Abstract
Phylogenetic reconstruction of three highly conserved proteins involved in bacterial conjugation (relaxase, coupling protein and a type IV secretion system ATPase) allowed the classification of transmissible elements in relaxase MOB families and mating pair formation MPF groups. These evolutionary studies point to the existence of a limited number of module combinations in transmissible elements, preferentially associated with specific genetic or environmental backgrounds. A practical protocol based on the MOB classification was implemented to detect and assort transmissible plasmids and integrative elements from γ-Proteobacteria. It was called “Degenerate Primer MOB Typing” or DPMT. It resulted in a powerful technique that discovers not only backbones related to previously classified elements (typically by PCR-based replicon typing or PBRT), but also distant new members sharing a common evolutionary ancestor. The DPMT method, conjointly with PBRT, promises to be useful to gain information on plasmid backbones and helpful to investigate the dissemination routes of transmissible elements in microbial ecosystems.
Keywords: relaxase, plasmid classification, degenerate primer MOB typing, conjugation, ICE, transmissible elements
Traditional Trends in Plasmid Classification
Plasmid biology emerged as a discipline with the study of fertility and resistance factors present in Enterobacteria, Pseudomonas and Staphylococci. At that emerging point, plasmids were assigned to different incompatibility groups (Inc), based on their ability to coexist in the same cell.1 Incompatibility was generally (but not always!) an indicator of high similarity in the replication and/or partition modules of members included in the same Inc and of large differences among members from different Inc.2,3 Many exceptions were found, as discussed previously.4 Since replication is an essential plasmid property, Inc grouping continued to base plasmid classification in spite of the previous caveats. Subsequent experimental approaches moved to molecular biology techniques, such as DNA hybridization,5 PCR (refs. 6–9, among others), and/or straight sequencing,10,11 which provided direct information on the replicon DNA sequence.
Plasmid incompatibility testing was finally abandoned due to its technical drawbacks. Nevertheless, the tradition of grouping plasmids by Inc persisted, probably motivated by the fact that much of plasmid biology research during the following decades concentrated on a few plasmid backbones encoding virulence and antibiotic-resistance determinants, hosted in the “classical” bacterial families, mostly Enterobacteriaceae, where Inc testing was developed. Molecular approaches inevitably led to further Inc subdivisions, which split some Inc groups (IncQ,12 IncH,13 IncP-1,14 IncF15 and IncX plasmids16) and allowed the discovery of new ones (PromA,17 IncR,18 GR groups from Acinetobacter baumannii,19 FIIK from Klebsiella pneumoniae, FIIY from Yersinia, and FIIS from Salmonella15). For IncN, IncHI1 and IncI1 groups, a plasmid MultiLocus Sequence Tying (pMLST) approach was implemented to identify closely plasmid variants spread recently and assort them in numerous profiles (http://pubmlst.org/plasmid/). At the end, the picture got even more inextricable and begged for the eclosion of a novel classification system.
Conjugation Modules and Types of Transmissible Elements
A classical plasmid backbone encompasses not only its replication module, but also partition and other maintenance systems as well as transfer functions. Transfer through conjugation is the main way used by plasmids for their dissemination among bacterial populations20 and it is one of the strategies that contribute to their maintenance at a population level.21,22 Hence, research attention focused on conjugation mainly due to its involvement in antibiotic resistance spreading. Details of the biochemical mechanism of conjugation and its ecological implications have been reviewed by references 23 and 24.
