Abstract
Using transgenic mouse models of breast cancer that ablate Src homology and collagen A (ShcA) expression or oncogene-coupled ShcA signaling, we previously showed that this adaptor is critical for mammary tumor onset and progression. We now provide the first evidence that ShcA regulates mammary tumorigenesis, in part, through its ability to regulate the adaptive immune response. Inactivation of ShcA signaling within tumor cells results in extensive CD4+ T-cell infiltration and induction of a humoral immune response in mammary tumors. This is associated with a robust CTL response in preneoplastic lesions that are deficient in ShcA signaling. Moreover, mammary tumor progression of ShcA-deficient hyperplasias is accelerated in a T cell–deficient background. We also uncover a clinically relevant correlation between high ShcA expression and low CTL infiltration in human breast cancers. Finally, we define a novel ShcA-regulated immune signature that functions as an independent prognostic marker of survival in human epidermal growth factor receptor 2+ and basal breast cancers. We reveal a novel role for tumor cell–derived ShcA in the establishment and maintenance of an immunosuppressive state.
Introduction
Src homology and collagen A (ShcA) couples to receptor and cytoplasmic tyrosine kinases and relays extracellular signals that govern cell proliferation, survival, invasion, and angiogenesis (1). Moreover, clinical studies show that elevated ShcA signaling correlates with increased nodal status, advanced stage, and relapse in breast cancer patients (2).
Transgenic mouse models have defined important roles for ShcA during breast cancer progression (3–5). Expression of a mutant polyomavirus middle T (MT) oncogene lacking the ShcA binding site (MT-Y250F) results in the delayed induction of mammary epithelial hyperplasia and a prolonged latency period before tumor formation (5). Conversely, restoration of the ShcA binding site to a mutant ErbB2 lacking its COOH terminal tyrosine phosphorylation sites is sufficient to restore transforming activity in vivo (3). We recently showed that loss of ShcA in the mammary epithelial compartment abrogates tumor development (4). These observations indicate that oncogene-coupled ShcA signaling is critical for mammary tumorigenesis.
Although ShcA functions in an autonomous fashion (1), it also regulates paracrine signaling between cell types. ShcA loss in myocytes impairs synaptic conductivity in Ia sensory neurons, leading to a spindle cell defect (6). In fibroblasts, engagement of ShcA downstream of receptor tyrosine kinases is sufficient to stimulate vascular endothelial growth factor production and elicit an angiogenic response (7). Finally, impaired ShcA signaling disrupts the ability of breast cancer cells to induce vasculature recruitment (4). These observations suggest that ShcA signaling in cancer cells is important for the establishment of a protumorigenic microenvironment.
To better understand the importance of tumor cell–derived ShcA on breast cancer progression, we modulated ShcA expression and signaling in well-characterized mouse models. Our work defines a novel role for ShcA in establishing an immunosuppressive state. Moreover, analysis of tissue microarrays and published gene expression data sets from human breast cancers reveal that elevated ShcA signaling is associated with the establishment of an immunosuppressive state and poor patient outcome. These observations provide the first evidence that ShcA signaling plays a critical role in modulating the immune response to favor a protumorigenic microenvironment.
Materials and Methods
Transgenic mice
Transgenic mouse models expressing polyomavirus MT [mouse mammary tumor virus (MMTV)/MT], a mutant MT lacking the ShcA binding site (MMTV/MT-250F) or a bicistronic transcript encoding an oncogenic ErbB2 allele coupled to Cre (MMTV/NIC), have been described (4, 5, 8). Mammary-specific deletion of ShcA in MMTV/NIC mice was achieved using ShcAfl/fl animals (6). FVB and athymic (nu/nu) mice were purchased from Charles River Laboratories. NIC/ShcAfl/fl multiparous females were bred from 8 weeks of age. All NIC/ShcAfl/fl tumors are described (Supplementary Table S1). Animals were monitored for tumor onset by weekly physical palpation and necropsied 6 weeks postpalpation. Mammary epithelial cells were isolated from 5-week-old mice as described (9) and injected (250,000 cells) into the fourth mammary gland of mice. Animal studies were approved by the Animal Resources Centre at McGill University and comply with guidelines set by the Canadian Council of Animal Care.
Microarray analysis
Total RNA was isolated from eight NIC/ShcAfl/fl tumors, representing five independent animals, and hybridized against a NIC pool containing equal amounts of RNA from five NIC tumors onto Agilent whole genome oligo microarrays (4 × 44 K) as previously described (10). Duplicate hybridizations were done on half of the samples using reverse dye labeling. Raw probe intensities were background corrected within and between arrays normalized using the normexp, loess, and quantile methods in Bioconductor (11). Differential expression was evaluated using LIMMA. The NIC pool identified genes that were commonly differentially expressed in NIC/ShcAfl/fl tumors. Subsequent validation used RNA from 10 independent NIC tumors, five of which comprised the NIC pool. The microarray data are deposited on GEO (GSE18996).
ShcA-regulated immune signature
The ShcA-regulated immune signature (SRIS) contains B cell– and T cell–specific genes that are differentially expressed in at least six of eight NIC/ShcAfl/fl tumors (P ≤ 0.01). The average fold change (log2) for each gene over all tumors or in those displaying differential expression is shown (Supplementary Table S3). Three genes were included, although they were not differentially expressed in six tumors, because their overexpression was validated in NIC/ShcAfl/fl tumors. Human orthologues were examined in 12 publicly available data sets comprising 2,481 patients (Supplementary Table S5). In each data set, hierarchical clustering using Euclidean distance and Wards algorithm separated the patients into two classes: those that increased (SRIS-high) and decreased (SRIS-low) expression of the signature. The SRIS-high patients were determined by cutting the top node. In the van de Vijver data set, a lower cluster was deemed more appropriate.
Molecular subtype was determined using correlation to the PAM50 centroids as described (12). In data sets where human epidermal growth factor receptor 2 (HER2) status was unavailable, it was determined by clustering expression of genes in the HER2 amplicon (Neurod2, Ppp1r1b, Stard3, Tcap, Pnmt, Perld1, ErbB2, C17orf37, C17orf37, Grb7, and Ikzf3). Patients were classified as ShcA-high or ShcA-low based on whether ShcA levels were above or below the mean for each data set. Analysis used R/Bioconductor (11), and Sweave documentation is available on request. Data sets containing overall survival time or time to relapse were combined in the statistical analysis as time to outcome. Kaplan-Meier analyses also show that a high SRIS is also associated with good outcome in the majority of individual data sets (Supplementary Fig. S13). In the multivariate analyses, estrogen receptor and HER2 positivity is based on immunohistochemical staining and was included given the variability in distribution of patients based on molecular or immunohistochemical subtyping.
