Abstract
Protein structures evolved through a complex interplay of cooperative interactions and it is still very challenging to design new protein folds de novo. Here, we present a strategy to design self-assembling polypeptide nanostructured polyhedra, based on modularization using orthogonal dimerizing segments. We designed end experimentally demonstrated formation of the tetrahedron that self-assembles from a single polypeptide chain comprising 12 concatenated coiled-coil-forming segments separated by flexible peptide hinges. Path of the polypeptide chain is guided by the defined order of segments that traverse each of the 6 edges of the tetrahedron exactly twice, forming coiled-coil dimers with their corresponding partners. Coincidence of the polypeptide termini in the same vertex is demonstrated by reconstitution of the split fluorescent protein by the polypeptide with the correct tetrahedral topology, while polypeptides with a deleted or scrambled segment order fail to self-assemble correctly. This design platform provides the basis for construction of new topological polypeptide folds based on the set of orthogonal interacting polypeptide segments.
Base complementarity of DNA can now be used to rationally design artificial nanostructures that can adopt versatile two and three-dimensional shapes, such as different polyhedra1–4. In contrast, the design of new polypeptide folds is significantly more complicated, due to contribution of many cooperative and long-range interactions. Yet in nature polypeptides perform most complex tasks, because they can form intricate tertiary structures with versatile chemical properties and functionalities. De novo protein fold design has been, with few exceptions, successful only for folds similar to those that exist in nature5–8. Self-assembled symmetrical polypeptide assemblies have been designed based on natural protein oligomerization domains9,10 and the interacting surfaces of natural trimerizing protein domains were optimized for the self-assembly into tetrahedral and octahedral nanostructures11,12. Peptides have also been used for the self-assembly of fibrils, nanotubes and nanospheres (reviewed in 13). Use of modular building elements could facilitate the design of new protein folds. Such a type of modular element is the coiled-coil dimer, which is composed of intertwined helices, first proposed in 195214. Interactions that govern the specificity of coiled-coil dimerization are fairly well understood15, as coiled coils are characterized by a heptad repeat that forms two turns of a helix spanning approximately 1 nm. Formation of coiled-coil dimers is governed by hydrophobic and electrostatic interactions, involving residues at positions a, d of the periodic heptad repeat and e, g, respectively (Supplementary Results, Supplementary Fig. 4). The specificity and orthogonality of the desired coiled-coil combination can be further improved by the negative design such as by introduction of an Asn residue at hydrophobic coiled-coil interaction position16. Staggered arrangement of self-complementary coiled-coil-forming segments has been used to prepare polypeptide nanofibrils17–19. However, the combination of 2 types of interacting coiled-coil elements can only lead to one-dimensional fibrillar assemblies18 and at least three different elements are required for two- or three-dimensional assemblies.
We surmised that the concatenated orthogonal polypeptide interacting segments could be used to guide the self-assembly of complex polypeptide polyhedra. This strategy was tested on the self-assembly of a tetrahedral polypeptide fold designed from 12 coiled-coil-forming peptides interspersed with short flexible peptide linkers and validated by the determination of the hydrodynamic diameter, stability, TEM and AFM imaging and by the confirmation of the coincidence of the N- and C-terminal segment.
RESULTS
Design of polyhedral fold from a single chain
We reasoned that it should be possible to self-assemble the designed 3-dimensional objects from a single polypeptide chain that is composed of concatenated coiled-coil-forming segments. This strategy relies on the ability of those segments to pair in a selected orientation with their complementary interacting segments within the same polypeptide chain, thus driving the self-assembly. Each of those coiled-coil-forming segments is, in isolation, unstructured and forms a coiled-coil helix only when it dimerizes with the corresponding complementary segment. Each edge of the constructed polyhedron should be traversed by a polypeptide chain exactly twice forming a rigid coiled-coil dimer. The topology of the self-assembled polypeptide chain should therefore define the orientation of each coiled-coil pair and their sequential arrangement. Tetrahedron is the simplest 3-dimensional geometric object consisting of 6 edges; thus, a tetrahedron could be constructed from 6 pairs i.e. 12 concatenated coiled-coil dimer-forming segments. In topological terms, the polypeptide path along the edges of a polyhedron corresponds to an Eulerian trail, an oriented path that connects each of the two vertices by exactly two connections. Such a trail is guaranteed to exist by the graph theory, because all vertices of a double tetrahedral graph have an even degree. We formally proved in Supplementary Note 1 that polyhedra are, in principle, realizable by a single multisegmental polypeptide chain approach. Additional constraints on the topology of the polypeptide trail were imposed to ensure the stability of the polyhedron by interlocking the edges at each vertex (Supplementary Note 1, Supplementary Fig. 1). Unexpectedly, we proved by graph theory analysis that a tetrahedron cannot be constructed exclusively from either parallel or antiparallel dimeric segments but only by a combination of both types. The ability of the polypeptide chain to form either parallel- or antiparallel-oriented dimers is therefore an additional advantage in comparison to nucleic acid-based designed nanostructures, since nucleic acid duplex can only assume antiparallel orientation. We showed that the tetrahedron could self-assemble from a single polypeptide chain that comprises either 3 parallel and 3 antiparallel coiled-coil pairs or 4 parallel and 2 antiparallel pairs, altogether encompassing 3 distinct topological solutions (Supplementary Note 1, Supplementary Figs. 2 and 3). The number of different path topologies increased rapidly with the number of polyhedral edges; however the number of parallel or antiparallel only topologies is very sparse (Supplementary Table 1). Additionally, we note that for all polyhedra traversed by a single chain, the beginning and end of the polypeptide trail must coincide at the same vertex, which can be tested experimentally.
