PIF3 recruits HDA15 to repress chlorophyll biosynthesis and photosynthesis gene expression in etiolated seedlings, suggesting that epigenetic regulation is critical for phytochrome- and PIF3-regulated light signal transduction. This study provides insight into the molecular mechanism underlying photomorphogenesis in plants.
Abstract
PHYTOCHROME INTERACTING FACTOR3 (PIF3) is a key basic helix-loop-helix transcription factor of Arabidopsis thaliana that negatively regulates light responses, repressing chlorophyll biosynthesis, photosynthesis, and photomorphogenesis in the dark. However, the mechanism for the PIF3-mediated transcription regulation remains largely unknown. In this study, we found that the REDUCED POTASSIUM DEPENDENCY3/HISTONE DEACETYLASE1-type histone deacetylase HDA15 directly interacted with PIF3 in vivo and in vitro. Genome-wide transcriptome analysis revealed that HDA15 acts mainly as a transcriptional repressor and negatively regulates chlorophyll biosynthesis and photosynthesis gene expression in etiolated seedlings. HDA15 and PIF3 cotarget to the genes involved in chlorophyll biosynthesis and photosynthesis in the dark and repress gene expression by decreasing the acetylation levels and RNA Polymerase II–associated transcription. The binding of HDA15 to the target genes depends on the presence of PIF3. In addition, PIF3 and HDA15 are dissociated from the target genes upon exposure to red light. Taken together, our results indicate that PIF3 associates with HDA15 to repress chlorophyll biosynthetic and photosynthetic genes in etiolated seedlings.
INTRODUCTION
Light is one of the most important environmental factors that govern plant growth and development. Plants can detect almost all facets of light, such as direction, duration, quantity, and wavelength using three major classes of photoreceptors: phytochromes (PHY), cryptochromes, and phototropins (Fankhauser and Chory, 1997). In Arabidopsis thaliana, red/far-red (R/FR) light receptors, phytochromes (PHYA to PHYE), regulate various light responses by initiating the transcriptional cascades that alter the expression of 10 to 30% of the entire transcriptome (Ma et al., 2001; Tepperman et al., 2001; Jiao et al., 2005). Light-regulated gene expression has served as a paradigm to understand transcriptional regulatory mechanisms in plants (Bertrand et al., 2005). Light-responsive cis-elements, which commonly are present in light-regulated promoters, are essential for light-controlled transcriptional activity (Arguello-Astorga and Herrera-Estrella, 1998). A number of light-responsive transcription factors have been identified through screening for light-responsive cis-element binding proteins and through genetic analyses of mutants that are deficient in their response to specific types of light (Jiao et al., 2007). These factors include the bZIP proteins ELONGATED HYPOCOTYL5 (HY5) and HY5 HOMOLOG (Oyama et al., 1997; Hardtke et al., 2000; Holm et al., 2002); the MYB proteins CIRCADIAN CLOCK ASSOCIATED1, LATE ELONGATED HYPOCOTYL, and LONG AFTER FAR-RED LIGHT1 (Schaffer et al., 1998; Wang and Tobin, 1998; Ballesteros et al., 2001); and the bHLH proteins PHYTOCHROME INTERACTING FACTOR1 (PIF1), PIF3, PIF4, and PIF5 (Ni et al., 1998; Huq and Quail, 2002; Huq et al., 2004; Khanna et al., 2007).
PIF3 is one of the most extensively characterized light-responsive transcription factors to date. Phenotypic and genetic evidence indicate that PIF3 mainly acts as a negative regulator in light responses, repressing light-mediated cotyledon expansion, opening, and chlorophyll biosynthesis in the dark (Kim et al., 2003; Shin et al., 2007, 2009; Stephenson et al., 2009; Sentandreu et al., 2011; Soy et al., 2012). Recent studies have revealed that PIF3 represses the majority of chlorophyll biosynthetic and photosynthetic genes in etiolated seedlings (Kim et al., 2003; Shin et al., 2009; Stephenson et al., 2009). In vitro binding assays showed that PIF3 binds specifically to cis-acting regulatory elements, such as G-boxes and E-boxes, in promoters of a variety of light-responsive genes (Martínez-García et al., 2000; Shin et al., 2007). However, the detailed mechanism of how PIF3 acts in light-regulated gene transcription remains obscure.
A fundamental mechanism controlling gene expression is the ability of many transcription factors to access the eukaryotic genome. Posttranslational modifications, including acetylation, methylation, phosphorylation, and ubiquitination of histones, play a key role in modulating dynamic chromatin structures and access to the DNA (Reyes et al., 2002; Wu et al., 2008). Among different types of histone modifications, reversible histone acetylation and deacetylation of histone tails appears as a key switch for interconversion between permissive and repressive states of chromatin domains (Kim et al., 2009). Hyperacetylation of histones relaxes chromatin structure and is associated with transcriptional activation, whereas hypoacetylation of histones induces chromatin compaction and gene repression (Marmorstein and Roth, 2001; Berger, 2007). Histone acetylation is catalyzed by intrinsic histone acetyltransferases (HATs) and histone deacetylases (HDACs). Based on primary homology with their yeast and mammalian counterparts, the Arabidopsis HDACs are classified into three types, the REDUCED POTASSIUM DEPENDENCY3/HISTONE DEACETYLASE1 (RPD3/HDA1), Silent Information Regulator2 (SIR2), and HISTONE DEACETYLASE2 (HD2) types, whereas the Arabidopsis HATs are grouped into four types, the GCN5-RELATED N-TERMINAL ACETYLTRANSFERASES (GNAT), MOZE YBF2/SAS3 SAS2 TIP60 (MYST), CREB-BINDING PROTEIN (CBP), and TATA-BINDING PROTEIN-ASSOCIATED FACTOR II 250 (TAFII250) types (Pandey et al., 2002).
Studies from genetic analyses of HAT and HDAC mutants in Arabidopsis suggest a role for histone acetylation in light-activated gene expression and development processes. Loss of function of the HAT HAF2 in Arabidopsis induces decreased chlorophyll accumulation and light-responsive mRNA levels (Bertrand et al., 2005). Mutations of the HAT GCN5 results in a long-hypocotyl phenotype and reduced light-inducible gene expression, whereas mutations of the HDAC HDA19 induce the opposite effects (Benhamed et al., 2006). Furthermore, quantitative trait locus mapping revealed that HDA6 is a positive regulator of light-controlled chromatin compaction (Tessadori et al., 2009). In addition, a recent genome-wide histone modification analysis revealed that the activation of photosynthetic genes correlates with dynamic acetylation changes in response to light (Charron et al., 2009). Collectively, these findings suggest a requirement for HATs and HDACs in regulating light-responsive gene transcription.
In this study, we found that PIF3 physically interacts with HDA15, a RPD3/HDA1-type HDAC. HDA15 has HDAC activity and represses chlorophyll biosynthesis and photosynthesis in etiolated seedlings. Furthermore, PIF3 is required to recruit HDA15 to the promoters of chlorophyll biosynthetic and photosynthetic genes and repress their transcription by histone deacetylation.
RESULTS
HDA15 Directly Interacts with PIF3
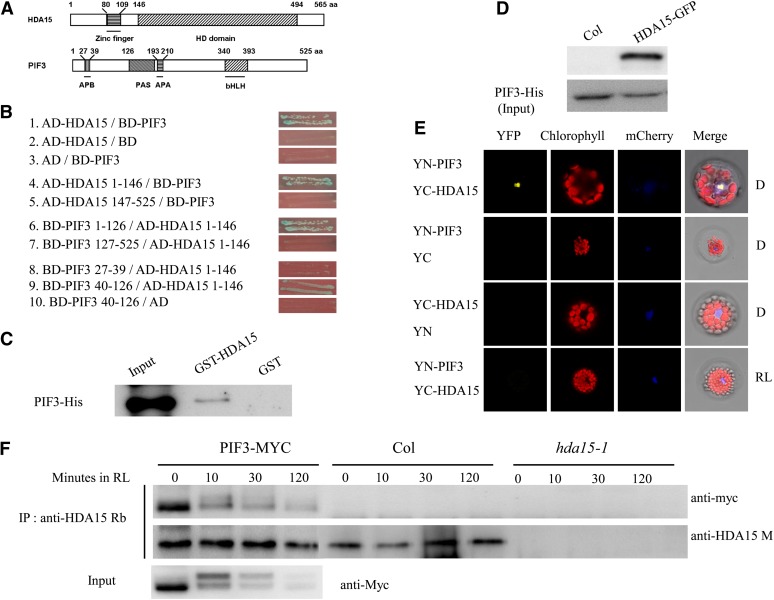
It was reported that PIF3 may act as a negative regulator in the gene expression involved in chlorophyll biosynthesis and photosynthesis (Shin et al., 2009; Stephenson et al., 2009). Many transcription repressors were found to regulate their target genes by recruiting HDACs (Yang and Grégoire, 2005; Luo et al., 2012; Zhou et al., 2013). Therefore, we examined whether PIF3 could directly interact with the HDACs by yeast two-hybrid assays. We found that PIF3 directly interacted with a RPD3/HDA1-type HDAC, HDA15, but not other HDACs in yeast cells (Figures 1A and 1B; see Supplemental Figure 1A online). Further deletion analyses identified that the N-terminal domain of HDA15 (amino acids 1 to 146) and the N-terminal domain of PIF3 (amino acids 40 to 126) were responsible for their interaction in yeast cells (Figures 1A and 1B). By contrast, HDA15 did not interact with PIF4 and PIF5 in yeast cells (see Supplemental Figure 1B online). These data indicate that HDA15 specifically interacts with PIF3.
