Abstract
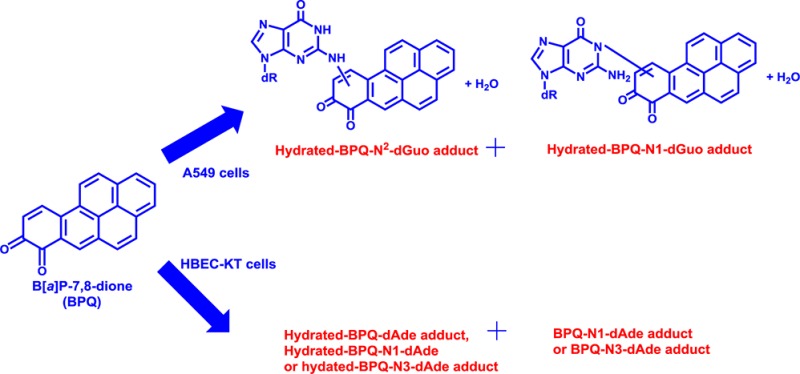
Metabolic activation of the proximate carcinogen benzo[a]pyrene-7,8-trans-dihydrodiol (B[a]P-7,8-trans-dihydrodiol) by aldo-keto reductases (AKRs) leads to B[a]P-7,8-dione that is both electrophilic and redox-active. B[a]P-7,8-dione generates reactive oxygen species resulting in oxidative DNA damage in human lung cells. However, information on the formation of stable B[a]P-7,8-dione-DNA adducts in these cells is lacking. We studied stable DNA adduct formation of B[a]P-7,8-dione in human lung adenocarcinoma A549 cells, human bronchoalveolar H358 cells, and immortalized human bronchial epithelial HBEC-KT cells. After treatment with 2 μM B[a]P-7,8-dione, the cellular DNA was extracted from the cell pellets subjected to enzyme hydrolysis and subsequent analysis by LC-MS/MS. Several stable DNA adducts of B[a]P-7,8-dione were only detected in A549 and HBEC-KT cells. In A549 cells, the structures of stable B[a]P-7,8-dione-DNA adducts were identified as hydrated-B[a]P-7,8-dione-N2-2′-deoxyguanosine and hydrated-B[a]P-7,8-dione-N1-2′-deoxyguanosine. In HBEC-KT cells, the structures of stable B[a]P-7,8-dione-DNA adducts were identified as hydrated-B[a]P-7,8-dione-2′-deoxyadenosine, hydrated-B[a]P-7,8-dione-N1- or N3-2′-deoxyadenosine, and B[a]P-7,8-dione-N1- or N3-2′-deoxyadenosine. In each case, adduct structures were characterized by MSn spectra. Adduct structures were also compared to those synthesized from reactions of B[a]P-7,8-dione with either deoxyribonucleosides or salmon testis DNA in vitro but were found to be different.
Introduction
Polycyclic aromatic hydrocarbons (PAHs) are ubiquitous environmental pollutants and suspect human carcinogens.1 PAHs are products of incomplete combustion and are emitted into the air we breathe, they are also present in first-hand and second hand smoke, and many are lung tumorigens in experimental animals.2 PAHs themselves are biologically inert, and their carcinogenic effects require metabolic activation to biologically reactive intermediates, which covalently modify deoxyribonucleic acid (DNA), to form DNA adducts that result in mutation.3 Three pathways of metabolic activation of benzo[a]pyrene (B[a]P), a representative PAH, have been proposed as the formation of radical cations,4 the formation of anti-diol-epoxides,5−7 and the formation of ortho-quinones.8,9
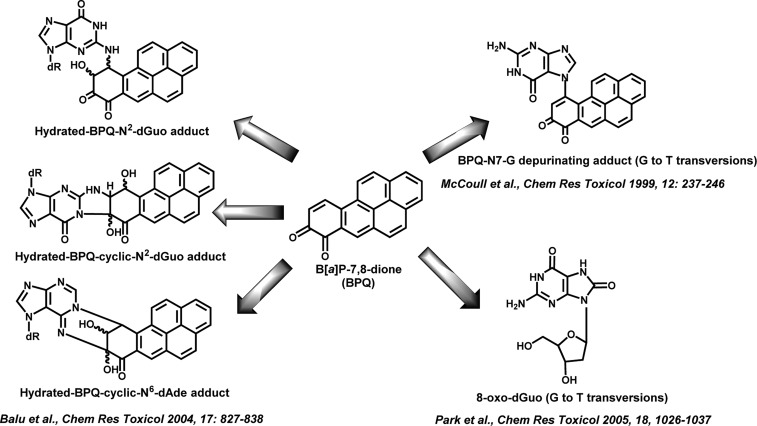
In the third pathway, benzo[a]pyrene-7,8-dione (B[a]P-7,8-dione), is produced by the oxidation of (±)-B[a]P-7,8-trans-dihydrodiol, which is catalyzed by aldo-keto reductases (AKRs).8,9 B[a]P-7,8-dione is both electrophilic and redox-active. The electrophilic B[a]P-7,8-dione can undergo 1,4- or 1,6-Michael addition with glutathione (GSH), and N-acetyl-l-cysteine (NAC) in vitro.10 It can also react with deoxyguanosine and deoxyadenosine to form a hydrated N2-deoxyguanosine adduct, a hydrated cyclic N2-deoxyguanosine adduct, a hydrated cyclic N6-deoxyadenosine adduct, or a N7-guanine depurinating adduct in vitro (Figure 1).11−14 B[a]P-7,8-dione can also be enzymatically and nonenzymatically reduced back to the catechol and can establish futile redox cycles that result in the amplification of reactive oxygen species (ROS) until cellular reducing equivalents (e.g., NADPH) are depleted.15,16 ROS formation can give rise to the mutagenic lesion 8-oxo-2′-deoxyguanosine (8-oxo-dGuo) in A549 and H358 cells (Figure 1).17,18 Both the N7-guanine depurinating adduct and 8-oxo-dGuo can give rise to G to T transversions, which are the dominant mutations found in K-ras and p53 in lung cancer.
Figure 1.
DNA adducts formed from B[a]P-7,8-dione and their mutations.
Human lung cells are major sites of inhalation exposure to B[a]P, and generation of the reactive metabolite B[a]P-7,8-dione from B[a]P has been previously demonstrated in H358 cells.19,20 The downstream metabolism of B[a]P-7,8-dione in human lung cells has also been reported.21 However, to our knowledge, information relevant to the formation of stable B[a]P-7,8-dione-DNA adducts in any human cell line is lacking. The objective of this study was to identify the stable B[a]P-7,8-dione-DNA adducts in human lung cells. A549 (human lung adenocarcinoma cells), H358 (human bronchoalveolar cells), and HBEC-KT (immortalized human bronchial epithelial cells) were selected as three human lung cell lines to investigate the comparative profiles of stable B[a]P-7,8-dione-DNA adducts.
