Abstract
Sindbis virus subgenomic mRNA is efficiently translated in infected vertebrate cells whereas host translation is shut-off. Deletions in the 5′ UTR of the subgenomic mRNA were made to investigate its role in viral gene expression. Deletion of nucleotides 1-10 and 11-20 caused a small plaque phenotype, reduced levels of subgenomic mRNA and structural proteins, and increased expression of nonstructural proteins. Whereas deletion 1-10 virus inhibited cellular protein synthesis, deletion 11-20 did so inefficiently. A large plaque revertant of deletion 11-20, possessing a duplication of the subgenomic promoter region, produced subgenomic mRNA at WT levels and restored inhibition of host protein synthesis. Further analysis of the mutant and revertant 5′UTR sequences showed the ability to shut-off host cell translation correlated with the efficiency of translation of subgenomic mRNA. We propose that the translational efficiency and quantity of the subgenomic mRNA play a role in inhibition of host cell translation.
INTRODUCTION
Sindbis Virus (SINV) is member of the Togaviridae family, and as the type species of the Alphavirus genus, it is well characterized at the genetic and molecular levels (Strauss and Strauss 1994). The virus is transmitted by mosquitoes and maintained in an enzootic cycle with an avian amplifying host. The outcome of infection in the vertebrate and arthropod host differs significantly. Infection of vertebrate cells results in inhibition of host cell gene expression at both the transcriptional and translational levels and ultimately cell death. In arthropod cells infection results in the establishment of a non-cytolytic, persistent infection with no global disruption of host cell gene expression.
The SINV genome is a single stranded positive sense RNA that is 11.7 kb in length, capped, and polyadenylated (Strauss and Strauss 1994). The 5′ two thirds of the genome encodes the nonstructural proteins that function to synthesize viral RNA and the 3′ one third encodes the structural proteins, which are translated from a subgenomic mRNA (SG mRNA) and function to assemble virus particles. The nonstructural proteins (nsP1-4) are translated as two polyproteins: P123, and read through of an opal stop codon produces P1234. The polyproteins are processed by the nsP2-associated proteinase activity to nsP1, nsP2, nsP3, and nsP4 (Ding and Schlesinger 1989; Hahn, Strauss et al. 1989; Hardy and Strauss 1989). The nonstructural proteins and the polyprotein intermediates function to replicate the genome and produce the SG mRNA (Lemm, Rumenapf et al. 1994; Shirako and Strauss 1994).
The SG mRNA is co-linear with the 3′ end of the genome and is transcribed from an internal promoter in the minus-strand replication intermediate (Cancedda, Villa-Komaroff et al. 1975) located -19/+5 in reference to the initiation site (Levis, Schlesinger et al. 1990; Raju and Huang 1991). The SG mRNA is a capped and polyadenylated 4.1 kb mRNA that has a 49 nt 5′UTR and a 322 nt 3′UTR (Strauss and Strauss 1994). Several factors are known to regulate transcription of the SG mRNA. In addition to the promoter region referenced above, Wielgosz et al. identified elements from -40/-20 and +6/+14 that separately modulated transcription (Wielgosz, Raju et al. 2001). Distinct amino acids in nsP4 (the viral RNA dependent RNA polymerase) were found to differentially effect promoter recognition and synthesis of G and SG RNAs, suggesting different mechanisms for generation of the two RNAs (Li and Stollar 2007). These mechanisms possibly alter nsP4 binding to the RNA or modulate activity of the RNA synthetic complex by altering its composition. Additionally, in work with Semliki Forest Virus (SFV) temperature sensitive mutants of nsP2 were found to decrease transcription of the 26S SG mRNA, and it was proposed that nsP2 interacts with the subgenomic promoter as a transcription factor (Suopanki, Sawicki et al. 1998). In addition to viral proteins, the host protein hnRNP K has been shown to bind to SG mRNA and may also modulate SG mRNA gene expression (Burnham, Gong et al. 2007).
Translation of the SG mRNA differs from translation of cellular mRNAs and even the SINV genomic RNA in that there are reduced requirements for specific initiation factors. The presence of a translational enhancer element in the 5′ proximal coding region of the SG mRNA allows for efficient translation in the absence of eIF2 (Frolov and Schlesinger 1994; Frolov and Schlesinger 1996; Sanz, Castello et al. 2009) in vertebrate cells. PKR activation as a consequence of virus replication and dsRNA detection results in the phosphorylation of eIF2α and consequently a shut-off of host cell translation. Thus the presence of the translational enhancer allows translation of viral structural proteins in an eIF2-limited environment. It has been observed that there is a reduced requirement for intact eIF4F components during SG mRNA translation in vertebrate cells (Sanz, Castello et al. 2009; Garcia-Moreno, Sanz et al.). It has also been reported that the related Semliki Forest virus SG mRNA bound eIF-4B and eIF-4E with greater affinity than cellular messages, potentially utilizing the initiation factors at lower concentrations (van Steeg, Thomas et al. 1981). This reduced requirement for translation factors may account for the ability of the SG mRNA to be efficiently translated in an environment in which cellular translation is inhibited.
The inhibition of cellular gene expression during SINV infection of vertebrate cells is the result of two indepedndent mechanisms: inhibition of transcription, and inhibition of translation (Gorchakov, Frolova et al. 2005). It is well established that nsP2 inhibits cellular transcription (Frolov, Agapov et al. 1999; Gorchakov, Frolova et al. 2005; Garmashova, Gorchakov et al. 2006); however, the mechanism by which translation of cellular mRNA is inhibited is not well understood. Recognition of dsRNA by PKR and consequent phosphorylation of eIF2α plays a role, but, as was demonstrated by Gorchakov et.al, this does not fully account for the inhibition of translation (Gorchakov, Frolova et al. 2004). Mutations in nsP2 and nsP4 were shown to prevent effective inhibition of host cell translation (Gorchakov, Frolova et al. 2004; Mayuri, Geders et al. 2008; Rupp, Jundt et al. 2011). Nevertheless, it should be noted that in each of these cases the mutations in the nonstructural protein also resulted in decreased virus RNA synthesis, particularly SG mRNA. These observations imply a role for the SG mRNA in inhibition of host cell translation.
Sequence and structural features of the SG mRNA implicated in its translation include the 5′UTR, the 3′UTR and the translational enhancer element located in the capsid coding region. While the role of the translational enhancer element has been clearly established in translation of the SG mRNA (Frolov and Schlesinger 1994; Frolov and Schlesinger 1996; Garcia-Moreno, Sanz et al. 2012; Ventoso 2012), the role of the 5′UTR in SG mRNA translation has not been extensively described. Through deletion analysis we began to assess the role of the 5′UTR during SG mRNA translation. A deletion of nucleotides 11-20 of the SG mRNA 5′UTR produced virus that was impaired for SG mRNA transcription, produced higher levels of nsP2, but failed to efficiently shutoff host cell translation. In contrast a deletion of nucleotides 1-10 of the SG mRNA 5'UTR was able to inhibit host cell translation while producing low levels of SG mRNA and high levels of nsP2. A plaque-purified revertant of Del11-20 contained a duplication of the subgenomic promoter that led to transcription of two SG mRNAs: that of the parental mutant and a larger SG mRNA possessing a 112 base extension of the 5′UTR. The revertant virus transcribed wild type levels of SG mRNA, efficiently inhibited host translation early in infection, but continued translation of nonstructural proteins late in infection. Examination of the effect of the WT, Del1-10, Del11-20 and revertant subgenomic 5'UTR on reporter gene expression in infected cells revealed that the efficiency with which the 5′UTR programmed translation correlated directly with the ability of the virus to inhibit host cell translation. On the basis of these data we propose that translational capacity, and to a degree, the quantity of the SG mRNA serves as a crucial factor in inhibiting host cell translation during alphavirus infection.