Figure 1.
HDA15 Interacts with PIF3 Both in Vitro and in Vivo.
(A) Schematic structures of HDA15 and PIF3 protein domains. aa, amino acids.
(B) HDA15 interacts with PIF3 in yeast cells. Different regions of HDA15 and PIF3 were fused with AD and BD vectors, respectively, and cotransformed into the yeast strain AH109. The transformants were plated on SD/-Leu-Trp-His plus 3-amino-1,2,4-triazole; 0.1 mM 3-amino-1,2,4-triazole was added to inhibit the autoactivation of PIF3 bait.
(C) HDA15 interacts with PIF3 in vitro. GST-HDA15 or GST was incubated with PIF3-His and GST resin, and the bound proteins were then eluted from resin and probed with the anti-His antibody.
(D) The interaction between HDA15 and PIF3 was detected by semi–in vivo pull-down assay. Recombinant PIF3-His protein pulls down HDA15-GFP from total protein extracts. Total proteins were extracted from 2-d-etiolated Col and 35S:HDA15-GFP lines. PIF3-His was used as loading control.
(E) HDA15 interacts with PIF3 in Arabidopsis protoplasts in darkness. HDA15 and PIF3 fused with the C terminus (YC) or the N terminus (YN) of YFP were cotransformed into protoplasts and then incubated in the dark (D) or 10 µmol m−2 s−1 RL for 12 h. mCherry was used as a nuclear maker. As a negative control, PIF3 and HDA15 fused with YC or YN and empty vectors were also cotransformed into protoplasts.
(F) Coimmunoprecipitation analysis of interaction between HDA15 and PIF3. Total proteins were extracted from Col, hda15-1, and PIF3-MYC seedlings grown in the dark for 2 d and exposed to RL for 0, 10 min, 30 min, or 2 h. PIF3-MYC is a transgenic line expressing myc-tagged PIF3 (Shin et al., 2007). A rabbit polyclonal anti-HDA15 antibody (anti-HDA15 Rb) was used for immunoprecipitation and further detected after SDS-PAGE with an anti-myc antibody. Endogenous HDA15 protein was detected with a mouse polyclonal anti-HDA15 antibody (anti-HDA15 M) after immunoprecipitation with anti-HDA15 Rb. Input PIF3-MYC protein was detected with an anti-myc antibody.
We further analyzed the interaction between HDA15 and PIF3 by pull-down assays. Purified PIF3-His recombinant protein was incubated with glutathione S-transferase (GST)-HDA15 fusion protein. PIF3-His was pulled down by GST-HDA15 (Figure 1C). Furthermore, PIF3-His recombinant protein was also incubated with HDA15-GFP (for green fluorescent protein) from total protein extracts of etiolated seedlings. In vivo HDA15-GFP protein was also pulled down by recombinant PIF3-His (Figure 1D). These data also support that HDA15 interacts with PIF3.
The interaction of HDA15 and PIF3 was further studied in vivo by the bimolecular fluorescence complementation (BiFC) assay. HDA15 and PIF3 were fused to the N-terminal 174–amino acid portion of yellow fluorescent protein (YFP) in the pEarleyGate201 vector and C-terminal 66–amino acid portion of YFP in the pEarleyGate202 vector (Lu et al., 2010), respectively. The constructs were codelivered into Arabidopsis protoplasts and then incubated in darkness or under red light (RL) for 12 h. Strong YFP signal was observed in the nucleus of dark-incubated Arabidopsis protoplasts, while no obvious signal was detected in the protoplasts incubated under RL conditions, suggesting that HDA15 and PIF3 interact in dark conditions only (Figure 1E). The corresponding fusion gene constructs were also codelivered into tobacco (Nicotiana tabacum) leaves by infiltration with Agrobacterium tumefaciens (GV3101). The direct interaction between HDA15 and PIF3 was also observed in the nuclei of tobacco epidermal cells in dark (see Supplemental Figure 2 online).
We also performed coimmunoprecipitation assays in 35S:PIF3-MYC transgenic plants (Shin et al., 2007). PIF3-MYC was coimmunoprecipitated with endogenous HDA15 in 2-d-old etiolated seedlings (Figure 1F). However, this interaction became weaker when the seedlings were exposed to RL for 10 min and almost disappeared after 2 h RL exposure. Taken together, our data suggest that HDA15 interacts with PIF3 only in dark conditions in Arabidopsis.
HDA15 Is a HDAC Localized in Nuclei
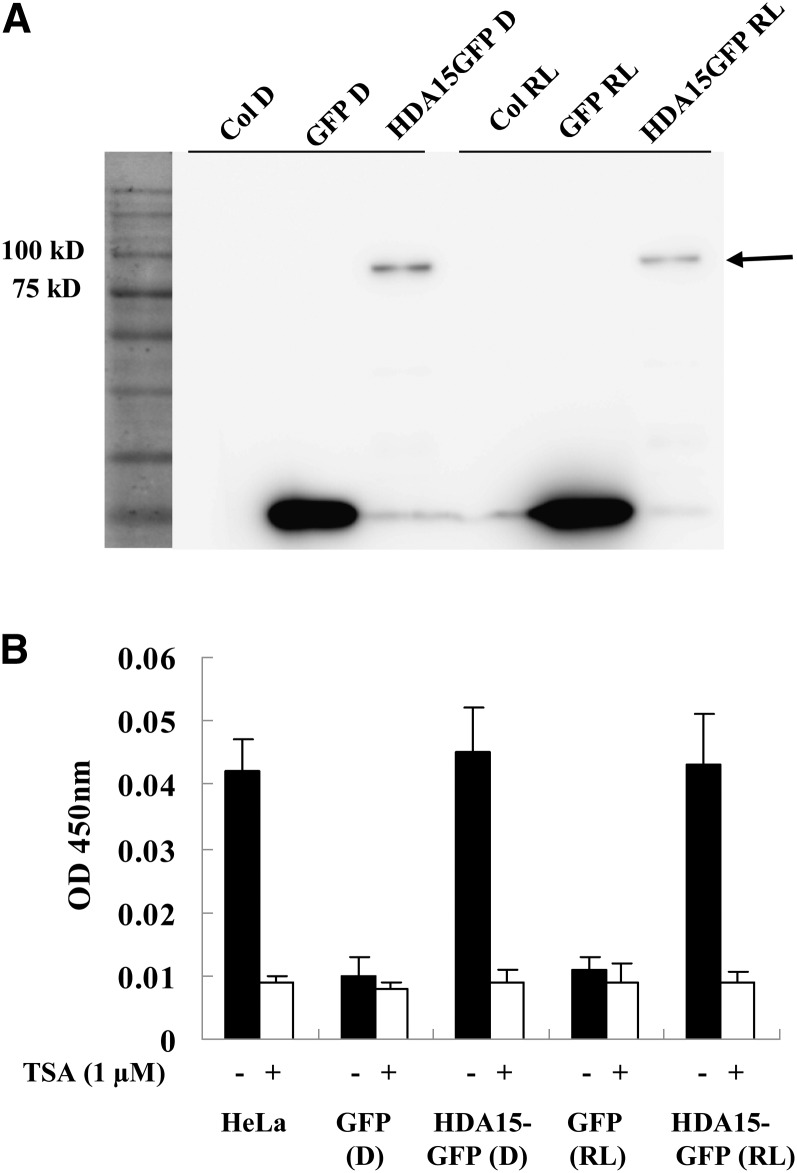
Recombinant HDA15 produced in Escherichia coli did not show measurable HDAC activity. Therefore, we purified GFP-tagged HDA15 from 2-d-old etiolated seedlings or seedlings transferred to RL light for 2 h using transgenic Arabidopsis expressing the HDA15-GFP chimeric gene driven by the cauliflower mosaic virus 35S promoter (Figure 2A). A prominent polypeptide migrated on a SDS-PAGE gel at the predicted size of 86 kD and was absent in nontransgenic and 35S:GFP plants (Figure 2A). HDAC activities were evaluated using a colorimetric assay. The HeLa nuclear extract that contains a blend of HDACs served as the positive control, while the HDAC inhibitor trichostatin A (TSA) was also added to demonstrate the specificity of deacetylation activities. As shown in Figure 2B, purified HDA15-GFP protein from both etiolated seedlings and seedlings exposed to RL clearly showed the HDAC activity. These data suggest that HDA15 has HDAC activity and that the activity is not affected by light.