Design of a single chain polypeptide tetrahedron
For the experimental test of the design of the tetrahedral topology we selected to construct the topology comprising 4 parallel and 2 antiparallel coiled-coil dimeric edges (Fig. 1), which is the topology with the largest number of parallel segments. Although both parallel and antiparallel coiled-coil dimeric arrangements are represented in nature20, significantly higher number of isolated parallel coiled-coil dimers have been characterized and designed15,21,22. We selected the pairs for the formation of 3 edges of the polyhedron from the orthogonal coiled-coil-forming pairs P3-P4, P5-P6 and P7-P8. Peptides P3 to P8, composed of 4 heptad repeats (see Online Methods for amino acid sequences), were designed on the basis of known coiled-coil stability and selectivity principles21 and the orthogonality of the peptide pairs was demonstrated (Supplementary Fig. 6). None of these peptides shares 3 or more consecutive heptads with the same interacting motif at positions a, d, e, and g. Besides the designed parallel heterodimers, we used one parallel homodimer (based on GCN423) and two antiparallel homodimers (the APH24 and Bcr peptide25) (see Online Methods for amino acid sequences) in order to demonstrate that besides heterodimers, homodimers can also be used for the self-assembly of polypeptide structures (Supplementary Fig. 6b). The tetrapeptide SGPG was selected as the flexible linker to connect the consecutive coiled-coil-forming segments. This linker introduces strong helix-interrupting residues proline and glycine to prevent formation of an extended helix spanning several coiled-coil-forming segments and to provide the hinge flexibility at the vertices, which is required for the formation of acute angles between the edges of a tetrahedron. The synthetic gene for the designed polypeptide, TET12 (Fig. 1a and Supplementary Table 2), was codon-optimized for E. coli, the polypeptide was expressed in bacteria, purified (Supplementary Fig. 7a), and refolded by dialysis. The secondary structure of the self-assembled polypeptide, assessed by the circular dichroism (Fig. 2a), was consistent with the designed molecular model with high helical content. Each of the 6 isolated peptide pairs that form the edges of a tetrahedron unfolded between 0.5 M and 1.8 M GdnHCl (Fig. 2b). Although the cooperativity of the designed tetrahedral fold is provided only by the sequential order of concatenated coiled-coil-forming elements rather than by the hydrophobic core, as in the native protein folds, the annealed tetrahedral polypeptide exhibited significantly greater stability than its coiled-coil segment constituents, with a midpoint of unfolding at 3 M GdnHCl (Fig. 2b). Self-assembled polypeptide particles had an average hydrodynamic diameter of 6.9 ± 0.4 nm, determined by DLS (Fig. 2c), which is close to the calculated dimensions26 of the molecular model of the designed tetrahedron (Supplementary Fig. 5). Its diameter was also significantly more compact than TET12 polypeptide denatured in 6 M GdnHCl which measured 20.0 ± 2.9 nm (Fig. 2c). At a polypeptide concentration above 4 μM in the refolding mixture, coiled-coil-forming elements of the TET12 formed intermolecular interactions with the complementary segments from other molecules during the refolding process, which led to formation of aggregates with hydrodynamic size above 80 nm (Supplementary Fig. 8).
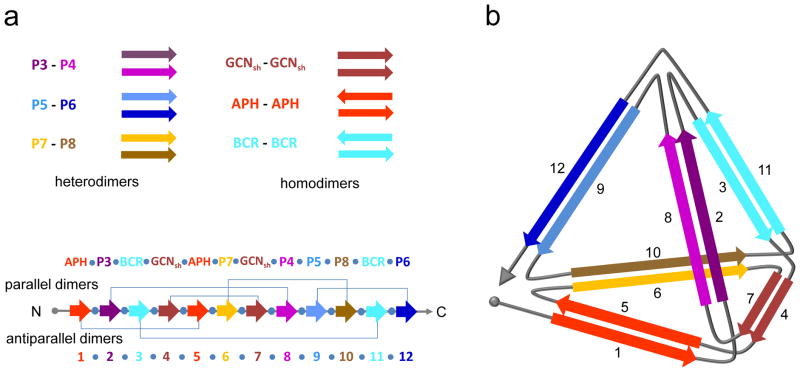
Figure 1. Design of the self-assembling polypeptide tetrahedron.
(a) The designed orthogonal peptide pairs P3-P4, P5-P6 and P7-P8, and homodimeric peptides APH, BCR and GCNsh are used to construct a tetrahedron-forming polypeptide chain, TET12. Twelve coiled-coil-forming elements are concatenated in a defined order, separated by the tetrapeptide linker SGPG (
 ), wherein the coiled-coil-forming segments marked 1 to 12 form parallel heterodimers (2–8, 6–10, 9–12), a parallel homodimer (4–7), and antiparallel homodimers (1–5, 3–11). (b) Schematic representation of the polypeptide path forming a tetrahedron. Arrows denote the orientation of the interacting coiled-coil-forming elements in the assembly.
), wherein the coiled-coil-forming segments marked 1 to 12 form parallel heterodimers (2–8, 6–10, 9–12), a parallel homodimer (4–7), and antiparallel homodimers (1–5, 3–11). (b) Schematic representation of the polypeptide path forming a tetrahedron. Arrows denote the orientation of the interacting coiled-coil-forming elements in the assembly.