We found that several stable B[a]P-7,8-dione-DNA adducts could be detected in A549 and HBEC-KT cells. In each case, adduct structures were characterized by MSn spectra. We found that A549 cells produced predominately 2′-deoxyguanosine (dGuo) adducts, and HBEC-KT cells produced predominately 2′-deoxyadenosine (dAde) adducts. Adduct structures were also compared to those synthesized from reactions of B[a]P-7,8-dione with dGuo, dAde and salmon testis DNA in vitro but were found to be different.
Materials and Methods
Caution:
All PAHs are potentially hazardous and should be handled in accordance with the NIH Guidelines for the Laboratory Use of Chemical Carcinogens.
Chemicals and Reagents
Cell culture medium and reagents were all obtained from Invitrogen Co. (Carlsbad, CA) except for fetal bovine serum (FBS) which was purchased from Hyclone (Logan, UT). B[a]P-7,8-Dione was synthesized according to published methods.22 2′-Deoxyguanosine (dGuo), 2′-deoxyadenosine (dAde), salmon testis DNA, DNase, nuclease P1, and shrimp alkaline phosphatase (SAP) were purchased from Sigma-Aldrich Co. (St. Louis, MO). All other chemicals used were of the highest grade available, and all solvents were HPLC grade.
Cell Culture
A549 (human lung adenocarcinoma) cells were obtained from American Type Culture Collection (ATCC #CCL-185) and maintained in F-12K nutrient mixture (Kaighn’s modification) with 10% heat-inactivated FBS, 2 mM l-glutamine, 100 units/mL penicillin, and 100 μg/mL streptomycin. H358 (human bronchoalveolar) cells were obtained from the American Type Culture Collection (ATCC #CRL-5807) and maintained in RPMI 1640 nutrient mixture with 10% heat-inactivated FBS, 2 mM l-glutamine, 100 units/mL penicillin, and 100 μg/mL streptomycin. HBEC-KT (immortalized human bronchial epithelial) cells originated from a patient without lung cancer were a gift from Dr. John Minna at University of Texas Southwestern Medical Center and maintained in keratinocyte serum free medium with 0.1–0.2 ng/mL recombinant epidermal growth factor (rEGF), 20–30 μg/mL bovine pituitary extract (BPE), and 2 mM l-glutamine. Cells were incubated at 37 °C in a humidified atmosphere containing 5% CO2 and were passaged every 3 days at a 1:6 dilution. Cultured cells with a passage number of 10–20 were used in the experiments to reduce variability due to cell culture conditions.
Identification of DNA Adducts of B[a]P-7,8-dione in Human Lung Cells
The cells (∼5 × 106) were treated with B[a]P-7,8-dione (2 μM, 0.2% DMSO) in HBSS buffer containing 1 mM sodium pyruvate as an energy source. The cell pellets were collected by scraping at 0 and 24 h. The DNA from the cell pellets was extracted with a Wako DNA extraction kit (Wako Chemicals USA, Inc., Richmond, VA) and was subsequently dissolved in 300 μL of 20 mM phosphate buffer at pH 7.0.
The solution containing cellular DNA (300 μL) was subjected to enzyme hydrolysis. Thirty microliters of 0.1 M MgCl2 and 400 units of DNase was added, followed by 1 h of incubation at 37 °C. A 5 μL solution containing 50 mM NaOAc and 5 mM ZnCl2, pH 5.0, was then added along with 7.5 units of nuclease P1. The sample was incubated for 1.5 h at 37 °C. At the end of the incubation, 40 μL of 0.5 M Tris-HCl and 50 mM MgCl2, pH 8.5, was added along with 20 units of shrimp alkaline phosphatase (SAP), and the sample was incubated for 1.5 h at 37 °C. Ethanol (900 μL) was added to the digestion solution, and the sample was chilled at 0 °C for 10 min. The sample solution was centrifuged at 14000 rpm for 10 min at 4 °C. The supernatant was transferred to another tube and evaporated under vacuum. The residue was redissolved in 100 μL methanol plus H2O (1:1) and analyzed by the ion trap LC-MS/MS. The injection volume was 20 μL. At the end of digestion, RP-HPLC showed that the digestion was complete since only the four deoxyribonucleosides were detected.
Data were acquired using a Waters Alliance 2690 HPLC system (Waters Corporation) coupled to a Finnigan LTQ spectrometer (Thermo Fisher Scientific, San Jose, CA). Separations were accomplished on a reverse-phase (RP) column (Zorbax-ODS C18, 5 μm, 4.6 mm × 250 mm) (DuPont Co., Wilmington, DE) with a guard column at ambient temperature. The mobile phase consisted of 5 mM ammonium acetate and 0.02% formic acid (v/v) in H2O (solvent A) and acetonitrile (solvent B) and was delivered at a flow rate of 0.5 mL/min. The linear gradient elution program was as follows: 20% to 80% B over 40 min, 80% to 95% B over 1 min, followed by an isocratic hold at 95% B for another 4 min. At 45 min, B was returned to 20% in 1 min, and the column was equilibrated for 14 min before the next injection. The total run time for each analysis was 60 min. During LC-MS/MS analysis, up to 6 min of the initial flow was diverted away from the mass spectrometer before the evaluation of eluants. The mass spectrometer was operated in the positive ion mode with an electrospray ionization (ESI) source. Eluants were monitored on the LTQ using product ion scan (MS2) and subsequent MS/MS/MS (MS3) modes. The mass spectrometry parameters, including spray voltage (4 kV), sheath gas flow rate (35 arbitrary units), auxiliary gas flow rate (18 arbitrary units), capillary temperature (220 °C), capillary voltage (9 V), and tube lens (80 V), were automatically optimized with a B[a]P-7,8-dione standard solution in methanol. An isolation width of three bracketed around the m/z of interest, activation Q of 0.25, and activation time of 30 ms were used for data acquisition. Xcalibur software, version 2.0 (Thermo Fisher Scientific), was used to control the LC-MS/MS system and to process data.
Synthesis of B[a]P-7,8-Dione-dGuo and B[a]P-7,8-Dione-dAde Adducts
The B[a]P-7,8-dione-dGuo and B[a]P-7,8-dione-dAde adducts were synthesized according to the method reported previously13 and subsequently analyzed by ion trap LC-MS/MS. The identity of the hydrated-B[a]P-7,8-dione-N2-dGuo, hydrated-B[a]P-7,8-dione-cyclic N2-dGuo and hydrated-B[a]P-7,8-dione-cyclic N6-dAde adducts were validated using pseudo-selected reaction monitoring (SRM) and MS2 modes in the same manner as that described above.
