Abstract
The molecular era of telomere biology began with the discovery that telomeres usually consist of G-rich simple repeats and end with 3′ single-stranded tails. Enormous progress has been made in identifying the mechanisms that maintain and replenish telomeric DNA and the proteins that protect them from degradation, fusions, and checkpoint activation. Although telomeres in different organisms (or even in the same organism under different conditions) are maintained by different mechanisms, the disparate processes have the common goals of repairing defects caused by semiconservative replication through G-rich DNA, countering the shortening caused by incomplete replication, and postreplication regeneration of G tails. In addition, standard DNA repair mechanisms must be suppressed or modified at telomeres to prevent their being recognized and processed as DNA double-strand breaks. Here, we discuss the players and processes that maintain and regenerate telomere structure.
Specialized processes (e.g., nonreciprocal recombination) have evolved to maintain telomeres. If a telomere is lost, it is recognized as a double-strand break, causing checkpoint arrest, telomere fusions, and degradation.
The ends of chromosomes or telomeres are comprised of telomeric DNA and its associated proteins. In most eukaryotes, telomeric DNA is highly repetitive and rich in guanosine clusters in the strand running 5′ to 3′ toward the chromosome end, such as 5′-GGGTTA-3′ (humans), 5′-G1–3T-3′ (budding yeast, Saccharomyces cerevisiae), and 5′- G2–8TTACA-3′ (fission yeast, Schizosaccharomyces pombe) (Fig. 1). The amount of telomeric DNA per end varies from ∼300 bp in yeasts to up to 150 kb in some mouse strains (see Table 1 for a list of abbreviations used in this article) (Blasco et al. 1997).
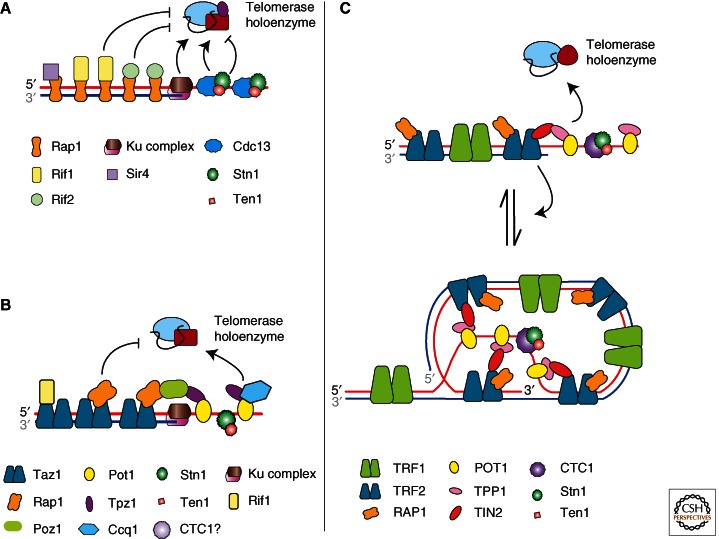
Figure 1.
Model structures and associated proteins of budding yeast, fission yeast, and human telomeres. (A) DNA structure and associated proteins of budding yeast telomeres. Arrows and blunt arrows denote up-regulation and down-regulation of telomerase recruitment, respectively (see Table 1 for abbreviations). (B) DNA structure and associated proteins of fission yeast telomeres (3′ end fold back and double-stranded DNA [dsDNA] invasion to form a t-loop is a potential alternate structure that is not shown [Tomaska et al. 2004]). (C) DNA structure and associated proteins of human telomeres.
Table 1.
Abbreviations used
| Abbreviation | Full name | Abbreviation | Full name |
|---|---|---|---|
| Nomenclature | Proteins | ||
| ALT | Alternative lengthening of telomeres | 53BP1 | p53-binding protein 1 |
| bp CB |
Base pair Cajal bodies |
ATM | Ataxia telangiectasia mutated; homolog of budding yeast Tel1 |
| ChIP CHO |
Chromatin immunoprecipitation Chinese hamster ovary |
ATR | Ataxia telangiectasia and rad3-related |
| DSB Est |
Double-stranded break Ever-shortening telomere |
Ccq1 | Coiled-coil protein quantitatively enriched 1 |
| G4 | G-quadruplex | Cdc13 | Cell division cycle 13 |
| H | Human | CST | Cdc13-Stn1-Ten1 |
| HAATI HTT |
Heterochromatin amplification-mediated and telomerase-independent Collective name for Drosophila telomere retrotransposons:HeT-A, TART, and Tahre |
CTC1 Est1, Est2, Est3 PARP1 |
Conserved telomere maintenance component 1 Ever-shortening telomere (1,2,3) Polyadenosine diphosphate ribose polymerase 1 |
| HR | Homologous recombination | Pot1 | Protection of telomeres 1 |
| kb | Kilobase | Rap1 | Repressor activator protein 1 |
| LINE-1 | Long interspersed element-one | Rif1, Rif2 | Rap1-interacting factors 1 and 2 |
| MEF | Mouse embryonic fibroblast | RPA | Replication protein A |
| NHEJ | Nonhomologous end joining | RTEL1 | Regulator of telomere length 1 |
| PAL-mechanism | Palindrome-dependent mechanism end maintenance | Sir4 SSB |
Silent information regulator 4 Single-stranded binding protein |
| RNP | Ribonucleoprotein particle | Stn1 | Suppressor of cdc thirteen 1 |
| RT ss |
Reverse transcriptase Single-stranded |
Taz1 | Telomere associated in Schizosaccharomyces pombe |
| t-Loop | Telomere-loop | TEBPα/β | Telomere end-binding protein α/β |
| UFB | Ultrafine bridges | Ten1 | Telomeric pathways with STN1 |
| TIN2 | TRF1-interacting nuclear protein 2 | ||
In addition to duplex telomeric DNA, telomeres end in G-rich overhangs or G-tails that are usually present at both ends of the chromosome. G-tails also vary in length: Human G-tails are between 100 and 280 nucleotides throughout the cell cycle (Makarov et al. 1997; Wright et al. 1997), whereas budding yeast G-tails are only ∼12–14 nucleotides (Larrivee et al. 2004) except in late S phase when they are 50–100 nucleotides (Wellinger et al. 1993). Long G-tails can fold back and invade the duplex region to form a specialized displacement loop called a t-loop (telomere loop) (Fig. 1C) that is thought to provide a protective cap (Griffith et al. 1999).
In addition to t-loops, G-strand telomeric DNA from most organisms forms G-quadruplex (G4) structures in vitro, a stable DNA secondary structure that is held together by multiple guanine–guanine base pairs (reviewed in Bochman et al. 2012). These structures can form within a single strand of DNA or from two or four strands and can be parallel or antiparallel. G4 DNA may contribute to telomere capping (see, e.g., Smith et al. 2011), and, depending on its exact structure, can inhibit or promote telomerase in vitro (Zahler et al. 1991; Zaug et al. 2005; Oganesian et al. 2006). The best evidence for G4 structures at telomeres in vivo comes from ciliates (Paeschke et al. 2005). An antibody that recognizes only antiparallel G4 structures comprised of G4T4 Oxytricha telomeric DNA binds telomeres in a cell-cycle-specific manner, and this binding depends on the telomere end-binding proteins, TEBPα/β (see section on Telomere Proteins) (Paeschke et al. 2005, 2008).
TELOMERE PROTEINS
Proteins that bind directly to telomeric DNA can be divided into single-stranded (ss) and double-stranded (ds) DNA-binding proteins (see Table 2 for a list of proteins discussed in this article and their names in yeasts and mammals.) (Fig. 1). The Oxytricha nova ss-binding heterodimeric TEBPα/β was the first ss telomere DNA-binding activity to be identified (Gottschling and Zakian 1986). Cdc13 is the G-tail-binding subunit of a heterotrimeric complex called CST (Cdc13-Stn1-Ten1) in budding yeast (Lin and Zakian 1996; Nugent et al. 1996; Hughes et al. 2000). The CST complex, which has so far been found only at telomeres, interacts with both Est1, a telomerase subunit (Qi and Zakian 2000; Pennock et al. 2001; Wu and Zakian 2011) and DNA Pol α, which is required to start DNA synthesis on both leading and lagging strands (Qi and Zakian 2000; Grossi et al. 2004).
Table 2.
List of major factors discussed
| Budding yeast | Fission yeast | Mammals | Others | |
|---|---|---|---|---|
| Duplex sequence-specific binding proteins | Rap1 | Taz1 | TRF1, TRF2 | HOAP (Drosophila) |
| Single-strand sequence-specific binding proteins | Cdc13 | Pot1 | POT1 (POT1a, POT1b) | TEBPα, TEBPβ (ciliates) Verrocchio (Drosophila) |
| Protein–protein-interacting proteins | ||||
| Conserved | Rif1 | Rif1 | RIF1 | |
| Species-specific | Rif2, Sir4 | Ccq1 | TPP1, TIN2 | |
| CST complex | Cdc13, Stn1, Ten1 | Stn1, Ten1 | CTC1, Stn1, Ten1 | |
| Checkpoint proteins with roles at telomeres | ||||
| ATR checkpoint kinase | Mec1 | Rad3 | ATR | |
| ATM checkpoint kinase | Tel1 | Tel1 | ATM | |
| Checkpoint proteins | MRX | MRN | MRN | |
| Telomerase holoenzyme components | ||||
| Catalytic subunit | Est2 | Trt1 | TERT | |
| RNA template | TLC1 | TER1 | TR, TERC | |
| Conserved accessory subunit | Est1 | Est1 | EST1A, EST1B | |
| Species-specific accessory subunit | Est3 | |||
| Helicases | ||||
| RecQ helicase | Sgs1 | Rqh1 | WRN, BLM | |
| Pif helicase | Pif1, Rrm3 | Pfh1 | PIF1 | |
| Others | RTEL, FANCJ | |||
Some factors are listed under multiple headings. WRN, Werner; BLM, Bloom.
In fission yeast, mammals, and some plants, many of the telomere functions of CST are performed by Pot1, a homolog of the ciliate TEBPα (Baumann and Cech 2001; Shakirov et al. 2005; Hockemeyer et al. 2006; Wu et al. 2006). However, vertebrates and plants also have a CST complex that protects telomeres and/or functions in telomere replication and telomerase regulation (Miyake et al. 2009; Surovtseva et al. 2009; reviewed in Price et al. 2010; Chen et al. 2012). In these organisms, Cdc13 is replaced by another protein, CTC1, whose mutation leads to inherited human disease, such as dyskeratosis congenita (Armanios 2012; Keller et al. 2012). Although fission yeast encodes Stn1 and Ten1-like proteins, a S. pombe CTC1/Cdc13-like protein has not been identified (reviewed in Subramanian and Nakamura 2010). Unlike budding yeast CST, mammalian CST is not telomere limited but has more global roles in DNA replication (Gu et al. 2012; Stewart et al. 2012). The various G-strand-binding proteins are not highly conserved in primary sequence, but all share OB (oligonucleotide/oligosaccharide binding) fold domain(s) that bind tightly to telomeric ssDNA with high sequence specificity (Horvath et al. 1998; Mitton-Fry et al. 2002; Lei et al. 2003).
The ds telomeric DNA is also bound by sequence-specific DNA-binding proteins. Again, the sequences of these proteins vary but they have in common that they bind DNA via a conserved Myb domain. ds telomere-binding proteins include budding yeast Rap1 (Conrad et al. 1990; Lustig et al. 1990), fission yeast Taz1 (Cooper et al. 1997), and mammalian TRF1 and TRF2 (Chong et al. 1995; Broccoli et al. 1997). Budding yeast Rap1 is not telomere limited but is also a transcriptional modulator at many genes (Lieb et al. 2001).
Proteins that bind telomeres via protein–protein interactions form a third class of telomeric proteins. For example, in budding yeast Rif1, Rif2, and Sir4 associate with telomeres via their interactions with the carboxyl terminus of Rap1, whereas Stn1 and Ten1 interact with telomeric ssDNA by association with Cdc13 (Grandin et al. 1997, 2001b). Another example is human POT1, which binds to G-tails via an OB fold, but whose stable telomere association relies on its interaction with TPP1, which strengthens its binding to telomeric ssDNA (Ye et al. 2004; Wang et al. 2007; Xin et al. 2007). TPP1 also interacts with TIN2, and TIN2 also interacts with TRF1. Thus, TIN2 forms a bridge between the ds and ss regions of the telomere (O’Connor et al. 2006). Finally, TRF2 binds RAP1, a non-DNA-binding homolog of budding yeast Rap1, which like its yeast counterpart, has both telomeric and transcriptional roles (Martinez et al. 2010). Collectively, POT1, TPP1, TRF1, TRF2, RAP1, and TIN2 form a six-member complex known as “shelterin” (de Lange 2005) that performs multiple telomeric functions. Fission yeast telomeres are also bound by a six-member shelterin-like complex (Miyoshi et al. 2008). Like mammalian shelterin, the fission yeast complex extends between Pot1 bound to ss telomeric DNA and the dsDNA binder Taz1.
TELOMERE FUNCTIONS
Telomeres are essential to provide a protective “cap” that shields chromosomes from degradation, fusions, and checkpoint recognition. If the cap is lost, telomeres can no longer be distinguished from a double-stranded break (DSB), which causes checkpoint arrest, telomere fusions, and rampant degradation. End protection or capping is due largely to telomere-binding proteins, but in almost all organisms, the protein cap is assembled only on telomeric DNA. Thus, for telomeres to carry out their essential capping functions, duplex telomeric DNA and G-tails must be maintained.
Most of the chromosome, including most of telomeric DNA, is replicated by standard semiconservative replication. However, chromosomal DNA cannot provide a template for replication of the very ends of linear DNA molecules (Watson 1972). This problem arises from the properties of DNA polymerases, which require a primer, usually RNA, to start DNA synthesis and replicate DNA only in the 5′ to 3′ direction. Removal of the terminal RNA primer at the 5′ ends of newly replicated strands leaves a gap that cannot be filled by a standard DNA polymerase. As a result, successive rounds of DNA replication result in gradual telomere attrition and, ultimately, loss of genetic information. A second replication problem derives from the importance of G-tails for telomere integrity (Lingner et al. 1995). After conventional semiconservative DNA replication, the leading strand DNA polymerase will generate a blunt-ended terminus at one end of each chromosome, and this blunt end cannot bind G-tail-binding proteins. G-tails are regenerated by degradation of the C-rich strand followed by C-strand resynthesis (see section Another Replication Problem: G-Tail Regeneration) (Wellinger et al. 1996).
Three major strategies are used to solve the first replication problem: telomerase (see section Telomerase), recombination (see section Recombinational Maintenance of Telomeric DNA), and transposition (see section Retrotransposons as Telomeres). Each of these mechanisms maintains chromosome ends by repairing the damage owing to incomplete replication. Thus, a study of telomere maintenance is really a study of telomere repair. The second “end replication problem,” G-tail regeneration, is solved by a variation of classical DSB repair.