Different genetic modules encode the proteins involved in conjugation. The relaxase, sometimes helped by accessory proteins, recognizes a specific sequence of the transmissible element named origin of transfer (oriT), forming a relaxosome.24 The relaxase cleaves one of the DNA strands at the nic site, resulting in a nucleoprotein complex that would be the translocated substrate.24 The mating pair formation system (MPF) is the exit way of the conjugative substrate from donor to recipient cells25 and an entry door for filamentous phages.26,27 By linking the relaxosome to the transport channel, the coupling protein (T4CP) recognizes the substrate,28-30 establishes contacts with the MPF28,31 and pumps the substrate by its ATPase activity.32
Transmissible elements are classified according to their transfer ability in conjugative (those that code all components needed for transfer) and mobilizable (those that encode the MOB but not the MPF transfer functions and hence require a helper to be transferred). According to the genetic location of the transfer genes, conjugation-transmissible elements can be plasmids or integrative elements hosted in the chromosome. The mobile repertoire is thus composed of conjugative and mobilizable plasmids (CP and MP), integrative and conjugative elements (ICE), and integrative and mobilizable elements (IME) (Fig. 1A). The relaxase is the only common gene to all transmissible elements. Eight relaxase families (MOB families) were identified in a global survey of transmissible elements.34 Phylogenetic relationships among relaxases constitute the rationale behind the MOB classification.33,35,36 Based on the presence or absence of specific MPF genes and the phylogeny of the most conserved MPF protein (VirB4/TraU-like), seven MPF were detected.33,34,37 Each MOB type is unequally distributed among plasmids of different sizes and different taxonomic classes. The association of a relaxase with a T4CP, as well as with each MPF type, is usually specific for each MOB family,37 suggesting that the genes involved in conjugative transfer have evolved into specific sets co-adapted to specific physiological and ecological contexts. Thus, MOB types provide not only a classification tool but also a valuable resource to predict the transfer characteristics of a plasmid and to follow its propagation routes in complex ecosystems.38
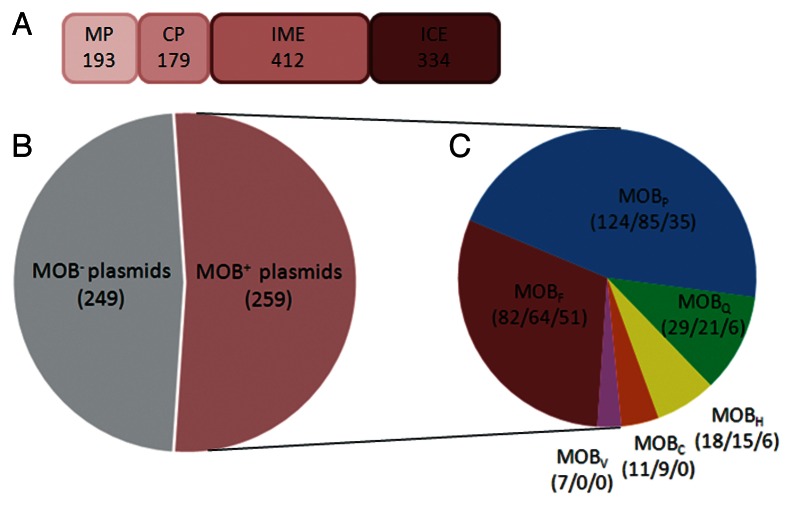
Figure 1. Bestiary of mobility elements. (A) Abundance of the different elements transmissible by conjugation. The number of conjugative plasmids (CP), mobilizable plasmids (MP), integrative and conjugative elements (ICE) and integrative and mobilizable elements (IME), identified in completely sequenced prokaryotes (1,207 chromosomes and 891 plasmids associated with chromosomes) is indicated for each element type and is proportional to each box width. Data were obtained from Dataset S1 in reference 33. (B) Proportion of transmissible (MOB+) and non-transmissible (MOB−) γ-Proteobacterial plasmids. (C) MOB family distribution in γ-Proteobacterial plasmids. For each MOB family which has representatives in γ-Proteobacteria the number of members is indicated by the first figure between parentheses. The second and third figures indicate the number of relaxases that can be detected by DPMT probes and the number of transmissible plasmids that can be detected by PBRT probes, respectively. Data presented in (B and C) were obtained from Supplementary Table 1 in reference 4.
According to a genomic survey,33 mobilizable elements outnumber conjugative elements both within integrated elements and within plasmids. Besides, the repertoire of ICE practically doubles that of CP (Fig. 1A). Furthermore, phylogenetic analyses indicate that CP and ICE show strikingly conserved patterns of conjugative genes. Exchanges of CP to and from ICE have been frequent along their evolutionary history. These findings suggest that CP often become ICE, and/or vice-versa, arguing for a unitary vision of the evolutionary dynamics of conjugative elements. For example, MOBH12 relaxases are encoded in both, conjugative plasmids (i.e., IncA/C) and ICE (i.e., IncJ elements).4
A Practical Approach to Classify Transmissible Elements According to their Relaxase: Pros and Cons
Most knowledge about conjugation comes from the study of plasmids and ICE hosted in Proteobacteria. The transfer systems of this phylum can be all grouped in six MOB and four MPF families.37 In class γ-Proteobacteria, which includes a significant amount of genera involved in infectious diseases, five MOB relaxase families include more than 95% of the elements (MOBF, MOBP, MOBQ, MOBH and MOBC) (Fig. 1B and C). A recently published screening method called “Degenerate Primer MOB Typing” (acronym DPMT) was developed to detect and classify relaxase genes carried by γ-Proteobacterial plasmids.4
Protein alignments of well-resolved clades in the five mentioned MOB phylogenies were analyzed to find blocks of residues with high global homology. Such blocks generally corresponded to catalytic motifs that contain sequence signatures of each MOB (sub)family.35,36 Following the CODEHOP strategy,39 degenerate primers, hybridizing to coding sequences of conserved amino acid motifs, were designed to amplify related relaxase genes. Such primers contained two regions: (1) a 3′ core sequence (around 12-mer) that hybridized with the codons that determine the block of conserved amino acids and, therefore, was degenerate to encompass different codon usages; and (2) a 5′ non-degenerate clamp sequence of variable length that contained a consensus of the most represented base at each position. The maximum degeneracy allowed in the oligonucleotide set was 24 (for a single primer) and 32 (for the sum of degeneracies of both oligonucleotides of a primer pair).
