Abstract
DNAJB6 is a constitutively expressed member of the HSP40 family. It has been described as a negative regulator of breast tumor progression and a regulator of epithelial phenotype. Expression of DNAJB6 is reported to be compromised with tumor progression. However, factors responsible for its down-regulation are still undefined. We used a knowledge-based screen for identifying miRNAs capable of targeting DNAJB6. In this work, we present our findings that hsa-miR-632 (miR-632) targets the coding region of DNAJB6. Invasive and metastatic breast cancer cells express high levels of miR-632 compared to mammary epithelial cells. Analysis of RNA from breast tumor specimens reveals inverse expression patterns of DNAJB6 transcript and miR-632. In response to exogenous miR-632 expression, DNAJB6 protein levels are down regulated and the resultant cell population shows significantly increased invasive ability. Silencing endogenous miR-632 abrogates invasive ability of breast cancer cells and promotes epithelial like characteristics noted by E-cadherin expression with concomitant decrease in mesenchymal markers such as Zeb2 and Slug. Thus, miR-632 is a potentially important epigenetic regulator of DNAJB6 which contributes to the down regulation of DNAJB6 and plays a supportive role in malignant progression.
Keywords: DnaJB6, hsa-miR-632
Introduction
Breast cancer is the leading cancer type for the estimated new cancer cases and second only to lung cancer in the estimated deaths for 2010 in women in the United States (1). Years of extensive research has implicated several intrinsic and extrinsic etiologic factors for the initiation and progression of breast cancer. Micro-RNAs are 21–23 nucleotide regulatory RNAs that can target mRNA and cause either transcription or translation repression (2, 3). In the recent years micro-RNAs have been emerging as critical epigenetic regulators which modulate almost all stages of breast cancer progression such as proliferation, angiogenesis, invasion, migration, chemoresistance and stem cell renewal (4–9).
The focus of our study is DNAJB6 (Mammalian Relative of DNAJ, Mrj), a constitutively expressed member of HSP 40 family (10). DNAJB6 gene codes for 2 spliced variants. The longer spliced variant is referred to as DNAJB6 a [Mrj(L)] and the shorter is DNAJB6 b [Mrj(S)] (11). Studies on murine DNAJB6 have revealed that it plays a critical role in embryonic development, specifically during chorioallantoic fusion (12). This gene product has also been implicated as a player in regulation of cell-cycle and mitosis (13). Mutations of DNAJB6 are implicated as a potential cause of limb-girdle muscular dystrophy (14, 15). We have previously reported that DNAJB6 protein expression is significantly compromised in advanced breast cancers (11). Re-expression of DNAJB6a abrogated tumor growth and metastasis in mammary-xenograft studies (11). DNAJB6b has been demonstrated to interact with uPAR to increase uPAR mediated cell adhesion to vitronectin (16). Other diseases such as neuro-degenerative disorders involving polyglutamine aggregate may also display a negative outcome if expression or function of DNAJB6 is compromised (17–19). However, factors that regulate the expression of this critical protein are yet to be understood. Hence we undertook an investigation to find out if micro-RNAs may play a role in downregulating DNAJB6 expression. In this study we describe that microRNA, hsa-miR-632 (referred here after as miR-632), downregulates DNAJB6 expression and evaluate its clinical significance.
Materials and Methods
Cell culture and reagents
MCF10A, MCF10AT, MCF10CA1d.cl.1 (obtained from Barbara Ann Karamanos Cancer Center, Detroit, MI, USA) were grown in DMEM/F12 (Invitrgen, Carslbad, USA) supplemented with 5% heat inactivated horse serum, 100ng/ml cholera toxin (Calbiochem, San Diego, CA), 10ug/ml insulin (Sigma, St. Louis, MO), 25ng/ml EGF (Sigma) and 500 ng/ml hydrocortisone (Sigma). SUM159 cells (purchased from Asterand plc, Detroit, MI, USA) were cultured in Ham's F-12 with 5% fetal bovine serum supplemented with insulin (5 mg/ml) and hydrocortisone (1 mg/ml). MDA-MB-231was grown as described previously (11). All cells were maintained in a humidified 5% CO2 environment at 37° C.
DNAJB6 rabbit polyclonal (MO1) antibody (Abnova Corp., Taipei City, Taiwan) was used (1:5000) with 5% milk in PBS containing 0.2% Tween 20). Horseradish peroxidase-β-actin (Sigma-Aldrich, St. Louis, MO) was used at (1:50,000). x-miR:has-mir-632 (cat # X-miR-8710R) and control mir were obtained from oligoengine™ (oligoengine, Seattle, WA).