Results
NIC/ShcAfl/fl mammary tumors emerge with reduced penetrance and delayed onset
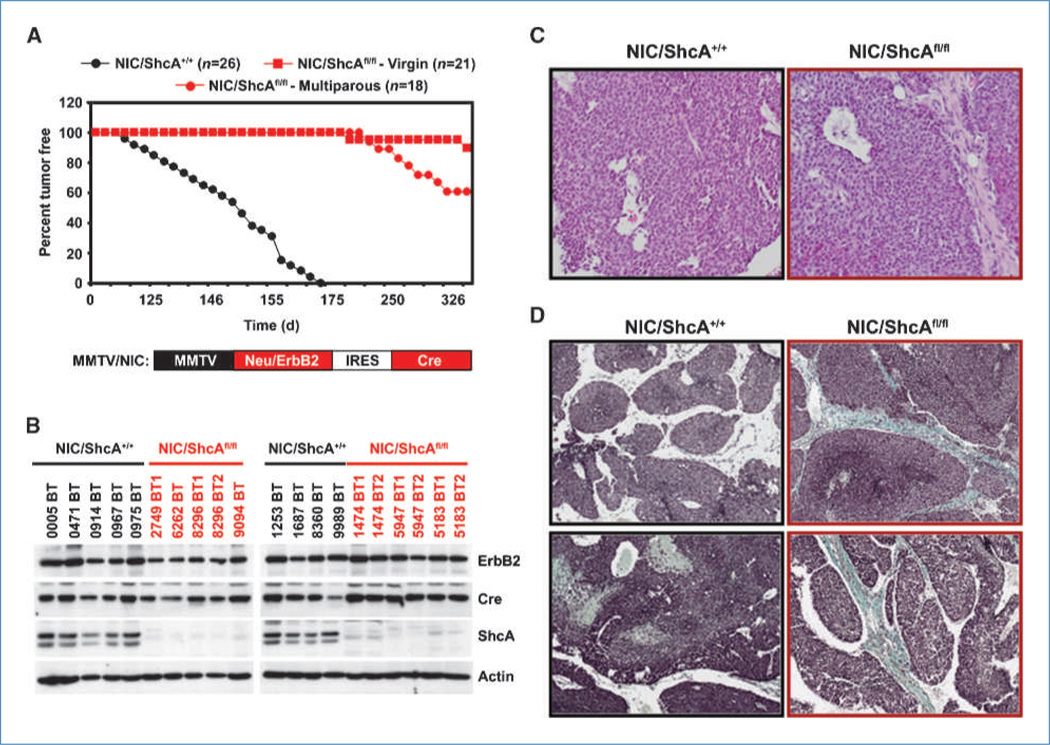
Using a breast cancer mouse model that coexpresses Neu/ErbB2 and Cre from a bicistronic transcript (MMTV/NIC; Fig. 1A), we showed that mammary epithelial-derived ShcA is critical for tumor induction (4). In an expanded cohort, we observed tumors in 9% of NIC/ShcAfl/fl virgin mice, albeit with a long latency period (Fig. 1A). Previous studies show that parity facilitates mammary tumor formation (13). With parity, we observed a 4-fold increase in the percentage of tumor-bearing NIC/ShcAfl/fl mice. These focal tumors arose after a long latency period and lacked ShcA (Fig. 1A and B; Supplementary Fig. S1A). NIC and NIC/ShcAfl/fl tumors were histologically similar, resembling the solid phenotype that is typical of ErbB2-driven tumors (Fig. 1C). Finally, NIC/ShcAfl/fl tumors displayed more collagen deposition (Fig. 1D) but metastasized at comparable rates relative to NIC tumors (Supplementary Fig. S2).
Figure 1.
NIC/ShcAfl/fl tumors emerge with parity. A, percentage of tumor-free mice over time for the indicated genotypes. B, immunoblot analysis on tumor cell lysates using the indicated antibodies. C, H&E staining of representative NIC and NIC/ShcAfl/fl tumors. D, trichrome-staining of NIC and NIC/ShcAfl/fl tumors. Blue staining shows collagen deposition.
A mechanism by which ShcA-null tumors could arise is through elevated expression of other Shc family members (14, 15). However, neither ShcB nor ShcC was differentially expressed between NIC and NIC/ShcAfl/fl tumors (Supplementary Fig. S1A and B). Whereas ShcD transcripts were elevated in NIC/ShcAfl/fl tumors (Supplementary Fig. S1B), ShcD protein levels were low and unaltered regardless of ShcA status (Supplementary Fig. S1C). Therefore, other Shc family members are not overexpressed in NIC/ShcAfl/fl tumors.
Further characterization revealed a modest increase in the proliferative index of NIC/ShcAfl/fl tumors (Supplementary Fig. S3A). This likely reflects the activation of compensatory pathways in these tumors, which contribute to their emergence. However, the apoptotic potential and vascular density of tumors were comparable regardless of ShcA status (Supplementary Fig. S3B and D). Moreover, isolated NIC and NIC/ShcAfl/fl mammary tumor cells similarly induced CD31+ cell recruitment, showing that neither is debilitated in its ability to elicit an angiogenic response (Supplementary Fig. S3E). Finally, whereas we observed variable levels of extracellular signal-regulated kinase activation, the p38 mitogen-activated protein kinase, c-Jun NH2 terminal kinase, and AKT pathways were unaltered between NIC and NIC/ShcAfl/fl tumors (Supplementary Fig. S4). Therefore, NIC/ShcAfl/fl tumors have adopted alternative strategies to activate signaling pathways, which are normally dependent on ShcA, to support tumor cell proliferation, survival, and angiogenesis.
Loss of ShcA signaling increases T-cell infiltration
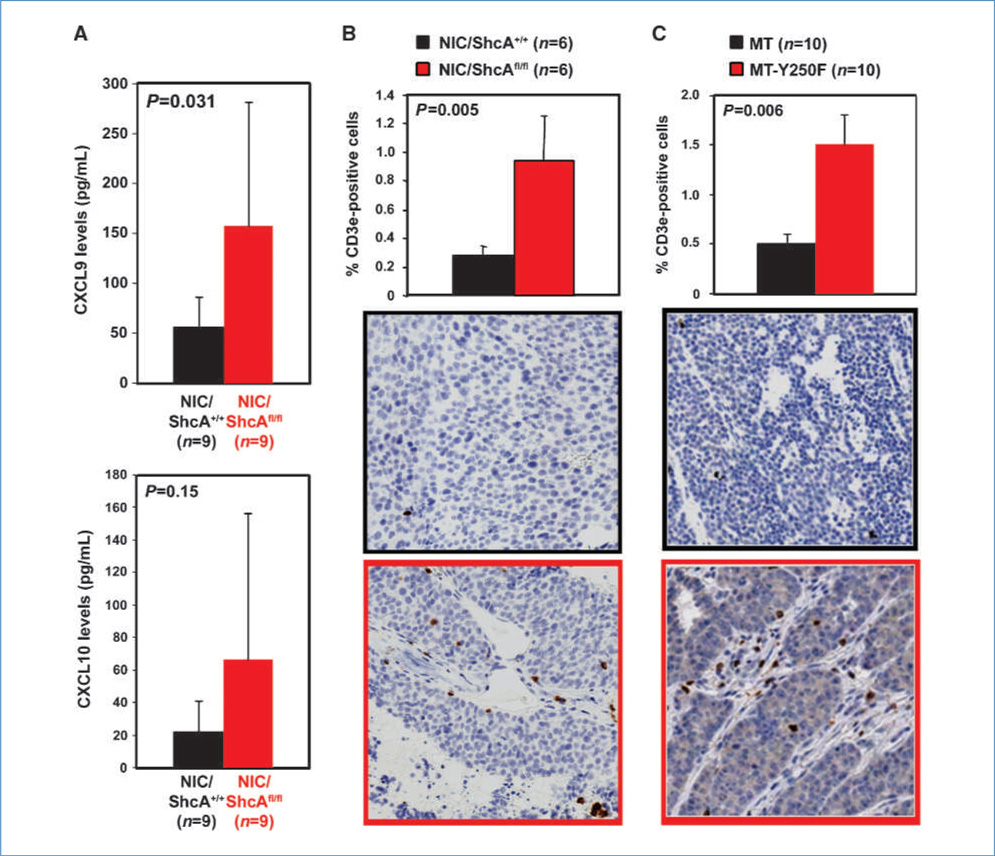
We used gene expression profiling to investigate the mechanism by which mammary tumors circumvent the requirement for ShcA. Bioinformatics analysis revealed a large number of differentially expressed chemokines and their receptors between NIC and NIC/ShcAfl/fl tumors (Supplementary Table S2). Several of these chemokines recruit and activate macrophages. However, we neither observed differences in their expression nor in macrophage recruitment between NIC and NIC/ShcAfl/fl tumors (Supplementary Fig. S5). Thus, we focused on CXCL9 and CXCL10, which are overexpressed in NIC/ShcAfl/fl tumors (Fig. 2A) and are chemotactic for CXCR3-expressing T cells and natural killer (NK) cells (16). Indeed, NIC/ShcAfl/fl tumors overexpress CXCR3 (Supplementary Fig. S7A) and exhibit a 3.5-fold increase in T-cell infiltration (Fig. 2B). We also used MMTV/MT-Y250F mice, which express a mutant MT protein that cannot recruit ShcA due to mutation of the ShcA binding site (5). MT-Y250F tumors also displayed a 3-fold increase in T-cell recruitment (Fig. 2C) and increased CXCL9 levels in MT-Y250F hyperplastic tissue (Supplementary Fig. S6). Thus, loss of ShcA signaling downstream of the transforming oncogene, in a tumor cell that retains ShcA, is sufficient to augment T-cell recruitment.
