Summary
Genomic sequencing reveals similar but limited numbers of protein-coding genes in different genomes, which begs the question of how organismal diversities are generated. Alternative pre-mRNA splicing, a widespread phenomenon in higher eukaryotic genomes, is thought to provide a mechanism to increase the complexity of the proteome and introduce additional layers for regulating gene expression in different cell types and during development. Among a large number of factors implicated in the splicing regulation are the SR protein family of splicing factors and SR protein-specific kinases. Here, we summarize the rules for SR proteins to function as splicing regulators, which depends on where they bind in exons versus intronic regions, on alternative exons versus flanking competing exons, and on cooperative as well as competitive binding between different SR protein family members on many of those locations. We review the importance of cycles of SR protein phosphorylation/dephosphorylation in the splicing reaction with emphasis on the recent molecular insight into the role of SR protein phosphorylation in early steps of spliceosome assembly. Finally, we highlight recent discoveries of SR protein-specific kinases in transducing growth signals to regulate alternative splicing in the nucleus and the connection of both SR proteins and SR protein kinases to human diseases, particularly cancer.
Keywords: SR splicing factors, SR protein-specific kinases, Splice-site selection, Alternative Splicing, Phosphorylation, Signal Transduction
Introduction
The completion of the human genome and genomes of virtually all model organisms has revealed the striking fact that the complexity of organisms is not correlated with the number of genes each genome encodes. Instead, the expression of individual genes in different cell types or during development is subjected to complex regulation via enhancer networks. In addition, gene expression in both quantity and quality is also known to subject to regulation by a variety of post-transcriptional mechanisms.
Pre-mRNA splicing removes intervening sequences from primary transcripts, a process essential for gene expression in eukaryotic cells. In addition to constitutive activities, higher eukaryotic cells produce a large number of mRNA isoforms as result of alternative splicing. Estimation based on unbiased transcriptome analysis suggests that up to 90% genes in humans undergo alternative splicing (Pan et al., 2008; Wang et al., 2008a), which may directly contribute to species and organ specificities during evolution and development (Barbosa-Morais et al., 2012; Merkin et al., 2012). Alternative splicing comes in all imaginable combinatory uses of RNA segments from primary transcripts, including the use of alternative promoters that are coupled to a downstream exon, cassette exons that can be included or skipped, alternative uses of the 5′ or 3′ splice sites that include or exclude specific exonic regions, intron retentions, and alternative polyadenylation events.
It has been generally believed that alternative splicing contributes to the complexity of the proteome and to differential stability of alternatively spliced transcripts though coupling with other post-transcriptional mechanisms, such as nonsense-mediated mRNA decay [NMD] or microRNA-induced mRNA degradation. Both non-coding RNA and the non-coding part of protein-coding transcripts (e.g. 5′UTR and 3′UTR) are also known to subject to alternative splicing. Since alternative splicing is so prevalent, it has been challenging to assign functions to individual mRNA isoforms. Some isoforms may constitute background noises from gene expression and imperfection of the splicing machinery. However, many mRNA isoforms clearly encode for functionally distinct proteins in stem cell pluripotency and reprogramming (Gabut et al., 2011), targeting critical signaling molecules to different cellular compartments (Xu et al., 2005), regulating cell proliferation versus death (Moore et al., 2010), switching metabolic pathways (Christofk et al., 2008), etc. Many alternative splicing events have been linked to various human diseases, including cancer. Readers are referred to several recent outstanding reviews on these topics (Cooper et al., 2009; David and Manley, 2010; Kalsotra and Cooper, 2011; Nilsen and Graveley, 2009). Overall, these findings point to a vital role for splicing in cellular regulation.
One of the major goals in the field is to understand how splicing is regulated. In general, splicing regulation has to be executed by specific RNA binding proteins via their interactions with cis-acting regulatory elements on primary transcripts, which is evident from a recent genome-wide analysis of alternative splicing in mammalian tissues (Merkin et al., 2012). A recent proteomic analysis indicates an alarmingly large number of proteins that may be directly or indirectly associated with mRNA (Castello et al., 2012). In reality, however, we know little about how many proteins can directly bind RNA and/or communicate with the splicing machinery. Furthermore, most splicing reactions appear to take place co-transcriptionally (Carrillo Oesterreich et al., 2011; Han et al., 2011b), and therefore, the transcription machinery and chromatin states may have profound influence on splice site selection (Luco et al., 2011; Pandit et al., 2008).
Approaching such a complex problem requires the definition of RNA elements recognized by specific RNA binding proteins and linking specific binding events to functional consequences. This has become more approachable by using the latest genomics technologies coupled with traditional functional dissection on minigenes. Indeed, several recent works have revised our general impression on simple division of RNA binding splicing regulators into splicing enhancers or repressors, as most, if not all, well-studied splicing regulators appear to exhibit position-dependent effects on splicing outcomes (Huelga et al., 2012; Pandit et al., 2013; Witten and Ule, 2011).
In this review, we “narrowly” focus on the family of SR proteins and SR protein-specific kinases with specific attention paid to their roles in the regulation of alternative splicing and regulation of splicing regulators in response to cellular signaling. In order for us to focus on major conceptual issues and recent advances, readers are referred to early comprehensive reviews on SR proteins (Lin and Fu, 2007; Long and Caceres, 2009) and SR protein kinases (Ghosh and Adams, 2011; Giannakouros et al., 2011).
SR proteins as the family of alternative splicing regulators
The SR protein family of splicing factors
SR proteins are part of a large superfamily of RNA binding proteins that share common RNA binding motifs (Figure 1A), which among others include heterogeneous nuclear ribonucleoparticle [hnRNP] proteins (Busch and Hertel, 2012; Tang et al., 2012b). The unique structure that distinguishes SR proteins from other RNA binding protein is the presence of an Arg/Ser-rich domain [called the RS domain, see Figure 1A)]. This signature RS domain has also been found in other proteins carrying several other types of protein domains, such as a Zn finger domain or an RNA helicase domain, which are generally known as SR-related proteins (Fu, 1995; Zhong et al., 2009b; Long and Caceres, 2009). An important feature of SR proteins is their extensive auto-regulation and cross-regulation to control the expression of individual SR proteins in the cell (Lareau et al., 2007; Ni et al., 2007).
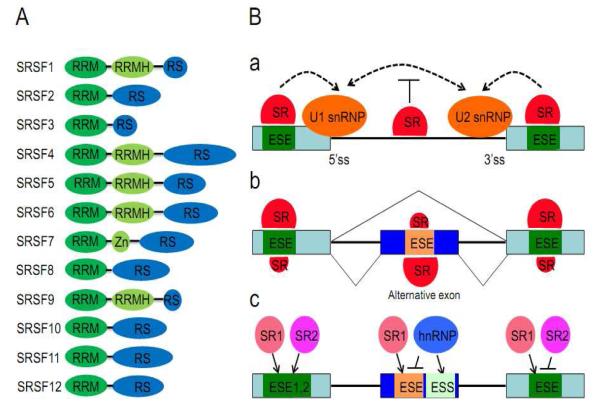
Figure 1.
Domain structures of SR proteins and rules for the regulation of alternative splicing by SR proteins. (A) The domain structure of core SR proteins. RRM: RNA recognition motif; RRMH: RRM homology; RS: arginine/serine-rich domain; Zn: Zinc knuckle. (B, panel a) Positional dependent effects of SR proteins on splice site selection. SR protein binding to exonic splicing enhancer (ESE) stimulates the recognition of the nearby 5′ and 3′ splice site by U1 and U2 snRNP, respectively. On the other hand, SR protein binding to intronic regions inhibits splicing, likely through interfering with the communication between the functional 5′ and 3′ splice sites. (B, panel b) SR protein-dependent exon inclusion or skipping. SR protein binding to the internal alternative exon promotes exon inclusion, whereas SR protein binding to the flanking competing exon(s) causes exon skipping. (B, panel c) Cooperative and competitive binding of SR proteins. Different SR proteins may bind to adjacent ESEs either independently or cooperatively, which will result in additive or synergistic effects on splice site selection. Different SR proteins may also compete on related ESEs. SR protein binding to ESEs is also known to compete with the interaction of other RNA binding proteins with adjacent exonic splicing silencer (ESS). SR proteins may therefore compete with hnRNP proteins in splice site selection via both cis- and trans-mechanisms.
Because multiple names have been given to some of the same SR proteins during the course of their discovery, a new standardized nomenclature has been proposed for the “core” SR protein family that consists of 12 relatively well-characterized members (Manley and Krainer, 2010). For example, SF2/ASF or ASF/SF2 becomes SRSF1; SC35 is renamed as SRSF2; and SRp20, SRp75, SRp40, SRp55, and 9G8 are now known as SRSF3 to SRSF7, respectively. The splicing community has been gradually adopting this new nomenclature, although it is possible that other structurally and functionally related proteins, such as human Tra2α/β and RNPS1, may eventually join this core, once their activities in splicing are further characterized.
Essential roles of SR proteins in constitutive splicing
The founding members of the SR protein family, such as SRSF1 and SRSF2, were discovered for their essential roles in constitutive splicing (Fu and Maniatis, 1990; Ge et al., 1991; Krainer et al., 1991; Zahler et al., 1992; 1993). These SR proteins were found to promote U1 snRNP binding to the 5′ splice site and U2 snRNP binding to the 3′ splice site, and they also bridge the communication between these initial splice site recognition events in the pre-spliceosome and the mature spliceosome (Cho et al., 2011; Fu and Maniatis, 1992; Kohtz et al., 1994; Roscigno and Garcia-Blanco, 1995). Even though such activities have been demonstrated, several widely held assumptions remain to be supported by direct experimental evidence. For example, a key assumption is that the RS domain in SR proteins mediates a protein-protein interaction network to facilitate U1-U2 communication during exon definition, cross intron interactions, and the eventual formation of higher order spliceosome. However, such interactions were mainly postulated based on yeast two-hybrid interactions or in vitro pull-down assays (Hertel and Graveley, 2005; Wu and Maniatis, 1993). Whether these interactions occur and how an SR protein specifically interacts with another SR protein or an RS-domain containing protein during spliceosome assembly awaits direct biochemical and structural evidence. Another key issue is concerned with the ability of the RS domain to directly interact with substrate RNA through its positive charges, which is detectable under specific experimental conditions (Shen and Green, 2004, 2007; Shen et al., 2004), but it is unclear whether this is an obligatory function of SR proteins in splicing.
Function and mechanism of SR proteins in regulated splicing
Perhaps the best-known function of SR proteins is their activities in regulated splicing. Numerous biochemical studies have established the ability of SR proteins to promote splice-site selection by binding to exonic splicing enhancers [ESEs] (Cavaloc et al., 1999; Liu et al., 2000; Liu et al., 1998; Schaal and Maniatis, 1999; Tacke and Manley, 1995). Multiple SR proteins may bind to a set of ESEs to exert additive effects in splice-site selection. Recent studies revealed that SR proteins not only promote exon inclusion, but also induce exon skipping, depending on they interact with pre-mRNA. As illustrated in Figure 1B, penal a, SR protein binding to an exon is positive for it inclusion, but its binding to an intronic sequence has the opposite effect(Dembowski et al., 2012; Erkelenz et al., 2013). Because alternative splicing is the choice of splice sites in competition, it has also been demonstrated (depicted in Figure 1B, panel b) that SR protein binding to the alternative exon promotes exon inclusion whereas SR protein binding to a flanking exon causes skipping of the internal alternative exon (Han et al., 2011a; Sanford et al., 2009). This pair of rules may explain antagonizing activities of different SR proteins in some specific alternative splicing events (Gallego et al., 1997; Ghigna et al., 2005; Lemaire et al., 1999; Solis et al., 2008), which is consistent with recent genome-wide analyses (Pandit et al., 2013; Sanford et al., 2009).
Despite extensive studies on model minigenes, a series of mechanistic issues remain to be addressed. Although it is possible to predict potential binding sites for several important SR proteins in mammalian genomes based on in vitro deduced binding consensus motifs (Cartegni et al., 2003) and in vivo CLIP-seq analysis (Anko et al., 2012; Pandit et al., 2013; Sanford et al., 2009), SR protein interaction with RNA at a specific location is known to subject to influence by multiple other determining factors, including RNA secondary structure and competition with other RNA binding proteins, such as various hnRNP proteins (Lin and Fu, 2007; Long and Caceres, 2009). Therefore, the RNA binding specificity of SR proteins, especially with respect to their binding landscape in mammalian transcriptomes, remains to be fully established. Recent studies illustrate that this critical issue may effectively be approached by structural analysis of SR protein in complex with RNA (Daubner et al., 2012), coupled with genome-wide protein-RNA mapping (Pandit et al., 2013). Accurate definition of actual binding sites of individual SR proteins is essential for eventual elucidation of the “splicing code” in mammalian genomes.