TELOMERASE
Introduction to Telomerase
Telomerase is a telomere-dedicated reverse transcriptase (Fig. 2A). At its core, telomerase consists of the reverse transcriptase subunit called Est2 (budding yeast), Trt1 (fission yeast), or TERT (mammals) and a tightly associated RNA molecule called TLC1 (budding yeast), TER1 (fission yeast), or TR (human). Telomerase RNAs contain a short segment that is complementary to the G-strand of telomeric DNA and serves as the template for telomere elongation.
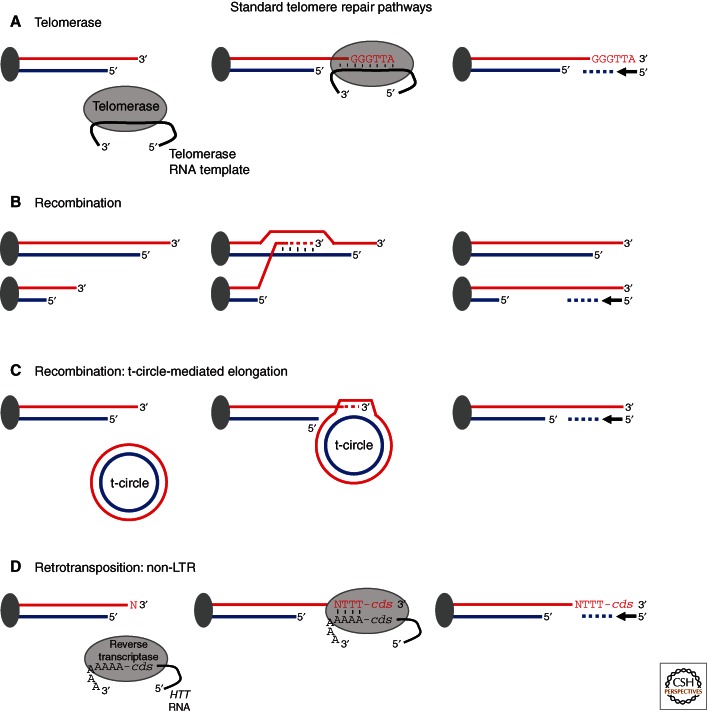
Figure 2.
Standard telomere repair pathways. Pathways are represented from left to right with DNA strands in red and blue. Centromeres are shown as dark ovals and relevant proteins and complexes are shown as labeled ovals. (A) Telomerase, a telomere-dedicated reverse transcriptase mechanism used in most eukaryotes. (B) Nonreciprocal recombination, used by a minority of eukaryotes, but is used as a survival strategy by most cells that survive without telomerase. (C) Recombination: t-circle-mediated elongation, a method of elongation that is best understood in yeasts in which circular DNA is used to template the addition of a new sequence to the end of the telomere. The new sequence can spread to other telomeres by nonreciprocal recombination. (D) Retrotransposition: tandem arrays of non-LTR (long terminal repeat) retrotransposons are added to the ends of telomeres. Retrotransposition is targeted only to telomeres probably because the transposon does not encode an endonuclease.
Although the catalytic and RNA subunits are necessary and sufficient in vitro for elongation of telomeric primers, other protein subunits are required for telomere lengthening in vivo. These accessory factors were first identified in budding yeast by their “ever-shortening telomere” (est) phenotype (Lundblad and Szostak 1989; Lendvay et al. 1996). For example, Est1, which has homologs in humans (Reichenbach et al. 2003; Snow et al. 2003) and fission yeast (Beernink et al. 2003), is necessary for the recruitment and/or activation of budding yeast telomerase (Evans and Lundblad 1999; Taggart et al. 2002; Chan et al. 2008; DeZwaan and Freeman 2009) owing to its binding both telomerase RNA (Livengood et al. 2002; Seto et al. 2002) and Cdc13 (Qi and Zakian 2000; Wu and Zakian 2011). Fission yeast Est1 interacts with the shelterin component Ccq1 to bring telomerase to telomeres (Moser et al. 2011; Webb and Zakian 2012; Yamazaki et al. 2012). Not all key telomerase subunits are conserved. For example, budding yeast Est3 is essential for telomerase activity in vivo (Lendvay et al. 1996), yet it is absent from fission yeast and human genomes, although it is structurally related to mammalian TPP1 (Lee et al. 2008a; Yu et al. 2008).
Cell-Cycle Regulation of Telomerase
Although telomerase activity can be detected in budding yeast extracts prepared from cells throughout the cell cycle (Diede and Gottschling 1999), telomeres are only extended during late S phase (Marcand et al. 2000). Likewise, although the catalytic core of telomerase is telomere associated throughout most of the cell cycle, the telomerase holoenzyme is telomere associated only in late S/G2 phase (Taggart et al. 2002; Chan et al. 2008; Tuzon et al. 2011). In fission yeast, telomerase is telomere associated only in late S phase (Moser et al. 2009a; Webb and Zakian 2012). Thus, in yeasts, telomerase action is cell-cycle regulated.
Cell-cycle regulation of budding yeast telomerase is due in part to regulated degradation of Est1 (Osterhage et al. 2006). Est1 peaks in late S phase (Taggart et al. 2002), and is required to recruit Est3 to telomeres (Tuzon et al. 2011). Thus, cell-cycle-regulated abundance of Est1 is, in principle, sufficient to explain the cell-cycle-regulated activity of telomerase. However, even if Est1 is at high levels in G1 phase and is incorporated along with Est3 into the holoenzyme, telomerase is not active in G1 phase (Osterhage et al. 2006). The telomerase inhibitory proteins Rif1 and Rif2 help sequester telomeres from telomerase in G1 phase (Gallardo et al. 2011). In addition, the long G tails, which are probably the preferred substrate for telomerase, are only present in late S phase (Wellinger et al. 1993). Indeed, removal of Rif proteins allows telomerase to act in G1 phase, perhaps because they protect telomeres from degradation (Bonetti et al. 2010b; Gallardo et al. 2011). In addition, Cdc13 is multiply phosphorylated in a cell-cycle-dependent manner, and several of these modifications affect telomere length and/or Est1 recruitment (Li et al. 2009; Wu et al. 2013).
In fission yeast, levels of telomerase subunits (Est1 and Trt1) (Dehe et al. 2012; Webb and Zakian 2012) and their regulator (Ccq1) (Tomita and Cooper 2008) are not cell-cycle affected. Taz1 enforces telomerase-telomere recruitment to late S phase, as evidenced by the larger temporal window of Trt1- and Est1-telomere association in taz1Δ cells (Dehe et al. 2012).
Telomerase is not active in most human somatic cells because transcription of TERT, the catalytic subunit, is repressed by several tumor suppressor pathways (Lin and Elledge 2003). In addition, the WNT signaling pathway, which is important for stem cell identity, affects TERT expression. β-catenin, a central player in WNT signaling, acts directly to activate TERT transcription in embryonic and adult stem cells as well as in human cancer cells (Hoffmeyer et al. 2012). The c-Myc oncogene also positively regulates TERT transcription (Wu et al. 1999).
In HeLa cells, which express high levels of telomerase, telomerase RNA is telomere associated in S phase (Jady et al. 2006) when telomeres are lengthened and telomerase activity is the highest (Zhu et al. 1996). At this time, Cajal bodies (CB), nuclear bodies composed primarily of the protein coilin that functions in RNP (ribonucleoprotein particle) modification/assembly, also associate with telomeres (Jady et al. 2006). CBs are important for both telomerase maturation and for recruiting telomerase to telomeres (reviewed in Londono-Vallejo and Wellinger 2012; Stern et al. 2012).
TRF1 is a negative regulator of telomere length (van Steensel and de Lange 1997). The checkpoint kinase ATM phosphorylates TRF1 on S367 resulting in release of TRF1 from telomeric DNA. Phosphorylated TRF1 forms non-telomere-associated nuclear foci during S and G2 phase that overlap with proteasome centers. Moreover, TRF1-S367D does not inhibit telomere lengthening (McKerlie et al. 2012). These findings implicate TRF1 as a major player in enforcing cell-cycle-regulated telomere extension in human cells.
Regulation of Telomerase by Telomere Length
In budding yeast, an assay that determines the amount of yeast telomeric DNA added per telomere in a single cell cycle reveals that only ∼10% of telomeres are lengthened in a given S phase (Teixeira et al. 2004). Moreover, the likelihood of telomere extension is inversely correlated with telomere length with short telomeres (∼100 bp) being about six times more likely to be lengthened than wild-type length telomeres (∼300 bp). Thus, unlike conventional DNA replication, telomerase does not act on every telomere in a given cell cycle, reinforcing the idea that telomerase is more akin to repair than replication.
Chromatin immunoprecipitation (ChIP) in budding yeast reveals that short telomeres bind more telomerase than wild-type telomeres (Bianchi and Shore 2007; Sabourin et al. 2007). This elevated binding is achieved by two nonoverlapping mechanisms. Short telomeres preferentially bind MRX (Mre11/Rad50/Xrs2), which in turn recruits Tel1 to short telomeres (Bianchi and Shore 2007; Hector et al. 2007; Sabourin et al. 2007; McGee et al. 2010). Tel1 kinase activity is required to promote preferential binding of telomerase to short telomeres (Hector et al. 2007; Sabourin et al. 2007). There are data for (Tseng et al. 2006) and against (Gao et al. 2010b; Wu et al. 2013) Cdc13 being the relevant telomere target of the Tel1 kinase in terms of telomerase recruitment. Alternatively, Tbf1, a transcription factor that binds at specific sites in subtelomeric DNA (Fourel et al. 1999), can recruit telomerase to short telomeres in the absence of Tel1 (Arneric and Lingner 2007).
The ability of Tel1 to distinguish short from wild-type length telomeres is dependent on Rif2, which is more abundant on long telomeres (Sabourin et al. 2007; McGee et al. 2010). Rif2 inhibits Tel1 binding because both proteins (Sabourin et al. 2007) interact with the carboxyl end of Xrs2 and thus compete with each other for MRX association (Hirano et al. 2009). Likewise, Tbf1 can inhibit MRX binding to telomeres (Fukunaga et al. 2012). In most situations, Tel1 affects only the frequency of telomere lengthening, but at short telomeres it also increases telomerase processivity (Chang et al. 2007).
Preferential lengthening of short telomeres is seen in some contexts in mammalian cells. For example, when telomerase minus mice are crossed to heterozygous telomerase plus mice, the average length of telomeres is not very different in the telomerase plus and telomerase minus progeny, but the telomerase plus animals no longer contain very short telomeres (Hemann et al. 2001). This result can be explained if mouse telomerase acts preferentially on short telomeres. However, overexpression of telomerase in primary cells and cancer cell lines lengthens both short and long telomeres, suggesting that human telomerase is targeted to short telomeres only when telomerase is limiting (Cristofari and Lingner 2006). Consistent with this possibility, every telomere is equivalently elongated in a given cell cycle in human cancer cell lines, such as H1299 and HeLa (Zhao et al. 2011).
TERRA, a Novel Telomerase Regulator
In yeasts and human cells, transcription is repressed in subtelomeric regions by a phenomenon called telomere position effect (TPE) (Gottschling et al. 1990; Nimmo et al. 1994; Baur et al. 2001; reviewed in Mondoux and Zakian 2006). Thus, the discovery of TERRA, nuclear noncoding telomeric repeat containing RNA, was unexpected (Azzalin et al. 2007; Luke et al. 2008; Schoeftner and Blasco 2008). TERRA, which has been described in mammals and yeasts, is transcribed from within the subtelomeric region toward the telomere. TERRA inhibits telomerase in at least two ways. The budding yeast Rat1 nuclease, which has general roles in mRNA processing, also degrades TERRA. When Rat1 levels are low, telomeres shorten. This shortening can be reversed by overexpressing RNaseH, which degrades RNA in RNA–DNA hybrids, suggesting that TERRA inhibits yeast telomerase by hybridizing to telomeric DNA (Luke et al. 2008). In humans, TERRA inhibits telomerase by interacting directly with telomerase (Redon et al. 2010). In addition, telomerase-mediated telomere lengthening inhibits TERRA production in human cell lines while at the same time increasing the length of TERRA transcripts, suggesting that telomerase action and TERRA feed back on each other (Arnoult et al. 2012).
RECOMBINATIONAL MAINTENANCE OF TELOMERIC DNA
Telomere Recombination in Telomerase Minus Cells
Most eukaryotes use telomerase to counter the loss of telomeric DNA that occurs during semiconservative replication. However, some organisms solve the end replication problem using nonreciprocal recombination (Fig. 2B). In some cases, recombination and telomerase are both active, whereas in other organisms, only recombination is used (reviewed in McEachern et al. 2000). However, most studies on recombinational maintenance of telomeres are from organisms that normally rely on telomerase, such as yeasts and humans. In these cases, recombination is a bypass mechanism that maintains telomeres when telomerase is not expressed. These bypass telomere maintenance pathways are called ALT (alternative lengthening of telomeres).
In budding yeast, most telomerase minus cells die. However, survivors, which come in two types, types I and II, arise in all telomerase-deficient cultures (Lundblad and Blackburn 1993; Teng and Zakian 1999). Type I telomeres are composed of long tandem arrays of the subtelomeric Y′ element capped by very short tracts of telomeric repeats (Lundblad and Blackburn 1993). Type II telomeres are dependent on the MRX complex and have very heterogeneous length telomeres, some very short and others as long as 12 kb (Teng and Zakian 1999; Teng et al. 2000). Telomerase minus fission yeast can maintain telomeres by either ALT (Nakamura et al. 1998) or chromosome circularization (Fig. 3A) (Nakamura et al. 1998).
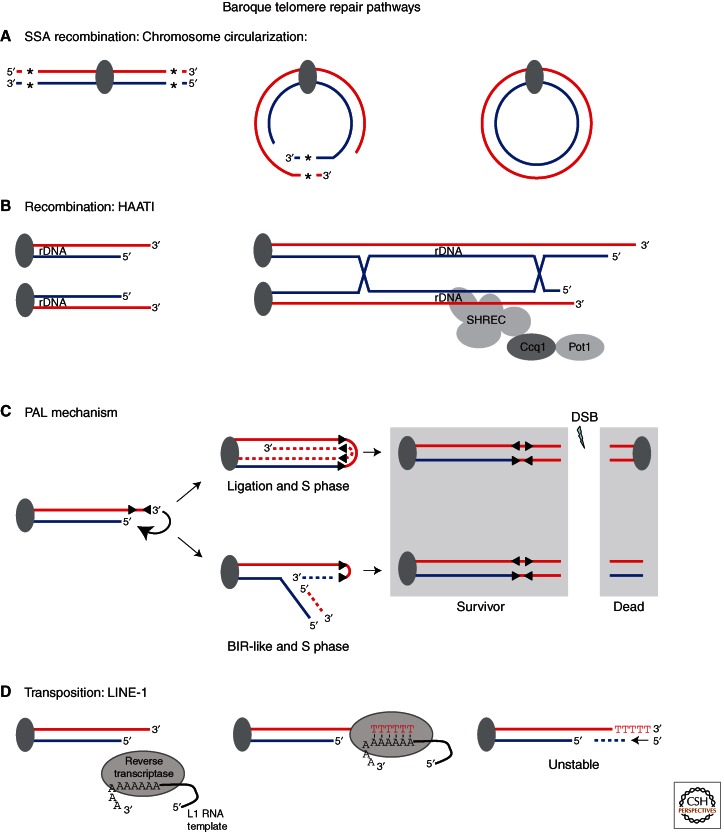
Figure 3.