Figure 2.
Increased T-cell infiltration in NIC/ShcAfl/fl and MT-Y250F tumors. A, relative CXCL9 and CXCL10 levels, as determined by ELISA, in lysates from NIC and NIC/ShcAfl/fl tumors. B and C, immunohistochemical staining of tumors with a CD3e-specific antibody. Data are representative of 6 (B) or 10 (C) tumors (10–20 fields per tumor) and are depicted as percentage of CD3e+ cells ± SE.
Infiltrating T cells in NIC/ShcAfl/fl tumors also upregulate costimulatory markers, including CD28 (2.6-fold) and ICOS (2.9-fold; Supplementary Fig. S7A). Significantly, ICOS transcripts are only expressed in antigen-activated T cells (17), and a statistically significant increase (1.8-fold) in ICOS transcripts is retained within NIC/ShcAfl/fl tumors following normalization to CXCR3 levels (Supplementary Fig. S7A). Thus, the increase in ICOS is not solely attributed to enhanced T-cell recruitment. We also show that interleukin-2 (IL-2), which is secreted by antigen-activated T cells (18), is increased in NIC/ShcAfl/fl tumors (Supplementary Fig. S7B). Finally, a higher percentage of proliferating T cells was observed in NIC/ShcAfl/fl tumors (Supplementary Fig. S7C).
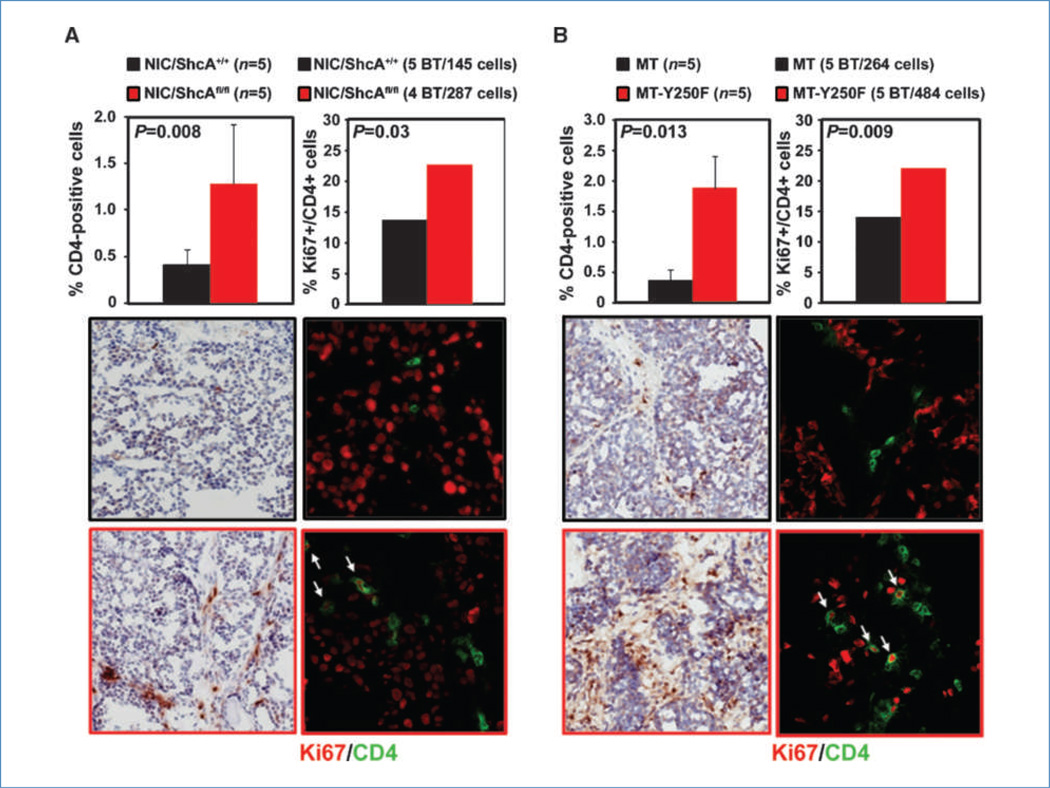
To specify the nature of the T-cell infiltrate, we assessed the extent of CTL and T helper (Th) cell recruitment into NIC/ShcAfl/fl and MT-Y250F tumors. The numbers of granzyme B+ and CD8+ cells were exceedingly low in tumors (Supplementary Fig. S8). In contrast, NIC/ShcAfl/fl tumors displayed a 3.1-fold increase in CD4+ cell recruitment and a 1.7-fold increase in the percentage of Ki67+/CD4+ T cells relative to controls (Fig. 3A). We also observed a 4.9-fold increase in CD4+ T-cell recruitment and a 1.6-fold increase in the percentage of Ki67+/CD4+ cells in MT-Y250F tumors compared with MT controls (Fig. 3B). These data suggest that impaired ShcA signaling in tumor cells results in increased recruitment of CD4+ T cells, a subset of which is proliferating and may be antigen activated.
Figure 3.
Increased infiltration of proliferating CD4+ T cells in NIC/ShcAfl/fl and MT-Y250F tumors. A, left, immunohistochemical staining of NIC and NIC/ShcAfl/fl tumors with a CD4-specific antibody. Data are representative of five tumors (10–20 fields per tumor) and are depicted as percentage of CD4+ cells ± SE. Right, immunofluorescent staining of NIC and NIC/ShcAfl/fl tumors with CD4 (green) and Ki67 (red) antibodies. The percentage of proliferating CD4+ cells were calculated from five NIC and four NIC/ShcAfl/fl tumors and include 145 and 287 CD4+ cells, respectively. B, immunohistochemical and immunofluorescent staining of MT and MT-Y250F tumors with a CD4-specific antibody. A total of 264 (MT) and 484 (MT-Y250F) CD4+ cells were counted.
CD4+ Th cells differentiate into numerous lineages that perform distinct biological functions, including induction of a CTL response (Th1), activation of humoral immunity (Th2), immunosuppression (Treg), and proinflammatory signaling (Th17; ref. 19). Cancer cells co-opt Th responses to favor Treg- and Th2-regulated immunity, which establishes a chronic inflammatory, protumorigenic state (20). The T-cell infiltrate in NIC/ShcAfl/fl or MT-Y250F tumors does not reflect a strong Th1 response given the virtual absence of CD8+ cells (Supplementary Fig. S8). Moreover, we observe a comparable percentage of FoxP3+ cells between NIC and NIC/ShcAfl/fl tumors (Supplementary Fig. S9A).