A set of 19 primer pairs was selected for its specificity and sensitivity using a collection of 33 reference relaxases. They represent more than 95% of the diversity of γ-Proteobacterial plasmids and are distributed in 16 MOB subfamilies with robust phylogenetic support. Once validated, DPMT was used to test two enterobacterial plasmid collections (originating from ESBL-resistant and pivmecillinam-resistant clinical isolates, respectively). In 93% of transconjugants, at least one MOB relaxase was detected by DPMT. The method detected not only relaxase genes identical to those already reported but also new MOB members ranging from 60 to 95% identity to the closer MOB homolog. These new branches, which populate known or new MOB subfamilies, will help to improve the MOB phylogenies and refine the DPMT set of primers. This fact underscores the power of DPMT to detect and classify plasmids that are undetected by other currently used methods (Figs. 1 and 2), singularly PBRT. Thus, DPMT is suitable for studying global plasmid diversity and finding deviant plasmids from well-studied backbones or those carried by a large number of taxonomic families. Other studies have also successfully used DPMT primer pairs conjointly with PBRT for identifying plasmids4,40 and ICE41 from clinical strains of Enterobacteria. ICE are gaining research momentum because of increasing evidence of their involvement in the dissemination of antibiotic resistance.42 So, the ability of DPMT to detect them is valuable. The philosophy that guided the development of the γ-Proteobacteria MOB primer set has been extended to encompass relaxases of other taxonomical groups of bacteria, such as Enterococci.43
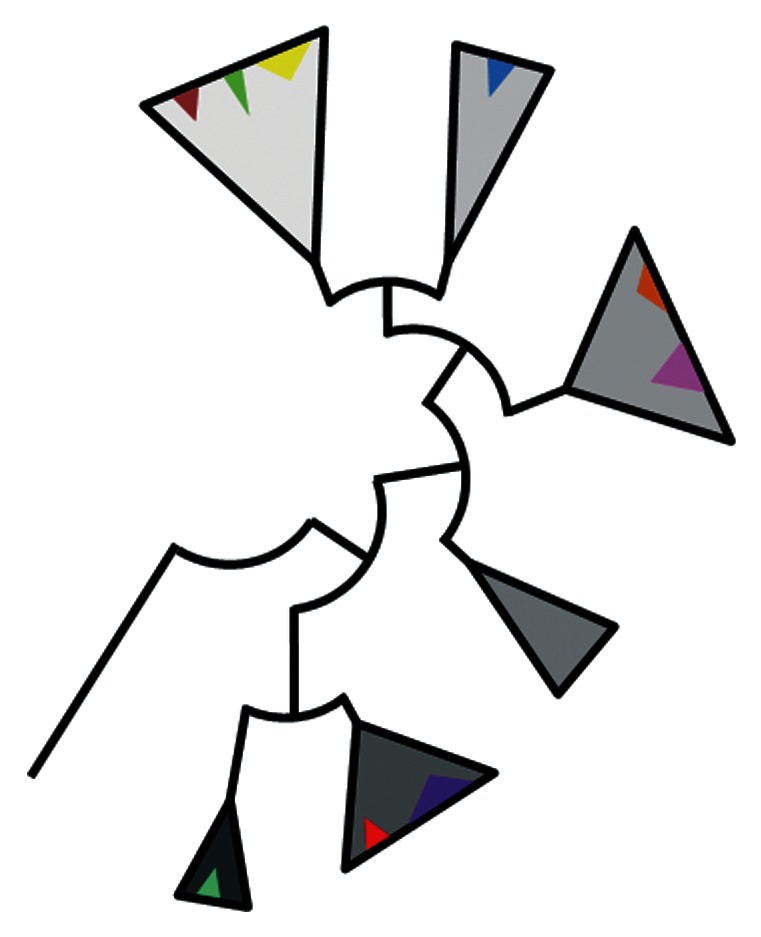
Figure 2. Phylogenetic depth of relaxase typing (DPMT) vs. Inc typing (PBRT). Schematic representation of an idealized relaxase family phylogeny. Grey-filled triangles represent monophyletic clades that constitute the different MOB subfamilies within a given family, and are thus detectable by a specific DPMT oligonucleotide primer pair. Smaller, colored triangles group relaxases belonging to a specific Inc plasmid group (which can be detected by a specific PBRT oligonucleotide primer pair). The width of the triangles is proportional to the number of relaxases they contain, while their height reflects the phylogenetic depth from the taxa to the last common ancestor of the group. Details on the correlation between Inc/REP types and MOB subfamilies for major plasmids of γ-Proteobacteria are provided in Figure 5 and Table 1 of reference 38, and Figure 8 and Supplementary Table S1 of reference 4.