RESULTS
Mutations in subgenomic 5′UTR affect virus growth
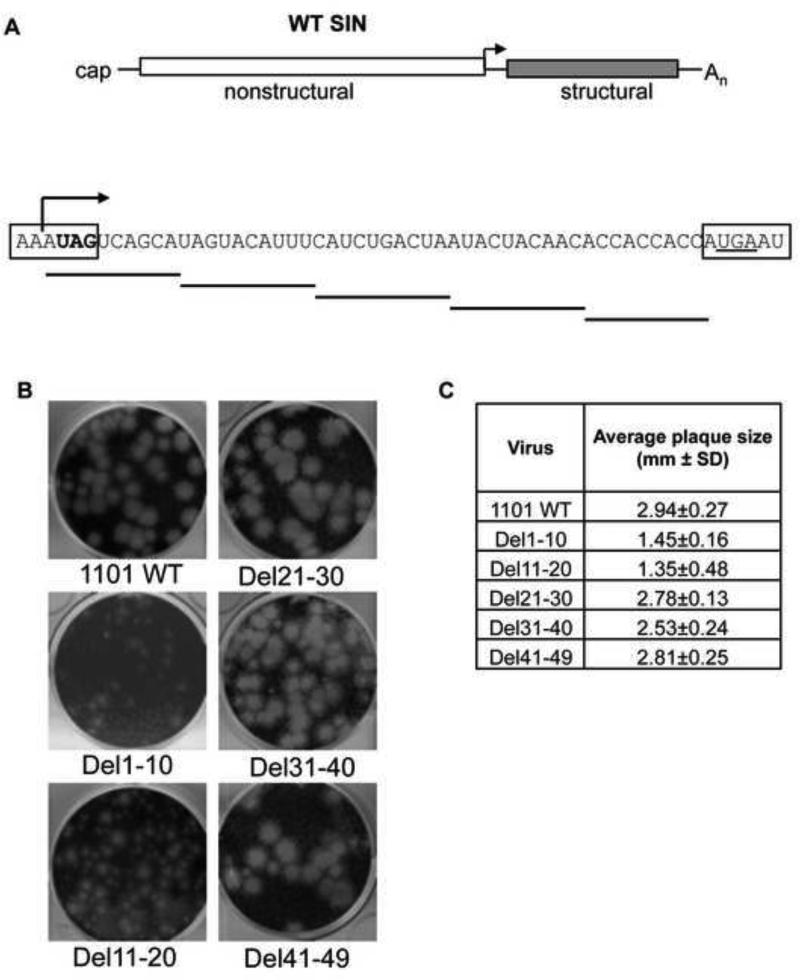
To assess the role of the subgenomic 5′UTR on virus gene expression, scanning deletions were made in the infectious clone of the viral genome corresponding to this region of the SG mRNA. Virus was generated by transfection of full-length in vitro transcribed RNA (Figure 1A). Virus was collected after 48 hours and characterized on the basis of plaque phenotype. Deletion of nucleotides 1-10 (Del1-10) of the subgenomic 5′UTR produced virus with a small plaque size as would be predicted based on the disruption of the minimal promoter for transcription of the subgenome and/or the addition to the C-terminus of nsP4. Deletion of nucleotides 11-20 (Del11-20) of the SG 5′UTR produced virus with a mixed plaque phenotype (Figure 1B). Deletions of nucleotides 21-30, 31-40 and 41-49 led to the production of virus with WT plaque size (Figure 1B and C).
Figure 1. Plaque size of SG 5′untranslated region mutants.
(A) Junction region of SINV genome and scanning deletions of the SG 5′UTR. Boxed sequence indicates coding regions of nonstructural and structural regions. Stop codon for nsP4 (bold) is disrupted by del1-10, and translation stops at the opal stop codon (underlined). The site of transcription of the subgenomic mRNA is indicated by the black arrow. (B) Plaque morphology of SIN WT and deletion viruses. Plaques visualized at 40 hours post infection. (C) Average plaque size for mutant viruses. The average plaque size of the Del11-20 virus is inclusive of the small and big plaques leading to higher standard deviation. Size determined by ImageJ software (NIH).
Deletion 1-10 and 11-20 in the 5′UTR of SG mRNA differentially affect host cell protein synthesis
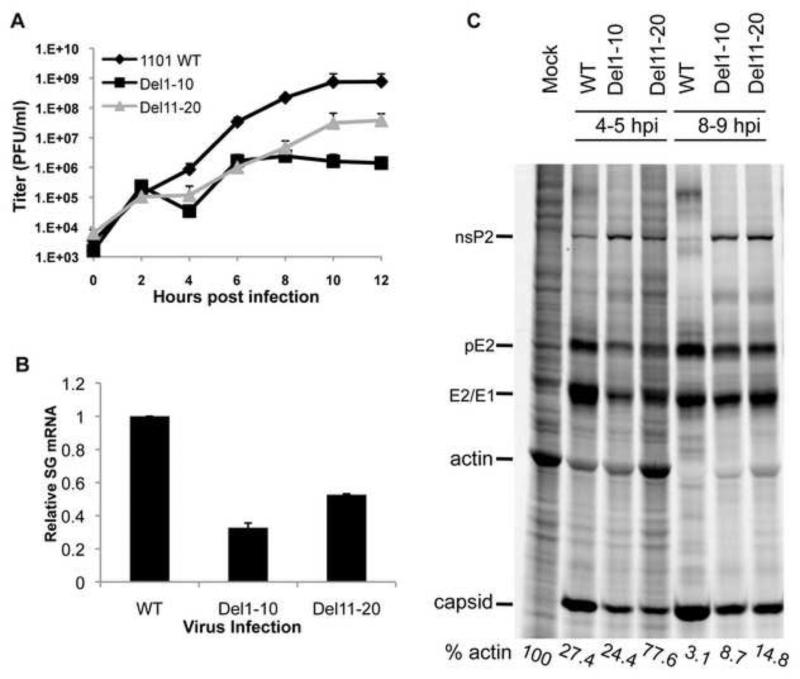
Single-step growth curves were performed to determine whether mutant viruses Del1-10 and Del11-20 displayed defects in growth kinetics as determine by single-step growth curve (Figure 2A). Del1-10 and Del11-20 were impaired for virus production starting at 4 hours post-infection (hpi) and continuing throughout the time course. Both mutant viruses showed a decrease in virus titer of greater than one log at 12 hpi when compared to wt.
Figure 2. Differences between Del1-10 and Del11-20 at the level of host translation.
(A) Growth curves indicating viral yield over time of WT, Del1-10, and Del11-20 viruses in BHK cells. The cells were infected with the respective viruses at an MOI of 10 PFU/cell. After infection cells were rinsed in PBS and media was collected and replaced every two hours. The viral titers are expressed in plaque forming units or PFU/ml. Titers are average from two independent experiments. (B) qRT-PCR quantification of subgenomic mRNA. Quantity of SG mRNA is shown as fold change over WT. Error bars represent SD for three independent experiments. (C) Protein synthesis in WT, Del1-10 and Del11-20 virus infected cells. Uninfected and infected cells were radiolabeled with [35S]-methionine-cysteine for one hour and visualized as described in Material and Methods. Cellular protein actin and viral proteins nsP2, pE2, E1/E2, and capsid are marked. Actin level in each lane is shown as a percentage of that in uninfected cells, and was determined by phosphorimagery.
The decreased virus titers were expected as both mutations extend into the optimal promoter sequence for synthesis of the SG mRNA. Analysis of SG mRNA levels by qRT-PCR demonstrated that both mutations caused a 50-70% decrease in the amount of SG mRNA in cells as compared to WT virus (Figure 2B). While it is possible that these mutations could decrease the stability of the SG mRNA, given the location of the deletions and their overlap with the optimal SG promoter it seems more likely that synthesis is reduced.
When the pattern of protein expression in infected cells was examined by metabolic labeling, differences between the two mutant viruses were observed. Expression of viral structural proteins (pE2, E1/E2, and capsid, Figure 2C) were reduced in Del1-10 as would be expected due to reduced SG mRNA levels, and nonstructural protein expression was increased as indicated by nsP2 levels. Interestingly, Del1-10 virus inhibited host cell protein synthesis at levels similar to WT virus early in infection (exemplified by actin levels), but this inhibition was less complete at later times post-infection. Del11-20 also, had reduced levels of structural protein synthesis and increased nonstructural protein synthesis. Nevertheless, this virus failed to inhibit host cell translation at early time post-infection, and inhibition at later times was less efficient than Del1-10 virus (Figure 2C). This implied a change in the interaction of the virus with the host cell. In order to further investigate this interaction, revertant viruses were isolated from the Del11-20 mutant virus following passage in BHK-21 cells.
A Del11-20 revertant virus contains a duplication of the subgenomic promoter
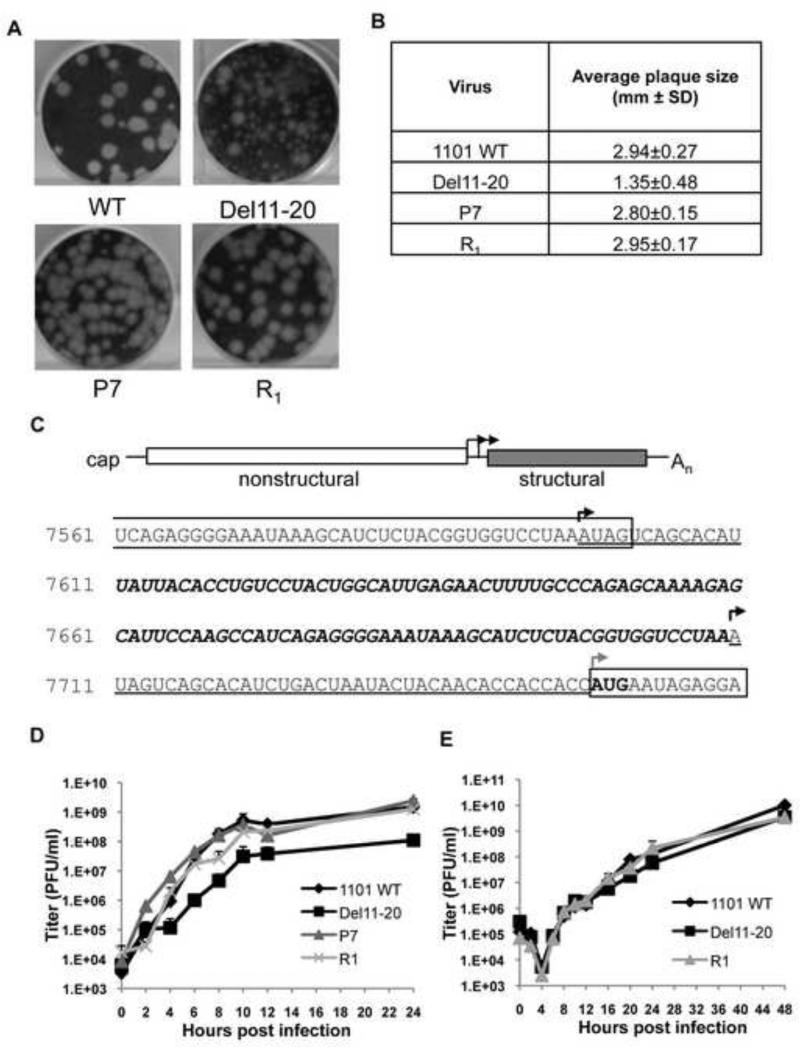
A deletion of bases 11-20 of the subgenomic 5′UTR reduced virus yield and caused a mixed plaque phenotype. We took advantage of the mixed plaque phenotype and sequenced large plaque revertants to identify mutations responsible for the large plaque size. Individual large plaques were selected at random and passaged as separate virus stocks. A number of these large plaque viruses displayed increased growth kinetics over the parental mutant, but failed to reach WT levels. Isolated large plaque virus, designated P7, was similar to SINV WT in plaque size (Figure 3A and 3B), its growth kinetics and final virus yield (Figure 3D).