Given that Th2 cells are required for humoral immunity, we characterized the extent of immunoglobulin synthesis in these tumors. NIC/ShcAfl/fl tumors displayed 15-fold and 10-fold increases in IgHM and IgJ transcript levels, the latter representing the joining chain for IgM assembly (Supplementary Fig. S10A). This correlated with a robust increase in IgM protein levels within NIC/ShcAfl/fl tumors (Supplementary Fig. S10B). In addition, a 10-fold increase in IgHG1 and IgHG2a transcripts (Supplementary Fig. S10A) and significantly elevated IgHG1, IgHG2a, IgHG2b, and IgHG3 protein levels (Supplementary Fig. S10B) were observed in NIC/ShcAfl/fl tumors. This was associated with a 2-fold increase in CD20+ B-cell recruitment into NIC/ShcAfl/fl mammary tumors (Supplementary Fig. S10C). Moreover, IgG was also elevated in MT-Y250F hyperplastic mammary glands and tumors (Supplementary Fig. S11). These data suggest that sustained loss of ShcA signaling within mammary tumors polarizes the immune response to favor increased Th2 and B-cell recruitment.
Loss of ShcA signaling relieves immunosuppression
Differentiated Th cells secrete unique cytokines to regulate diverse biological functions (20, 21). IFNγ is produced from Th1 cells and initiates an antitumor immune response. IL-4 is a Th2-derived cytokine that stimulates B-cell differentiation and chronic inflammation in cancer cells. IL-10 is secreted from Treg and Th2 cells to mediate immunosuppression (22). End-stage tumors derived from NIC or NIC/ShcAfl/fl mice expressed comparable IFNγ, IL-4, and IL-10 levels, suggesting that tumors must adjust the steady-state expression of these cytokines to levels that permit outgrowth (Supplementary Fig. S9B).
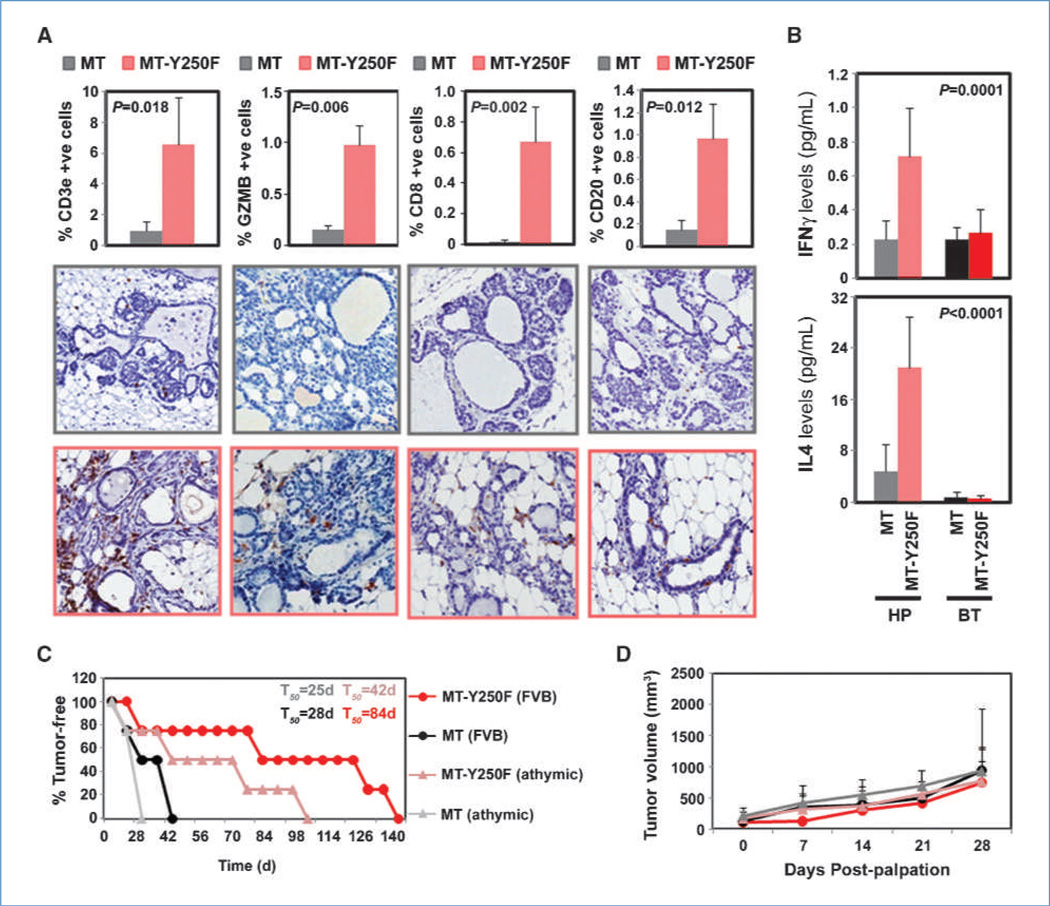
We wished to establish the importance of ShcA signaling in regulating T-cell recruitment early during cancer progression. Mammary glands from tumor-bearing and tumor-free NIC/ShcAfl/fl animals were histologically normal (data not shown). This is not surprising given the long tumor latency and reduced penetrance observed in virgin and multiparous NIC/ShcAfl/fl mice (Fig. 1). This suggests that ShcA signaling is required early during mammary tumorigenesis, making it difficult to isolate preneoplastic NIC/ShcAfl/fl lesions. However, the MT mouse model is ideally suited to study mammary tumor progression (23). Therefore, we focused on the MT and MT-Y250F mouse models, which uniformly develop hyperplasias before overt tumor development (5, 23). MT-Y250F hyperplastic mammary glands displayed a robust increase in CD3e+ T-cell infiltration relative to MT controls (Fig. 4A). We also examined the cytokine profiles of hyperplastic mammary glands and tumors from MT and MT-Y250F animals. All tumors possessed low and comparable IFNγ and IL-4 levels (Fig. 4B). In contrast, MT-Y250F hyperplastic glands exhibited 3.2-fold and 4.4-fold increases in IFNγ and IL-4 levels, respectively (Fig. 4B). This was associated with a large and significant increase in the recruitment of granzyme B+, CD8+, and CD20+ cells into MT-Y250F hyperplastic structures (Fig. 4A). However, these cell types were virtually absent in early preneoplastic MT lesions (Fig. 4A). This suggests that loss of ShcA signaling relieves an immunosuppressive state early during the transformation process.
Figure 4.
Increased CD8+ and CD20+ cell recruitment into hyperplastic MT-Y250F mammary glands. A, CD3e, granzyme B, CD8, and CD20 immunohistochemical staining of mammary glands from MT and MT-Y250F mice. The percentage of positivity was determined from hyperplastic lesions retaining a hollow lumen. Data are representative of four to six animals for each genotype, scoring the following number of hyperplasias: CD3: 176, MT and 158, MT-Y250F; granzyme B: 138, MT and 103, MT-Y250F; CD8: 125, MT and 143, MT-Y250F; and CD20: 124, MT and 112, MT-Y250F. Data are depicted as percentage of positive cells ± SE. B, relative IFNγ and IL-4 levels, as determined by ELISA, in lysates from MT and MT-Y250F mammary glands and tumors (n = 9). Cytokine levels were normalized to tubulin levels, as determined by fluorescent immunoblotting. C, isolated mammary epithelial cells from 5-week-old MT and MT-Y250F animals were injected into the mammary fat pads of FVB or athymic mice (n = 4 each). Mammary tumor onset was monitored by weekly physical palpation, and the percentage of tumor-free mice over time is indicated. D, following first palpation, tumor volumes were determined by caliper measurements.
To functionally examine the contribution of adaptive immunity to delayed tumor progression in MT-Y250F mice, we exploited the fact that mammary glands of 5-week-old MT females only contain hyperplasias and/or adenomas (23). Therefore, isolated mammary epithelial cells from 5-week-old MT and MT0Y250F animals were injected into the mammary fat pads of immunocompetent (FVB) or immunocompromised (athymic) mice that lack T cells. We show that MT tumors emerged with similar kinetics, regardless of the immune status of recipient animals (Fig. 4C). In contrast, we observed a 2-fold decrease in the latency of MT-Y250F tumor onset in athymic mice (Fig. 4C). However, once formed, the growth kinetics of MT and MT-Y250F tumor cells was comparable in both FVB and athymic mice (Fig. 4D). This suggests that an intact T-cell response contributes to the delayed tumor onset observed in MT-Y250F animals but cannot impair tumor outgrowth following progression to carcinoma.