DPMT potency is achieved by a combination of phylogenetic support and the use of degenerate primers to uncover most codon variants of the relaxase signatures. As shown in Figure 2, DPMT finds and classifies backbones that share a common relaxase ancestor (“zoom out” strategy), while Inc classification and currently available PBRT and pMLST probes are useful at a lower phylogenetic depth, that is, at detecting practically identical backbones that carry different cargo genes (“zoom in” strategy). In a single MOB subfamily detected by DPMT, relaxases from different Inc sets can be grouped. On the other hand, plasmids of such Inc groups do not contain relaxases dispersed in different MOB subfamilies. Very few exceptions are observed to this statement, which can usually be explained by events of recombination, plasmid cointegration, and deletions of secondary replicons. In practical terms, only a few dozens of plasmid backbones are repeatedly detected in clinical and environmental isolates of γ-Proteobacteria, associated or not with antibiotic resistance genes.23,44 Most of them are MOB+. Therefore, a multiplex PCR MOB typing (MPMT) that uses a set of non-degenerate oligonucleotide primers is being presently developed to uncover the relaxases of those backbones, complementing PBRT in faster plasmid screenings (Fig. 3).
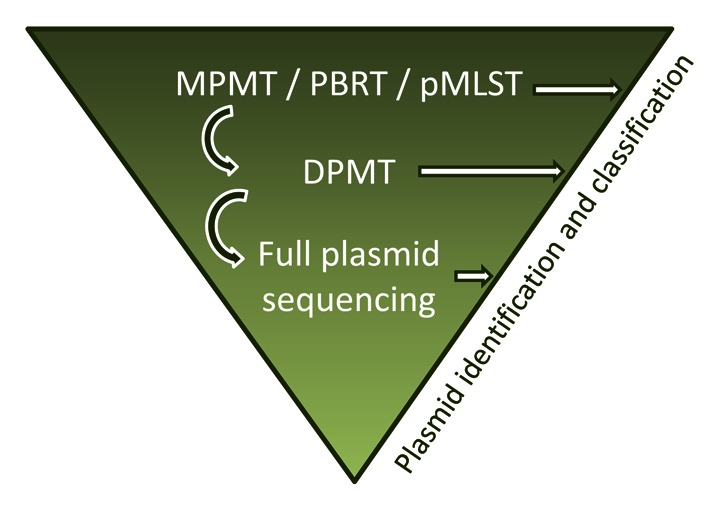
Figure 3. Workflow proposal for plasmid identification and classification. Plasmid DNA samples are first subjected to multiplex PCR MOB typing (MPMT) and/or PCR-based replicon typing (PBRT) and/or plasmid MultiLocus Sequence Typing (pMLST). Negative hits are subjected to DPMT-based MOB typing. If DPMT does not result in plasmid identification, full plasmid sequencing is performed. Positive hits (horizontal white arrows) represent plasmids that can be directly identified and classified. Negative hits (bent arrows), are subjected to the next classification protocol.
The relaxase-based plasmid classification was criticized by reference 45 with the argument that MOB and RepABC replicon phylogenies were not fully congruent. The main line of defense against that criticism is to mirror the problems of plasmid classification with those of bacterial classification. Even the concept of bacterial species is still controversial because of non-coherent phylogenies for some bacterial clades.46,47 Nevertheless, new data emerged from metagenomic studies (instead of just 16S rRNA sequences) point to the existence of sequence-discrete populations, microbial communities predominantly organized in genetically and ecologically discernible populations, which possess the attributes expected for species.48 Some degree of recombination between plasmids can be expected because more than one plasmid with homologous modules can coexist into the same cell. This is aggravated by the fact that plasmids are evolutionary units that propagate and replicate in different host structures, in turn influencing the selection of other units generated by introgressive descent.49 By being part of the horizontal gene pool, plasmids explore many genomic landscapes where potential homologs can be found. The effect of recombination can be higher in the case of plasmids hosted in bacteria capable of natural competence for transformation, such as Firmicutes, α-, β- and ε-Proteobacteria.50-53 Evidence suggests that there is not a single plasmid module that can escape from recombination. Even within the repABC family, horizontal gene transfer events of individual genes within the rep operon were detected. Besides, a particular phylogeny was found for each member of this operon, suggesting that each has its own evolutionary dynamics.54 This fact does not necessarily invalidate a classification scheme, it only establishes the intervals of confidence in the inference of coevolution of different backbone modules.