Figure 2.
HDA15 Has HDAC Activity.
(A) Affinity purification of HDA15-GFP expressed in transgenic plants. HDA15-GFP was purified with GFP-Trap magnetic particles from extracts of 2-d-etiolated HDA15-GFP seedlings or seedlings exposed to RL (10 µmol m−2 s−1) for 2 h, and a GFP antibody was used for immunoblotting analysis. D, dark.
(B) Colorimetric HDAC activity assay of purified HDA15-GFP. HeLa nuclear extracts that contain a blend of HDACs served as the positive control. TSA was added to samples to demonstrate the specificity of deacetylation activities. The HDAC activity was expressed as OD as measured by a spectrophotometer at 405 nm. Transgenic plants expressing GFP were used as a control. The values are shown as means ± sd.
We also used 35S:HDA15-GFP transgenic plants to examine the subcellular localization of HDA15 (see Supplemental Figures 3A and 3B online). We observed that HDA15-GFP protein was localized in the nucleus of 4-d-old etiolated seedlings both in dark and white light conditions (see Supplemental Figure 3A online). When etiolated HDA15-GFP seedlings were transferred to RL for 10, 30, or 120 min, the HDA15-GFP proteins were also localized in the nucleus (see Supplemental Figure 3B online). These data indicated that HDA15 is constitutively localized in the nucleus in both dark and light conditions in Arabidopsis seedlings. A previous study indicated that HDA15 can undergo the nuclear-cytoplasm shuttling driven by light in Arabidopsis protoplasts using a transient assay (Alinsug et al., 2012), suggesting that HDA15 may also be localized in the cytoplasm under different conditions.
HDA15 and PIF3 Repress Chlorophyll Biosynthesis in the Dark
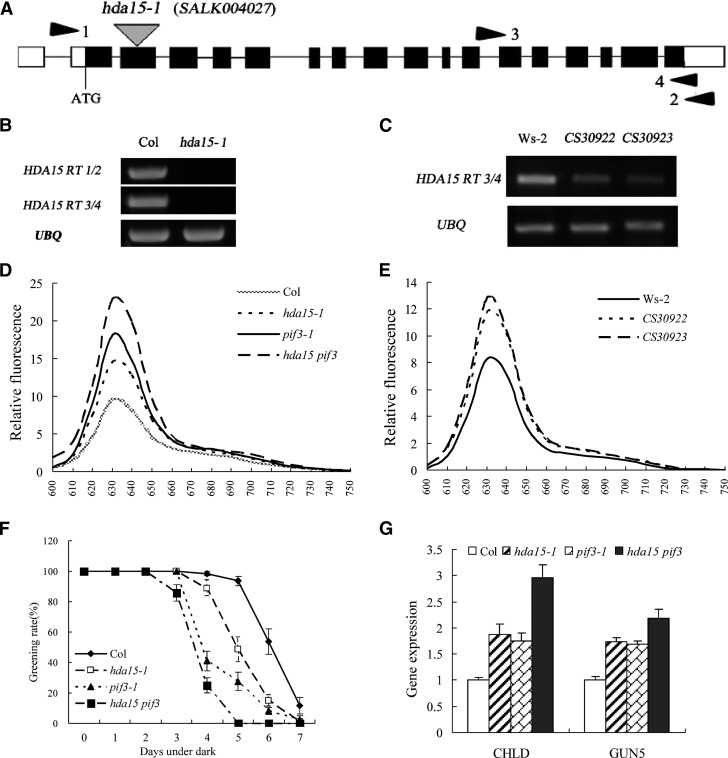
To further investigate the function of HDA15, a T-DNA insertion mutant, hda15-1 (SALK_004027), and two RNA interference (RNAi) lines (CS30922 and CS30923) were analyzed. The T-DNA was inserted in the second exon of HDA15, and no HDA15 transcript was detected in the homozygous hda15-1 plants (Figures 3A and 3B). Furthermore, the expression of HDA15 mRNA in the two RNAi lines CS30922 and CS30923 was markedly decreased compared with the wild type (Figure 3C). We also generated the hda15 pif3 double mutants by crossing hda15-1 and pif3-1 (Kim et al., 2003) to investigate the genetic interaction between HDA15 and PIF3.
Figure 3.
HDA15 Negatively Regulates Chlorophyll Biosynthetic and Photosynthetic Genes in the Dark.
(A) Exon-intron structure of HDA15. White boxes represent untranslated regions, black boxes represent coding regions, lines represent introns, and triangles indicate the T-DNA insertion in hda15-1 (SALK_004027). Positions of primers used for RT-PCR in (B) and (C) are indicated by arrowheads.
(B) and (C) RT-PCR analysis of HDA15 mRNA in hda15-1 and HDA15 RNAi lines (CS30922 and CS30923). UBQ10 was used as an internal control. Ws-2, Wassilewskija-2.
(D) and (E) Relative fluorescence of protochlorophyllide in etiolated hda15-1, pif3-1, hda15 pif3, and HDA15 RNAi seedlings. The protochlorophyllide was extracted from 4-d-old etiolated seedlings.
(F) Greening rates of seedlings grown in the dark for various periods of time followed by 2 d of continuous white light exposure (100 µmol m−2 s−1). The values are shown as means ± sd.
(G) qRT-PCR analysis of gene expression related to chlorophyll biosynthesis genes, CHLD and GUN5, in 2-d-old etiolated hda15-1, pif3-1, and hda15 pif3 seedlings. The values are shown as means ± sd.
The protochlorophyllide contents in etiolated hda15-1 and RNAi plants were measured (Shin et al., 2009). Etiolated hda15-1, hda15 pif3, and HDA15 RNAi seedlings accumulated a relative higher amount of protochlorophyllide compared with the wild type (Figures 3D and 3E), suggesting that HDA15 may negatively regulate protochlorophyllide biosynthesis in the dark. Etiolated seedlings that accumulate abnormally high levels of chlorophyll intermediates are bleached when transferred to the light (Huq et al., 2004). We grew Columbia (Col) wild type, hda15-1, pif3-1, and hda15 pif3 for several days in the dark and then transferred into white light. The greening rate of hda15-1 was lower than that of the wild type after 4 d or longer of dark treatment (Figure 3F). The hda15 pif3 double mutant showed an even lower greening rate compared with hda15-1 and pif3-1 single mutants, with almost all the seedlings became bleached when grew 5 d or longer in the dark (Figure 3F). These data suggest that hda15 mutants accumulate a higher level of chlorophyll intermediates than the wild type.
The observed phenotype prompted us to test whether HDA15 regulates the expression of the genes involved in the chlorophyll biosynthetic pathway (see Supplemental Figure 4 online). CHLD and GUN5 encode the CHL D and CHL H subunit of the Mg2+-chelatase enzyme complex, the first enzyme in the chlorophyll biosynthetic branch in the tetrapyrrole pathway (Mochizuki et al., 2001), whereas CRD1 encodes a putative di-iron enzyme that is required for the biosynthesis of protochlorophyllide from Mg-protoporphyrin IX ME (Tanaka and Tanaka, 2007). The expression of CHLD, GUN5, and CRD was upregulated (>1.5-fold to Col) in the 2-d-etiolated seedlings of hda15 mutants compared with the wild type (see Supplemental Figure 4 online), suggesting that HDA15 may repress the expression of the chlorophyll biosynthetic genes. The upregulation of CHLD, GUN5, and CRD1 in hda15-1 indicated that HDA15 may repress the conversion from protoporphyrin IX to protochlorophyllide in the chlorophyll biosynthetic pathway.
