Abstract
Huntington’s disease (HD) is an autosomal dominant hereditary disease caused by a trinucleotide repeat mutation in the huntingtin gene that results in an increased number of glutamine residues in the N terminus of huntingtin protein. Mutant huntingtin leads to progressive impairment of motor function, cognitive dysfunction, and neuropsychiatric disturbance. There are no disease-modifying treatments available. During the past decade, Sirtuin-1 (SIRT1) has been the focus of intense investigation and discussion because it regulates longevity in multiple organisms and has shown beneficial effects in a variety of models of neurodegenerative disorders. Studies in different animal models provide convincing evidence that SIRT1 protects neurons in mouse models of HD well as in Caenorhabditis. elegans, although controversial results were reported in a fly model. Indeed, many connections exist between the deacetylation function of SIRT1 and its role in neuroprotection. As a result, pharmacological interventions targeting SIRT1 might become promising strategies to combat HD. This review summarizes recent progress in SIRT1 research, with a focus on the specificity of this protein as a potential therapeutic target for HD, as well as existing challenges for developing SIRT1 modulators for clinical use.
Keywords: Huntington’s disease, SIRT1, Sirtuins, Therapeutics
1. Introduction
Huntington’s disease (HD) is an autosomal dominant neurodegenerative disease that is caused by a CAG triplet repeat expansion in the huntingtin gene, which encodes an expanded polyglutamine stretch in the huntingtin (Htt) protein [1]. The disease is inherited with age-dependent penetrance, and repeat CAG lengths of 40 or more are associated with nearly full penetrance by age 65 years [2]. The prevalence of HD is 7–10/100,000 in the Western world [3], with many more people at risk of the disease. Longer CAG repeats predict earlier onset, accounting for up to 50–70% of variance in age of onset, with the remainder likely to be due to modifying genes and the environment [4, 5].
Clinical features of HD include progressive involuntary movement disorders, psychiatric signs, cognitive decline, and a shortened lifespan. Currently, there is no therapy that modifies the disease progression. Thus, identification of new targets, strategies for drug discovery and therapeutic approaches are now becoming a critical point.
Htt is a large protein predicted to consist mainly of repeated units of about 50 amino acids, termed HEAT repeats [3], this protein is truncated and gives rise to toxic N-terminal fragments, and also undergoes extensive post-translational modification[4]. The cellular functions of Htt are still not completely understood. Defects in energy metabolism and mitochondrial respiratory enzymes have been identified in postmortem brain tissues from HD cases as well as in HD models [6–9]. Mutant Htt affects mitochondria and cellular metabolism in multiple ways. For example, mutant Htt could have direct or indirect effects on mitochondria [4], impair the mitochondrial disulfide relay system [9], and compromise energy metabolism, and increase oxidative damage [6, 10]. Moreover, mutant Htt alters transcription of PPARGC1A, which encodes a transcription factor peroxisome proliferator-activated receptor-gamma coactivator 1α (PGC1α), which in turn controls transcription of many nuclear-encoded proteins necessary for mitochondrial function and cellular energy metabolism [11, 12]. Abnormalities in mitochondrial function and bioenergetics contribute to cell death in HD-affected individuals, in both central and peripheral tissues [13–16]. Energy deficits thus are recognized as important pathogenic pathways in HD [17, 18]. Notably, the onset of energy-related manifestations at the presymptomatic stage indicates that energy deficits are likely to be an early phenomenon in the cascade of events leading to HD pathogenesis [19–22]. These findings highlight the importance of disturbed energy metabolism in HD pathogenesis.
Our previous study showed that calorie restriction could ameliorate the motor phenotype and extend survival of N171-82Q HD mice [7], indicating that pathways related to energy metabolism can modify disease progression in HD. Calorie restriction increases mitochondrial biogenesis by inducing endothelial nitric oxide synthase (eNOS), and NO can activate the SIRT1 gene [23, 24] which is the mammalian ortholog of yeast Sir2, and a highly conserved NAD+-dependent protein deacetylase. Moreover, SIRT1 has been suggested to mediate some beneficial effects of calorie restriction [25–28]. It has been demonstrated that SIRT1 enhances the ability of cells to counter oxidative stress: first, SIRT1 may offer protection against oxidative stress through the modulation of FOXOs [29]. Second, SIRT1 protects cells against oxidative stress by increasing the activity of catalase [30, 31]. Third, SIRT1 induces the antioxidant enzyme MnSOD [32]. Lastly, SIRT1 deacetylates its substrate PGC-1α and enhances its transcriptional activity, thereby preventing oxidative stress [33, 34].
SIRT1 is a nuclear protein, that is predominantly expressed in neurons [35]; it has thus emerged as a key regulator for energy metabolism of neurons [20]. SIRT1 is highly expressed in the mouse brain during embryogenesis [36], as well as in the adult brain, including important metabolic centers of the brain, such as the hypothalamus [35]. During aging, SIRT1 expression is decreased in specific nuclei of the hypothalamus of mice [37–39]. Although the precise functions of SIRT1 in neurons are still unclear, they seem to be important players in neurodegenerative disorders. The subcellular localization of SIRT1 likely depends on cell type, stress status, and molecular interactions [40].
Despite a recent controversy on the role of SIRT1(Sir2) in extension of life-span in eukaryotes [41], SIRT1 in mammalian system seems to be beneficial in many neurodegenerative diseases, including Wallerian degeneration (wlds) [42], Alzheimer’s disease (AD), amyotrophic lateral sclerosis (ALS), Parkinson’s disease (PD), and HD [43–50], suggesting that the modulation of SIRT1 activity might be important in treatment of these neurodegenerative diseases. Nevertheless, the connection between SIRT1 and calorie restriction, the precise normal function of SIRT1, and its role in neurodegeneration warrant further investigation.