We recently showed that such competition also occurs between different SR proteins, and as result, binding of one SR protein may either enhance or repress binding ofother SR proteins (Pandit et al., 2013). Because multiple SR proteins function on a set of ESEs, which are abundantly present in both alternative and constitutive exons, our recent genome-wide binding and functional analysis on the two founding members of SR proteins SRSF1 and SRSF2 paint a complex picture for SR protein-regulated alternative splicing in vivo, emphasizing a combinatory control that depends on [1] specific binding by individual SR proteins, [2] the locations of individual binding events, [3] synergistic as well as competitive interactions among different SR proteins, and [4]competition with other RNA binding splicing regulators [Figure1B, panel c]. Therefore, the specific regulatory outcomes likely result from the actions of multiple SR proteins in conjunction with other splicing regulators. Such regulatory networks are likely subjected to transcriptional and post-posttranscriptional controls during development and cell differentiation, leading to the establishment of tissue-specific and cell type-specific splicing programs in mammals.
Activities of SR proteins beyond splicing
It is worth mentioning that SR proteins have roles both before and after splicing, including interactions with chromatin (Loomis et al., 2009), coupling with the transcription machinery (Das et al., 2007; Lin et al., 2008), mRNA export out of the nucleus (Huang and Steitz, 2005), regulation of RNA stability via the nonsense mediated RNA decay (Zhang and Krainer, 2004), and translational control by shuttling SR proteins in the cytoplasm (Michlewski et al., 2008; Sanford et al., 2004). Readers are referred to a recent review on these topics (Zhong et al., 2009b) as well as key literature information herein cited. Therefore, studying SR proteins is a gateway to understand a large range of cellular activities in gene expression in mammalian cells.
Regulation of SR proteins by post-translational modifications
Different types of post-translational modifications on SR proteins
At least three types of post-transcriptional modifications are known to occur on SR proteins, including methylation, acetylation, and phosphorylation. Arginine methylation has been detected on many RNA binding proteins, particularly hnRNP proteins (Liu and Dreyfuss, 1995). Arginine methylation in SR proteins and SR-related proteins was first reported on Npl3p in budding yeast (Siebel and Guthrie, 1996). The methylation site was mapped near the end of the protein, adjacent to the phosphorylation site. Interestingly, the relative stable arginine methylation blocks phosphorylation, the latter of which is required for Npl3p to interact with its nuclear import receptor Mtr10p, and as a result, hyper-methylation of Npl3p showed a defective nuclear import phenotype (Yun and Fu, 2000). Because Npl3p has recently been found to play important roles in coupling between transcription and splicing in the nucleus (Kress et al., 2008; Moehle et al., 2012), it will be interesting to determine how such activities might be regulated at the level of nuclear import. More recently, three methylated arginine residues were identified between the two RRMs in the mammalian SR protein SRSF1 (Sinha et al., 2010). Such modification appears to play a positive role in promoting nuclear import of the SR protein, although the modification does not seem to affect the interaction of the SR protein with its nuclear import receptor. Importantly, defective SRSF1 nuclear import could be linked to enhanced activities of the SR protein in translation in the cytoplasm, but attenuated activities in the nucleus, such as the regulation of alternative splicing and coupling with nonsense-mediated mRNA decay (Sinha et al., 2010).
Large-scale proteomic analysis has also revealed extensive lysine acetylation in both SR proteins and SR protein-specific kinases, which may represent an important class of non-histone substrates recognized by various histone actyltransferases [HATs] in mammalian cells (Choudhary et al., 2009). Indeed, the HAT Tip60 was subsequently reported to specifically modify SRSF2, thereby regulating the turnover of the SR protein, and such effect appears to be subject to the counter regulation by the deacetylase HDAC6 (Edmond et al., 2011). These findings raise an intriguing possibility that different SR proteins and regulators of SR proteins may be under control by different combinations of HATs and HDACs, thus constituting a potentially critical regulatory network to modulate splice site selection in mammalian cells.
Perhaps the best understood regulation of SR proteins is through phosphorylation catalyzed by multiple kinases, all of which belong to the CMGC family of kinases (Kannan and Neuwald, 2004). The discovery of SR protein phosphorylation was in fact co-incident with the discovery of the SR protein family because initial antibodies raised against endogenous SR proteins all recognize a phosphoepitope(s) in their RS domains, which dramatically retard the migration of native SR proteins in SDS-PAGE (Roth et al., 1991; Zahler et al., 1992). Because the phosphorylation/dephosphorylation cycle of SR proteins has been linked to all of their activities in the cell [see below], identification and characterization of specific kinases and phosphatases involved have been pursued by many laboratories, including ours, since the discovery of SR proteins two decades ago.
SR protein specific kinases
Our group discovered the first SR protein-specific kinase through purification of a cell cycle activity responsible for the redistribution of SR proteins from highly localized speckled domains in interphase cells to the nucleoplasm when cells enter mitosis (Gui et al., 1994a; Gui et al., 1994b). This activity was found to correspond to a serine kinase specific for the RS domain present in all SR proteins, thus named as SR protein specific kinase 1 or SRPK1 (Gui et al., 1994a). Based on the homology search, SRPK2 and SRPK3 as well as their alternatively spliced products were subsequently discovered in humans and mice (Kuroyanagi et al., 1998; Nakagawa et al., 2005; Nikolakaki et al., 2001; Wang et al., 1998b). These kinases constitute a unique family of kinases characterized by a long divergent spacer sequence that separate the kinase domains into two lobes, a feature common among tyrosine kinases, but rare in serine/threonine kinases (Giannakouros et al., 2011; Nolen et al., 2004).
Interestingly, the SRPK family is conserved in all eukaryotic cells from plant to animals, all the way down to budding yeast [see comprehensive review of all SRPK family members in Giannakouroset al., 2011], even though the genome of budding yeast does not seem to encode for any typical SR protein essential for pre-mRNA splicing, as in higher eukaryotic cells. However, the only yeast SRPK1 family, known as Sky1p, has been shown to phosphorylate several SR protein-related proteins, including Npl3p and Hrb1p (Porat et al., 2006; Siebel et al., 1999). At least in the case of Npl3p, Sky1p-mediated phosphorylation appears to regulate its cellular localization and facilitate protein-protein and protein-RNA interactions, functions that are highly related to those mediated by SRPKs in mammalian cells (Gilbert et al., 2001; Yeakley et al., 1999). Interestingly, while SRPK1 is ubiquitously expressed, SRPK2 is expressed mainly in the nervous system and the expression of SRPK3 is largely confined in muscle cells (Nakagawa et al., 2005; Wang et al., 1998a), indicating that individual SRPK family members may have unique functions in different cell types or during development.
Besides SRPKs, several other kinases or kinase activities have been shown to be able to transfer phosphates to SR proteins in vitro, including cAMP-dependent protein kinase A [PAK] and protein kinase C [PKC] (Colwill et al., 1996b), Akt (Blaustein et al., 2005; Patel et al., 2005), Topoisomerase I(Rossi et al., 1996), dual-specificity tyrosine phosphorylation-regulated kinases [DYRKs] (Aranda et al., 2011; de Graaf et al., 2004), and cyclin-dependent like kinases [Clk1-4] (Colwill et al., 1996a; Colwill et al., 1996b; Duncan et al., 1998). However, only SRPKs and Clks have been directly shown to be responsible for SR protein phosphorylation in vivo through either genetic ablation or chemical inhibition (Fukuhara et al., 2006; Hayes et al., 2006; Yomoda et al., 2008; Zhong et al., 2009a). Clk-1 was initially linked to SR proteins in a yeast two-hybrid screen, and like SRPKs, overexpression of a Clk was able to induce redistribution of SR proteins from nuclear speckles to the nucleoplasm (Colwill et al., 1996b). A similar activity was also reported with DYRK1A (Alvarez et al., 2003). Although SRPKs seem to be highly specific for SR proteins and SR protein-related splicing factors, these kinases clearly have other substrates implicated in other cellular functions(Giannakouros et al., 2011).
A major distinction between the SRPK and Clk families of kinases is their subcellular localization. SRPKs are detected in both the cytoplasm and the nucleus with overexpressed kinase largely anchored in the cytoplasm (Wang et al., 1998a). SRPK cellular partition is regulated, at least in part, by the unique spacer domain in each SRPK family member, as deletion of the spacer shifts the kinase to the nucleus (Ding et al., 2006; Ngo et al., 2008; Nolen et al., 2001). Initial evidence suggests that the spacer has little effect on the kinase activity, but a more recent study indicates that it can enhance the catalysis by increasing the rate-liming ADP release step in the kinase reaction (Aubol et al., 2012). In contrast, all Clk family members are constitutively localized in the nucleus and show extensive colocalization with SR proteins in nuclear speckles (Colwill et al., 1996b; Duncan et al., 1998). These differential localization patterns suggest that the two families of SR protein kinases may participate in a “relay” in phosphorylating SR proteins to synergistically regulate their activities in the cell (Ngo et al., 2005). For example, cytoplasmic SRPKs may be responsible for initial phosphorylation of SR proteins to facilitate their nuclear import, whereas nuclear Clks may fine-tune the phosphorylation state of SR proteins in the nucleus. Therefore, the two families of kinases may function in a synergistic and/or complementary fashion to regulate splicing, RNA export, and other processes of RNA metabolism in mammalian cells.
Unlike most other kinases that transfer phosphate to their substrate by a “hit and run” mechanism, both SRPK and Clk families of kinases appear to act like a “polymerizing kinase” (Ghosh and Adams, 2011). In other words, once the kinase binds to its substrate, it continues to transfer multiple phosphates to adjacent Ser/Arg dipeptides without dissociating from the substrate after each round of the kinase reaction. This processive mode of action has been demonstrated on SRPK1 in catalyzing a chain of reactions within the RS domain of SRSF1 (Aubol et al., 2003). The structural basis for this unusual property of SRPK1 is due to a MAPK insert in the large lobe of the kinase, which serves as the docking site for an SR protein substrate to bind (Hagopian et al., 2008; Ngo et al., 2008). For further mechanistic details, readers are referred to a recent comprehensive review on SRPK1-catalyzed phosphorylation reaction(Ghosh and Adams, 2011).
Regulation of splicing by phosphorylation
Requirement of the phosphorylation/dephosphorylation cycle for splicing
The requirement for reversible phosphorylation in the splicing reaction was first demonstrated using various phosphatase inhibitors, showing that inhibition of PP2A blocked the second step catalysis whereas inhibition of PP1 and PP2A together blocked both steps in nuclear extracts, indicating that dephosphorylation is required for splicing activity, but not for splicing complex formation (Mermoud et al., 1992). Conversely, while purified PP1 blocked the formation of the pre-spliceosome as well as its conversion to the full spliceosome, it showed no effect on splicing after spliceosome assembly (Mermoud et al., 1994; Stojdl and Bell, 1999). These data suggest that phosphorylation is required for spliceosome assembly, but dephosphorylation is critical for catalysis within the assembled spliceosome. Reversible SR protein phosphorylation appears to underlie this phosphorylation/dephosphorylation requirement for spliceosome progression from assembly to catalysis because purified SR proteins from insert cells (which are known to be properly phosphorylated) were able to restore the spliceosome formation and splicing activity to PP1-inhibited extracts (Mermoud et al., 1994). A thiophosphorylated SR protein, which is resistant to phosphatases, could complement S100 extracts (the cytosolic fraction generated during the preparation of nuclear extracts; this fraction, contains all essential spliceosomal components except SR proteins) for spliceosome assembly, but the assembled spliceosome was unable to become splicing competent (Cao et al., 1997; Roscigno and Garcia-Blanco, 1995; Xiao and Manley, 1997, 1998). These observations led to a general model in which SR protein phosphorylation is necessary for assembly of spliceosomal components whereas dephosphorylation is essential for splicing catalysis.