Baroque telomere repair pathways. Pathways are represented from left to right with DNA strands in red and blue. Centromeres are shown as dark ovals and relevant proteins and complexes are shown as labeled ovals. (A) Single-strand annealing recombination: chromosome circularization, a telomerase minus cell bypass mechanism found in fission yeast telomerase minus cells that is dependent on regions of homology, which are used to prime DNA synthesis and fuse the two ends of a single chromosome. *Denotes regions of homology. (In the absence of Taz1 nonhomologous end joining is used to fuse the ends [not shown] [Ferreira and Cooper 2001; Wang and Baumann 2008].) (B) Recombination: HAATI, a mode of telomere maintenance in fission yeast telomerase minus cells that maintains chromosome ends by recombination between rDNA arrays and recruitment of protective factors by heterochromatic interactions. (C) PAL-mechanism, a bypass mechanism in telomerase and C-strand degradation deficient budding yeast cells that uses palindromic sequence to generate a telomeric hairpin structure, which can undergo two pathways that both generate a single survivor after cell division. Palindromic sequences are depicted as mirrored black triangles. (D) Transposition: LINE-1, a process so far only observed in Chinese hamster ovary cells in which dysfunctional telomeres serve as substrates for LINE-1 endonuclease-independent retrotransposition. cds, coding sequence; SSA, single-strand annealing; SHREC, Snf2/Hdac-containing repressor complex; BIR, break-induced recombination (also see Table 1).
Telomerase is activated in ∼90% of tumors, but 5%–10% of human tumors use ALT (Bryan et al. 1997; Dunham et al. 2000). As in type II ALT in budding yeast, telomere length is highly heterogeneous in human ALT. Extrachromosomal circles of telomeric DNA, known as t-circles, are associated with ALT in yeasts and mammals (Natarajan and McEachern 2002; Natarajan et al. 2003; Cesare and Griffith 2004; Wang et al. 2004). The mechanistic importance of t-circles to ALT is best documented in the yeast Kluyveromyces lactis in which t-circles have been shown to provide the template for recombinational telomere lengthening (Fig. 2C) (Natarajan and McEachern 2002). t-circles in human ALT cells are similar in size to t-loops leading to the proposal that t-circles are generated by HR-mediated deletion of t-loops (Cesare and Griffith 2004).
Recombination in Telomerase-Proficient Cells
Recombination can regulate telomere length even in telomerase plus cells. For example, generation of t-circles by recombination is used in telomerase-proficient cells to shorten abnormally long telomeres. This MRX-dependent process, called telomere rapid deletion (TRD) was discovered in budding yeast, in which it can reduce exceptionally long telomeres to wild-type length in a single cell cycle (Li and Lustig 1996). A similar trimming mechanism occurs in mammals. For example, in mouse cells lacking the amino-terminal basic domain of TRF2, t-loop-sized telomeric circles are excised from leading strand telomeres via homologous recombination (HR) (Wang et al. 2004); t-circles also play a role in telomere trimming in human cancer cells that overexpress telomerase (Pickett et al. 2009).
Given that telomeres are repetitive, it is probably not surprising that HR can contribute to their maintenance. This type of event was first documented in budding yeast, in which heterologous telomere repeats can recombine by events that initiate at the boundary of unique and telomeric DNA and transfer a telomere length’s worth of DNA in a single recombination event (Pluta and Zakian 1989; Wang and Zakian 1990). In mice, Rad51D localizes to mitotic and meiotic chromosomes and its depletion (in combination with reduced p53) results in short telomeres and high levels of telomere fusions (Tarsounas et al. 2004), as does depletion of Rad54 (Jaco et al. 2003). The tumor suppressor and recombination facilitator BRCA2 binds HeLa telomeres in S and G2 phase, and this binding promotes telomere binding of RAD51, another Rad51 paralog (Badie et al. 2010). In mice, conditional loss of BRCA2 results in short and fragile telomeres, whereas human brca2−/− tumors have short telomeres.
Although the above data suggest that HR contributes to telomere maintenance, several lines of evidence indicate that HR is normally suppressed at telomeres. For example, in budding yeast, telomere deprotection in cdc13–1 mec3–2 cells induces recombination-dependent telomere hyperelongation (Grandin et al. 2001a). In fission yeast, Taz1 inhibits telomere hyper-recombination (Miller et al. 2006; Rog et al. 2009). In mouse cells, elevated telomere sister chromatid exchange is only seen when cells are defective for two telomere proteins (e.g., TRF2 and Ku70) (Celli et al. 2006; Sfeir et al. 2010). These results suggest that Ku and TRF2 act in parallel to suppress HR at mammalian telomeres.
RETROTRANSPOSONS AS TELOMERES
Although telomerase is an ancient and widespread activity, some organisms lost telomerase at some point in evolution. The fruit fly is the best-studied example of an organism that naturally lacks telomerase. Instead of simple repeats, the ends of fly chromosomes bear tandem arrays of three different non-LTR (long terminal repeat) retrotransposons: HeT-A, TART, and Tahre (collectively called HTT elements) (Fig. 2D) (reviewed in Mason et al. 2008). Internal to HTT are middle repetitive sequences similar to those found internal to the telomerase-generated repeats in organisms with canonical telomeric DNA. HTT elements are replenished by a combination of transposition and HR. The 5′ regions in all HTTs encode Gag genes and in TART and Tahre, also encode a reverse transcriptase (RT) (reviewed in Pardue and DeBaryshe 2008).
HTT elements are telomere limited because they transpose only to DNA ends. This limitation is thought to result from their having lost the endonuclease gene that is found in most retrotransposons (Mason et al. 2008). Its absence is proposed to prevent insertion of the transposon into internal regions of the chromosome. A single targeted transposition places several kb of dispensable DNA at a chromosome end. Although HTT-bearing ends are eroded by incomplete DNA replication, the rate of terminal loss is very slow, ∼50–100 bp/telomere/fly generation (Levis 1989). A model of telomere-targeted HTT transposition followed by terminal sequence loss is supported by the structure of HTT arrays at individual chromosome ends: The tandem copies of HTT elements are positioned in head-to-tail arrays with their 3′ poly(A) ends pointing toward the centromere (Levis et al. 1993). The sequence at the 5′ end is variable, reflecting different extents of sequence loss before a new transposon arrives to form the most distal part of the chromosome.
At first glance, fly telomeres appear to be unrelated to telomerase-generated ends. However, HET-A has the same nucleotide strand bias as canonical telomeres with G residues enriched in the strand running 5′ to 3′ toward the chromosome end (Abad and Villasante 1999). Second, the sequence of the telomerase catalytic subunit has higher sequence similarity to the TART-encoded RT than to many other RTs (Nakamura et al. 1997). Third, many proteins that function at telomerase-generated telomeres, such as ATM and MRN, also have telomere functions in flies (Gao et al. 2010a). HOAP is a heterotrimeric complex that binds Drosophila telomeres and is required for telomere capping. Remarkably, Verrocchio, one of the three proteins in this complex, has an OB fold, suggesting that the complex contacts DNA in the same manner as canonical G-tail-binding proteins (Raffa et al. 2009, 2010). However, unlike yeast and mammalian telomere-binding proteins, fly telomere proteins bind in a sequence-independent manner (Fanti et al. 1998; Cenci et al. 2003), similar to the mammalian CST complex (Price et al. 2010).
Even more amazing than the surprising discovery that transposons can act as telomeres, in certain circumstances, telomere function in flies can be supplied by an epigenetic mechanism that involves the assembly of telomere proteins onto unique sequence DNA. These terminal deletions are readily isolated in mu-2 mutant flies but can also be recovered on P-element mobilization (reviewed in Pardue and Debaryshe 2011). The occurrence of sequence nonspecific telomeres is reminiscent of neocentromeres.
Even organisms that normally rely on telomerase may use transposons in certain situations. For example, LINE-1, the most abundant class of retrotransposons in mammals, has been reported to move to dysfunctional telomeres in CHO (Chinese hamster ovary) cells. This novel pathway (Fig. 3D) seems to be linked to loss of the LINE-1 endonuclease. Transposition to telomeres is increased in cells expressing a dominant–negative allele of TRF2 that disrupts telomere capping (Morrish et al. 2007). Although rare, the budding yeast subtelomeric Y′ element can move via an RNA-mediated process in telomerase-deficient cells (Maxwell et al. 2004).
EVEN MORE BIZARRE MECHANISMS THAT MAINTAIN LINEAR CHROMOSOMES
Approximately half of the samples of the most common soft tissue sarcoma (liposarcomas) from two different patient panels did not show characteristics of either telomerase activation or ALT (Costa et al. 2006; Johnson et al. 2007). These findings suggest that there are as-yet-undiscovered mechanisms to maintain linear chromosomes in human tumors. One possibility is that LINE-1 provides telomere function in these tumors. Even more baroque mechanisms have been described in yeasts. For example, in budding yeast, rare survivors arise in telomerase and recombination-defective cultures, if the strain also lacks either the Exo1 nuclease or the Sgs1 helicase (Maringele and Lydall 2004; Lee et al. 2008b), two proteins involved in C-strand degradation in telomerase-proficient cells (see section Another Replication Problem: G-Tail Regeneration). These cells maintain chromosome ends via a palindrome-dependent mechanism (PAL-mechanism) that uses a break-induced replication-like strategy to generate large palindromic regions at chromosome ends (Fig. 3C). In fission yeast, HAATI (heterochromatin amplification mediated and telomerase independent) provides a telomerase bypass mechanism in which ribosomal or subtelomeric repeats provide end function, but only if the repeats are heterochromatic (Fig. 3B) (Jain et al. 2010).
SEMICONSERVATIVE REPLICATION OF TELOMERIC DNA
Consideration of telomere replication problems usually focuses on mechanisms that compensate for incomplete replication. However, even semiconservative replication of telomeric DNA is a problem for the replication machinery and can lead to telomere damage. The G-rich and repetitive nature of telomeric DNA increases chances of fork slippage and formation of stable secondary structures, such as G4 DNA. Secondary structures are particularly likely given that the G strand is the template for lagging-strand synthesis, which is transiently single stranded during the time required for the leading strand replisome to expose enough DNA to start a new Okazaki fragment. As G4-forming sequences slow fork progression in nontelomeric contexts (London et al. 2008; Lopes et al. 2011; Paeschke et al. 2011), it would not be surprising if they also do so at telomeres. Finally, telomeres are coated with proteins, and this “telosomal” chromatin structure (Wright and Zakian 1995) can create “roadblocks” for replication (Fig. 1). Fork slowing is probably owing to the sequence and/or chromatin structure of telomeric DNA (not its position) as even internal tracts of telomeric DNA slow fork progression in budding and fission yeasts (Ivessa et al. 2002; Miller et al. 2006).
Fragile sites are chromosomal regions that are difficult to replicate especially during conditions of replication stress, such as low nucleotide pools (Sfeir et al. 2009). They often occur at repetitive DNA and/or at stable protein complexes, and their breakage promotes chromosome translocations, such as those seen in many cancers. Although fragile sites have been known for years, the idea that telomeres are fragile sites is recent.
Telomeres were first shown to slow replication forks in budding yeast. By two-dimensional gel analysis and DNA polymerase occupancy, replication forks slow as they move through subtelomeric and telomeric DNA (Ivessa et al. 2002; Azvolinsky et al. 2009). In wild-type cells, this pausing is decreased in the absence of certain telomere-binding proteins, such as Sir4 (Ivessa et al. 2003), suggesting that the non-nucleosomal protein structure of the telomere impedes fork progression. However, as first shown in fission yeast, duplex telomere-binding proteins that bind directly to telomeric DNA actually promote replication through these sequences (Miller et al. 2006). Thus, deletion of fission yeast Taz1 results in replication fork stalling within telomeres, especially at subtelomeric–telomeric borders. Deletion of Taz1 also causes profound telomerase-mediated telomere elongation (Nakamura et al. 1998). However, fork stalling is not simply a consequence of very long telomeres as deletion of the Taz1-interacting protein Rap1 also results in long telomeres but not in slow telomere replication (Miller et al. 2006). When the replication machinery stalls in taz1Δ cells, it recruits telomerase to the subtelomeric/telomeric junction, which can explain the extremely long telomeres in taz1Δ cells (Dehe et al. 2012). Perhaps stalled forks regress into chicken foot structures, which are then degraded to produce a 3′ overhang that recruits telomerase (Dehe et al. 2012).
Deletion of TRF1 in mouse embryo fibroblasts results in fragile telomeres that appear in mitotic spreads as multiple discrete telomeric signals at a single chromosome end (Martinez et al. 2009; Sfeir et al. 2009). In addition, single molecule analysis reveals that replication forks stall at a low, but significant frequency, especially at the subtelomeric–telomeric boundary in these cells (Sfeir et al. 2009). By cytological criteria, simultaneous removal of TRF1 and either of two DNA helicases, BLM or the RTEL, which interact with TRF1 and produce fragile telomeres on individual shRNA knockdown, does not increase the number of fragile telomeres as compared to cells without TRF1 alone. This epistatic relationship suggests a model in which TRF1 at the subtelomere–telomere boundary recruits helicase(s) to stalled replication forks. Fragile telomeres are also seen in mammalian cells depleted for CTC1 (Gu et al. 2012) or the nonsense-mediated decay (NMD) protein UPF1 (which interacts with both hTPP1 and hEST1A) (Fukuhara et al. 2005; Chawla et al. 2011).
DNA HELICASES WITH SPECIALIZED ROLES IN TELOMERE REPAIR
The advent of whole genome sequencing revealed that eukaryotes encode a surprisingly large number of helicases. For example, budding yeast has ∼140 open reading frames with helicase motifs (Shiratori et al. 1999). Unexpectedly many of these helicases have nonoverlapping functions in telomere maintenance.
The 5′-3′Pif1 family helicases are present in some bacteria and almost all eukaryotes (Bochman et al. 2011), and mutations in the single human PIF helicase are associated with increased breast cancer risk (Chisholm et al. 2012). Although most eukaryotes encode only one Pif1 helicase, budding yeast encodes two, Pif1 and Rrm3. Pif1 affects telomeres by inhibiting telomerase at both telomeres and DSBs (Schulz and Zakian 1994; Zhou et al. 2000; Myung et al. 2001). In its absence, telomerase binding is higher, telomerase is more processive, telomeres are longer, and de novo telomere formation at DSBs is sharply increased. Pif1 inhibits telomerase by releasing telomerase from both telomeres and DSBs (Boule et al. 2005). Pif1 probably interacts directly with Est2, the telomerase catalytic subunit, as mutations in the Est2 finger domain result in long telomeres that are Pif1 insensitive (Eugster et al. 2006).
