Abstract
Legionella pneumophila is a bacterial pathogen present in aquatic environments that can cause a severe pneumonia called Legionnaires’ disease. Soon after its recognition, it was shown that Legionella replicates inside amoeba, suggesting that bacteria replicating in environmental protozoa are able to exploit conserved signaling pathways in human phagocytic cells. Comparative, evolutionary, and functional genomics suggests that the Legionella–amoeba interaction has shaped this pathogen more than previously thought. A complex evolutionary scenario involving mobile genetic elements, type IV secretion systems, and horizontal gene transfer among Legionella, amoeba, and other organisms seems to take place. This long-lasting coevolution led to the development of very sophisticated virulence strategies and a high level of temporal and spatial fine-tuning of bacteria host–cell interactions. We will discuss current knowledge of the evolution of virulence of Legionella from a genomics perspective and propose our vision of the emergence of this human pathogen from the environment.
Legionella can infect many eukaryotic cells, including amoeba, ciliates, and human cells where it causes Legionnaires’ disease. A large mobile gene pool combined with a highly dynamic genome seems to underlie its success.
Legionellosis or Legionnaires’ disease is a severe pneumonia caused by bacteria belonging to the genus Legionella. The first member of this genus, Legionella pneumophila, was recognized as a human pathogen during an outbreak of severe pneumonia in 1976 in Philadelphia, PA (Fraser et al. 1977; McDade et al. 1977). Since then, outbreaks occur every year, in particular in the industrialized world, and legionellosis remains an up-to-date health problem. Thirty-five years after the discovery of Legionella pneumophila, the genus Legionella has grown considerably as it now contains 59 species (http://old.dsmz.de/microorganisms/bacterial_nomenclature_info.php?genus=Legionella), all of which are ubiquitous, environmental bacteria. Although 24 different species of Legionella have been isolated at least once from humans, L. pneumophila is the major cause of Legionnaires’ disease worldwide as it causes more than 90% of the diagnosed cases. Legionella longbeachae is the second cause of legionellosis, responsible for 3.9% of the cases (Yu et al. 2002; Newton et al. 2010). Interestingly, the epidemiology is different in Australia and New Zealand, where L. pneumophila accounts for only 45.7% but L. longbeachae for 30.4% of the cases. Thus, research focuses mainly on L. pneumophila and L. longbeachae. In contrast, little or nothing is known about the other Legionella species with respect to their environmental distribution, ecology, or the capacity to trigger human disease. All Legionella species, except L. longbeachae, which is present mainly in compost and potting mix, are found in freshwater environments (Whiley and Bentham 2011).
Legionella are intracellular bacteria whose natural hosts are aquatic protozoa, in which these bacteria replicate and are protected from harsh environmental conditions (Rowbotham 1980). Legionella are able to parasitize at least 20 different species of amoeba, two species of ciliated protozoa, and one slime mold (Lau and Ashbolt 2009), but are associated most frequently with amoeba belonging to the genera Acanthamoeba, Hartmanella, and Naegleria (Fields 1996). Interestingly, not all Legionella will grow in the same amoebal host (Rowbotham 1986) and certain amoeba seem also to be resistant to Legionella infection (Atlan et al. 2012). Thus, Legionella are broad host-range protozoan parasites but some species, in particular L. pneumophila, are also pathogens of humans. The vehicle of transmission to humans is aerosols loaded with Legionella. Changes in the human lifestyle in the last decade led to an increased number of artificial water systems such as air conditioning units, cooling towers, showers, and fountains, where water is given off in a fine spray providing the bacterium with the possibility to reach humans as an accidental host. However, although Legionella is able to infect humans, human infection is a dead end for Legionella replication, as person-to-person transmission has never been reported. Therefore, the evolution of virulence traits in L. pneumophila and L. longbeachae has resulted from the interaction with environmental protozoa.
The capacity of Legionella to infect its natural hosts, amoeba or ciliated protozoa but also human macrophages, indicates that the mechanisms developed by this bacterium to infect low eukaryotes can also be used for infection of cells of high eukaryotes like humans as was proposed the first time by Rowbotham in 1980 (Rowbotham 1980). This also suggests that virulence factors of Legionella probably target conserved eukaryotic pathways, allowing Legionella to infect many phagocytic eukaryotic cells like different aquatic protozoa, ciliates, and human cells. Therefore, the identification of what are these factors, how they have been acquired, and what are their functions, is fundamental to decipher host–pathogen interactions and the virulence strategies used by intracellular pathogens.
In recent years, it has been shown that several intracellular bacterial pathogens are able to enter and replicate in environmental amoebae, similar to what is known for Legionella (Molmeret et al. 2005). This observation has led to the idea that free-living amoebae are the “Trojan horses” of the microbial world, as they have a role in survival, replication, and transmission of bacteria (Winiecka-Krusnell and Linder 1999). The interaction between both organisms provides bacteria with the capacity to infect human cells and to become human pathogens. Furthermore, genome analyses and comparative genomics of amoeba-associated bacteria revealed that common virulence factors are present in bacteria living with amoebae (Schmitz-Esser et al. 2010). This finding suggested that the genes coding for these virulence factors may be interchanged and that they may thus be part of mobile genetic elements. The complete set of mobile genetic elements of a genome is commonly called “mobilome” (Siefert 2009), a term we will use hereafter. Particularly in bacteria living with amoebae, this mobilome seems to be highly dynamic. Because L. pneumophila is probably the best-studied amoeba-associated bacterium, it has become a model to study the role of amoebal hosts in bacterial pathogenesis and the acquisition of virulence factors from this host–pathogen relationship.
In this review, we will present the state-of- the-art of the evolution of virulence in Legionella from a genomics perspective and how current data increased our knowledge about Legionella virulence evolution and host–pathogen interaction and acquisition of virulence factors in general.
THE Legionella GENOMES ARE GENETICALLY DIVERSE
L. pneumophila strains Philadelphia, Paris, and Lens were the first Legionella genomes sequenced (Cazalet et al. 2004; Chien et al. 2004). Each strain contains one circular chromosome that is characterized by an average G + C content of about 38% and by a size between 3.3 and 3.5 Mb. In each strain ∼3000 protein-coding genes are predicted, representing a coding capacity of 88% (Table 1A). Comparative genomics revealed that ∼300 genes (∼10%) are specific for each strain. Taking into account that these strains belong to the same species and serogroup, the genomic diversity among them is relatively high. Further analysis of the gene content of 217 L. pneumophila strains using a Legionella DNA array confirmed that the gene content of the L. pneumophila genome is highly diverse (Cazalet et al. 2008). Both studies suggested that the main source of diversity among the Legionella genomes is mobile genetic elements and horizontal gene transfer among L. pneumophila strains, but also among strains belonging to different Legionella species, other bacterial species, and most surprisingly, probably also between Legionella and their eukaryotic hosts (Cazalet et al. 2004, 2008).
Table 1.