Figure 3. Revertant plaque purified virus, P7 is similar to WT in titer and plaque size and contains a duplication of the subgenomic promoter.
(A) Plaque morphology of the WT, Del11-20, P7 and recombinant R1 viruses. (B) Average plaque size for WT, Del11-20, P7 and recombinant R1 viruses as determined by ImageJ software (NIH). (C) Identification of the duplicated promoter region in revertant P7 virus. P7 contains a duplication of 112 bases: 13 bases of the 5′ SG mRNA Del11-20 untranslated region (underlined), followed by of 99 bases of 3′ nsP4 sequence (bold italics), followed by another copy of SG mRNA del11-20 5′ UTR (underlined). Boxed areas indicate protein-coding regions. Black arrows indicate potential initiation sites for subgenomic mRNA transcription. The gray arrow indicates the site of translational initiation of the structural proteins. Growth curves indicating viral titers over time for WT, Del11-20, P7 and R1 viruses in BHK cells (D) and C636 cells (E). Cells were infected with the respective viruses at an MOI of 10 PFU/cell. The viral titers are expressed in plaque forming units or PFU/ml. Titers are average from two independent experiments.
Sequencing the genome of revertant P7 identified a duplication of the subgenomic promoter (Figure 3C). The 3′ coding region of nsP4 was maintained as was the authentic UAG amber stop codon (first boxed sequence). NsP4 sequence was followed by 13 bases of Del11-20 5′UTR sequence (underlined), a duplication of 99 bases of 3′ nsP4 coding sequence (bold italics), and the parental Del11-20 mutant SG mRNA 5′UTR (underlined). The duplication reconstitutes the minimal promoter (-19/+5) and 96% of the optimal subgenomic promoter (-98/+14). This duplication of the promoter region could lead to transcription of two SG mRNAs, one with a 39 base 5′UTR (parental Del11-20) and one with 151 base 5′UTR (P7), both of which would begin coding for capsid at the authentic start codon. No other mutations were found in the plaque-purified virus P7. To confirm the role of the duplicated subgenomic promoter in restoration of the large plaque phenotype, the junction region of the revertant virus was cloned into a pToto1101 SINV WT background. Recombinant 1101-P7 (R1) virus was generated by transfection of an in vitro transcribed RNA. A P0 stock of R1 was compared to plaque purified virus P7 and found to be nearly identical in plaque size, growth kinetics, and virus yield (Figure 3A, 3B and 3D) demonstrating that the duplication of the subgenomic promoter is responsible for the revertant phenotype. P0 stocks of recombinant R1 were used for the remaining experiments.
Since SINV infects both vertebrate and mosquito cells we determined whether the defect in Del 11-20 was host-species specific. Single-step growth curves were performed in mosquito cells (C6/36), and in contrast to the vertebrate cells no significant difference in the viral titers in the WT, Del11-20 and R1 viruses was observed (Figure 3E). This indicates that the cause of Del11-20 decreased growth in vertebrate cells is host cell specific rather than a fundamental problem with structural protein expression levels.
Del11-20 and revertant virus, R1 change levels of viral and host protein synthesis
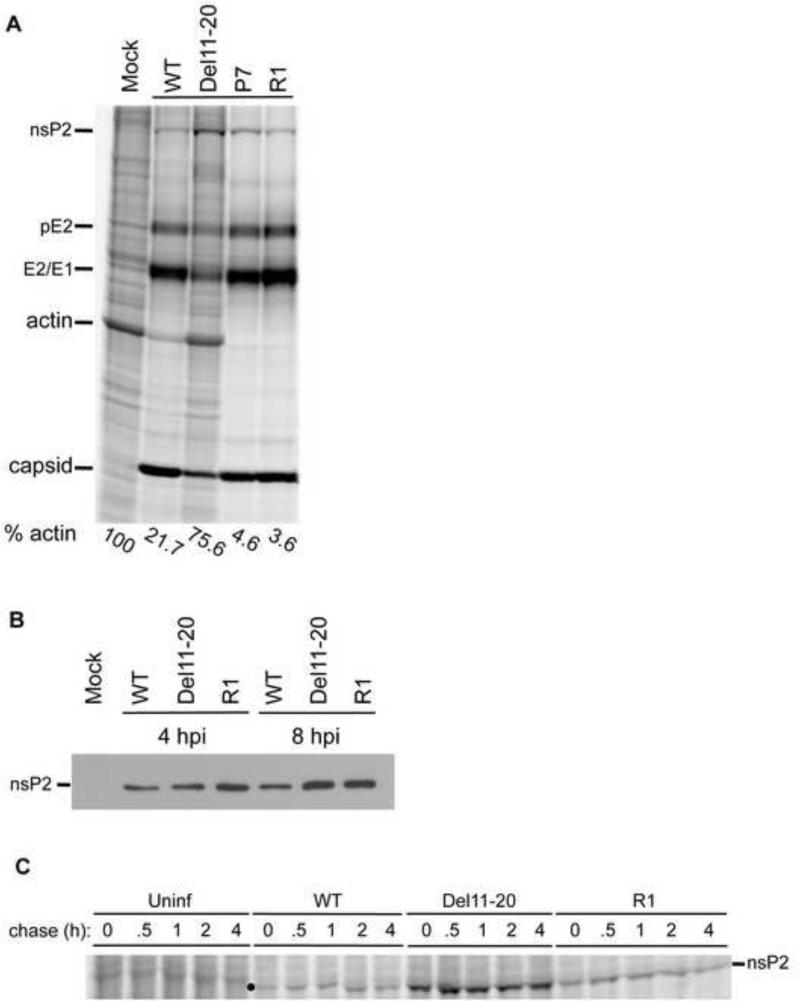
Del11-20 exhibited over a log decrease in virus yield while R1 demonstrated phenotypic reversion to WT plaque size, growth kinetics and virus yield. To determine what effect the duplication of the UTR had on viral and cellular protein expression we radiolabeled proteins in uninfected and infected cells with [35S]-methionine/cysteine. Mock and virus infected (MOI=10) cells were radiolabeled with [35S]-methionine/cysteine for one hour at 4 hpi. Cells were harvested and proteins separated by SDS-PAGE and visualized by phosphorimaging (Figure 4A). WT virus efficiently inhibited host cell translation (shown by actin levels) by 4 hpi, whereas viral structural proteins (capsid and glycoproteins) continue to be efficiently translated. As was observed previously Del11-20 failed to efficiently inhibit host translation and altered viral protein synthesis. Viral structural protein synthesis remained lower than SINV WT whereas nonstructural protein production (as exemplified by nsP2 labeling) was higher than WT. Similarly, actin translation was higher in Del11-20 and was more similar to uninfected cells at 4 hpi (Figure 4A).
Figure 4. Altered levels of viral and host protein synthesis in Del11-20 and R1 viruses.
(A) Protein synthesis in WT, Del11-20, P7 and R1 virus infected cells. Uninfected and infected cells were radiolabeled with [35S]-methionine-cysteine at 4 hours post-infection for one hour and visualized as described in Material and Methods. Cellular protein actin and viral proteins nsP2, pE2, E1/E2, and capsid are marked. Actin level in each lane is shown as a percentage of that in uninfected cells, and was determined by phosphorimagery. (B) Western Blot indicating accumulated levels of nsP2 in uninfected and infected cells. (C) Pulse chase analysis in uninfected, WT, Del11-20 and R1 virus infected cells. Uninfected and infected cells were starved and radiolabeled with [35S]-methionine-cysteine for 30 minutes. Cells were washed twice in PBS and chased with regular media supplemented with 10 mM methionine and 5 mM cysteine. At time points indicated cells were harvested in lysis buffer and visualized as described in Materials and Methods. The location of nsP2 migration is marked by ● on the gel image.
Recombinant R1 virus restored plaque size and titer to WT levels. Similarly levels of viral structural proteins were restored to WT levels (Figure 4A). Additionally, the nsP2 synthesis phenotype was partially restored with levels being lower than for Del11-20, but slightly higher than observed in WT infection. R1 appeared to inhibit actin translation better than WT. A similar translation profile was observed in P7 virus infected cells, further confirming that the duplication of the junction region was responsible for the revertant phenotype.