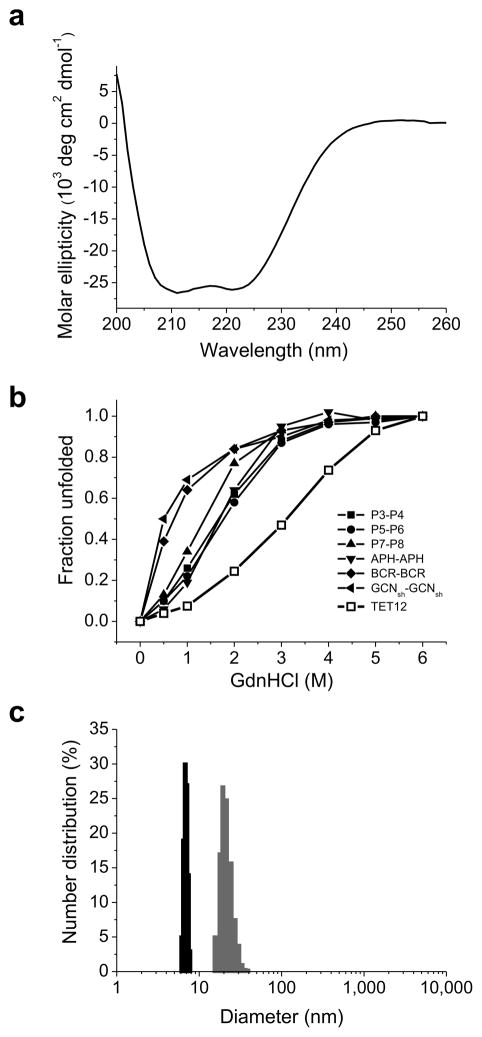
Figure 2. Self-assembly of the tetrahedron-forming polypeptide TET12.
(a) Analysis of the secondary structure of self-assembled TET12 polypeptide was performed by a circular dichroism measurement. The average curve of three scans of the representative sample is shown. The analysis of this far-UV CD spectrum indicates 82% of α-helical structure. (b) Increased thermodynamic stability of TET12 in comparison to its dimeric coiled-coil peptide building elements, determined by CD signal at 222 nm depending on the concentration of GdnHCl. The representative curves are shown. (c) The hydrodynamic diameter of the TET12 unfolded in 6 M GdnHCl (gray) and self-assembled TET12 (black), 20.0 ± 2.9 nm and 6.9 ± 0.4 nm, respectively, were determined by DLS. The mean values ± s.d. of three independent experiments are calculated. The representative histogram graph of one measurement is shown.
Visualization of the assembled structures by microscopy
Atomic force microscopy (AFM) was used to visualize the self-assembled polypeptide structures that were bound to the mica substrate. We found small, discrete, homogeneous objects that were produced under the slow annealing conditions by dialysis at low polypeptide concentrations (100 nM), wherein structural features were arranged in a triangular shape with dimensions below 25 nm (Fig. 3a). AFM images of polypeptide particles were similar to the images of DNA tetrahedra formed from trigonal DNA star components3. In both polynucleotide and our polypeptide tetrahedra collapse of the tetrahedral three-dimensional structure was observed probably due to interactions with mica and dehydration. Additionally, the inherent characteristic of AFM is a tip-sample convolution, which makes features appear wider and is especially prominent when features are of similar size as the tip diameter. Further, we used positive uranyl staining to obtain the sufficient contrast of small polypeptide particles for TEM visualization. The observed particles exhibited triangular shape or other projections of a tetrahedron whose inner edges measured approximately 5 nm (Supplementary Fig. 9a), in agreement with hydrodynamic size measurement (Fig. 2c) and consistent with the length of the designed coiled-coil dimers (Supplementary Fig. 5). Even more convincing topological arrangement of the vertices of the particles was observed by a prolonged uranyl staining procedure, resulting in an increased concentration of the stain at vertices, where the three linkers converge (Fig. 3b and Supplementary Fig. 9b). In those images we observed the arrangement of vertices corresponding to different projections of a tetrahedron. In addition to polypeptide refolding by dialysis, slow temperature annealing also produced objects of similar size and shape (Supplementary Fig. 9e). Annealing at the higher, aggregation-prone 10 μM concentration led to formation of a connected network (Supplementary Fig. 10). Polypeptide TET12 comprised an N-terminal hexahistidine polypeptide tag that can bind Ni2+ ions complexed with nitrilotriacetic acid (NTA). This allowed us to probe the location of the polypeptide N-terminus by the addition of 1.8-nm nanogold beads coated with Ni-NTA to the self-assembled polypeptide TET12. Clearly recognizable triangles and other tetrahedral projections were visible by TEM, with a dense nanogold sphere attached to one vertex (Fig. 3c,d and Supplementary Fig. 9c,d), which improved the contrast and provided an additional evidence of the dimensions of the formed nanostructures, corroborating the success of the design.
Figure 3. Imaging of the assembled TET12 reveals tetrahedral topology.