General features of the Legionella genome sequences published
| (A) L. pneumophila | ||||||||
|---|---|---|---|---|---|---|---|---|
| Paris | Lens | Philadelphia | Corby | Alcoy | 130b | Lorraine | HL06041035 | |
| Chromosome size (Mb) | 3.5 | 3.3 | 3.4 | 3.6 | 3.5 | 3.5 | 3.5 | 3.5 |
| G + C content (%) | 38 | 38 | 38 | 38 | 38 | 38 | 38 | 38 |
| Number of genes | 3178 | 3034 | 3083 | 3290 | 3197 | 3293 | 3170 | 3184 |
| Number of protein-coding genes | 3079 | 2921 | 2999 | 3193 | 3191 | 3288 | 3080 | 3079 |
| Pseudogenes | 45 | 59 | 32 | 44 | ? | ? | 37 | 53 |
| tRNA | 43 | 43 | 43 | 44 | ? | 42 | 44 | 43 |
| 16S/23S/5S | 3/3/3 | 3/3/3 | 3/3/3 | 3/3/3 | 3/3/3 | 3/3/3 | 3/3/3 | 3/3/3 |
| Coding density (%) | 87 | 87 | 88 | 87 | 86 | 87 | 87 | |
| Plasmids | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| (B) L. longbeachae | |||||
|---|---|---|---|---|---|
| NSW 150# | D-4968+ | ATCC39462+ | 98072+ | C-4E7+ | |
| Chromosome size (Kb) | 4.0 | 4.0 | 4.1 | 4.0 | 3.9 |
| G + C content (%) | 37.1 | 37.0 | 37.0 | 37.0 | 37 |
| Number of genes | 3660 | 3557 | – | – | – |
| 16S/23S/5S | 4/4/4 | 4/4/4 | 4/4/4 | 4/4/4 | 4/4/4 |
| No. of contigs > 0.5–300 kb | Complete | 13 | 64 | 65 | 63 |
| N50 contig sizea | Complete | – | 138 kb | 129 kb | 134 kb |
| Percentage of coverageb | 100% | 96.3 | 96.3 | 93.4 | 93.1 |
| Number of SNP with NSW150 | – | 1900 | 1611 | 16 853 | 16 820 |
| Plasmids | 1 | 1 | 0 | 1 | 1 |
aN50 contig size, calculated by ordering all contig sizes and adding the lengths (starting from the longest contig) until the summed length exceeds 50% of the total length of all contigs.
bFor SNP detection: #, completed sequence; +, draft sequence, not completed; SNP, single nucleotide polymorphism.
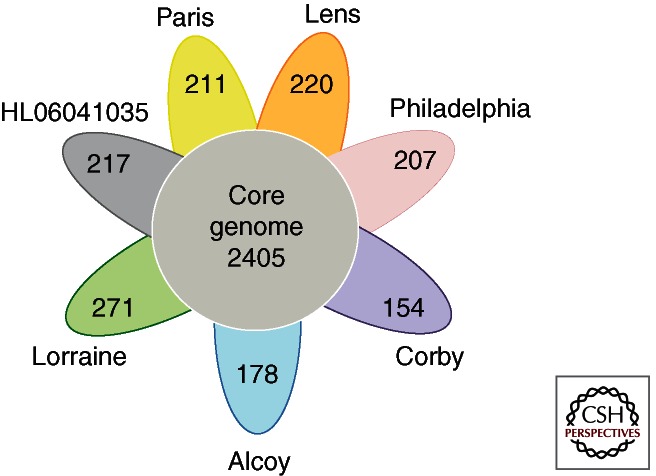
Recent years have seen five new L. pneumophila genomes sequenced providing the possibility to study their diversity and evolution in depth (Table 1A) (Steinert et al. 2007; D’Auria et al. 2010; Schroeder et al. 2010; Gomez-Valero et al. 2011b). Comparative analyses of the eight available genomes showed that the core genome contains 2405 orthologous groups of genes, and each genome when compared with the seven others contains between 154 and 271 strain-specific genes (Fig. 1). Furthermore, more than 1000 genes are not present in all eight genomes and thus also belong to the flexible gene content or accessory genome. In contrast, the L. longbeachae genomes, five of which have been sequenced to date (Cazalet et al. 2010; Kozak et al. 2010), have a slightly larger genome size of 3.9–4.1 Mb and show much lower diversity among them (Table 1B). When comparing two strains of L. longbeachae belonging to different serogroups (Sg), the overall DNA polymorphism is less than 0.4%, whereas this polymorphism is ∼2% between L. pneumophila strains of the same Sg (Cazalet et al. 2010). Interestingly, despite the fact that L. pneumophila and L. longbeachae cause the same human disease, high divergence of these two species was observed. Only 2290 (65.2%) L. longbeachae genes are orthologous to L. pneumophila genes, whereas 1222 (34.8%) are L. longbeachae specific with respect to L. pneumophila Paris, Lens, Philadelphia, and Corby (Cazalet et al. 2010; Gomez-Valero et al. 2011b). The different genome analyses, when combined, revealed that the accessory genome of Legionella, containing approximately 3500 genes when eight L. pneumophila and five L. longbeachae genomes are analyzed, is large and, most interestingly, the main part seems to be transferable, thus constituting the mobilome of the genus.
Figure 1.
Shared and specific gene content of eight L. pneumophila genomes. Each petal represents a genome with an associated color. The number in the center of the diagram represents the orthologous groups of genes shared by all the genomes. The number inside of each individual petal corresponds to the specific genes of each genome with nonorthologous genes in any of the other genomes. The orthologous groups were defined using the program PanOCT (Fouts et al. 2012) (the parameters used were e-value ≤1 × 10−5, percent identity ≥35, and length of the match ≥75).
THE MOBILOME OF Legionella HAS A DIVERSE ORIGIN
The Legionella mobilome comprises plasmids, integrative conjugative elements, insertion sequences, and genomic-island-like regions like most of the prokaryotic genomes. However, it also has a particular feature, which is a large array of so-called eukaryotic-like genes that were identified during the L. pneumophila genome analyses of strains Paris and Lens in 2004 (Cazalet et al. 2004). These genes code for proteins that show the strongest similarity, not to prokaryotic but to eukaryotic proteins, or encode motifs known to be implicated in protein–protein interactions, which are present only or primarily in eukaryotes. The eukaryotic-like proteins were predicted to mimic host proteins to allow intracellular replication of Legionella and are thus good candidates for being implicated in host–pathogen interactions (Cazalet et al. 2004). Interestingly, many of these potential virulence factors never had been identified in a prokaryotic genome before the discovery in L. pneumophila, suggesting that L. pneumophila may have developed specific mechanisms to cross talk with its eukaryotic hosts (Bruggemann et al. 2006). The presence of a high number of eukaryotic-like proteins in the genus Legionella was further confirmed when analyzing different L. pneumophila and L. longbeachae genomes (de Felipe et al. 2005; Cazalet et al. 2010; Kozak et al. 2010; Schroeder et al. 2010; Gomez-Valero et al. 2011b). Thus, the Legionella genomes reflect the interaction of this bacterium with amoeba, which is probably the driving force in the evolution of Legionella pathogenicity.