In addition to its role in transcriptional shut-off, nsP2 has also been implicated in the inhibition of host translation. Therefore a simple assumption would be that increased levels of nsP2 should result in more efficient shut-off of host cell translation. Since this was not the case with Del11-20 we decided to examine further the levels of accumulated nsP2 as well as the stability of nsP2 in virus infected cells to determine whether excess nsP2 was being degraded, potentially altering its effect on host translation. As shown in Figure 4B, Del11-20 infected cells nsP2 accumulated to levels equivalent to those seen in WT infected cells at 4 hpi, and accumulated to higher levels by 8hpi. To determine the nsP2 produced in Del11-20 infected cells was stable, pulse-chase experiments were performed with WT, Del11-20, and R1 infected cells at 4 hpi. Cells were labeled for 30 minutes and chased up to 4 hours in the presence of unlabeled methionine and cysteine (Figure 4C). At 0 hr post-labeling, levels of nsP2 synthesis were similar to previous results, with Del11-20 and R1 infected cells producing elevated nsP2. There was no visible loss of labeled nsP2 in cells infected with any of the three viruses at any time post-labeling, indicating nsP2 was stable during the chase. This shows that excess nsP2 is stable in Del11-20 or R1 infected cells and indicates that nsP2 turnover is not responsible for reduced inhibition of host translation in Del11-20 infected cells. These data suggest a mechanism of inhibition of host cell gene expression that is independent of absolute nsP2 levels, and, given the restoration of shut-off in the R1 virus infected cells, may be related to the SG mRNA 5′UTR.
Del11-20 does not alter shut-off of host cell transcription
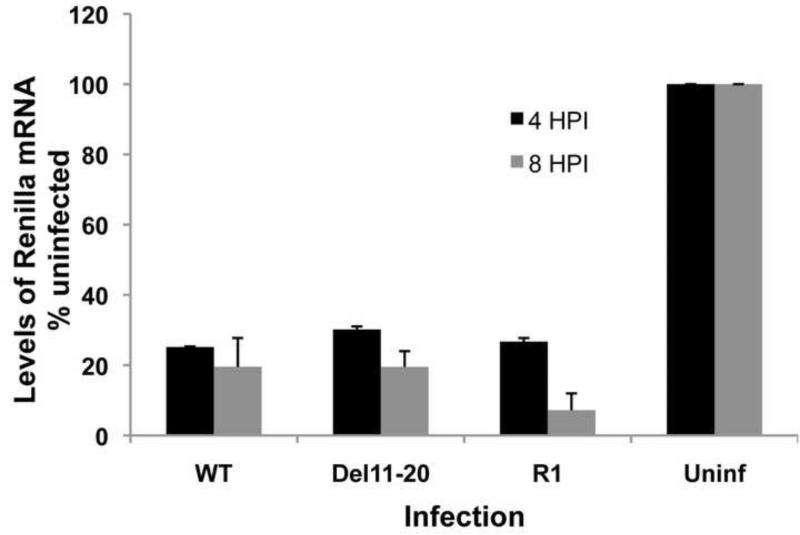
Del11-20 reduced the levels of SG mRNA and structural proteins produced, led to a decrease in inhibition of host shut-off of translation, and increased accumulation of nsP2, a known modulator of cellular transcription and translation. Increased levels of host translation could result from decreased inhibition of transcription leading to more mRNAs available for translation. To test this possibility, we performed a transcription assay where infected and uninfected cells were transfected with a plasmid encoding Renilla luciferase under the control of a CMV (RNA polII-dependent) promoter. Renilla mRNA levels were quantified using qRT-PCR (Figure 5). Four hours post-infection Renilla luciferase mRNA levels in WT, Del11-20 and R1 infected cells were similarly reduced between 70% and 75% as compared to uninfected cells with no significant differences between samples. Later in infection (8 hpi), Renilla mRNA levels from WT SINV and Del11-20 infected cells were reduced by 80% while transcription by R1 infected cells was reduced by over 90%. This indicates early viral mechanisms of transcription inhibition were functioning similarly in SINV WT, Del11-20, and R1 infections. These data demonstrate that the WT, Del11-20, and R1 viruses are equally capable of efficient inhibition of host cell transcription, and therefore lack of transcriptional inhibition is not the reason for the loss of inhibition of host cell protein synthesis. Furthermore, these data indicate that nsP2 is functional with regards to transcriptional shutoff.
Figure 5. Inhibition of host cell transcription by WT, Del11-20 and R1 viruses.
(A) Renilla luciferase is expressed from a CMV promoter on the pRL-CMV DNA vector. (B) Replicate qPCR analysis of Renilla mRNA levels after transfection of infected and uninfected cells with the pRL-CMV DNA vector. Cells were transfected with the plasmid DNA at 2 or 6 hpi and harvested after an additional 2 h incubation. Levels of Renilla mRNA are shown as a percentage of levels in uninfected cells. Error bars represent standard deviation for three independent experiments.
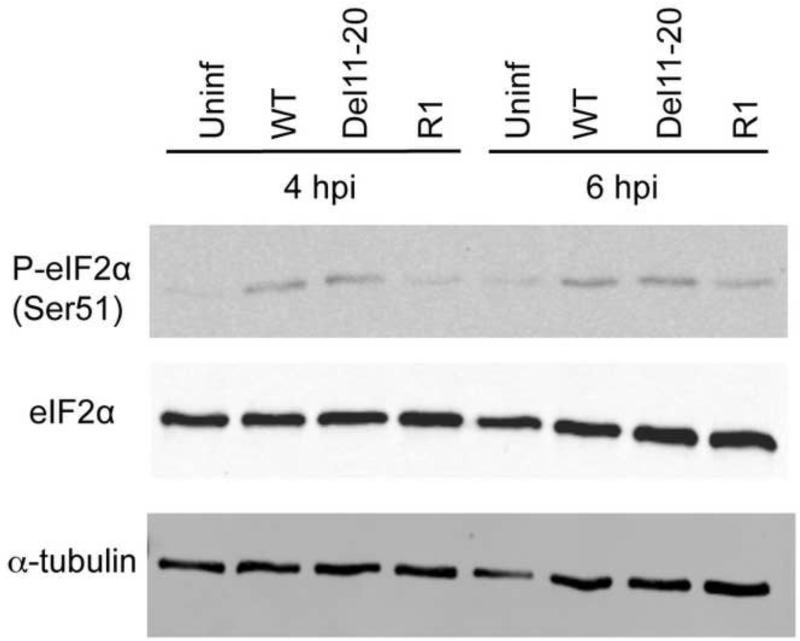
Shut-off of host translation is independent of eIF2 phosphorylation
Virus replication and hence accumulation of dsRNA molecules is known to lead to activation of the kinase, PKR (Green and Mathews 1992; Romano, Garcia-Barrio et al. 1998; Dar, Dever et al. 2005; Weber, Wagner et al. 2006). Activated PKR leads to phosphorylation of the translation initiation factor 2 α (eIF2α) subunit that, in turn leads to down-regulation of viral as well as host translation (Balachandran, Roberts et al. 2000; Stojdl, Abraham et al. 2000; Dever 2002; Dar, Dever et al. 2005). SINV is known to activate PKR leading to phosphorylation of eIF2α (Gorchakov, Frolova et al. 2004). It is also known to possess a cis-acting secondary structure in the 26S RNA that facilitates translation of structural proteins in the absence of eIF2 (Ventoso, Sanz et al. 2006; Toribio and Ventoso 2010). Since the WT and R1 viruses inhibit host translation whereas the Del11-20 virus fails to do so completely, we examined whether this was as a result of differential activation of PKR. BHK-21 cells were infected with all three viruses, and whole cell lysates were prepared at 4 and 6 hpi. Lysates were then probed for levels of phosphorylated as well as total eIF2α (Figure 6). At 4 and 6 hpi, eIF2α was phosphorylated at the same levels in WT and Del11-20 virus whereas R1 showed a decreased level of phosphorylation. The total levels of eIF2α are similar in all samples. The same effect was observed when this was repeated at 4 hpi in another mammalian cell line, HEK 293 cells (data not shown). Similar levels of eIF2α phosphorylation in WT as well Del11-20 virus infected cells indicates that the difference in host translation inhibition is not due to differential PKR activation and eIF2α phosphorylation. Also, reduced levels of eIF2α phosphorylation in R1, which restores WT levels of host shut-off, further underscores this conclusion.
Figure 6. Phosphorylation of eIF2 in WT and mutant virus infected cells.
Western Blot analysis of uninfected and infected BHK cells 4 and 6hpi using antibodies to phospho-eIF2α and total eIF2α. Cells were mock infected or infected with an MOI of 10 PFU/cell and whole cells lysates were made. αTubulin was used as the loading control.