Synthesis of DNA Adducts of B[a]P-7,8-Dione in Salmon Testis DNA
Incubations of B[a]P-7,8-dione with salmon testis DNA were performed in a final volume of 0.3 mL containing 25 μg of B[a]P-7,8-dione, 75 μg of salmon testis DNA, and 20 mM phosphate buffer at pH 7.0. Reactions were performed at 37 °C for 24 h in the dark. Unreacted B[a]P-7,8-dione was removed by repetitive extraction with cold H2O-saturated ethyl acetate. The DNA pellet was precipitated from the aqueous layer and was subsequently hydrolyzed by stepwise addition of DNase, nuclease P1, and SAP. Ethanol was added to the digestion solution, and the supernatant was dried and subjected to the analysis by ion trap LC-MS/MS. The identity of the DNA adducts of B[a]P-7,8-dione were validated using pseudo-selected reaction monitoring (SRM), MS2, and subsequent MS3 modes in the same manner as that described above. At the end of digestion, RP-HPLC showed that the digestion was complete since only the four deoxyribonucleosides were detected.
Results
Strategy
A549, H358, and HBEC-KT cells were treated with 2 μM B[a]P-7,8-dione to detect and identify the potential DNA adducts using ion trap LC-MS/MS. Previous disposition studies with radiolabeled 1,3-[3H2]-B[a]P-7,8-dione in these three cell lines indicated that there was incorporation of radioactivity into the cell pellet that reached a plateau at 6 h and then stayed constant for up to 24 h. Subfractionation of the cell pellet indicated that the radioactivity was in the protein and DNA fractions with the largest portion being covalently bound to protein.21 Thus, in our studies we elected to detect covalent DNA adducts following a 24 h treatment with B[a]P-7,8-dione.
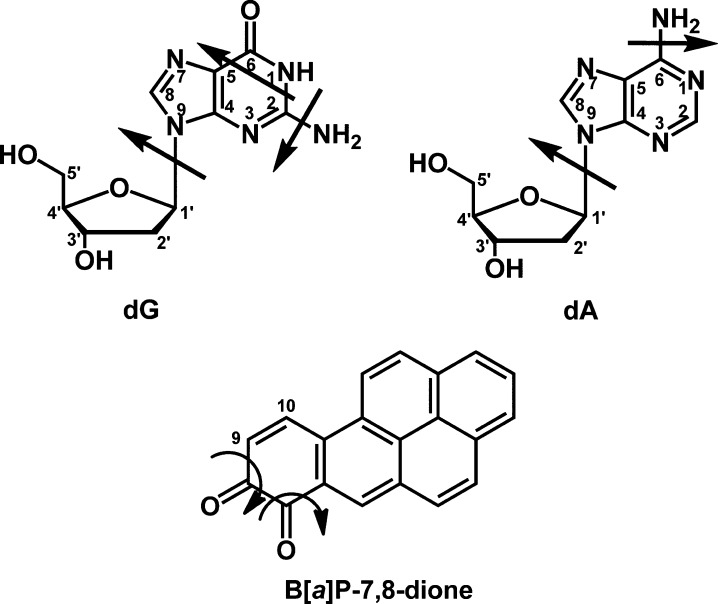
The structures of the potential DNA adducts were compared to authentic synthesized standards obtained by reacting B[a]P-7,8-dione with either deoxyribonucleosides as described by Balu, N. et al.13 or salmon testis DNA. The common fragmentation patterns of dGuo, dAde, and B[a]P-7,8-dione have been reported previously19,23−26 and are shown in Figure 2. The most generally characteristic ions in deoxyribonucleoside mass spectra in the positive ion mode are those representing the base: b (loss of 117 amu), b + H (loss of 116 amu), and b + 2H (loss of 115 amu).23,25,26 The loss of 115 amu results from the loss of dR with the abstraction of one hydrogen atom from the sugar to the base, to yield the aglycone ion. B[a]P-7,8-Dione can theoretically react with the N1, N2, or N3 positions of dGuo to yield three possible adducts, but addition by the N2-exocyclic amino group was expected. Similarly, B[a]P-7,8-dione can theoretically react with the N1, N3, or N6 positions of dAde to yield three possible adducts, but addition by the N6-exocyclic amino group was expected. Although covalent binding at the N7 positions of dGuo and dAde is also possible, these reactions would give rise to unstable depurinating adducts due to the cleavage of the N-glycosidic bond. On the basis of these known fragmentation patterns, the MSn spectra of potential DNA adducts can be interpreted to obtain more structural information as to their identity.
Figure 2.
Fragmentation patterns of dGuo, dAde and B[a]P-7,8-dione.
Identification of B[a]P-7,8-Dione-dGuo Adducts in A549 Cells
Hydrated-B[a]P-7,8-dione-dGuo adducts were detected in DNA digested from A549 cells following treatment with 2 μM B[a]P-7,8-dione for 24 h. Two peaks with retention times of 11.40 and 13.85 min were detected by monitoring the MS3 chromatograms (m/z 566→450→) at 0 h (Figure 3A) and 24 h (Figure 3B). The corresponding MS2 spectra (m/z 566) of these two adducts showed the loss of deoxyribose from the hydrated molecular ion (Figure 3C). The MS3 spectra (m/z 566→450→) of the adduct with a retention time of 11.40 min (Figure 3D) and the adduct with a retention time of 13.85 min (Figure 3E) showed reasonable fragmentation patterns from both the guanine nucleobase and B[a]P-7,8-dione (loss of CO). In particular, the loss of HNCO (43 amu) in Figure 3D and NH3 (17 amu) in Figure 3E are characteristic of the fragmentation of the guanine ring.24 For the adduct with a retention time of 11.40 min, the loss of HNCO (43 amu) shown in Figure 3D strongly indicates that the covalent binding did not occur at the N1 position of dGuo. Covalent binding at the N3 position of dGuo could also be ruled out since the loss of NH3 (17 amu) was not observed. Thus, Michael addition followed by hydration could account for the formation of the hydrated-B[a]P-7,8-dione-N2-dGuo adduct with a retention time of 11.40 min as expected. For the adduct with a retention time of 13.85 min, the loss of NH3 (17 amu) shown in Figure 3E strongly indicates that the covalent binding did not occur at the N2 exocyclic amino group of dGuo. Covalent binding at the N3 position of dGuo could also be ruled out since the loss of HNCO (43 amu) was not observed in Figure 3E. Thus, Michael addition by the enol-imidazole tautomer of guanine followed by hydration could account for the formation of the hydrated-B[a]P-7,8-dione-N1-dGuo adduct with a retention time of 13.85 min. The specific position by which guanine attacks B[a]P-7,8-dione and the specific position of B[a]P-7,8-dione hydration could not be ascertained based on mass spectrometry data only. On the basis of the known addition chemistry for B[a]P-7,8-dione,10 it is speculated that for both adducts guanine addition might occur at the C-10 position of B[a]P-7,8-dione followed by subsequent hydration of the quinone as proposed by N. Balu et al.13
Figure 3.