While it is clear that SR proteins need to be dephosphorylated for RNA catalysis in the mature spliceosome, we have little clue about when and how such dephosphorylation is triggered. Purified spliceosomes do not seem to contain a stoichiometric amount of a phosphatase, indicating that PP1 and/or PP2A may act as diffusible regulators, rather than as built-in components of the spliceosome. In contrast, SRPKs are assembled into the spliceosome with SRPK1 being specifically associated with U1 snRNP and SRPK2 with U4/6-U5 tri-snRNP containing complexes (Mathew et al., 2008). These findings suggest that different SRPK family members may have both overlapping [i.e. phosphorylating SR proteins] and distinct functions [i.e. phosphorylating other RS domain-containing splicing factors] in splicing, although it is currently unclear whether any spliceosome complexes are associated with SRPK3 or Clk. In addition, the spliceosome-associated SRPK1 or SRPK2 may continue to catalyze phosphorylation of some RS domain-containing proteins [such as Prp28] to ensure spliceosome assembly in a orderly fashion (Mathew et al., 2008). The presence of SRPKs in the spliceosome may also provide a timing device for specific phosphatases to act on specific substrates for the spliceosome to progress into the active form. These possibilities are in line with the observed genetic interaction of the SRPK family member [Sky1p] with several second step splicing factors [Prp8 and Prp17/Slu4] in budding yeast (Dagher and Fu, 2001). Thus, while the evidence casts SRPKs as critical players in spliceosomal control, we still have much to learn about their specific roles in these events.
Distinct functions of differentially phosphorylated SR proteins in splicing
The requirement for phosphorylated SR proteins to facilitate spliceosome assembly is at least two-fold. One is to prevent non-specific interaction of their highly positively charged RS domains with RNA and the other is to allow phosphorylation-dependent interaction of SR proteins with the U1 70K protein and likely other RS domain-containing proteins to establish network interactions critical for spliceosome assembly (Tacke et al., 1997; Xiao and Manley, 1998; Yeakley et al., 1999). The requirement for neutralizing positive charges by phosphorylation in the RS domain agrees with the current model where SR proteins bind via their RRMs toexonic splicing enhancers and subsequently promote protein-protein interactions through their RS domains, but appears incompatible to the proposed charge-based interactions of the RS domain with the 3′ splice site in the early phases of spliceosome assembly (Shen and Green, 2004; Shen et al., 2004). However, such seemly incompatible requirements might be satisfied by progressive phosphorylation of SR proteins.
A more recent study provides evidence for such a scenario, suggesting that SR proteins may undergo progressive phosphorylation in early steps of spliceosome assembly. In this study, only partially phosphorylated SRSF1 [in its RS1 subdomain] was found to bind with high affinity to its target RNA, compared to both unphosphorylated and fully phosphorylated SRSF1 [in both RS1 and RS2 subdomains] (Cho et al., 2011). This study further demonstrated several previously unrecognized interactions between the RRM of SRSF1 and the RRM of the U1 70K protein and between the RRM of the SR protein and its RS domain. Interestingly, these protein-protein interactions appear to involve an RRM interface distinct from that responsible for binding to RNA in each case. Importantly, only fully phosphorylated SRSF1 could enhance the formation of a ternary complex containing SRSF1, target RNA, and the U1 70K protein (Cho et al., 2011). These findings therefore suggest a new model, as illustrated in Figure 2, where partially phosphorylated SRSF1 binds to specific splicing enhancers and further phosphorylation subsequently induces a switch from intra- to intermolecular interactions to promote the formation of the ternary complex with U1 snRNP. The presence of SRPK1 in the U1 complex may thus engineer such progressive phosphorylation of SR proteins to facilitate the switch and thus the progression of spliceosome assembly. It remains to be determined whether other SR proteins use this two-step mechanism to promote U1 snRNP binding to a functional 5′ splice site.
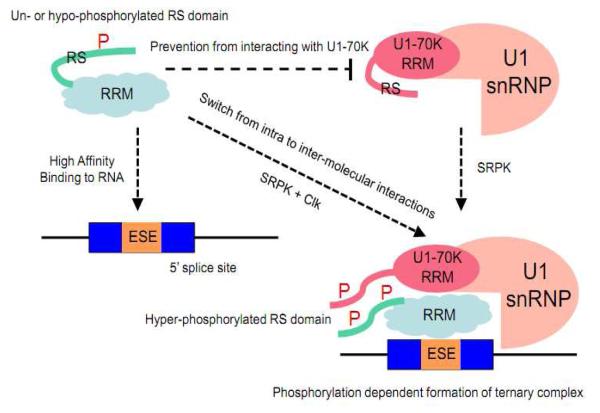
Figure 2.
Phosphorylation regulation of SR proteins in spliceosome assembly. The RS domains of both the SR protein SRSF1 and the SR-related protein U1-70K can engage in intra- and inter-molecular interactions. In un- or hypo-phosphorylated state, the RS domain is engaged in an intra-molecular interaction with one interface of the SRSF1 RRM domain, which the other interface binds RNA with high affinity. However this prevents the SRSF1 from interacting with the U1 70K protein. Hyper-phosphorylation of the RS domain of SRSF1 by SRPK1 plus Clk is proposed to switch this intra-molecular interactions to inter-molecule interactions through protein-protein interactions via both the RS domains and RRMs between SRSF1 and U1-70K, allowing the formation of the ternary complex containing the SR protein, U1 snRNP, and the RNA containing a functional 5′ splice site.
The study described above on SRSF1 illustrates that SR proteins may require a specific phosphorylation state to function properly in splicing. This has been well illustrated earlier with SRSF10 [previously known as SRp38], which does not seem to have any activity in splicing in its fully phosphorylated state, but acts as a splicing activator when it is partially dephosphorylated (Shin et al., 2004; Shin and Manley, 2002). Interestingly, the cell appears to use this mechanism to repress splicing during mitosis or in response to heat shock. These studies illustrate that an SR protein may have completely distinct functions in different phosphorylation states, although SRSF10 appears to exhibit some unique properties and thus possess a particular sensitivity to such differential regulation, compared to other SR family members (Shin et al., 2005). These findings are fully consistent with dynamic phosphorylation of SR proteins as a functional consequence of constant competition between kinases and phosphatases in the cell to regulate their functions in splicing(Shi and Manley, 2007; Zhou et al., 2012).
Phosphorylation of SR protein recycling in the cell
Besides the importance of the phosphorylation/dephosphorylation cycle for SR proteins to function in the splicing reaction, phosphorylation has also been demonstrated to regulate SR protein recycling in the cell. As shown in Figure 3, SRPK1-mediated phosphorylation plays an important role in facilitating nuclear import of SR proteins(Kataoka et al., 1999; Lai et al., 2000; Lai et al., 2001; Yun and Fu, 2000),although not all SR proteins have to enter the nucleus in such a phosphorylation dependent fashion(Hamelberg et al., 2007; Yun et al., 2003).After entering the nucleus, SR proteins in a proper phosphorylation state may be directly recruited to nascent pre-mRNA for co-transcriptional splicing (Misteli et al., 1998).
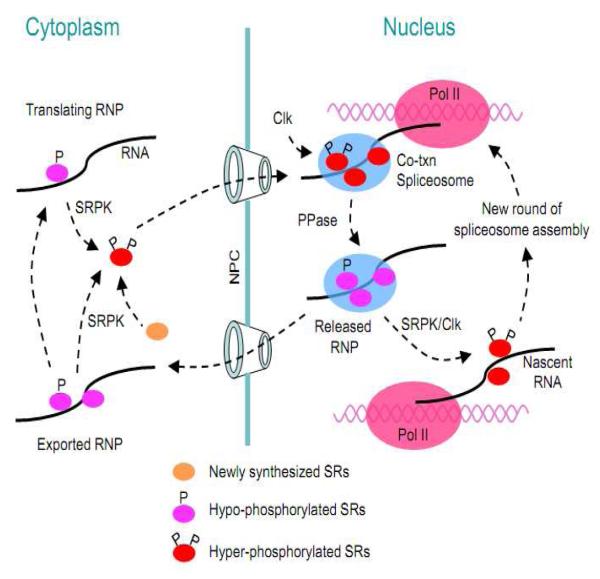
Figure 3.
Phosphorylation regulation of intercellular trafficking of SR proteins. SR proteins are initially phosphorylated by the SRPK family of kinases, which promotes nuclear import of SR proteins. After entering the nucleus, SR proteins may be further phosphorylated by the Clk family of kinases. Hyper-phosphorylated SR proteins are recruited to nascent pre-mRNA for co-transcriptional splicing. The splicing reaction requires dephosphorylation of SR proteins within the spliceosome. SR proteins associated with mRNA appear to have two alternative recycling pathways. One is through re-phosphorylation, which may release the SR protein from spliced mRNA, allowing them to recycle within the nucleus. Alternatively, hypo-phosphorylated SR proteins on spliced mRNA may be exported out of the nucleus, which functions to enhance mRNA export and stimulate translation in the cytoplasm. Re-phosphorylation in the cytoplasm may facilitate the release of the SR proteins from spliced mRNA and then promote their re-import back to the nucleus.
At the end of splicing, there are probably multiple routines for SR proteins to recycle in the cell. The first is for SR proteins to become re-phosphorylated within the nucleus to participate in the next round of the splicing reaction. This is consistent with the ability of overexpressed SR protein kinases to release SR proteins from post-splicing complexes, which tend to become aggregated in the speckled nuclear domains (Colwill et al., 1996b; Gui et al., 1994a; Ngo et al., 2005; Wang et al., 1998a).
Alternatively, dephosphorylated SR proteins may remain associated with spliced mRNA, thereby escorting spliced mRNA to the mRNA export machinery, because only hypo-phosphorylated SRSF1 is able to bind TAP, a key co-factor required for mRNA export (Huang et al., 2003; Huang et al., 2004; Lai and Tarn, 2004). A recent study showed that multiple SR proteins are part of the exon junction complex [EJC] on spliced mRNA (Singh et al., 2012), consistent with roles of SR proteins in mRNA export (Reed and Cheng, 2005)and non-sense mediated RNA decay (Zhang and Krainer, 2004). This leads a fraction of SR proteins to travel with spliced mRNA to the cytoplasm where they enhance the translation of the mRNA (Sanford et al., 2005; Sanford et al., 2004). Re-phosphorylation of SR proteins in the cytoplasm may serve to dissociate SR proteins from spliced mRNA and facilitate their re-import back to the nucleus, although this has only been demonstrated in budding yeast on the SR-like protein Npl3p (Gilbert et al., 2001). These activities may contribute to the shuttling property associated with all SR proteins with the exception of SRSF2 (Caceres et al., 1998; Sapra et al., 2009).
It is currently unknown what determines the recycling of SR proteins within the nucleus versus through the cytoplasm. In light of the observation that purified PP1 can also disrupt post-splicing complexes in nuclear speckles (Misteli and Spector, 1996), it is conceivable that SR protein kinases may direct them to recycle within the nucleus while SR protein phosphatases may promote them to recycle through the cytoplasm. This may explain why SRSF2 does not shuttle because it is highly resistant to dephosphorylation by phosphatases (Lin et al., 2005), which may be the underlying mechanism for the identified nuclear retention signal in the RS domain of this unique SR protein (Cazalla et al., 2002). Importantly, such different routes of SR protein recycling may help create different pools of SR proteins in the cell and differential distribution of SR proteins in different cellular compartments may affect SR protein-regulated splicing because all SR proteins are known to affect alternative splicing in a dosage dependent manner. This concept has been proposed for hnRNP A1 in response to a stress signaling (van der Houven van Oordt et al., 2000).
It is also important to point out that shuttling of SR proteins out of the nucleus does not seem to be essential for cell viability as indicated by the functional complementation assay performed on SRSF1-depleted cells (Lin et al., 2005). We recently show that SRSF2 binds to both pre-mRNA and spliced mRNA in the cell (Pandit et al., 2013). Because SRSF2 does not shuttle between the nucleus and the cytoplasm, this observation implies that this non-shuttling SR protein has to be removed from spliced mRNA prior to export, which may constitute a new regulatory step in mRNA export. The non-shuttling property of SRSF2 is also in line with its involvement in transcription elongation (Lin et al., 2008).
SR protein kinases in signaling
Signal-induced splicing
The term “regulation” has been broadly used in the splicing field to refer to any changes in splicing that are under the control of cis-acting elements or trans-acting factors. Strictly speaking, however, regulation should be an inducible event for cells to respond to an internal cue or an external signal. Although various splicing regulators are targets of signaling molecules and impact alternative splicing as highlighted in Figure 4, little is known about how these signals are transduced to the nucleus to regulate splicing either through existing or novel signaling pathways.