While it is common to find plasmids with more than one replicon, plasmids with more than one relaxase gene are the exceptions, a fact that helps in the univocal classification of transmissible plasmids based on DPMT. On the other hand, only eight MOB families uncover the complete diversity of conjugative elements, while a higher and unknown amount of replication initiation protein families exists. There are even some plasmids (i.e., ColE1-like) that do not code for their own initiator protein. A tour de force was made 15 years ago by reference 55 to summarize the strategies the circular bacterial plasmids use to initiate replication and to control their copy number. Evolutionary studies of some replicon families have been already performed, such as: repFIB56 and repFIIA57 present in Enterobacteria, those related to RepA of plasmid R388,58 repABC from α-Proteobacteria,54,59,60 DnaA-like from Rhodobacterales61 and 36 rep-families from Enterococci and Staphylococci.8,9,62 Nevertheless, a phylogenetic analysis of the complete diversity of initiator proteins is still missing. Once achieved, such analysis, in combination with those performed for conjugative systems and other backbone regions such as the partition63 and addiction systems,64,65 would serve to evaluate the role played by recombination in the shaping of plasmid backbones in different phyla. Then, the common elements of the backbones of each family and the phylogenetic depth of stable assemblies of plasmid modules could be better analyzed. The sum of all these studies will hopefully provide a more accurate and operational classification based on a deeper knowledge of plasmid diversity.
Acknowledgments
This work was supported by Spanish Ministry of Education (BFU2011-26608), and grants nº 248919/FP7-ICT-2009-4 and 282004/FP7-HEALTH.2011.2.3.1-2 from the European VII Framework Program to FdlC. MPBG was funded by a JAE-Doc_2009 postdoctoral contract from Consejo Superior de Investigaciones Científicas (CSIC) which was cofinanced by the European Science Foundation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/mge/article/24263
References
- 1.Datta N, Hedges RW. Compatibility groups among fi - R factors. Nature. 1971;234:222–3. doi: 10.1038/234222a0. [DOI] [PubMed] [Google Scholar]
- 2.Novick RP. Plasmid incompatibility. Microbiol Rev. 1987;51:381–95. doi: 10.1128/mr.51.4.381-395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Austin S, Nordström K. Partition-mediated incompatibility of bacterial plasmids. Cell. 1990;60:351–4. doi: 10.1016/0092-8674(90)90584-2. [DOI] [PubMed] [Google Scholar]
- 4.Alvarado A, Garcillán-Barcia MP, de la Cruz F. A degenerate primer MOB typing (DPMT) method to classify gamma-proteobacterial plasmids in clinical and environmental settings. PLoS One. 2012;7:e40438. doi: 10.1371/journal.pone.0040438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Couturier M, Bex F, Bergquist PL, Maas WK. Identification and classification of bacterial plasmids. Microbiol Rev. 1988;52:375–95. doi: 10.1128/mr.52.3.375-395.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Götz A, Pukall R, Smit E, Tietze E, Prager R, Tschäpe H, et al. Detection and characterization of broad-host-range plasmids in environmental bacteria by PCR. Appl Environ Microbiol. 1996;62:2621–8. doi: 10.1128/aem.62.7.2621-2628.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63:219–28. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 8.Jensen LB, Garcia-Migura L, Valenzuela AJ, Løhr M, Hasman H, Aarestrup FM. A classification system for plasmids from enterococci and other Gram-positive bacteria. J Microbiol Methods. 2010;80:25–43. doi: 10.1016/j.mimet.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 9.Lozano C, García-Migura L, Aspiroz C, Zarazaga M, Torres C, Aarestrup FM. Expansion of a plasmid classification system for Gram-positive bacteria and determination of the diversity of plasmids in Staphylococcus aureus strains of human, animal, and food origins. Appl Environ Microbiol. 2012;78:5948–55. doi: 10.1128/AEM.00870-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiesner M, Calva E, Fernández-Mora M, Cevallos MA, Campos F, Zaidi MB, et al. Salmonella Typhimurium ST213 is associated with two types of IncA/C plasmids carrying multiple resistance determinants. BMC Microbiol. 