Duplication of the junction region in revertant virus leads to transcription of two SG mRNAs
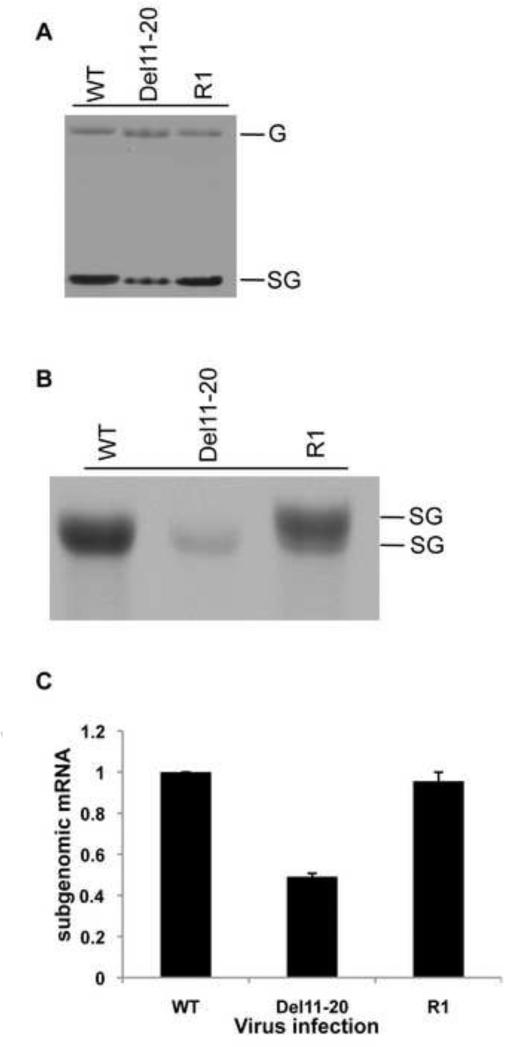
Revertant virus R1 contained a duplication of the subgenomic promoter and restored levels of structural proteins to WT levels indicating that a change in the levels of SG mRNA may have occurred. The presence of two functional promoters provides the potential for the production of two SG mRNAs. To determine if two species of SG mRNA were transcribed from the revertant R1 and to determine their levels of expression, we analyzed viral RNA synthesis by radiolabeling infected cells with [3H]-uridine in the presence of actinomycin-D (Figure 7A). RNA was harvested 8 hpi following a one-hour label. Infection resulted in synthesis of plus-strand genomic (G) and subgenomic (SG) RNAs. Del11-20 produced similar levels of genomic RNA compared to WT but had reduced levels of SG mRNA (Figure 7A). R1 virus synthesized genome and SG mRNA at WT levels. Slight changes in the migration of the R1 genome and SG mRNA compared to Del 11-20 were observable. When allowed to migrate further in the gel two SG mRNAs were seen from R1 infected cells, indicating that the duplication in the junction region resulted in two functional and active subgenomic promoters restoring SG mRNA levels close to WT (Figure 7B).
Figure 7. Analysis of SG mRNA levels.
(A) Agarose-phosphate gel of viral RNA synthesis labeled with [3H]-uridine. Thirty minutes prior to labeling cells were treated with Actinomycin D at 10ug/ml and labeled with 50uCi/ml [5′-3H]-uridine for one hour at 7 hours post infection. RNA was isolated and visualized as described in Materials and Methods. (B) Identification of two subgenomic RNAs. [3H]-uridine labeled RNA was separated on an agarose phosphate gel. (C) qPCR quantification of SG mRNA. Levels of SG mRNA are shown as fold change over WT. Error bars represent SD for three independent experiments.
To determine the total cumulative amount of SG mRNA transcribed by each virus, levels of SG mRNA were quantified by qRT-PCR (Figure 7C). RNA was isolated from infected cells, reverse transcribed with a primer specific to the structural protein coding sequence, and the resulting cDNA was amplified in a qPCR reaction to quantify SG mRNA levels. Levels of SG mRNA were calculated after subtraction of genome levels determined by qRT-PCR using primers to the nonstructural protein coding sequence. This data confirmed the radiolabeling data showing that the duplication of the junction region in the R1 virus led to restoration of SG mRNA levels to that of WT. These data indicated that the level of SG mRNA might play a role in the inhibition of host cell proteins synthesis. However, we had previously observed that Del1-10 virus had a similar effect on host shut-off as the WT despite producing lower amounts of SG mRNA and hence we decided examine the effect the different 5′UTR sequences had on translational efficiency of the SG mRNA.
Changes in the 5′UTR alter efficiency of SG mRNA translation
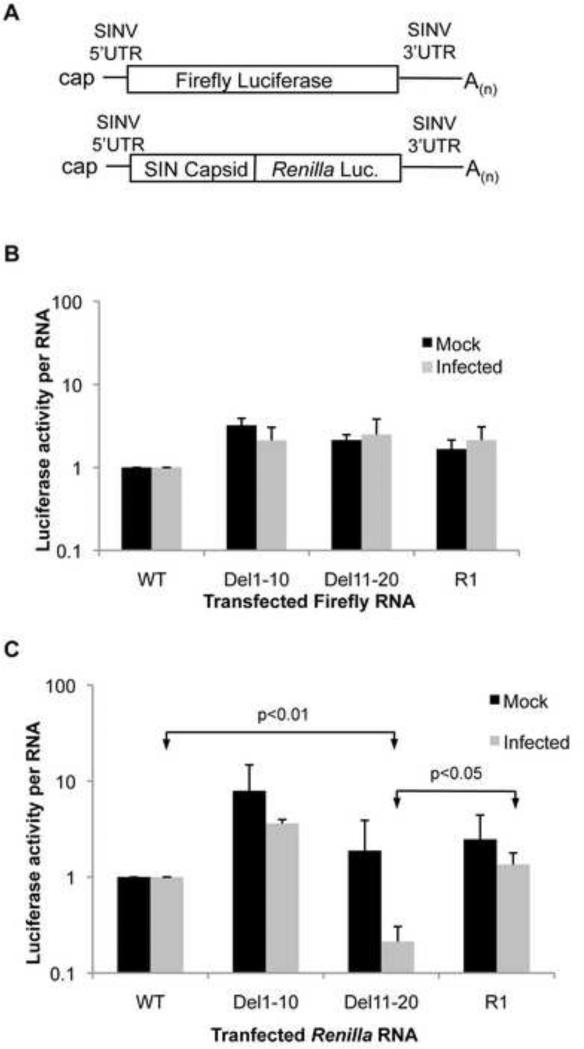
In order to assess the effect of mutations in the 5′UTR and the revertant sequence on translation efficiency of the SG mRNA the 5′UTR sequences were fused upstream of firefly luciferase followed by the authentic viral 3′UTR and polyA tail (Figure 8A). RNAs were generated in vitro and transfected into uninfected cells or infected cells at 4 hpi. Cells were incubated at 37°C for two additional hours then harvested. Luciferase activity was standardized to transfected RNA levels as determined by qRT-PCR. Upon examination of the translation efficiency of each RNA it was apparent that changes in the 5′UTR alone had little effect on how well the mRNA was translated in either uninfected or infected cells when compared to mRNA possessing the viral WT 5′UTR (Figure 8B).
Figure 8. Altered 5′UTRs alter efficiency of translation.
(A) Diagrams of luciferase encoding RNAs. (B) Firefly luciferase activity from reporter RNAs possessing the SG 5′UTR from WT, Del1-10, Del11-20 and R1 viruses fused to firefly luciferase and followed by the SINV 3′UTR and poly(A) tail. (C) Renilla luciferase activity from a reporter RNA possessing the SG 5′UTR from WT, Del1-10, Del11-20 and R1 viruses fused in frame with the viral capsid coding sequence followed by Renilla luciferase, the viral 3′UTR and poly(A) tail. All RNAs were transfected into mock infected and WT SINV infected BHK-21 cells at 4hpi and incubated for an additional 2 hours before harvesting. Luciferase activity was determined and normalized to transfected RNA levels as determined by qRT-PCR analysis and shown as fold change over WT. Data shown is representative of three independent experiments. Error bars represent SD.
Previous reports have clearly demonstrated the importance of the translational enhancer located within the 5′ coding region of the capsid gene in the SG mRNA (Frolov and Schlesinger 1994; Frolov and Schlesinger 1996). Therefore, the 5′UTRs were fused to capsid sequence followed by Renilla luciferase, the viral 3′UTR and poly(A) tail (Figure 8A). M-fold predictions indicate that the translational enhancer structure is maintained with each of the 5′UTR sequences tested (data not show). In both infected and uninfected cells it was apparent that the 5′UTR when combined with the translational enhancer had a significant effect on the translation efficiency of the RNA. The Del1-10 5′UTR increased the translation efficiency when compared to the WT 5′UTR in both uninfected and infected cells (Figure 8C). In contrast translation of mRNA possessing the Del11-20 5′UTR was significantly less efficient in infected cells (20% of WT, p<0.01). The R1 5′UTR sequence led to translation that was at least as efficient as that seen for the WT sequence in both uninfected and infected cells. These data strongly indicate that the 5′UTR and the enhancer element function in concert to facilitate translation of the SG mRNA and that deletion of nucleotides 11-20 of the 5′UTR have a detrimental effect on translation efficiency of the mRNA in an infected cell. It is also apparent that the ability of the 5′UTR to program translation correlates directly with the ability of the virus to inhibit cellular translation.
It should be noted that in all cases the translation of transfected RNA was significantly lower in infected cells than in uninfected cells. This has been reported previously and may indicate a requirement for viral transcription of the RNA in order for efficient translation in infected cells (Sanz, Castello et al. 2007). Nevertheless, it is apparent that changes to the 5′UTR, in combination with the translation enhancer, do alter the efficiency of translation in both infected and uninfected cells, and that Del11-20 has a particularly detrimental effect on translation in infected cells possibly indicating a reduced ability to compete for factors necessary for translation in the infected cell.