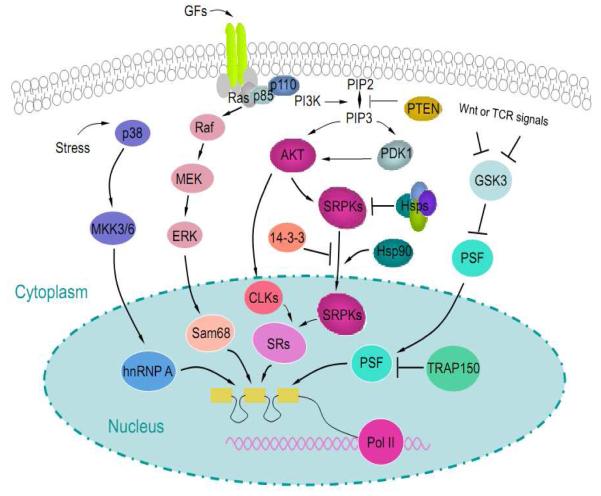
Figure 4.
Regulation of alternative splicing by signaling. The p38 kinase transduces stress signals to hnRNP A1 by the MAPK pathway. The Wnt signaling pathway, by regulating GSK3, phosphorylates and potentiates the activity of PSF by releasing the splicing regulator from the inhibitory complex with TRAP150. Growth factor signals activate both the Raf-MEK-ERK pathway to modify Sam68 and thePI3K-Akt pathway. Activated Akt binds to SRPKs and induces their release from the inhibitory heat shock complexes. This triggers nuclear translocation of SRPKs, which is antagonized by 14-3-3 proteins and enhanced by Hsp90. In the nucleus, SRPKs act in synergy with Clk to phosphorylate SR proteins. There is evidence suggesting that activated Akt may also regulate Clk in the nucleus. In addition, it is likely that signal transduction pathways highlighted here may induce post-translational modification of multiple splicing regulators, which may function in a combinatorial fashion to modulate splice site selection in the nucleus. The unique spectrum of splicing regulators, coupled with distinct activities of individual signaling pathways, may be responsible for specific splicing programs in different cell types or during development.
Several studies have illustrated common experimental approaches for studying signal-dependent splicing regulation by connecting a signaling molecule to a specific splicing regulator, and then to a particular alternative splicing event, which has been extensively reviewed (Heyd and Lynch, 2011; Lynch, 2007; Shin and Manley, 2004). For example, in response to osmotic stress, activated MKK-p38 modifies hnRNP A1 to induce its accumulation in the cytoplasm (van der Houven van Oordt et al., 2000). As hnRNP A1 is a general splicing repressor, its restriction in the cytoplasm may de-repress many alternative exons, but this hypothesis has remained to be directly tested by global analysis. In activated T cells, ERK [extracellular signal-regulated kinase] phosphorylates SAM68 [SRC-associated in mitosis, 68 kDa] and alters the activity of this RNA binding protein in the regulation of alternative splicing of the CD44 gene (Matter et al., 2002). Alternative splicing of this important cell surface receptor appears to also subject to regulation by the Wnt/β-catenin and Akt pathways via different SR proteins (Goncalves et al., 2008; 2009). Insulin has been found to alter alternative splicing of the PKCβII gene via induced phosphorylation of the SR protein SRSF5 [formerly known as SRp40] in response to the activation of the PI3K/Akt pathway (Chalfant et al., 1995; Patel et al., 2004; Patel et al., 2005). However, it has been unclear whether PKCβII splicing is solely regulated by SRSF5 because growth factor signaling has also been shown to affect the phosphorylation state of SRSF1 and SRSF7 via the Akt pathway (Blaustein et al., 2005). It is also unclear whether increased phosphorylation of various SR proteins in growth factor-treated is mediated directly by activated Akt or indirectly via an SR protein kinase or both (Jiang et al., 2009).
The studies described above and many other examples illustrate the regulation of specific alternative splicing events by individual signaling molecules. However, we clearly lack the global view on splicing regulation in response to a specific signaling event in terms of [1] how many splicing regulators are induced or modified, [2] how these regulators may act together to change the splicing program in the cell, and [3] how such splicing program is responsible for some specific biological outputs. These efforts are essential to understand the regulation of the transcriptome at the splicing level in development and disease.
SR protein-specific kinases as transducers of growth signals
A recent effort in our lab illustrates the use of some modern global approaches for dissecting a critical signal transduction pathway via SR protein-specific kinases (Zhou et al., 2012). As shown in Figure 4, the localization of the splicing kinases in the cytoplasm appears to be particularly suitable for them to serve as signal transducers for splicing regulation. Previous studies showed that a large spacer insert domain anchors SRPKs in the cytoplasm of both yeast and mammals (Ding et al., 2006; Siebel et al., 1999; Takeuchi and Yanagida, 1993; Tang et al., 2012c; Wang et al., 1998b). In response to a signal, these SR protein kinases translocate to the nucleus to phosphorylate SR proteins (Zhong et al., 2009a). The Ye lab was the first to show that activated Akt could directly transfer phosphates to two specific sites in SRPK2, which induces SRPK2 nuclear translocation, leading to the phosphorylation of several SR and SR-related proteins, including SRSF2 and Acinus (Jang et al., 2009; Jang et al., 2008). In one of these studies, enhanced SRSF2 phosphorylation was further linked to the induction of cyclin D in a p53 dependent manner, thus forcing neurons to re-enter mitosis, resulting in mitotic catastrophe and neuronal death. Interestingly, this pathway appears to play a key part in a mouse brain stroke model (Jang et al., 2009).
The study on SRPK2 in neurons suggests that the Akt pathway may regulate other SRPKs. This appears to be the case in EGF-treated mammalian cells where SRPK1 phosphorylation was induced (Zhou et al., 2012). However, unlike SRPK2, there is no consensus motif for Akt-mediated phosphorylation in SRPK1. Instead, upon activation, Akt appears to form a stable complex with SRPK1 that induces SRPK1 autophosphorylation. This releases SRPK1 from heat shock protein complexes, allowing the activated SRPK1 to translocate to the nucleus and trigger SR protein hyper-phosphorylation [Figure 4]. By surveying a large set of alternative splicing events using an mRNA isoform-sensitive technology, EGF was found to induce a large splicing program that could be blocked by inhibitors against specific components of the PI3K/Akt pathway, but not by inhibitors against the JAK/STAT, MEKK/ERK or PKC pathways (Zhou et al., 2012). Most importantly, the entire EGF-induced splicing program was diminished when SRPK1, SRPK2, or both were inactivated by RNAi. Furthermore, Rapamycin inhibition of the mTOR pathway, which is a major signal branch in the Akt pathway for transcriptional and translational control in mammals, was found to have little effect on the EGF-invoked splicing program. This is somewhat surprising, at least initially, because mTOR has been shown to play a key role in SRSF1 overexpression-induced cellular transformation (Karni et al., 2007; Karni et al., 2008). A more recent study suggests that mTOR activities could be modulated by SRSF1-induced splicing events, indicating that mTOR is a target, rather than a signal transducer, for regulated splicing (Ben-Hur et al., 2012). One may further imagine various feed-back and/or feed-forward loops in which growth factors may transduce signals via Akt and SRPKs to regulate splicing in the nucleus and some specific splicing events may in turn reinforce the growth signals in synergy with activated mTORs to regulate cell proliferation and transformation in mammalian cells.
Molecular chaperones involved in modulating splicing regulators
Molecular chaperones have long been known to function not only in assisting folding of newly synthesized proteins but also in controlling various signal transduction events. This also applies to splicing regulation via both SR proteins and SR protein kinases. As mentioned earlier, SRSF10 dephosphorylation was rapidly induced by heat shock, which converts it from a splicing activator to repressor. The heat shock protein Hsp27 appears to confer a “thermotolerance” to the cell against a mild heat shock by preventing SRSF10 dephosphorylation (Shi et al., 2011) or by facilitating its rephosphorylation after heat shock (Marin-Vinader et al., 2006).
The major heat shock proteins Hsp70 and Hsp90 have been found to associate with SRPKs via their co-chaperones, which is responsible for anchoring the splicing kinases in the cytoplasm (Zhong et al., 2009a). As illustrated in Figure 4, Akt-dependent activation of SRPK1 not only induces the release of the kinase from the Hsp70-containing complex, but also triggers the re-connection of the kinase with Hsp90, and this rearrangement with molecular chaperones appears to play a critical role in facilitating SRPK1 nuclear translocation (Zhou et al., 2012). Finally, the abundant 14-3-3 family members have also been implicated in controlling SR protein phosphorylation in the cell. These proteins appear to bind and protect SRSF10 from dephosphorylation (Shi and Manley, 2007) as well as limit the amount of Akt-activated SRPKs that can translocate to the nucleus (Jang et al., 2009; Zhou et al., 2012). These findings suggest that molecular chaperones and 14-3-3 proteins provide check and balance for SR proteins and SR protein kinases to function properly in signal-induced splicing responses.
SR proteins and SR protein kinases in development and disease
Essential role of SR proteins and SR proteins kinases in development
Genetic studies published to date indicate that SR proteins are each essential for animal development (Ding et al., 2004; Feng et al., 2009; Jumaa et al., 1999; Wang et al., 2001). Tissue-specific ablation of individual SR proteins has also been shown to cause aberrant splicing in different tissues or cell types (Sen et al., 2013; Wang et al., 2001; Xu et al., 2005). Our unpublished studies indicate that both SRPK1 and SRPK2 are also essential for mouse development. This is consistent with the essential role of a Drosophila SRPK homologue in spindle microtubule assembly during meiosis (Loh et al., 2012) and with the functional requirement of the C. elegance SRPK homologue for germline development (Kuroyanagi et al., 2000). However, it has been unclear whether SRPKs function through SR proteins and/or other substrates, such as the Lamin B receptor and P1 protamine (Papoutsopoulou et al., 1999a; Papoutsopoulou et al., 1999b). It is also plausible that SRPKs may phosphorylate a variety of RS domain-containing proteins that are critical for cell cycle progression. This includes SON, an SR-like RNA binding protein that has been demonstrated to play a key role in mitosis by regulating the splicing efficiency of microtube and other critical components of the cell cycle machinery (Ahn et al., 2011; Sharma et al., 2011).
SR proteins and SR protein kinases in cancer
SR proteins have been implicated in cancer. In particular, SRSF1 and SRSF3 appear to function as oncoproteins in human cancer because their overexpression is able to promote anchorage-independent cell growth in vitro and tumor growth in nude mice (Jia et al., 2010; Karni et al., 2007). In fact, multiple other SR proteins have been found to regulate cell proliferation and apoptosis in diverse cancer cell types via other critical splicing events (Cohen-Eliav et al., 2013; Gautrey and Tyson-Capper, 2012; Jia et al., 2010; Stickeler et al., 1999; Tang et al., 2012a). Most strikingly, a burst of recent studies demonstrated that specific mutations in SRSF2 are tightly linked to specific leukemia in patients (Lasho et al., 2012; Meggendorfer et al., 2012;Patnaik et al., 2013; Yoshida et al., 2011). These findings have strongly implicated SR proteins as specific disease genes.
At the molecular level, SRSF1 has been found to regulate alternative splicing of multiple signaling molecules, including MNK2 and S6K1, to promote MAK-independent phosphorylation of eIF4E, thereby enhancing protein synthesis in the cytoplasm (Karni et al., 2007; Karni et al., 2008). More recent studies from the Krainer lab demonstrated that SRSF1 is a direct target of c-Myc (Das et al., 2012), and SRSF1 and Myc act in synergy to induce cellular transformation (Anczukow et al., 2012). These studies have linked aberrant SR protein expression to some established oncogenic pathways. In addition, SR proteins are also known to affect genome instability in both chicken and human cells (Li and Manley, 2005; Xiao et al., 2007), which may contribute to cancer progression.
SR protein kinases have also been implicated in cancer. The SRPK family of kinases has been reported to be overexpressed in various human cancers (Hayes et al., 2006; Hayes et al., 2007). In particular, SRPK2 overexpression could enhance cell proliferation of leukemia cells, indicating a direct role in tumorigenesis (Jang et al., 2008). Similarly, all Clk kinase family members appeared overexpressed during erythroleukemia cell differentiation (Garcia-Sacristan et al., 2005). More recently, both SRPK1 and SRPK2 were found to be overexpressed in non-small cell lung cancer and their overexpression correlated with hyper-phosphorylation of SRSF1 and SRSF2 (Gout et al., 2012). Curiously, genetic ablation of the SRPK homolog SKY1 conferred a cisplatin-resistant phenotype in budding yeast (Schenk et al., 2001) and SRPK1 reduction was also linked to several cisplatin-resistant human tumors(Krishnakumar et al., 2008; Odunsi et al., 2012; Schenk et al., 2002; Schenk et al., 2004). These findings suggest that both over- and under regulation of SRPKs may contribute to different aspects of human cancer. Although the mechanism has remained elusive in most of these cases, the tumorigenic activities of both SR proteins (Li and Manley, 2005; Wang et al., 2008b; Xiao et al., 2007)and SR protein kinases (Paulsen et al., 2009) are likely linked to their functions in maintaining genome stability and generating aberrant protein isoforms by changing splicing patterns (Goncalveset al., 2008; 2009; Christofk et al., 2008).