2011;11:9. doi: 10.1186/1471-2180-11-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shearer JE, Wireman J, Hostetler J, Forberger H, Borman J, Gill J, et al. Major families of multiresistant plasmids from geographically and epidemiologically diverse staphylococci. G3 (Bethesda) 2011;1:581–91. doi: 10.1534/g3.111.000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rawlings DE, Tietze E. Comparative biology of IncQ and IncQ-like plasmids. Microbiol Mol Biol Rev. 2001;65:481–96. doi: 10.1128/MMBR.65.4.481-496.2001. [Table of contents]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilmour MW, Thomson NR, Sanders M, Parkhill J, Taylor DE. The complete nucleotide sequence of the resistance plasmid R478: defining the backbone components of incompatibility group H conjugative plasmids through comparative genomics. Plasmid. 2004;52:182–202. doi: 10.1016/j.plasmid.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 14.Schlüter A, Szczepanowski R, Pühler A, Top EM. Genomics of IncP-1 antibiotic resistance plasmids isolated from wastewater treatment plants provides evidence for a widely accessible drug resistance gene pool. FEMS Microbiol Rev. 2007;31:449–77. doi: 10.1111/j.1574-6976.2007.00074.x. [DOI] [PubMed] [Google Scholar]
- 15.Villa L, García-Fernández A, Fortini D, Carattoli A. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother. 2010;65:2518–29. doi: 10.1093/jac/dkq347. [DOI] [PubMed] [Google Scholar]
- 16.Johnson TJ, Bielak EM, Fortini D, Hansen LH, Hasman H, Debroy C, et al. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid. 2012;68:43–50. doi: 10.1016/j.plasmid.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Van der Auwera GA, Król JE, Suzuki H, Foster B, Van Houdt R, Brown CJ, et al. Plasmids captured in C. metallidurans CH34: defining the PromA family of broad-host-range plasmids. Antonie Van Leeuwenhoek. 2009;96:193–204. doi: 10.1007/s10482-009-9316-9. [DOI] [PubMed] [Google Scholar]
- 18.García-Fernández A, Fortini D, Veldman K, Mevius D, Carattoli A. Characterization of plasmids harbouring qnrS1, qnrB2 and qnrB19 genes in Salmonella. J Antimicrob Chemother. 2009;63:274–81. doi: 10.1093/jac/dkn470. [DOI] [PubMed] [Google Scholar]
- 19.Bertini A, Poirel L, Mugnier PD, Villa L, Nordmann P, Carattoli A. Characterization and PCR-based replicon typing of resistance plasmids in Acinetobacter baumannii. Antimicrob Agents Chemother. 2010;54:4168–77. doi: 10.1128/AAC.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halary S, Leigh JW, Cheaib B, Lopez P, Bapteste E. Network analyses structure genetic diversity in independent genetic worlds. Proc Natl Acad Sci U S A. 2010;107:127–32. doi: 10.1073/pnas.0908978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lundquist PD, Levin BR. Transitory derepression and the maintenance of conjugative plasmids. Genetics. 1986;113:483–97. doi: 10.1093/genetics/113.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fox RE, Zhong X, Krone SM, Top EM. Spatial structure and nutrients promote invasion of IncP-1 plasmids in bacterial populations. ISME J. 2008;2:1024–39. doi: 10.1038/ismej.2008.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heuer H, Schmitt H, Smalla K. Antibiotic resistance gene spread due to manure application on agricultural fields. Curr Opin Microbiol. 2011;14:236–43. doi: 10.1016/j.mib.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 24.de la Cruz F, Frost LS, Meyer RJ, Zechner EL. Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol Rev. 2010;34:18–40. doi: 10.1111/j.1574-6976.2009.00195.x. [DOI] [PubMed] [Google Scholar]
- 25.Llosa M, Gomis-Rüth FX, Coll M, de la Cruz Fd F. Bacterial conjugation: a two-step mechanism for DNA transport. Mol Microbiol. 2002;45:1–8. doi: 10.1046/j.1365-2958.2002.03014.x. [DOI] [PubMed] [Google Scholar]
- 26.Bradley DE, Sirgel FA, Coetzee JN, Hedges RW, Coetzee WF. Phages C-2 and J: IncC and IncJ plasmid-dependent phages, respectively. J Gen Microbiol. 1982;128:2485–98. doi: 10.1099/00221287-128-10-2485. [DOI] [PubMed] [Google Scholar]
- 27.Coetzee JN, Bradley DE, Fleming J, du Toit L, Hughes VM, Hedges RW. Phage pilH alpha: a phage which adsorbs to IncHI and IncHII plasmid-coded pili. J Gen Microbiol. 1985;131:1115–21. doi: 10.1099/00221287-131-5-1115. [DOI] [PubMed] [Google Scholar]