DISCUSSION
Virus infections frequently result in the inhibition of host cell gene expression. This inhibition facilitates virus replication by muting the antiviral response of the cell and also increasing the availability of cellular machinery to promote virus replication. Different viruses utilize different strategies to achieve the same ends, frequently disrupting cellular gene expression at the level of translation. Poliovirus inhibits cellular translation through cleavage of eIF4G but promotes viral expression through cap-independent IRES elements (Etchison, Milburn et al. 1982; Krausslich, Nicklin et al. 1987; Gradi, Svitkin et al. 1998). Influenza virus snatches cap structures from cellular pre-mRNAs to both promote viral and inhibit host expression (Kash, Goodman et al. 2006). Additionally influenza and adenovirus infections lead to dephosphorylation of eIF4E inhibiting its function in the initiation of translation; however viral mRNAs are efficiently translated (Feigenblum and Schneider 1993; Zhang, Feigenblum et al. 1994). In old world alphaviruses nsP2 has been implicated in inhibition of host transcription and translation (Frolov and Schlesinger 1994; Gorchakov, Frolova et al. 2004; Gorchakov, Frolova et al. 2005; Garmashova, Gorchakov et al. 2006; Mayuri, Geders et al. 2008).
Our present study adds to our understanding of SINV-mediated inhibition of host cell gene expression. Two mutations in the 5′UTR (Del1-10 and Del11-20) of the SINV SG mRNA were observed to be detrimental to viral growth, reducing SG mRNA synthesis, and structural protein expression. While both mutant viruses displayed increased expression of nsP2, Del11-20 was unable to efficiently inhibit host cell translation. A suppressor mutant with a duplicated fragment of the promoter region restored levels of SG mRNA synthesis and shut-off host translation. These data suggested that the levels of SG mRNA play a role in inhibition of host cell translation, possibly by recruiting limiting amounts of translation factors. Yet the low levels of SG mRNA synthesized by the Del1-10 virus in combination with its ability to inhibit host translation indicated that high levels of SG mRNA were not required for inhibition. We found that shut-off of host translation correlated with the translational efficiency and to some degree the quantity of the SG mRNA bestowed by the 5′UTR in combination with the translation enhancer element. Elevated levels of intracellular nsP2 did not lead to translational inhibition. Our data indicate that efficient translation of the SG mRNA results in inhibition of host cell translation suggesting that the SG mRNA competes with cellular mRNAs for limiting factors in the infected cell essential for translation.
It has been previously reported that alphavirus SG mRNA has a reduced requirement for certain initiation factors like eIF4B (van Steeg, van Grinsven et al. 1981), eIF4G (Castello, Sanz et al. 2006), eIF4A (Garcia-Moreno, Sanz et al. 2012) and eIF2α (Gorchakov, Frolova et al. 2004; Ventoso, Sanz et al. 2006) in infected cells, in part owing to particular structural elements present in the mRNA which circumvent the need of the mRNA to bind to these initiation factors to initiate translation (Gale, Tan et al. 2000; Jang 2006). Elements within the SG mRNA have been shown to promote its translation, specifically the translational enhancer located in the capsid coding sequence. This element appears to function to reduce or eliminate the requirement for eIF2 in vertebrate cells but has little effect in mosquito cells (Ventoso 2012). Our data imply that elements in the 5′UTR act in combination with the translational enhancer for efficient translation in vertebrate cells, and we speculate that these elements are required to recruit a factor(s) necessary for translation in vertebrate cells. The ability of the Del1-10 5′UTR and the revertant (R1) 5′UTR to efficiently program translation in combination with their ability to inhibit host cell translation demonstrates a mechanism of translational inhibition that is not a direct effect of viral nsP2, the protein that has been previously suggested to be responsible for the inhibition of translation (Gorchakov, Frolova et al. 2004; Mayuri, Geders et al. 2008).
Prior reports examining inhibition of host cell translation have focused on the carboxy-terminal domain of nsP2 (Gorchakov, Frolova et al. 2004; Mayuri, Geders et al. 2008). This region of the protein is similar in structure to a methyltransferase domain while lacking the specific residues in the appropriate location for methyltransferase activity (Russo, White et al. 2006). Mutations in this region of the protein have been associated with decreased ability to inhibit both host cell transcription and translation (Frolov, Agapov et al. 1999; Frolova, Fayzulin et al. 2002; Gorchakov, Frolova et al. 2004; Mayuri, Geders et al. 2008). It is noteworthy that in the studies examining inhibition of translation the mutations in nsP2 also impaired viral RNA synthesis thus reducing levels of SG mRNA during infection. Interestingly, previously published observations from our lab showed that mutations in the amino-terminal region of the viral polymerase, nsP4, resulted in a decrease in viral RNA synthesis and a persistence of host cell translation throughout infection (Rupp, Jundt et al. 2011). Additionally reports by Gorchakov et al indicate that virus producing nsP2 only in the form of a polyprotein rather than mature nsP2 was capable of inhibition of host cell translation (Gorchakov, Frolova et al. 2008). This suggests a mechanism for host cell translational inhibition that is independent of nsP2. Our finding that reversions restoring SG mRNA synthesis correlated with restored inhibition of host cell translation support the idea of an nsP2-independent mechanism of host translation inhibition.
Re-examination of prior reports in combination with findings presented here suggest that inhibition of host cell translation is, at least in part, contingent on SG mRNA competing for cellular factors necessary for efficient translation. The Del11-20 revertant sequence (R1) examined here achieves this in two ways: first, by increasing the amount of SG mRNA being made through the duplication of the subgenomic promoter, and secondly by enhancing the efficiency of translation of the SG mRNA possessing the 5′UTR extension. The observation that the Del1-10 5′UTR can efficiently promote translation (Figure 8C) of the SG mRNA implies that in WT virus nucleotides 11-20 of the 5′UTR are responsible for enhancing translation and possibly binding a required factor. We hypothesize that the R1 reversion provides a structure that functionally substitutes for nucleotides 11-20 in enhancing translation.
The factor(s) bound by the 5′UTR are currently unknown and identifying these is the focus of future studies. One possibility is that the 5′UTR cooperates with the translational enhancer element to facilitate the independence from, or reduced requirement for eIF2 (Ventoso 2012). PKR-dependent phosphorylation of eIF2α in infected cells reduces the amount of active eIF2 for initiation of translation. It is possible that the 5′UTR and translational enhancer function to efficiently bind the limiting amounts of eIF2 thus promoting SG mRNA translation and completely inhibiting host cell translation. Examining such hypotheses and identifying cognate binding partners for the SG mRNA will further elucidate our understanding of the influence of alphavirus on host cell gene expression.
MATERIALS AND METHODS
Cells, plasmids, and viruses
BHK-21 (Baby Hamster Kidney) and Mosquito C6/36 cells were obtained from the American Type Culture Collection. These cells were grown at 37°C in 5% CO2 in MEM Alpha (Invitrogen) and supplemented with 5% fetal bovine serum, glutamine, and anti-biotic/anti-mycotic. pToto1101 plasmid containing a cDNA copy of the SIN genome was a gift from Charlie Rice (Rice, Levis et al. 1987). pRL-CMV was obtained from Promega. SINV was generated by transfection of BHK-21 cells with infectious RNA in vitro transcribed from XhoI linearized pToto1101 (Rice, Levis et al. 1987). Del1-10, Del11-20, Del21-30, Del31-40, and Del41-49 deletion of the SG 5′UTR were generated using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies). Mutations were confirmed by sequencing. Mutant viruses were generated from transfection of an infectious RNA in vitro transcribed from linearized mutant plasmid. pToto1101-R1 was constructed from a ligation of HpaI/AatII digested pToto1101 and a RT-PCR product of plaque purified virus P7 amplified with primers TM040-(6910) 5′-CTATGGCGTTAACCGGTCTGATG and TM149-(8062) 5′-CGATGGTTCCTTGCACGTGCAGAG. All infections were done for one hour at a MOI of 10.
Virus growth kinetics
BHK-21 cells were infected with virus at an m.o.i of 10. One-hour after adding the inoculum cells were washed with PBS and fresh medium was added and an aliquot removed to represent time 0. At 2 hr intervals growth medium samples were taken and stored at -80 °C. Cultures were washed and fresh medium was added. This process was repeated up to 12 hpi. A final sample was taken at 24 hpi. For growth in mosquito cells C6/36 cells were infected as described above and samples of medium were taken every 4 h up to 24 hpi. A final sample was taken at 48 hpi. Titers from each sample at each time point were determined on BHK-21 cells and plotted against time.
[35S] Protein labeling and quantification
Thirty minutes prior to labeling, infected and uninfected cells were washed in phosphate buffered saline (PBS) and incubated in media lacking methionine or cysteine. Media was removed and replaced with methionine and cysteine depleted media supplemented with [35S]-methionine and cysteine (EXPRESS35S Protein Labeling Mix, Perkin Elmer) at a final concentration of 50 mCi/ml. Cells were incubated for one hour at 37°C and harvested in ice cold lysis buffer (10 mM Tris HCl pH 7.4, 20 mM NaCl, 1 mM EDTA, 0.4% sodium deoxycholate, 1% NP40). Insoluble material was removed by a high-speed spin at 16,000 × g for 1 minute. Equal volumes of lysate were loaded and proteins were separated by SDS-PAGE (10% acrylamide), dried, exposed to a phosphor screen, imaged on a Typhoon 9200 imager (GE Healthcare), and processed in ImageQuant software (Molecular Dynamics). For pulse chase experiments, cells were starved of methionine and cysteine and radiolabeled with depleted media supplemented with [35S]-methionine and cysteine at 50 mCi/ml for 30 minutes. Cells were rinsed twice with PBS and chased with regular media supplemented with 10 mM methionine and 5 mM cysteine. Cells were harvested and proteins visualized as previously described.