Although few eukaryotic-like genes were then also identified in other bacteria like Rickettsia belli or Wolbachia (Ogata et al. 2006; Walker et al. 2007), Legionella is the bacterium with the widest variety of eukaryotic-like proteins and eukaryotic domain-carrying proteins; a finding that still holds true today, despite the hundreds of different bacterial genome sequences that have been sequenced and analyzed since then. In 2004, more than 60 of these proteins were described in the genomes of strains Paris and Lens (Cazalet et al. 2004) and in 2005, in strain Philadelphia (de Felipe et al. 2005). Later, the analysis of the distribution of these genes among more than 200 L. pneumophila strains using DNA arrays showed that the conservation of certain eukaryotic-like genes within the species L. pneumophila was high, indicating strong environmental selection pressures for their preservation (Cazalet et al. 2008).
Recently, the sequencing and analyses of five additional L. pneumophila genomes revealed that over half of the eukaryotic-like proteins are present in all eight L. pneumophila strains (Gomez-Valero et al. 2011a,b). We thus propose that these conserved eukaryotic-like proteins have probably allowed the ancestor of L. pneumophila to better adapt to the intracellular environment, but now most of them are evolving as part of the core genome. Interestingly, approximately 30% of the proteins containing eukaryotic motifs are also present in the species L. longbeachae (Cazalet et al. 2010; Gomez-Valero et al. 2011a). These proteins might be part of the core of eukaryotic-like proteins essential for the genus Legionella for its intracellular replication. However, simultaneously, a flexible pool of eukaryotic-like proteins is present in the Legionella genomes. It represents more than half of the eukaryotic-like proteins identified to date in different L. pneumophila strains. The fact that even phylogenetically very closely related strains like the L. pneumophila strains HL06041035 and Paris present some differences in their repertoire of eukaryotic-like proteins suggests that the acquisition and loss of these genes is an ongoing process that has taken place multiple times and further underlines the high dynamics and plasticity of the Legionella genomes. In addition to horizontal gene transfer a process of convergent evolution might be important in the evolution of Legionella as, although many of the proteins containing eukaryotic motifs are not conserved between the species L. pneumophila and L. longbeachae, the same eukaryotic motifs like F-box, U-box, ankyrin, or serine thereonine protein kinase (STPK) motifs are found in both species (Cazalet et al. 2010). A similar observation has been made when the genome sequence of the amoeba symbiont “Candidatus Amoebophilus asiaticus” has been analyzed. Interestingly, many of the eukaryotic motifs identified in Legionella are also present in this bacterium (Schmitz-Esser et al. 2010). Most interestingly, an enrichment analysis comparing the fraction of all functional protein domains among 514 bacterial proteomes revealed that all genomes of bacteria replicating in amoeba were enriched in protein domains that are predominantly found in eukaryotic proteins (Schmitz-Esser et al. 2010). The most highly enriched domains were ANK repeats, LRR, SEL1 repeats, and F- and U-box domains, all of which are also present in L. pneumophila and L. longbeachae. Taken together, genome analyses of different bacteria associated with amoeba undertaken since the L. pneumophila genome sequence had been published in 2004, showed that bacteria that can exploit amoebae as hosts share a set of eukaryotic domain proteins that are most probably important for bacteria–host–cell interactions, despite their different lifestyles and their large phylogenetic diversity. Thus, a global, convergent adaptation mechanism to intracellular parasitism seems to exist in amoeba-associated bacteria.
EUKARYOTIC-LIKE PROTEINS OF Legionella ARE VIRULENCE FACTORS THAT MIMIC HOST PROTEINS
One important prediction from the genome analysis of strains Paris and Lens was that the eukaryotic-like proteins of Legionella are secreted virulence factors able to interfere with many different host-signaling pathways by mimicking host proteins (Cazalet et al. 2004; Bruggemann et al. 2006). Therefore, many groups started functional studies focusing on these proteins and within a short period of time the importance of the eukaryotic-like proteins for the virulence of Legionella was shown (de Felipe et al. 2008; Nora et al. 2009; Hubber and Roy 2010). The main secretion system of Legionella, essential for intracellular growth, is the type IVB Dot/Icm secretion system (Marra and Shuman 1992; Berger and Isberg 1993). Using many different methods it has been shown that at least 275 L. pneumophila proteins are secreted by this system (Campodonico et al. 2005; de Felipe et al. 2005; Shohdy et al. 2005; de Felipe et al. 2008; Burstein et al. 2009; Heidtman et al. 2009; Zhu et al. 2011), which represents nearly 10% of the protein-coding genes predicted in the L. pneumophila genomes. At least 75 of these are eukaryotic-like proteins or carry eukaryotic domains and, for certain of these, it has also been shown experimentally that they interfere with host–cell signaling pathways (Table 2).


The diverse motifs identified, like ankyrin repeats, F-box and U-box domains, STPK domains, coil-coiled domains, SET, and Sel1 or Sec-7 domains indicate also very diverse functional roles of these proteins. This fact is, for example, reflected in the very high number of different posttranslational modifications Legionella is able to induce in its hosts. To date, it has been shown that L. pneumophila proteins are able to ubiquitinate, phosphorylate, lipidate, glycosylate, AMPylate, DeAMPylate, phosphocholinate, and dephosphoryl-cholinate specific host proteins to modulate multiple host pathways to the pathogen's advantage (Rolando and Buchrieser 2012). Recently, a new posttranslational modification induced by L. pneumophila was reported. The Legionella-secreted effector, RomA, was shown to be a SET domain-containing methyltransferase that uniquely modifies host chromatin to repress gene expression and promote efficient intracellular bacterial replication (Rolando et al. 2013). Interestingly, a combination of eukaryotic domains was predicted in several of the eukaryotic-like proteins, leading to a modular protein structure. One example is AnkB/LegAU13/Ceg27/Lpg2144/Lpp2082 that contains three different eukaryotic protein domains: an F-box domain, an ankyrin domain, and a CaaX motif (Table 2). The F-box domain interacts with the SKP1 component of the SCF1 ubiquitin ligase complex and allows Legionella to exploit the host ubiquitination machinery, whereas the ankyrin motif, a widespread protein–protein interaction motif, probably interacts with the cellular target like Parvin B described for strain Paris (Price et al. 2009; Lomma et al. 2010). The F-box domain, as well as the two ankyrin protein–protein interaction domains, is essential for the biological function of AnkB. The CaaX motif exploits the host prenyltransferase machinery to facilitate membrane localization of L. pneumophila effector proteins (Ivanov et al. 2010; Price et al. 2010). Another modular protein is SdhA/Lpg0376, a Dot/Icm-secreted virulence factor that contains a GRIP and a coiled-coil domain (Table 2). SdhA is required for growth within macrophages and protection from host–cell death (Laguna et al. 2006). Furthermore, the Legionella-containing vacuole is actively stabilized by the SdhA protein during intracellular replication, which affords the bacterium protection from cytosolic host factors that degrade bacteria and initiate immune responses (Creasey and Isberg 2012). AnkY/LegA9/Lpg0402 is also a modular protein as it contains an STPK domain and an ankyrin motif. However, to date, its functional role and putative implication in virulence has not been shown.