Plasmid Constructs
Oligos designed to encompass hsa-miR-632 target site in the ORF of DNAJB6 mRNA 5'agcttttctttgggaatcgaaggggtccccgaggaagcagaagccgagggacggggtcgttttta---3'and 3'- tgatctaaaaacgaccccgtccctcggcttctgcttcctcggggaccccttcgattcccaaagaa---5' were annealed and cloned into HindIII and SpeI sites of pMIR-Report vector (Ambion, Austin, TX) to generate pMIR-Report-DNAJB6. The hsa-miR-632 expression construct was generated in pIRES2-EGFP (Clontech, Madison. WI) by annealing commercially synthesized oligos corresponding to the mature miRNA hsa-miR-632 5'ctcgagacggctaccaccacgtcccacaggaagcagacacaaaatggccgacggcctcgttccccgctccgcctcccgtcaagcactgcggtaggaggcg- 3' and 5'tgccgatggtggtgcagggtgtccttcgtctgtgttttaccggctgccggagcaaggggcgaggcggagggcagttcgtgacgccatcctccgcgaattc- 3' and then cloning them into XhoI and EcoRI sites of pIRES2-EGFP.
Transfection
Cells were transfected with miRNA inhibitor scrambled control clone pEZX-AM01 (CmiR-AN0001-AM01, GeneCopoeia Inc, Rockville, MD, USA ) or miRNA inhibitor against hsa-miR-632-pEZX-AM01 (HmiR-AN0742-AM01) using lipofectamine 2000. MCF10AT cells were tansfected with pIRES2EGFP - vector or hsa-miR- 632-pIRES2EGFP using Fugene 6 (Roche, In, USA).
Transfection with anti-miR-632
To inhibit endogenous miRNA, 50 or 100nM x-miR-632 or control mir (oligoengine™) was transfected into cells using Lipofectamine 2000. Cells were assayed for knockdown 48 hrs post transfection.
Western Blots
Cells were transfected with miR-632-pIRES2EGFP or pIRES2EGFP vector alone using Lipofectamine 2000 according to manufacturer's intructions (Invitrogen). Cells were harvested 42 hrs post transfection in NP-40 lysis buffer. The lysates (20ug) were resolved on SDS-PAGE and transferred onto PVDF membranes. The immunoblot was developed using relevant primary and secondary antibodies as per the respective manufacturer's instructions.
Luciferase Reporter Assay
Cells were transfected with 50 ng pMIR-Report or pMIR-Report-DNAJB6 in combination with either 100 ng pIRES2EGFP vector alone or pIRES2EGFP-miR-632 construct and 25ng of β-gal plasmid using with Lipofectamine 2000 (Invitrogen) as stated in manufacturer's protocol. The assay was terminated 36 hrs. post-transfection. Luciferase activity was measured using Turner 20/20 luminometer (Turner Biosystems , Sunnyvale, CA). β-galactosidase activity was quantitated using Synergy 4 ™ plate reader (BioTek, Winooski, VT, USA)was used as normalization control.
Quantitative-RT-PCR
To measure mRNA levels in cell lines or tissues, RNA isolated from cell lines or tissue samples were subjected to qRT-PCR. 1ug of total RNA was used to synthesize cDNA (High Capacity Reverse Transcription Kit, Applied Biosystems, Foster City, CA). PCR was performed using 40ng of cDNA with respective Taqman primer probe using BioRad iQ5 Real Time Detection system (BioRad, Hercules, CA). The gene expression Ct values of mRNAs from each sample were calculated by normalizing with endorse control, glyceraldehyde-3-phosphate dehydrogenase and relative quantitation values were plotted using GraphPad Prism (La Jolla, CA).
miRNA levels were also analyzed by real time as follows: cDNA was generated using microRNA Reverse Transcription kit (Applied Biosystems). Total RNA was used to generate cDNA using primers specific to U6 (control) or has-miR-632. PCR was performed using both U6 (control) or has-miR-632 Taqman primer probes and Taqman Universal Master Mix, No Amperase UNG (Applied Biosystems). The cycling conditions were initial step of 95°C for 10 min followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 min. has-miR-632 miRNA levels were normalized to U6 levels.
Invasion assay
Invasion assays were conducted using 8 uM polyethylene terpthalate filters (BD Pharmingen), as described earlier (20). Cells (transfected with vector or hsa-miR-632) were allowed to invade through matrigel coated filters for 18 hrs. in a transwell. Cells invaded to the lower sides of the transwell, were stained using 0.05% crystal violet, and the cell number was counted as described before (11).