The only other mutations known to increase telomere addition at DSBs in yeast are those that prevent DSB resection, as in sgs1Δ exo1Δ mutant cells (Chung et al. 2010). These data suggest that HR and telomere addition are in competition at DSBs with Pif1 pushing the DSB toward HR. Because de novo telomere addition results in terminal deletions that generate aneuploidy for the affected region, it is better for the cell to repair DSBs by HR, and indeed, telomere addition in budding yeast is repressed by Mec1 (ATR), and this inhibition involves Mec1-mediated Pif1 phosphorylation (Makovets and Blackburn 2009). Pif1 phosphorylation promotes its activity at DSBs but not at telomeres, thus increasing the probability that DSBs are channeled to HR, not telomerase.
Budding yeast Rrm3 also affects telomeres but in a different manner from Pif1. The replication pausing that occurs at telomeres and subtelomeric regions in wild-type cells is increased about 10-fold in the absence of Rrm3 (Ivessa et al. 2002; Azvolinsky et al. 2009). However, Rrm3 promotes replication at many discrete sites throughout the genome, such as tRNA genes and centromeres, not just at telomeres (Ivessa et al. 2003). Likewise, the fission yeast Pif1 protein Pfh1 does not inhibit telomerase (Zhou et al. 2002; Pinter et al. 2008) but rather promotes fork progression at many sites (Sabouri et al. 2012; Steinacher et al. 2012), including telomeres (N Sabouri, in prep.). In the absence of Rrm3 or Pfh1, breaks are detected in telomeric regions, indicating that yeast telomeres are fragile sites when they are under replication stress (Ivessa et al. 2002; N Sabouri, in prep.). The effects of mouse and human PIF1 on telomeres and telomerase have also been examined: Although telomere length is normal in mice lacking mPIF1, mPIF1 and hPIF1 coimmunoprecipitate with TERT (Mateyak and Zakian 2006; Snow et al. 2007), and hPIF1 overexpression results in telomere shortening (Zhang et al. 2006).
Like Pif1 family helicases, the 3′-5′ RecQ helicases are widespread, and their mutation causes genome instability to manifest as replication and recombination defects. Humans encode five RecQ helicases, and mutations in three of these—Bloom’s (BLM), Werner’s (WRN), and RecQ4—cause inherited predisposition to cancer and/or premature aging (reviewed in Paeschke et al. 2010). In addition to their genome-wide functions, many RecQ helicases have telomeric roles.
Although budding and fission yeasts encode two RecQ helicases, only the BLM homologs—Sgs1 in budding yeast and Rqh1 in fission yeast—have been analyzed for telomere functions. Deletion of Sgs1 or Rqh1 has little or no effect on telomere length. However, Sgs1 promotes the telomeric C-strand resection that occurs in late S phase (see section Another Replication Problem: G-Tail Regeneration) and is required to generate type II survivors in telomerase null cells (see section Telomere Recombination in Telomerase Minus Cells) (Azam et al. 2006; Lee et al. 2007). Rqh1 is needed for telomere maintenance in certain genetic backgrounds that compromise telomeres. For example, Rqh1 is required to disentangle telomere recombination intermediates that arise in telomerase-deficient cells (Ukimori et al. 2012).
Mammalian RecQ DNA helicases also affect telomeres, and these effects probably contribute to the disease phenotypes associated with their mutation. For example, mutations in the WRN helicase cause Werner syndrome (WS), a premature aging disease. Reduced life span is recapitulated when cells from WS patients are placed in culture, as WS cells senesce after far fewer population doublings than cells from healthy individuals. Remarkably, this reduced division potential is suppressed simply by expressing TERT in WS cells (Wyllie et al. 2000). Thus, even though WS cells have genome-wide defects, the critical problem that results in life span reduction can be explained by its telomere role(s).
The first hint that WRN acts at telomeres came from Southern blot analysis that showed that telomeric DNA is lost faster from WS telomeres than from controls (Schulz et al. 1996). This finding can be explained by the occurrence of fragile telomeres in WS cells (Crabbe et al. 2004). Suppression of telomere fragility requires the helicase but not the nuclease activity of WRN. By using co-FISH, a method that distinguishes leading and lagging strands, telomere defects occur only during lagging-strand synthesis, which uses the G-rich strand of the telomere as a template. As with replicative senescence, introduction of telomerase suppresses telomere fragility. Given that WRN associates with telomeres during S phase, it likely has a direct role in telomere replication (Opresko et al. 2004). Finally, in vitro studies show that WRN can unwind t-loops (Opresko et al. 2004) and G4 structures (Mohaghegh et al. 2001).
BLM affects telomeres by promoting ALT (Stavropoulos et al. 2002). However, BLM also has telomere functions in normal human fibroblasts: It localizes to telomeres and its shRNA knockdown increases the number of telomere free ends in these cells (Barefield and Karlseder 2012). BLM acts at a late stage in the replication of hard-to-replicate sites, such as centromeres and telomeres, probably by decatenating sister chromatids. BLM coats DNA strands in ultrafine bridges (UFB), structures that connect sister loci at the very end of S phase (Chan et al. 2009). The number of UFBs increases in BLM-deficient cells (Chan et al. 2007) leading to the hypothesis that BLM resolves replication intermediates that are otherwise manifest as fragile sites (Chan et al. 2007; Barefield and Karlseder 2012). In vitro data also support telomeric roles for BLM as its ability to unwind telomeric duplexes and t-loops is stimulated by POT1 (Opresko et al. 2005).
RTEL1 (regulator of telomere length 1) helicases, which are not found in yeasts, were first identified because a gain-of-function allele in mouse RTEL1 has a dominant effect on telomere length (Zhu et al. 1998; Ding et al. 2004). RTEL1 helicases are among a small number of helicases with an iron–sulfur (FeS) domain that is essential for their in vivo functions. Mutations in human versions of known FeS helicases, such as FANCJ, to which RTEL1 is related, cause inherited genome instability (Cantor and Guillemette 2011), and we anticipate that disease-causing alleles of human RTEL1 will be found.
Mice with RTEL1 knockouts die after 10–11.5 d of gestation. Cells derived from RTEL1-deficient embryos have very short, unstable telomeres and high levels of chromosome fusions (Ding et al. 2004). RTEL1 is implicated in the resolution of telomere DNA secondary structures. For example, t-circles accumulate in cycling RTEL1-deficient mouse cells, leading the investigators to suggest that RTEL1 processes t-loops to allow telomere replication through the end of the chromosome (Vannier et al. 2012). RTEL1’s ability to suppress t-circle formation was presaged by the in vitro demonstration that D-loop recombination intermediates are resolved by RTEL1 (Barber et al. 2008). RTEL1 might also unwind G4 telomeric structures, as treatment of RTEL-deficient mouse cells with a compound that stabilizes G4 structures increases telomere fragility (Vannier et al. 2012). Collectively, these results suggest that RTEL1 assists telomere replication by removing t-loops and G4 structures.
ANOTHER REPLICATION PROBLEM: G-TAIL REGENERATION
As first shown in ciliated protozoa (Klobutcher et al. 1981), most eukaryotic chromosomes have G-tails on both telomeres. G-tails are critical for the capping functions of telomeres and are also a better substrate than blunt-ended molecules for telomerase. Nonetheless, G-tails are not universal. About half of the telomeres in angiosperm plants are blunt ended (the other half has extremely short G-tails) (Kazda et al. 2012), whereas half of Caenorhabditis elegans chromosomes end in C-tails (Raices et al. 2008). C-tails are also detected in both dividing and postreplicative mammalian cells (Oganesian and Karlseder 2011).
Even if telomeric G-tails are not ubiquitous, they are widespread, which raises the question of how they are generated. Although G-tails could be telomerase generated, G-tails are not reduced in length or abundance in telomerase null yeast or mice (Wellinger et al. 1996; Hemann and Greider 1999). At the lagging-strand telomere, a short G-tail can arise by incomplete replication. However, yeast and mammalian G-tails are longer than the 8–12 nucleotide G-tails predicted from removal of the terminal RNA primer, and the product of the leading strand polymerase is expected to be a blunt end. Thus, G-tails are not a by-product of conventional DNA replication.
In budding yeast, G-tails are generated by postreplication degradation of the C-rich strand (Wellinger et al. 1996). This degradation is strikingly similar to the 5′ degradation that occurs at DSBs before their activating an ATR/Mec1-mediated checkpoint response and HR. The difference between the two processes lies in the extent of nucleolytic processing: In budding yeast, DSB resection can extend as much as 25 kb from the site of a DSB (Vaze et al. 2002), whereas the maximum length of G-tails is ∼100 nucleotides (Wellinger et al. 1993).
C-strand degradation and 5′-3′ resection of DSBs are both cell-cycle regulated owing to their dependence on the kinase activity of CDK1 (Wellinger et al. 1993; Ira et al. 2004; Frank et al. 2006; Vodenicharov and Wellinger 2006). Moreover, the same enzymes are involved in DSB and telomere processing. The MRX complex and the Sae2 nuclease (homolog of human CtIP) are the first to arrive at DSBs followed by Tel1, which is recruited by MRX (Lisby et al. 2004). These activities initiate DSB resection by removing short stretches of nucleotides from the DSB (Cejka et al. 2010). Exo1 and Sgs1/Dna2 define two downstream pathways that lead to extensive degradation of the 5′-terminated strand to generate the long 3′ overhangs that are required for efficient homology search (Mimitou and Symington 2008; Nimonkar et al. 2011). Similarly, MRX, SAE2, SGS1, and, to a lesser extent, EXO1 all affect telomere C-strand degradation (Larrivee et al. 2004; Bonetti et al. 2010a,b; Longhese et al. 2010). The processes of DSB resection and C-strand degradation are conserved from yeasts to humans. As in budding yeast, the mammalian MRN complex, CtIP, BLM (Sgs1 homolog), and ExoI are all involved in C-strand degradation once telomeres are deprotected by loss of shelterin subunits (Sfeir and de Lange 2012). In mammals, C-strand degradation is controlled at another level such that ∼80% of the C strands terminate in 3′-CCAATC-5′, whereas the last base of the G strand is not precise (Sfeir et al. 2005).
Deprotection of mammalian telomeres by removing shelterin subunits and/or other proteins, such as the Apollo nuclease, leads to fusions that occur preferentially at the leading strand (Deng et al. 2009; Dimitrova and de Lange 2009; Lam et al. 2010; Wu et al. 2010). These results suggest that the structure at the very ends of the two nascent strands is different. In telomerase-negative human cancer cells, lagging-strand G-tails are much longer than those at the leading strand (Zhao et al. 2008), and telomerase expression increases the overhang length only at leading, not lagging, strand ends (Chai et al. 2006). Furthermore, the timing of G-tail maturation appears to be different at the two ends. The lagging strand G tail reaches its final length in early S phase shortly after semiconservative replication of telomeric DNA, whereas the leading-strand end reaches its mature length in late S phase (Zhao et al. 2009; Chow et al. 2012). Thus, in both yeast and normal mammalian cells, G-tails are generated differently at leading- and lagging-strand telomeres.
PREVENTING TELOMERES FROM BEING SEEN AS DSBs
Distinguishing telomeres from DSBs is a major challenge. The solution is not simply one of threshold (i.e., the cell being able to tolerate a certain number of DSBs before triggering a DNA damage checkpoint) because even a single DSB triggers checkpoint arrest in budding yeast (Sandell and Zakian 1993) and mammals (Huang et al. 1996). Distinguishing telomeres from DSBs is critical because events that occur normally at DSBs, including extensive degradation, nonhomologous end-joining (NHEJ)-mediated fusions, and signaling that results in a checkpoint-activated cell-cycle arrest, are disastrous for the cell if they occur at telomeres. Yet telomeres look very much like DSBs, and counterintuitively, many of the proteins that recognize and repair DSBs have telomere functions (Nakamura et al. 2002; Subramanian and Nakamura 2010). In addition, the same enzymes that resect DSBs carry out C-strand degradation, a normal event in telomere metabolism. Thus, the activities of these proteins must be differently regulated at DSBs and telomeres. Two evolutionarily distinct telomere complexes, CST and shelterin, impose these differences.
Protecting Telomeres from Extensive Resection
At DSBs, extensive DNA degradation results in a long stretch of 3′-ssDNA that is coated with replication protein A (RPA), the generic single-strand-binding (SSB) protein that binds ssDNA with minimal sequence specificity. RPA binding to ssDNA further enhances nucleolytic degradation (Cejka et al. 2010; Nimonkar et al. 2011) and also recruits the checkpoint kinase ATR/Mec1, which sets into motion a checkpoint-mediated cell-cycle response. At budding yeast telomeres, C-strand degradation generates ssTG1–3 telomeric DNA, which is bound by CST (Taggart et al. 2002), which unlike RPA, does not recruit Mec1 (McGee et al. 2010; Hector et al. 2012). Budding yeast CST limits C-strand degradation as loss of any of the three CST subunits results in rapid and extensive C-strand resection, activation of the checkpoint response, and S/G2 arrest (Garvik et al. 1995; Grandin et al. 1997, 2001b; Vodenicharov and Wellinger 2006; Paschini et al. 2012).
Deletion of other budding yeast telomere-binding proteins is less catastrophic. For example, the Ku complex protects telomeres from Exo1 degradation (Bertuch and Lundblad 2004), especially in G1 phase (Bonetti et al. 2010a). As a result, ykuΔ cells have long G-tails throughout the cell cycle (Boulton and Jackson 1996; Gravel et al. 1998; Polotnianka et al. 1998). Nonetheless, telomere function is not lost in ykuΔ cells. Likewise, Rif2 and to a lesser extent Rif1, which associate with telomere-bound Rap1 in the duplex portion of telomeres, inhibit MRX/Sae2/Sgs1-mediated degradation, yet telomere capping is still maintained in cells lacking either protein (Negrini et al. 2007; Bonetti et al. 2009, 2010a,b; Ribeyre and Shore 2012).
In fission yeast, the shelterin-like complex prevents telomere degradation. Removal of Taz1 or its binding partner Rap1 results in accumulation of long G overhangs but not cell-cycle arrest (Miller et al. 2005). However, loss of Pot1 results in telomere loss within a single cell cycle (Baumann and Cech 2001; Pitt and Cooper 2010). Stn1 and Ten1 also prevent rapid telomere loss and end-to-end fusions (Martin et al. 2007).
In mammals, multiple proteins contribute in a redundant manner to prevent telomere degradation. Even when mouse telomeres are completely stripped of shelterin and Ku, dramatic telomere degradation is not observed unless 53BP1, a DNA damage response and checkpoint mediator protein, is also depleted (Sfeir and de Lange 2012). hPOT1 and mPOT1b both suppress telomere resection (Hockemeyer et al. 2006; He et al. 2009; Palm et al. 2009), whereas CST does not play a major role in telomere protection (Miyake et al. 2009).