Hydrated-B[a]P-7,8-dione-dGuo adducts detected from A549 cell pellets. (A) MS3 chromatogram at 0 h. (B) MS3 chromatogram at 24 h. (C) MS2 spectrum. (D) MS3 spectrum at 11.40 min. (E) MS3 spectrum at 13.85 min. A549 cells (∼5 × 106) were treated with B[a]P-7,8-dione (2 μM, 0.2% (v/v) DMSO) in HBSS buffer containing 1 mM sodium pyruvate. The cell pellets were collected at 0 and 24 h. The DNA from cell pellets was extracted and subsequently hydrolyzed before analysis on an ion trap LC-MS/MS.
Identification of B[a]P-7,8-Dione-dAde Adducts in HBEC-KT Cells
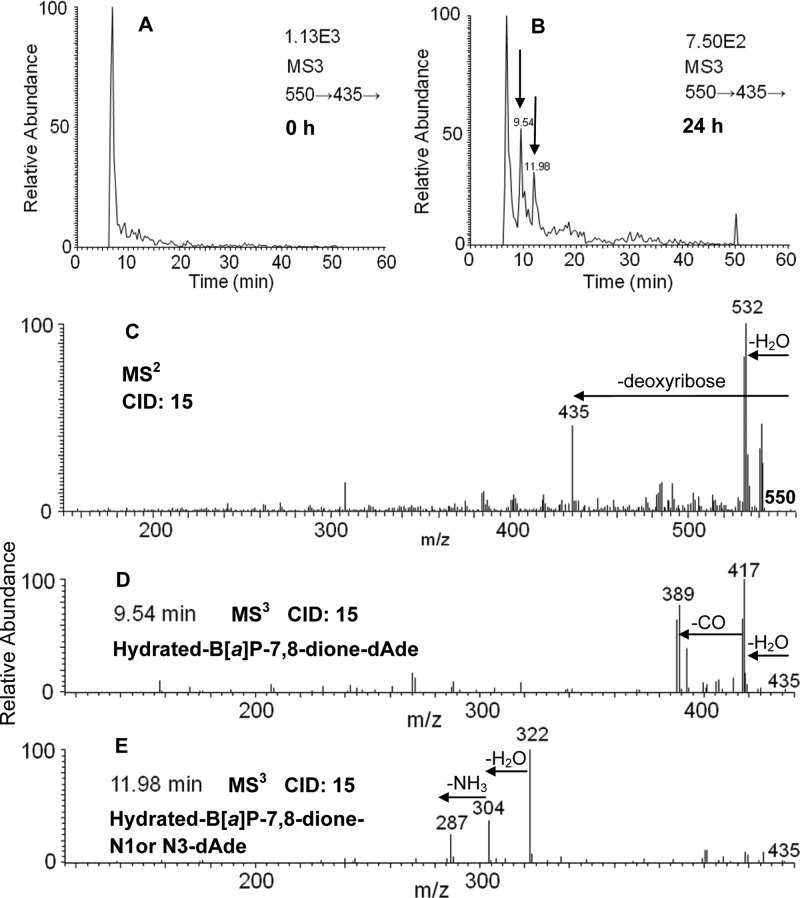
Hydrated-B[a]P-7,8-dione-dAde adducts were detected in DNA digested from HBEC-KT cells following treatment with 2 μM B[a]P-7,8-dione for 24 h. Two peaks with retention times of 9.54 and 11.98 min were detected by monitoring the MS3 chromatograms (m/z 550→435→) at 0 h (Figure 4A) and 24 h (Figure 4B). The corresponding MS2 spectra (m/z 550) of these two adducts showed the loss of deoxyribose from the hydrated molecular ion (Figure 4C). The MS3 spectra (m/z 550→435→) of the adduct with a retention time of 9.54 min (Figure 4D) and the adduct with a retention time of 11.98 min (Figure 4E) showed reasonable fragmentation patterns from the adenine nucleobase and B[a]P-7,8-dione (loss of CO). In particular, the loss of NH3 (17 amu) in Figure 4E is characteristic of the fragmentation of the adenine ring.24 The loss of NH3 (17 amu) shown in Figure 4E strongly indicates that the covalent binding did not occur at the N6 exocyclic amino group of dAde as expected. Thus, Michael addition followed by hydration could account for the formation of the hydrated-B[a]P-7,8-dione-N1- or N3-dAde adduct with a retention time of 11.98 min. The adduct with a retention time of 9.54 min gave no fragmentation patterns assignable to adenine in Figure 4D, and thus, this adduct could be a hydrated-B[a]P-7,8-dione-N1, N3, or N6-dAde adduct. The specific position by which adenine attacks B[a]P-7,8-dione and the specific position of B[a]P-7,8-dione hydration could not be ascertained based on mass spectrometry data only. On the basis of the known addition chemistry for B[a]P-7,8-dione,10 it is speculated that a 1,4-Michael addition of adenine might occur at the C-10 position of B[a]P-7,8-dione followed by subsequent hydration of the quinone.
Figure 4.
Hydrated-B[a]P-7,8-dione-dAde adducts detected from HBEC-KT cells. (A) MS3 chromatogram at 0 h. (B) MS3 chromatogram at 24 h. (C) MS2 spectrum. (D) MS3 spectrum at 9.54 min. (E) MS3 spectrum at 11.98 min. HBEC-KT cells (∼5 × 106) were treated with B[a]P-7,8-dione (2 μM, 0.2% (v/v) DMSO) in HBSS buffer containing 1 mM sodium pyruvate. The cell pellets were collected at 0 and 24 h. The DNA from cell pellets was extracted and subsequently hydrolyzed before analysis on an ion trap LC-MS/MS.
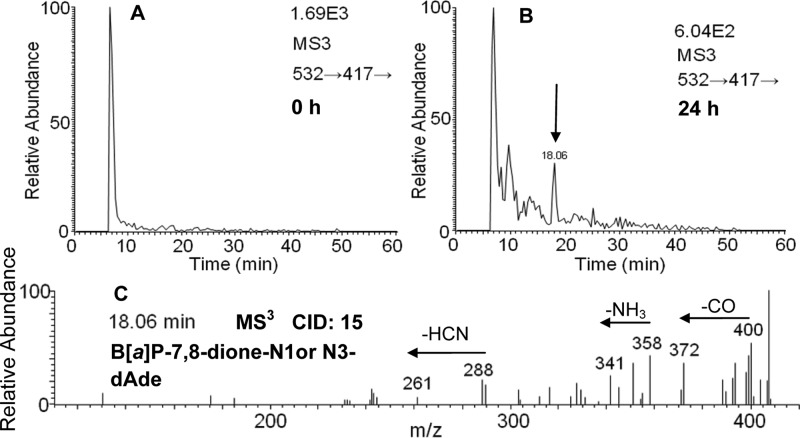
Comparison of MS3 chromatograms (m/z 532→417→) of the digested DNA from HBEC-KT cells at 0 h (Figure 5A) and 24 h (Figure 5B) showed that one peak with a retention time of 18.06 min was a potential B[a]P-7,8-dione-dAde adduct. The corresponding MS3 spectra (m/z 532→417→) of this adduct (Figure 5C) showed reasonable fragmentation patterns from both B[a]P-7,8-dione (loss of CO) and adenine. In particular, the loss of NH3 (17 amu) and HCN (27 amu) in Figure 5C is characteristic of the fragmentation of the adenine ring.24 The loss of NH3 (17 amu) shown in Figure 5C strongly indicates that the covalent binding did not occur at the N6 position of dAde. Thus, Michael addition in the absence of hydration likely accounts for the formation of the B[a]P-7,8-dione-N1- or N3-dAde adduct with a retention time of 18.06 min. The specific position by which adenine attacks B[a]P-7,8-dione could not be ascertained based on mass spectrometry data only. On the basis of the known addition chemistry for B[a]P-7,8-dione,10 it is speculated that a 1,4-Michael addition of adenine might occur at the C-10 position of B[a]P-7,8-dione.