- 28.Llosa M, Zunzunegui S, de la Cruz F. Conjugative coupling proteins interact with cognate and heterologous VirB10-like proteins while exhibiting specificity for cognate relaxosomes. Proc Natl Acad Sci U S A. 2003;100:10465–70. doi: 10.1073/pnas.1830264100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mihajlovic S, Lang S, Sut MV, Strohmaier H, Gruber CJ, Koraimann G, et al. Plasmid r1 conjugative DNA processing is regulated at the coupling protein interface. J Bacteriol. 2009;191:6877–87. doi: 10.1128/JB.00918-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lang S, Zechner EL. General requirements for protein secretion by the F-like conjugation system R1. Plasmid. 2012;67:128–38. doi: 10.1016/j.plasmid.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Paz HD, Larrea D, Zunzunegui S, Dehio C, de la Cruz F, Llosa M. Functional dissection of the conjugative coupling protein TrwB. J Bacteriol. 2010;192:2655–69. doi: 10.1128/JB.01692-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cabezon E, Lanza VF, Arechaga I. Membrane-associated nanomotors for macromolecular transport. Curr Opin Biotechnol. 2012;23:537–44. doi: 10.1016/j.copbio.2011.11.031. [DOI] [PubMed] [Google Scholar]
- 33.Guglielmini J, Quintais L, Garcillán-Barcia MP, de la Cruz F, Rocha EP. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet. 2011;7:e1002222. doi: 10.1371/journal.pgen.1002222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guglielmini J, de la Cruz F, Rocha EP. Evolution of conjugation and type IV secretion systems. Mol Biol Evol. 2013;30:315–31. doi: 10.1093/molbev/mss221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Francia MV, Varsaki A, Garcillán-Barcia MP, Latorre A, Drainas C, de la Cruz F. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol Rev. 2004;28:79–100. doi: 10.1016/j.femsre.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 36.Garcillán-Barcia MP, Francia MV, de la Cruz F. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol Rev. 2009;33:657–87. doi: 10.1111/j.1574-6976.2009.00168.x. [DOI] [PubMed] [Google Scholar]
- 37.Smillie C, Garcillán-Barcia MP, Francia MV, Rocha EP, de la Cruz F. Mobility of plasmids. Microbiol Mol Biol Rev. 2010;74:434–52. doi: 10.1128/MMBR.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcillán-Barcia MP, Alvarado A, de la Cruz F. Identification of bacterial plasmids based on mobility and plasmid population biology. FEMS Microbiol Rev. 2011;35:936–56. doi: 10.1111/j.1574-6976.2011.00291.x. [DOI] [PubMed] [Google Scholar]
- 39.Rose TM, Schultz ER, Henikoff JG, Pietrokovski S, McCallum CM, Henikoff S. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 1998;26:1628–35. doi: 10.1093/nar/26.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valverde A, Cantón R, Garcillán-Barcia MP, Novais A, Galán JC, Alvarado A, et al. Spread of bla(CTX-M-14) is driven mainly by IncK plasmids disseminated among Escherichia coli phylogroups A, B1, and D in Spain. Antimicrob Agents Chemother. 2009;53:5204–12. doi: 10.1128/AAC.01706-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mata C, Miró E, Alvarado A, Garcillán-Barcia MP, Toleman M, Walsh TR, et al. Plasmid typing and genetic context of AmpC β-lactamases in Enterobacteriaceae lacking inducible chromosomal ampC genes: findings from a Spanish hospital 1999-2007. J Antimicrob Chemother. 2012;67:115–22. doi: 10.1093/jac/dkr412. [DOI] [PubMed] [Google Scholar]
- 42.Toleman MA, Walsh TR. Combinatorial events of insertion sequences and ICE in Gram-negative bacteria. FEMS Microbiol Rev. 2011;35:912–35. doi: 10.1111/j.1574-6976.2011.00294.x. [DOI] [PubMed] [Google Scholar]
- 43.Goicoechea P, Romo M, Coque TM, Baquero F, de la Cruz F, Martínez-Martínez L, et al. Identification of enterococcal plasmids by Multiplex-PCR-based relaxase typing. 18th European Congress of Clinical Microbiology and Infectious Diseases 2008. [Google Scholar]
- 44.Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother. 2009;53:2227–38. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petersen J. Phylogeny and compatibility: plasmid classification in the genomics era. Arch Microbiol. 2011;193:313–21. doi: 10.1007/s00203-011-0686-9. [DOI] [PubMed] [Google Scholar]
- 46.Hanage WP, Fraser C, Spratt BG. Fuzzy species among recombinogenic bacteria. BMC Biol. 2005;3:6. doi: 10.1186/1741-7007-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Konstantinidis KT, Ramette A, Tiedje JM. The bacterial species definition in the genomic era. Philos Trans R Soc Lond B Biol Sci. 2006;361:1929–40. doi: 10.1098/rstb.2006.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caro-Quintero A, Konstantinidis KT. Bacterial species may exist, metagenomics reveal. Environ Microbiol. 2012;14:347–55. doi: 10.1111/j.1462-2920.2011.02668.x. [DOI] [PubMed] [Google Scholar]