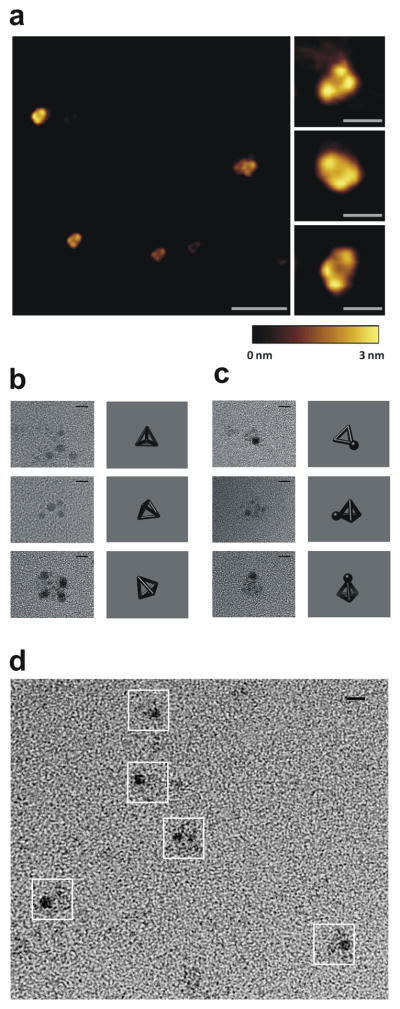
(a) A representative AFM image of the self-assembled TET12 polypeptide (scale bar represents 100 nm) and close-up view (right; scale bars, 20 nm). A height scale bar is shown at the bottom. (b–d) TEM images of self-assembled TET12 tetrahedral structures. Representative particles from TEM images and projections of a tetrahedron in the matching orientation are shown, where the samples on grids were stained either with uranyl for 3 min (b) or first with 1.8-nm-Ni-NTA-nanogold beads followed by 1 min uranyl staining (c,d). White boxes in (d) indicate the identified particles. Scale bars, 5 nm.
Verification of a closed polypeptide topological path
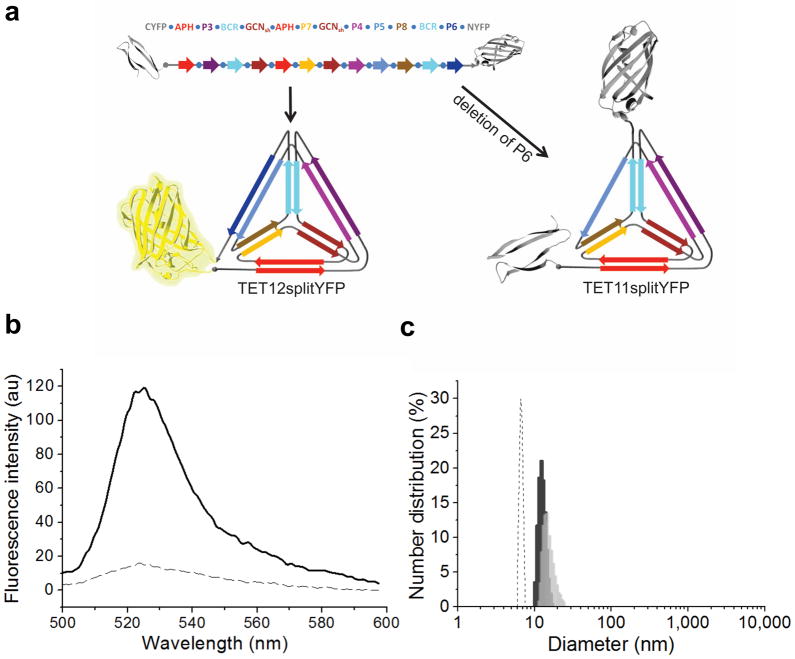
The N- and C-terminus of the TET12 polypeptide must by design coincide at the same vertex of the self-assembled tetrahedron (Supplementary Note 1 and Fig. 1b). We confirmed this property by grafting an N- and C-terminal segment of a split yellow fluorescent protein (YFP) to each terminus of the polypeptide. Therefore, the YFP fluorescence could only be reconstituted by the correct self-assembly (Fig. 4a). This is a strong topological constraint, because the first and the last segments of TET12 do not interact directly and the proximity of both termini of the self-assembled tetrahedron is a consequence of the complex global topology, encoded in the sequential order of coiled-coil-forming segments (Fig. 1a,b). The recombinant fusion polypeptide TET12splitYFP, isolated from bacteria (Supplementary Fig. 7b), contained a high content of α-helices (data not shown); yet, the fluorescence was not observed, suggesting misfolding of the tetrahedron in bacteria, presumably due to kinetically trapped intermediates or formation of intermolecular aggregates. This finding demonstrates that YFP does not reconstitute in nonspecific aggregates. However, dialysis of the fusion polypeptide TET12splitYFP solubilized in 6 M GdnHCl resulted in high fluorescence intensity of the reconstituted split YFP (Fig. 4b), confirming formation of the correct circular path of the chain and the coincidence of both termini of the polypeptide chain at the same vertex of the structure. TEM analysis additionally confirmed the formation of tetrahedron-like particles from TET12splitYFP polypeptide (Supplementary Fig. 9f).
Figure 4. Determination of the sequence-dependent topology of the self-assembled polypeptide.
(a) Scheme of the polypeptide chains TET12splitYFP and TET11splitYFP, used to establish the coincident N- and C-termini of the polypeptide chain and requirement for the defined order of segments for the assembly of a tetrahedron. (b) Reconstituted YFP fluorescence measurements reveal the correct self-assembly of TET12splitYFP (—), whereas the deletion of a single building element (TET11splitYFP) prevents formation of complete tetrahedron and YFP reconstitution (---). The average curves of three scans of the representative samples are shown. (c) DLS measurements show increased hydrodynamic diameters of the assembled polypeptides TET11 with a single deleted segment (13.9 ± 1.1 nm, dark gray) and TET12scr with two swapped segments (15.0 ± 1.5 nm, light gray) in comparison to the diameter of the assembled TET12 (6.9 ± 0.4 nm, dashed). The mean values ± s.d. of three independent experiments arecalculated. The representative histogram graph of one measurement is shown.