The role of SIRT1 in HD
The first report demonstrating the connection between SIRT1 and HD came from studies by Parker and colleagues, who found that overexpression of Sir2.1 (Mammalian Sir2 orthology is called SIRT1) or treatment with resveratrol rescues neuronal dysfunction phenotypes induced by mutant polyglutamine in Caenorhabditis elegans [51]. In contrast to the neuroprotective effect of Sir2.1 in C. elegans, Pallos and co-workers reported that 50% reduction of Sir2 extended survival and preserved neurons containing photoreceptor in flies expressing mutant Htt [52]. Interestingly, in the fly model system, overexpression of Sir2 does not reduce the lethality or the level of neuronal degeneration caused by mutant Htt. Studies in both C. elegans and Drosophila suggest that complete loss of Sir2 is deleterious in the worm [51] and is deleterious compared with heterozygous loss in mutant Htt-challenged flies [52]. Although heterozygous loss of Sir2 is protective in flies, heterozygous loss of Sir2 in worms was not examined. Nevertheless, reduction of Sir2 neither alters the life-span of flies not expressing Htt nor siblings expressing Htt. There is controversial in several aspects of the role of sirtuins in lifespan in C. elegans and Drosophila, and studies have indicated that Sir2 overexpression did not increase lifespan and dietary restriction increased fly lifespan independently of dSir2 [53]. However, overexpression of Sir2 increases the longevity of normal flies and the longevity of diseased flies is slightly increased by elevated Sir2 [52]. The precise mechanism of these controversial results remains to be clarified. The different results might be due to the amount of Sir2, its activation status, and different downstream targets involved. Whatever the mechanism, these controversial results warrant further investigation of the role of SIRT1 in mammalian systems.
Indeed, two independent studies done by our group [54] and Krainc’s group [55] demonstrated that modulating the levels of SIRT1 has therapeutic benefit in three different HD mouse models, and putative downstream targets of SIRT1 involved in improved disease outcomes are also identified. Krainc’s group showed that brain-specific knockout of SIRT1 results in exacerbation of neuropathology in an R6/2 mouse model of HD, whereas overexpression of SIRT1 by knock-in it to the endogenous β-actin locus improves survival of R6/2 HD mice, ameliorates neuropathology and mutant Htt aggregation in the R6/2 model [55]. By comparison, our group used a different truncated HD mouse model, N171-82Q mice, and a full-length HD model in which mutant Htt was expressed from a bacteria artificial chromosome (BACHD mouse model), we showed that overexpression of SIRT1 driven by a prion protein promoter attenuates brain atrophy, preserves endogenous dopamine- and cyclic AMP-regulated phosphoprotein, Mr=32000 (DARPP32), a marker of medium spiny neurons that are selectively degenerated in HD, and improves motor function in both HD mouse models [54].
Reduced transcription of brain derived neurotrophic factor (BDNF) gene and axonal transport of proteins have been implicated as a key pathogenesis resulting in selective neurodegeneration and neuronal dysfunction in HD [56–58]. Increasing BDNF levels by genetic manipulation or with agents that promote production of endogenous BDNF improves motor function, attenuates brain atrophy, and/or extends survival in HD mice [59, 60]. To gain further understanding of the mechanism of protection by SIRT1 in HD, our group demonstrated that BDNF protein levels were preserved by SIRT1. Consistent with our findings on BDNF, Krainc’s group demonstrated that SIRT1 transactivates BDNF expression at a promoter region that is also regulated by the cyclic AMP response element binding (CREB) transcription factor. Moreover, Krainc’s group demonstrated that transducer of regulated CREB activity 1 (TORC1), a transcriptional coactivator known to enhance CREB function, is involved in SIRT1-mediated regulation of BDNF transcription. Through a comprehensive biochemical analysis, the group determined that SIRT1-mediated deacetylation of TORC1 at certain lysine residues promotes the interaction of TORC1 with CREB, and that mutant Htt interferes with SIRT1 through a physical interaction that may be dependent on polyglutamine length, which requires the presence of TORC1 [55].
We had previously implicated BDNF in the rescue of the HD phenotypes [61–64], and therefore we examined the BDNF receptor and found that SIRT1 overexpression favored the phosphorylation and consequent activation of the BDNF receptor TrkB. We also evaluated the effects of FOXO3a, which is a well-known SIRT1 target and candidate neuroprotective factor, and found that restoration of ATP levels by SIRT1 in cultured HD striatal-like neurons depends upon FOXO3a, and that FOXO3a overexpression is associated with the recovery of BDNF and DARPP32 expression in HD cells [55]. In addition, our results and Krainc’s findings indicate that SIRT1 deacetylase activity is required for neuroprotection in HD cells. Moreover, we found that mutant Htt inhibits SIRT1 deacetylase activity [55]. Taken together, these two independent studies provide solid evidence supporting the view that SIRT1 modifies disease phenotypes in mouse models of HD.
In contrast, there is a disconnection between the observed effects of SIRT1 on aggregation of mutant Htt protein: SIRT1 overexpression or knockout modulates inclusion formation in the brain of the R6/2 mice [55], whereas SIRT1 has no effect on mutant Htt aggregation in N171-82Q mice [54]. This difference may be because aggregation in R6/2 is more pronounced and less variable than it is in N171-82Q mice. In addition, several published reports implicate insulin signaling in the regulation of protein aggregation, including in HD [65]. It is therefore possible that altered insulin sensitivity in these HD mice contributes to the observed differences, but further studies will be required to address this in more detail. Nonetheless, this difference in aggregation does not minimize the key finding that SIRT1 rescues neuronal loss in different HD-like mouse models.
There is also discrepancy in the effects of SIRT1 in HD mouse survival. SIRT1 overexpression extended survival of R6/2 mice, but not N171-82Q mice. Two important points may provide some clarification on this issue. First, we showed that N171-82Q mice develop insulin resistance that was not seen in the R6/2 model. Published reports describe a complicated relation between insulin signaling and neurodegeneration. While some papers suggest that reduced insulin signaling is protective in CNS, others suggest the opposite [65]. Similarly, the role of SIRT1 in insulin signaling is complex and primarily depends on context. In relation to TORC1 discussed in Krainc’s paper, recent work showed that increased insulin signaling inhibits TORC1 activity [66]. It is therefore possible that insulin resistance in N171-82Q mice contributes to differences in survival upon SIRT1 rescue. Further studies will be required to resolve this issue. Second, it is not precisely known why HD-like mice die. Studies by La Spada [11] showed that Htt fragment models (R6/2 and N171-82Q) develop profound hypothermia that presumably contributes to death in these animals (since hypothermia was not observed in HD-like mice that have normal life span). Since humans have very little brown fat compared to rodents, however; it is not likely that hypothermia plays a similar role in humans with HD [11]. Of note, patients with HD most often die from complications of pneumonia. Therefore it can be argued that survival as an outcome measure in HD-like mice, although very useful, does not always directly inform about human disease. Nonetheless, it is widely accepted that neuropathological changes observed in HD-like mice strongly resemble those in human disease, and on this key measure, two papers are in agreement. Specifically, both papers establish that SIRT1 mediated rescue of striatal atrophy, the hallmark of HD. Importantly, the groups used different methods to achieve the same conclusion.