Hiding Telomeres from Checkpoints
The two major DNA damage checkpoint pathways, ATR/Mec1 and ATM/Tel1, are both important in telomere maintenance. In budding yeast, Tel1 kinase activity plays a major role in recruiting telomerase to short telomeres (Chang et al. 2007; Hector et al. 2007; Sabourin et al. 2007), whereas the role of Mec1 is minor and only clearly evident in the absence of TEL1 (Ritchie et al. 1999). The roles of ATR and ATM in telomere regulation in fission yeast are reversed, with the ATR homolog Rad3 playing a major role and the ATM homolog Tel1 a minor role (Naito et al. 1998; Nakamura et al. 2002; Moser et al. 2009b). Human telomeres from patients with ataxia telangiectasia, a disease attributable to mutations in ATM, shorten faster than ATM+ telomeres (Metcalfe et al. 1996). However, elongation of short telomeres in mouse is not dependent on ATM (Feldser et al. 2006) or ATR (Verdun and Karlseder 2006; McNees et al. 2010).
At processed DSB breaks and stalled replication forks, RPA-coated ssDNA activates the ATR/Mec1 checkpoint response (Zou and Elledge 2003). At telomeres, resection is much more limited than at DSBs, and the exposed ssDNA is bound by telomeric DNA sequence-specific SSBs that exclude RPA. Removal of either Cdc13 or POT1 leads to rapid Mec1/ATR checkpoint activation and permanent cell-cycle arrest (Garvik et al. 1995; Denchi and de Lange 2007; Guo et al. 2007; Hirano and Sugimoto 2007; Gong and de Lange 2010). In budding yeast, a single short telomere, which binds high levels of Tel1, does not cause cell-cycle arrest (Sabourin et al. 2007) but does result in Rad53 phosphorylation (Baldo et al. 2008) suggesting that early steps in checkpoint activation, but not later steps, occur. In telomerase-minus primary human fibroblasts, ATR and ATM are both recruited to telomeres around the time of telomere replication (Verdun and Karlseder 2006). Because telomeres do not elicit a checkpoint-mediated cell-cycle arrest, there must be mechanisms that allow ATR/ATM telomere binding without triggering their kinase activity, and/or activation of their downstream effectors must be attenuated.
Given that RPA is much more abundant than telomere ss-binding proteins, it is puzzling that telomeric G-tails do not bind RPA, which would elicit a checkpoint response. For example, in budding yeast, there is 10 times more RPA than Cdc13 (Ghaemmaghami et al. 2003; Wu and Zakian 2011) and about three orders of magnitude more RPA than POT1 in mammalian cells (Takai et al. 2011). Remarkably, the affinity of mammalian RPA for telomeric ssDNA in vitro is comparable to that of POT1-TPP1 (Flynn et al. 2011; Takai et al. 2011). Thus, mechanisms other than sequence-specific binding are needed to exclude RPA from telomeres and prevent their activating a checkpoint. In budding yeast, the Rap1-interacting protein Rif1 possesses “anticheckpoint” activity. Rif1 limits RPA binding to deprotected telomeres generated in cdc13–1 cells grown at nonpermissive temperatures. It also inhibits RPA binding to a DSB made adjacent to a short stretch of telomeric DNA without affecting accumulation of ssDNA at the break (Xue et al. 2011; Ribeyre and Shore 2012).
In mammals, two mechanisms aid POT1-TPP1 exclusion of RPA from G-tails. First, hnRNPA1 (heterogeneous nuclear ribonucleoprotein A1), an abundant nuclear protein that binds to both hnRNA and to telomeric ssDNA with high affinity (McKay and Cooke 1992; Ding et al. 1999), can efficiently displace RPA, but not POT1, from telomeric ssDNA (Flynn et al. 2011). TERRA binds hnRNPA1 and thereby prevents it from binding telomeric ssDNA. TERRA levels are high in early G1 and low in late S (Porro et al. 2010). Thus, during early S phase, when RPA is required for semiconservative replication, TERRA levels are high enough to sequester hnRNPA1. During late S phase after telomere replication, TERRA levels are low freeing hnRNPA1 to bind telomeric ssDNA to facilitate POT1 assembly on telomeres. The second mechanism involves POT1 tethering to the shelterin complex through an interaction between TIN2 and TPP1 (Takai et al. 2011). This tethering mechanism is probably the more critical one for keeping POT1 telomere bound, as the TPP1-binding domain, but not the DNA-binding domain of POT1 is required for POT1 telomere localization and protection (Xin et al. 2007).
Even when RPA accumulates at telomeres and ATR is activated, the cell has still another mechanism to prevent an ATR checkpoint response. In fission yeast, for example, RPA (Rad11) and ATR (Rad3) both accumulate at deprotected telomeres in taz1Δ cells (Carneiro et al. 2010), and telomeres are quickly lengthened by telomerase in a Rad3-dependent manner (Nakamura et al. 2002). Despite this Rad3 activation, its downstream effector Chk1 is not activated and cell-cycle progression is not perturbed. Fission yeast Pot1 and, to a lesser extent, Ccq1 suppress the Rad3 checkpoint pathway (Miyoshi et al. 2008; Tomita and Cooper 2008) by inhibiting telomere association of the Set9 methylase, which modifies lysine 20 of histone H4. The lack of H4K20me2 modification in telomeric chromatin prevents stable recruitment of the 53BP1 homolog, Crb2, which functions upstream of Chk1 (Carneiro et al. 2010). Similarly, the human POT1 and mouse POT1a suppress the ATR checkpoint (Denchi and de Lange 2007; Guo et al. 2007).
Inhibition of Standard and Alternative NHEJ
In budding yeast Rap1, Rif2, and Sir4 prevent formation of NHEJ-mediated telomere fusions (Marcand et al. 2008). Similarly, fission yeast Rap1 cooperates with its binding partner Taz1 to inhibit these events (Miller et al. 2005). TRF2 plays the major role in suppressing NHEJ at mammalian telomeres, perhaps because of its ability to promote t-1oop formation (Griffith et al. 1999; Stansel et al. 2001), as t-loops are thought to sequester telomeres from NHEJ. In addition, the TRF2-interacting protein SNM1B/Apollo, a 5′ exonuclease, processes the end of newly synthesized leading-strand telomeres, preventing fusions between telomere sister chromatids (Lam et al. 2010; Wu et al. 2010). TRF2 also inhibits ATM activation at least in part via SNM1B/Apollo interaction (Lam et al. 2010; Wu et al. 2010), a prerequisite for efficient NHEJ (Denchi and de Lange 2007). The ATM target MDC1 promotes telomere fusions in TRF2−/− mouse cells by recruiting 53BP1 to deprotected telomeres (Dimitrova and de Lange 2006). 53BP1 enhances the local mobility of dysfunctional telomeres, which increases their chances of finding another telomere with which to fuse (Dimitrova et al. 2008). The nuclease activity of Mre11 also affects NHEJ activation at a step downstream from ATM. Because the fusion events in TRF2−/− Mre11−/− cells primarily involve fusion of two leading-strand telomeres, Mre11 likely contributes to leading-strand telomere processing, which generates unsuitable substrates for NHEJ (Deng et al. 2009).
Even though Ku is essential for NHEJ and promotes end-to-end fusion of deprotected telomeres, paradoxically Ku is telomere associated in many organisms (Gravel et al. 1998; Hsu et al. 1999, 2000; Nakamura et al. 2002; Fisher et al. 2004). Ku promotes telomere maintenance in several ways. In budding yeast, Ku functions in telomerase recruitment (Fisher et al. 2004; Chan et al. 2008) and telomere protection (Bertuch and Lundblad 2004; Bonetti et al. 2010a). In mammals, Ku coordinates with shelterin to block telomere access to alt-NHEJ pathway, a PARP1 and ligase 3-dependent pathway (Wang et al. 2009; Sfeir and de Lange 2012).
FINAL THOUGHTS
Telomere maintenance is essential for chromosome integrity yet it presents multiple challenges. Because the very ends of chromosomes cannot be replicated by the standard replication machinery, specialized repairlike processes have evolved to compensate for the loss of DNA arising from incomplete replication, such as telomerase, a telomere-dedicated RT; recombination, especially in telomerase-deficient cells; and retrotransposition, so far documented in only a small number of organisms, and even more baroque mechanisms, for example, PAL. Budding yeast uses all of these mechanisms, depending on the specific situation or genetic context, and we would not be surprised if a similar diversity of end maintenance activities is found in human cancers.
ACKNOWLEDGMENTS
We thank Carolyn Price for her thoughtful comments on the manuscript and the National Institutes of Health for support.
Footnotes
Editors: Errol C. Friedberg, Stephen J. Elledge, Alan R. Lehmann, Tomas Lindahl, and Marco Muzi-Falconi
Additional Perspectives on DNA Repair, Mutagenesis, and Other Responses to DNA Damage available at www.cshperspectives.org
REFERENCES
- Abad JP, Villasante A 1999. The 3′ non-coding region of the Drosophila melanogaster HeT-A telomeric retrotransposon contains sequences with propensity to form G-quadruplex DNA. FEBS Lett 453: 59–62 [DOI] [PubMed] [Google Scholar]
- Armanios M 2012. An emerging role for the conserved telomere component 1 (CTC1) in human genetic disease. Pediatr Blood Cancer 59: 209–210 [DOI] [PubMed] [Google Scholar]
- Arneric M, Lingner J 2007. Tel1 kinase and subtelomere-bound Tbf1 mediate preferential elongation of short telomeres by telomerase in yeast. EMBO Rep 8: 1080–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoult N, Van Beneden A, Decottignies A 2012. Telomere length regulates TERRA levels through increased trimethylation of telomeric H3K9 and HP1α. Nat Struct Mol Biol 19: 948–956 [DOI] [PubMed] [Google Scholar]
- Azam M, Lee JY, Abraham V, Chanoux R, Schoenly KA, Johnson FB 2006. Evidence that the S. cerevisiae Sgs1 protein facilitates recombinational repair of telomeres during senescence. Nucleic Acids Res 34: 506–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azvolinsky A, Giresi PG, Lieb JD, Zakian VA 2009. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol Cell 34: 722–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J 2007. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318: 798–801 [DOI] [PubMed] [Google Scholar]
- Badie S, Escandell JM, Bouwman P, Carlos AR, Thanasoula M, Gallardo MM, Suram A, Jaco I, Benitez J, Herbig U, et al. 2010. BRCA2 acts as a RAD51 loader to facilitate telomere replication and capping. Nat Struct Mol Biol 17: 1461–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldo V, Testoni V, Lucchini G, Longhese MP 2008. Dominant TEL1-hy mutations compensate for Mec1 lack of functions in the DNA damage response. Mol Cell Biol 28: 358–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber LJ, Youds JL, Ward JD, McIlwraith MJ, O’Neil NJ, Petalcorin MI, Martin JS, Collis SJ, Cantor SB, Auclair M, et al. 2008. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell 135: 261–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barefield C, Karlseder J 2012. The BLM helicase contributes to telomere maintenance through processing of late-replicating intermediate structures. Nucleic Acids Res 40: 7358–7367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P, Cech TR 2001. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292: 1171–1175 [DOI] [PubMed] [Google Scholar]
- Baur JA, Zou Y, Shay JW, Wright WE 2001. Telomere position effect in human cells. Science 292: 2075–2077 [DOI] [PubMed] [Google Scholar]
- Beernink HT, Miller K, Deshpande A, Bucher P, Cooper JP 2003. Telomere maintenance in fission yeast requires an Est1 ortholog. Curr Biol 13: 575–580 [DOI] [PubMed] [Google Scholar]
- Bertuch AA, Lundblad V 2004. EXO1 contributes to telomere maintenance in both telomerase-proficient and telomerase-deficient Saccharomyces cerevisiae. Genetics 166: 1651–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A, Shore D 2007. Increased association of telomerase with short telomeres in yeast. Genes Dev 21: 1726–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW 1997. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91: 25–34 [DOI] [PubMed] [Google Scholar]
- Bochman ML, Judge CP, Zakian VA 2011. The Pif1 family in prokaryotes: What are our helicases doing in your bacteria? Mol Biol Cell 22: 1955–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman ML, Paeschke K, Zakian VA 2012. DNA secondary structures: Stability and function of G-quadruplex structures. Nat Rev Genet 13: 770–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetti D, Martina M, Clerici M, Lucchini G, Longhese MP 2009. Multiple pathways regulate 3′ overhang generation at S. cerevisiae telomeres. Mol Cell 35: 70–81 [DOI] [PubMed] [Google Scholar]
- Bonetti D, Clerici M, Anbalagan S, Martina M, Lucchini G, Longhese MP 2010a. Shelterin-like proteins and Yku inhibit nucleolytic processing of Saccharomyces cerevisiae telomeres. PLoS Genet 6: e1000966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetti D, Clerici M, Manfrini N, Lucchini G, Longhese MP 2010b. The MRX complex plays multiple functions in resection of Yku- and Rif2-protected DNA ends. PLoS ONE 5: e14142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boule JB, Vega LR, Zakian VA 2005. The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature 438: 57–61 [DOI] [PubMed] [Google Scholar]
- Boulton SJ, Jackson SP 1996. Identification of a Saccharomyces cerevisiae Ku80 homologue: Roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res 24: 4639–4648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broccoli D, Smogorzewska A, Chong L, de Lange T 1997. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat Genet 17: 231–235 [DOI] [PubMed] [Google Scholar]
- Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR 1997. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med 3: 1271–1274 [DOI] [PubMed] [Google Scholar]
- Cantor SB, Guillemette S 2011. Hereditary breast cancer and the BRCA1-associated FANCJ/BACH1/BRIP1. Future Oncol 7: 253–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro T, Khair L, Reis CC, Borges V, Moser BA, Nakamura TM, Ferreira MG 2010. Telomeres avoid end detection by severing the checkpoint signal transduction pathway. Nature 467: 228–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, Kowalczykowski SC 2010. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature 467: 112–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli GB, Denchi EL, de Lange T 2006. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat Cell Biol 8: 885–890 [DOI] [PubMed] [Google Scholar]
- Cenci G, Siriaco G, Raffa GD, Kellum R, Gatti M 2003. The Drosophila HOAP protein is required for telomere capping. Nat Cell Biol 5: 82–84 [DOI] [PubMed] [Google Scholar]
- Cesare AJ, Griffith JD 2004. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol Cell Biol 24: 9948–9957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai W, Du Q, Shay JW, Wright WE 2006. Human telomeres have different overhang sizes at leading versus lagging strands. Mol Cell 21: 427–435 [DOI] [PubMed] [Google Scholar]
- Chan KL, North PS, Hickson ID 2007. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J 26: 3397–3409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A, Boule JB, Zakian VA 2008. Two pathways recruit telomerase to Saccharomyces cerevisiae telomeres. PLoS Genet 4: e1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, Palmai-Pallag T, Ying S, Hickson ID 2009. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol 11: 753–760 [DOI] [PubMed] [Google Scholar]