- 49.Bapteste E, Lopez P, Bouchard F, Baquero F, McInerney JO, Burian RM. Evolutionary analyses of non-genealogical bonds produced by introgressive descent. Proc Natl Acad Sci U S A. 2012;109:18266–72. doi: 10.1073/pnas.1206541109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen I, Dubnau D. DNA uptake during bacterial transformation. Nat Rev Microbiol. 2004;2:241–9. doi: 10.1038/nrmicro844. [DOI] [PubMed] [Google Scholar]
- 51.Hanage WP, Fraser C, Spratt BG. The impact of homologous recombination on the generation of diversity in bacteria. J Theor Biol. 2006;239:210–9. doi: 10.1016/j.jtbi.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 52.González V, Bustos P, Ramírez-Romero MA, Medrano-Soto A, Salgado H, Hernández-González I, et al. The mosaic structure of the symbiotic plasmid of Rhizobium etli CFN42 and its relation to other symbiotic genome compartments. Genome Biol. 2003;4:R36. doi: 10.1186/gb-2003-4-6-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crossman LC, Castillo-Ramírez S, McAnnula C, Lozano L, Vernikos GS, Acosta JL, et al. A common genomic framework for a diverse assembly of plasmids in the symbiotic nitrogen fixing bacteria. PLoS One. 2008;3:e2567. doi: 10.1371/journal.pone.0002567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Castillo-Ramírez S, Vázquez-Castellanos JF, González V, Cevallos MA. Horizontal gene transfer and diverse functional constrains within a common replication-partitioning system in Alphaproteobacteria: the repABC operon. BMC Genomics. 2009;10:536. doi: 10.1186/1471-2164-10-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.del Solar G, Giraldo R, Ruiz-Echevarría MJ, Espinosa M, Díaz-Orejas R. Replication and control of circular bacterial plasmids. Microbiol Mol Biol Rev. 1998;62:434–64. doi: 10.1128/mmbr.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gibbs MD, Spiers AJ, Bergquist PL. RepFIB: a basic replicon of large plasmids. Plasmid. 1993;29:165–79. doi: 10.1006/plas.1993.1020. [DOI] [PubMed] [Google Scholar]
- 57.Osborn AM, da Silva Tatley FM, Steyn LM, Pickup RW, Saunders JR. Mosaic plasmids and mosaic replicons: evolutionary lessons from the analysis of genetic diversity in IncFII-related replicons. Microbiology. 2000;146:2267–75. doi: 10.1099/00221287-146-9-2267. [DOI] [PubMed] [Google Scholar]
- 58.Fernández-López R, Garcillán-Barcia MP, Revilla C, Lázaro M, Vielva L, de la Cruz F. Dynamics of the IncW genetic backbone imply general trends in conjugative plasmid evolution. FEMS Microbiol Rev. 2006;30:942–66. doi: 10.1111/j.1574-6976.2006.00042.x. [DOI] [PubMed] [Google Scholar]
- 59.Cevallos MA, Cervantes-Rivera R, Gutiérrez-Ríos RM. The repABC plasmid family. Plasmid. 2008;60:19–37. doi: 10.1016/j.plasmid.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 60.Petersen J, Brinkmann H, Pradella S. Diversity and evolution of repABC type plasmids in Rhodobacterales. Environ Microbiol. 2009;11:2627–38. doi: 10.1111/j.1462-2920.2009.01987.x. [DOI] [PubMed] [Google Scholar]
- 61.Petersen J, Brinkmann H, Berger M, Brinkhoff T, Päuker O, Pradella S. Origin and evolution of a novel DnaA-like plasmid replication type in Rhodobacterales. Mol Biol Evol. 2011;28:1229–40. doi: 10.1093/molbev/msq310. [DOI] [PubMed] [Google Scholar]
- 62.Garcia-Migura L, Liebana E, Jensen LB. Transposon characterization of vancomycin-resistant Enterococcus faecium (VREF) and dissemination of resistance associated with transferable plasmids. J Antimicrob Chemother. 2007;60:263–8. doi: 10.1093/jac/dkm186. [DOI] [PubMed] [Google Scholar]
- 63.Gerdes K, Møller-Jensen J, Bugge Jensen R. Plasmid and chromosome partitioning: surprises from phylogeny. Mol Microbiol. 2000;37:455–66. doi: 10.1046/j.1365-2958.2000.01975.x. [DOI] [PubMed] [Google Scholar]
- 64.Leplae R, Geeraerts D, Hallez R, Guglielmini J, Drèze P, Van Melderen L. Diversity of bacterial type II toxin-antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Res. 2011;39:5513–25. doi: 10.1093/nar/gkr131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guglielmini J, Van Melderen L. Bacterial toxin-antitoxin systems: Translation inhibitors everywhere. Mob Genet Elements. 2011;1:283–90. doi: 10.4161/mge.18477. [DOI] [PMC free article] [PubMed] [Google Scholar]