These findings underscore the complexity inherent in working with alternative HD models and various SIRT1 mouse models in which SIRT1 modulation is accomplished in different ways. These two studies provide compelling support to the view that SIRT1 provides beneficial effects in HD mouse models, however, and also raise important questions. It is possible that the contradictory results on the effects of SIRT1 in models of HD might be explained by different effector pathways or mechanisms and by context-dependent effects or different levels of SIRT1 activation. SIRT1 has numerous targets, and different models of HD display different phenotypes as a result of activation of various targets and mechanisms. Therefore, it is not surprising to encounter contradictory data, especially in different species and different models.
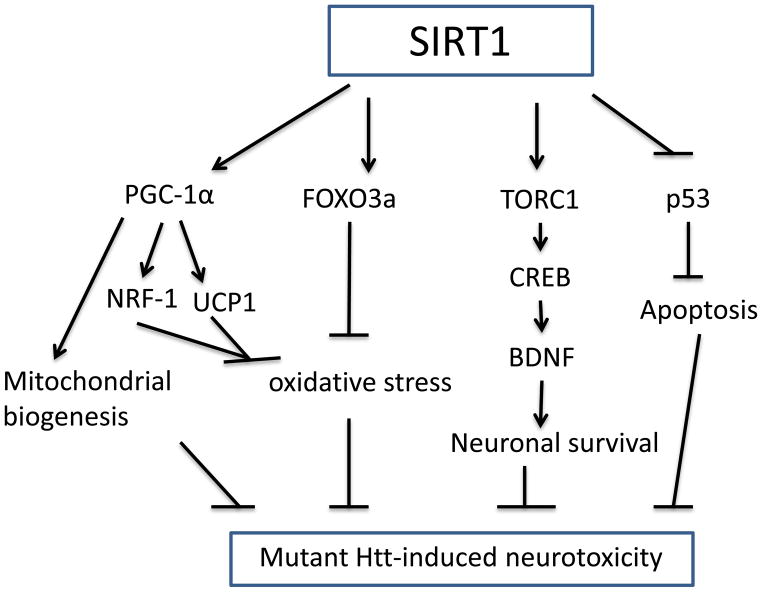
As already described, the role of SIRT1 in HD has been debated, and conflicting reports have argued that either SIRT1 activation or inhibition can be neuroprotective [52]. Although there are substrates that are critical for mediating the biological effects of SIRT1 (Figure 1), the mechanism(s) underlying these controversial effects of SIRT1 on mutant Htt between mammalian system and drosophila are not known. SIRT1 might deacetylate different substrates and subsequently has different consequence. The downstream signaling pathways reveal important differences between mammals and flies. For example, drosophila activates the NF-κB precursor Relish by IKK- and caspase-dependent endoproteolytic cleavage. On the other hand, mammalian NF-κBs are activated by the proteasome-mediated destruction of IκB proteins, and/or the proteasome-mediated degradation of the C-terminal inhibitory domain of NF-κB precursors p100 and p105 [67]. Despite this caveat, SIRT1 deserves careful consideration as a therapeutic target for HD. Hence, although many questions remain to be answered, these two studies in mammalian HD models suggest that SIRT1 should receive even more attention, particularly in seeking treatments for HD.
Figure 1. Schematic diagram of potential neuroprotective molecular mechanisms activated by SIRT1 in Huntington’s disease.
SIRT1 deacetylates its multiple targets, including PGC-1α, FOXO3a, TORC1, and p53, which are altered the activity by mutant huntingtin (Htt). SIRT1 increases PGC-1α transcriptional activity and thereby activates mitochondrial factors NRF-1, and UCP1 which prevent oxidative stress; at the same time, PGC-1α also regulates other mitochondrial genes and promotes mitochondrial biogenesis. Deacetylation of FOXO3a promotes its anti-oxidant activity. TORC1 is a coactivator of transcription factor CREB that promotes transcription of brain derived neurotrophic factor (BDNF), SIRT1 deacetylates TORC1 and increases its binding to CREB. The transcription activity of proapoptotic factor p53 can be inhibited by SIRT1 deacetylation.
2. The application of small-molecule SIRT1 modulators in HD
2.1. SIRT1 activators in HD
Resveratrol, a natural polyphenolic compound, was found to be an activator of SIRT1 in vitro [68, 69]. It has been reported that resveratrol produces changes associated with longer life-span, including increased insulin sensitivity, reduced insulin-like growth factor-1 (IGF-I) levels, increased AMP-activated protein kinase (AMPK) and PGC-1α activity, increased mitochondrial number, and improved motor function [69, 70]. The first report of the effect of resveratrol in HD came from Parker and co-workers [51], showed that resveratrol rescued mutant polyglutamine-induced neuronal death in striatal cells derived from HdhQ111 knock-in mice. Later, resveratrol exhibited protective effect in 3-nitropropionic acid (3-NP)-induced HD mouse model and genetic HD models [71, 72]. Pallos and co-worker showed that resveratrol dose-dependently increased neuronal survival in mutant Htt-challenged flies [52].
Whether the biological effects observed for resveratrol administration are causally and mechanistically associated with direct activation of SIRT1 is currently a matter of debate. In addition, whether or not resveratrol can penetrate the blood-brain barrier and act directly on the striatal neurons is not known. Recently, Ho and co-workers reported that resveratrol treatment increased expression of PGC-1α, and its downstream targets, nuclear respiratory factor-1 (NRF-1) and uncoupling protein-1 (UCP-1) in brown adipose tissue (BAT), but resveratrol had no effect on PGC-1α, or NRF-1 in the striatum, and provided no significant improvement in motor performance, survival, or striatal atrophy in N171-82Q HD mice[68]. These results suggest that resveratrol may not cross the blood-brain barrier, and some beneficial effects of resveratrol might be due to its effects on peripheral energy metabolism. On the other hand, SIRT1 regulates liver kinase b1 (LKB1), an upstream regulator of adenine monophosphate-activated protein kinase (AMPK) [73, 74]. Thus the effects of resveratrol may be mediated by inhibition of LKB1. Indeed, knockdown of LKB1 reduces the ability of resveratrol to protect cells from mitochondrial dysfunction [74]. Importantly, resveratrol has not improved mitochondrial function in animals lacking SIRT1, and SIRT1 plays an essential role in the ability of moderate doses of resveratrol to activate AMPK and improve mitochondrial function [75].