Tetrahedral self-assembly depends on the segment order
Polypeptide self-assembly into the tetrahedral topology depended on the order of coiled-coil-forming segments within the polypeptide, just as folding of the native protein relies on the precise amino acid sequence. Thus, the self-assembly should be sensitive to the disruption of the order of coiled-coil building elements. To this end, we tested 2 additional polypeptide constructs: one in which a single coiled-coil-forming segment was deleted (TET11) and another, scrambled construct that retained all coiled-coil-forming segments but in which 2 segments were swapped (TET12Scr, with the segment order 4,5,1,2,3,6,7,8,9,10,11,12) (see Online Methods for amino acid sequences). We predicted that both polypeptides would fail to adopt the tetrahedral topology. Indeed, both constructs exhibited increased aggregation during refolding, had increased hydrodynamic size of the soluble monomers, 13.9 ± 1.1 nm and 15.0 ± 1.5 nm for TET11 and TET12Scr, respectively (Fig. 4c), and exhibited decreased stability (Supplementary Fig. 11). In contrast to TET12, we did not observe formation of any regular structures by TEM in either the truncated or scrambled constructs (data not shown). Moreover, in the TET11 polypeptide, which had a deletion of a single coiled-coil-forming segment (P6), the N- and C-terminus no longer coincided and, as expected, the tetrahedral polypeptide (TET11splitYFP) could not reconstitute YFP, in contrast to the integral tetrahedral construct TET12splitYFP (Fig. 4b).
DISCUSSION
We have demonstrated that the rational design of tertiary structures from natural polymers, as pioneered by the DNA-based self-assembly, can be extended to polypeptides, based on the specificity of pairwise interactions between the coiled-coil building elements. Modularization of the design, based on the well understood building blocks, allowed us to use the topological design rather than full atomistic modeling. The ability of coiled-coil-forming peptide segments to adopt either parallel or antiparallel orientation expands the number of reachable topological arrangements and facilitated the formation of a complex polypeptide scaffold. This principle enabled us to design a tetrahedral topology that self-assembles from a single polypeptide chain, while the tetrahedron based on DNA origami has to be composed of several chains or include a four-stranded edge27. The described method allowed us to create a new topological protein fold, which has not been observed in nature. The design based on concatenated coiled-coil-forming segments can produce asymmetric structures, which is difficult to achieve by the assembly of protein domains or for other nanoparticles composed of organic or inorganic building blocks. We demonstrated attachment of nanogold or protein domain cargo to the N- and/or C-termini of the polypeptide, and suggest other vertices of the polypeptide structures could similarly be decorated with selected functional groups. Due to their variable length and high length: width aspect ratio coiled-coil dimers seem to be an almost ideal type of building elements, allowing formation of large cavities within the polypeptide polyhedra in contrast to interacting protein oligomerization domains, that occupy a significant fraction of the volume. Interactions between coiled-coil segments can also be used to combine several building blocks into larger assemblies28. The presented technology could therefore be used for a range of applications, such as utilizing designed cavities and arrangement of the amino acid side chains for the drug delivery or creation of artificial catalytic sites. Polypeptides can be produced cost-effectively in the recombinant form, which is a clear advantage for the technological implementation of designed polypeptide structures. The current experimental limitation is the number of available orthogonal coiled-coil-forming segments. More than 2000 coiled-coil-containing proteins in the human proteome29 and hundreds of natural coiled-coils30,31 represent the source of potential building elements, while the rational design of new orthogonal sets may extend the variability in size, oligomerization state or other features. The principles of “polypeptide origami” demonstrated on the design of a tetrahedron could be applied to construct other polyhedra, e.g. listed in the Supplementary Table 1 or, in general, any shapes that can be tessellated.
ONLINE METHODS
Peptide and linker sequences
The sequences of the coiled-coil-forming peptides used in the TET12 are artificial sequences P3 to P8 and APH, as well as natural BCR and a homodimer-forming peptide based on GCN, GCNsh.
P3:SPEDEIQQLEEEIAQLEQKNAALKEKNQALKYG; P4:SPEDKIAQLKQKIQALKQENQQLEEENAALEYG; P5:SPEDENAALEEKIAQLKQKNAALKEEIQALEYG; P6:SPEDKNA ALKEEIQALEEENQALEEKIAQLKYG; P7: SPEDEIQALEEKNAQLKQEIAALEEKNQAL KYG; P8:SPEDKIAQLKEENQQLEQKIQALKEENAALEYG; APH:MKQLEKELKQLEKEL QAIEKQLAQLQWKAQARKKKLAQLKKKLQA; BCR:DIEQELERAKASIRRLEQEVNQERSRMAYLQTLLAK; GCNsh:QLEDKVEELLSKNYHLENEVARLKKLVG
The tetrapeptide linker SGPG was positioned between every two consecutive coiled-coil-forming peptide segments to interrupt the sequence with helical propensity and enable the flexibility at the vertices.