A recent study suggested that PIF3 may act as a negative regulator of chlorophyll biosynthesis by repressing genes required for chlorophyll biosynthesis, such as CHLD and GUN5 in the dark (Shin et al., 2009). We further compared the expression levels of CHLD and GUN5 in etiolated hda15, pif3, and hda15 pif3 mutants. Compared with the wild type, the transcript levels of CHLD and GUN5 were all increased in 2-d-etiolated hda15, pif3, and double mutants (Figure 3G). The hda15 pif3 double mutants showed highest expression of CHLD and GUN5, which is in accordance with the relatively high amount of protochlorophyllide in etiolated hda15 pif3 seedlings (Figure 3F). These data indicate that both HDA15 and PIF3 repress the gene expression involved in chlorophyll biosynthetic pathways.
HDA15 Is a Repressor of Photosynthetic Genes in the Dark
To further explore the roles of HDA15 in etiolated seedlings, we analyzed the global gene expression profiles of 2-d-old etiolated hda15-1 seedlings using an Affymetrix microarray (ATH1). Compared with the wild type, 520 genes were upregulated but only 192 genes were downregulated in hda15-1 seedlings (see Supplemental Data Sets 1A and 1B online), suggesting that HDA15 may act mainly as a transcriptional repressor in etiolated seedlings. The genes affected by the hda15 mutation are involved in a variety of biological processes, including the following Gene Ontology categories: metabolic process, transcription, response to stimulus, photosynthesis, transport, protein phosphorylation, biosynthetic process, developmental process, translation, cellular process, and nucleic acid metabolic process (see Supplemental Data Sets 1A and 1B and Supplemental Figures 5A and 5B online), indicating that HDA15 plays important roles in regulation of growth and developmental processes in Arabidopsis.
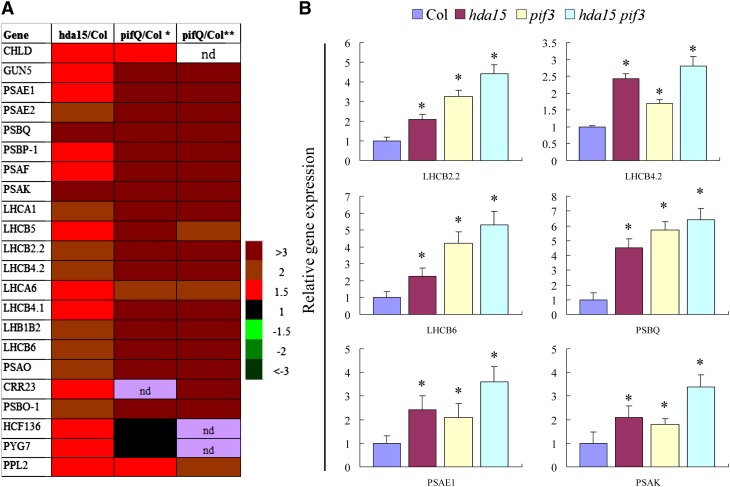
We compared the gene expression profiles of the hda15 and pif1 pif3 pif4 pif5 quadruple (pifQ) mutants (Leivar et al., 2009; Shin et al., 2009). Of the 520 induced genes in hda15, 62 (∼12%) genes were also upregulated in the pifQ mutant (see Supplemental Table 1 online). Most of the light-responsive genes regulated by HDA15 were also upregulated in etiolated pifQ mutants (Figure 4A). The transcripts of a large subset of photosynthetic genes were elevated in hda15, including PSAE1, PSAE2, PSAF, PSAK, PSAO, LHCB2.2, LHCA6, LHCB5, LHCB4.2, LHCB4.1, and LHB1B2 (Figure 4A; see Supplemental Table 1 online). PSAE1, PSAE2, PSAF, PSAK, PSBQ, and PSAO encode the subunits of photosystem I, whereas the others encode the subunits of light harvesting complexes of photosystems I and II. The expression patterns of these genes were further confirmed by quantitative RT-PCR (qRT-PCR) (see Supplemental Figure 5C online).
Figure 4.
HDA15 Represses a Subset of Light-Responsive Genes in Etiolated Seedlings.
(A) Two-day-old etiolated hda15 and pifQ seedlings show similar transcriptional profiles of chlorophyll biosynthetic and photosynthetic genes. *Data analyzed by Shin et al. (2009); **data analyzed by Leivar et al. (2009). The color code indicates relative transcript levels. nd, not determined.
(B) qRT-PCR analysis of genes related to photosynthesis in etiolated Col, hda15, pif3, and hda15 pif3 seedlings; UBQ10 was used as an internal control. The values are shown as means + sd (t test: *P < 0.05, difference from the wild type).
We also analyzed the transcript levels of some of the HDA15-regulated light-responsive genes in etiolated pif3-1 and hda15 pif3 seedlings. Similar to hda15-1, the expression of LHCB2.2, LHCB4.2, LHCB6, PSBQ, PSAE1, and PSAK was also upregulated in pif3-1 and hda15 pif3 seedlings (Figure 4B). Compared with the single mutants, increased expression of LHB1B2, LHCB6, PSAE2, PSAK, and PSBQ in hda15 pif3 double mutants was observed, suggesting that HDA15 and PIF3 may act synergistically in the regulation of gene expression. Taken together, these data suggested that both HDA15 and PIF3 function as repressors of photosynthetic genes in etiolated seedlings.
HDA15 and PIF3 Repress Histone Acetylation and RNAPII-Mediated Transcription of Target Genes
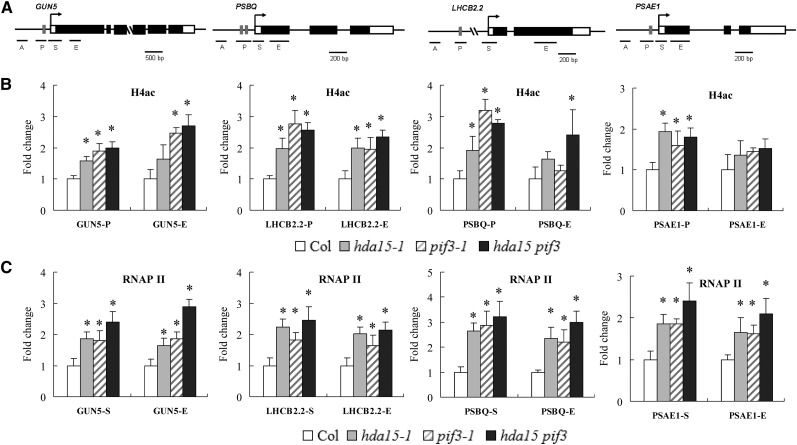
To evaluate whether the high expression of GUN5, LHCB2.2, PSBQ, and PSAE1 in hda15-1 and pif3-1 seedlings is related to histone modifications, we determined the histone acetylation levels of these genes by ChIP assays. The histone H4 acetylation levels of the promoters and exons of GUN5, LHCB2.2, PSBQ, and PSAE1 were elevated in hda15, pif3, and hda15 pif3 mutants (Figures 5A and 5B). By contrast, the nuclear occupancy of these genes was not changed (see Supplemental Figure 6 online). These data suggest that both HDA15 and PIF3 may repress gene expression by deceasing histone acetylation levels.
Figure 5.
HDA15 and PIF3 Repress the Histone Acetylation and RNAPII-Mediated Transcription of Chlorophyll Biosynthetic and Photosynthetic Genes.
(A) Schematic diagram of the HDA15 and PIF3 coregulated genes GUN5, PSBQ, LHCB2.2, and PSAE1. Gray boxes indicate G-box elements (CACGTG). A and P indicate promoter regions, and S and E indicate transcriptional starting site and exon.
(B) ChIP analysis of H4ac levels at the promoters (P) and exons (E) of GUN5, LHCB2.2, PSBQ, and PSAE1 in Col, hda15, pif3, and hda15 pif3 mutants.
(C) ChIP analysis of RNAPII enrichment at the transcription starting sites (S) and exons (E) of GUN5, LHCB2.2, PSBQ, and PSAE1 in Col, hda15, pif3, and hda15 pif3 plants. The amounts of DNA after ChIP were quantified and normalized to ACTIN2. The values are shown as means + sd (t test: *P < 0.05, compared with the wild type).