3D culture
3D cultures were grown following the protocol by Debnath et al. (21). Briefly, eight well-chambered cover glass slides (Thermo Scientific, Waltham, MA, USA) were placed on ice and coated with 50 ml of 3D Culture Matrix Basement Membrane Extract Reduced Growth Factor (phenol red free) from Trevigen (Gaithersburg, MD, USA). The slide was incubated for 30 min in 37 °C incubator. Cells (5000/well, 36 hrs post transfection) were plated in growth media containing 2% reduced growth factor basement membrane extract (Trevigen). Media was changed every 4 days and the morphology was documented digitally using a Nikon microscope (Nikon, Tokyo, Japan) using the 20 × objective. Laminin V was stained using MAB-D4B5 (Chemicon, USA)
Patient samples Processing
Upon obtaining signed consent, surgically excised breast tumor specimens were flash frozen in liquid nitrogen with minimal lag time. Patient tissues were banked per Mitchell Cancer Institute-BioBank protocol (IRB approval #03-092), prior to being procured for use (IRB approval #09-287). All specimens were coded and de-identified. Frozen tissue was grinded using mortar and pestle. Total RNA was isolated using miRNAeasy kit (Qiagen, Valencia, CA) as recommended in manufacturer's protocol.
RESULTS
Identification of potential miRNA regulators of DNAJB6
miRNAs predicted to target DNAJB6 transcript (3' UTR as well as coding region) were cataloged using the Sanger data base (Table 1). Eleven distinct miRNAs are predicted to target DNAJB6. Out of these 5 miRNAs are predicted to target the coding region and 6 are predicted to target the non-coding region (3'UTR). The outcome of this search was manually queried for miRNAs that were reported to be overexpressed in malignancies. Four miRNAs (mir-197, 632, 376a and 628-3p) targeting the coding region and one (mir-424) targeting the 3'UTR were cited in literature to be overexpressed in certain malingnancies.
Table 1.
Micro-RNAs predicted to target DNAJB6 transcript are catalogued using the Sanger data base. The highlighted miRNAs are predicted to target the coding region of DNAJB6 and the rest are predicted to target the non-coding region (3'UTR).
| Over Expressed | Mechanism | Reference | |
|---|---|---|---|
| miR-197 | SCLCs and NSCLCs Male breast cancer Follicular thyroid carcinoma |
Speculated Oncogenic | Du L et al. Mol Cancer Res. 2009 Aug;7(8): 1234–43. Lehmann U. et al. BMC Cancer. 2010 Mar 23;10:109. Weber F J et al. Clin Endocrinol Metab. 2006 Sep;91(9):3584–91. |
| miR-632 | Myelodysplastic syndrome | Unknown | Erdogan B., et al. Experimental Hematology 2011 September, 39 (9), 2011 915–926.e2 |
| miR-376a | Pancreatic cancer | Unknown | Batra et al. Cancer Letters 292(1), 1 June 2010 |
| miR-628-3p | Myc induced | Unknown | Mestdagh P. et al. Oncopene 2010;29:1394–1404. |
| miR-29b-2* | No data found | - | - |
| miR-369 | No data found | - | - |
| miR-29c | No data found | - | - |
| miR-597 | No data found | - | - |
| Mir-591 | No data found | - | - |
| miR-801 | No data found | - | - |
| miR-424 | Under expressed Ovarian cancer | Dahiya N,, et al. 2008. PLoS ONE 3(6): e2436. |
The expression of DNAJB6 has been reported to be highly compromised in invasive (grade III) and metastatic (lymph nodes) breast cancer (11). Hence we queried a set of breast cancer cell lines (viz. MCF10CA1d.cl.1, SUM159 and MDA-MB-231) with documented highly invasive and metastatic properties in xenograft studies (22–24) for the expression of these miRNAs in comparison with MCF10A, an immortalized mammary epithelial cell line (Figure 1A). Tumorigenic but non-invasive and non-metastatic MCF10AT cell line was also used in this comparison. mir-376a and mir-628-3p were not detectable in these cells. Of the three that showed detectable levels, mir-197 expression did not show a distinct trend; if at all there was a slight reduction in expression. However, mir-424 and miR-632 expression showed a distinct increase in the three invasive and metastatic cell lines in comparison with MCF10A and MCF10AT. These two miRNAs were evaluated for their capacity to reduce DNAJB6 protein levels by transiently tranfecting the respective miRNAs in MCF10A (cell line with high level of DNAJB6). The results showed that miR-632 was able to cause a noticeable reduction in the protein levels of both the spliced variants of DNAJB6 (viz. DNAJB6a and b) (Figure 1B). Digital densitometry analysis shows that there is 74% reduction in DNAJB6a and 55% reduction in DNAJB6b (Supplemental Figure 1).
Figure 1. Screening for miRNAs that can potentially target DNAJB6.