Molecular modeling of a TET12 tetrahedron
A molecular model of the self-assembled TET12 was constructed using the MODELLER comparative modeling software, version 9.10 (http://salilab.org/modeller/)32,33. Structural features, such as atom-atom distances and dihedral angles, were extracted from known three-dimensional structures of coiled-coil dimers (Protein Data Bank codes 1zik for parallel dimers, and 1k1f for anti-parallel dimers). Candidate structures of the designed sequence were restrained to reproduce these features along each tetrahedron edge, as well as correct stereochemistry, excluded volume, and known properties of proteins such as main chain and side chain dihedral angle distributions. To reduce the conformational search space, the linkers were initially placed at the tetrahedron vertices and the remaining residues simply interpolated between them. 2000 candidate models were generated and the model that best satisfied all restraints was selected. The structure was verified using MolProbity34.
Construction of plasmids
The plasmid BB-NIC-II-HisN-TET12 used for the production of TET12 polypeptide was constructed by ligation of the NgoMIV and SpeI digested synthetic gene (1392 bp) encoding polypeptide TET12 synthesized by Geneart (Regensburg, GE) into plasmid BB-NIC-II-HisN previously cut with NgoMIV and SpeI (2261 bp, http://partsregistry.org/wiki/index.php?title=Part:BBa_K245005 ). BB-NIC-II-HisN contains the T7 promoter and a His-tag coding sequence upstream of the gene. TET12scr was constructed with ligation of three PCR products. Regions encoding GCNsh-APH, APH-P3-BCR and P7-GCNsh-P4-P5-P8-BCR-P6 were amplified from the TET12 gene and first ligated into BB-NIC-II-HisN separately through NgoMIV and SpeI restriction sites. Afterwards BB-NIC-II-HisN-APH-P3-BCR was cut with EcoRI and BspEI and ligated as front insert into EcoRI and NgoMIV digested BB-NIC-II-HisN-P7-GCNsh-P4-P5-P8-BCR-P6. BB-NIC-II-HisN-GCNsh-APH was cut with EcoRI and BspEI and ligated as front insert into BB-NIC-II-HisN-APH-P3-BCR-P7-GCNsh-P4-P5-P8-BCR-P6 previously cut with EcoRI and NgoMIV. The resulting order of the coiled-coil-forming segments was GCNsh-APH-APH-P3-BCR-P7-GCNsh-P4-P5-P8-BCR-P6 (TET12scr). TET11 was obtained by PCR amplification of the APH-P3-BCR- GCNsh -APH-P7- GCNsh-P4-P5-P8-BCR encoding region from the TET12 gene. The PCR product was then digested by NgoMIV and SpeI and subcloned into the BB-NIC-II-HisN plasmid previously cut with NgoMIV and SpeI. Split YFP fusions were constructed by joining the C-terminal segment of YFP (CYFP) at the 5′-end and the segment encoding the N-terminal portion of YFP (NYFP) to the 3′ of the TET12 or TET11 coding regions. First, BB-NIC-II-HisN-CYFP (this plasmid was obtained by amplifying CYFP from a synthetic gene synthesized by Geneart and subcloning it into BB-NIC-II-HisN) with EcoRI and BspEI and the resulting insert was ligated into EcoRI and NgoMIV digested BB-NIC-II-HisN-TET12 or BB-NIC-II-HisN-TET11. Afterwards BB-NIC-II-HisN-CYFP-TET12 or BB-NIC-II-HisN-CYFP-TET11 was cut with EcoRI and BspEI and the resulting front insert ligated into BB-NIC-II-HisN-NYFP (this plasmid was obtained by amplifying NYFP from a synthetic gene synthesized by Geneart and subcloning it into BB-NIC-II-HisN) previously cut with EcoRI and NgoMIV. All constructs were verified by DNA sequencing.
Polypeptide production and isolation
Peptides for testing the orthogonality of the building blocks were synthesized by small scale solid state chemical synthesis the Keck Biology Resource Lab of the Yale school of medicine; mass spectra of the purified products were in agreement with expected values. E. coli BL21(DE3)pLysS cultures transformed with proper plasmids were grown at 37 °C and 160 rpm in LB medium containing ampicillin (0.1 mg/ml). After induction of polypeptide production with 1 mM IPTG, incubation proceeded for additional 4 hours. Cell pellets were resuspended in the lysis buffer (10 mM Tris pH 8.0, 0.1 % deoxycholate) and sonicated. Bacterial inclusion bodies were solubilized in 6 M guanidine hydrochloride (GdnHCl) and purified by chelating chromatography (Ni-NTA agarose column, Quiagen, GE) followed by size exclusion chromatography (SEC, Agilent, USA) on Proteema 300Å 5μM, 8ξ300 mm column (PSS,GE). Eluted fractions that contained polypeptides were dialyzed Milli Q water and the precipitate was dissolved in 6 M GdnHCl.
Polypeptide self-assembly
Purified polypeptides were diluted to a concentration of 100 nM in 6 M GdnHCl in 20 mM Tris buffer, pH 8.5, 150 mM NaCl. Refolding proceeded by dialysis against 20 mM Tris buffer, pH 8.5, 150 mM NaCl for 20 h at 4 °C. After dialysis the samples were prepared for TEM and AFM characterization. Then the sample was concentrated with a 30-kDa MWCO concentrator (Amicon, USA) and the solution of 4 μM concentration was used for CD, DLS and fluorescence measurements. Slow temperature annealing was performed by cooling the sample from 90 °C to 20 °C over 20 hours.