We further analyzed RNA polymerase II (RNAPII) occupancy on GUN5, LHCB2.2, PSBQ, and PSAE1 in etiolated hda15, pif3, and hda15 pif3 mutants by ChIP assays using an anti-RNAPII antibody. A relatively higher enrichment of RNAPII binding to the transcriptional starting sites and exons was observed in hda15, pif3, and hda15 pif3 mutants compared with the wild type (Figure 5C), suggesting that HDA15 and PIF3 repress RNAPII-associated transcription in etiolated seedlings. Taken together, our data indicate that both HDA15 and PIF3 repress the transcription of chlorophyll biosynthetic and photosynthetic genes by decreasing histone acetylation and RNAPII-associated transcription in etiolated seedlings.
HDA15 and PIF3 Cotarget to the Promoters of Chlorophyll Biosynthetic and Photosynthetic Genes in Etiolated Seedlings
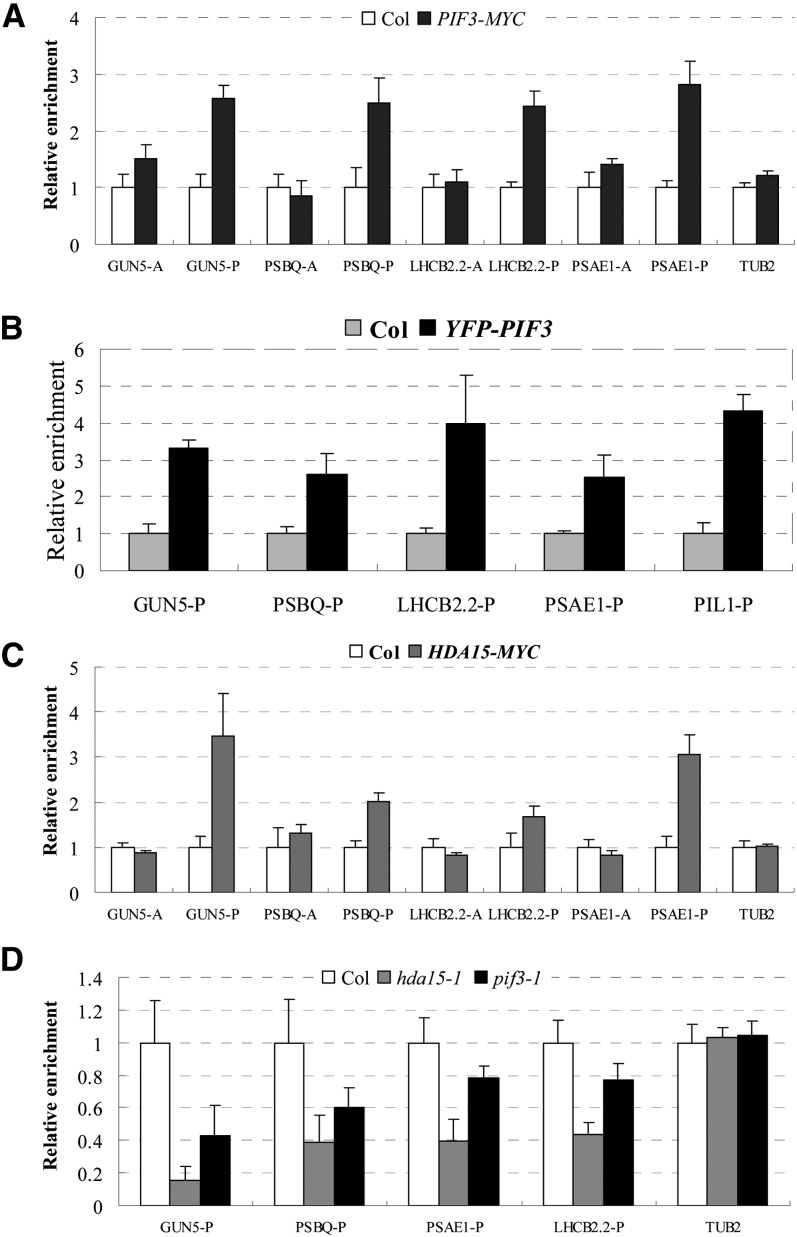
Genome-wide transcription and ChIP analysis revealed that HDA15 and PIF3 coregulate a large subset of chlorophyll biosynthetic and photosynthetic genes. To examine whether these genes are direct targets of HDA15 and PIF3, ChIP assays were performed using transgenic lines expressing YFP-tagged PIF3 (YFP-PIF3) (Al-Sady et al., 2006), MYC-tagged PIF3 (PIF3-MYC) (Shin et al., 2007), and MYC-tagged HDA15 (HDA15-MYC) (see Supplemental Figures 7A and 7B online). Overexpression of 35S:HDA15-MYC in hda15-1 can complement the protochlorophyllide defect phenotype (see Supplemental Figure 7C online), suggesting that the HDA15-MYC protein is functional in vivo. ChIP analyses in etiolated PIF3-MYC and YFP-PIF3 seedlings showed that PIF3 specifically bound to the G-box regions of GUN5, LHCB2.2, PSBQ, and PSAE1 in etiolated seedlings (Figures 6A and 6B). Similar to a previous study (Soy et al., 2012), we also found that PIF3 bound to the promoter of PIL1 (Figure 6B). Furthermore, the G-box regions of GUN5, LHCB2.2, PSBQ, and PSAE1 were also direct binding sites of HDA15 (Figure 6C). As a control, we analyzed SAG20 and CSD1, two unrelated genes upregulated in hda15 based on the microarray and qRT-PCR analysis (see Supplemental Data Set 1A and Supplemental Figure 8A online). SAG20 and CSD1 were not direct binding targets of HDA15 (see Supplemental Figure 8B online). These data suggest that HDA15 and PIF3 may specifically cotarget to the promoters of chlorophyll biosynthetic and photosynthetic genes in etiolated seedlings.
Figure 6.
HDA15 and PIF3 Cotarget to the Promoters of Chlorophyll Biosynthetic and Photosynthetic Genes in the Dark.
(A) ChIP-qPCR analysis of PIF3-MYC DNA fragments coimmunoprecipitated with the anti-myc antibody relative to Col DNA. PIF3-MYC is a transgenic line expressing myc-tagged PIF3 (Shin et al., 2007). TUB2 was used as a control. The values are shown as means ± sd.
(B) ChIP-qPCR analysis of enrichment of YFP-PIF3 to the promoters of the light-responsive genes in etiolated Col and YFP-PIF3 seedlings. An anti-GFP antibody was used for the immunoprecipitation. YFP-PIF3 is a transgenic line expressing YFP-tagged PIF3 under the control of PIF3 native promoter (Al-Sady et al., 2006). PIL1 was used as a positive control as described by Soy et al. (2012). The values are shown as means ± sd.
(C) ChIP-qPCR analysis of DNA fragments coimmunoprecipitated with the anti-myc antibody relative to Col. HDA15-MYC is a transgenic line expressing a myc-tagged HDA15. TUB2 was used as a control. The values are shown as means ± sd.
(D) ChIP analysis with an anti-HDA15 antibody in 2-d-old etiolated Col, hda15-1, and pif3-1 seedlings. The amounts of DNA after ChIP were quantified and normalized to ACTIN2. A and P are promoter regions as indicated in Figure 5A. The values are shown as means ± sd.
To further investigate whether the binding of HDA15 to the target genes is dependent on the presence of PIF3, we performed ChIP assays with an anti-HDA15 antibody using 2-d-old etiolated Col, hda15-1, and pif3-1 seedlings. Much lower enrichment of HDA15 protein in the promoters of GUN5, LHCB2.2, PSBQ, and PSAE1 was detected in hda15-1 seedlings compared with the wild type (Figure 6D), confirming that these genes are the direct targets of HDA15. Furthermore, relatively lower enrichment of HDA15 protein in the promoters of GUN5, LHCB2.2, PSBQ, and PSAE1 was also detected in pif3-1 (Figure 6D), suggesting that the association of HDA15 with the promoters of light-responsive genes is dependent on PIF3.