ShcA expression is associated with reduced CTL infiltration and poor outcome in breast cancer patients
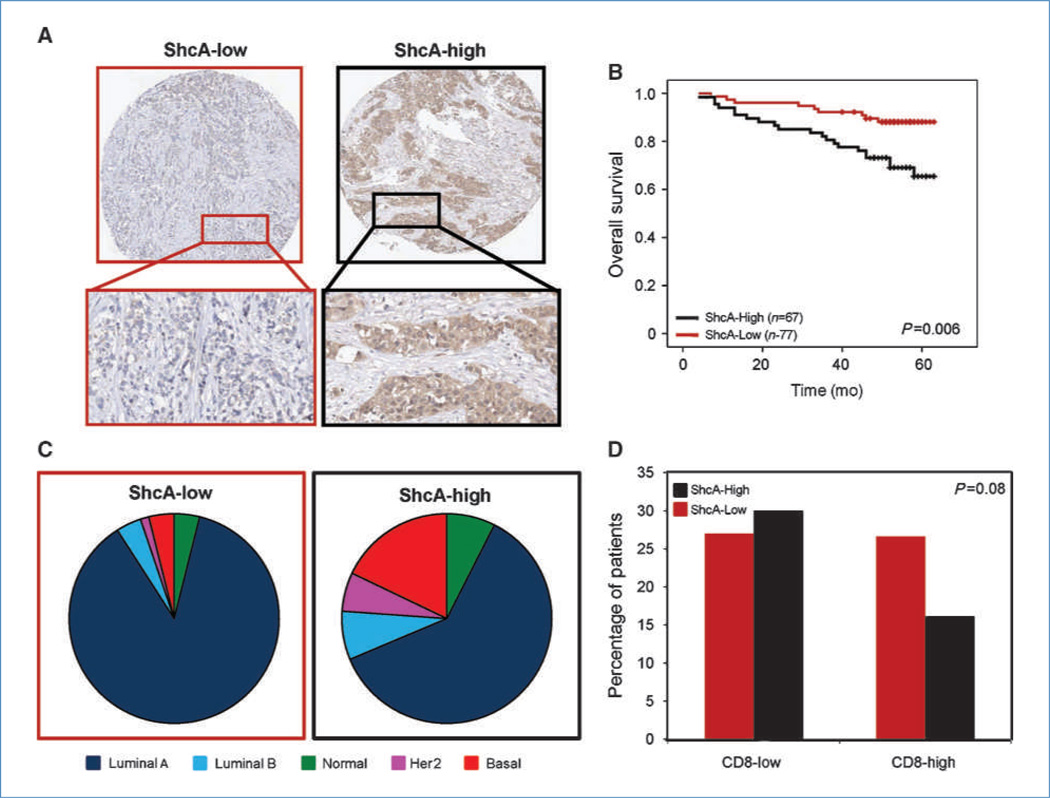
To understand the clinical significance of ShcA signaling and tumor immunity, we stained a tissue microarray, containing 179 invasive breast carcinomas, with CD4-, CD8-, FoxP3-, or ShcA-specific antibodies. The tumors were classified into two groups (ShcA-high and ShcA-low) based on cytoplasmic staining intensity and percentage of positively stained cells (Fig. 5A). We observed significantly reduced overall survival (Fig. 5B) and an increased percentage of basal and HER2+ tumors within the ShcA-high group (Fig. 5C). Finally, high ShcA expression within breast tumors correlated with reduced CD8+ T-cell infiltration compared with the ShcA-low group (Fig. 5D). However, we did not observe any differences in CD4 or FoxP3 positivity within ShcA-low versus ShcA-high tumors (data not shown). These observations suggest that high ShcA levels are associated with the HER2 and basal subtypes, reduced survival, and diminished CTL recruitment in breast tumors.
Figure 5.
High ShcA expression is associated with diminished CD8 infiltration, basal subtype, and reduced survival in primary breast cancers. A tissue microarray was stained with a ShcA-specific antibody. A, the tumors were classified into two groups: ShcA-low (n = 77) and ShcA-high (n = 67). B, Kaplan-Meier analysis of each group with respect to overall survival. C, distribution of ShcA-low and ShcA-high patients within the five molecular subtypes (P = 0.008). D, the tissue microarray was also stained with a CD8-specific antibody. Tumors within the ShcA-low group are associated with increased CD8+ T-cell infiltration.
A SRIS predicts good outcome within basal and HER2 breast cancer patients
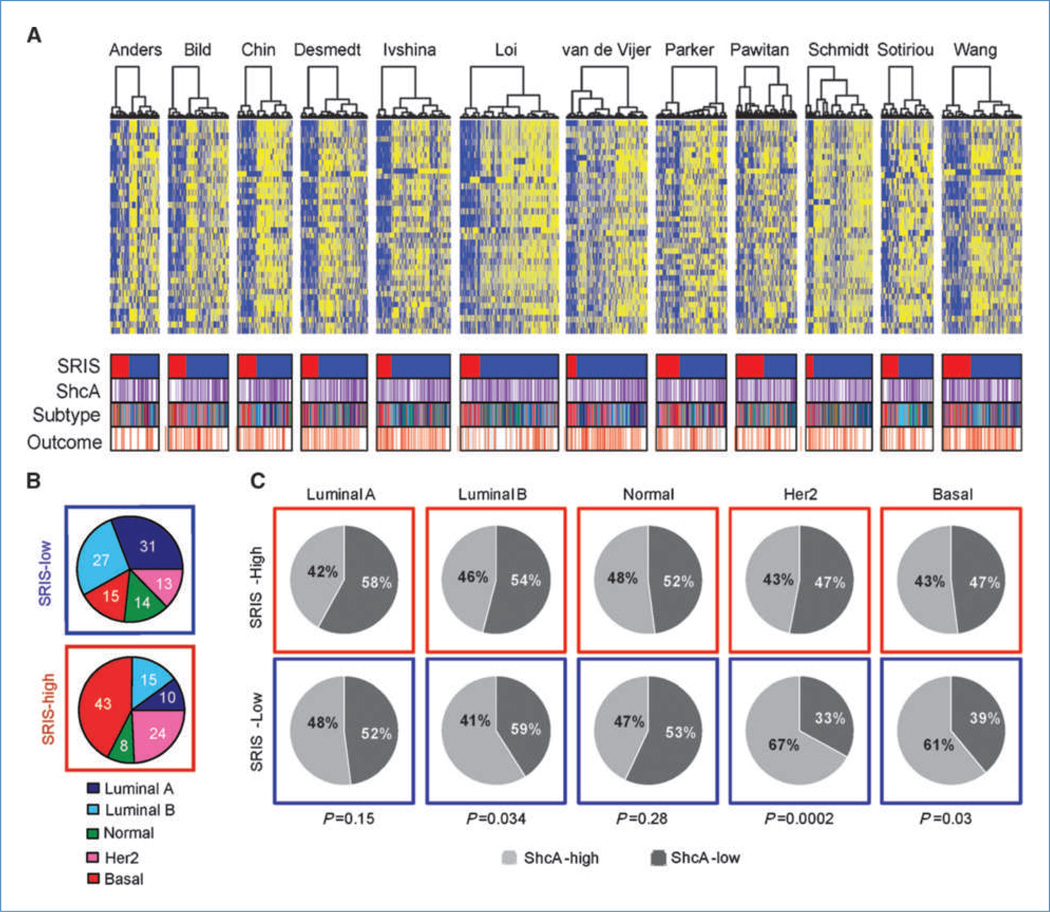
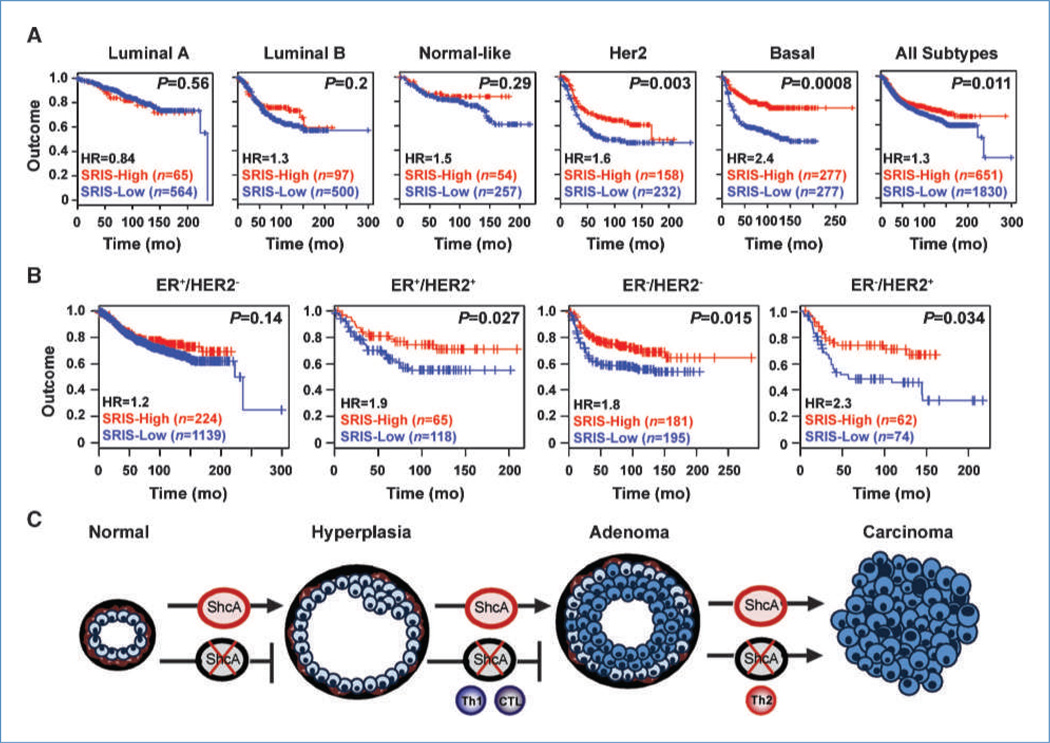
To further explore the clinical significance of ShcA expression in human breast cancer, we developed a 43-gene SRIS, representing those genes linked to the B-cell or T-cell response, which were differentially expressed by gene expression profiling between NIC and NIC/ShcAfl/fl tumors (Supplementary Table S3). We used the SRIS to cluster 2,481 patients from 12 publicly available data sets. Hierarchical clustering consistently segregated the patients into groups that either overexpressed (ShcA-high; n = 651) or repressed (ShcA-low; n = 1,830) the signature (Fig. 6A). Importantly, the SRIS discriminated the stromal, but not epithelial, contribution of the tumor (refs. 24, 25; Supplementary Fig. S12). Molecular subtype analyses revealed that the SRIS-high group was associated with a statistically significant increase in the percentage of patients within the HER2 and basal subtypes (P < 2.2 × 10−16; Fig. 6B). Consistent with a dependency of the SRIS on ShcA, higher ShcA levels were specifically observed within the SRIS-low group of HER2 and basal breast cancer patients (Fig. 6C). Moreover, a high SRIS was associated with good outcome within these subtypes but had no predictive power for luminal patients (Fig. 7A). The SRIS also stratified poor and good outcome patients within the HER2 and triple-negative subtypes based on immunohistochemical classification (Fig. 7B). Finally, we show that the SRIS predicted survival in HER2+ and basal breast cancer patients independently of grade and lymph node status (Supplementary Table S4). These data suggest that ShcA signaling favors immunosuppression in HER2 and basal breast cancers, which may contribute to poor patient outcome. Low ShcA levels are associated with the acquisition of an immune signature that predicts survival and may represent a novel prognostic marker within these poor outcome subtypes.
