Abstract
Acute promyelocytic leukemia is characterized by a chromosomal translocation involving the retinoic acid receptor alpha gene. To identify co-operating pathways to leukemogenesis, we crossed MRP8-PML/RARA transgenic mice with BXH-2 mice which harbor an endogenous murine leukemia virus that causes acute myeloid leukemia. Approximately half of the leukemias that arose in this cross showed features of acute promyelocytic leukemia. We identified 22 proviral insertion sites in acute promyelocytic-like leukemias and focused our analysis on insertion at Sox4, a HMG box transcription factor. Using a transplant model, co-operation between PML-RARα and Sox4 was confirmed with increased penetrance and reduced latency of disease. Interestingly, karyotypic analysis revealed cytogenetic changes suggesting that the factors combined to initiate but not complete leukemic transformation. The cooperation between these transcription factors is consistent with the paradigm of multiple routes to the disease and reinforces the concept that transcription factor networks are important therapeutic targets in myeloid leukemias.
Introduction
While acute myeloid leukemia (AML) presents with a heterogeneous phenotype and genotype, somatic chromosomal aberrations are a common molecular abnormality. The t(15;17) PML-RARα fusion product is characteristic of a subtype of AML, namely acute promyelocytic leukemia (APL), which is recapitulated by the MRP8-PML/RARA transgenic model.1 This has modeled the common, secondary chromosomal aberrations in human APL2 as well as co-operating pathways during leukemic transformation.3,4 Current data support the idea that PML-RARα serves as the initiating event in APL. The conceptual model of co-operation between receptor tyrosine kinase mutations (Class-I) and mutations targeting transcription factors (Class-II) has led to significant advances in our knowledge of myeloid leukemogenesis.5 However, other routes clearly exist, such as co-operative transformation mediated by alterations in more than one transcription factor. Replication-competent murine leukemia viruses (that lack oncogenic sequences) and murine strains with high levels of endogenous retroviral activity6 have been invaluable tools for the identification of co-operating events.
The Sry-related high mobility group (HMG) box 4 (Sox4) transcription factor binds directly to the minor groove of DNA within a conserved region.7,8 On the one hand, ablation of Sox4 leads to embryogenic lethality due to cardiac defects,9 whereas there is mounting evidence that overexpression of Sox4 contributes to tumorigenesis. Directly relevant to myeloid leukemia, the Sox4 locus has been previously identified as a viral integration target in co-operation with the Evi1 transcription factor.10 We describe here the novel co-operation of PML-RARα with Sox4 during myeloid leukemogenesis, highlighting collaborative changes in transcription factors as one route to myeloid leukemia, and emphasizing the importance of disrupting transcription factor abnormalities for treatment of hematopoietic neoplasia.11,12
Design and Methods
Mice
Mice were bred and maintained at the University of California in San Francisco in accordance with institutional Animal Care and Use Committee guidelines. FVB/n MRP8-PML/RARA mice have been previously described.1 Animals were sacrificed when moribund or when physical examination and blood cell counts indicated likely rapid progression of illness. Daily inspection of mice included assessment for general signs of illness. Blood was obtained via retro-orbital bleeding of animals presenting with raised criteria for malaise or moribund.
Retroviral transduction and transplantation
The ecotropic BOSC23 packaging line was transfected with retroviral constructs and virus harvested as previously described3 using the MSCV control plasmid13 or one carrying the mouse Sox4 cDNA. Transduced cells were washed, counted with trypan blue and injected into the tail vein of lethally (9 Gy) irradiated recipient FVB/n mice.