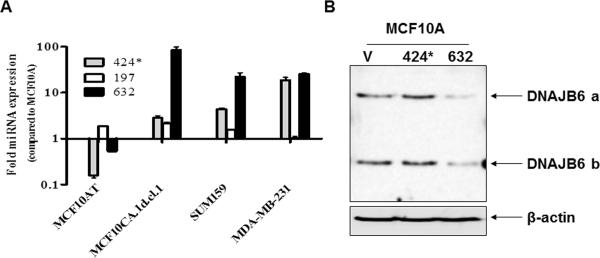
(A) Levels of each of the miRNAs, miR-197, miR-424 and miR-632 from human Breast cancer cell lines MCF10AT, MCF10CA1d.cl.1, SUM159 and MDA-MB-231 were compared to immortalized human mammary epithelial cell line MCF10A using real time-quantitive-PCR. The graph is presented in logarithmic scale for Y-axis. Reactions were performed in triplicate and the experiment was performed twice.
(B) mir-424 and miR-632 were transiently expressed in MCF10A. Total protein extract (20μg) was resolved using SDS-PAGE and subjected to western blot analysis for DNAJB6 levels. β-actin level was determined to confirm equal loading.
A putative miR-632 binding site exists in the DNAJB6 ORF
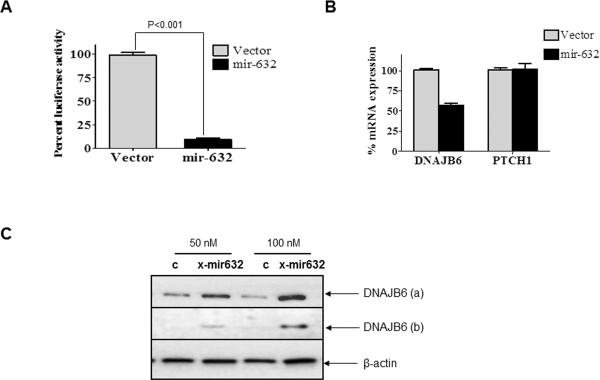
To validate the miR-632 targeting site from DNAJB6, we cloned the putative binding site of miR-632 (as per Sanger miRNA search software prediction) from the open reading frame (ORF) of DNAJB6 in pMIR-REPORT™ vector to obtain pMIR-REPORT-DNAJB6. This reporter was co-transfected with a construct that constitutively overexpresses mature miR-632 (miR-632-pIRES2EGFP). We found a 90% reduction in the activity of pMIR-Report-DNAJB6 in MCF10A cells (Figure 2 A). These observations provided additional strength to the proposed role of miR-632 in targeting DNAJB6.
Figure 2. miR-632 targets DNAJB6 for degradation.
(A) pMIR-REPORT-DNAJB6 (containing putative binding site of miR-632 from the ORF of DNAJB6) was co-transfected with miR-632 expression vector, miR-632-pIRES2EGFP or empty vector control. The assay was performed in triplicate and the experiment was performed twice. The luciferase activity readings were normalized with activity from a co-transfected β-gal expressing control. The error bars represent standard error of mean (SEM)
(B) Levels of DNAJB6 and PTCH1 transcript were evaluated from MCF10A cells treated with miR-632-pIRES2EGFP or empty vector control. GAPDH was used as endorse control. The error bars represent standard error of mean (SEM). The reactions were performed in triplicate and the experiment was performed three times.
(C) MDA-MB-231 cells were treated with X-miR-632 or control (50 and 100 nM). Total protein extract (30μg) was resolved using SDS-PAGE and subjected to western blot analysis for DNAJB6 levels. β-actin levels were determined as loading control.
miRNAs have several predicted targets and are known to alter expression at transcription or translation or both levels. We tested effect of miR-632 on DNAJB6 transcript in MCF10A cells. Compared to the non-targeting control, the transcript levels of DNAJB6 in miR-632 transfected cells were reduced by about 40%. Patched 1 (PTCH1) is another predicted target of miR-632. However, the transcript levels of PTCH1 were unaltered in the DNAJB6 expressors (Figure 2 B). This endorsed the specificity of miR-632 in targeting DNAJB6.
To further evaluate the validity of our findings, we used (X-miR-632) to silence the expression of miR-632 from MDA-MB-231 cells. Western blot analysis for DNAJB6 showed that at 50 nm as well as 100 nm concentration of X-miR-632 there was a noticeable increase in levels of both DNAJB6 varients (Figure 2 C and Supplementary Figure 2). Cumulatively these observations confirm the ability of miR-632 to downregulate DNAJB6 expression.