Figure 6.
The SRIS derived from NIC/ShcAfl/fl tumors is enriched within the Her2 and basal subtypes. A, heatmaps of SRIS in 12 publicly available breast cancer data sets, encompassing 2,481 patients. The SRIS segregates the patients into two main clusters: those that display increased (n = 766; blue) or decreased (n = 2,269; yellow) expression of the signature. B, pie charts illustrating the distribution of molecular subtypes within the SRIS-high and SRIS-low patients. C, pie charts illustrating the relative ShcA abundance, at the transcript level, between SRIS-high and SRIS-low patients within each molecular subtype.
Figure 7.
The SRIS predicts good outcome within the Her2 and basal subtypes. Kaplan-Meier analysis within the SRIS-low (blue) and SRIS-high (red) groups for each of the molecular (A) or immunohistochemical (B) subtypes. The number of patients within each subtype is shown. HR, hazards ratio. C, ShcA-deficient mammary epithelial cells are debilitated in their ability to progress from hyperplasia. Loss of ShcA signaling relieves immunosuppression in early hyperplastic mammary glands, resulting in the activation of an antitumorigenic Th1/CTL response. Progressively growing lesions, however, polarize the immune response to favor Th2-mediated immunity, which establishes a chronic inflammatory state to favor transition to carcinoma.
Discussion
We show a novel role for tumor cell autonomous ShcA in limiting the T-cell response within the microenvironment. Reduced ShcA signaling may increase the expression of chemokines that promote T-cell infiltration. Indeed, CXCL9 and CXCL10 proteins and CXCL9, CXCL10, and CXCL11 transcripts were overexpressed in NIC/ShcAfl/fl tumors. They are chemotactic for CXCR3-expressing T cells and NK cells (16), which may influence tumor cell cytotoxicity. Previous studies show that overexpression of CXCL9 in cancer cells increased T-cell infiltration, decreased tumor outgrowth, and prolonged survival of immunocompetent, but not immunocompromised, animals (26).
It is equally plausible that reduced ShcA signaling indirectly establishes an activated stroma that permits T-cell infiltration. The density of infiltrating leukocytes, including B cells, T cells, and macrophages, progressively increases during breast cancer progression in clinical samples and mouse models (22, 27). Moreover, ovalbumin-tagged breast cancer cells that regress following adoptive transfer of ovalbumin-specific T cells show increased T-cell infiltration and a proportionally denser, more collagen-rich stroma compared with progressively growing tumors (28). Finally, PTEN loss in stromal fibroblasts induces collagen deposition and innate immune cell infiltration (29). NIC/ShcAfl/fl and MT-Y250F (5) tumors contain a denser, more collagen-rich stroma and exhibit increased T-cell recruitment. This raises the possibility that matrix remodeling provides a conduit for immune cell recruitment and that tumor cells regulate the expression of chemokines that dictate the nature of the immune infiltrate.
NIC tumors were derived from virgin females, whereas NIC/ShcAfl/fl tumors were obtained from virgin and multiparous mice (Supplementary Table S1), raising the question of whether parity influenced the observed gene expression patterns. However, NIC/ShcAfl/fl tumors from virgin and multiparous mice showed similar increases in collagen deposition, immunoglobulin synthesis, and B-cell and T-cell recruitment. Moreover, these results were recapitulated within MT-Y250F tumors, which were derived from virgin females. Although we cannot exclude the possibility that some differentially expressed genes are affected by pregnancy, regulation of immune-related genes is likely to be ShcA dependent and independent of parity.