Even though there are multiple pharmacological effects of resveratrol in various model systems, the appropriate dosage is also a critical factor [73, 74, 76]; relatively high concentrations are required to achieve desirable pharmacological effects. This has prompted a search for more potent SIRt1 activators. Improved SIRT1 agonists are beginning to emerge. High-throughput screening of small molecules led to the identification of three SIRT1-activating molecules, SRT1460, SRT1720, and SRT2183, which are structurally unrelated to and 1000-fold more potent than resveratrol in assay in vitro [77]. These molecules, when used in multiple rodent animal studies, including high-fat-diet-induced obese mice, ob/ob mice, and Zucker fa/fa rats, were able to normalize glucose homeostasis by improvements in insulin sensitivity [77]. Whether SRT1460, SRT1720, and SRT2183 act as direct SIRT1 activators is currently debated, however, a recent study attributed activation of SIRT1 by these drugs to binding to the fluorphore, Fluor-de-Lys biochemical assay, which is a commonly used assay for SIRT1 activity, and thus may be an artifact. These results cast doubts on the specificity of these molecules, and whether their pharmacological action might be ascribed to one or more of their demonstrated off-target activities [78].
It is well known that increased intracellular levels of NAD+ activate sirtuin-dependent metabolic control. Thus, compounds that modulate NAD+/NADH ratios are likely to affect SIRT1-mediated metabolic control. Tests of brain-penetrable specific SIRT1 activators in HD mouse models will provide important insight on whether activation of SIRT1 can be a therapeutic target in HD. There is an ongoing effort to develop specific SIRT1 activators that can penetrate the brain. Sirtuis, a GSK company, is currently developing this kind of compounds. Test of brain penetrable Sirt1 activators in HD mouse model is undergoing, hope the results will be released in near future.
3.2 SIRT1 inhibitors in HD
Whether SIRT1 should be inhibited instead of activated in HD is controversial. The catalytic activity of SIRT1 is physiologically inhibited by nicotinamide (NAM). Mechanistically, NAM binds to a conserved region in the SIRT1 catalytic site and favors a Base Exchange reaction instead of deacetylation. Hathorn and co-workers reported that NAM improves motor function and upregulates PGC-1α and BDNF expression in an HD mouse model [79]. NAM is also a substrate for NAD+ biosynthesis, however, and it is also believed to promote improvements in energy production because of its role as a precursor of NAD, an important molecule involved in energy metabolism. Increasing NAM concentrations increases the available NAD molecules that are involved in energy metabolism, thus increasing the amount of energy available in the cell. NAM can also increase cellular energy by inhibiting poly-ADP-ribose polymerase. When poly-ADP-ribose polymerase is activated, it depletes the supply of NAD by transferring poly-ADP-ribose subunits from NAD to various DNA repair enzymes. Depletion of NAD leads to the depletion of ATP owing to decreased activity in both glycolysis and the Krebs cycle. When NAM inhibits the poly-ADP ribose polymerase, it essentially prevents the NAD molecules from becoming depleted, administration of NAM to mice may actually result in increased NAD+ biosynthesis and SIRT1 activation. Therefore, the effects of NAM in HD mice may result from activation of SIRT1 instead of its inhibition.
The current challenge is that no specific SIRT1 inhibitors are available, and the first generation of sirtuin deacetylase inhibitors such as sirtinol [80], identified in high-throughput screening, had low potency and selectivity [81]. Subsequently, potency and selectivity were greatly improved by rational drug design [82]. These redesigned sirtinol derivatives were found to be more potent, but they remain non-selective inhibitors of both SIRT1 and SIRT2. Further high-throughput screening has led to the finding that indoles are potent and selective SIRT1 inhibitors [83]. Interestingly, kinetic analyses suggest that these inhibitors bind after the release of nicotinamide from the enzyme, and prevent the release of deacetylated peptide and 2′-O-acetyl-ADP-ribose, the products of enzyme-catalyzed deacetylation. These newly discovered SIRT1 inhibitors seem to be cell-permeable, orally bioavailable, and metabolically stable.
Suramin was reported as a SIRT1 inhibitor, but it has also been shown to be a weak inhibitor of SIRT5 deacetylase. Efforts to design improved derivatives of the SIRT1 inhibitor suramin have led to the discovery of highly potent and selective inhibitors of SIRT1 and SIRT2 [84]. The therapeutic application of the sirtuin inhibitors has just begun and the further efficacy trials in different animal models will be crucial for evaluating therapeutic potentials of new SIRT1 inhibitors for HD. Testing specific SIRT1 inhibitors in mammalian HD models will no doubt reveal the role of SIRT1 in HD.
Sirtinol has been shown to be beneficial in flies expressing mutant Htt [52]. Since sirtinol also inhibits SIRT2, it is not possible to exclude the notion that protection is due to inhibition of SIRT2, which has recently been shown to be protective in HD mouse models [85]. An ongoing European trial called PADDINGTON is testing a drug designed to reduce the effect of SIRT1. On the basis of their own experiments, including studies in HD mice, scientists at Sienna believe that reducing SIRT1 activity might help cells remove the mutant Htt that causes HD. This is consistent with studies in the fly model that have shown that reducing levels of Sir2 in fruit flies protects them from damage caused by mutant Htt [52]. A Phase I clinical trial is underway to treat HD with the specific SIRT1 inhibitor EX-527 [86, 87]. When released, the results should provide insight into whether SIRT1 inhibitors will be therapeutically helpful for this disease.