miR-632 targeting of DNAJB6 is conserved across species
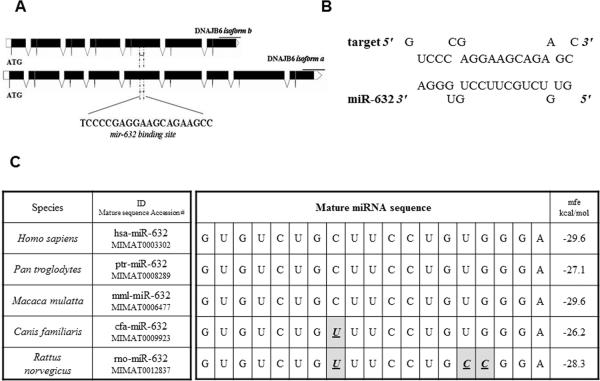
Analysis of the predicted target site of miR-632 in the context of the genomic DNA and mRNA sequence of human DNAJB6 gene revealed that it targets a region in exon 5 of the DNAJB6 gene (Figure 3 A). As seen in from the complementarity between the seed sequence and the mRNA depicted in Figure 3 B, this region is common to both the spliced variants of DNAJB6. Cross-species sequence comparison revealed that mature miR-632 sequence remains fairly unaltered. In fact, it is identical in the primates and deviates minimally in other mammals such as dogs and rodents. Importantly, the mfe (minimum-free energy) of the respective miR-632 hybrid with respective DNAJB6 target sequence remains reasonably comparable (Figure 3 C). These observations imply that the relationship of the miR632 in targeting DNAJB6 may be conserved throughout evolution and hence suggestive of the importance of miR-632 as a regulator of DNAJB6.
Figure 3. Analysis of miR-632 binding site from DNAJB6.
(A) Diagrammatic representation of miR-632 binding site. Solid black boxes represent exons.
(B) Diagrammatic representation of binding of miR-632 seed sequence to its target site in DNAJB6.
(C) Cross species sequence comparison miR-632. Highlighted boxes with bold italicized underlined letters depict deviation from human miR-632 sequence. mfe = minimum-free energy.
hsa-miR-632 and DNAJB6 levels show inverse trends in breast cancer tissues
We analyzed miR-632 and DNAJB6 levels from RNA harvested from normal breasts tissue and 15 breast tumors. The data shows decreased expression of DNAJB6 in cancerous tissues compared to normal breast tissue where as miR-632 levels were found to be higher in cancerous tissues as compared to the non-cancerous tissues (Figure 4). Thirteen out of 15 tumor specimens showed decreased DNAJB6 transcript and out of those, twelve specimens showed high miR-632 levels relative to normal breast tissue (exact binomial test, p < 0.0001).
Figure 4. hsa-miR-632 and DNAJB6 levels show inverse trends in breast cancer tissues.
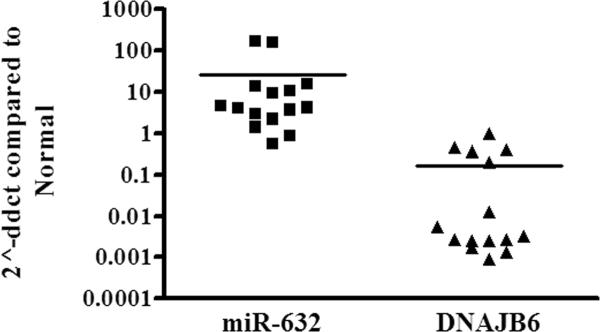
Total RNA from 15 breast tumors was analyzed by qRT-PCR for miR-632 and DNAJB6 levels from. The levels were compared with average of normal levels as calibrator, designated as 1. The graph is presented in logarithmic scale for Y-axis.
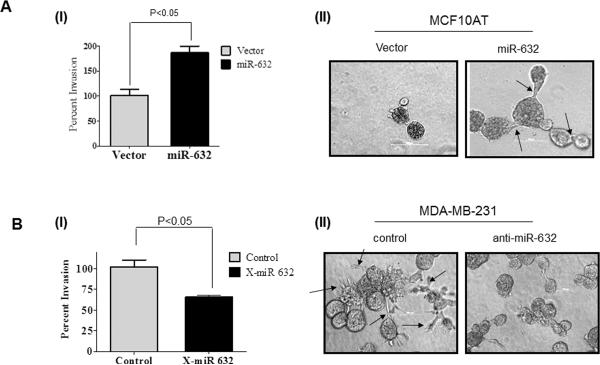
miR-632 expression enhances the invasive properties
We have previously reported that DNAJB6 enhances epithelial attributes and inhibits the mesenchymal phenotype (11, 25). The mesenchymal phenotype confers invasive ability upon breast cancer cells (26). Hence we analyzed the impact of expression of miR-632 on invasion and levels of mesenchymal markers. MCF10AT breast cancer cells are tumorigenic but non-invasive in nude mice xenograft studies. These cells express very low levels of miR-632 and are known to have high levels of DNAJB6. miR-632 was over expressed in these cells by transiently transfecting miR-632-pIRES2EGFP. Our results showed that expression of miR-632 caused about a 2-fold increase in the invasive ability of the cells (Figure 5 A [I]). Conversely we tested effect of silencing expression of miR-632 in the invasive and metastatic MDA-MB-231 breast cancer cell line. Our observations reveal a 35% decrease in the invasive ability of these cells upon silencing miR-632 compared to the control treated cells (Figure 5 B [I]).
Figure 5. miR-632 expression enhances the invasive properties.