Figure 5.
B[a]P-7,8-dione-dAde adducts detected from HBEC-KT cells. (A) MS3 chromatogram at 0 h. (B) MS3 chromatogram at 24 h. (C) MS3 spectrum at 18.06 min. HBEC-KT cells (∼5 × 106) were treated with B[a]P-7,8-dione (2 μM, 0.2% (v/v) DMSO) in HBSS buffer containing 1 mM sodium pyruvate. The cell pellets were collected at 0 and 24 h. The DNA from cell pellets was extracted and subsequently hydrolyzed before analysis on an ion trap LC-MS/MS.
Synthesis of Adducts by Reacting B[a]P-7,8-Dione with Deoxyribonucleosides
B[a]P-7,8-Dione was reacted with dGuo under conditions similar to those previously published,13 and three peaks with the retention time of 17.34, 17.61, and 19.69 min were observed corresponding to the mass transition of a protonated hydrated-B[a]P-7,8-dione-dGuo (m/z 566) undergoing the loss of deoxyribose (Figure S-1A, Supporting Information). Comparison of the fragmentation patterns of these peaks with the corresponding hydrated-B[a]P-7,8-dione-dGuo adducts reported previously13 showed a good concordance (Table 1). The structures of these adducts were previously confirmed by nuclear magnetic resonance (NMR)13 and are shown in Figure S-1A (Supporting Information). However, these adducts were different from stable B[a]P-7,8-dione-dGuo adducts detected in A549 cells based on the retention times (Table 1).
Table 1. Mass Spectrometric Properties of Stable DNA Adducts of B[a]P-7,8-Dione.
| reaction system | B[a]P-7,8-dione-dGuo | B[a]P-7,8-dione-dAde |
|---|---|---|
| dN | hydrated-B[a]P-7,8-dione-dGuo (17.34 min, 17.61 min, 19.69 min, 566 [M+H]+, 450 [M+H-deoxyribose]+) | hydrated-B[a]P-7,8-dione-dAde (18.69 min, 550 [M+H]+, 434 [M+H-deoxyribose]+) |
| salmon testis DNA | hydrated-B[a]P-7,8-dione-N3-dGuo (13.93 min, 566 [M+H]+, 451 [M+2H-deoxyribose]+), B[a]P-7,8-dione-dGuo (19.11 min, 548 [M+H]+, 433 [M+2H-deoxyribose]+) | hydrated-B[a]P-7,8-dione-N1 or N3-dAde (10.29 min, 13.79 min, 550 [M+H]+, 435 [M+2H-deoxyribose]+), B[a]P-7,8-dione-dAde (20.54 min, 532 [M+H]+, 417 [M+2H-deoxyribose]+) |
| A549 | hydrated-B[a]P-7,8-dione-N2-dGuo (11.40 min, 566 [M+H]+, 450 [M+H-deoxyribose]+), hydrated-B[a]P-7,8-dione-N1-dGuo (13.85 min, 566 [M+H]+, 450 [M+H-deoxyribose]+) | not detected |
| HBEC-KT | not detected | hydrated-B[a]P-7,8-dione-dAde (9.54 min, 550 [M+H]+, 435 [M+2H-deoxyribose]+), hydrated-B[a]P-7,8-dione-N1 or N3-dAde (11.98 min, 550 [M+H]+, 435 [M+2H-deoxyribose]+), B[a]P-7,8-dione-N1 or N3-dAde (18.06 min, 532 [M+H]+, 417 [M+2H-deoxyribose]+) |
B[a]P-7,8-Dione was also reacted with dAde under conditions similar to those previously published,13 and one peak with a retention time of 18.69 min was observed corresponding to the mass transition of a protonated hydrated-B[a]P-7,8-dione-dAde (m/z 550) undergoing the loss of deoxyribose (Figure S-1B, Supporting Information). Comparison of the fragmentation patterns of this peak with the corresponding hydrated-B[a]P-7,8-dione-dAde adducts reported previously13 showed a good concordance (Table 1). The structure of this adduct was previously confirmed by NMR13 and is shown in Figure S-1B (Supporting Information). However, this adduct was different from stable B[a]P-7,8-dione-dAde adducts detected in HBEC-KT cells based on the retention times (Table 1).
Detection of DNA Adducts by Reacting B[a]P-7,8-Dione with Salmon Testis DNA
Comparison of MS3 chromatograms (m/z 566→450→) of the digested DNA obtained from the reaction system of B[a]P-7,8-dione with salmon testis DNA at 0 and 24 h showed that one peak with a retention time of 13.93 min was a potential hydrated-B[a]P-7,8-dione-dGuo adduct (Figure S-2, Supporting Information). The MS3 spectra (m/z 566→450→) of this adduct showed reasonable fragmentation patterns from both guanine and B[a]P-7,8-dione (Figure S-2, Supporting Information). The loss of NH3 (17 amu) indicates that the covalent binding did not occur at the N2 position of dGuo. The loss of HNCO (43 amu) indicates that the covalent binding did not occur at the N1 position of dGuo. Thus, Michael addition followed by hydration could account for the formation of the hydrated-B[a]P-7,8-dione-N3-dGuo adduct with a retention time of 13.93 min. Comparison of pseudo-SRM chromatograms (m/z 548→432) of the digested DNA obtained from the reaction system of B[a]P-7,8-dione with salmon testis DNA at 0 and 24 h showed that one peak with a retention time of 19.11 min was a potential B[a]P-7,8-dione-dGuo adduct (Figure S-3, Supporting Information). The specific position by which guanine attacks B[a]P-7,8-dione and the specific position of B[a]P-7,8-dione hydration could not be ascertained based on mass spectrometry data only. These dGuo adducts were different from stable B[a]P-7,8-dione-dGuo adducts detected in A549 cells based on retention time (Table 1).