We hypothesize that loss of ShcA signaling within preneoplastic lesions relieves an immunosuppressive state. The observation that tumor onset is significantly delayed in MT-Y250F (5) and NIC/ShcAfl/fl animals and MT-Y250–induced tumor onset is accelerated in a T cell–deficient background supports this model. It has been shown that MT-induced tumor formation and outgrowth proceeds normally in CD4- and CD8-deficient backgrounds, but the degree of lung metastasis is severely reduced in CD4−/− mice (27). However, ShcA signaling is intact in these tumors, which establishes an immunosuppressive state and negates any role for the adaptive immune response during cancer progression. We observed lung metastases in 70% and 75% of NIC and NIC/ShcAfl/fl mice, respectively. Given this high metastatic potential, it may be difficult to see enhanced lung metastases.
There is strong evidence for activation of Th1/CTL-mediated antitumor immunity in preneoplastic NIC/ShcAfl/fl and MT-250F lesions. Significantly increased numbers of infiltrating CD8+ and granzyme B+ cells surround MT-Y250F hyperplastic lesions relative to MT controls. IgG2a expression is elevated in NIC/ShcAfl/fl tumors, and IgG2a class switch recombination is dependent on IFNγ signaling (30). In addition, IFNγ levels are significantly increased in MT-Y250F hyperplastic mammary glands but reduced to comparable levels in MT and MTY250F tumors. The emergence of MT-Y250F, but not MT, mammary tumors is accelerated in a T cell–deficient background. These data suggest that a Th1/CTL-driven immune response is recruited to ShcA-deficient preneoplastic lesions and likely contributes to delayed progression to carcinoma. However, established MT-Y250F and NIC/ShcAfl/fl tumors lack CD8+ T cells, suggesting that a Th1/CTL response is progressively lost during tumor progression and is replaced by Th2/humoral immunity (Fig. 7C). We hypothesize that this Th1-to-Th2 switch is an adaptive response that permits emerging tumors to escape cytotoxicity and establishes a chronic inflammatory state that facilitates tumorigenesis.
Elevated IL-4 levels and increased B-cell recruitment also characterize MT-Y250F preneoplastic lesions. Moreover, NIC/ShcAfl/fl tumors overexpress CXCL13 (Supplementary Table S2), which is chemotactic for B cells. These tumors display increased B-cell recruitment and elevated immunoglobulin levels, suggesting that humoral immunity is engaged and sustained throughout tumor progression. Indeed, B cells are most abundant in DCIS lesions of breast cancer patients but remain elevated in invasive carcinomas (22). Moreover, in the acute phase, B cells may elicit antitumor immune responses, whereas chronically activated B cells secrete cytokines and immunoglobulins that exert proinflammatory functions (22). Thus, sustained B-cell recruitment and elevated immunoglobulin synthesis in MT-Y250F and NIC/ShcAfl/fl tumors may contribute to the tumorigenic process.
Reduced ShcA signaling in primary breast cancers is associated with increased CTL infiltration. However, Th cells predominate in ShcA-deficient mouse mammary tumors. Several reasons can account for this discrepancy. Although CD4 levels do not correlate with ShcA levels in primary breast cancers, this marker cannot discriminate between Th1, Th2, and Treg cells, which exert opposing functions during cancer progression. Moreover, in the transgenic mouse models, ShcA expression or oncogene-coupled ShcA signaling is completely ablated. In contrast, ShcA levels are diminished, but never absent, in primary breast cancers. This suggests that a greater selective pressure is exerted on ShcA-deficient mouse mammary tumors to favor a polarized, Th2-dependent protumorigenic response. Reduced ShcA levels in human breast cancers are sufficient to permit progression to carcinoma but may also allow engagement of antitumor immune responses, which would increase patient survival.
We observe a strong association between ShcA expression with the HER2 and basal subtypes. Paradoxically, primary tumors overexpressing genes within the SRIS express lower ShcA levels and yet are also classified within the HER2 and basal subtypes. This reflects the fact that high ShcA levels select for poor outcome HER2 and basal breast cancer patients, whereas the SRIS strongly discerns good prognosis patients within these subtypes. Moreover, the SRIS has no predictive value within the luminal subtypes in a cohort encompassing over 1,400 patients. This suggests that the SRIS is not a general predictor but is limited to HER2 and basal breast cancers. The inverse correlation between ShcA expression, acquisition of the SRIS signature, and outcome within these subtypes suggests that regulation of ShcA signaling is critical for disease progression in human breast cancers. ShcA signals downstream of numerous receptor tyrosine kinases in breast cancer cells (1). Indeed, HER2 and basal breast cancers are associated with amplification, overexpression, and activation of receptor tyrosine kinases (RTK), including ErbB2, ErbB3, epidermal growth factor receptor, c-Kit, and Met (31–36), each of which couple to ShcA (37–41). We provide the first observation that RTK signaling is important for tumor immunosuppression. Therefore, combining RTK inhibitors with immune-based therapies may have significant therapeutic potential in the treatment of breast cancer patients.
Supplementary Material
Acknowledgments
We thank Peter Siegel and Robert Annan for the critical reading of the manuscript, Cynthia Lavoie for mammary fat pad injections, Elitza Germanov and Vi Minh Tri Su for their histologic support, and Julie Livingstone and Indrani Vasudeva Murthy for their bioinformatics assistance.
Grant Support
Terry Fox team grant 020002, Canadian Breast Cancer Research Alliance operating grant 019491, and Canadian Institutes of Health Research (CIHR) operating grants 6849, 13466, 89727, and 67211. Funding is acknowledged for the Genomics Core from Terry Fox and CIHR team grants, along with Enterprise Ireland and Science Foundation Ireland. W.J. Muller is supported by a CRC chair in Molecular Oncology, T. Pawson is a CIHR distinguished scientist, D. DeNardo was supported by the American Cancer Society, and L.M. Coussens acknowledges support from NIH/National Cancer Institute and DOD BCRP Era of Hope Scholar Award W81XWH-06-1-0416. M. Park holds the Diane and Sal Guerrera chair in Cancer Genetics at McGill University.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Ursini-Siegel J, Muller WJ. The ShcA adaptor protein is a critical regulator of breast cancer progression. Cell Cycle. 2008;7:1936–1943. doi: 10.4161/cc.7.13.6205. [DOI] [PubMed] [Google Scholar]
- 2.Davol PA, Bagdasaryan R, Elfenbein GJ, Maizel AL, Frackelton AR., Jr Shc proteins are strong, independent prognostic markers for both node-negative and node-positive primary breast cancer. Cancer Res. 2003;63:6772–6783. [PubMed] [Google Scholar]