(A) (I) MCF10AT cells were transiently transfected miR-632-pIRES2EGFP and were allowed to invade through matrigel coated filters for 18 hrs. Invaded cells were visualized using crystal violet, and the cell number was counted and compared to invasion of cells transfected with empty vector control. (II) MCF10AT cells were transiently transfected with empty pIRES2EGFP vector or miR-632-pIRES2EGFP and their growth in 3-D was assessed. Arrows point the invasive outgrowth. Experiments were performed in triplicate and the experiment was performed two times.
(B) (I) MDA-MB-231 cells were treated with X-miR-632 (100 nM) or the control and the invasion assay was performed as described before. (II) MDA-MB-231 cells were transfected with miRNA inhibitor scrambled control clone pEZX-AM01 (control) or anti-miR-632 pIRES2EGFP and their growth in 3-D was assessed. Arrows point the invasive outgrowth.. Experiments were performed in triplicate and the experiment was performed two times.
Growth in 3-D matrix assays has been accepted to more accurately recapitulates the epithelial organization seen in breast ducts in vivo (27). MCF10AT cells transfected with empty vector formed well circumscribed, acinar structures. Notably, consistent with the results of the invasion assay, when miR-632 was expressed in these cells they formed branched outgrowths indicated by arrows in Figure 5A(II). Cells embedded in 3-D growth environment can produce basement membrane molecules such as laminin 5 (28). The miR-632 expressing MCF10AT structures retained the ability to deposit basement membrane but exhibited broken (indicated by arrows) laminin 5 around the invasive edge (Supplementary Figure 3).
MDA-MB-231 cells are known to show stellate 3-D morphology with projected invasive growths that can link several cell colonies (29). Our observations presented in Figure 5B(II) indicate that consistent with the results of the invasion assay, upon introduction of anti-mir-632, majority of the cells tend to lose the stellate projections and appear more rounded in 3-D culture. Notably, the anti-mir-632 transfected MDA-MB-231 cells showed the presence of intact laminin 5 which was broken (indicated by arrows) in the control anti-mir transfected cells (Supplementary Figure 3).
Cumulatively these observations indicate a gain in invasive-mesenchymal like cellular morphology in the presence of miR-632. Expression analysis using qRT-PCR revealed that X-miR-632 treatment down regulated mesenchymal markers Zeb2 and SLUG whereas epithelial marker, E-cadherin was up-regulated. These observations are consistent with our previous report of EMT reversal seen in DNAJB6 expressors are consistent with our previous report of EMT reversal seen in DNAJB6 expressors (Supplementary Figure 4).
Discussion
Differential regulation of miRNA in cancers of diverse types is known. Many human miRNA genes are located at genomic loci associated with cancer suggesting the role of miRNAs in cancer (30). The role of miRNAs in breast cancer cell growth and metastasis has been widely accepted (7, 31–35). miRNAs have emerged as novel candidates markers and treatment targets in breast cancer and have a huge potential of evolving into effective biomarkers because of their stability and availability for detection (36–38).
However, newer miRNAs are still being discovered and validated for their activity. To date the role of miR-632 in cancer has not been described. Also no specific target been validated for this miRNA. Notably, the importance of miR-632 is getting realized. Recent interesting studies by Erdogan et al revealed that expression level of miR-632 is positively associated with myelodysplastic syndromes (MDS) and has high discrimination ability compared to normal (39). Cancer for the most part is associated with aging; hence it is interesting to note that an aging-associated disorder such as MDS, shows up-regulation of miR-632. One other detailed study involving analysis of exosomes secreted by immune cells revealed high miR-632 levels in the exosomes (40). This study shows that miRNAs transferred during immune synapsis are able to modulate gene expression in recipient cells. Thus it is conceivable that DNAJB6 expression in the breast epithelial cells could also be modulated by miR-632 delivered through exosomes in the microenvironment.
The phenomenon of Epithelial-Mesenchymal-transition (EMT) has been implied as a determinant of the invasiveness of solid tumors (41–43). Mesenchymal characteristics are responsible for the invasive ability of breast cancer cells (44–46). Findings from our laboratory have indicated that loss of DNAJB6 expression is concurrent with tumor progression and may contribute to mesenchymal like properties (11, 25). Our work presented here identifies miR-632 as a potential negative regulator of DNAJB6 expression. Overexpression of miR-632 in MCF10AT lead to increased invasion, conversely, silencing miR-632 reduced invasive ability and mesenchymal markers. These observations are consistent with its action of reducing DNAJB6 expression. Our observations show that miR-632 is capable of silencing both spliced variants of DNAJB6. However, the extent of silencing is variable and this may reflect as differential interactions of miR-632 with the spliced variants. Interestingly immunohistochemical analysis of DNAJB6 in IDC grade III and in lymph node metastasis of breast cancer shows a total loss of DNAJB6 protein signal, which is expected if both the spliced variants are abrogated. There could be multiple negative regulators of DNAJB6 expression that may be responsible for abrogating DNAJB6 expression and based on observation presented here, we propose that miR-632 is one of the negative regulators of DNAJB6 expression.