Angiogenesis has long been linked to alternative splicing of VEGF, which produces both pro- and anti-angiogenic isoforms, and SR proteins are known to play a critical role in regulated VEGF splicing (Qiu et al., 2009). Recent studies revealed that inactivation of the tumor suppressor WT1 gene caused derepression ofSRPK1 and overexpression of this splicing kinase, stimulated nuclear import ofSRSF1, leading to the increased production of the pro-angiogenic isoform of VEGF (Nowak et al., 2010). The small molecule inhibitor of SRPK1 or SRPK1 RNAi was found to effectively block angiogenesis, thus retarding tumor growth in nude mice (Amin et al., 2011). These findings suggest that SRPK1 may serve as a potential therapeutic target for cancer treatment (Oltean et al., 2012). However, it has also been reported that SRSF2 appears to play an opposite role in VEGF splicing by favoring the production of the anti-angiogenic isoform of the VEGF gene (Merdzhanova et al., 2010). Considered together, these observations indicate that different SR proteins may compete with one another in the regulation of VEGF splicing. It will be particularly interesting to determine in future studies how different SR protein kinases are involved in modulating nuclear import and splicing activities of various SR proteins in different cell types to regulate angiogenesis.
Conclusions
Pre-mRNA splicing is an essential step in gene expression in eukaryotic cells and alternative splicing generates transcriptome diversity, which not only alters the proteome but also introduces additional layers of regulation. SR proteins are unique RNA binding proteins involved in both constitutive and regulated splicing. A major challenge is to understand how SR proteins mediate protein-RNA and protein-protein interaction networks in the spliceosome, which may be ultimately elucidated by structural analysis of splicing complexes at different stages of assembly. As an important class of splicing regulators, SR proteins are widely involved in modulating splice-site selection. Because of their overlapping and distinct activities in RNA binding and protein-protein interactions, we have much to learn about how splicing is regulated through both collaborative and competitive effects of different SR proteins and SR-regulated splicing regulators in mammalian genomes. SR proteins are also known to play diverse roles in coupling splicing with multiple other steps in gene expression. Therefore, despite their discovery two decades ago, it appears that we have a long way to go before a clear picture of their biological functions and action mechanisms in development and disease comes into focus.
Like most other regulators of gene expression, SR proteins are extensively modified at the post-translational level. Phosphorylation catalyzed by specific SR protein kinases plays a considerable role in controlling SR protein activities. Recent evidence suggests that SR proteins are not simply activated by phosphorylation and inactivated by dephosphorylation in their RS domains. Instead, site-specific or region-specific phosphorylation of SR proteins appears to modulate their functions in different stages of RNA processing as well as recycling in the cell. Furthermore, phosphorylation and other modes of post-translational modification of SR proteins have the potential to connect SR proteins to diverse signaling networks in the cell. The recent discovery of SRPKs as the major branch of the Akt pathway has linked growth signals to regulated splicing, which may directly contribute to the function of SR proteins in cellular transformation and other oncogenic processes. This discovery opens new doors for understanding the regulation of RNA metabolism in mammalian cells and developing SR protein-specific kinases as a new class of therapeutic targets for treating cancer and other human diseases.
Acknowledgements
The authors thank Drs. J. Adams and G. Ghosh for critical comments on the manuscript. Works in the authors’ laboratory are supported by NIH grants [GM049369 and GM052872] to X-D.F.
References
- Ahn EY, DeKelver RC, Lo MC, Nguyen TA, Matsuura S, Boyapati A, Pandit S, Fu XD, Zhang DE. SON controls cell-cycle progression by coordinated regulation of RNA splicing. Mol Cell. 2011;42:185–198. doi: 10.1016/j.molcel.2011.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez M, Estivill X, de la Luna S. DYRK1A accumulates in splicing speckles through a novel targeting signal and induces speckle disassembly. J Cell Sci. 2003;116:3099–3107. doi: 10.1242/jcs.00618. [DOI] [PubMed] [Google Scholar]
- Amin EM, Oltean S, Hua J, Gammons MV, Hamdollah-Zadeh M, Welsh GI, Cheung MK, Ni L, Kase S, Rennel ES, et al. WT1 mutants reveal SRPK1 to be a downstream angiogenesis target by altering VEGF splicing. Cancer Cell. 2011;20:768–780. doi: 10.1016/j.ccr.2011.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anczukow O, Rosenberg AZ, Akerman M, Das S, Zhan L, Karni R, Muthuswamy SK, Krainer AR. The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat Struct Mol Biol. 2012;19:220–228. doi: 10.1038/nsmb.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anko ML, Muller-McNicoll M, Brandl H, Curk T, Gorup C, Henry I, Ule J, Neugebauer KM. The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol. 2012;13:R17. doi: 10.1186/gb-2012-13-3-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda S, Laguna A, de la Luna S. DYRK family of protein kinases: evolutionary relationships, biochemical properties, and functional roles. FASEB J. 2011;25:449–462. doi: 10.1096/fj.10-165837. [DOI] [PubMed] [Google Scholar]
- Aubol BE, Chakrabarti S, Ngo J, Shaffer J, Nolen B, Fu XD, Ghosh G, Adams JA. Processive phosphorylation of alternative splicing factor/splicing factor 2. Proc Natl Acad Sci U S A. 2003;100:12601–12606. doi: 10.1073/pnas.1635129100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubol BE, Plocinik RM, McGlone ML, Adams JA. Nucleotide release sequences in the protein kinase SRPK1 accelerate substrate phosphorylation. Biochemistry. 2012;51:6584–6594. doi: 10.1021/bi300876h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, Kim T, Misquitta-Ali CM, Wilson MD, Kim PM, Odom DT, Frey BJ, Blencowe BJ. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- Ben-Hur V, Denichenko P, Siegfried Z, Maimon A, Krainer A, Davidson B, Karni R. S6K1 alternative splicing modulates its oncogenic activity and regulates mTORC1. Cell Rep. 2012;3:103–115. doi: 10.1016/j.celrep.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaustein M, Pelisch F, Tanos T, Munoz MJ, Wengier D, Quadrana L, Sanford JR, Muschietti JP, Kornblihtt AR, Caceres JF, et al. Concerted regulation of nuclear and cytoplasmic activities of SR proteins by AKT. Nat Struct Mol Biol. 2005;12:1037–1044. doi: 10.1038/nsmb1020. [DOI] [PubMed] [Google Scholar]
- Busch A, Hertel KJ. Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip Rev RNA. 2012;3:1–12. doi: 10.1002/wrna.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres JF, Screaton GR, Krainer AR. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 1998;12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W, Jamison SF, Garcia-Blanco MA. Both phosphorylation and dephosphorylation of ASF/SF2 are required for pre-mRNA splicing in vitro. RNA. 1997;3:1456–1467. [PMC free article] [PubMed] [Google Scholar]
- Carrillo Oesterreich F, Bieberstein N, Neugebauer KM. Pause locally, splice globally. Trends Cell Biol. 2011;21:328–335. doi: 10.1016/j.tcb.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaloc Y, Bourgeois CF, Kister L, Stevenin J. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA. 1999;5:468–483. doi: 10.1017/s1355838299981967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- Cazalla D, Zhu J, Manche L, Huber E, Krainer AR, Caceres JF. Nuclear export and retention signals in the RS domain of SR proteins. Mol Cell Biol. 2002;22:6871–6882. doi: 10.1128/MCB.22.19.6871-6882.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfant CE, Mischak H, Watson JE, Winkler BC, Goodnight J, Farese RV, Cooper DR. Regulation of alternative splicing of protein kinase C beta by insulin. J Biol Chem. 1995;270:13326–13332. doi: 10.1074/jbc.270.22.13326. [DOI] [PubMed] [Google Scholar]
- Cho S, Hoang A, Sinha R, Zhong XY, Fu XD, Krainer AR, Ghosh G. Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly. Proc Natl Acad Sci U S A. 2011;108:8233–8238. doi: 10.1073/pnas.1017700108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Cohen-Eliav M, Golan-Gerstl R, Siegfried Z, Andersen CL, Thorsen K, Orntoft TF, Mu D, Karni R. The splicing factor SRSF6 is amplified and is an oncoprotein in lung and colon cancers. J. Patho. 2013;l229:630–639. doi: 10.1002/path.4129. [DOI] [PubMed] [Google Scholar]
- Colwill K, Feng LL, Yeakley JM, Gish GD, Caceres JF, Pawson T, Fu XD. SRPK1 and Clk/Sty protein kinases show distinct substrate specificities for serine/arginine-rich splicing factors. J Biol Chem. 1996a;271:24569–24575. doi: 10.1074/jbc.271.40.24569. [DOI] [PubMed] [Google Scholar]
- Colwill K, Pawson T, Andrews B, Prasad J, Manley J, Bell J, Duncan P. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distriubtion. EMBO J. 1996b;15:265–275. [PMC free article] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagher SF, Fu XD. Evidence for a role of Sky1p-mediated phosphorylation in 3′ splice site recognition involving both Prp8 and Prp17/Slu4. RNA. 2001;7:1284–1297. doi: 10.1017/s1355838201016077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell. 2007;26:867–881. doi: 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- Das S, Anczukow O, Akerman M, Krainer AR. Oncogenic splicing factor SRSF1 is a critical transcriptional target of MYC. Cell Rep. 2012;1:110–117. doi: 10.1016/j.celrep.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubner GM, Clery A, Jayne S, Stevenin J, Allain FH. A syn-anti conformational difference allows SRSF2 to recognize guanines and cytosines equally well. EMBO J. 2012;31:162–174. doi: 10.1038/emboj.2011.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 2010;24:2343–2364. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf K, Hekerman P, Spelten O, Herrmann A, Packman LC, Bussow K, Muller-Newen G, Becker W. Characterization of cyclin L2, a novel cyclin with an arginine/serine-rich domain: phosphorylation by DYRK1A and colocalization with splicing factors. J Biol Chem. 2004;279:4612–4624. doi: 10.1074/jbc.M310794200. [DOI] [PubMed] [Google Scholar]
- Dembowski JA, An P, Scoulos-Hanson M, Yeo G, Han J, Fu XD, Grabowski PJ. Alternative Splicing of a Novel Inducible Exon Diversifies the CASK Guanylate Kinase Domain. J Nucleic Acids. 2012;2012:816237. doi: 10.1155/2012/816237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding JH, Xu X, Yang D, Chu PH, Dalton ND, Ye Z, Yeakley JM, Cheng H, Xiao RP, Ross J, et al. Dilated cardiomyopathy caused by tissue-specific ablation of SC35 in the heart. EMBO J. 2004;23:885–896. doi: 10.1038/sj.emboj.7600054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding JH, Zhong XY, Hagopian JC, Cruz MM, Ghosh G, Feramisco J, Adams JA, Fu XD. Regulated cellular partitioning of SR protein-specific kinases in mammalian cells. Mol Biol Cell. 2006;17:876–885. doi: 10.1091/mbc.E05-10-0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan PI, Stojdl DF, Marius RM, Scheit KH, Bell JC. The Clk2 and Clk3 dual-specificity protein kinases regulate the intranuclear distribution of SR proteins and influence pre-mRNA splicing. Exp Cell Res. 1998;241:300–308. doi: 10.1006/excr.1998.4083. [DOI] [PubMed] [Google Scholar]