One possible explanation for the conflict in compounds targeting SIRT1 is that, unlike adding or removing copies of genes, a drug that is aimed at a particular target often hits several others as well, or a drug may inhibit some forms of a protein but not others. Irrespective of the results, accumulating data suggest that SIRT1 is an attractive target for intervention in HD.
3. Concluding remarks
SIRT1 offers an excellent opportunity for investigating the connection between brain aging and neurodegenerative diseases, with the aim of uncovering new molecular pathways and target molecules for drug development. Further investigation into the targets and functions of SIRT1 and other members of the sirtuin family will help in the development of new strategies for protection against neurodegenerative diseases.
Neurodegenerative disorders are complex diseases with many underlying pathways, and SIRT1 is a molecule with numerous different substrates in the cell. Therefore, it is not surprising that SIRT1 is involved in neurodegenerative disorders. Although exciting discoveries have been made in SIRT1 neurobiology, important questions still remain to be answered. For example, why SIRT1 behaves differently in different tissues needs to be precisely elucidated. As the numbers of SIRT1 substrates identified have increased, the situation has become more complex, because the pathways involved overlap with other proteins.
HD mouse models actually show non-neural phenotypes, including metabolic abnormalities, and reduced body weight has been observed in several HD mouse models as well as perturbed glucose metabolism and insulin pathway regulation. As SIRT1 elicits multiple divergent effects in the central nervous system and periphery, different outcomes might occur, depending on how, when, and where SIRT1 is activated.
Decades of biomedical progress and vast improvements in our living conditions allow us to lead productive lives into old age. Although pharmacological or genetic activation of SIRT1 resembles the beneficial effects of caloric restriction, making it an attractive drug target, we should not forget that SIRT1 acts on many different downstream targets, which are involved in numerous biological activities. Drugs that can specifically activate SIRT1 and inhibit SIRT1 need to be tested in HD mammalian models, in order to identify whether the benefits of these drugs are sustained. If so, clinical trials will be necessary to follow up and to address whether SIRT1 modulators have beneficial effects in HD patients.
Acknowledgments
The authors acknowledge grants NS074196 (W.D), NS 072344 (W.D) from the National Institutes of Health, CHDI A3875 (W.D), Hereditary Disease foundation (W.D) which supported preparation of this manuscript and some of the research reported herein.
Footnotes
The author declares no conflicts of interest in writing this article.
References
- 1.A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell. 1993 Mar 26;72(6):971–83. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.Langbehn DR, Brinkman RR, Falush D, Paulsen JS, Hayden MR. A new model for prediction of the age of onset and penetrance for Huntington’s disease based on CAG length. Clin Genet. 2004 Apr;65(4):267–77. doi: 10.1111/j.1399-0004.2004.00241.x. [DOI] [PubMed] [Google Scholar]
- 3.Hoppitt T, Calvert M, Pall H, Rickards H, Sackley C. Huntington’s disease. Lancet. 2010 Oct 30;376(9751):1463–4. doi: 10.1016/S0140-6736(10)61989-7. [DOI] [PubMed] [Google Scholar]
- 4.Ross CA, Tabrizi SJ. Huntington’s disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 2011 Jan;10(1):83–98. doi: 10.1016/S1474-4422(10)70245-3. [DOI] [PubMed] [Google Scholar]
- 5.Wexler NS, Lorimer J, Porter J, Gomez F, Moskowitz C, Shackell E, et al. Venezuelan kindreds reveal that genetic and environmental factors modulate Huntington’s disease age of onset. Proc Natl Acad Sci U S A. 2004 Mar 9;101(10):3498–503. doi: 10.1073/pnas.0308679101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Browne SE. Mitochondria and Huntington’s disease pathogenesis: insight from genetic and chemical models. Ann N Y Acad Sci. 2008 Dec;1147:358–82. doi: 10.1196/annals.1427.018. [DOI] [PubMed] [Google Scholar]
- 7.Gu M, Gash MT, Mann VM, Javoy-Agid F, Cooper JM, Schapira AH. Mitochondrial defect in Huntington’s disease caudate nucleus. Ann Neurol. 1996 Mar;39(3):385–9. doi: 10.1002/ana.410390317. [DOI] [PubMed] [Google Scholar]
- 8.Mann VM, Cooper JM, Javoy-Agid F, Agid Y, Jenner P, Schapira AH. Mitochondrial function and parental sex effect in Huntington’s disease. Lancet. 1990 Sep 22;336(8717):749. doi: 10.1016/0140-6736(90)92242-a. [DOI] [PubMed] [Google Scholar]
- 9.Napoli E, Wong S, Hung C, Ross-Inta C, Bomdica P, Giulivi C. Defective mitochondrial disulfide relay system, altered mitochondrial morphology and function in Huntington’s disease. Hum Mol Genet. 2013 Mar 1;22(5):989–1004. doi: 10.1093/hmg/dds503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imarisio S, Carmichael J, Korolchuk V, Chen CW, Saiki S, Rose C, et al. Huntington’s disease: from pathology and genetics to potential therapies. Biochem J. 2008 Jun 1;412(2):191–209. doi: 10.1042/BJ20071619. [DOI] [PubMed] [Google Scholar]
- 11.Weydt P, Pineda VV, Torrence AE, Libby RT, Satterfield TF, Lazarowski ER, et al. Thermoregulatory and metabolic defects in Huntington’s disease transgenic mice implicate PGC-1alpha in Huntington’s disease neurodegeneration. Cell Metab. 2006 Nov;4(5):349–62. doi: 10.1016/j.cmet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 12.Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006 Oct 6;127(1):59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 13.Turner C, Schapira AH. Mitochondrial matters of the brain: the role in Huntington’s disease. J Bioenerg Biomembr. 2010 Jun;42(3):193–8. doi: 10.1007/s10863-010-9290-y. [DOI] [PubMed] [Google Scholar]