Circular dichroism (CD)
CD measurements were performed on a Chirascan CD spectrometer (Applied Photophysics, Leatherhead, UK) in far-UV in a 1-mm quartz cell (Hellma, Mullheim, Germany) at 20 °C using 1 nm step, 1 nm bandwidth and 1 s sampling. Secondary structure of 4 μM polypeptide solution was analyzed by scan measurement from 200 nm to 260 nm. A chemical denaturation study was performed at different concentrations of GdnHCl from 0 M to 6 M, and the thermodynamic stability of the structures was determined from the ellipticity at 222 nm. The sample measurements were performed in triplicate and the mean values ± s.d. were calculated.
Dynamic light scattering (DLS)
The size of self-assembled structures in samples of 4 μM concentration was measured on a Zetasizer Nano (Malvern, UK) at 20 °C using an angle of 173° and 633-nm laser. The size distribution of particles was recorded, and a hydrodynamic diameter was calculated using the software provided by the manufacturer.
Fluorescence measurements
Fluorescence measurements were performed with an LS55 spectrofluorimeter (Perkin Elmer Life Sciences, UK) with a double monochromator. All measurements were done at 20 °C in 3-mm × 3-mm quartz cell. A slit width of 5 nm was used for both excitation and emission. The samples of 4 μM concentration were excited at 485 nm and the emission spectra from 500 nm to 600 nm were recorded.
Transmission electron microscopy (TEM)
Polypeptide solutions of 100 nM concentration were applied to holey carbon-coated copper grids (SPI, West Chester, USA). The grid was placed on a droplet of sample solution (15 μL) for 2 min. Then, the grid was rinsed 3 times with distilled water. The uranyl staining was performed using 2 % (w/v) uranyl acetate or formate by incubating the grid on a drop of stain solution for 1 min (short time staining) or for extended time of 3 min (long time staining). Excess solution was removed by rinsing the grid with water. Alternatively, before positive staining 1.8-nm-NiNTA-nanogold beads (Nanoprobes, NY, USA) were added to the samples, according to manufacturer’s instructions, to form a complex with the hexa-His-tag of the polypeptide. The structures were observed on TEM (JOEL JEM 2100 LaB6 microscope, Japan), operated at 180 kV. Images were acquired on a Gatan Orius CCD camera in underfocus.
Atomic force microscopy (AFM)
A drop (10 μl) of 100 nM polypeptide solution was spotted onto freshly cleaved mica surface and held for 15 s to achieve strong adsorption. The sample drop was then washed twice with filtered Milli-Q water and dried under a nitrogen stream. Protein samples were imaged in the acoustic alternating current mode on an Agilent Technologies 5500 scanning probe microscope using silicon cantilevers (Arrow-NCR-50) with a force constant of 42 N/m.
Supplementary Material
Acknowledgments
This research was supported by grants from the Slovenian Research Agency (J2-2131, P4-0176 to RJ) and Centre of Excellence EN-FIST, financed in part by the EU structural funds. Ben Webb and Andrej Šali acknowledge NIH grants R01 GM083960 and R01 GM54762 to AS. We thank the rest of the members of the 2009 Slovenian iGEM Team (students Marko Verce, Anja Lukan, Nika Debeljak, Špela Miklavič and Urška Jelerčič, and mentors Ota Fekonja, Jelka Pohar, Robert Bremšak and Mojca Benčina) for their pioneering work on the development of concatenated coiled-coil based nanostructures (http://2009.igem.org/Team:Slovenia), underlying the development of the polypeptide polyhedra; Robert Bremšak for excellent technical support, Jernej Rus for calculation of the Supplementary Table 1, Slovenian Electron microscopy center for the use of electron microscopes and Kristina Djinovič Carugo for comments on the manuscript.
Footnotes
Author contributions
R.J. conceived the idea and designed the tetrahedral polypeptide, participated in the mathematical analysis of the polyhedral topology, coordinated the project, discussed the results and wrote the manuscript; H.G. performed TEM, DLS, CD and fluorescence experiments, coordinated the project, discussed the results and wrote the manuscript; S.B. prepared, purified, and analyzed polypeptides, performed TEM, CD and fluorescence experiments, discussed the results and wrote the manuscript; T.D. prepared, purified, and analyzed polypeptides, performed AFM measurements, discussed the results and wrote the manuscript; D.V. performed TEM measurements; I.H.-B. performed AFM measurements; A.M. performed DLS measurements; B.W. prepared the molecular model; A.Š. prepared the molecular model and wrote the manuscript; S.K. solved the mathematical analysis of the polyhedral topology and wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
Supplementary information is available in the online version of the paper.