- Edmond V, Moysan E, Khochbin S, Matthias P, Brambilla C, Brambilla E, Gazzeri S, Eymin B. Acetylation and phosphorylation of SRSF2 control cell fate decision in response to cisplatin. EMBO J. 2011;30:510–523. doi: 10.1038/emboj.2010.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkelenz S, Mueller WF, Evans MS, Busch A, Schoneweis K, Hertel KJ, Schaal H. Position-dependent splicing activation and repression by SR and hnRNP proteins rely on common mechanisms. RNA. 2013;19:96–102. doi: 10.1261/rna.037044.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Valley MT, Lazar J, Yang AL, Bronson RT, Firestein S, Coetzee WA, Manley JL. SRp38 regulates alternative splicing and is required for Ca(2+) handling in the embryonic heart. Dev Cell. 2009;16:528–538. doi: 10.1016/j.devcel.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XD. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- Fu XD, Maniatis T. Factor required for mammalian spliceosomeassembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Fu XD, Maniatis T. The 35-kDa mammalian splicing factor SC35 mediates specific interactions between U1 and U2 small nuclear ribonucleoprotein particles at the 3′ splice site. Proc Natl Acad Sci U S A. 1992;89:1725–1729. doi: 10.1073/pnas.89.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara T, Hosoya T, Shimizu S, Sumi K, Oshiro T, Yoshinaka Y, Suzuki M, Yamamoto N, Herzenberg LA, Herzenberg LA, et al. Utilization of host SR protein kinases and RNA-splicing machinery during viral replication. Proc Natl Acad Sci U S A. 2006;103:11329–11333. doi: 10.1073/pnas.0604616103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabut M, Samavarchi-Tehrani P, Wang X, Slobodeniuc V, O’Hanlon D, Sung HK, Alvarez M, Talukder S, Pan Q, Mazzoni EO, Nedelec S, Wichterle H, Woltjen K, Hughes TR, Zandstra PW, Nagy A, Wrana JL, Blencowe BJ. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell. 2011;147:132–146. doi: 10.1016/j.cell.2011.08.023. [DOI] [PubMed] [Google Scholar]
- Gallego ME, Gattoni R, Stevenin J, Marie J, Expert-Bezancon A. The SR splicing factors ASF/SF2 and SC35 have antagonistic effects on intronic enhancer-dependent splicing of the beta-tropomyosin alternative exon 6A. EMBO J. 1997;16:1772–1784. doi: 10.1093/emboj/16.7.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sacristan A, Fernandez-Nestosa MJ, Hernandez P, Schvartzman JB, Krimer DB. Protein kinase clk/STY is differentially regulated during erythroleukemia cell differentiation: a bias toward the skipped splice variant characterizes postcommitment stages. Cell Res. 2005;15:495–503. doi: 10.1038/sj.cr.7290319. [DOI] [PubMed] [Google Scholar]
- Gautrey HL, Tyson-Capper AJ. Regulation of Mcl-1 by SRSF1 and SRSF5 in Cancer Cells. PLoS One. 2012;7:e51497. doi: 10.1371/journal.pone.0051497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H, Zuo P, Manley JL. Primary structure of the humansplicing factor ASF reveals similarities with Drosophilaregulators. Cell. 1991;66:373–382. doi: 10.1016/0092-8674(91)90626-a. [DOI] [PubMed] [Google Scholar]
- Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, Comoglio PM, Green MR, Riva S, Biamonti G. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell. 2005;20:881–890. doi: 10.1016/j.molcel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- Ghosh G, Adams JA. Phosphorylation mechanism and structure of serine-arginine protein kinases. FEBS J. 2011;278:587–597. doi: 10.1111/j.1742-4658.2010.07992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakouros T, Nikolakaki E, Mylonis I, Georgatsou E. Serine-arginine protein kinases: a small protein kinase family with a large cellular presence. FEBS J. 2011;278:570–586. doi: 10.1111/j.1742-4658.2010.07987.x. [DOI] [PubMed] [Google Scholar]
- Gilbert W, Siebel CW, Guthrie C. Phosphorylation by Sky1p promotes Npl3p shuttling and mRNA dissociation. RNA. 2001;7:302–313. doi: 10.1017/s1355838201002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves V, Matos P, Jordan P. The beta-catenin/TCF4 pathway modifies alternative splicing through modulation of SRp20 expression. RNA. 2008;14:2538–2549. doi: 10.1261/rna.1253408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves V, Matos P, Jordan P. Antagonistic SR proteins regulate alternative splicing of tumor-related Rac1b downstream of the PI3-kinase and Wnt pathways. Hum Mol Genet. 2009;18:3696–3707. doi: 10.1093/hmg/ddp317. [DOI] [PubMed] [Google Scholar]
- Gout S, Brambilla E, Boudria A, Drissi R, Lantuejoul S, Gazzeri S, Eymin B. Abnormal expression of the pre-mRNA splicing regulators SRSF1, SRSF2, SRPK1 and SRPK2 in non small cell lung carcinoma. PLoS One. 2012;7:e46539. doi: 10.1371/journal.pone.0046539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui JF, Lane WS, Fu XD. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature. 1994a;369:678–682. doi: 10.1038/369678a0. [DOI] [PubMed] [Google Scholar]
- Gui JF, Tronchere H, Chandler SD, Fu XD. Purification and characterization of a kinase specific for the serine- and arginine-rich pre-mRNA splicing factors. Proc Natl Acad Sci U S A. 1994b;91:10824–10828. doi: 10.1073/pnas.91.23.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagopian JC, Ma CT, Meade BR, Albuquerque CP, Ngo JC, Ghosh G, Jennings PA, Fu XD, Adams JA. Adaptable molecular interactions guide phosphorylation of the SR protein ASF/SF2 by SRPK1. J Mol Biol. 2008;382:894–909. doi: 10.1016/j.jmb.2008.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamelberg D, Shen T, McCammon JA. A proposed signaling motif for nuclear import in mRNA processing via the formation of arginine claw. Proc Natl Acad Sci U S A. 2007;104:14947–14951. doi: 10.1073/pnas.0703151104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Ding JH, Byeon CW, Kim JH, Hertel KJ, Jeong S, Fu XD. SR proteins induce alternative exon skipping through their activities on the flanking constitutive exons. Mol Cell Biol. 2011a;31:793–802. doi: 10.1128/MCB.01117-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Xiong J, Wang D, Fu XD. Pre-mRNA splicing: where and when in the nucleus. Trends Cell Biol. 2011b;21:336–343. doi: 10.1016/j.tcb.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes GM, Carrigan PE, Beck AM, Miller LJ. Targeting the RNA splicing machinery as a novel treatment strategy for pancreatic carcinoma. Cancer Res. 2006;66:3819–3827. doi: 10.1158/0008-5472.CAN-05-4065. [DOI] [PubMed] [Google Scholar]
- Hayes GM, Carrigan PE, Miller LJ. Serine-arginine protein kinase 1 overexpression is associated with tumorigenic imbalance in mitogen-activated protein kinase pathways in breast, colonic, and pancreatic carcinomas. Cancer Res. 2007;67:2072–2080. doi: 10.1158/0008-5472.CAN-06-2969. [DOI] [PubMed] [Google Scholar]
- Hertel KJ, Graveley BR. RS domains contact the pre-mRNA throughout spliceosome assembly. Trends Biochem Sci. 2005;30:115–118. doi: 10.1016/j.tibs.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Heyd F, Lynch KW. Degrade, move, regroup: signaling control of splicing proteins. Trends Biochem Sci. 2011;36:397–404. doi: 10.1016/j.tibs.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Gattoni R, Stevenin J, Steitz JA. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol Cell. 2003;11:837–843. doi: 10.1016/s1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- Huang Y, Steitz JA. SRprises along a messenger’s journey. Mol Cell. 2005;17:613–615. doi: 10.1016/j.molcel.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Huang Y, Yario TA, Steitz JA. A molecular link between SR protein dephosphorylation and mRNA export. Proc Natl Acad Sci U S A. 2004;101:9666–9670. doi: 10.1073/pnas.0403533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelga SC, Vu AQ, Arnold JD, Liang TY, Liu PP, Yan BY, Donohue JP, Shiue L, Hoon S, Brenner S, et al. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep. 2012;1:167–178. doi: 10.1016/j.celrep.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, Liu X, Fu H, Rees H, Yepes M, Levey A, Ye K. Interaction of Akt-phosphorylated SRPK2 with 14-3-3 mediates cell cycle and cell death in neurons. J Biol Chem. 2009;284:24512–24525. doi: 10.1074/jbc.M109.026237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang SW, Yang SJ, Ehlen A, Dong S, Khoury H, Chen J, Persson JL, Ye K. Serine/arginine protein-specific kinase 2 promotes leukemia cell proliferation by phosphorylating acinus and regulating cyclin A1. Cancer Res. 2008;68:4559–4570. doi: 10.1158/0008-5472.CAN-08-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia R, Li C, McCoy JP, Deng CX, Zheng ZM. SRp20 is a proto-oncogene critical for cell proliferation and tumor induction and maintenance. Int J Biol Sci. 2010;6:806–826. doi: 10.7150/ijbs.6.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K, Patel NA, Watson JE, Apostolatos H, Kleiman E, Hanson O, Hagiwara M, Cooper DR. Akt2 regulation of Cdc2-like kinases (Clk/Sty), serine/arginine-rich (SR) protein phosphorylation, and insulin-induced alternative splicing of PKCbetaII messenger ribonucleic acid. Endocrinology. 2009;150:2087–2097. doi: 10.1210/en.2008-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumaa H, Wei G, Nielsen PJ. Blastocyst formation is blocked in mouse embryos lacking the splicing factor SRp20. Curr Biol. 1999;9:899–902. doi: 10.1016/s0960-9822(99)80394-7. [DOI] [PubMed] [Google Scholar]
- Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet. 2011;12:715–729. doi: 10.1038/nrg3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan N, Neuwald AF. Evolutionary constraints associated with functional specificity of the CMGC protein kinases MAPK, CDK, GSK, SRPK, DYRK, and CK2alpha. Protein Sci. 2004;13:2059–2077. doi: 10.1110/ps.04637904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. 2007;14:185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karni R, Hippo Y, Lowe SW, Krainer AR. The splicing-factor oncoprotein SF2/ASF activates mTORC1. Proc Natl Acad Sci U S A. 2008;105:15323–15327. doi: 10.1073/pnas.0801376105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N, Bachorik JL, Dreyfuss G. Transportin-SR, a nuclear import receptor for SR proteins. J Cell Biol. 1999;145:1145–1152. doi: 10.1083/jcb.145.6.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohtz JD, Jamison SF, Will CL, Zuo P, Luhrmann R, Garcia-Blanco MA, Manley JL. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- Krainer AR, Mayeda A, Kozak D, Binns G. Functionalexpression of cloned human splicing factor SF2: Homology toRNA-binding proteins, U1 70K, and Drosophila splicingregulators. Cell. 1991;66:383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- Kress TL, Krogan NJ, Guthrie C. A single SR-like protein, Npl3, promotes pre-mRNA splicing in budding yeast. Mol Cell. 2008;32:727–734. doi: 10.1016/j.molcel.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar S, MOhan A, Kandalam M, Ramkumar HL, Venkatesa N, Das RR. SRPK1: A cisplatin sensitive protein expressed in retinoblastoma. Pediatr Blood Cancer. 2008;50:402–406. doi: 10.1002/pbc.21088. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi H, Kimura T, Wada K, Hisamoto N, Matsumoto K, Hagiwara M. SPK-1, a C. elegans SR protein kinase homologue, is essential for embryogenesis and required for germline development. Mech Dev. 2000;99:51–64. doi: 10.1016/s0925-4773(00)00477-9. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi N, Onogi H, Wakabayashi T, Hagiwara M. Novel SR-protein-specific kinase, SRPK2, disassembles nuclear speckles. Biochem Biophys Res Commun. 1998;242:357–364. doi: 10.1006/bbrc.1997.7913. [DOI] [PubMed] [Google Scholar]
- Lai MC, Lin RI, Huang SY, Tsai CW, Tarn WY. A human importin-beta family protein, transportin-SR2, interacts with the phosphorylated RS domain of SR proteins. J Biol Chem. 2000;275:7950–7957. doi: 10.1074/jbc.275.11.7950. [DOI] [PubMed] [Google Scholar]
- Lai MC, Lin RI, Tarn WY. Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc Natl Acad Sci U S A. 2001;98:10154–10159. doi: 10.1073/pnas.181354098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MC, Tarn WY. Hypophosphorylated ASF/SF2 binds TAP and is present in messenger ribonucleoproteins. J Biol Chem. 2004;279:31745–31749. doi: 10.1074/jbc.C400173200. [DOI] [PubMed] [Google Scholar]
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- Lasho TL, Jimma T, Finke CM, Patnaik M, Hanson CA, Ketterling RP, Pardanani A, Tefferi A. SRSF2 mutations in primary myelofibrosis: significant clustering with IDH mutations and independent association with inferior overall and leukemia-free survival. Blood. 2012;120:4168–4171. doi: 10.1182/blood-2012-05-429696. [DOI] [PubMed] [Google Scholar]