Quantitative-PCR
Oligo-dTprimed cDNA samples were prepared from total RNA isolated by means of Trizol extraction (Invitrogen, Carlsbad, CA, USA) using Superscript (Invitrogen,Carlsbad, CA, USA). PCR was conducted in triplicate with 20 μL reaction volumes of 1X Taqman buffer (1X Applied Biosystems PCR buffer, 20% glycerol, 2.5% gelatin, 60nM Rox as a passive reference), 5.5 mM MgCl2, 0.5 mM each primer, 0.2 μM each deoxynucleotide triphosphate (dNTP), 200 nM probe, and 0.025 unit/μL AmpliTaq Gold (Applied Biosystems) with 5 ng cDNA. PCR was conducted on the ABI 7900HT (Applied Biosystems) using the following cycle parameters: 1 cycle at 95° for 10 min and 40 cycles at 95° for 15 s, and at 60° for 1 min. Analysis was carried out using the SDS v.2.3 software supplied with the ABI 7900HT to determine the Ct values of each reaction. GapDH (Rodent): Forward TGC ACC ACC AAC TGC TTA G; Reverse GGA TGC AGG GAT GAT GTT C; Probe 5’-FAM- CAG AAG ACT GTG GAT GGC CCC TC-BHQ1-3’. Sfpi1 (Mouse): Forward CGT CCT CGA TAC TCC CAT GGT; Reverse AAG GTT TGA TAA GGG AAG CAC ATC; Probe 5’-FAM-TCA GTC ACC AGG TTT CCT ACA TGC CCC-BHQ1-3’. Further information is available in the Online Supplementary Appendix.
Statistical analysis
Ct values were determined for three test and three reference reactions in each sample, averaged, and subtracted to obtain the ACt [ACt = Ct (test locus) – Ct (control locus)]. PCR efficiencies were measured for all custom assays and were greater than or equal to 90%. Therefore, relative fold difference was calculated for each primer/probe combination as 2−ΔCt × 100.
Results and Discussion
With the aim of identifying co-operating events with PML-RARα during myeloid leukemogenesis, we first back-crossed MRP8-PML/RARA transgenic mice with the BXH-2 mice three times and aged mice for leukemia development. BXH-2 strain mice harbor an endogenous retrovirus that can induce AML.14 Although latency of disease was similar in BXH-2 mice with and without MRP8-PML/RARA, leukemias with features of APL were only observed when PML-RARα was present (data not shown). Twenty-two insertion sites were identified in eight leukemias with a promyelocytic morphology; with nine being reported as common insertion sites in the Mouse Retroviral Tagged Cancer Gene Database (RTCGD; http://rtcgd.ncifcrf.gov) which included Sox4, Meis1, Lck, Sfpi1, Nfil3 and Cbl-b. We focused our analysis on an insertion identified in one animal in the disease relevant HMG box transcription factor Sox4. Previously published sample data sets using Affymetrix Human Genome U133 show significantly increased levels of Sox4 transcript from APL patients versus normal human promyelocytes (P<0.005)5 (Online Supplementary Figure S1).
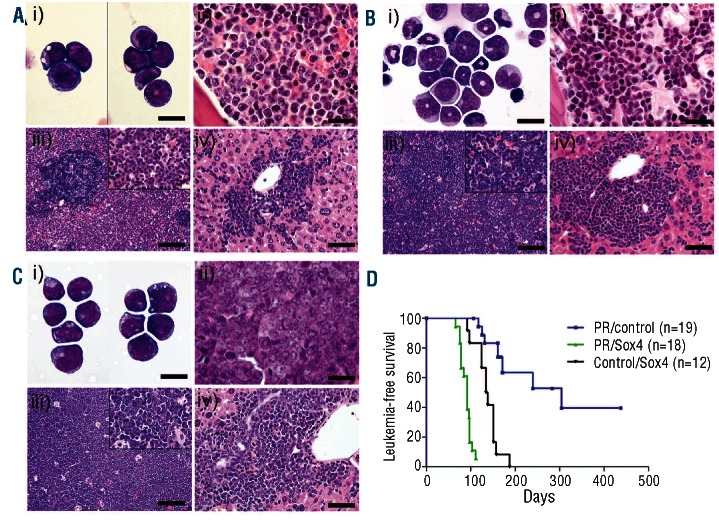
To assess the potential co-operation between PML-RARα and Sox4 we utilized a bone marrow transplant assay using retroviral transduction of Sox4 into control or MRP8-PML/RARA cells. Representative pathology at necropsy from PML-RARα animals is consistent with previous observations,1 where promyelocytes are observed in marrow cytology with immature cells observed by marrow histopathology (Figure 1A). Marrow cytology (Table 1) and histopathology from PML-RARα + Sox4 mice showed more differentiated cells, predominantly with features of