- 3.Dankort D, Maslikowski B, Warner N, et al. Grb2 and Shc adapter proteins play distinct roles in Neu (ErbB-2)-induced mammary tumorigenesis: implications for human breast cancer. Mol Cell Biol. 2001;21:1540–1551. doi: 10.1128/MCB.21.5.1540-1551.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ursini-Siegel J, Hardy WR, Zuo D, et al. ShcA signalling is essential for tumour progression in mouse models of human breast cancer. EMBO J. 2008;27:910–920. doi: 10.1038/emboj.2008.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Webster MA, Hutchinson JN, Rauh MJ, et al. Requirement for both Shc and phosphatidylinositol 3′ kinase signaling pathways in polyomavirus middle T-mediated mammary tumorigenesis. Mol Cell Biol. 1998;18:2344–2359. doi: 10.1128/mcb.18.4.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hardy WR, Li L, Wang Z, et al. Combinatorial ShcA docking interactions support diversity in tissue morphogenesis. Science. 2007;317:251–256. doi: 10.1126/science.1140114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saucier C, Khoury H, Lai KM, et al. The Shc adaptor protein is critical for VEGF induction by Met/HGF and ErbB2 receptors and for early onset of tumor angiogenesis. Proc Natl Acad Sci U S A. 2004;101:2345–2350. doi: 10.1073/pnas.0308065101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White DE, Kurpios NA, Zuo D, et al. Targeted disruption of β1-integrin in a transgenic mouse model of human breast cancer reveals an essential role in mammary tumor induction. Cancer Cell. 2004;6:159–170. doi: 10.1016/j.ccr.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 10.Schade B, Lam SH, Cernea D, et al. Distinct ErbB-2 coupled signaling pathways promote mammary tumors with unique pathologic and transcriptional profiles. Cancer Res. 2007;67:7579–7588. doi: 10.1158/0008-5472.CAN-06-4724. [DOI] [PubMed] [Google Scholar]
- 11.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Polyak K. Pregnancy and breast cancer: the other side of the coin. Cancer Cell. 2006;9:151–153. doi: 10.1016/j.ccr.2006.02.026. [DOI] [PubMed] [Google Scholar]
- 14.Jones N, Hardy WR, Friese MB, et al. Analysis of a Shc family adaptor protein, ShcD/Shc4, that associates with muscle-specific kinase. Mol Cell Biol. 2007;27:4759–4773. doi: 10.1128/MCB.00184-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Bryan JP, Songyang Z, Cantley L, Der CJ, Pawson T. A mammalian adaptor protein with conserved Src homology 2 and phosphotyrosine-binding domains is related to Shc and is specifically expressed in the brain. Proc Natl Acad Sci U S A. 1996;93:2729–2734. doi: 10.1073/pnas.93.7.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vandercappellen J, Van Damme J, Struyf S. The role of CXC chemokines and their receptors in cancer. Cancer Lett. 2008;267:226–244. doi: 10.1016/j.canlet.2008.04.050. [DOI] [PubMed] [Google Scholar]
- 17.Watts TH, DeBenedette MA. T cell co-stimulatory molecules other than CD28. Curr Opin Immunol. 1999;11:286–293. doi: 10.1016/s0952-7915(99)80046-6. [DOI] [PubMed] [Google Scholar]
- 18.Waldmann TA. The biology of interleukin-2 and interleukin-15: implications for cancer therapy and vaccine design. Nat Rev Immunol. 2006;6:595–601. doi: 10.1038/nri1901. [DOI] [PubMed] [Google Scholar]
- 19.Zhu J, Paul WE. CD4 T cells: fates, functions, and faults. Blood. 2008;112:1557–1569. doi: 10.1182/blood-2008-05-078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johansson M, Denardo DG, Coussens LM. Polarized immune responses differentially regulate cancer development. Immunol Rev. 2008;222:145–154. doi: 10.1111/j.1600-065X.2008.00600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glimcher LH, Murphy KM. Lineage commitment in the immune system: the T helper lymphocyte grows up. Genes Dev. 2000;14:1693–1711. [PubMed] [Google Scholar]
- 22.DeNardo DG, Coussens LM. Inflammation and breast cancer. Balancing immune response: crosstalk between adaptive and innate immune cells during breast cancer progression. Breast Cancer Res. 2007;9:212–221. doi: 10.1186/bcr1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin EY, Jones JG, Li P, et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol. 2003;163:2113–2126. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boersma BJ, Reimers M, Yi M, et al. A stromal gene signature associated with inflammatory breast cancer. Int J Cancer. 2008;122:1324–1332. doi: 10.1002/ijc.23237. [DOI] [PubMed] [Google Scholar]
- 25.Finak G, Bertos N, Pepin F, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 26.Walser TC, Ma X, Kundu N, Dorsey R, Goloubeva O, Fulton AM. Immune-mediated modulation of breast cancer growth and metastasis by the chemokine Mig (CXCL9) in a murine model. J Immunother. 2007;30:490–498. doi: 10.1097/CJI.0b013e318031b551. [DOI] [PubMed] [Google Scholar]
- 27.DeNardo DG, Barreto JB, Andreu P, et al. CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell. 2009;16:91–102. doi: 10.1016/j.ccr.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin ML, Wall EM, Sandwith E, et al. Density of tumour stroma is correlated to outcome after adoptive transfer of CD4(+) and CD8 (+) T cells in a murine mammary carcinoma model. Breast Cancer Res Treat. 2010;121:753–763. doi: 10.1007/s10549-009-0559-y. [DOI] [PubMed] [Google Scholar]
- 29.Trimboli AJ, Cantemir-Stone CZ, Li F, et al. Pten in stromal fibroblasts suppresses mammary epithelial tumours. Nature. 2009;461:1084–1091. doi: 10.1038/nature08486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng SL, Szabo SJ, Glimcher LH. T-bet regulates IgG class switching and pathogenic autoantibody production. Proc Natl Acad Sci U S A. 2002;99:5545–5550. doi: 10.1073/pnas.082114899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheang MC, Voduc D, Bajdik C, et al. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res. 2008;14:1368–1376. doi: 10.1158/1078-0432.CCR-07-1658. [DOI] [PubMed] [Google Scholar]
- 32.Graveel CR, DeGroot JD, Su Y, et al. Met induces diverse mammary carcinomas in mice and is associated with human basal breast cancer. Proc Natl Acad Sci U S A. 2009;106:12909–12914. doi: 10.1073/pnas.0810403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nalwoga H, Arnes JB, Wabinga H, Akslen LA. Expression of EGFR and c-kit is associated with the basal-like phenotype in breast carcinomas of African women. APMIS. 2008;116:515–525. doi: 10.1111/j.1600-0463.2008.01024.x. [DOI] [PubMed] [Google Scholar]
- 34.Ponzo MG, Lesurf R, Petkiewicz S, et al. Met induces mammary tumors with diverse histologies and is associated with poor outcome and human basal breast cancer. Proc Natl Acad Sci U S A. 2009;106:12903–12908. doi: 10.1073/pnas.0810402106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 36.Wiseman SM, Makretsov N, Nielsen TO, et al. Coexpression of the type 1 growth factor receptor family members HER-1, HER-2, and HER-3 has a synergistic negative prognostic effect on breast carcinoma survival. Cancer. 2005;103:1770–1777. doi: 10.1002/cncr.20970. [DOI] [PubMed] [Google Scholar]
- 37.Batzer AG, Blaikie P, Nelson K, Schlessinger J, Margolis B. The phosphotyrosine interaction domain of Shc binds an LXNPXY motif on the epidermal growth factor receptor. Mol Cell Biol. 1995;15:4403–4409. doi: 10.1128/mcb.15.8.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dankort DL, Wang Z, Blackmore V, Moran MF, Muller WJ. Distinct tyrosine autophosphorylation sites negatively and positively modulate neu-mediated transformation. Mol Cell Biol. 1997;17:5410–5425. doi: 10.1128/mcb.17.9.5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lennartsson J, Blume-Jensen P, Hermanson M, Ponten E, Carlberg M, Ronnstrand L. Phosphorylation of Shc by Src family kinases is necessary for stem cell factor receptor/c-kit mediated activation of the Ras/MAP kinase pathway and c-fos induction. Oncogene. 1999;18:5546–5553. doi: 10.1038/sj.onc.1202929. [DOI] [PubMed] [Google Scholar]
- 40.Schaeper U, Gehring NH, Fuchs KP, Sachs M, Kempkes B, Birchmeier W. Coupling of Gab1 to c-Met, Grb2, and Shp2 mediates biological responses. J Cell Biol. 2000;149:1419–1432. doi: 10.1083/jcb.149.7.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vijapurkar U, Cheng K, Koland JG. Mutation of a Shc binding site tyrosine residue in ErbB3/HER3 blocks heregulin-dependent activation of mitogen-activated protein kinase. J Biol Chem. 1998;273:20996–21002. doi: 10.1074/jbc.273.33.20996. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.