Supplementary Material
Acknowledgements
Grant support: USPHS grants CA140472 (R. S. Samant) and CA138850 (L. A. Shevde).
J. Rostas is a recipient of the American Medical Association Seed Grant 2011.
R. S. Samant is the recipient of the Mayer Mitchell Award for Excellence in Cancer Research and acknowledges the support.
References
- 1.Jemal A, Siegel R, Xu J, et al. Cancer statistics. CA Cancer J Clin. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5(10):827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dumont N, Tlsty TD. Reflections on miR-ing effects in metastasis. Cancer Cell. 2009;16(1):3–4. doi: 10.1016/j.ccr.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 5.Rameshwar P. Potential novel targets in breast cancer. Curr Pharm Biotechnol. 2009;10(2):148–153. doi: 10.2174/138920109787315024. [DOI] [PubMed] [Google Scholar]
- 6.Wright JA, Richer JK, Goodall GJ. microRNAs and EMT in mammary cells and breast cancer. J Mammary Gland Biol Neoplasia. 15(2):213–223. doi: 10.1007/s10911-010-9183-z. [DOI] [PubMed] [Google Scholar]
- 7.O'Day E, Lal A. MicroRNAs and their target gene networks in breast cancer. Breast Cancer Res. 12(2):201. doi: 10.1186/bcr2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu Z, Baserga R, Chen L, et al. microRNA, cell cycle, and human breast cancer. Am J Pathol. 176(3):1058–1064. doi: 10.2353/ajpath.2010.090664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66(15):7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 10.Mitra A, Shevde LA, Samant RS. Multi-faceted role of HSP40 in cancer. Clin Exp Metastasis. 2009;26(6):559–567. doi: 10.1007/s10585-009-9255-x. [DOI] [PubMed] [Google Scholar]
- 11.Mitra A, Fillmore RA, Metge BJ, et al. Large isoform of MRJ (DNAJB6) reduces malignant activity of breast cancer. Breast Cancer Res. 2008;10(2):R22. doi: 10.1186/bcr1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunter PJ, Swanson BJ, Haendel MA, et al. Mrj encodes a DnaJ-related co-chaperone that is essential for murine placental development. Development. 1999;126(6):1247–1258. doi: 10.1242/dev.126.6.1247. [DOI] [PubMed] [Google Scholar]
- 13.Dey S, Banerjee P, Saha P. Cell cycle specific expression and nucleolar localization of human J-domain containing co-chaperone Mrj. Mol Cell Biochem. 2009;322(1–2):137–142. doi: 10.1007/s11010-008-9950-y. [DOI] [PubMed] [Google Scholar]
- 14.Sarparanta J, Jonson PH, Golzio C, et al. Mutations affecting the cytoplasmic functions of the co-chaperone DNAJB6 cause limb-girdle muscular dystrophy. Nat Genet. 44(4):450–455. doi: 10.1038/ng.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harms MB, Sommerville RB, Allred P, et al. Exome sequencing reveals DNAJB6 mutations in dominantly-inherited myopathy. Ann Neurol. 71(3):407–416. doi: 10.1002/ana.22683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Bock CE, Lin Z, Mekkawy AH, et al. Interaction between urokinase receptor and heat shock protein MRJ enhances cell adhesion. Int J Oncol. 36(5):1155–1163. doi: 10.3892/ijo_00000598. [DOI] [PubMed] [Google Scholar]
- 17.Fayazi Z, Ghosh S, Marion S, et al. A Drosophila ortholog of the human MRJ modulates polyglutamine toxicity and aggregation. Neurobiol Dis. 2006;24(2):226–244. doi: 10.1016/j.nbd.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 18.Chuang JZ, Zhou H, Zhu M, et al. Characterization of a brain-enriched chaperone, MRJ, that inhibits Huntingtin aggregation and toxicity independently. J Biol Chem. 2002;277(22):19831–19838. doi: 10.1074/jbc.M109613200. [DOI] [PubMed] [Google Scholar]
- 19.Durrenberger PF, Filiou MD, Moran LB, et al. DnaJB6 is present in the core of Lewy bodies and is highly up-regulated in parkinsonian astrocytes. J Neurosci Res. 2009;87(1):238–245. doi: 10.1002/jnr.21819. [DOI] [PubMed] [Google Scholar]
- 20.Menezes ME, Mitra A, Shevde LA, et al. DNAJB6 governs a novel regulatory loop determining Wnt/beta-catenin signaling activity. Biochem J. doi: 10.1042/BJ20120205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30(3):256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 22.Morrow KA, Das S, Metge BJ, et al. Loss of Tumor Suppressor Merlin in Advanced Breast Cancer Is due to Post-translational Regulation. J Biol Chem. 286(46):40376–40385. doi: 10.1074/jbc.M111.250035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris LG, Pannell LK, Singh S, et al. Increased vascularity and spontaneous metastasis of breast cancer by hedgehog signaling mediated upregulation of cyr61. Oncogene. doi: 10.1038/onc.2011.