- Lemaire R, Winne A, Sarkissian M, Lafyatis R. SF2 and SRp55 regulation of CD45 exon 4 skipping during T cell activation. Eur J Immunol. 1999;29:823–837. doi: 10.1002/(SICI)1521-4141(199903)29:03<823::AID-IMMU823>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol. 2008;15:819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Fu XD. SR proteins and related factors in alternative splicing. Adv Exp Med Biol. 2007;623:107–122. doi: 10.1007/978-0-387-77374-2_7. [DOI] [PubMed] [Google Scholar]
- Lin S, Xiao R, Sun P, Xu X, Fu XD. Dephosphorylation-dependent sorting of SR splicing factors during mRNP maturation. Mol Cell. 2005;20:413–425. doi: 10.1016/j.molcel.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Liu HX, Chew SL, Cartegni L, Zhang MQ, Krainer AR. Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Mol Cell Biol. 2000;20:1063–1071. doi: 10.1128/mcb.20.3.1063-1071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HX, Zhang M, Krainer AR. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Dreyfuss G. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol Cell Biol. 1995;15:2800–2808. doi: 10.1128/mcb.15.5.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh BJ, Cullen CF, Vogt N, Ohkura H. The conserved kinase SRPK regulates karyosome formation and spindle microtubule assembly in Drosophila oocytes. J Cell Sci. 2012;125:4457–4462. doi: 10.1242/jcs.107979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- Loomis RJ, Naoe Y, Parker JB, Savic V, Bozovsky MR, Macfarlan T, Manley JL, Chakravarti D. Chromatin binding of SRp20 and ASF/SF2 and dissociation from mitotic chromosomes is modulated by histone H3 serine 10 phosphorylation. Mol Cell. 2009;33:450–461. doi: 10.1016/j.molcel.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch KW. Regulation of alternative splicing by signal transduction pathways. Adv Exp Med Biol. 2007;623:161–174. doi: 10.1007/978-0-387-77374-2_10. [DOI] [PubMed] [Google Scholar]
- Manley JL, Krainer AR. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins) Genes Dev. 2010;24:1073–1074. doi: 10.1101/gad.1934910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Vinader L, Shin C, Onnekink C, Manley JL, Lubsen NH. Hsp27 enhances recovery of splicing as well as rephosphorylation of SRp38 after heat shock. Mol Biol Cell. 2006;17:886–894. doi: 10.1091/mbc.E05-07-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Hartmuth K, Mohlmann S, Urlaub H, Ficner R, Luhrmann R. Phosphorylation of human PRP28 by SRPK2 is required for integration of the U4/U6-U5 tri-snRNP into the spliceosome. Nat Struct Mol Biol. 2008;15:435–443. doi: 10.1038/nsmb.1415. [DOI] [PubMed] [Google Scholar]
- Matter N, Herrlich P, Konig H. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature. 2002;420:691–695. doi: 10.1038/nature01153. [DOI] [PubMed] [Google Scholar]
- Meggendorfer M, Roller A, Haferlach T, Eder C, Dicker F, Grossmann V, Kohlmann A, Alpermann T, Yoshida K, Ogawa S, Koeffler HP, Kern W, Haferlach C, Schnittger S. SRSF2 mutations in 275 cases with chronic myelomonocytic leukemia (CMML) Blood. 2012;120:3080–3088. doi: 10.1182/blood-2012-01-404863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdzhanova G, Gout S, Keramidas M, Edmond V, Coll JL, Brambilla C, Brambilla E, Gazzeri S, Eymin B. The transcription factor E2F1 and the SR protein SC35 control the ratio of pro-angiogenic versus antiangiogenic isoforms of vascular endothelial growth factor-A to inhibit neovascularization in vivo. Oncogene. 2010;29:5392–5403. doi: 10.1038/onc.2010.281. [DOI] [PubMed] [Google Scholar]
- Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science. 2012;338:1593–1599. doi: 10.1126/science.1228186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermoud JE, Cohen P, Lamond AI. Ser/Thr-specific protein phosphatases are required for both catalytic steps of pre-mRNA splicing. Nucleic Acids Res. 1992;20:5263–5269. doi: 10.1093/nar/20.20.5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermoud JE, Cohen PT, Lamond AI. Regulation of mammalian spliceosome assembly by a protein phosphorylation mechanism. EMBO J. 1994;13:5679–5688. doi: 10.1002/j.1460-2075.1994.tb06906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michlewski G, Sanford JR, Caceres JF. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol Cell. 2008;30:179–189. doi: 10.1016/j.molcel.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Misteli T, Caceres JF, Clement JQ, Krainer AR, Wilkinson MF, Spector DL. Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J Cell Biol. 1998;143:297–307. doi: 10.1083/jcb.143.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Spector DL. Serine/threonine phosphatase 1 modulates the subnuclear distribution of pre-mRNA splicing factors. Mol Biol Cell. 1996;7:1559–1572. doi: 10.1091/mbc.7.10.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehle EA, Ryan CJ, Krogan NJ, Kress TL, Guthrie C. The Yeast SR-Like Protein Npl3 Links Chromatin Modification to mRNA Processing. PLoS Genet. 2012;8:e1003101. doi: 10.1371/journal.pgen.1003101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Wang Q, Kennedy CJ, Silver PA. An alternative splicingnetwork links cell-cycle control to apoptosis. Cell. 2010;142:625–636. doi: 10.1016/j.cell.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa O, Arnold M, Nakagawa M, Hamada H, Shelton JM, Kusano H, Harris TM, Childs G, Campbell KP, Richardson JA, et al. Centronuclear myopathy in mice lacking a novel muscle-specific protein kinase transcriptionally regulated by MEF2. Genes Dev. 2005;19:2066–2077. doi: 10.1101/gad.1338705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo JC, Chakrabarti S, Ding JH, Velazquez-Dones A, Nolen B, Aubol BE, Adams JA, Fu XD, Ghosh G. Interplay between SRPK and Clk/Sty kinases in phosphorylation of the splicing factor ASF/SF2 is regulated by a docking motif in ASF/SF2. Mol Cell. 2005;20:77–89. doi: 10.1016/j.molcel.2005.08.025. [DOI] [PubMed] [Google Scholar]
- Ngo JC, Giang K, Chakrabarti S, Ma CT, Huynh N, Hagopian JC, Dorrestein PC, Fu XD, Adams JA, Ghosh G. A sliding docking interaction is essential for sequential and processive phosphorylation of an SR protein by SRPK1. Mol Cell. 2008;29:563–576. doi: 10.1016/j.molcel.2007.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O’Brien G, Shiue L, Clark TA, Blume JE, Ares M., Jr. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated. Genes Dev. 2007;21:808–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolakaki E, Kohen R, Hartmann AM, Stamm S, Georgatsou E, Giannakouros T. Cloning and characterization of an alternatively spliced form of SR protein kinase 1 that interacts specifically with scaffold attachment factor-B. J Biol Chem. 2001;276:40175–40182. doi: 10.1074/jbc.M104755200. [DOI] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2009;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen B, Taylor S, Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- Nolen B, Yun CY, Wong CF, McCammon JA, Fu XD, Ghosh G. The structure of Sky1p reveals a novel mechanism for constitutive activity. Nat Struct Biol. 2001;8:176–183. doi: 10.1038/84178. [DOI] [PubMed] [Google Scholar]
- Nowak DG, Amin EM, Rennel ES, Hoareau-Aveilla C, Gammons M, Damodoran G, Hagiwara M, Harper SJ, Woolard J, Ladomery MR, et al. Regulation of vascular endothelial growth factor (VEGF) splicing from pro-angiogenic to anti-angiogenic isoforms: a novel therapeutic strategy for angiogenesis. J Biol Chem. 2010;285:5532–5540. doi: 10.1074/jbc.M109.074930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odunsi K, Mhawech-Fauceglia P, Andrews C, Beck A, Amuwo O, Lele S, Black JD, Huang RY. Elevated expression of the serine-arginine protein kinase 1 gene in ovarian cancer and its role in Cisplatin cytotoxicity in vitro. PLoS One. 2012;7:e51030. doi: 10.1371/journal.pone.0051030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oltean S, Gammons M, Hulse R, Hamdollah-Zadeh M, Mavrou A, Donaldson L, Salmon AH, Harper SJ, Ladomery MR, Bates DO. SRPK1 inhibition in vivo: modulation of VEGF splicing and potential treatment for multiple diseases. Biochem Soc Trans. 2012;40:831–835. doi: 10.1042/BST20120051. [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Pandit S, Wang D, Fu XD. Functional integration of transcriptional and RNA processing machineries. Curr Opin Cell Biol. 2008;20:260–265. doi: 10.1016/j.ceb.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit S, Zhou Y, Shiue L, Coutinho-Mansfield G, Li H, Qiu J, Huang J, Yeo G, Ares M, Jr., Fu X-D. Genome-wide analysis reveals SR protein cooperation and competition in regulated splicing. Mol Cell. 2013 doi: 10.1016/j.molcel.2013.03.001. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papoutsopoulou S, Nikolakaki E, Chalepakis G, Kruft V, Chevaillier P, Giannakouros T. SR protein-specific kinase 1 is highly expressed in testis and phosphorylates protamine 1. Nucleic Acids Res. 1999a;27:2972–2980. doi: 10.1093/nar/27.14.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papoutsopoulou S, Nikolakaki E, Giannakouros T. SRPK1 and LBR protein kinases show identical substrate specificities. Biochem Biophys Res Commun. 1999b;255:602–607. doi: 10.1006/bbrc.1999.0249. [DOI] [PubMed] [Google Scholar]
- Patel NA, Apostolatos HS, Mebert K, Chalfant CE, Watson JE, Pillay TS, Sparks J, Cooper DR. Insulin regulates protein kinase CbetaII alternative splicing in multiple target tissues: development of a hormonally responsive heterologous minigene. Mol Endocrinol. 2004;18:899–911. doi: 10.1210/me.2003-0391. [DOI] [PubMed] [Google Scholar]
- Patel NA, Kaneko S, Apostolatos HS, Bae SS, Watson JE, Davidowitz K, Chappell DS, Birnbaum MJ, Cheng JQ, Cooper DR. Molecular and genetic studies imply Akt-mediated signaling promotes protein kinase CbetaII alternative splicing via phosphorylation of serine/arginine-rich splicing factor SRp40. J Biol Chem. 2005;280:14302–14309. doi: 10.1074/jbc.M411485200. [DOI] [PubMed] [Google Scholar]
- Patnaik MM, Lasho TL, Finke CM, Hanson CA, Hodnefield JM, Knudson RA, Ketterling RP, Pardanani A, Tefferi A. Spliceosome mutations involving SRSF2, SF3B1, and U2AF35 in chronic myelomonocytic leukemia: Prevalence, clinical correlates, and prognostic relevance. Am J Hematol. 2013;88:201–206. doi: 10.1002/ajh.23373. [DOI] [PubMed] [Google Scholar]
- Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell. 2009;35:228–239. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porat Z, Erez O, Kahana C. Cellular localization and phosphorylation of Hrb1p is independent of Sky1p. Biochim Biophys Acta. 2006;1763:207–213. doi: 10.1016/j.bbamcr.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Hoareau-Aveilla C, Oltean S, Harper SJ, Bates DO. The anti-angiogenic isoforms of VEGF in health and disease. Biochem Soc Trans. 2009;37:1207–1213. doi: 10.1042/BST0371207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R, Cheng H. TREX, SR proteins and export of mRNA. Curr Opin Cell Biol. 2005;17:269–273. doi: 10.1016/j.ceb.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Roscigno RF, Garcia-Blanco MA. SR proteins escort the U4/U6.U5 tri- snRNP to the spliceosome. RNA. 1995;1:692–706. [PMC free article] [PubMed] [Google Scholar]
- Rossi F, Labourier E, Forne T, Divita G, Derancourt J, Riou JF, Antoine E, Cathala G, Brunel C, Tazi J. Specific phosphorylation of SR proteins by mammalian DNA topoisomerase I. Nature. 1996;381:80–82. doi: 10.1038/381080a0. [DOI] [PubMed] [Google Scholar]