Light Dissociates the Binding of PIF3 and HDA15 from Their Target Genes
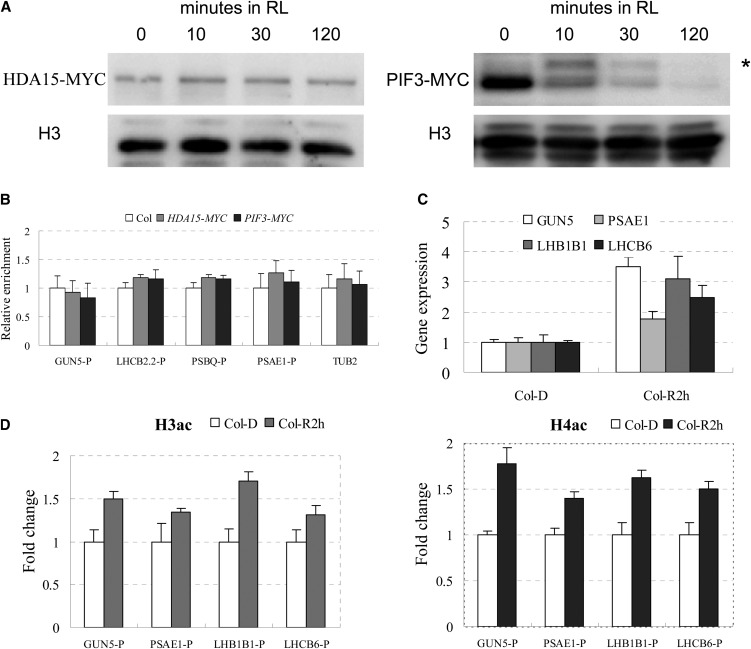
Previous studies showed that light triggers activated forms of phytochromes and induces rapid phosphorylation and degradation of PIF3 (Bauer et al., 2004; Park et al., 2004; Shen et al., 2005; Al-Sady et al., 2006), indicating that PIF3 and its interacting proteins may be disassociated from their target genes upon exposure to light. We analyzed the binding of PIF3 and HDA15 to their target genes during seedling deetiolation. We first checked the protein levels of HDA15-MYC and PIF3-MYC in the transgenic lines grown for 2 d in darkness and then exposed to RL. The protein level of HDA15 was not changed (Figure 7A), suggesting that HDA15 protein was stable during dark-to-light conversion. In PIF3-MYC transgenic plants, a band of a larger molecular mass appeared immediately after Rp exposure, which may represent the phosphorylated form of PIF3 (Bauer et al., 2004; Park et al., 2004; Shen et al., 2005; Al-Sady et al., 2006). The phosphorylated PIF3 was degraded 2 h after transfer to RL (Figure 7A).
Figure 7.
RL Dissociates HDA15 and PIF3 from Target Genes and Promotes Gene Expression.
(A) Immunoblot analysis of epitope-tagged HDA15 and PIF3 in HDA15-MYC and PIF3-MYC seedlings grown for 2 d in the dark and then maintained in darkness or exposed to RL at 10 µmol m−2 s−1 before extraction. Blotted samples were probed with an anti-myc antibody. Histone H3 was used as a loading control. Asterisk indicates the phosphorylated form of PIF3.
(B) ChIP analysis of the binding of HDA15 and PIF3 to the G-box regions of the promoters in the light-responsive genes when dark-grown seedlings were exposed to RL. P indicates G-box–containing regions in the promoters (as indicated in Figure 5A). The values are shown as means ± sd.
(C) Expression levels of light-responsive genes in Col grown for 2 d in dark and then exposed to RL for 2 h (Col-R2 h) compared with those in dark-grown seedlings (Col-D). The values are shown as means ± sd.
(D) ChIP analysis of histone H3 and H4 acetylation levels of light-responsive genes in Col grown for 2 d in dark and then maintained in darkness or exposed to RL for 2 h. P indicates promoter region. ACTIN2 was used as an internal control. The values are shown as means ± sd.
We used ChIP assays to examine whether PIF3 and HDA15 could still bind to GUN5, LHCB2.2, PSAE1, and PSBQ when etiolated seedlings were exposed to RL. Compared with the strong binding of PIF3 and HDA15 to the promoters of the target genes (Figure 6) in dark-grown seedlings, there was no difference in the relative enrichment of PIF3 and HDA15 among the transgenic plants (PIF3-MYC and HDA15-MYC) and wild-type plants after 2 h RL exposure (Figure 7B), suggesting that PIF3 and HDA15 proteins were dissociated from the target genes after light exposure.
We also detected the expression levels of the early light-inducible genes, GUN5, PSAE1, LHB1B1, and LHCB6, in wild-type seedlings after RL exposure. The transcription of these genes was upregulated after transfer to RL for 2 h (Figure 7C). ChIP analysis showed that H3 and H4 acetylation levels in the promoters of these genes were enriched after 2 h RL (Figure 7D). Collectively, our data suggest that light dissociates HDA15 and PIF3 from their target genes, resulting in increased histone acetylation and gene expression.
HDA15 and PIF3 Act Independently in the Regulation of Hypocotyl Elongation
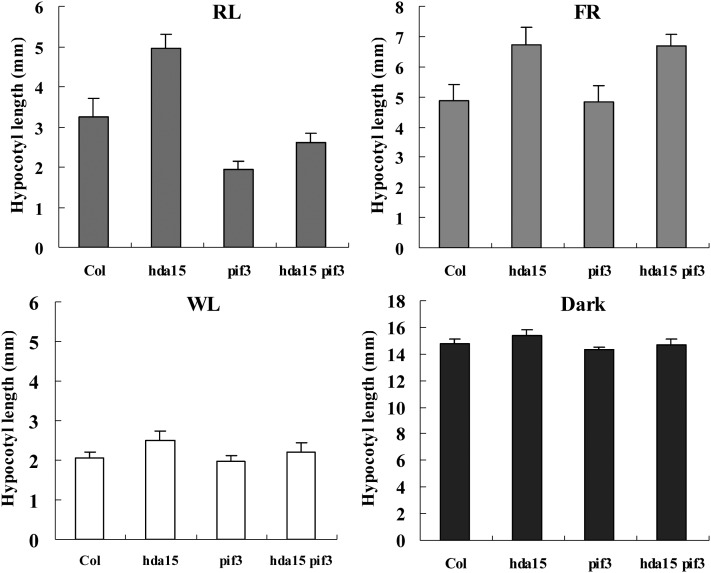
We further studied the phenotype of hda15 mutants under various light conditions. Phenotypic analysis showed that hda15-1 and HDA15 RNAi seedlings exhibited relative longer hypocotyls than the wild type under RL and FR conditions (Figure 8; see Supplemental Figure 9 online), indicating that HDA15 might positively regulate PHYA- and PHYB-mediated inhibition of hypocotyl elongation. Previous studies demonstrated that PIF3 is a negative regulator of PHYB- but not PHYA-mediated inhibition of hypocotyl elongation (Kim et al., 2003). We further studied whether HDA15 and PIF3 interact genetically to regulate hypocotyl elongation under various light conditions. Similar to the results from previous studies (Kim et al., 2003), the pif3 mutants had shorter hypocotyls under RL compared with the wild type. hda15 pif3 double mutants showed an intermediate hypocotyl length compared with hda15-1 and pif3-1 seedlings under RL (Figure 8), suggesting that HDA15 might act independently of PIF3 in regulation of PHYB-mediated inhibition of hypocotyl elongation. When grown under FR, the hypocotyl length of pif3 was similar to that of the wild type, whereas the hypocotyl length of hda15 pif3 was similar to that of hda15. Taken together, these data suggest that HDA15 and PIF3 may independently regulate hypocotyl elongation in seedlings growth.
Figure 8.
HDA15 and PIF3 Act Independently in the Regulation of Hypocotyl Elongation.
Quantification of hypocotyl length phenotypes of 4-d-old Col, hda15, pif3, and hda15 pif3 seedlings under various light conditions. RL, 5 µmol m−2 s−1; FR, 5 µmol m−2 s−1; white light (WL), 100 µmol m−2 s−1. The values are shown as means ± sd.
A previous study showed that mutations of HDA19 in Arabidopsis resulted in a short-hypocotyl phenotype under RL and FR (Benhamed et al., 2006), indicating a negative role of HDA19 in PHYA- and PHYB-mediated inhibition of hypocotyl elongation. We generated hda15 hda19 double mutants by genetic crossing hda15-1 and hda19-3 (Kim et al., 2008). hda15 hda19 seedlings showed intermediate hypocotyl lengths compared with hda15 and hda19 single mutants under RL and FR conditions (see Supplemental Figure 10 online), suggesting that HDA15 and HDA19 may act antagonistically in regulation of hypocotyl elongation. These data suggest that different HDACs may have distinct functions in hypocotyl growth.