A specific example for the amazing temporal and spatial fine-tuning Legionella has evolved in its interactions with the host cell is the Dot/Icm-secreted effector LubX/Lpg2830. LubX (for Legionella U-box) is a protein containing two U-box domains (U-box1 and U-box2) similar to eukaryotic E3 ubiquitin ligases that function as a ubiquitin ligase in conjunction with host UbcH5a or UbcH5c E2 enzymes and mediates polyubiquitination of cellular Clk1 (Cdc2-like kinase) (Kubori et al. 2008). U-box1 seems to be critical for ubiquitin ligation, whereas the U-box2 domain interacts with the substrate. Recently, it was shown that the secreted L. pneumophila effector protein SidH is a second target of LubX within host cells. LubX directly binds and polyubiquitinates SidH in vitro and mediates its proteasomal degradation in infected cells. LubX is the first example of a bacterial metaeffector that regulates spatially and temporally the expression level of another effector (Kubori et al. 2010). The identification of this mechanism points to the fact that there might be many different levels of regulation among the large number of Legionella effectors as well as between the effectors and their host targets.
EUKARYOTIC-LIKE PROTEINS WITNESS THE LONG-LASTING COEVOLUTION OF Legionella WITH EUKARYOTIC CELLS
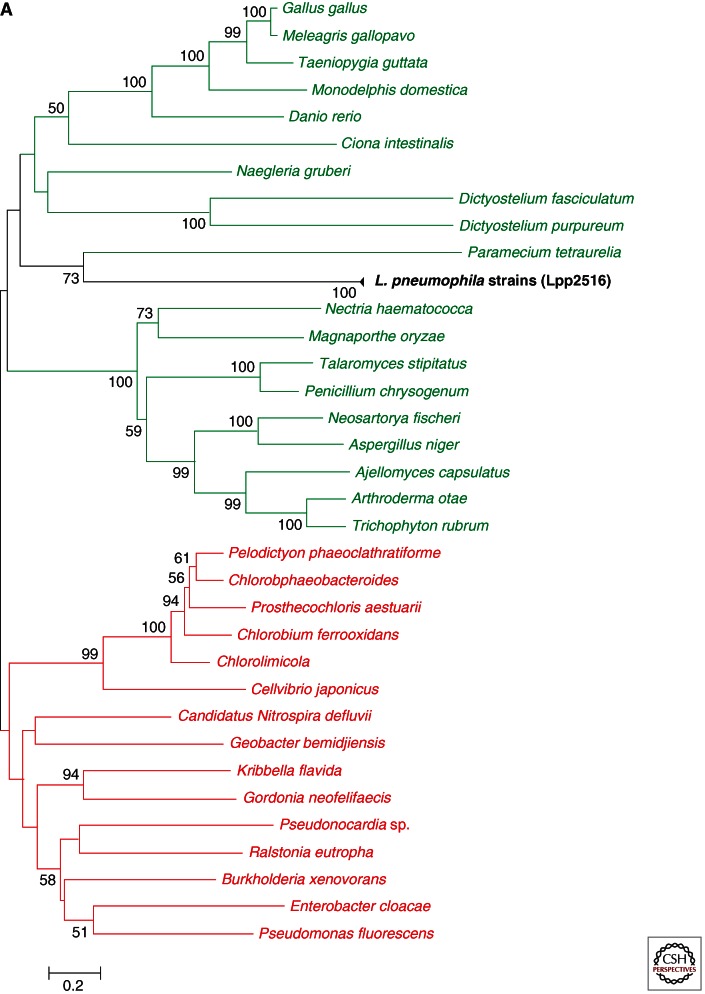
The presence of these many eukaryotic-like proteins in the genome of amoeba-associated bacteria raises the question of the origin of this part of the genome. Are these eukaryotic-like genes laterally acquired or are they the result of adaptive changes generating motifs similar to the eukaryotic ones? The latter is known as “convergent evolution” in contrast to horizontal gene transfer (HGT). Although both can lead to similar proteins, similarity resulting from HGT is detectable at the primary sequence level, whereas convergent evolution more frequently leads to similar tridimensional structures but not necessarily to similar amino acid sequences. Legionella possesses all necessary factors for acquiring genes laterally as these bacteria are naturally competent and a complete recombination machinery is present, thus it is conceivable that eukaryotic-like proteins have been acquired by HGT. To analyze this question, different phylogenetic analyses of some of these eukaryotic-like proteins have been performed revealing a clustering of the Legionella eukaryotic-like proteins with eukaryotic sequences, further supporting the hypothesis that these were acquired by HGT from eukaryotic organisms like amoeba (de Felipe et al. 2005; Nora et al. 2009; Lurie-Weinberger et al. 2010; Gomez-Valero et al. 2011a). An example is the enzyme arylamine N-acetyltransferase (NAT) encoded by lpp2516 in strain Paris. The L. pneumophila NAT is an atypical member of the arylamine N-acetyltransferase family that allows L. pneumophila to detoxify aromatic amine chemicals and to grow in their presence (Kubiak et al. 2012). This enzyme probably allows L. pneumophila that is exposed to many chemicals in its natural and man-made environments, to better adapt and survive in these environments (Kubiak et al. 2012). Phylogenetic reconstruction shown in Figure 2A indicates that horizontal acquisition of this gene from ciliates is the most probable origin of this enzyme in Legionella (Kubiak et al. 2012).
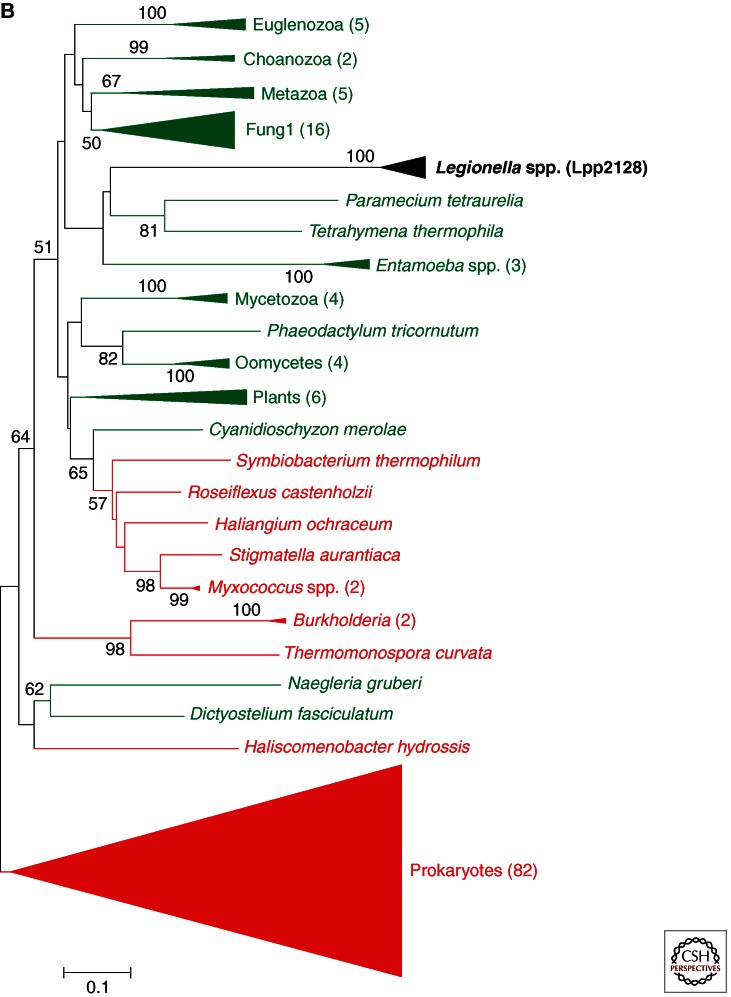
Figure 2.