- Roth MB, Zahler AM, Stolk JA. A conserved family of nuclear phosphoproteins localized to sites of polymerase II transcription. J Cell Biol. 1991;115:587–596. doi: 10.1083/jcb.115.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JR, Ellis JD, Cazalla D, Caceres JF. Reversible phosphorylation differentially affects nuclear and cytoplasmic functions of splicing factor 2/alternative splicing factor. Proc Natl Acad Sci U S A. 2005;102:15042–15047. doi: 10.1073/pnas.0507827102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JR, Gray NK, Beckmann K, Caceres JF. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 2004;18:755–768. doi: 10.1101/gad.286404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JR, Wang X, Mort M, Vanduyn N, Cooper DN, Mooney SD, Edenberg HJ, Liu Y. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res. 2009;19:381–394. doi: 10.1101/gr.082503.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapra AK, Anko ML, Grishina I, Lorenz M, Pabis M, Poser I, Rollins J, Weiland EM, Neugebauer KM. SR protein family members display diverse activities in the formation of nascent and mature mRNPs in vivo. Mol Cell. 2009;34:179–190. doi: 10.1016/j.molcel.2009.02.031. [DOI] [PubMed] [Google Scholar]
- Schaal TD, Maniatis T. Selection and characterization of pre-mRNA splicing enhancers: identification of novel SR protein-specific enhancer sequences. Mol Cell Biol. 1999;19:1705–1719. doi: 10.1128/mcb.19.3.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk PW, Boersma AW, Brandsma JA, den Dulk H, Burger H, Stoter G, Brouwer J, Nooter K. SKY1 is involved in cisplatin-induced cell kill in Saccharomyces cerevisiae, and inactivation of its human homologue, SRPK1, induces cisplatin resistance in a human ovarian carcinoma cell line. Cancer Res. 2001;61:6982–6986. [PubMed] [Google Scholar]
- Schenk PW, Boersma AW, Brok M, Burger H, Stoter G, Nooter K. Inactivation of the Saccharomyces cerevisiae SKY1 gene induces a specific modification of the yeast anticancer drug sensitivity profile accompanied by a mutator phenotype. Mol Pharmacol. 2002;61:659–666. doi: 10.1124/mol.61.3.659. [DOI] [PubMed] [Google Scholar]
- Schenk PW, Stoop H, Bokemeyer C, Mayer F, Stoter G, Oosterhuis JW, Wiemer E, Looijenga LH, Nooter K. Resistance to platinum-containing chemotherapy in testicular germ cell tumors is associated with downregulation of the protein kinase SRPK1. Neoplasia. 2004;6:297–301. doi: 10.1593/neo.03406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen S, Jumaa H, Webster NJ. Splicing factor SRSF3 is crucial for hepatocyte differentiation and metabolic function. Nat Commun. 2013;4:1336. doi: 10.1038/ncomms2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Markey M, Torres-Munoz K, Varia S, Kadakia M, Bubulya A, Bubulya PA. Son maintains accurate splicing for a subset of human pre-mRNAs. J Cell Sci. 2011;124:4286–4298. doi: 10.1242/jcs.092239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Green MR. A pathway of sequential arginine-serine-rich domain-splicing signal interactions during mammalian spliceosome assembly. Mol Cell. 2004;16:363–373. doi: 10.1016/j.molcel.2004.10.021. [DOI] [PubMed] [Google Scholar]
- Shen H, Green MR. RS domain-splicing signal interactions in splicing of U12-type and U2-type introns. Nat Struct Mol Biol. 2007;14:597–603. doi: 10.1038/nsmb1263. [DOI] [PubMed] [Google Scholar]
- Shen H, Kan JL, Green MR. Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol Cell. 2004;13:367–376. doi: 10.1016/s1097-2765(04)00025-5. [DOI] [PubMed] [Google Scholar]
- Shi Y, Manley JL. A complex signaling pathway regulates SRp38 phosphorylation and pre-mRNA splicing in response to heat shock. Mol Cell. 2007;28:79–90. doi: 10.1016/j.molcel.2007.08.028. [DOI] [PubMed] [Google Scholar]
- Shi Y, Nishida K, Campigli Di Giammartino D, Manley JL. Heat shock-induced SRSF10 dephosphorylation displays thermotolerance mediated by Hsp27. Mol Cell Biol. 2011;31:458–465. doi: 10.1128/MCB.01123-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin C, Feng Y, Manley JL. Dephosphorylated SRp38 acts as a splicing repressor in response to heat shock. Nature. 2004;427:553–558. doi: 10.1038/nature02288. [DOI] [PubMed] [Google Scholar]
- Shin C, Kleiman FE, Manley JL. Multiple properties of the splicing repressor SRp38 distinguish it from typical SR proteins. Mol Cell Biol. 2005;25:8334–8343. doi: 10.1128/MCB.25.18.8334-8343.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin C, Manley JL. The SR protein SRp38 represses splicing in M phase cells. Cell. 2002;111:407–417. doi: 10.1016/s0092-8674(02)01038-3. [DOI] [PubMed] [Google Scholar]
- Shin C, Manley JL. Cell signalling and the control of pre-mRNA splicing. Nat Rev Mol Cell Biol. 2004;5:727–738. doi: 10.1038/nrm1467. [DOI] [PubMed] [Google Scholar]
- Siebel CW, Feng L, Guthrie C, Fu XD. Conservation in budding yeast of a kinase specific for SR splicing factors. Proc Natl Acad Sci U S A. 1999;96:5440–5445. doi: 10.1073/pnas.96.10.5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel CW, Guthrie C. The essential yeast RNA binding protein Np13p is methylated. Proc Natl Acad Sci U S A. 1996;93:13641–13646. doi: 10.1073/pnas.93.24.13641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, Weng Z, Moore MJ. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell. 2012;151:750–764. doi: 10.1016/j.cell.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha R, Allemand E, Zhang Z, Karni R, Myers MP, Krainer AR. Arginine methylation controls the subcellular localization and functions of the oncoprotein splicing factor SF2/ASF. Mol Cell Biol. 2010;30:2762–2774. doi: 10.1128/MCB.01270-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis AS, Peng R, Crawford JB, Phillips JA, 3rd, Patton JG. Growth hormone deficiency and splicing fidelity: two serine/arginine-rich proteins, ASF/SF2 and SC35, act antagonistically. J Biol Chem. 2008;283:23619–23626. doi: 10.1074/jbc.M710175200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stickeler E, Kittrell F, Medina D, Berget SM. Stage-specific changes in SR splicing factors and alternative splicing in mammary tumorigenesis. Oncogene. 1999;18:3574–3582. doi: 10.1038/sj.onc.1202671. [DOI] [PubMed] [Google Scholar]
- Stojdl DF, Bell JC. SR protein kinases: the splice of life. Biochem Cell Biol. 1999;77:293–298. [PubMed] [Google Scholar]
- Tacke R, Chen Y, Manley JL. Sequence-specific RNA binding by an SR protein requires RS domain phosphorylation: creation of an SRp40-specific splicing enhancer. Proc Natl Acad Sci U S A. 1997;94:1148–1153. doi: 10.1073/pnas.94.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacke R, Manley JL. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi M, Yanagida M. A mitotic role for a novel fission yeast protein kinase dsk1 with cell cycle stage dependent phosphorylation and localization. Mol Biol Cell. 1993;4:247–260. doi: 10.1091/mbc.4.3.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Horikawa I, Ajiro M, Robles AI, Fujita K, Mondal AM, Stauffer JK, Zheng ZM, Harris CC. Downregulation of splicing factor SRSF3 induces p53beta, an alternatively spliced isoform of p53 that promotes cellular senescence. Oncogene. 2012a doi: 10.1038/onc.2012.288. doi:10.1038/onc.2012.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YH, Han SP, Kassahn KS, Skarshewski A, Rothnagel JA, Smith R. Complex Evolutionary Relationships Among Four Classes of Modular RNA-Binding Splicing Regulators in Eukaryotes: The hnRNP, SR, ELAV-Like and CELF Proteins. J Mol Evol. 2012b;75:214–228. doi: 10.1007/s00239-012-9533-0. [DOI] [PubMed] [Google Scholar]
- Tang Z, Luca M, Taggart-Murphy L, Portillio J, Chang C, Guven A, Lin RJ, Murray J, Carr A. Interacting factors and cellular localization of SR protein-specific kinase Dsk1. Exp Cell Res. 2012c;318:2071–2084. doi: 10.1016/j.yexcr.2012.05.020. [DOI] [PubMed] [Google Scholar]
- van der Houven van Oordt W, Diaz-Meco MT, Lozano J, Krainer AR, Moscat J, Caceres JF. The MKK(3/6)-p38-signaling cascade alters the subcellular distribution of hnRNP A1 and modulates alternative splicing regulation. J Cell Biol. 2000;149:307–316. doi: 10.1083/jcb.149.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008a;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H-Y, Lin W, Dyck JA, Yeakley JM, Songyang Z, Cantley LC, Fu XD. SRPK2: A differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J Cell Biol. 1998a;140:737–750. doi: 10.1083/jcb.140.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HY, Lin W, Dyck JA, Yeakley JM, Songyang Z, Cantley LC, Fu XD. SRPK2: a differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J Cell Biol. 1998b;140:737–750. doi: 10.1083/jcb.140.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HY, Xu X, Ding JH, Bermingham JR, Jr., Fu XD. SC35 plays a role in T cell development and alternative splicing of CD45. Mol Cell. 2001;7:331–342. doi: 10.1016/s1097-2765(01)00181-2. [DOI] [PubMed] [Google Scholar]
- Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, Kim S, Xu X, Zheng Y, Chilton B, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008b;14:312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witten JT, Ule J. Understanding splicing regulation through RNA splicing maps. Trends Genet. 2011;27:89–97. doi: 10.1016/j.tig.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JY, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- Xiao R, Sun Y, Ding JH, Lin S, Rose DW, Rosenfeld MG, Fu XD, Li X. Splicing regulator SC35 is essential for genomic stability and cell proliferation during mammalian organogenesis. Mol Cell Biol. 2007;27:5393–5402. doi: 10.1128/MCB.00288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao SH, Manley JL. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- Xiao SH, Manley JL. Phosphorylation-dephosphorylation differentially affects activities of splicing factor ASF/SF2. EMBO J. 1998;17:6359–6367. doi: 10.1093/emboj/17.21.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Yang D, Ding JH, Wang W, Chu PH, Dalton ND, Wang HY, Bermingham JR, Jr., Ye Z, Liu F, et al. ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell. 2005;120:59–72. doi: 10.1016/j.cell.2004.11.036. [DOI] [PubMed] [Google Scholar]
- Yeakley JM, Tronchere H, Olesen J, Dyck JA, Wang HY, Fu XD. Phosphorylation regulates in vivo interaction and molecular targeting of serine/arginine-rich pre-mRNA splicing factors. J Cell Biol. 1999;145:447–455. doi: 10.1083/jcb.145.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yomoda J, Muraki M, Kataoka N, Hosoya T, Suzuki M, Hagiwara M, Kimura H. Combination of Clk family kinase and SRp75 modulates alternative splicing of Adenovirus E1A. Genes Cells. 2008;13:233–244. doi: 10.1111/j.1365-2443.2008.01163.x. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- Yun CY, Fu XD. Conserved SR protein kinase functions in nuclear import and its action is counteracted by arginine methylation in Saccharomyces cerevisiae. J Cell Biol. 2000;150:707–718. doi: 10.1083/jcb.150.4.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun CY, Velazquez-Dones AL, Lyman SK, Fu XD. Phosphorylation-dependent and -independent nuclear import of RS domain-containing splicing factors and regulators. J Biol Chem. 2003;278:18050–18055. doi: 10.1074/jbc.M211714200. [DOI] [PubMed] [Google Scholar]
- Zahler AM, Lane WS, Stolk JA, Roth MB. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- Zahler AM, Neugebauer KM, Lane WS, Roth MB. Distinct functions of SR proteins in alternative pre-mRNA splicing. Science. 1993;260:219–222. doi: 10.1126/science.8385799. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Krainer AR. Involvement of SR proteins in mRNA surveillance. Mol Cell. 2004;16:597–607. doi: 10.1016/j.molcel.2004.10.031. [DOI] [PubMed] [Google Scholar]
- Zhong XY, Ding JH, Adams JA, Ghosh G, Fu XD. Regulation of SR protein phosphorylation and alternative splicing by modulating kinetic interactions of SRPK1 with molecular chaperones. Genes Dev. 2009a;23:482–495. doi: 10.1101/gad.1752109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong XY, Wang P, Han J, Rosenfeld MG, Fu XD. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol Cell. 2009b;35:1–10. doi: 10.1016/j.molcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Qiu J, Liu W, Zhou Y, Plocinik RM, Li H, Hu Q, Ghosh G, Adams JA, Rosenfeld MG, et al. The Akt-SRPK-SR axis constitutes a major pathway in transducing EGF signaling to regulate alternative splicing in the nucleus. Mol Cell. 2012;47:422–433. doi: 10.1016/j.molcel.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]