- 14.Oliveira JM. Mitochondrial bioenergetics and dynamics in Huntington’s disease: tripartite synapses and selective striatal degeneration. J Bioenerg Biomembr. 2010 Jun;42(3):227–34. doi: 10.1007/s10863-010-9287-6. [DOI] [PubMed] [Google Scholar]
- 15.Quintanilla RA, Johnson GV. Role of mitochondrial dysfunction in the pathogenesis of Huntington’s disease. Brain Res Bull. 2009 Oct 28;80(4–5):242–7. doi: 10.1016/j.brainresbull.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beal MF. Mitochondria and neurodegeneration. Novartis Found Symp. 2007;287:183–92. doi: 10.1002/9780470725207.ch13. discussion 92–6. [DOI] [PubMed] [Google Scholar]
- 17.Mochel F, Haller RG. Energy deficit in Huntington disease: why it matters. J Clin Invest. 2011 Feb;121(2):493–9. doi: 10.1172/JCI45691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenstock TR, Duarte AI, Rego AC. Mitochondrial-Associated Metabolic Changes and Neurodegeneration in Huntington’s Disease - From Clinical Features to the Bench. Curr Drug Targets. 2010 Jul 1; doi: 10.2174/1389450111007011218. [DOI] [PubMed] [Google Scholar]
- 19.Browne SE, Beal MF. Oxidative damage in Huntington’s disease pathogenesis. Antioxid Redox Signal. 2006 Nov-Dec;8(11–12):2061–73. doi: 10.1089/ars.2006.8.2061. [DOI] [PubMed] [Google Scholar]
- 20.Chen HI, Liou SH, Ho SF, Wu KY, Sun CW, Chen MF, et al. Oxidative DNA damage estimated by plasma 8-hydroxydeoxyguanosine (8-OHdG): influence of 4, 4′-methylenebis (2-chloroaniline) exposure and smoking. J Occup Health. 2007 Sep;49(5):389–98. doi: 10.1539/joh.49.389. [DOI] [PubMed] [Google Scholar]
- 21.Sorolla MA, Reverter-Branchat G, Tamarit J, Ferrer I, Ros J, Cabiscol E. Proteomic and oxidative stress analysis in human brain samples of Huntington disease. Free Radic Biol Med. 2008 Sep 1;45(5):667–78. doi: 10.1016/j.freeradbiomed.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 22.Santamaria A, Perez-Severiano F, Rodriguez-Martinez E, Maldonado PD, Pedraza-Chaverri J, Rios C, et al. Comparative analysis of superoxide dismutase activity between acute pharmacological models and a transgenic mouse model of Huntington’s disease. Neurochem Res. 2001 Apr;26(4):419–24. doi: 10.1023/a:1010911417383. [DOI] [PubMed] [Google Scholar]
- 23.Nisoli E, Tonello C, Cardile A, Cozzi V, Bracale R, Tedesco L, et al. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005 Oct 14;310(5746):314–7. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- 24.Haigis MC, Guarente LP. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006 Nov 1;20(21):2913–21. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 25.Chalkiadaki A, Guarente L. Sirtuins mediate mammalian metabolic responses to nutrient availability. Nat Rev Endocrinol. 2012 May;8(5):287–96. doi: 10.1038/nrendo.2011.225. [DOI] [PubMed] [Google Scholar]
- 26.Shimokawa I, Trindade LS. Dietary restriction and aging in rodents: a current view on its molecular mechanisms. Aging Dis. 2010 Oct;1(2):89–107. [PMC free article] [PubMed] [Google Scholar]
- 27.Canto C, Auwerx J. Caloric restriction, SIRT1 and longevity. Trends Endocrinol Metab. 2009 Sep;20(7):325–31. doi: 10.1016/j.tem.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wakeling LA, Ions LJ, Ford D. Could Sirt1-mediated epigenetic effects contribute to the longevity response to dietary restriction and be mimicked by other dietary interventions? Age (Dordr) 2009 Dec;31(4):327–41. doi: 10.1007/s11357-009-9104-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maiese K, Chong ZZ, Shang YC. OutFOXOing disease and disability: the therapeutic potential of targeting FoxO proteins. Trends Mol Med. 2008 May;14(5):219–27. doi: 10.1016/j.molmed.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hasegawa K, Wakino S, Yoshioka K, Tatematsu S, Hara Y, Minakuchi H, et al. Sirt1 protects against oxidative stress-induced renal tubular cell apoptosis by the bidirectional regulation of catalase expression. Biochem Biophys Res Commun. 2008 Jul 18;372(1):51–6. doi: 10.1016/j.bbrc.2008.04.176. [DOI] [PubMed] [Google Scholar]
- 31.Chong ZZ, Maiese K. Enhanced tolerance against early and late apoptotic oxidative stress in mammalian neurons through nicotinamidase and sirtuin mediated pathways. Curr Neurovasc Res. 2008 Aug;5(3):159–70. doi: 10.2174/156720208785425666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanno M, Kuno A, Yano T, Miura T, Hisahara S, Ishikawa S, et al. Induction of manganese superoxide dismutase by nuclear translocation and activation of SIRT1 promotes cell survival in chronic heart failure. J Biol Chem. 2010 Mar 12;285(11):8375–82. doi: 10.1074/jbc.M109.090266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsunemi T, Ashe TD, Morrison BE, Soriano KR, Au J, Roque RA, et al. PGC-1alpha rescues Huntington’s disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci Transl Med. 2012 Jul 11;4(142):142ra97. doi: 10.1126/scitranslmed.3003799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.La Spada AR. PPARGC1A/PGC-1alpha, TFEB and enhanced proteostasis in Huntington disease: defining regulatory linkages between energy production and protein-organelle quality control. Autophagy. 2012 Dec;8(12):1845–7. doi: 10.4161/auto.21862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramadori G, Lee CE, Bookout AL, Lee S, Williams KW, Anderson J, et al. Brain SIRT1: anatomical distribution and regulation by energy availability. J Neurosci. 2008 Oct 1;28(40):9989–96. doi: 10.1523/JNEUROSCI.3257-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987 May;116(1):9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guarente L. Sirtuins, aging, and metabolism. Cold Spring Harb Symp Quant Biol. 2011;76:81–90. doi: 10.1101/sqb.2011.76.010629. [DOI] [PubMed] [Google Scholar]
- 38.Guarente L, Franklin H. Epstein Lecture: Sirtuins, aging, and medicine. N Engl J Med. 2011 Jun 9;364(23):2235–44. doi: 10.1056/NEJMra1100831. [DOI] [PubMed] [Google Scholar]