Phylogenetic tree of selected eukaryotic-like proteins of Legionella spp. (A) Phylogeny of the enzyme arylamine N-acetyltransferase (NAT) (Lpp2516) of L. pneumophila. (B) Phylogeny of the sphingosine-1 phosphate lyse Lpp2128 of L. pneumophila. Homolog sequences were recruited from a database containing only completed genome sequences. Selected representatives of all eukaryotic groups and one representative of each bacterial species are included in the analyses. After blastp only significant hits were recruited (e-value <10 × 10−4), and only one hit for each species was retained. The alignments were performed with T-coffee (expresso) for Lpp2616 and Muscle for Lpp2128, and followed by manual curation. Phylogenies were reconstructed using a distance method (NJ) with 1000 bootstrap replicates. The corresponding support values are shown in each node (values lower than 50 are not represented). Bars represent 20% and 10% of estimated phylogenetic divergence, respectively. (Legend continues on following page.)
Another convincing example is the eukaryote-like sphingosine-1-phosphate lyase of L. pneumophila that is encoded by lpp2128 in strain Paris. It has been shown that the L. pneumophila Spl is a Dot/Icm-secreted effector that is able to complement the sphingosine-sensitive phenotype of Saccharomyces cervisiae and that colocalizes with host–cell mitochondria (Degtyar et al. 2009). Figure 2B shows a phylogenetic reconstruction of Spl sequences that we have undertaken. The L. pneumophila Spl protein sequence clearly falls into the eukaryotic clade of SPL sequences and those of aquatic protozoa are the closest phylogenetic relatives, suggesting acquisition of this gene by HGT from an amoeba host. Most interestingly, Legionella also encodes proteins not present in any prokaryote sequenced to date, as amoebae are the only organisms in which we identified sequences with significant similarity. An example is Lpg1684 that codes a protein of unknown function and belongs to the eukaryotic D123 protein family. Eukaryotic-like proteins may have been acquired also from other eukaryotic organisms like viruses. For example, the eukaryotic-like protein Lpg2416 is an ankyrin-containing protein whose best homolog is present in the Acanthamoeba polyphaga mimivirus, a giant virus infecting Acanthamoeba that are also the hosts of Legionella (Lurie-Weinberger et al. 2010). Furthermore, recent studies have shown that these eukaryotic-like proteins can also be exchanged among different amoeba-related bacteria, adding complexity to the possible evolutionary scenarios. For example, the sequencing and analysis of the genome of L. drancourtii have identified numerous cases of exchange between the intracellular bacteria of the order Legionellales and Chlamydiales (Gimenez et al. 2011). However, for several eukaryotic-like proteins, a clear description of their phylogenetic history remains difficult, as the acquisition events might have been too ancient for the phylogenetic signal in the sequences to be preserved. Moreover, in some of these proteins, the eukaryotic sequence is restricted to a motif and therefore to a very short region, for which phylogenetic analyses are less evident. Furthermore, only few genome sequences of aquatic amoeba and other protozoa that are most probably the main donors are currently present in the databases. Only once the gap of genome sequences of aquatic protozoa in the databases is filled will we be able to fully understand the origin and history of these eukaryotic-like proteins.
Independently of the origin of these eukaryotic-like genes, their mechanism of integration remains unknown. How are these eukaryotic genes without any similarity to prokaryotic sequences integrated in the Legionella genome? Many eukaryotic genes have introns, but their prokaryotic counterparts do not. How can these introns be processed or how did they “disappear?” It is tempting to propose that this happened not via HGT but via “horizontal RNA transfer.” Indeed, the L. pneumophila genomes encode a reverse transcriptase that could be implicated in this process. However, the integration mechanism remains to be discovered. Moreover, there is evidence for the presence of a secretion signal in the carboxy terminal of the Dot/Icm substrates (Nagai et al. 2005; Kubori et al. 2008; Huang et al. 2010); therefore, acquisition of this signal is also a challenge for these proteins to be secreted. Thus, although we start to discern how these eukaryotic proteins have appeared in the Legionella genomes, many questions with respect to the detailed mechanism of acquisition and integration remain unanswered.
THE Legionella ACCESSORY GENES ARE PART OF THE MOBILOME
The most evident component of the Legionella accessory genome is the plasmids. The strains Paris, Lens, and Lorraine each contain a plasmid of 132, 60, and 150 kb, respectively (Cazalet et al. 2004; Chien et al. 2004). Likewise, L. longbeachae strain NSW150 possesses a plasmid of 72 kb, and also strain D-4968 seems to contain a plasmid (Kozak et al. 2010). These replicons show a heterogeneous distribution among the different genomes, as the same plasmids or partially similar plasmids are present in different strains (Cazalet et al. 2008). Likewise, a comparative analysis of these plasmids revealed that the 30.4 kb Tra region present in the plasmid of L. pneumophila strain Paris shows much higher identity with the Tra region located on the L. longbeachae plasmid, than with those of the other L. pneumophila strains sequenced (Gomez-Valero et al. 2011b). Interestingly, regions coding proteins homologous to Tra proteins probably coding for a conjugative machinery are present in all Legionella plasmids. They are similar to F-type T4SS that allow the synthesis of a long and flexible pilus for conjugation in liquid and solid media (Lawley et al. 2003). However, such a system is also found in a chromosomal localization in the L. pneumophila strain Philadelphia and in L. longbeachae strain NSW150. In both strains, this region is inserted in a tRNA gene next to an integrase and it is bordered by flanking repeats. The presence of these elements suggests that these T4SSs are mobile and that their heterogeneous distribution is the result of the lateral movement of these plasmids. Furthermore, these conjugative elements can be found in different plasmids or can be completely or partially present in the chromosome, indicating that they might have the capacity to integrate and excise from the Legionella genomes. Indeed, this has been shown for the region carrying the Lvh genes. Lvh is a genomic-island-like region that encodes a T4ASS implicated in conjugation and in virulence-related phenotypes under laboratory conditions mimicking the spread of Legionnaires’ disease from environmental niches (Ridenour et al. 2003; Bandyopadhyay et al. 2007). This region can be integrated in the chromosome but can also excise in a site-specific manner to exist as a low copy number plasmid that is undoubtedly transferable among strains (Doleans-Jordheim et al. 2006). Indeed, it is present in five of the eight sequenced L. pneumophila strains, in L. longbeachae strain D-4968 (Kozak et al. 2010), and according to hybridization results also partly in Legionella rubrilucens (Cazalet et al. 2008). Interestingly, in the Legionella strains where the Lvh T4SS is not present, another Tra system, a P-type T4SS that codes for short and rigid pili that allow surface mating for conjugation, is present in the same chromosomal position (Gomez-Valero et al. 2011b). This remarkable diversity and mobility of conjugative elements probably coding T4SSs in Legionella and their presence in all strains, suggests a role in the bacterial adaptation to the different environments and hosts.