metamyelocytes (Figure 1B). Cells appeared most blastic in the leukemias that developed in Control/Sox4 animals (Figure 1C). Leukemic infiltration into other tissues including spleen, liver and lymph nodes was also detected in leukemic animals in all three groups (Figure 1A-C and data not shown). Mice that received cells with both PML-RARα and Sox4 had consistently increased white blood cell counts (Online Supplementary Figure S2) but with similar immunophenotypic profiles as compared to single expressors (using the Mac-1/Gr-1/c-Kit/CD34 markers; data not shown) and increased leukemia-associated mortality, with median survival of 92 days as compared to 136 days or 304 days in mice with Sox4 or PML-RARα alone, respectively (Figure 1D). Interestingly, peripheral platelet counts were somewhat higher in PML-RARα + Sox4 leukemic animals as compared to PML-RARα alone leukemic mice, suggesting that thrombocytopenia does not track the course of the leukemic progression in the PML-RARα+ Sox4 model (Online Supplementary Figure S2). Additionally, interpretable SKY analysis of leukemia cells from 7 PML-RARα + Sox4 animals revealed 3 with no clonal abnormalities, 2 with loss of a sex chromosome only, and 2 with loss of a sex chromosome together with a gain of chromosome 8. While the cytogenetic pattern of leukemia cells from Sox4-only animals were not obtained, the specific karyotypic abnormalities seen in PML-RARα + Sox4 animals were consistent with that of our previously published observations from the PML-RARα animals with a high frequency of loss of a sex chromosome (57% vs. 55%); however the overall frequency of abnormalities appeared reduced in the PML-RARα + Sox4 cohort (57% vs. 93%)3 (Online Supplementary Table S1). Our findings indicate that although the combination of Sox4 and PML-RARα is neither sufficient to complete leukemogenesis nor able to fundamentally change the karyotypic pathway to progression, the proteins can work cooperatively to strongly initiate leukemic transformation.
Figure 1.
Sox4 co-operates with PML-RARα to induce AML. Representative pathology of: (i) marrow cytology, (ii) marrow histopathology, (iii) spleen (or lymph node in the case of Sox4) and (iv) and liver for (A) PML-RARα, (B) PML-RARα + Sox4 and (C) Sox4 mice. Although all groups represent similar degrees of organ infiltration of blast cells, PML-RARα + Sox4 group show a more pronounced involvement of metamyelocytes. Scale bar represents 13.3 micrometers for marrow cytology, 20 micrometers for marrow histopathology, 100 micrometers for spleen/lymph node and 40 micrometers for spleen/lymph node inset, and 50 micrometers for liver. Marrow cytology: Wright’s Giemsa stain. Histopathology: Hematoxylin & Eosin. (D) Kaplan-Meier survival curves of animals in days from transplant censored for non-leukemic deaths with similar incidences between the groups. These represent significantly reduced survival in PML-RARα + Sox4 mice as compared to the PML-RARα or Sox4 alone animals (P<0.0001). Non-leukemic deaths were at 116 days (euthanized due to fighting), 161 days (T-cell lymphoma phenotype), 293 days (lung tumor) and 431 days (euthanized due to age without disease).
Table 1.
Differential cell counts of Wright’s Giemsa stained bone marrow cells from PML-RARα24 and PML-RARα + Sox4 leukemias.