DISCUSSION
Involvement of HDACs in Light-Regulated Gene Expression
Previous work showed that light-induced expression of the pea (Pisum sativum) plastocyanin gene (PetE) in tobacco was associated with hyperacetylation of both histones H3 and H4 (Chua et al., 2001, 2003), suggesting that histone acetylation of chromatin acts as an important regulatory switch to integrate light signals to control gene expression. Further phenotypic analysis of HAT and HDAC mutants in Arabidopsis revealed a role of histone acetylation in light-regulated development processes. Mutations of the HAT GCN5 resulted in reduced light-inducible gene expression (Benhamed et al., 2006), whereas loss of function of HDA15 leads to upregulation of a number of chlorophyll biosynthetic and photosynthetic genes. These findings suggest that HDACs and HATs may act antagonistically in plant light responses. Similar to the previous finding (Charron et al., 2009), we also found that activation of photosynthetic genes was accompanied with an increase of H3 acetylation. Increased H4 acetylation in the chlorophyll biosynthetic and photosynthetic genes was also observed in the hda15 mutant, suggesting that HDA15 may be the major HDAC that contributes to the regulation of H4 acetylation.
Accurate initiation of gene transcription requires multiple factors, including transcription cofactors (coactivators or corepressors) (Bertrand et al., 2005). In vitro studies have shown that transcription cofactors are usually associated with chromatin remodeling and modification factors, such as HDACs and HATs, which determine the level of acetylation of specific Lys residues within the core histone tails (Eberharter and Becker, 2002). Previous studies indicated that GCN5 directly binds to the promoters of RBCS-1A and LHCB1.1 (Benhamed et al., 2006). Further ChIP-on-chip analyses revealed that GCN5 binds to 38% of the light-regulated genes (Benhamed et al., 2008). In this study, we found that several chlorophyll biosynthetic and photosynthetic genes are the direct targets of HDA15. These results suggest a direct involvement of HATs and HDACs in transcriptional regulation of light-responsive genes in Arabidopsis. Phenotypic observation and gene expression analyses of HAT mutants, haf1, taf1, and gcn5, have suggested a genetic association of these HATs with HY5, which encodes a transcriptional activator of photomorphogenesis (Bertrand et al., 2005; Benhamed et al., 2006). It is still unclear whether HY5 interacts directly or indirectly with these HAT proteins to promote light responses. In this study, we present evidence indicating that HDA15 physically interacts with PIF3, a transcriptional repressor of chlorophyll and photosynthesis genes. Therefore, HATs and HDACs may associate with different transcription factors to modulate light-regulated gene expression.
PIF3 Represses Chlorophyll Biosynthetic and Photosynthetic Genes via Association with HDA15
PIF3 was initially proposed as a positive component in light-regulated chloroplast development. This conclusion was mainly supported by the data that the pif3 mutant showed a reduced level of chlorophyll compared with wild-type seedlings upon transfer to RL after 4 d in the dark (Monte et al., 2004). However, recent studies support that PIF3 acts as a negative regulator in chloroplast development based on the findings that PIF3 decreases the expression of chlorophyll biosynthetic and photosynthetic genes and functions additively with PIF1 to repress chlorophyll biosynthesis in 2-d-old dark-grown seedlings (Shin et al., 2009; Stephenson et al., 2009). However, the molecular mechanism of how PIF3 regulates chlorophyll biosynthesis was still unclear. Our study reveals that the chlorophyll biosynthetic and photosynthetic genes are the direct targets of PIF3. These findings further support the hypothesis that PIF3 acts a negative regulator in chloroplast development by directly repressing chlorophyll biosynthetic and photosynthetic genes.
More recent studies indicated that transcription repressors can recruit HDACs to regulate gene expression in plants. Arabidopsis AGL15, a MADS transcription factor, modulates target gene expression via recruitment of a repression complex containing RPD3/HDA1-type HDACs, HDA6 and HDA19 (Hill et al., 2008). The AP2/EREBP-type transcription factor ERF7 interacts with SIN3, a global corepressor of transcription, which in turn interacts with HDA19 to repress abscisic acid–mediated gene expression (Song et al., 2005). In addition, AS1 and HSL2 recruit HDA6 and HDA19 to regulate leaf and seed development, respectively (Luo et al., 2012; Zhou et al., 2013). In mammalian cells, the bHLH-ZIP transcription factor MAD recruits HDACs together with SIN3 to prevent cell cycle–related gene expression (Laherty et al., 1997). These findings indicate that transcription factors, including bHLH proteins, can recruit HDAC complexes to regulate target gene expression. In this study, we showed that PIF3 physically associated with a RPD3/HDA1-type HDAC, HDA15, suggesting that they act in the same protein complex. A correlation between light-responsive gene expression and histone acetylation was observed, supporting the involvement of HDACs in light responses (Guo et al., 2008; Charron et al., 2009). Increased levels of histone H4 acetylation were observed at chlorophyll biosynthetic and photosynthetic genes in etiolated hda15, pif3, and pif3 hda15 seedlings, indicating that both HDA15 and PIF3 regulate expression of these genes through histone deacetylation. Furthermore, purified HDA15 demonstrated HDAC activity in etiolated seedlings, supporting an involvement of histone deacetylation in repression of the chlorophyll development genes. The binding of HDA15 to the target genes depended on the presence of PIF3. Taken together, these findings suggest that PIF3 represses chlorophyll biosynthetic and photosynthetic genes via recruiting HDA15 to decrease their histone acetylation levels.
Similar to PIF3, other PIFs, such as PIF1, PIF4, and PIF5, are also required to repress photomorphogenic development in the dark. pif1 mutant seedlings accumulate excess free protochlorophyllides, suggesting a negative role of PIF1 in chlorophyll development (Huq et al., 2004). A recent work demonstrated that PIF1 also directly binds to the promoter of PSY, which encodes the main enzyme of the carotenoid biosynthesis pathway, resulting in PSY repression in the dark (Toledo-Ortiz et al., 2010). Genome-wide expression analysis showed that genes involved in chlorophyll biosynthesis and photosynthesis were highly upregulated in the etiolated pifQ mutant (Shin et al., 2009; Stephenson et al., 2009), indicating that these PIFs may function redundantly and additively in repression of chlorophyll development. We found that HDA15 specifically interacted with PIF3. It remains to be determined whether other PIFs may associate with different HDACs to repress chlorophyll biosynthetic genes in etiolated seedlings.
Moreover, recent studies revealed a key role of DELLA proteins in chlorophyll and carotenoid biosynthesis (Cheminant et al., 2011). Since DELLAs promote protochlorophyllide biosynthesis by reducing the binding of PIFs to the promoters of target genes (Cheminant et al., 2011), DELLAs might regulate the chlorophyll biosynthetic genes by inhibiting the occupancy of the HDA15-PIF3 repression complex at the chlorophyll biosynthetic genes. Furthermore, DELLA proteins also promote the expression of POR genes independent of PIFs, thus preventing photobleaching (Cheminant et al., 2011). In this study, we showed that the expression of POR genes was not obviously altered in etiolated hda15 mutants. The photooxidative phenotype of hda15-1 mutant might due to accumulation of a higher amount of protochlorophyllide.
Light Dissociates PIF3 and HDA15 from Their Target Genes
The degradation of transcription factors is important in the regulation of light signaling (Henriques et al., 2009). Previous studies showed that PIF3 protein is stable in the dark, but it is rapidly degraded upon exposure to light (Bauer et al., 2004). This light-induced degradation is promoted by phyA, phyB, and phyD and is mediated by the ubiquitin-26S proteasome pathway (Park et al., 2004; Al-Sady et al., 2006). In this study, we showed that PIF3 was dissociated from the target genes after exposure to RL for 2 h, which may be due to rapid degradation of PIF3 by RL (Bauer et al., 2004). Although the protein level of HDA15 did not change during dark–light transition, HDA15 dissociated from the target genes after light exposure, suggesting that PIF3 is required for the binding of HDA15 to the target genes. Due to the degradation of PIF3 protein, no obvious interaction between HDA15 and PIF3 was observed under RL. Taken together, these data support the notion that phytochromes relieve repression of light responses via degradation of PIF3 and dissociation of HDA15 from their target genes.