- 39.Nakagawa T, Guarente L. Sirtuins at a glance. J Cell Sci. 2011 Mar 15;124(Pt 6):833–8. doi: 10.1242/jcs.081067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J Biol Chem. 2007 Mar 2;282(9):6823–32. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 41.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–95. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004 Aug 13;305(5686):1010–3. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- 43.Srivastava S, Haigis MC. Role of sirtuins and calorie restriction in neuroprotection: implications in Alzheimer’s and Parkinson’s diseases. Curr Pharm Des. 2011;17(31):3418–33. doi: 10.2174/138161211798072526. [DOI] [PubMed] [Google Scholar]
- 44.Donmez G, Wang D, Cohen DE, Guarente L. SIRT1 suppresses beta-amyloid production by activating the alpha-secretase gene ADAM10. Cell. 2010 Jul 23;142(2):320–32. doi: 10.1016/j.cell.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Polito L, Kehoe PG, Forloni G, Albani D. The molecular genetics of sirtuins: association with human longevity and age-related diseases. Int J Mol Epidemiol Genet. 2010;1(3):214–25. [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J, Fivecoat H, Ho L, Pan Y, Ling E, Pasinetti GM. The role of Sirt1: at the crossroad between promotion of longevity and protection against Alzheimer’s disease neuropathology. Biochim Biophys Acta. 2010 Aug;1804(8):1690–4. doi: 10.1016/j.bbapap.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 47.Kim D, Nguyen MD, Dobbin MM, Fischer A, Sananbenesi F, Rodgers JT, et al. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer’s disease and amyotrophic lateral sclerosis. EMBO J. 2007 Jul 11;26(13):3169–79. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Donmez G, Arun A, Chung CY, McLean PJ, Lindquist S, Guarente L. SIRT1 protects against alpha-synuclein aggregation by activating molecular chaperones. J Neurosci. 2012 Jan 4;32(1):124–32. doi: 10.1523/JNEUROSCI.3442-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Jiang M, Wang J, Fu J, Du L, Jeong H, West T, et al. Neuroprotective role of Sirt1 in mammalian models of Huntington’s disease through activation of multiple Sirt1 targets. Nat Med. 2012 Jan;18(1):153–8. doi: 10.1038/nm.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jeong H, Cohen DE, Cui L, Supinski A, Savas JN, Mazzulli JR, et al. Sirt1 meiates neuroprotection from mutant huntingtin by activation of the TORC1 and CREB transcriptional pathway. Nat Med. 2012 Jan;18(1):159–65. doi: 10.1038/nm.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parker JA, Arango M, Abderrahmane S, Lambert E, Tourette C, Catoire H, et al. Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nat Genet. 2005 Apr;37(4):349–50. doi: 10.1038/ng1534. [DOI] [PubMed] [Google Scholar]
- 52.Pallos J, Bodai L, Lukacsovich T, Purcell JM, Steffan JS, Thompson LM, et al. Inhibition of specific HDACs and sirtuins suppresses pathogenesis in a Drosophila model of Huntington’s disease. Hum Mol Genet. 2008 Dec 1;17(23):3767–75. doi: 10.1093/hmg/ddn273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burnett C, Valentini S, Cabreiro F, Goss M, Somogyvari M, Piper MD, et al. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 2011 Sep 22;477(7365):482–5. doi: 10.1038/nature10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang M, Wang J, Fu J, Du L, Jeong H, West T, et al. Neuroprotective role of Sirt1 in mammalian models of Huntington’s disease through activation of multiple Sirt1 targets. Nat Med. Jan;18(1):153–8. doi: 10.1038/nm.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jeong H, Cohen DE, Cui L, Supinski A, Savas JN, Mazzulli JR, et al. Sirt1 mediates neuroprotection from mutant huntingtin by activation of the TORC1 and CREB transcriptional pathway. Nat Med. Jan;18(1):159–65. doi: 10.1038/nm.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zuccato C, Liber D, Ramos C, Tarditi A, Rigamonti D, Tartari M, et al. Progressive loss of BDNF in a mouse model of Huntington’s disease and rescue by BDNF delivery. Pharmacol Res. 2005 Aug;52(2):133–9. doi: 10.1016/j.phrs.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 57.Zuccato C, Ciammola A, Rigamonti D, Leavitt BR, Goffredo D, Conti L, et al. Loss of huntingtin-mediated BDNF gene transcription in Huntington’s disease. Science. 2001 Jul 20;293(5529):493–8. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- 58.Zuccato C, Cattaneo E. Role of brain-derived neurotrophic factor in Huntington’s disease. Prog Neurobiol. 2007 Apr;81(5–6):294–330. doi: 10.1016/j.pneurobio.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 59.Xie Y, Hayden MR, Xu B. BDNF overexpression in the forebrain rescues Huntington’s disease phenotypes in YAC128 mice. J Neurosci. 2010 Nov 3;30(44):14708–18. doi: 10.1523/JNEUROSCI.1637-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Giralt A, Carreton O, Lao-Peregrin C, Martin ED, Alberch J. Conditional BDNF release under pathological conditions improves Huntington’s disease pathology by delaying neuronal dysfunction. Mol Neurodegener. 2011;6(1):71. doi: 10.1186/1750-1326-6-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Duan W, Guo Z, Jiang H, Ware M, Li XJ, Mattson MP. Dietary restriction normalizes glucose metabolism and BDNF levels, slows disease progression, and increases survival in huntingtin mutant mice. Proc Natl Acad Sci U S A. 2003 Mar 4;100(5):2911–6. doi: 10.1073/pnas.0536856100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Duan W, Peng Q, Masuda N, Ford E, Tryggestad E, Ladenheim B, et al. Sertraline slows disease progression and increases neurogenesis in N171-82Q mouse model of Huntington’s disease. Neurobiol Dis. 2008 Jun;30(3):312–22. doi: 10.1016/j.nbd.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peng Q, Masuda N, Jiang M, Li Q, Zhao M, Ross CA, et al. The antidepressant sertraline improves the phenotype, promotes neurogenesis and increases BDNF levels in the R6/2 Huntington’s disease mouse model. Exp Neurol. 2008 Mar;210(1):154–63. doi: 10.1016/j.expneurol.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Duan W, Guo Z, Jiang H, Ware M, Mattson MP. Reversal of behavioral and metabolic abnormalities, and insulin resistance syndrome, by dietary restriction in mice deficient in brain-derived neurotrophic factor. Endocrinology. 2003 Jun;144(6):2446–53. doi: 10.1210/en.2002-0113. [DOI] [PubMed] [Google Scholar]