The fact that PML-RARα + Sox4 leads to myeloid leukemia is conceptually consistent with observations of co-operation between transcriptions factors, such as Sox4/PU.1,16 Sox4/CREB,17 Sox4/Evi1,10 Hoxa9/Meis1a18 and that of PML-RARα/Myc426. PML-RARα can promote increased proliferation of hematopoietic progenitors19 and Sox4 can enhance proliferation and decrease differentiation.10,20,21 Co-operative expansion of myeloid progenitors by these proteins might provide a platform for the cumulative acquisition of additional genetic changes that complete leukemic transformation. Interestingly, in the context of another myeloid leukemia model, Sox4 has been shown to transactivate the AKV LTR and up-regulate provirally activated Evi1.10 The cumulative impact of transcription factor changes in this model and in our model is the dysregulation of differentiation program and interference with the normal control of proliferation and survival.
Our analyses of published array data concur with the recently described inverse correlation between SOX4 and PU.1 (encoded by the SPI1 gene) in human AMLs15,17 (data not shown). However, when APL patient samples were specifically analyzed, the relationship is less robust and a range of SPI1 expression levels was observed even when there was significant expression of SOX4 (Online Supplementary Figure S3). Furthermore, in the context of our in vivo mouse model, exogenous expression of Sox4 on the background of PML-RARα was associated with a relative increase in Sfpi1 levels, the murine homolog of SPI1 (Online Supplementary Figure S4). Hence, although downregulation of SPI1 levels by SOX4 appears to be one mechanism by which SOX4 contributes to AML, this does not appear predominant in APL: relatively higher rather than lower SPI1/Sfpi1 levels are observed both in a subset of human APLs and in our murine model. Given the observations that PML-RARα + Sox4 leukemias showed greater neutrophilic differentiation than PML-RARα leukemias (Table 1), it was possible that the higher Sfpi1 levels seen in a subset of these leukemias (Online Supplementary Figure S4) reflected this increased maturation. To assess this possibility PML-RARα, Sox4, and PML-RARα + Sox4 leukemias were transplanted into secondary recipients and Sfpi1 levels were assessed in both leukemic spleens and in sorted promyelocytes. Sfpi1 levels in all three leukemias were higher than in normal promyelocytes and Sfpi1 appeared enriched in sorted promyelocytes as compared to unsorted leukemic spleens for both PML-RARα and PML-RARα + Sox4 leukemias (Online Supplementary Figure S5). The increased Sfpi1 in sorted promyelocytes compared to total leukemic cells suggests that the high levels of Sfpi1 in a subset of PML-RARα + Sox4 leukemias do not simply reflect the presence of maturing neutrophilic cells. Hence, the range of SPI1/Sfpi1 levels measured in human and mouse APL (Online Supplementary Figures S3 and S4) appears to reflect true biological variation. Interestingly, for the PML-RARα + Sox4 leukemia studied in Online Supplementary Figure S5, Sfpi1 levels were reduced relative to the PML-RARα alone cells, consistent with previous observations.16 Altogether, our findings support the idea that inhibition of SPI1/Sfpi1 expression plays a role in some, but not all, SOX4 expressing leukemias.
Increased expression of SOX4 has been reported in many cancers including medulloblastoma, as well as cancers of the prostate, bladder, colon and lung (non-small cell).22 Our models described here provide robust systems in which gene expression profile analyses may reveal additional PML-RARα targets alone or in combination with Sox4 and as comparators to human APL. Furthermore, our finding that PML-RARα + Sox4 co-operate in leukemogenesis is consistent with the observation that therapeutic levels of retinoic acid result in reduced levels of SOX4 in the context of APL.23 To further examine the therapeutic potential of ATRA, leukemia-free survival of secondary recipients with placebo or ATRA was determined. Interestingly, ATRA is able to significantly increase the leukemia-free survival of PML-RARα + Sox4, as well as the expected PML-RARα cohort, as compared to their respective placebo treated cohorts (P=0.0038 and P=0.0052). Sox4 alone animals were insensitive to the ATRA treatment (Figure 2). Given the leukemic potency of Sox4 we observed, this protein is an attractive target for therapy of malignancies in which it is over-expressed whose efficacy may be exponentially raised in concert with retinoic acid in the presence of the PML-RARα lesion.