An intriguing feature of these probably mobile regions is their association with genes coding for homologs of a putative phage repressor protein (PrpA) and for homologs of LvrA, LvrB, and LvrC, first described for the Lvh region of L. pneumophila. LvrC is a homolog of CsrA, an RNA-binding protein crucial for the regulation of the switch between the replicative and transmissive phases of L. pneumophila (Molofsky and Swanson 2004). It is tempting to assume that the CsrA homolog encoded on each of these regions is implicated in the regulation of the mobility and/or the integration or excision of these islands. Perhaps under certain conditions it is advantageous for Legionella to have multiple copies of these plasmids to achieve a higher expression of these genes or for promoting high frequencies of DNA interchange to rapidly adapt to new conditions. This idea is further underlined by the observation that the presence of homologous Lvr loci is not only limited to the Lvh or Tra regions, but they are also present in other genomic locations that seem to be mobile. Thus, large parts of the accessory genome of Legionella are mobile, and it seems that their mobility is regulated by a particular mechanism that still remains elusive.
CONCLUSIONS: THE Legionella ACCESSORY GENOME IS PART OF A GLOBAL MOBILOME CONFERRING VERSATILITY
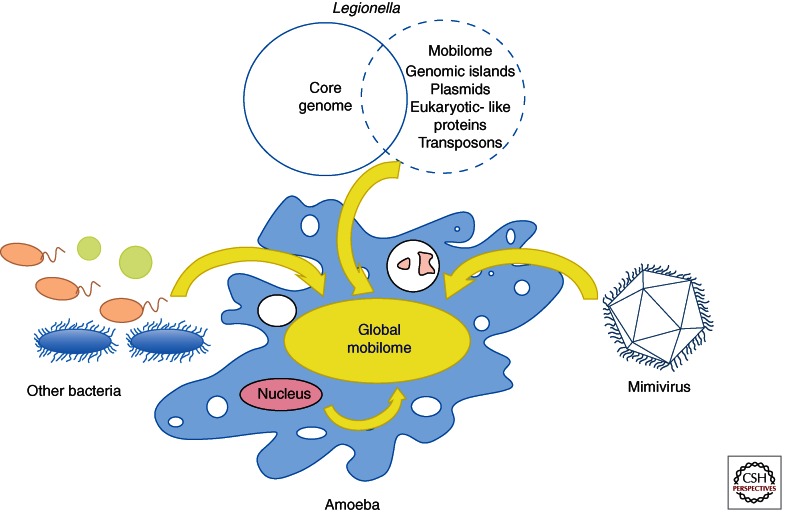
One of the best-known characteristics of obligate, intracellular bacteria is a reduction of the genome size as many genes become dispensable given the stability of the intracellular environment they inhabit. In addition, there is a minor probability of DNA exchange as they live in a closed niche (Moya et al. 2008). Extreme examples are the symbiontic bacteria Buchnera aphidicola, an insect endosymbiont found in the aphid Cinaria cedri with a genome size of 416 Kb coding solely for 357 protein-coding genes (Pérez-Brocal et al. 2006) or the obligate intracellular parasite Rickettsia prowazekii, the causative agent of epidemic typhus with a 1.1 Mb genome (Andersson et al. 1998). However, this does not hold true for amoeba-associated bacteria like Legionella, as no signs that they follow a reductive evolutionary path are present in their genomes, although Legionella are probably replicating exclusively intracellularly. Furthermore, Legionella have highly dynamic genomes and a large flexible gene pool is present in the genus comprising plasmids, mobile genetic elements, and genes probably transferred from their hosts. This large mobilome confers to these bacteria their high versatility and capacity to adapt to many different environmental conditions and to infect diverse hosts like amoeba, protists, probably algae, and humans. Thus, the intracellular environment provided by amoeba is different from the environment in which symbionts like (e.g., Buchneria or Rickettsia live), probably because amoeba may host different bacteria and viruses at the same time, allowing exchange of genetic material among a variety of organisms. We thus propose that amoeba and aquatic protozoa form communities with a large pool of exchangeable genetic material, and have a large common accessory genome, derived from the host and from the diverse intracellular organisms that inhabit them as parasites or symbionts (Fig. 3). This big “global mobilome” allows the constant acquisition of DNA and provides a large repertoire of diverse functions for adapting to changing environmental conditions. This large mobile gene pool combined with the high genome dynamics provide the basis of versatility and adaptability in Legionella and allow successful intracellular parasitism in many different hosts.
Figure 3.
Model of the genetic flow inside amoeba. The accessory genome of Legionella, other intracellular bacteria, mimivirus and amoeba are contributing to a “global mobilome” that is accessible to the different organisms living inside amoeba to adapt to different conditions and allows versatility in the host–pathogen interactions.
ACKNOWLEDGMENTS
This work received financial support from the Institut Pasteur, the Centre National de la Recherche (CNRS), the Institut Carnot-Pasteur MI, the Fondation pour la Recherche Médicale (FRM), grant No. DEQ20120323697, ANR-10-LABX-62-IBEID, and the ANR-10-PATH-004 project “MobilGenomics,” in the frame of ERA-NET PathoGenoMics. We are most grateful to Philippe Glaser for critical reading of the manuscript and for fruitful discussions.