- 65.Cohen E, Dillin A. The insulin paradox: aging, proteotoxicity and neurodegeneration. Nat Rev Neurosci. 2008 Oct;9(10):759–67. doi: 10.1038/nrn2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang B, Goode J, Best J, Meltzer J, Schilman PE, Chen J, et al. The insulin-regulated CREB coactivator TORC promotes stress resistance in Drosophila. Cell Metab. 2008 May;7(5):434–44. doi: 10.1016/j.cmet.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Silverman N, Maniatis T. NF-kappaB signaling pathways in mammalian and insect innate immunity. Genes Dev. 2001 Sep 15;15(18):2321–42. doi: 10.1101/gad.909001. [DOI] [PubMed] [Google Scholar]
- 68.Ho DJ, Calingasan NY, Wille E, Dumont M, Beal MF. Resveratrol protects against peripheral deficits in a mouse model of Huntington’s disease. Exp Neurol. 2010 Sep;225(1):74–84. doi: 10.1016/j.expneurol.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 69.Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006 Nov 16;444(7117):337–42. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baur JA, Sinclair DA. Therapeutic potential of resveratrol: the in vivo evidence. Nat Rev Drug Discov. 2006 Jun;5(6):493–506. doi: 10.1038/nrd2060. [DOI] [PubMed] [Google Scholar]
- 71.Kumar P, Padi SS, Naidu PS, Kumar A. Effect of resveratrol on 3-nitropropionic acid-induced biochemical and behavioural changes: possible neuroprotective mechanisms. Behav Pharmacol. 2006 Sep;17(5–6):485–92. doi: 10.1097/00008877-200609000-00014. [DOI] [PubMed] [Google Scholar]
- 72.Maher P, Dargusch R, Bodai L, Gerard PE, Purcell JM, Marsh JL. ERK activation by the polyphenols fisetin and resveratrol provides neuroprotection in multiple models of Huntington’s disease. Hum Mol Genet. 2011 Jan 15;20(2):261–70. doi: 10.1093/hmg/ddq460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Scott E, Steward WP, Gescher AJ, Brown K. Resveratrol in human cancer chemoprevention--choosing the ‘right’ dose. Mol Nutr Food Res. 2012 Jan;56(1):7–13. doi: 10.1002/mnfr.201100400. [DOI] [PubMed] [Google Scholar]
- 74.Stocco B, Toledo K, Salvador M, Paulo M, Koyama N, Torqueti Toloi MR. Dose-dependent effect of resveratrol on bladder cancer cells: chemoprevention and oxidative stress. Maturitas. 2012 May;72(1):72–8. doi: 10.1016/j.maturitas.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 75.Price NL, Gomes AP, Ling AJ, Duarte FV, Martin-Montalvo A, North BJ, et al. SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab. 2012 May 2;15(5):675–90. doi: 10.1016/j.cmet.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Girbovan C, Morin L, Plamondon H. Repeated resveratrol administration confers lasting protection against neuronal damage but induces dose-related alterations of behavioral impairments after global ischemia. Behav Pharmacol. 2012 Feb;23(1):1–13. doi: 10.1097/FBP.0b013e32834eafa3. [DOI] [PubMed] [Google Scholar]
- 77.Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007 Nov 29;450(7170):712–6. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pacholec M, Bleasdale JE, Chrunyk B, Cunningham D, Flynn D, Garofalo RS, et al. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J Biol Chem. 2010 Mar 12;285(11):8340–51. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hathorn T, Snyder-Keller A, Messer A. Nicotinamide improves motor deficits and upregulates PGC-1alpha and BDNF gene expression in a mouse model of Huntington’s disease. Neurobiol Dis. 2011 Jan;41(1):43–50. doi: 10.1016/j.nbd.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov. 2008 Oct;7(10):854–68. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- 81.Grozinger CM, Chao ED, Blackwell HE, Moazed D, Schreiber SL. Identification of a class of small molecule inhibitors of the sirtuin family of NAD-dependent deacetylases by phenotypic screening. J Biol Chem. 2001 Oct 19;276(42):38837–43. doi: 10.1074/jbc.M106779200. [DOI] [PubMed] [Google Scholar]
- 82.Mai A, Massa S, Lavu S, Pezzi R, Simeoni S, Ragno R, et al. Design, synthesis, and biological evaluation of sirtinol analogues as class III histone/protein deacetylase (Sirtuin) inhibitors. J Med Chem. 2005 Dec 1;48(24):7789–95. doi: 10.1021/jm050100l. [DOI] [PubMed] [Google Scholar]
- 83.Napper AD, Hixon J, McDonagh T, Keavey K, Pons JF, Barker J, et al. Discovery of indoles as potent and selective inhibitors of the deacetylase SIRT1. J Med Chem. 2005 Dec 15;48(25):8045–54. doi: 10.1021/jm050522v. [DOI] [PubMed] [Google Scholar]
- 84.Trapp J, Meier R, Hongwiset D, Kassack MU, Sippl W, Jung M. Structure-activity studies on suramin analogues as inhibitors of NAD+-dependent histone deacetylases (sirtuins) ChemMedChem. 2007 Oct;2(10):1419–31. doi: 10.1002/cmdc.200700003. [DOI] [PubMed] [Google Scholar]
- 85.Chopra V, Quinti L, Kim J, Vollor L, Narayanan KL, Edgerly C, et al. The sirtuin 2 inhibitor AK-7 is neuroprotective in Huntington’s disease mouse models. Cell Rep. 2012 Dec 27;2(6):1492–7. doi: 10.1016/j.celrep.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang F, Wang S, Gan L, Vosler PS, Gao Y, Zigmond MJ, et al. Protective effects and mechanisms of sirtuins in the nervous system. Prog Neurobiol. 2011 Nov;95(3):373–95. doi: 10.1016/j.pneurobio.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Donmez G, Outeiro TF. SIRT1 and SIRT2: emerging targets in neurodegeneration. EMBO Mol Med. 2013 Feb 18; doi: 10.1002/emmm.201302451. [DOI] [PMC free article] [PubMed] [Google Scholar]