Figure 2.
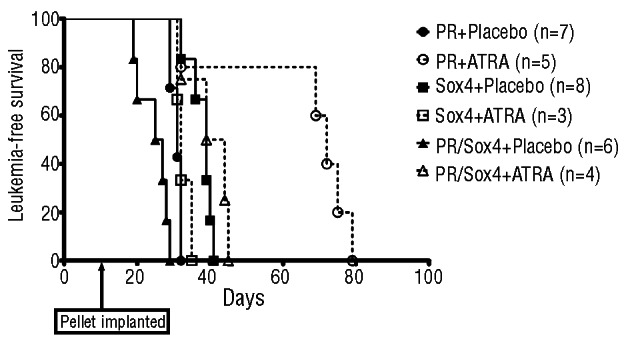
ATRA is able to extend survival of PML-RARα and PML-RARα + Sox4 secondary transplanted mice. Kaplan-Meier survival curves of secondary transplanted animals with placebo or ATRA pellet implantation. Compared to their respective placebo controls, ATRA is able to significantly extend survival in PML-RARα mice (2.3fold difference in median survival; P=0.0052) as well as in PML-RARα + Sox4 mice (1.6-fold difference in median survival; P=0.0038).
Supplementary Material
Acknowledgments
We thank The Mouse Pathology Core of the UCSF Helen Diller Family Comprehensive Cancer Center; Kirsten Copren and Lillian Edmondson from the Genome Analysis Core Facility, Helen Diller Family Comprehensive Cancer Center, University of California, San Francisco; Noel Lenny and James Downing from St Jude Children’s Research Hospital Memphis; Neal Copeland and Nancy Jenkins for providing the Sox4 expressing retroviral vector; Tim Ley for helpful discussion; and Elizabeth M. Davis from the University of Chicago for expert assistance with the SKY analysis.
Footnotes
Funding
The project described was supported by Award Number R01CA095274 from the National Cancer Institute, and LLS-SCOR (7015-09).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. SCK was a scholar of the Leukemia and Lymphoma Society. NO is funded by Leukaemia and Lymphoma Research (UK). BY is supported by award number 2011CB933501 from the National Basic Research Program of China (973 Program).
Authorship and Disclosures
Information on authorship, contributions, and financial and other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Brown D, Kogan S, Lagasse E, Weissman I, Alcalay M, Pelicci PG, et al. A PMLRARalpha transgene initiates murine acute promyelocytic leukemia. Proc Natl Acad. Sci USA. 1997;94(6):2551–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Le Beau MM, Bitts S, Davis EM, Kogan SC. Recurring chromosomal abnormalities in leukemia in PML-RARA transgenic mice parallel human acute promyelocytic leukemia. Blood. 2002;99(8):2985–91 [DOI] [PubMed] [Google Scholar]
- 3.Jones L, Wei G, Sevcikova S, Phan V, Jain S, Shieh A, et al. Gain of MYC underlies recurrent trisomy of the MYC chromosome in acute promyelocytic leukemia. J Exp Med. 2010;207(12):2581–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le Beau MM, Davis EM, Patel B, Phan VT, Sohal J, Kogan SC. Recurring chromosomal abnormalities in leukemia in PML-RARA transgenic mice identify cooperating events and genetic pathways to acute promyelocytic leukemia. Blood. 2003;102(3):1072–4 [DOI] [PubMed] [Google Scholar]
- 5.Dash A, Gilliland DG. Molecular genetics of acute myeloid leukaemia. Best. Pract Res Clin Haematol. 2001;14(1):49–64 [DOI] [PubMed] [Google Scholar]
- 6.Jenkins NA, Copeland NG, Taylor BA, Bedigian HG, Lee BK. Ecotropic murine leukemia virus DNA content of normal and lymphomatous tissues of BXH-2 recombinant inbred mice. J Virol. 1982;42(2):379–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van de Wetering M, Clevers H. Sequence-specific interaction of the HMG box proteins TCF-1 and SRY occurs within the minor groove of a Watson-Crick double helix. EMBO J. 1992;11(8):3039–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harley VR, Jackson DI, Hextall PJ, Hawkins JR, Berkovitz GD, Sockanathan S, et al. DNA binding activity of recombinant SRY from normal males and XY females. Science. 1992;255(5043):453–6 [DOI] [PubMed] [Google Scholar]