- Chang M, Arneric M, Lingner J 2007. Telomerase repeat addition processivity is increased at critically short telomeres in a Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev 21: 2485–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla R, Redon S, Raftopoulou C, Wischnewski H, Gagos S, Azzalin CM 2011. Human UPF1 interacts with TPP1 and telomerase and sustains telomere leading-strand replication. EMBO J 30: 4047–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LY, Redon S, Lingner J 2012. The human CST complex is a terminator of telomerase activity. Nature 488: 540. [DOI] [PubMed] [Google Scholar]
- Chisholm KM, Aubert SD, Freese KP, Zakian VA, King MC, Welcsh PL 2012. A genomewide screen for suppressors of Alu-mediated rearrangements reveals a role for PIF1. PLoS ONE 7: e30748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong L, van Steensel B, Broccoli D, Erdjument-Bromage H, Hanish J, Tempst P, de Lange T 1995. A human telomeric protein. Science 270: 1663–1667 [DOI] [PubMed] [Google Scholar]
- Chow TT, Zhao Y, Mak SS, Shay JW, Wright WE 2012. Early and late steps in telomere overhang processing in normal human cells: The position of the final RNA primer drives telomere shortening. Genes Dev 26: 1167–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WH, Zhu Z, Papusha A, Malkova A, Ira G 2010. Defective resection at DNA double-strand breaks leads to de novo telomere formation and enhances gene targeting. PLoS Genet 6: e1000948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad MN, Wright JH, Wolf AJ, Zakian VA 1990. RAP1 protein interacts with yeast telomeres in vivo: Overproduction alters telomere structure and decreases chromosome stability. Cell 63: 739–750 [DOI] [PubMed] [Google Scholar]
- Cooper JP, Nimmo ER, Allshire RC, Cech TR 1997. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 385: 744–747 [DOI] [PubMed] [Google Scholar]
- Costa A, Daidone MG, Daprai L, Villa R, Cantu S, Pilotti S, Mariani L, Gronchi A, Henson JD, Reddel RR, et al. 2006. Telomere maintenance mechanisms in liposarcomas: Association with histologic subtypes and disease progression. Cancer Res 66: 8918–8924 [DOI] [PubMed] [Google Scholar]
- Crabbe L, Verdun RE, Haggblom CI, Karlseder J 2004. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science 306: 1951–1953 [DOI] [PubMed] [Google Scholar]
- Cristofari G, Lingner J 2006. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J 25: 565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehe PM, Rog O, Ferreira MG, Greenwood J, Cooper JP 2012. Taz1 enforces cell-cycle regulation of telomere synthesis. Mol Cell 46: 797–808 [DOI] [PubMed] [Google Scholar]
- de Lange T 2005. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev 19: 2100–2110 [DOI] [PubMed] [Google Scholar]
- Denchi EL, de Lange T 2007. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 448: 1068–1071 [DOI] [PubMed] [Google Scholar]
- Deng Y, Guo X, Ferguson DO, Chang S 2009. Multiple roles for MRE11 at uncapped telomeres. Nature 460: 914–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZwaan DC, Freeman BC 2009. The conserved Est1 protein stimulates telomerase DNA extension activity. Proc Natl Acad Sci 106: 17337–17342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diede SJ, Gottschling DE 1999. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases α and δ. Cell 99: 723–733 [DOI] [PubMed] [Google Scholar]
- Dimitrova N, de Lange T 2006. MDC1 accelerates nonhomologous end-joining of dysfunctional telomeres. Genes Dev 20: 3238–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, de Lange T 2009. Cell cycle-dependent role of MRN at dysfunctional telomeres: ATM signaling-dependent induction of nonhomologous end joining (NHEJ) in G1 and resection-mediated inhibition of NHEJ in G2. Mol Cell Biol 29: 5552–5563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, Chen YC, Spector DL, de Lange T 2008. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 456: 524–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Hayashi MK, Zhang Y, Manche L, Krainer AR, Xu RM 1999. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev 13: 1102–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, Schertzer M, Wu X, Gertsenstein M, Selig S, Kammori M, Pourvali R, Poon S, Vulto I, Chavez E, et al. 2004. Regulation of murine telomere length by Rtel: An essential gene encoding a helicase-like protein. Cell 117: 873–886 [DOI] [PubMed] [Google Scholar]
- Dunham MA, Neumann AA, Fasching CL, Reddel RR 2000. Telomere maintenance by recombination in human cells. Nat Genet 26: 447–450 [DOI] [PubMed] [Google Scholar]
- Eugster A, Lanzuolo C, Bonneton M, Luciano P, Pollice A, Pulitzer JF, Stegberg E, Berthiau AS, Forstemann K, Corda Y, et al. 2006. The finger subdomain of yeast telomerase cooperates with Pif1p to limit telomere elongation. Nat Struct Mol Biol 13: 734–739 [DOI] [PubMed] [Google Scholar]
- Evans SK, Lundblad V 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286: 117–120 [DOI] [PubMed] [Google Scholar]
- Fanti L, Giovinazzo G, Berloco M, Pimpinelli S 1998. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol Cell 2: 527–538 [DOI] [PubMed] [Google Scholar]
- Feldser D, Strong MA, Greider CW 2006. Ataxia telangiectasia mutated (Atm) is not required for telomerase-mediated elongation of short telomeres. Proc Natl Acad Sci 103: 2249–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MG, Cooper JP 2001. The fission yeast Taz1 protein protects chromosomes from Ku-dependent end-to-end fusions. Mol Cell 7: 55–63 [DOI] [PubMed] [Google Scholar]
- Fisher TS, Taggart AK, Zakian VA 2004. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat Struct Mol Biol 11: 1198–1205 [DOI] [PubMed] [Google Scholar]
- Flynn RL, Centore RC, O’Sullivan RJ, Rai R, Tse A, Songyang Z, Chang S, Karlseder J, Zou L 2011. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature 471: 532–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourel G, Revardel E, Koering CE, Gilson E 1999. Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J 18: 2522–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank CJ, Hyde M, Greider CW 2006. Regulation of telomere elongation by the cyclin-dependent kinase CDK1. Mol Cell 24: 423–432 [DOI] [PubMed] [Google Scholar]
- Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E 2005. SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell 17: 537–547 [DOI] [PubMed] [Google Scholar]
- Fukunaga K, Hirano Y, Sugimoto K 2012. Subtelomere-binding protein Tbf1 and telomere-binding protein Rap1 collaborate to inhibit localization of the Mre11 complex to DNA ends in budding yeast. Mol Biol Cell 23: 347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo F, Laterreur N, Cusanelli E, Ouenzar F, Querido E, Wellinger RJ, Chartrand P 2011. Live cell imaging of telomerase RNA dynamics reveals cell cycle-dependent clustering of telomerase at elongating telomeres. Mol Cell 44: 819–827 [DOI] [PubMed] [Google Scholar]
- Gao G, Walser JC, Beaucher ML, Morciano P, Wesolowska N, Chen J, Rong YS 2010a. HipHop interacts with HOAP and HP1 to protect Drosophila telomeres in a sequence-independent manner. EMBO J 29: 819–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Toro TB, Paschini M, Braunstein-Ballew B, Cervantes RB, Lundblad V 2010b. Telomerase recruitment in Saccharomyces cerevisiae is not dependent on Tel1-mediated phosphorylation of Cdc13. Genetics 186: 1147–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvik B, Carson M, Hartwell L 1995. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol 15: 6128–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS 2003. Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Gong Y, de Lange T 2010. A Shld1-controlled POT1a provides support for repression of ATR signaling at telomeres through RPA exclusion. Mol Cell 40: 377–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling DE, Zakian VA 1986. Telomere proteins: Specific recognition and protection of the natural termini of Oxytricha macronuclear DNA. Cell 47: 195–205 [DOI] [PubMed] [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA 1990. Position effect at S. cerevisiae telomeres: Reversible repression of Pol II transcription. Cell 63: 751–762 [DOI] [PubMed] [Google Scholar]
- Grandin N, Reed SI, Charbonneau M 1997. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev 11: 512–527 [DOI] [PubMed] [Google Scholar]
- Grandin N, Damon C, Charbonneau M 2001a. Cdc13 prevents telomere uncapping and Rad50-dependent homologous recombination. EMBO J 20: 6127–6139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandin N, Damon C, Charbonneau M 2001b. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J 20: 1173–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Larrivee M, Labrecque P, Wellinger RJ 1998. Yeast Ku as a regulator of chromosomal DNA end structure. Science 280: 741–744 [DOI] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T 1999. Mammalian telomeres end in a large duplex loop. Cell 97: 503–514 [DOI] [PubMed] [Google Scholar]
- Grossi S, Puglisi A, Dmitriev PV, Lopes M, Shore D 2004. Pol12, the B subunit of DNA polymerase α, functions in both telomere capping and length regulation. Genes Dev 18: 992–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu P, Min JN, Wang Y, Huang C, Peng T, Chai W, Chang S 2012. CTC1 deletion results in defective telomere replication, leading to catastrophic telomere loss and stem cell exhaustion. EMBO J 31: 2309–2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Deng Y, Lin Y, Cosme-Blanco W, Chan S, He H, Yuan G, Brown EJ, Chang S 2007. Dysfunctional telomeres activate an ATM-ATR-dependent DNA damage response to suppress tumorigenesis. EMBO J 26: 4709–4719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Wang Y, Guo X, Ramchandani S, Ma J, Shen MF, Garcia DA, Deng Y, Multani AS, You MJ, et al. 2009. Pot1b deletion and telomerase haploinsufficiency in mice initiate an ATR-dependent DNA damage response and elicit phenotypes resembling dyskeratosis congenita. Mol Cell Biol 29: 229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hector RE, Shtofman RL, Ray A, Chen BR, Nyun T, Berkner KL, Runge KW 2007. Tel1p preferentially associates with short telomeres to stimulate their elongation. Mol Cell 27: 851–858 [DOI] [PubMed] [Google Scholar]
- Hector RE, Ray A, Chen BR, Shtofman R, Berkner KL, Runge KW 2012. Mec1p associates with functionally compromised telomeres. Chromosoma 121: 277–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann MT, Greider CW 1999. G-strand overhangs on telomeres in telomerase-deficient mouse cells. Nucleic Acids Res 27: 3964–3969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann MT, Strong MA, Hao LY, Greider CW 2001. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 107: 67–77 [DOI] [PubMed] [Google Scholar]
- Hirano Y, Sugimoto K 2007. Cdc13 telomere capping decreases Mec1 association but does not affect Tel1 association with DNA ends. Mol Biol Cell 18: 2026–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano Y, Fukunaga K, Sugimoto K 2009. Rif1 and rif2 inhibit localization of tel1 to DNA ends. Mol Cell 33: 312–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Daniels JP, Takai H, de Lange T 2006. Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell 126: 63–77 [DOI] [PubMed] [Google Scholar]
- Hoffmeyer K, Raggioli A, Rudloff S, Anton R, Hierholzer A, Del Valle I, Hein K, Vogt R, Kemler R 2012. Wnt/β-catenin signaling regulates telomerase in stem cells and cancer cells. Science 336: 1549–1554 [DOI] [PubMed] [Google Scholar]
- Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC 1998. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell 95: 963–974 [DOI] [PubMed] [Google Scholar]
- Hsu HL, Gilley D, Blackburn EH, Chen DJ 1999. Ku is associated with the telomere in mammals. Proc Natl Acad Sci 96: 12454–12458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HL, Gilley D, Galande SA, Hande MP, Allen B, Kim SH, Li GC, Campisi J, Kohwi-Shigematsu T, Chen DJ 2000. Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes Dev 14: 2807–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LC, Clarkin KC, Wahl GM 1996. Sensitivity and selectivity of the DNA damage sensor responsible for activating p53-dependent G1 arrest. Proc Natl Acad Sci 93: 4827–4832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Weilbaecher RG, Walterscheid M, Lundblad V 2000. Identification of the single-strand telomeric DNA binding domain of the Saccharomyces cerevisiae Cdc13 protein. Proc Natl Acad Sci 97: 6457–6462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, et al. 2004. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431: 1011–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa AS, Zhou JQ, Schulz VP, Monson EK, Zakian VA 2002. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev 16: 1383–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA 2003. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol Cell 12: 1525–1536 [DOI] [PubMed] [Google Scholar]
- Jaco I, Munoz P, Goytisolo F, Wesoly J, Bailey S, Taccioli G, Blasco MA 2003. Role of mammalian Rad54 in telomere length maintenance. Mol Cell Biol 23: 5572–5580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady BE, Richard P, Bertrand E, Kiss T 2006. Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol Biol Cell 17: 944–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain D, Hebden AK, Nakamura TM, Miller KM, Cooper JP 2010. HAATI survivors replace canonical telomeres with blocks of generic heterochromatin. Nature 467: 223–227 [DOI] [PubMed] [Google Scholar]
- Johnson JE, Gettings EJ, Schwalm J, Pei J, Testa JR, Litwin S, von Mehren M, Broccoli D 2007. Whole-genome profiling in liposarcomas reveals genetic alterations common to specific telomere maintenance mechanisms. Cancer Res 67: 9221–9228 [DOI] [PubMed] [Google Scholar]
- Kazda A, Zellinger B, Rossler M, Derboven E, Kusenda B, Riha K 2012. Chromosome end protection by blunt-ended telomeres. Genes Dev 26: 1703–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller RB, Gagne KE, Usmani GN, Asdourian GK, Williams DA, Hofmann I, Agarwal S 2012. CTC1 mutations in a patient with dyskeratosis congenita. Pediatr Blood Cancer 59: 311–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klobutcher LA, Swanton MT, Donini P, Prescott DM 1981. All gene-sized DNA molecules in four species of hypotrichs have the same terminal sequence and an unusual 3′ terminus. Proc Natl Acad Sci 78: 3015–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YC, Akhter S, Gu P, Ye J, Poulet A, Giraud-Panis MJ, Bailey SM, Gilson E, Legerski RJ, Chang S 2010. SNMIB/Apollo protects leading-strand telomeres against NHEJ-mediated repair. EMBO J 29: 2230–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrivee M, LeBel C, Wellinger RJ 2004. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev 18: 1391–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Kozak M, Martin JD, Pennock E, Johnson FB 2007. Evidence that a RecQ helicase slows senescence by resolving recombining telomeres. PLoS Biol 5: e160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Mandell EK, Tucey TM, Morris DK, Lundblad V 2008a. The Est3 protein associates with yeast telomerase through an OB-fold domain. Nat Struct Mol Biol 15: 990–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Mogen JL, Chavez A, Johnson FB 2008b. Sgs1 RecQ helicase inhibits survival of Saccharomyces cerevisiae cells lacking telomerase and homologous recombination. J Biol Chem 283: 29847–29858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Podell ER, Baumann P, Cech TR 2003. DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature 426: 198–203 [DOI] [PubMed] [Google Scholar]
- Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V 1996. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 144: 1399–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis RW 1989. Viable deletions of a telomere from a Drosophila chromosome. Cell 58: 791–801 [DOI] [PubMed] [Google Scholar]
- Levis RW, Ganesan R, Houtchens K, Tolar LA, Sheen FM 1993. Transposons in place of telomeric repeats at a Drosophila telomere. Cell 75: 1083–1093 [DOI] [PubMed] [Google Scholar]
- Li B, Lustig AJ 1996. A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes Dev 10: 1310–1326 [DOI] [PubMed] [Google Scholar]
- Li S, Makovets S, Matsuguchi T, Blethrow JD, Shokat KM, Blackburn EH 2009. Cdk1-Dependent phosphorylation of Cdc13 coordinates telomere elongation during cell-cycle progression. Cell 136: 50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb JD, Liu X, Botstein D, Brown PO 2001. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet 28: 327–334 [DOI] [PubMed] [Google Scholar]
- Lin SY, Elledge SJ 2003. Multiple tumor suppressor pathways negatively regulate telomerase. Cell 113: 881–889 [DOI] [PubMed] [Google Scholar]
- Lin JJ, Zakian VA 1996. The Saccharomyces CDC13 protein is a single-strand TG1–3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc Natl Acad Sci 93: 13760–13765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Cooper JP, Cech TR 1995. Telomerase and DNA end replication: No longer a lagging strand problem? Science 269: 1533–1534 [DOI] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R 2004. Choreography of the DNA damage response: Spatiotemporal relationships among checkpoint and repair proteins. Cell 118: 699–713 [DOI] [PubMed] [Google Scholar]
- Livengood AJ, Zaug AJ, Cech TR 2002. Essential regions of Saccharomyces cerevisiae telomerase RNA: Separate elements for Est1p and Est2p interaction. Mol Cell Biol 22: 2366–2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- London TB, Barber LJ, Mosedale G, Kelly GP, Balasubramanian S, Hickson ID, Boulton SJ, Hiom K 2008. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J Biol Chem 283: 36132–36139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londono-Vallejo JA, Wellinger RJ 2012. Telomeres and telomerase dance to the rhythm of the cell cycle. Trends Biochem Sci 37: 391–399 [DOI] [PubMed] [Google Scholar]
- Longhese MP, Bonetti D, Manfrini N, Clerici M 2010. Mechanisms and regulation of DNA end resection. EMBO J 29: 2864–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes J, Piazza A, Bermejo R, Kriegsman B, Colosio A, Teulade-Fichou MP, Foiani M, Nicolas A 2011. G-quadruplex-induced instability during leading-strand replication. EMBO J 30: 4033–4046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J 2008. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell 32: 465–477 [DOI] [PubMed] [Google Scholar]
- Lundblad V, Blackburn EH 1993. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell 73: 347–360 [DOI] [PubMed] [Google Scholar]
- Lundblad V, Szostak JW 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57: 633–643 [DOI] [PubMed] [Google Scholar]
- Lustig AJ, Kurtz S, Shore D 1990. Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science 250: 549–553 [DOI] [PubMed] [Google Scholar]
- Makarov VL, Hirose Y, Langmore JP 1997. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell 88: 657–666 [DOI] [PubMed] [Google Scholar]
- Makovets S, Blackburn EH 2009. DNA damage signalling prevents deleterious telomere addition at DNA breaks. Nat Cell Biol 11: 1383–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S, Brevet V, Mann C, Gilson E 2000. Cell cycle restriction of telomere elongation. Curr Biol 10: 487–490 [DOI] [PubMed] [Google Scholar]
- Marcand S, Pardo B, Gratias A, Cahun S, Callebaut I 2008. Multiple pathways inhibit NHEJ at telomeres. Genes Dev 22: 1153–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maringele L, Lydall D 2004. Telomerase- and recombination-independent immortalization of budding yeast. Genes Dev 18: 2663–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin V, Du LL, Rozenzhak S, Russell P 2007. Protection of telomeres by a conserved Stn1-Ten1 complex. Proc Natl Acad Sci 104: 14038–14043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez P, Thanasoula M, Munoz P, Liao C, Tejera A, McNees C, Flores JM, Fernandez-Capetillo O, Tarsounas M, Blasco MA 2009. Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev 23: 2060–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez P, Thanasoula M, Carlos AR, Gomez-Lopez G, Tejera AM, Schoeftner S, Dominguez O, Pisano DG, Tarsounas M, Blasco MA 2010. Mammalian Rap1 controls telomere function and gene expression through binding to telomeric and extratelomeric sites. Nat Cell Biol 12: 768–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JM, Frydrychova RC, Biessmann H 2008. Drosophila telomeres: An exception providing new insights. Bioessays 30: 25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateyak MK, Zakian VA 2006. Human PIF helicase is cell cycle regulated and associates with telomerase. Cell Cycle 5: 2796–2804 [DOI] [PubMed] [Google Scholar]
- Maxwell PH, Coombes C, Kenny AE, Lawler JF, Boeke JD, Curcio MJ 2004. Ty1 mobilizes subtelomeric Y′ elements in telomerase-negative Saccharomyces cerevisiae survivors. Mol Cell Biol 24: 9887–9898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern MJ, Krauskopf A, Blackburn EH 2000. Telomeres and their control. Annu Rev Genet 34: 331–358 [DOI] [PubMed] [Google Scholar]
- McGee JS, Phillips JA, Chan A, Sabourin M, Paeschke K, Zakian VA 2010. Reduced Rif2 and lack of Mec1 target short telomeres for elongation rather than double-strand break repair. Nat Struct Mol Biol 17: 1438–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay SJ, Cooke H 1992. hnRNP A2/B1 binds specifically to single stranded vertebrate telomeric repeat TTAGGGn. Nucleic Acids Res 20: 6461–6464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKerlie M, Lin S, Zhu XD 2012. ATM regulates proteasome-dependent subnuclear localization of TRF1, which is important for telomere maintenance. Nucleic Acids Res 40: 3975–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNees CJ, Tejera AM, Martinez P, Murga M, Mulero F, Fernandez-Capetillo O, Blasco MA 2010. ATR suppresses telomere fragility and recombination but is dispensable for elongation of short telomeres by telomerase. J Cell Biol 188: 639–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalfe JA, Parkhill J, Campbell L, Stacey M, Biggs P, Byrd PJ, Taylor AM 1996. Accelerated telomere shortening in ataxia telangiectasia. Nat Genet 13: 350–353 [DOI] [PubMed] [Google Scholar]
- Miller KM, Ferreira MG, Cooper JP 2005. Taz1, Rap1 and Rif1 act both interdependently and independently to maintain telomeres. EMBO J 24: 3128–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Rog O, Cooper JP 2006. Semi-conservative DNA replication through telomeres requires Taz1. Nature 440: 824–828 [DOI] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitton-Fry RM, Anderson EM, Hughes TR, Lundblad V, Wuttke DS 2002. Conserved structure for single-stranded telomeric DNA recognition. Science 296: 145–147 [DOI] [PubMed] [Google Scholar]
- Miyake Y, Nakamura M, Nabetani A, Shimamura S, Tamura M, Yonehara S, Saito M, Ishikawa F 2009. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol Cell 36: 193–206 [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Kanoh J, Saito M, Ishikawa F 2008. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science 320: 1341–1344 [DOI] [PubMed] [Google Scholar]
- Mohaghegh P, Karow JK, Brosh RM Jr, Bohr VA, Hickson ID 2001. The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res 29: 2843–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondoux M, Zakian VA 2006. Telomere position effect: Silencing near the end. In Telomeres, 2nd ed. (ed. de Lange T, Lundblad V, Blackburn E). Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Morrish TA, Garcia-Perez JL, Stamato TD, Taccioli GE, Sekiguchi J, Moran JV 2007. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature 446: 208–212 [DOI] [PubMed] [Google Scholar]
- Moser BA, Subramanian L, Chang YT, Noguchi C, Noguchi E, Nakamura TM 2009a. Differential arrival of leading and lagging strand DNA polymerases at fission yeast telomeres. EMBO J 28: 810–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser BA, Subramanian L, Khair L, Chang YT, Nakamura TM 2009b. Fission yeast Tel1(ATM) and Rad3 (ATR) promote telomere protection and telomerase recruitment. PLoS Genet 5: e1000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser BA, Chang YT, Kosti J, Nakamura TM 2011. Tel1ATM and Rad3ATR kinases promote Ccq1-Est1 interaction to maintain telomeres in fission yeast. Nat Struct Mol Biol 18: 1408–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K, Chen C, Kolodner RD 2001. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411: 1073–1076 [DOI] [PubMed] [Google Scholar]
- Naito T, Matsuura A, Ishikawa F 1998. Circular chromosome formation in a fission yeast mutant defective in two ATM homologues. Nat Genet 20: 203–206 [DOI] [PubMed] [Google Scholar]
- Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR 1997. Telomerase catalytic subunit homologs from fission yeast and human. Science 277: 955–959 [DOI] [PubMed] [Google Scholar]
- Nakamura TM, Cooper JP, Cech TR 1998. Two modes of survival of fission yeast without telomerase. Science 282: 493–496 [DOI] [PubMed] [Google Scholar]
- Nakamura TM, Moser BA, Russell P 2002. Telomere binding of checkpoint sensor and DNA repair proteins contributes to maintenance of functional fission yeast telomeres. Genetics 161: 1437–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan S, McEachern MJ 2002. Recombinational telomere elongation promoted by DNA circles. Mol Cell Biol 22: 4512–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan S, Groff-Vindman C, McEachern MJ 2003. Factors influencing the recombinational expansion and spread of telomeric tandem arrays in Kluyveromyces lactis. Eukaryot Cell 2: 1115–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini S, Ribaud V, Bianchi A, Shore D 2007. DNA breaks are masked by multiple Rap1 binding in yeast: Implications for telomere capping and telomerase regulation. Genes Dev 21: 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo ER, Cranston G, Allshire RC 1994. Telomere-associated chromosome breakage in fission yeast results in variegated expression of adjacent genes. EMBO J 13: 3801–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC 2011. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev 25: 350–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent CI, Hughes TR, Lue NF, Lundblad V 1996. Cdc13p: A single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274: 249–252 [DOI] [PubMed] [Google Scholar]
- O’Connor MS, Safari A, Xin H, Liu D, Songyang Z 2006. A critical role for TPP1 and TIN2 interaction in high-order telomeric complex assembly. Proc Natl Acad Sci 103: 11874–11879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oganesian L, Karlseder J 2011. Mammalian 5' C-rich telomeric overhangs are a mark of recombination-dependent telomere maintenance. Mol Cell 42: 224–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oganesian L, Moon IK, Bryan TM, Jarstfer MB 2006. Extension of G-quadruplex DNA by ciliate telomerase. EMBO J 25: 1148–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opresko PL, Otterlei M, Graakjaer J, Bruheim P, Dawut L, Kolvraa S, May A, Seidman MM, Bohr VA 2004. The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol Cell 14: 763–774 [DOI] [PubMed] [Google Scholar]
- Opresko PL, Mason PA, Podell ER, Lei M, Hickson ID, Cech TR, Bohr VA 2005. POT1 stimulates RecQ helicases WRN and BLM to unwind telomeric DNA substrates. J Biol Chem 280: 32069–32080 [DOI] [PubMed] [Google Scholar]
- Osterhage JL, Talley JM, Friedman KL 2006. Proteasome-dependent degradation of Est1p regulates the cell cycle-restricted assembly of telomerase in Saccharomyces cerevisiae. Nat Struct Mol Biol 13: 720–728 [DOI] [PubMed] [Google Scholar]
- Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ 2005. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat Struct Mol Biol 12: 847–854 [DOI] [PubMed] [Google Scholar]
- Paeschke K, Juranek S, Simonsson T, Hempel A, Rhodes D, Lipps HJ 2008. Telomerase recruitment by the telomere end binding protein-β facilitates G-quadruplex DNA unfolding in ciliates. Nat Struct Mol Biol 15: 598–604 [DOI] [PubMed] [Google Scholar]
- Paeschke K, McDonald KR, Zakian VA 2010. Telomeres: Structures in need of unwinding. FEBS Lett 584: 3760–3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paeschke K, Capra JA, Zakian VA 2011. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell 145: 678–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, Hockemeyer D, Kibe T, de Lange T 2009. Functional dissection of human and mouse POT1 proteins. Mol Cell Biol 29: 471–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardue ML, DeBaryshe PG 2008. Drosophila telomeres: A variation on the telomerase theme. Fly (Austin) 2: 101–110 [DOI] [PubMed] [Google Scholar]
- Pardue ML, Debaryshe P 2011. Adapting to life at the end of the line: How Drosophila telomeric retrotransposons cope with their job. Mob Genet Elements 1: 128–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschini M, Toro TB, Lubin JW, Braunstein-Ballew B, Morris DK, Lundblad V 2012. A naturally thermolabile activity compromises genetic analysis of telomere function in Saccharomyces cerevisiae. Genetics 191: 79–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennock E, Buckley K, Lundblad V 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104: 387–396 [DOI] [PubMed] [Google Scholar]
- Pickett HA, Cesare AJ, Johnston RL, Neumann AA, Reddel RR 2009. Control of telomere length by a trimming mechanism that involves generation of t-circles. EMBO J 28: 799–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter SF, Aubert SD, Zakian VA 2008. The Schizosaccharomyces pombe Pfh1p DNA helicase is essential for the maintenance of nuclear and mitochondrial DNA. Mol Cell Biol 28: 6594–6608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt CW, Cooper JP 2010. Pot1 inactivation leads to rampant telomere resection and loss in one cell cycle. Nucleic Acids Res 38: 6968–6975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta AF, Zakian VA 1989. Recombination occurs during telomere formation in yeast. Nature 337: 429–433 [DOI] [PubMed] [Google Scholar]
- Polotnianka RM, Li J, Lustig AJ 1998. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr Biol 8: 831–834 [DOI] [PubMed] [Google Scholar]
- Porro A, Feuerhahn S, Reichenbach P, Lingner J 2010. Molecular dissection of telomeric repeat-containing RNA biogenesis unveils the presence of distinct and multiple regulatory pathways. Mol Cell Biol 30: 4808–4817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CM, Boltz KA, Chaiken MF, Stewart JA, Beilstein MA, Shippen DE 2010. Evolution of CST function in telomere maintenance. Cell Cycle 9: 3157–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Zakian VA 2000. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev 14: 1777–1788 [PMC free article] [PubMed] [Google Scholar]