[3H] RNA labeling
Mock or virus infected cells were treated with Actinomycin D (Sigma) at 10 mg/ml 30 minutes prior to labeling. At 7 hpi cells were labeled with 50 mCi/ml [5-3H]-uridine (Moravek) for one hour in the presence of 10 mg/ml Actinomycin D. RNA was Trizol (Invitrogen) extracted and subject to agarose-phosphate gel electrophoresis following glyoxal denaturation (Hardy and Rice 2005). The gel was visualized by fluorography.
RT-PCR of plaque purified virus
Thirty-five mm dishes were infected with plaque-purified virus and at 8 hpi total RNA was Trizol extracted and quantified. 500 ng total RNA was subject to reverse transcription with a plus sense virus specific primer (TM005-(10373), 5′-GACAATTCGACGTACGCCTCACTC) and Improm II Reverse Transcriptase (Promega). One μL of the RT reaction was added to a standard PCR reaction with pfu high-fidelity polymerase, and primer sets scanning the viral genome. PCR products were purified and sequenced.
Quantitative RT-PCR
Five hours post infection total RNA was Trizol extracted and treated with DNA-free (Ambion) following the manufacturer's directions. 500 ng RNA was reverse transcribed with oligo(dT) primer and Improm II Reverse Transcriptase (Promega). 50 ng cDNA was PCR amplified using Brilliant II SYBR Green qPCR master mix (Agilent Technologies) with the following primers: nsP1 forward997: 5′-GGTTACACACAATAGCGAGGGCTT, nsP1 reverse-1197: 5′-TGGTGTTCCTGTTAGTCCTACCGT-3′, capsid forward-9600: 5′-CGGAAAGACGGTCAGACAATTCACCGTCGACCGAG-3′, capsid reverse-9856: 5′-GGGATTACGGCTTGTGGGGCCAGGGCGTATGG-3′, E1 forward 5′-TCAGATGCACCACTGGTCTCAACA-3′, E1 reverse 5′-ATTGACCTTCGCGGTCGGATTCAT-3′. Serial dilutions of pToto1101 plasmid were used for generation of a standard curve, and absolute quantities of genome and subgenome were derived from Ct values fit to the standard curve (Sokoloski, Hayes et al. 2012). Molar levels of subgenome were calculated by subtraction of genome levels. For quantification of luciferase messages, primers Renilla forward 5′-CACGCTGAAAGTGTAGTAGATGTG and Renilla reverse 5′-CTCTTTGAATGGTTCAAGATATGC; or Firefly Forward 5′-TCAAAGAGGCGAACTGTGTG and Firefly Reverse 5′-GGTGTTGGAGCAAGATGGAT, were used in qPCR reactions with cDNA prepared as previously described.
Western blot analysis
Samples from whole-cell extracts of uninfected or infected BHK cells were separated by SDS-PAGE (8 to 12% polyacrylamide) and transferred onto a nitrocellulose membrane. Blots were blocked and probed in Tris-buffered saline (TBS) with 5% nonfat dry milk and 0.1% Tween 20. Blots were probed with rabbit anti-tubulin antibody (Sigma), rabbit anti-nsP2 polyclonal antiserum, rabbit phospho-eIF2α (Ser 51) and rabbit eIF2α (#9721, #9722-Cell Signaling Technology) followed by an anti-rabbit goat antiserum conjugated to horseradish peroxidase (HRP). Bands were visualized by chemiluminescence (SuperSignal West Pico; Pierce).
Luciferase constructs and assays
Firefly subgenomic 5′UTR reporters were generated by amplification of the junction region of pToto1101 WT, pToto1101 Del11-20, or pToto1101-R1 with primers containing BamHI (5′-CGTGGCGGATCCCCTGAAAAGG-3′) and NcoI (5′-CGTCTTCCATGGTGGTGGTGTTGTAGTATTAGTC-3′) sites. Digested products were ligated into a BamHI/NcoI digest of p5′SIN3′SIN (Frolov, Hardy et al. 2001). The reporter SG mRNA was then amplified with primers specific for each 5′UTR downstream of an SP6 promoter and a 3′ primer that incorporated the XhoI site and the 3′UTR and poly (A21) tail of the SINV genome. These products were ligated into a pUC19 cloning vector and used as template for linearization and transcription. Renilla luciferase subgenomic UTR translational enhancer reporters were made from a three part fusion PCR incorporating: subgenomic 5′UTR and capsid protein with three amino acids of E3 for proper cleavage, Renilla luciferase, and WT SINV 3′UTR with poly (A21) tail. The first fragment was amplified from pToto1101, pToto1101del11-20, or pToto1101-R1 with 5′ primer containing an SP6 promoter and sequence specific for the beginning of each SG mRNA and a 3′ Cap-Rluc fusion primer (5′-GGATCATAAACTTTCGAAGTCATTGCTGCGGACCACTCTTCTG-3′). A second fragment was amplified from pRL-CMV with 5′ Cap-Rluc fusion primer (5′-CAGAAGAGTGGTCCGCAGCAATGACTTCGAAAGTTTATGATCC-3′) and a 3′ Rluc-UTR fusion primer (5′-GGATCATTGGGGCGTAGCGGTCATTATTGTTCATTTTTGAGAACTCGCTCAACG-3′). The final fragment was amplified from pToto1101 with 5′ Rluc-UTR primer (5′-CGTTGAGCGAGTTCTCAAAAATGAACAATAATGACCGCTACGCCCCAATGATCC-3′) and a primer incorporating poly(A21) tail and XhoI restriction site. These products were separately amplified and gel extracted and equal molar quantities were used in a Phusion (Finnzymes) PCR reaction following the manufacturer's directions. The final product was ligated into a pUC19 cloning vector and used as template for linearization and transcription in presence of CAP analog. All reporter RNAs were in vitro transcribed and treated with RQ1 RNase free DNase (Promega). Prior to quantification RNA was purified by phenol/chloroform extraction and ethanol precipitation. For Firefly and Renilla luciferase subgenomic reporter assays, infected or uninfected cells were transfected with 1ug of reporter mRNA at 4 hpi and incubated at 37°C for an additional 2 hours. Cells were lysed and reporter activity was determined using Luciferase Assay System, or Renilla Luciferase Assay System (Promega) following the manufacturer's directions. Activity was read using a BioTek Synergy 2 (BioTek) plate reader. Luciferase encoding RNA in each sample was quantified by qRT-PCR and luciferase activity was standardized on the basis of RNA quantity. Results are the average of at least 3 independent biological experiments.
HIGHLIGHTS.
Mutations in the 5′UTR of Sindbis virus prevent inhibition of host cell translation
Levels of viral nsP2 do not correlate with host cell translational inhibition
Translational efficiency of subgenomic RNA is dependent on the 5′UTR
Host cell inhibition restored by increased translation efficiency of subgenomic RNA
ACKNOWLEDGMENTS
The authors wish to thank Pranav Danthi for critical reading of the paper and members of the Hardy, Mukhopadhyay, and Danthi labs for their help during manuscript preparation. This work was supported by grant MCB-0749482 from the National Science Foundation and grant R01 AI090077 from the National Institutes of Health both to R.W.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Balachandran S, Roberts PC, et al. Essential role for the dsRNA-dependent protein kinase PKR in innate immunitvto viral infection. Immunity. 2000;13(1):129–141. doi: 10.1016/s1074-7613(00)00014-5. [DOI] [PubMed] [Google Scholar]
- Burnham AJ, Gong L, et al. Heterogeneous nuclear ribonuclear protein K interacts with Sindbis virus nonstructural proteins and viral subgenomic mRNA. Virology. 2007 doi: 10.1016/j.virol.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Cancedda R, Villa-Komaroff L, et al. Initiation sites for translation of sindbis virus 42S and 26S messenger RNAs. Cell. 1975;6(2):215–222. doi: 10.1016/0092-8674(75)90012-4. [DOI] [PubMed] [Google Scholar]
- Castello A, Sanz MA, et al. Translation of Sindbis virus 26S mRNA does not require intact eukariotic initiation factor 4G. J Mol Biol. 2006;355(5):942–956. doi: 10.1016/j.jmb.2005.11.024. [DOI] [PubMed] [Google Scholar]
- Dar AC, Dever TE, et al. Higher-order substrate recognition of eIF2alpha by the RNA-dependent protein kinase PKR. Cell. 2005;122(6):887–900. doi: 10.1016/j.cell.2005.06.044. [DOI] [PubMed] [Google Scholar]
- Dever TE. Gene-specific regulation by general translation factors. Cell. 2002;108(4):545–556. doi: 10.1016/s0092-8674(02)00642-6. [DOI] [PubMed] [Google Scholar]
- Ding MX, Schlesinger MJ. Evidence that Sindbis virus NSP2 is an autoprotease which processes the virus nonstructural polvprotein. Virology. 1989;171(1):280–284. doi: 10.1016/0042-6822(89)90539-4. [DOI] [PubMed] [Google Scholar]
- Etchison D, Milburn SC, et al. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J Biol Chem. 1982;257(24):14806–14810. [PubMed] [Google Scholar]