Footnotes
Editors: Pascale Cossart and Stanley Maloy
Additional Perspectives on Bacterial Pathogenesis available at www.perspectivesinmedicine.org
REFERENCES
- Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Pontén T, Alsmark UC, Podowski RM, Näslund AK, Eriksson AS, Winkler HH, Kurland CG 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396: 133–140 [DOI] [PubMed] [Google Scholar]
- Atlan D, Coupat-Goutaland B, Risler A, Reyrolle M, Souchon M, Briolay J, Jarraud S, Doublet P, Pélandakis M 2012. Micriamoeba tesseris nov. gen. nov. sp: A new taxon of free-living small-sized Amoebae non-permissive to virulent Legionellae. Protist 163: 888–902 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay P, Liu S, Gabbai CB, Venitelli Z, Steinman HM 2007. Environmental mimics and the Lvh type IVA secretion system contribute to virulence-related phenotypes of Legionella pneumophila. Infect Immun 75: 723–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger KH, Isberg RR 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol 7: 7–19 [DOI] [PubMed] [Google Scholar]
- Bruggemann H, Cazalet C, Buchrieser C 2006. Adaptation of Legionella pneumophila to the host environment: Role of protein secretion, effectors and eukaryotic-like proteins. Curr Opin Microbiol 9: 86–94 [DOI] [PubMed] [Google Scholar]
- Burstein D, Zusman T, Degtyar E, Viner R, Segal G, Pupko T 2009. Genome-scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS Pathog 5: e1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campodonico EM, Chesnel L, Roy CR 2005. A yeast genetic system for the identification and characterization of substrate proteins transferred into host cells by the Legionella pneumophila Dot/Icm system. Mol Microbiol 56: 918–933 [DOI] [PubMed] [Google Scholar]
- Cazalet C, Rusniok C, Bruggemann H, Zidane N, Magnier A, Ma L, Tichit M, Jarraud S, Bouchier C, Vandenesch F, et al. 2004. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat Genet 36: 1165–1173 [DOI] [PubMed] [Google Scholar]
- Cazalet C, Jarraud S, Ghavi-Helm Y, Kunst F, Glaser P, Etienne J, Buchrieser C 2008. Multigenome analysis identifies a worldwide distributed epidemic Legionella pneumophila clone that emerged within a highly diverse species. Genome Res 18: 431–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalet C, Gomez-Valero L, Rusniok C, Lomma M, Dervins-Ravault D, Newton HJ, Sansom FM, Jarraud S, Zidane N, Ma L, et al. 2010. Analysis of the Legionella longbeachae genome and transcriptome uncovers unique strategies to cause Legionnaires’ disease. PLoS Genet 6: e1000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien M, Morozova I, Shi S, Sheng H, Chen J, Gomez SM, Asamani G, Hill K, Nuara J, Feder M, et al. 2004. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305: 1966–1968 [DOI] [PubMed] [Google Scholar]
- Creasey EA, Isberg RR 2012. The protein SdhA maintains the integrity of the Legionella-containing vacuole. Proc Natl Acad Sci 109: 3481–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Auria G, Jimenez-Hernandez N, Peris-Bondia F, Moya A, Latorre A 2010. Legionella pneumophila pangenome reveals strain-specific virulence factors. BMC Genomics 11: 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Felipe KS, Pampou S, Jovanovic OS, Pericone CD, Ye SF, Kalachikov S, Shuman HA 2005. Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J Bacteriol 187: 7716–7726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Felipe KS, Glover RT, Charpentier X, Anderson OR, Reyes M, Pericone CD, Shuman HA 2008. Legionella eukaryotic-like type IV substrates interfere with organelle trafficking. PLoS Pathog 4: e1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degtyar E, Zusman T, Ehrlich M, Segal G 2009. A Legionella effector acquired from protozoa is involved in sphingolipids metabolism and is targeted to the host cell mitochondria. Cell Microbiol 11: 1219–1235 [DOI] [PubMed] [Google Scholar]
- Doleans-Jordheim A, Akermi M, Ginevra C, Cazalet C, Kay E, Schneider D, Buchrieser C, Atlan D, Vandenesch F, Etienne J, et al. 2006. Growth-phase-dependent mobility of the lvh-encoding region in Legionella pneumophila strain Paris. Microbiology 152: 3561–3568 [DOI] [PubMed] [Google Scholar]
- Fields BS 1996. The molecular ecology of Legionellae. Trends Microbiol 4: 286–290 [DOI] [PubMed] [Google Scholar]
- Fouts DE, Brinkac L, Beck E, Inman J, Sutton G 2012. PanOCT: Automated clustering of orthologs using conserved gene neighborhood for pan-genomic analysis of bacterial strains and closely related species. Nucleic Acids Res 40: e172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser DW, Tsai TR, Orenstein W, Parkin WE, Beecham HJ, Sharrar RG, Harris J, Mallison GF, Martin SM, McDade JE, et al. 1977. Legionnaires’ disease: Description of an epidemic of pneumonia. N Engl J Med 297: 1189–1197 [DOI] [PubMed] [Google Scholar]
- Gimenez G, Bertelli C, Moliner C, Robert C, Raoult D, Fournier PE, Greub G 2011. Insight into cross-talk between intra-amoebal pathogens. BMC Genomics 12: 542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Valero L, Rusniok C, Cazalet C, Buchrieser C 2011a. Comparative and functional genomics of Legionella identified eukaryotic like proteins as key players in host–pathogen interactions. Front Microbiol 2: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Valero L, Rusniok C, Jarraud S, Vacherie B, Rouy Z, Barbe V, Medigue C, Etienne J, Buchrieser C 2011b. Extensive recombination events and horizontal gene transfer shaped the Legionella pneumophila genomes. BMC Genomics 12: 536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidtman M, Chen EJ, Moy MY, Isberg RR 2009. Large-scale identification of Legionella pneumophila Dot/Icm substrates that modulate host cell vesicle trafficking pathways. Cell Microbiol 11: 230–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Boyd D, Amyot WM, Hempstead AD, Luo ZQ, O’Connor TJ, Chen C, Machner M, Montminy T, Isberg RR 2010. The E Block motif is associated with Legionella pneumophila translocated substrates. Cell Microbiol 13: 227–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubber A, Roy CR 2010. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol 26: 261–283 [DOI] [PubMed] [Google Scholar]
- Ivanov SS, Charron G, Hang HC, Roy CR 2010. Lipidation by the host prenyltransferase machinery facilitates membrane localization of Legionella pneumophila effector proteins. J Biol Chem 285: 34686–34698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak NA, Buss M, Lucas CE, Frace M, Govil D, Travis T, Olsen-Rasmussen M, Benson RF, Fields BS 2010. Virulence factors encoded by Legionella longbeachae identified on the basis of the genome sequence analysis of clinical isolate D-4968. J Bacteriol 192: 1030–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubiak X, Dervins-Ravault D, Pluvinage B, Chaffotte AF, Gomez-Valero L, Dairou J, Busi F, Dupret JM, Buchrieser C, Rodrigues-Lima F 2012. Characterization of an acetyltransferase that detoxifies aromatic chemicals in Legionella pneumophila. Biochem J 445: 219–229 [DOI] [PubMed] [Google Scholar]
- Kubori T, Hyakutake A, Nagai H 2008. Legionella translocates an E3 ubiquitin ligase that has multiple U-boxes with distinct functions. Mol Microbiol 67: 1307–1319 [DOI] [PubMed] [Google Scholar]