- 9.Schilham MW, Oosterwegel MA, Moerer P, Ya J, de Boer PA, van de Wetering M, et al. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 1996;380(6576):711–4 [DOI] [PubMed] [Google Scholar]
- 10.Boyd KE, Xiao YY, Fan K, Poholek A, Copeland NG, Jenkins NA, et al. Sox4 cooperates with Evi1 in AKXD-23 myeloid tumors via transactivation of proviral LTR. Blood. 2006;107(2):733–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de The H, Chen Z. Acute promyelocytic leukaemia: novel insights into the mechanisms of cure. Nat Rev Cancer. 2010;10(11):775–83 [DOI] [PubMed] [Google Scholar]
- 12.Cerchietti LC, Ghetu AF, Zhu X, Da Silva GF, Zhong S, Matthews M, et al. A small-molecule inhibitor of BCL6 kills DLBCL cells in vitro and in vivo. Cancer Cell. 2010;17(4):400–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hawley RG, Fong AZ, Burns BF, Hawley TS. Transplantable myeloproliferative disease induced in mice by an interleukin 6 retro-virus. J Exp Med. 1992;176(4):1149–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bedigian HG, Johnson DA, Jenkins NA, Copeland NG, Evans R. Spontaneous and induced leukemias of myeloid origin in recombinant inbred BXH mice. J Virol. 1984;51(3):586–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Payton JE, Grieselhuber NR, Chang LW, Murakami M, Geiss GK, Link DC, et al. High throughput digital quantification of mRNA abundance in primary human acute myeloid leukemia samples. J Clin Invest. 2009;119(6):1714–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aue G, Du Y, Cleveland SM, Smith SB, Dave UP, Liu D, et al. Sox4 cooperates with PU.1 haploinsufficiency in murine myeloid leukemia. Blood. 2011;118(17):4674–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sandoval S, Kraus C, Cho EC, Cho M, Bies J, Manara E, et al. Sox4 cooperates with CREB in myeloid transformation. Blood. 2012;120(1):155–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17(13):3714–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du C, Redner RL, Cooke MP, Lavau C. Overexpression of wild-type retinoic acid receptor alpha (RARalpha) recapitulates retinoic acid-sensitive transformation of primary myeloid progenitors by acute promyelocytic leukemia RARalpha-fusion genes. Blood. 1999;94(2):793–802 [PubMed] [Google Scholar]
- 20.Du Y, Spence SE, Jenkins NA, Copeland NG. Cooperating cancer-gene identification through oncogenic-retrovirus-induced insertional mutagenesis. Blood. 2005;106(7):2498–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kvinlaug BT, Chan WI, Bullinger L, Ramaswami M, Sears C, Foster D, et al. Common and overlapping oncogenic pathways contribute to the evolution of acute myeloid leukemias. Cancer Res. 2011;71(12):4117–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Penzo-Mendez AI. Critical roles for SoxC transcription factors in development and cancer. Int. J. Biochem. Cell Biol. 2010;42(3):425–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meani N, Minardi S, Licciulli S, Gelmetti V, Coco FL, Nervi C, et al. Molecular signature of retinoic acid treatment in acute promyelocytic leukemia. Oncogene. 2005;24(20):3358–68 [DOI] [PubMed] [Google Scholar]
- 24.Kogan SC, Hong SH, Shultz DB, Privalsky ML, Bishop JM. Leukemia initiated by PMLRARalpha: the PML domain plays a critical role while retinoic acid-mediated transactivation is dispensable. Blood. 2000;95(5):1541–50 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.