- Feigenblum D, Schneider RJ. Modification of eukaryotic initiation factor 4F during infection by influenza virus. J Virol. 1993;67(6):3027–3035. doi: 10.1128/jvi.67.6.3027-3035.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov I, Agapov E, et al. Selection of RNA replicons capable of persistent noncytopathic replication in mammalian cells. J Virol. 1999;73(5):3854–3865. doi: 10.1128/jvi.73.5.3854-3865.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov I, Hardy R, et al. Cis-acting RNA elements at the 5′ end of Sindbis virus genome RNA regulate minus- and plus-strand RNA synthesis. RNA. 2001;7(11):1638–1651. doi: 10.1017/s135583820101010x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov I, Schlesinger S. Comparison of the effects of Sindbis virus and Sindbis virus replicons on host cell protein synthesis and cytopathogenicity in BHK cells. J Virol. 1994;68(3):1721–1727. doi: 10.1128/jvi.68.3.1721-1727.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov I, Schlesinger S. Translation of Sindbis virus mRNA: effects of sequences downstream of the initiating codon. J Virol. 1994;68(12):8111–8117. doi: 10.1128/jvi.68.12.8111-8117.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov I, Schlesinger S. Translation of Sindbis virus mRNA: analysis of sequences downstream of the initiating AUG codon that enhance translation. J Virol. 1996;70(2):1182–1190. doi: 10.1128/jvi.70.2.1182-1190.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova EI, Fayzulin RZ, et al. Roles of nonstructural protein nsP2 and Alpha/Beta interferons in determining the outcome of Sindbis virus infection. J Virol. 2002;76(22):11254–11264. doi: 10.1128/JVI.76.22.11254-11264.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale M, Jr., Tan SL, et al. Translational control of viral gene expression in eukaryotes. Microbiol Mol Biol Rev. 2000;64(2):239–280. doi: 10.1128/mmbr.64.2.239-280.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Moreno M, Sanz MA, et al. Requirements for eIF4A and eIF2 during translation of Sindbis virus subgenomic mRNA in vertebrate and invertebrate host cells1. Cell. Microbiol. 2012 doi: 10.1111/cmi.12079. doi: 10.1111/cmi.12079. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Garmashova N, Gorchakov R, et al. Sindbis virus nonstructural protein nsP2 is cytotoxic and inhibits cellular transcription. J Virol. 2006;80(12):5686–5696. doi: 10.1128/JVI.02739-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorchakov R, Frolova E, et al. Inhibition of transcription and translation in Sindbis virus-infected cells. J Virol. 2005;79(15):9397–9409. doi: 10.1128/JVI.79.15.9397-9409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorchakov R, Frolova E, et al. A new role for ns polyprotein cleavage in Sindbis virus replication. J Virol. 2008;82(13):6218–6231. doi: 10.1128/JVI.02624-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorchakov R, Frolova E, et al. PKR-dependent and -independent mechanisms are involved in translational shutoff during Sindbis virus infection. J Virol. 2004;78(16):8455–8467. doi: 10.1128/JVI.78.16.8455-8467.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradi A, Svitkin YV, et al. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc Natl Acad Sci U S A. 1998;95(19):11089–11094. doi: 10.1073/pnas.95.19.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green SR, Mathews MB. Two RNA-binding motifs in the double-stranded RNA-activated protein kinase, DAI. Genes Dev. 1992;6(12B):2478–2490. doi: 10.1101/gad.6.12b.2478. [DOI] [PubMed] [Google Scholar]
- Hahn YS, Strauss EG, et al. Mapping of RNA- temperature-sensitive mutants of Sindbis virus: assignment of complementation groups A, B, and G to nonstructural proteins. J Virol. 1989;63(7):3142–3150. doi: 10.1128/jvi.63.7.3142-3150.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy RW, Rice CM. Requirements at the 3′ end of the sindbis virus genome for efficient synthesis of minus-strand RNA. J Virol. 2005;79(8):4630–4639. doi: 10.1128/JVI.79.8.4630-4639.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy WR, Strauss JH. Processing the nonstructural polyproteins of sindbis virus: nonstructural proteinase is in the C-terminal half of nsP2 and functions both in cis and in trans. J Virol. 1989;63(11):4653–4664. doi: 10.1128/jvi.63.11.4653-4664.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang SK. Internal initiation: IRES elements of picornaviruses and hepatitis c virus. Virus Res. 2006;119(1):2–15. doi: 10.1016/j.virusres.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Kash JC, Goodman AG, et al. Hijacking of the host-cell response and translational control during influenza virus infection. Virus Res. 2006;119(1):111–120. doi: 10.1016/j.virusres.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Krausslich HG, Nicklin MJ, et al. Poliovirus proteinase 2A induces cleavage of eucaryotic initiation factor 4F polypeptide p220. J Virol. 1987;61(9):2711–2718. doi: 10.1128/jvi.61.9.2711-2718.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemm JA, Rumenapf T, et al. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 1994;13(12):2925–2934. doi: 10.1002/j.1460-2075.1994.tb06587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R, Schlesinger S, et al. Promoter for Sindbis virus RNA-dependent subgenomic RNA transcription. J Virol. 1990;64(4):1726–1733. doi: 10.1128/jvi.64.4.1726-1733.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ML, Stollar V. Distinct sites on the Sindbis virus RNA-dependent RNA polymerase for binding to the promoters for the synthesis of genomic and subgenomic RNA. J Virol. 2007;81(8):4371–4373. doi: 10.1128/JVI.02672-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayuri TW, Geders, et al. Role for conserved residues of sindbis virus nonstructural protein 2 methyltransferase-like domain in regulation of minus-strand synthesis and development of cytopathic infection. J Virol. 2008;82(15):7284–7297. doi: 10.1128/JVI.00224-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju R, Huang HV. Analysis of Sindbis virus promoter recognition in vivo, using novel vectors with two subgenomic mRNA promoters. J Virol. 1991;65(5):2501–2510. doi: 10.1128/jvi.65.5.2501-2510.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice CM, Levis R, et al. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J Virol. 1987;61(12):3809–3819. doi: 10.1128/jvi.61.12.3809-3819.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano PR, Garcia-Barrio MT, et al. Autophosphorylation in the activation loop is required for full kinase activity in vivo of human and yeast eukaryotic initiation factor 2 alpha kinases PKR and GCN2. Mol Cell Biol. 1998;18(4):2282–2297. doi: 10.1128/mcb.18.4.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp JC, Jundt N, et al. Requirment for the amino-terminal domain of Sindbis virus nsP4 during virus infection. J. Virol. 2011;85(7):3449–3460. doi: 10.1128/JVI.02058-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo AT, White MA, et al. The crystal structure of the Venezuelan equine encephalitis alphavirus nsP2 protease. Structure. 2006;14(9):1449–1458. doi: 10.1016/j.str.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Sanz MA, Castello A, et al. Viral translation is coupled to transcription in sindbis virus-infected cells. J Virol. 2007;81(13):7061–7068. doi: 10.1128/JVI.02529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz MA, Castello A, et al. Dual mechanism for the translation of subgenomic mRNA from Sindbis virus in infected and uninfected cells. PLoS ONE. 2009;4(3):e4772. doi: 10.1371/journal.pone.0004772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirako Y, Strauss JH. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J Virol. 1994;68(3):1874–1885. doi: 10.1128/jvi.68.3.1874-1885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokoloski KJ, Hayes CA, et al. Sindbis virus infectivity improves during the course of infection in both mammalian and mosquito cells. Virus Res. 2012;167:26–33. doi: 10.1016/j.virusres.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojdl DF, Abraham N, et al. The murine double-stranded RNA-dependent protein kinase PKR is required for resistance to vesicular stomatitis virus. J Virol. 2000;74(20):9580–9585. doi: 10.1128/jvi.74.20.9580-9585.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss JH, Strauss EG. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58(3):491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suopanki J, Sawicki DL, et al. Regulation of alphavirus 26S mRNA transcription by replicase component nsP2. J Gen Virol. 1998;79(Pt 2):309–319. doi: 10.1099/0022-1317-79-2-309. [DOI] [PubMed] [Google Scholar]
- Toribio R, Ventoso I. Inhibition of host translation by virus infection in vivo. Proc Natl Acad Sci U S A. 2010;107(21):9837–9842. doi: 10.1073/pnas.1004110107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steeg H, Thomas A, et al. Shutoff of neuroblastoma cell protein synthesis by Semliki Forest virus: loss of ability of crude initiation factors to recognize early Semliki Forest virus and host mRNA's. J Virol. 1981;38(2):728–736. doi: 10.1128/jvi.38.2.728-736.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steeg H, van Grinsven M, et al. Initiation of protein synthesis in neuroblastoma cells infected by Semliki Forest Virus. A decreased requirement of late viral mRNA for eIF-4B and cap binding protein. FEBS Lett. 1981;129(1):62–66. doi: 10.1016/0014-5793(81)80756-9. [DOI] [PubMed] [Google Scholar]
- Ventoso I. Adaptive changes in alphavirus mRNA translation allowed colonization of vertebrate hosts. J Virol. 2012;86(17):9484–9494. doi: 10.1128/JVI.01114-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventoso I, Sanz MA, et al. Translational resistance of late alphavirus mRNA to eIF2alpha phosphorylation: a strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev. 2006;20(1):87–100. doi: 10.1101/gad.357006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F, Wagner V, et al. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol. 2006;80(10):5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielgosz MM, Raju R, et al. Sequence requirements for Sindbis virus subgenomic mRNA promoter function in cultured cells. J Virol. 2001;75(8):3509–3519. doi: 10.1128/JVI.75.8.3509-3519.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Feigenblum D, et al. A late adenovirus factor induces eIF-4E dephosphorylation and inhibition of cell protein synthesis. J Virol. 1994;68(11):7040–7050. doi: 10.1128/jvi.68.11.7040-7050.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]