- Raffa GD, Siriaco G, Cugusi S, Ciapponi L, Cenci G, Wojcik E, Gatti M 2009. The Drosophila modigliani (moi) gene encodes a HOAP-interacting protein required for telomere protection. Proc Natl Acad Sci 106: 2271–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffa GD, Raimondo D, Sorino C, Cugusi S, Cenci G, Cacchione S, Gatti M, Ciapponi L 2010. Verrocchio, a Drosophila OB fold-containing protein, is a component of the terminin telomere-capping complex. Genes Dev 24: 1596–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raices M, Verdun RE, Compton SA, Haggblom CI, Griffith JD, Dillin A, Karlseder J 2008. C. elegans telomeres contain G-strand and C-strand overhangs that are bound by distinct proteins. Cell 132: 745–757 [DOI] [PubMed] [Google Scholar]
- Redon S, Reichenbach P, Lingner J 2010. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res 38: 5797–5806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenbach P, Hoss M, Azzalin CM, Nabholz M, Bucher P, Lingner J 2003. A human homolog of yeast Est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr Biol 13: 568–574 [DOI] [PubMed] [Google Scholar]
- Ribeyre C, Shore D 2012. Anticheckpoint pathways at telomeres in yeast. Nat Struct Mol Biol 19: 307–313 [DOI] [PubMed] [Google Scholar]
- Ritchie KB, Mallory JC, Petes TD 1999. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol Cell Biol 19: 6065–6075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rog O, Miller KM, Ferreira MG, Cooper JP 2009. Sumoylation of RecQ helicase controls the fate of dysfunctional telomeres. Mol Cell 33: 559–569 [DOI] [PubMed] [Google Scholar]
- Sabouri N, McDonald KR, Webb CJ, Cristea IM, Zakian VA 2012. DNA replication through hard-to-replicate sites, including both highly transcribed RNA Pol II and Pol III genes, requires the S. pombe Pfh1 helicase. Genes Dev 26: 581–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabourin M, Tuzon CT, Zakian VA 2007. Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol Cell 27: 550–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell LL, Zakian VA 1993. Loss of a yeast telomere: Arrest, recovery, and chromosome loss. Cell 75: 729–739 [DOI] [PubMed] [Google Scholar]
- Schoeftner S, Blasco MA 2008. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol 10: 228–236 [DOI] [PubMed] [Google Scholar]
- Schulz VP, Zakian VA 1994. The saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell 76: 145–155 [DOI] [PubMed] [Google Scholar]
- Schulz VP, Zakian VA, Ogburn CE, McKay J, Jarzebowicz AA, Edland SD, Martin GM 1996. Accelerated loss of telomeric repeats may not explain accelerated replicative decline of Werner syndrome cells. Hum Genet 97: 750–754 [DOI] [PubMed] [Google Scholar]
- Seto AG, Livengood AJ, Tzfati Y, Blackburn EH, Cech TR 2002. A bulged stem tethers Est1p to telomerase RNA in budding yeast. Genes Dev 16: 2800–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, de Lange T 2012. Removal of shelterin reveals the telomere end-protection problem. Science 336: 593–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir AJ, Chai W, Shay JW, Wright WE 2005. Telomere-end processing the terminal nucleotides of human chromosomes. Mol Cell 18: 131–138 [DOI] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T 2009. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, Kabir S, van Overbeek M, Celli GB, de Lange T 2010. Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science 327: 1657–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakirov EV, Surovtseva YV, Osbun N, Shippen DE 2005. The Arabidopsis Pot1 and Pot2 proteins function in telomere length homeostasis and chromosome end protection. Mol Cell Biol 25: 7725–7733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiratori A, Shibata T, Arisawa M, Hanaoka F, Murakami Y, Eki T 1999. Systematic identification, classification, and characterization of the open reading frames which encode novel helicase-related proteins in Saccharomyces cerevisiae by gene disruption and Northern analysis. Yeast 15: 219–253 [DOI] [PubMed] [Google Scholar]
- Smith JS, Chen Q, Yatsunyk LA, Nicoludis JM, Garcia MS, Kranaster R, Balasubramanian S, Monchaud D, Teulade-Fichou MP, Abramowitz L, et al. 2011. Rudimentary G-quadruplex-based telomere capping in Saccharomyces cerevisiae. Nat Struct Mol Biol 18: 478–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow BE, Erdmann N, Cruickshank J, Goldman H, Gill RM, Robinson MO, Harrington L 2003. Functional conservation of the telomerase protein Est1p in humans. Curr Biol 13: 698–704 [DOI] [PubMed] [Google Scholar]
- Snow BE, Mateyak M, Paderova J, Wakeham A, Iorio C, Zakian V, Squire J, Harrington L 2007. Murine Pif1 interacts with telomerase and is dispensable for telomere function in vivo. Mol Cell Biol 27: 1017–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansel RM, de Lange T, Griffith JD 2001. T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J 20: 5532–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavropoulos DJ, Bradshaw PS, Li X, Pasic I, Truong K, Ikura M, Ungrin M, Meyn MS 2002. The Bloom syndrome helicase BLM interacts with TRF2 in ALT cells and promotes telomeric DNA synthesis. Hum Mol Genet 11: 3135–3144 [DOI] [PubMed] [Google Scholar]
- Steinacher R, Osman F, Dalgaard JZ, Lorenz A, Whitby MC 2012. The DNA helicase Pfh1 promotes fork merging at replication termination sites to ensure genome stability. Genes Dev 26: 594–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern JL, Zyner KG, Pickett HA, Cohen SB, Bryan TM 2012. Telomerase recruitment requires both TCAB1 and Cajal bodies independently. Mol Cell Biol 32: 2384–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JA, Wang F, Chaiken MF, Kasbek C, Chastain PD 2nd, Wright WE, Price CM 2012. Human CST promotes telomere duplex replication and general replication restart after fork stalling. EMBO J 31: 3537–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian L, Nakamura TM 2010. To fuse or not to fuse: How do checkpoint and DNA repair proteins maintain telomeres? Front Biosci 15: 1105–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtseva YV, Churikov D, Boltz KA, Song X, Lamb JC, Warrington R, Leehy K, Heacock M, Price CM, Shippen DE 2009. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol Cell 36: 207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taggart AK, Teng SC, Zakian VA 2002. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297: 1023–1026 [DOI] [PubMed] [Google Scholar]
- Takai KK, Kibe T, Donigian JR, Frescas D, de Lange T 2011. Telomere protection by TPP1/POT1 requires tethering to TIN2. Mol Cell 44: 647–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarsounas M, Munoz P, Claas A, Smiraldo PG, Pittman DL, Blasco MA, West SC 2004. Telomere maintenance requires the RAD51D recombination/repair protein. Cell 117: 337–347 [DOI] [PubMed] [Google Scholar]
- Teixeira MT, Arneric M, Sperisen P, Lingner J 2004. Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell 117: 323–335 [DOI] [PubMed] [Google Scholar]
- Teng SC, Zakian VA 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol Cell Biol 19: 8083–8093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng SC, Chang J, McCowan B, Zakian VA 2000. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol Cell 6: 947–952 [DOI] [PubMed] [Google Scholar]
- Tomaska L, Willcox S, Slezakova J, Nosek J, Griffith JD 2004. Taz1 binding to a fission yeast model telomere: Formation of telomeric loops and higher order structures. J Biol Chem 279: 50764–50772 [DOI] [PubMed] [Google Scholar]
- Tomita K, Cooper JP 2008. Fission yeast Ccq1 is telomerase recruiter and local checkpoint controller. Genes Dev 22: 3461–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng SF, Lin JJ, Teng SC 2006. The telomerase-recruitment domain of the telomere binding protein Cdc13 is regulated by Mec1p/Tel1p-dependent phosphorylation. Nucleic Acids Res 34: 6327–6336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzon CT, Wu Y, Chan A, Zakian VA 2011. The Saccharomyces cerevisiae telomerase subunit Est3 binds telomeres in a cell cycle- and Est1-dependent manner and interacts directly with Est1 in vitro. PLoS Genet 7: e1002060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukimori S, Kawabata N, Shimada H, Imano R, Takahashi K, Yukawa M, Tsuchiya E, Ueno M 2012. A double mutant between fission yeast telomerase and RecQ helicase is sensitive to thiabendazole, an anti-microtubule drug. Biosci Biotechnol Biochem 76: 264–269 [DOI] [PubMed] [Google Scholar]
- Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MI, Ding H, Boulton SJ 2012. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell 149: 795–806 [DOI] [PubMed] [Google Scholar]
- van Steensel B, de Lange T 1997. Control of telomere length by the human telomeric protein TRF1. Nature 385: 740–743 [DOI] [PubMed] [Google Scholar]
- Vaze MB, Pellicioli A, Lee SE, Ira G, Liberi G, Arbel-Eden A, Foiani M, Haber JE 2002. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol Cell 10: 373–385 [DOI] [PubMed] [Google Scholar]
- Verdun RE, Karlseder J 2006. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell 127: 709–720 [DOI] [PubMed] [Google Scholar]
- Vodenicharov MD, Wellinger RJ 2006. DNA degradation at unprotected telomeres in yeast is regulated by the CDK1 (Cdc28/Clb) cell-cycle kinase. Mol Cell 24: 127–137 [DOI] [PubMed] [Google Scholar]
- Wang X, Baumann P 2008. Chromosome fusions following telomere loss are mediated by single-strand annealing. Mol Cell 31: 463–473 [DOI] [PubMed] [Google Scholar]
- Wang SS, Zakian VA 1990. Telomere-telomere recombination provides an express pathway for telomere acquisition. Nature 345: 456–458 [DOI] [PubMed] [Google Scholar]
- Wang RC, Smogorzewska A, de Lange T 2004. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119: 355–368 [DOI] [PubMed] [Google Scholar]
- Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M 2007. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature 445: 506–510 [DOI] [PubMed] [Google Scholar]
- Wang Y, Ghosh G, Hendrickson EA 2009. Ku86 represses lethal telomere deletion events in human somatic cells. Proc Natl Acad Sci 106: 12430–12435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD 1972. Origin of concatemeric T7 DNA. Nat New Biol 239: 197–201 [DOI] [PubMed] [Google Scholar]
- Webb CJ, Zakian VA 2012. Schizosaccharomyces pombe Ccq1 and TER1 bind the 14-3-3-like domain of Est1, which promotes and stabilizes telomerase-telomere association. Genes Dev 26: 82–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger RJ, Wolf AJ, Zakian VA 1993. Saccharomyces telomeres acquire single-strand TG1–3 tails late in S phase. Cell 72: 51–60 [DOI] [PubMed] [Google Scholar]
- Wellinger RJ, Ethier K, Labrecque P, Zakian VA 1996. Evidence for a new step in telomere maintenance. Cell 85: 423–433 [DOI] [PubMed] [Google Scholar]
- Wright JH, Zakian VA 1995. Protein-DNA interactions in soluble telosomes from Saccharomyces cerevisiae. Nucleic Acids Res 23: 1454–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright WE, Tesmer VM, Huffman KE, Levene SD, Shay JW 1997. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev 11: 2801–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zakian VA 2011. The telomeric Cdc13 protein interacts directly with the telomerase subunit Est1 to bring it to telomeric DNA ends in vitro. Proc Natl Acad Sci 108: 20362–20369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KJ, Grandori C, Amacker M, Simon-Vermot N, Polack A, Lingner J, Dalla-Favera R 1999. Direct activation of TERT transcription by c-MYC. Nat Genet 21: 220–224 [DOI] [PubMed] [Google Scholar]
- Wu L, Multani AS, He H, Cosme-Blanco W, Deng Y, Deng JM, Bachilo O, Pathak S, Tahara H, Bailey SM, et al. 2006. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell 126: 49–62 [DOI] [PubMed] [Google Scholar]
- Wu P, van Overbeek M, Rooney S, de Lange T 2010. Apollo contributes to G overhang maintenance and protects leading-end telomeres. Mol Cell 39: 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, DiMaggio P, Perlman D, Zakian V, Garcia B 2013. Novel phosphorylation sites in the S. cerevisiae Cdc13 protein reveal new targets for telomere length regulation. J Proteome Res 12: 316–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyllie FS, Jones CJ, Skinner JW, Haughton MF, Wallis C, Wynford-Thomas D, Faragher RG, Kipling D 2000. Telomerase prevents the accelerated cell ageing of Werner syndrome fibroblasts. Nat Genet 24: 16–17 [DOI] [PubMed] [Google Scholar]
- Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O’Connor MS, Songyang Z 2007. TPP1 is a homologue of ciliate TEBP-β and interacts with POT1 to recruit telomerase. Nature 445: 559–562 [DOI] [PubMed] [Google Scholar]
- Xue Y, Rushton MD, Maringele L 2011. A novel checkpoint and RPA inhibitory pathway regulated by Rif1. PLoS Genet 7: e1002417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H, Tarumoto Y, Ishikawa F 2012. Tel1(ATM) and Rad3(ATR) phosphorylate the telomere protein Ccq1 to recruit telomerase and elongate telomeres in fission yeast. Genes Dev 26: 241–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye JZ, Hockemeyer D, Krutchinsky AN, Loayza D, Hooper SM, Chait BT, de Lange T 2004. POT1-interacting protein PIP1: A telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev 18: 1649–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu EY, Wang F, Lei M, Lue NF 2008. A proposed OB-fold with a protein-interaction surface in Candida albicans telomerase protein Est3. Nat Struct Mol Biol 15: 985–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Williamson JR, Cech TR, Prescott DM 1991. Inhibition of telomerase by G-quartet DNA structures. Nature 350: 718–720 [DOI] [PubMed] [Google Scholar]
- Zaug AJ, Podell ER, Cech TR 2005. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc Natl Acad Sci 102: 10864–10869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DH, Zhou B, Huang Y, Xu LX, Zhou JQ 2006. The human Pif1 helicase, a potential Escherichia coli RecD homologue, inhibits telomerase activity. Nucleic Acids Res 34: 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Hoshiyama H, Shay JW, Wright WE 2008. Quantitative telomeric overhang determination using a double-strand specific nuclease. Nucleic Acids Res 36: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Sfeir AJ, Zou Y, Buseman CM, Chow TT, Shay JW, Wright WE 2009. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell 138: 463–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Abreu E, Kim J, Stadler G, Eskiocak U, Terns MP, Terns RM, Shay JW, Wright WE 2011. Processive and distributive extension of human telomeres by telomerase under homeostatic and nonequilibrium conditions. Mol Cell 42: 297–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Monson EK, Teng SC, Schulz VP, Zakian VA 2000. Pif1p helicase, a catalytic inhibitor of telomerase in yeast. Science 289: 771–774 [DOI] [PubMed] [Google Scholar]
- Zhou JQ, Qi H, Schulz VP, Mateyak MK, Monson EK, Zakian VA 2002. Schizosaccharomyces pombe pfh1+ encodes an essential 5′ to 3′ DNA helicase that is a member of the PIF1 subfamily of DNA helicases. Mol Biol Cell 13: 2180–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Kumar R, Mandal M, Sharma N, Sharma HW, Dhingra U, Sokoloski JA, Hsiao R, Narayanan R 1996. Cell cycle-dependent modulation of telomerase activity in tumor cells. Proc Natl Acad Sci 93: 6091–6095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Hathcock KS, Hande P, Lansdorp PM, Seldin MF, Hodes RJ 1998. Telomere length regulation in mice is linked to a novel chromosome locus. Proc Natl Acad Sci 95: 8648–8653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]