- Kubori T, Shinzawa N, Kanuka H, Nagai H 2010. Legionella metaeffector exploits host proteasome to temporally regulate cognate effector. PLoS Pathog 6: e1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguna RK, Creasey EA, Li Z, Valtz N, Isberg RR 2006. A Legionella pneumophila-translocated substrate that is required for growth within macrophages and protection from host cell death. Proc Natl Acad Sci 103: 18745–18750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau HY, Ashbolt NJ 2009. The role of biofilms and protozoa in Legionella pathogenesis: Implications for drinking water. J Appl Microbiol 107: 368–378 [DOI] [PubMed] [Google Scholar]
- Lawley TD, Klimke WA, Gubbins MJ, Frost LS 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol Lett 224: 1–15 [DOI] [PubMed] [Google Scholar]
- Lomma M, Dervins-Ravault D, Rolando M, Nora T, Newton HJ, Sansom FM, Sahr T, Gomez-Valero L, Jules M, Hartland EL, et al. 2010. The Legionella pneumophila F-box protein Lpp2082 (AnkB) modulates ubiquitination of the host protein parvin B and promotes intracellular replication. Cell Microbiol 12: 1272–1291 [DOI] [PubMed] [Google Scholar]
- Lurie-Weinberger MN, Gomez-Valero L, Merault N, Glockner G, Buchrieser C, Gophna U 2010. The origins of eukaryotic-like proteins in Legionella pneumophila. Int J Med Microbiol 300: 470–481 [DOI] [PubMed] [Google Scholar]
- Marra A, Shuman HA 1992. Genetics of Legionella pneumophila virulence. Annu Rev Genet 26: 51–69 [DOI] [PubMed] [Google Scholar]
- McDade JE, Shepard CC, Fraser DW, Tsai TR, Redus MA, Dowdle WR 1977. Legionnaires’ disease: Isolation of a bacterium and demonstration of its role in other respiratory disease. N Engl J Med 297: 1197–1203 [DOI] [PubMed] [Google Scholar]
- Molmeret M, Horn M, Wagner M, Santic M, Abu Kwaik Y 2005. Amoebae as training grounds for intracellular bacterial pathogens. Appl Environ Microbiol 71: 20–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AB, Swanson MS 2004. Differentiate to thrive: Lessons from the Legionella pneumophila life cycle. Mol Microbiol 53: 29–40 [DOI] [PubMed] [Google Scholar]
- Moya A, Pereto J, Gil R, Latorre A 2008. Learning how to live together: Genomic insights into prokaryote-animal symbioses. Nat Rev Genet 9: 218–229 [DOI] [PubMed] [Google Scholar]
- Nagai H, Cambronne ED, Kagan JC, Amor JC, Kahn RA, Roy CR 2005. A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc Natl Acad Sci 102: 826–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton HJ, Ang DK, van Driel IR, Hartland EL 2010. Molecular pathogenesis of infections caused by Legionella pneumophila. Clin Microbiol Rev 23: 274–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora T, Lomma M, Gomez-Valero L, Buchrieser C 2009. Molecular mimicry: An important virulence strategy employed by Legionella pneumophila to subvert host functions. Future Microbiol 4: 691–701 [DOI] [PubMed] [Google Scholar]
- Ogata H, La Scola B, Audic S, Renesto P, Blanc G, Robert C, Fournier PE, Claverie JM, Raoult D 2006. Genome sequence of Rickettsia bellii illuminates the role of amoebae in gene exchanges between intracellular pathogens. PLoS Genet 2: e:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Brocal V, Gil R, Ramos S, Lamelas A, Postigo M, Michelena JM, Silva FJ, Moya A, Latorre A 2006. A small microbial genome: The end of a long symbiotic relationship? Science 314: 312–313 [DOI] [PubMed] [Google Scholar]
- Price CT, Al-Khodor S, Al-Quadan T, Santic M, Habyarimana F, Kalia A, Kwaik YA 2009. Molecular mimicry by an F-box effector of Legionella pneumophila hijacks a conserved polyubiquitination machinery within macrophages and protozoa. PLoS Pathog 5: e1000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CT, Al-Quadan T, Santic M, Jones SC, Abu Kwaik Y 2010. Exploitation of conserved eukaryotic host cell farnesylation machinery by an F-box effector of Legionella pneumophila. J Exp Med 207: 1713–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridenour DA, Cirillo SL, Feng S, Samrakandi MM, Cirillo JD 2003. Identification of a gene that affects the efficiency of host cell infection by Legionella pneumophila in a temperature-dependent fashion. Infect Immun 71: 6256–6263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolando M, Buchrieser C 2012. Post-translational modifications of host proteins by Legionella pneumophila: A sophisticated survival strategy. Future Microbiol 7: 369–381 [DOI] [PubMed] [Google Scholar]
- Rolando M, Sanulli S, Rusniok C, Gomez-Valero L, Bertholet C, Sahr T, Margueron R, Buchrieser C 2013. Legionella pneumophila effector RomA uniquely modifies host chromatin to repress gene expression and promote intracellular bacterial replication. Cell Host Microbe 13: 395–405 [DOI] [PubMed] [Google Scholar]
- Rowbotham TJ 1980. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J Clin Pathol 33: 1179–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowbotham TJ 1986. Current views on the relationships between amoebae, legionellae and man. Isr J Med Sci 22: 678–689 [PubMed] [Google Scholar]
- Schmitz-Esser S, Tischler P, Arnold R, Montanaro J, Wagner M, Rattei T, Horn M 2010. The genome of the amoeba symbiont “Candidatus Amoebophilus asiaticus” reveals common mechanisms for host cell interaction among amoeba-associated bacteria. J Bacteriol 192: 1045–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder GN, Petty NK, Mousnier A, Harding CR, Vogrin AJ, Wee B, Fry NK, Harrison TG, Newton HJ, Thomson NR, et al. 2010. Legionella pneumophila strain 130b possesses a unique combination of type IV secretion systems and novel Dot/Icm secretion system effector proteins. J Bacteriol 192: 6001–6016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shohdy N, Efe JA, Emr SD, Shuman HA 2005. Pathogen effector protein screening in yeast identifies Legionella factors that interfere with membrane trafficking. Proc Natl Acad Sci 102: 4866–4871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siefert JL 2009. Defining the mobilome. Methods Mol Biol 532: 13–27 [DOI] [PubMed] [Google Scholar]
- Steinert M, Heuner K, Buchrieser C, Albert-Weissenberger C, Glockner G 2007. Legionella pathogenicity: Genome structure, regulatory networks and the host cell response. Int J Med Microbiol 297: 577–587 [DOI] [PubMed] [Google Scholar]
- Walker T, Klasson L, Sebaihia M, Sanders MJ, Thomson NR, Parkhill J, Sinkins SP 2007. Ankyrin repeat domain-encoding genes in the wPip strain of Wolbachia from the Culex pipiens group. BMC Biol 5: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiley H, Bentham R 2011. Legionella longbeachae and legionellosis. Emerg Infect Dis 17: 579–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winiecka-Krusnell J, Linder E 1999. Free-living amoebae protecting Legionella in water: The tip of an iceberg? Scand J Infect Dis 31: 383–385 [DOI] [PubMed] [Google Scholar]
- Yu VL, Plouffe JF, Pastoris MC, Stout JE, Schousboe M, Widmer A, Summersgill J, File T, Heath CM, Paterson DL, et al. 2002. Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired legionellosis: An international collaborative survey. J Infect Dis 186: 127–128 [DOI] [PubMed] [Google Scholar]
- Zhu W, Banga S, Tan Y, Zheng C, Stephenson R, Gately J, Luo ZQ 2011. Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila. PLoS ONE 6: e17638. [DOI] [PMC free article] [PubMed] [Google Scholar]