Comparison of MS3 chromatograms (m/z 550→435→) of the digested DNA obtained from the reaction system of B[a]P-7,8-dione with salmon testis DNA at 0 and 24 h showed that two peaks with retention times of 10.29 and 13.79 min were potential hydrated-B[a]P-7,8-dione-dAde adducts (Figure S-4, Supporting Information). The MS3 spectra (m/z 550→435→) of these adducts showed reasonable fragmentation patterns from both adenine and B[a]P-7,8-dione (Figure S-4, Supporting Information). The loss of NH3 (17 amu) indicates that the covalent binding did not occur at the N6 position of dAde. Thus, Michael addition followed by hydration could account for the formation of the hydrated-B[a]P-7,8-dione-N1- or N3-dAde adduct with retention times of 10.29 and 13.79 min. Comparison of pseudo-SRM chromatograms (m/z 532→417) of the digested DNA obtained from the reaction system of B[a]P-7,8-dione with salmon testis DNA at 0 and 24 h showed that one peak with a retention time of 20.54 min was a potential B[a]P-7,8-dione-dAde adduct (Figure S-5, Supporting Information). The specific position by which adenine attacks B[a]P-7,8-dione and the specific position of B[a]P-7,8-dione hydration could not be ascertained based on mass spectrometry data only. These dAde adducts were different from stable B[a]P-7,8-dione-dAde adducts detected in HBEC-KT cells based on retention time (Table 1).
Discussion
This study reports the novel discovery of the stable DNA adducts of B[a]P-7,8-dione in two different human lung cell lines: A549 and HBEC-KT. B[a]P-7,8-dione is the signature metabolite of the AKR pathway of B[a]P activation and has been shown to be mutagenic in vitro solely by generating ROS.15 The covalent binding of electrophilic B[a]P-7,8-dione with DNA nucleobases was previously reported in vitro,11−14 but there was a failure to detect these adducts in the A/J mouse lung model of B[a]P carcinogenesis, when animals were treated with B[a]P, B[a]P-7,8-trans-dihydrodiol, or B[a]P-7,8-dione.27 This is the first report of the ability of B[a]P-7,8-dione to form in vitro stable DNA adducts in human lung cells.
The covalent binding of electrophilic B[a]P metabolites (e.g., anti-diol-epoxides or radical cations) to DNA has been found to be a key step in the initiation of cancer.2,5,15 Formation of dGuo adducts or depurinating adducts from these respective metabolites can lead to G to T transversions.11−14 Formation of stable B[a]P-7,8-dione-DNA adducts leads to a loss of electrophilicity, but the adducts are still capable of redox-cycling to produce oxidative stress and oxidative DNA damage. Identification of the stable B[a]P-7,8-dione-DNA adducts in human lung cells will allow their mutagenicity to be assessed in lesion bypass assays using site-specifically modified oligonucleotides.
After treatment of A549, H358, and HBEC-KT cells with radiolabeled 1,3-[3H2]-B[a]P-7,8-dione, it was found that a small amount of radioactivity was distributed in the DNA extracted from all cell pellets (data not shown), which is consistent with the previous finding of radioactivity disposition in isolated rat hepatocytes.28 Among the three human lung cells, the amount of radioactivity distributed in the DNA extracted from H358 cell pellets was less than the other two (data not shown). In the present study, the B[a]P-7,8-dione-DNA adducts were only detected in A549 and HBEC-KT cells and not H358 cells, which could be accounted for by a lower formation of B[a]P-7,8-dione-DNA adducts in H358 cells. The B[a]P-7,8-dione-dGuo adducts were only detected in A549 cells, whereas the B[a]P-7,8-dione-dAde adducts were only detected in HBEC-KT cells. We hypothesize that these different DNA-adduct profiles may reflect differences in nucleotide excision repair in the two cell lines.
In the present study, a hydrated-B[a]P-7,8-dione-N2-dGuo adduct was detected in A549 cells. However, the specific position by which guanine attacks B[a]P-7,8-dione and the specific position of B[a]P-7,8-dione hydration could not be ascertained based on mass spectrometry data only. It has been previously reported that the in vitro reaction of B[a]P-7,8-dione with dGuo generated hydrated-B[a]P-7,8-dione-N2-dGuo, with 1,4-Michael addition occurring at C-10 position of B[a]P-7,8-dione and hydration occurring across the C-9 and C-10 positions of B[a]P-7,8-dione.13 These previous experiments were replicated to compare the properties of the synthetic hydrated-B[a]P-7,8-dione-N2-dGuo adduct with that formed in A549 cells. It was found that these two adducts differed based on retention time. Whereas the synthetic hydrated-B[a]P-7,8-dione-N2-dGuo adduct had a retention time of 19.69 min, the adduct found in A549 cells was more polar and gave a retention time of 11.40 min. One explanation for this discrepancy is that the specific position of guanine attack to B[a]P-7,8-dione and the specific position of B[a]P-7,8-dione hydration in a cell-based system are different from those occurring in the in vitro reaction.
This is the first time that a hydrated-B[a]P-7,8-dione-N1-dGuo adduct was detected in A549 cells. Michael addition by the enol-imidazole tautomer of guanine followed by hydration could account for the formation of this hydrated-B[a]P-7,8-dione-N1-dGuo adduct. However, the specific position of guanine attack to B[a]P-7,8-dione and the specific position of B[a]P-7,8-dione hydration could not be ascertained based on mass spectrometry data only. It has been previously reported that a N1-dGuo quinone methide adduct was observed along with a N2-dGuo adduct and a N7-guanine adduct in duplex DNA following alkylation with quinone methides.29,30 It was found that the N1-dGuo adduct was preferentially formed with a quinolinyl methide in vitro and that this adduct is favored thermodynamically. In this instance, the adduct was fully characterized by 2D-NMR.31
B[a]P-7,8-dione-3′-mononucleotide adducts were formed in vitro by reacting B[a]P-7,8-dione with calf thymus DNA using [32P]-postlabeling as the detection method.14 When B[a]P-7,8-dione-3′-mononucleotide adducts were subjected to dephosphorylation, they corresponded to B[a]P-7,8-dione-nucleoside adducts that were formed by reacting B[a]P-7,8-dione with dGuo and dAde.14 However, not all of the adducts detected in calf thymus DNA could be identified by comigration with authentic synthetic standards. We also reported that a B[a]P-7,8-dione-adenine adduct detected in the media of human lung cells following B[a]P-7,8-dione treatment was derived from the cellular nucleotide pool rather than DNA.21 We find that none of the stable B[a]P-7,8-dione-DNA adducts detected in A549 and HBEC-KT cells correspond to those previously identified. These cellular B[a]P-7,8-dione-DNA adducts were different from those formed with either deoxyribonucleoside or salmon testis DNA.