References
- 1.Chen JH, Seeman NC. Synthesis from DNA of a molecule with the connectivity of a cube. Nature. 1991;350:631–3. doi: 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]
- 2.Rothemund PW. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 3.He Y, et al. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature. 2008;452:198–201. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- 4.Han DR, et al. DNA Origami with Complex Curvatures in Three-Dimensional Space. Science. 2011;332:342–346. doi: 10.1126/science.1202998. [DOI] [PubMed] [Google Scholar]
- 5.Kuhlman B, et al. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–8. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 6.Regan L, DeGrado WF. Characterization of a helical protein designed from first principles. Science. 1988;241:976–8. doi: 10.1126/science.3043666. [DOI] [PubMed] [Google Scholar]
- 7.Fleishman SJ, et al. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science. 2011;332:816–21. doi: 10.1126/science.1202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hecht MH, Richardson JS, Richardson DC, Ogden RC. De novo design, expression, and characterization of Felix: a four-helix bundle protein of native-like sequence. Science. 1990;249:884–91. doi: 10.1126/science.2392678. [DOI] [PubMed] [Google Scholar]
- 9.Padilla JE, Colovos C, Yeates TO. Nanohedra: using symmetry to design self assembling protein cages, layers, crystals, and filaments. Proc Natl Acad Sci U S A. 2001;98:2217–21. doi: 10.1073/pnas.041614998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doles T, Bozic S, Gradisar H, Jerala R. Functional self-assembling polypeptide bionanomaterials. Biochemical Society Transactions. 2012;40:629–634. doi: 10.1042/BST20120025. [DOI] [PubMed] [Google Scholar]
- 11.King NP, et al. Computational design of self-assembling protein nanomaterials with atomic level accuracy. Science. 2012;336:1171–4. doi: 10.1126/science.1219364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai YT, Cascio D, Yeates TO. Structure of a 16-nm cage designed by using protein oligomers. Science. 2012;336:1129. doi: 10.1126/science.1219351. [DOI] [PubMed] [Google Scholar]
- 13.Lakshmanan A, Zhang S, Hauser CA. Short self-assembling peptides as building blocks for modern nanodevices. Trends Biotechnol. 2012;30:155–65. doi: 10.1016/j.tibtech.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Crick FH. Is alpha-keratin a coiled coil? Nature. 1952;170:882–3. doi: 10.1038/170882b0. [DOI] [PubMed] [Google Scholar]
- 15.Woolfson DN. The design of coiled-coil structures and assemblies. Adv Protein Chem. 2005;70:79–112. doi: 10.1016/S0065-3233(05)70004-8. [DOI] [PubMed] [Google Scholar]
- 16.Fong JH, Keating AE, Singh M. Predicting specificity in bZIP coiled-coil protein interactions. Genome Biol. 2004;5:R11. doi: 10.1186/gb-2004-5-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Potekhin SA, et al. De novo design of fibrils made of short alpha-helical coiled coil peptides. Chem Biol. 2001;8:1025–32. doi: 10.1016/s1074-5521(01)00073-4. [DOI] [PubMed] [Google Scholar]
- 18.Gribbon C, et al. MagicWand: a single, designed peptide that assembles to stable, ordered alpha-helical fibers. Biochemistry. 2008;47:10365–71. doi: 10.1021/bi801072s. [DOI] [PubMed] [Google Scholar]
- 19.Gauba V, Hartgerink JD. Self-assembled heterotrimeric collagen triple helices directed through electrostatic interactions. J Am Chem Soc. 2007;129:2683–90. doi: 10.1021/ja0683640. [DOI] [PubMed] [Google Scholar]
- 20.Moutevelis E, Woolfson DN. A periodic table of coiled-coil protein structures. J Mol Biol. 2009;385:726–32. doi: 10.1016/j.jmb.2008.11.028. [DOI] [PubMed] [Google Scholar]
- 21.Gradisar H, Jerala R. De novo design of orthogonal peptide pairs forming parallel coiled-coil heterodimers. J Pept Sci. 2011;17:100–6. doi: 10.1002/psc.1331. [DOI] [PubMed] [Google Scholar]
- 22.Bromley EH, Sessions RB, Thomson AR, Woolfson DN. Designed alpha-helical tectons for constructing multicomponent synthetic biological systems. J Am Chem Soc. 2009;131:928–30. doi: 10.1021/ja804231a. [DOI] [PubMed] [Google Scholar]
- 23.Lumb KJ, Carr CM, Kim PS. Subdomain folding of the coiled coil leucine zipper from the bZIP transcriptional activator GCN4. Biochemistry. 1994;33:7361–7. doi: 10.1021/bi00189a042. [DOI] [PubMed] [Google Scholar]
- 24.Gurnon DG, Whitaker JA, Oakley MG. Design and characterization of a homodimeric antiparallel coiled coil. J Am Chem Soc. 2003;125:7518–9. doi: 10.1021/ja0357590. [DOI] [PubMed] [Google Scholar]
- 25.Taylor CM, Keating AE. Orientation and oligomerization specificity of the Bcr coiled-coil oligomerization domain. Biochemistry. 2005;44:16246–56. doi: 10.1021/bi051493t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ortega A, Amoros D, Garcia de la Torre J. Prediction of hydrodynamic and other solution properties of rigid proteins from atomic- and residue-level models. Biophys J. 2011;101:892–8. doi: 10.1016/j.bpj.2011.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z, et al. A replicable tetrahedral nanostructure self-assembled from a single DNA strand. J Am Chem Soc. 2009;131:13093–8. doi: 10.1021/ja903768f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyle AL, et al. Squaring the circle in peptide assembly: from fibers to discrete nanostructures by de novo design. J Am Chem Soc. 2012;134:15457–67. doi: 10.1021/ja3053943. [DOI] [PubMed] [Google Scholar]
- 29.Rackham OJ, et al. The evolution and structure prediction of coiled coils across all genomes. J Mol Biol. 2010;403:480–93. doi: 10.1016/j.jmb.2010.08.032. [DOI] [PubMed] [Google Scholar]
- 30.Reinke AW, Grant RA, Keating AE. A synthetic coiled-coil interactome provides heterospecific modules for molecular engineering. J Am Chem Soc. 2010;132:6025–31. doi: 10.1021/ja907617a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newman JR, Keating AE. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science. 2003;300:2097–101. doi: 10.1126/science.1084648. [DOI] [PubMed] [Google Scholar]
- 32.Eswar N, et al. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics. 2006;Chapter 5(Unit 5):6. doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 34.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallographica Section D-Biological. Crystallography. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.