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das S, Harris LG, Metge BJ, et al. The hedgehog pathway transcription factor GLI1 promotes malignant behavior of cancer cells by up-regulating osteopontin. J Biol Chem. 2009;284(34):22888–22897. doi: 10.1074/jbc.M109.021949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitra A, Menezes ME, Shevde LA, et al. DNAJB6 induces degradation of beta-catenin and causes partial reversal of mesenchymal phenotype. J Biol Chem. 285(32):24686–24694. doi: 10.1074/jbc.M109.094847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guarino M, Rubino B, Ballabio G. The role of epithelial-mesenchymal transition in cancer pathology. Pathology. 2007;39(3):305–318. doi: 10.1080/00313020701329914. [DOI] [PubMed] [Google Scholar]
- 27.Jacks T, Weinberg RA. Taking the study of cancer cell survival to a new dimension. Cell. 2002;111(7):923–925. doi: 10.1016/s0092-8674(02)01229-1. [DOI] [PubMed] [Google Scholar]
- 28.Zahir N, Lakins JN, Russell A, et al. Autocrine laminin-5 ligates alpha6beta4 integrin and activates RAC and NFkappaB to mediate anchorage-independent survival of mammary tumors. J Cell Biol. 2003;163(6):1397–1407. doi: 10.1083/jcb.200302023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kenny PA, Lee GY, Myers CA, et al. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol Oncol. 2007;1(1):84–96. doi: 10.1016/j.molonc.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 32.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449(7163):682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 33.Hurst DR, Edmonds MD, Welch DR. Metastamir: the field of metastasis-regulatory microRNA is spreading. Cancer Res. 2009;69(19):7495–7498. doi: 10.1158/0008-5472.CAN-09-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pfeffer U, Romeo F, Noonan DM, et al. Prediction of breast cancer metastasis by genomic profiling: where do we stand? Clin Exp Metastasis. 2009;26(6):547–558. doi: 10.1007/s10585-009-9254-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi M, Liu D, Duan H, et al. Metastasis-related miRNAs, active players in breast cancer invasion, and metastasis. Cancer Metastasis Rev. 29(4):785–799. doi: 10.1007/s10555-010-9265-9. [DOI] [PubMed] [Google Scholar]
- 36.Andorfer CA, Necela BM, Thompson EA, et al. MicroRNA signatures: clinical biomarkers for the diagnosis and treatment of breast cancer. Trends Mol Med. 17(6):313–319. doi: 10.1016/j.molmed.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 37.Greene SB, Herschkowitz JI, Rosen JM. Small players with big roles: microRNAs as targets to inhibit breast cancer progression. Curr Drug Targets. 11(9):1059–1073. doi: 10.2174/138945010792006762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heneghan HM, Miller N, Kerin MJ. MiRNAs as biomarkers and therapeutic targets in cancer. Curr Opin Pharmacol. 10(5):543–550. doi: 10.1016/j.coph.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 39.Erdogan B, Facey C, Qualtieri J, et al. Diagnostic microRNAs in myelodysplastic syndrome. Exp Hematol. 39(9):915–926. e912. doi: 10.1016/j.exphem.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 40.Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen DX, Bos PD, Massague J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9(4):274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 42.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 43.May CD, Sphyris N, Evans KW, et al. Epithelial-mesenchymal transition and cancer stem cells: a dangerously dynamic duo in breast cancer progression. Breast Cancer Res. 13(1):202. doi: 10.1186/bcr2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Creighton CJ, Chang JC, Rosen JM. Epithelial-mesenchymal transition (EMT) in tumor-initiating cells and its clinical implications in breast cancer. J Mammary Gland Biol Neoplasia. 15(2):253–260. doi: 10.1007/s10911-010-9173-1. [DOI] [PubMed] [Google Scholar]
- 45.Blick T, Widodo E, Hugo H, et al. Epithelial mesenchymal transition traits in human breast cancer cell lines. Clin Exp Metastasis. 2008;25(6):629–642. doi: 10.1007/s10585-008-9170-6. [DOI] [PubMed] [Google Scholar]
- 46.Micalizzi DS, Farabaugh SM, Ford HL. Epithelial-mesenchymal transition in cancer: parallels between normal development and tumor progression. J Mammary Gland Biol Neoplasia. 15(2):117–134. doi: 10.1007/s10911-010-9178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.