Previously, B[a]P-7,8-dione was only found to be mutagenic in a yeast based system that reported p53 mutations when redox-cycling conditions were used.32 In those experiments, reaction of B[a]P-7,8-dione with p53 cDNA in vitro failed to form adducts that were mutagenic, and subsequent work focused on oxidative DNA lesions and their ability to cause mutation.32 It is likely that any covalent adducts that formed in these experiments would be similar to those observed in salmon testis DNA and differ from those formed in mammalian cells. This would suggest that it will be critical to obtain complete structural information on the cell based adducts so that their ability to cause mutations can be assessed.
In summary, we have identified the stable B[a]P-7,8-dione-DNA adducts in three human lung cells. The novel findings in this study are that several stable B[a]P-7,8-dione-DNA adducts were detected in A549 and HBEC-KT cells. In A549 cells, the structures of stable B[a]P-7,8-dione-DNA adducts were identified as hydrated-B[a]P-7,8-dione-N2-dGuo and hydrated-B[a]P-7,8-dione-N1-dGuo. In HBEC-KT cells, the structures of stable B[a]P-7,8-dione-DNA adducts were identified as hydrated-B[a]P-7,8-dione-dAde, hydrated-B[a]P-7,8-dione-N1- or N3-dAde, and B[a]P-7,8-dione-N1- or N3-dAde. In each case, adduct structures were characterized by MSn spectra. Adduct structures were also compared to those synthesized from reactions of B[a]P-7,8-dione with either deoxyribonucleosides or salmon testis DNA but were found to be different.
Glossary
Abbreviations
- AKR
aldo-keto reductase
- B[a]P
benzo[a]pyrene
- (+)-anti-B[a]PDE
(+)-7β,8α-dihydroxy-7,8,9,10-tetrahydro-9α,10α-oxo-benzo[a]pyrene
- (±)-B[a]P-7,8-trans-dihydrodiol
(−)-7β,8α-dihydroxy-7,8-dihydrobenzo[a]pyrene
- B[a]P-7,8-dione
benzo[a]pyrene-7,8-dione
- dAde
2′-deoxyadenosine
- dGuo
2′-deoxyguanosine
- DNA
deoxyribonucleic acid
- ESI
electrospray ionization
- FBS
fetal bovine serum
- HBSS
Hank’s balanced salt solution
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- 8-oxo-dGuo
8-oxo-2′-deoxyguanosine
- MH+
protonated molecular ion
- PAH
polycyclic aromatic hydrocarbon
- ROS
reactive oxygen species
- SAP
shrimp alkaline phosphatase
Supporting Information Available
Extracted ion chromatograms of pseudo-SRM transitions for hydrated-B[a]P-7,8-dione-dGuo adducts and hydrated-B[a]P-7,8-dione-dAde adducts prepared synthetically according to the method reported by Balu, N. et al.;12 MS3 chromatograms and spectra of the protonated hydrated-B[a]P-7,8-dione-dGuo adduct (m/z 566) and the protonated hydrated-B[a]P-7,8-dione-dAde adduct (m/z 550) prepared synthetically using B[a]P-7,8-dione and salmon testis DNA; extracted ion chromatograms of pseudo-SRM transitions for B[a]P-7,8-dione-dGuo adduct and B[a]P-7,8-dione-dAde adduct prepared synthetically using B[a]P-7,8-dione and salmon testis DNA at 0 h and 24 h. This material is available free of charge via the Internet at http://pubs.acs.org.
This work was supported by National Institutes of Health grants (1R01-CA39504 and P30-ES013508 to T.M.P.).
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Dipple A. (1983) Formation, metabolism, and mechanism of action of polycyclic aromatic hydrocarbons. Cancer Res. 43, 2422s–2425s. [PubMed] [Google Scholar]
- IARC (2010) Some Non-Heterocyclic Polycyclic Aromatic Hydrocarbons and Some Related Exposures, in IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, World Health Organization, Lyons, France. [PMC free article] [PubMed] [Google Scholar]
- Rothman N.; Poirier M. C.; Baser M. E.; Hansen J. A.; Gentile C.; Bowman E. D.; Strickland P. T. (1990) Formation of polycyclic aromatic hydrocarbon-DNA adducts in peripheral white blood cells during consumption of charcoal-broiled beef. Carcinogenesis 11, 1241–1243. [DOI] [PubMed] [Google Scholar]
- Cavalieri E. L.; Rogan E. G. (1995) Central role of radical cations in metabolic activation of polycyclic aromatic hydrocarbons. Xenobiotica 25, 677–688. [DOI] [PubMed] [Google Scholar]
- Conney A. H. (1982) Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons: G. H. A. Clowes Memorial Lecture. Cancer Res. 42, 4875–4917. [PubMed] [Google Scholar]
- Shimada T.; Gillam E. M.; Oda Y.; Tsumura F.; Sutter T. R.; Guengerich F. P.; Inoue K. (1999) Metabolism of benzo[a]pyrene to trans-7,8-dihydroxy-7,8-dihydrobenzo[a]pyrene by recombinant human cytochrome P450 1B1 and purified liver epoxide hydrolase. Chem. Res. Toxicol. 12, 623–629. [DOI] [PubMed] [Google Scholar]
- Shimada T.; Oda Y.; Gillam E. M.; Guengerich F. P.; Inoue K. (2001) Metabolic activation of polycyclic aromatic hydrocarbons and other procarcinogens by cytochromes P450 1A1 and P450 1B1 allelic variants and other human cytochromes P450 in Salmonella typhimurium NM2009. Drug Metab. Dispos. 29, 1176–1182. [PubMed] [Google Scholar]
- Palackal N. T.; Burczynski M. E.; Harvey R. G.; Penning T. M. (2001) The ubiquitous aldehyde reductase (AKR1A1) oxidizes proximate carcinogen trans-dihydrodiols to o-quinones: potential role in polycyclic aromatic hydrocarbon activation. Biochemistry 40, 10901–10910. [DOI] [PubMed] [Google Scholar]
- Palackal N. T.; Lee S. H.; Harvey R. G.; Blair I. A.; Penning T. M. (2002) Activation of polycyclic aromatic hydrocarbon trans-dihydrodiol proximate carcinogens by human aldo-keto reductase (AKR1C) enzymes and their functional overexpression in human lung carcinoma (A549) cells. J. Biol. Chem. 277, 24799–24808. [DOI] [PubMed] [Google Scholar]
- Murty V. S.; Penning T. M. (1992) Characterization of mercapturic acid and glutathionyl conjugates of benzo[a]pyrene-7,8-dione by two-dimensional NMR. Bioconjugate Chem. 3, 218–224. [DOI] [PubMed] [Google Scholar]
- Shou M.; Harvey R. G.; Penning T. M. (1993) Reactivity of benzo[a]pyrene-7,8-dione with DNA. Evidence for the formation of deoxyguanosine adducts. Carcinogenesis 14, 475–482. [DOI] [PubMed] [Google Scholar]
- McCoull K. D.; Rindgen D.; Blair I. A.; Penning T. M. (1999) Synthesis and characterization of polycyclic aromatic hydrocarbon o-quinone depurinating N7-guanine adducts. Chem. Res. Toxicol. 12, 237–246. [DOI] [PubMed] [Google Scholar]
- Balu N.; Padgett W. T.; Lambert G. R.; Swank A. E.; Richard A. M.; Nesnow S. (2004) Identification and characterization of novel stable deoxyguanosine and deoxyadenosine adducts of benzo[a]pyrene-7,8-quinone from reactions at physiological pH. Chem. Res. Toxicol. 17, 827–838. [DOI] [PubMed] [Google Scholar]
- Balu N.; Padgett W. T.; Nelson G. B.; Lambert G. R.; Ross J. A.; Nesnow S. (2006) Benzo[a]pyrene-7,8-quinone-3′-mononucleotide adduct standards for 32P postlabeling analyses: detection of benzo[a]pyrene-7,8-quinone-calf thymus DNA adducts. Anal. Biochem. 355, 213–223. [DOI] [PubMed] [Google Scholar]
- Penning T. M.; Burczynski M. E.; Hung C. F.; McCoull K. D.; Palackal N. T.; Tsuruda L. S. (1999) Dihydrodiol dehydrogenases and polycyclic aromatic hydrocarbon activation: generation of reactive and redox active o-quinones. Chem. Res. Toxicol. 12, 1–18. [DOI] [PubMed] [Google Scholar]
- Flowers-Geary L.; Bleczinki W.; Harvey R. G.; Penning T. M. (1996) Cytotoxicity and mutagenicity of polycyclic aromatic hydrocarbon ortho-quinones produced by dihydrodiol dehydrogenase. Chem.-Biol. Interact. 99, 55–72. [DOI] [PubMed] [Google Scholar]
- Park J. H.; Mangal D.; Tacka K. A.; Quinn A. M.; Harvey R. G.; Blair I. A.; Penning T. M. (2008) Evidence for the aldo-keto reductase pathway of polycyclic aromatic trans-dihydrodiol activation in human lung A549 cells. Proc. Natl. Acad. Sci. U.S.A. 105, 6846–6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangal D.; Vudathala D.; Park J. H.; Lee S. H.; Penning T. M.; Blair I. A. (2009) Analysis of 7,8-dihydro-8-oxo-2’-deoxyguanosine in cellular DNA during oxidative stress. Chem. Res. Toxicol. 22, 788–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H.; Gelhaus S. L.; Mangal D.; Harvey R. G.; Blair I. A.; Penning T. M. (2007) Metabolism of benzo[a]pyrene in human bronchoalveolar H358 cells using liquid chromatography-mass spectrometry. Chem. Res. Toxicol. 20, 1331–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D.; Harvey R. G.; Blair I. A.; Penning T. M. (2011) Quantitation of benzo[a]pyrene metabolic profiles in human bronchoalveolar (H358) cells by stable isotope dilution liquid chromatography-atmospheric pressure chemical ionization mass spectrometry. Chem. Res. Toxicol. 24, 1905–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M.; Liu X.; Basu S. S.; Zhang L.; Kushman M. E.; Harvey R. G.; Blair I. A.; Penning T. M. (2012) Metabolism and distribution of benzo[a]pyrene-7,8-dione (B[a]P-7,8-dione) in human lung cells by liquid chromatography tandem mass spectrometry: detection of an adenine B[a]P-7,8-dione adduct. Chem. Res. Toxicol. 25, 993–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey R. G.; Dai Q.; Ran C.; Penning T. M. (2004) Synthesis of the o-quinones and other oxidized metabolites of polycyclic aromatic hydrocarbons implicated in carcinogenesis. J. Org. Chem. 69, 2024–2032. [DOI] [PubMed] [Google Scholar]
- Biemann K.; McCloskey J. A. (1962) Application of mass spectrometry to structure problems. VI. Nucleosides. J. Am. Chem. Soc. 84, 2005–2007. [Google Scholar]
- Rice J. M.; Dudek G. O. (1967) Mass spectra of nucleic acid derivatives. II. Guanine, adenine, and related compounds. J. Am. Chem. Soc. 89, 2719–2725. [DOI] [PubMed] [Google Scholar]
- Shaw S. J.; Desideri D. M.; Tsuboyam K.; McCloskey J. A. (1970) Mass spectrometry of nucleic acid components - Analogs of adenosine. J. Am. Chem. Soc. 92, 2510–2522. [DOI] [PubMed] [Google Scholar]
- Lawson A. M.; Stillwell R. N.; Tacker M. M.; Tsuboyam K.; McCloskey J. A. (1971) Mass spectrometry of nucleic acid components - Trimethylsilyl derivatives of nucleotides. J. Am. Chem. Soc. 93, 1014–1023. [DOI] [PubMed] [Google Scholar]
- Nesnow S.; Nelson G.; Padgett W. T.; George M. H.; Moore T.; King L. C.; Adams L. D.; Ross J. A. (2010) Lack of contribution of covalent benzo[a]pyrene-7,8-quinone-DNA adducts in benzo[a]pyrene-induced mouse lung tumorigenesis. Chem.-Biol. Interact. 186, 157–165. [DOI] [PubMed] [Google Scholar]
- Flowers L.; Bleczinski W. F.; Burczynski M. E.; Harvey R. G.; Penning T. M. (1996) Disposition and biological activity of benzo[a]pyrene-7,8-dione. A genotoxic metabolite generated by dihydrodiol dehydrogenas. Biochemistry 35, 13664–13672. [DOI] [PubMed] [Google Scholar]
- Lewis M. A.; Yoerg D. G.; Bolton J. L.; Thompson J. A. (1996) Alkylation of 2’-deoxynucleosides and DNA by quinone methides derived from 2,6-di-tert-butyl-4-methylphenol. Chem. Res. Toxicol. 9, 1368–1374. [DOI] [PubMed] [Google Scholar]
- Veldhuyzen W. F.; Lam Y. F.; Rokita S. E. (2001) 2’-Deoxyguanosine reacts with a model quinone methide at multiple sites. Chem. Res. Toxicol. 14, 1345–1351. [DOI] [PubMed] [Google Scholar]
- Zhou Q.; Xu T.; Mangrum J. B. (2007) Selective N1-alkylation of 2’-deoxyguanosine with a quinolinyl quinone methide. Chem. Res. Toxicol. 20, 1069–1074. [DOI] [PubMed] [Google Scholar]
- Yu D.; Berlin J. A.; Penning T. M.; Field J. (2002) Reactive oxygen species generated by PAH o-quinones cause change-in-function mutations in p53. Chem. Res. Toxicol. 15, 832–842. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.