Gene activation markers, H3 and H4 acetylation, are enriched at the promoters of light-responsive genes after RL exposure, suggesting that HAT proteins may be recruited to these loci by transcriptional activators. HY5 is a bZIP transcription activator involved in promoting photomorphogenesis (Oyama et al., 1997; Lee et al., 2007; Kobayashi et al., 2012). HY5 binds to the G-box element and directly regulates key chlorophyll biosynthetic and photosynthetic genes (Oyama et al., 1997; Lee et al., 2007; Kobayashi et al., 2012). Both PIF3 and HY5 were shown to target to the promoters of anthocyanin biosynthetic genes, CHS, CHI, F3′H, and LDOX, at distinct cis-elements (Shin et al., 2007). These findings suggest a possibility that HY5 and PIF3 may act antagonistically to regulate genes involved in chlorophyll biosynthesis and photosynthesis. Therefore, HY5 may play a dominant role in transcription regulation after dissociation of PIF3 from the target promoters by light. Upon light exposure, HY5 may recruit GCN5 and/or other HATs to activate the expression of light-responsive genes by histone acetylation.
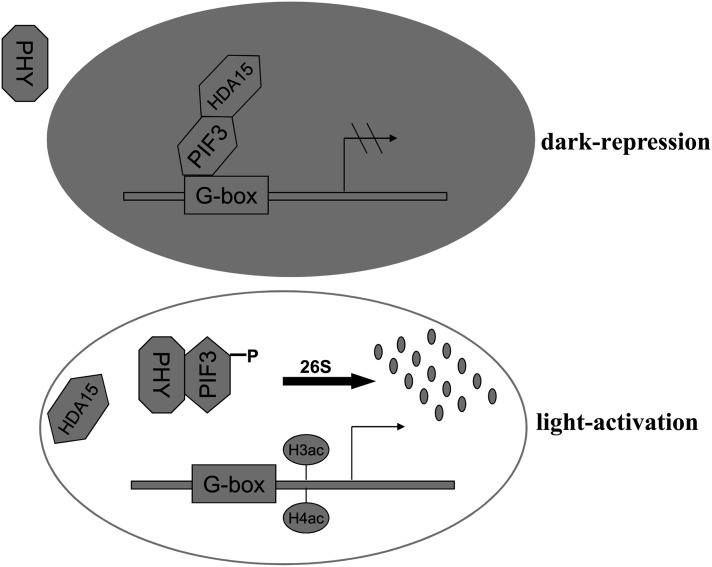
In summary, our results support a model of PIF3 and HDA15 interaction in chlorophyll biosynthesis and photosynthesis pathways (Figure 9). In the dark, PIF3 recruits HDA15 to the promoters of chlorophyll biosynthetic and photosynthetic genes and represses their expression via histone deacetylation. Upon light exposure, the active forms of phytochromes induce rapid degradation of PIF3 and dissociation of HDA15 from their target genes, resulting in derepression of the chlorophyll biosynthetic and photosynthetic genes.
Figure 9.
Proposed Model of HDA15 and PIF3 Function in Modulating Light-Responsive Gene Expression.
In the dark, PIF3 recruits HDA15 to the G-box elements of light-responsive genes to repress their expression. Upon light exposure, the active forms of phytochromes translocate into the nucleus and induce rapid phosphorylation and degradation of PIF3, resulting in the dissociation of HDA15 from the targets and activation of the light-responsive genes.
METHODS
Plant Materials and Growth Conditions
The hda15 T-DNA mutant hda15-1 (SALK_004027), HDA15 RNAi lines (CS30922 and CS30923), hda19-3 (SALK_139443), and pif3-1 (SALK_030753) were obtained from the ABRC (http://www.Arabidopsis.org/). hda15-1, hda19-3, and pif3-1 are in the Col background, whereas HDA15 RNAi lines are in the Wassilewskija background. 35S:PIF3-MYC and PIF3:YFP-PIF3 transgenic lines used in this study were described previously (Al-Sady et al., 2006; Shin et al., 2007). For 35S:HDA15-MYC and 35S:HDA15-GFP transgenic lines, the full-length cDNA of HDA15 was subcloned into pN-TAP (CD3-679) (Rubio et al., 2005) and PK7WGF2 (Karimi et al., 2005) binary vectors separately, and transgenic plants were generated using the floral dip method (Clough and Bent, 1998). Arabidopsis thaliana plants were grown under long-day conditions (16-h-light/8-h-dark cycle) at 22°C. For measurement of morphogenetic phenotypes, seeds were plated on half-strength Murashige-Skoog medium (Sigma-Aldrich) agar plates containing 0.3% Suc and imbibed for 3 d at 4°C in the dark. After germination was induced under white light for 6 h (100 µmol m−2 s−1), the seedlings were grown in various light conditions at 22°C for 4 d.
Production of Anti-HDA15 Antibodies
The full-length coding sequence of HDA15 was cloned into pGEX-4T-3 (GE Healthcare) and transformed into Escherichia coli strain BL21 (DE3). The recombinant proteins were affinity purified using Glutathione Sepharose 4B beads (GE Healthcare). GST fusion proteins were separated by SDS-PAGE. Bands of interest were excised and used as antigens for antibody production. Antibodies were produced in rabbit and mouse by Genomics BioSci and Tech.
RT-PCR and Real-Time RT-PCR Analysis
Total RNA was extracted with Trizol reagent (Invitrogen) according to the manufacturer’s protocol and used to synthesize cDNA. The gene-specific primers used for real-time PCR are listed in Supplemental Table 2 online. Each sample was quantified at least in triplicate and normalized using Ubiquitin10 (UBQ10) as an internal control.
Microarray Experiment
Surface-sterilized seeds of the wild-type Col and hda15-1 mutant were plated on half-strength Murashige and Skoog agar plates containing 0.3% Suc and imbibed for 3 d at 4°C in the dark. After germination was induced under white light for 6 h (100 µmol m−2 s−1), the seedlings were grown in darkness at 22°C for 48 h. Total RNA was extracted as described (Jiang et al., 2010). cDNA and cRNA synthesis and hybridization to 22K Affymetrix Gene Chips (ATH1) were performed according to the manufacturer’s instructions. Microarray assays were performed with triplicate biological samples. Raw data were processed with Affymetrix Microarray Suite 5.0, and the resulting CEL files were further analyzed using Genespring V11.5 (Agilent Technologies). We employed a false discovery rate set at 5% to identify the up- and downregulated genes. Genes with 1.5-fold expression difference between the wild type and hda15-1 mutant with statistically significance (P value < 0.05) were selected. The up- or downregulated genes were classified with Gene Ontology analysis.
Protochlorophyllide Determination and Greening Rate Assays
Protochlorophyllide content was measured as described (Shin et al., 2009). Briefly, 10 frozen 4-d-old etiolated seedlings were ground with a Mixer-mill, and pigments were extracted by gently agitating the seedling powder in 1 mL of cold 80% acetone in opaque tubes for 1 h in the dark at 4°C. After centrifugation at 4°C for 10 min, the supernatant was collected and the relative fluorescence was measured with a fluorescence spectrophotometer. The excitation wavelength was 440 nm, and the fluorescence emission spectra were recorded between 600 and 800 nm a bandwidth of 5 nm. To determine the greening rate, more than 50 seedlings were grown in darkness for the indicated number of days after germination and transferred into continuous white light (100 µmol m−2 s−1) for 2 d. The seedlings can be classified into two types: normal greening (with dark-green cotyledons) and impaired greening (with white, yellow, or light-green cotyledons). The greening rate was calculated by the percentage of normal greening seedlings (Zhong et al., 2009).
ChIP Assays
ChIP assays were performed as described (Gendrel et al., 2005; Yu et al., 2011; Liu et al., 2012). Chromatin was extracted from 2-d-old dark grown seedlings. After fixation with formaldehyde, the chromatin was sheared to an average length of 500 bp by sonication and then immunoprecipitated with specific antibodies including anti-c-myc (A-14) (catalog number sc-789; Santa Cruz), anti-GFP (catalog number 11814460001; Roche), anti-acetyl-histone H3 (catalog number 06-599; Millipore), anti-acetyl-histone H4 (catalog number 06-866; Millipore), anti-RNAPII (catalog number sc-33754; Santa Cruz), and anti-H4 (catalog number 05-858; Millipore). The cross-linking was then reversed, and the amount of each precipitated DNA fragment was determined by real-time PCR using specific primers (see Supplemental Table 2 online).
Yeast Two-Hybrid Assays
Yeast two-hybrid assays were performed according to the instructions for the Matchmaker GAL4-based two-hybrid system 3 (Clontech). Constructs were generated by cloning the full-length or deletion fragments of HDA15 and PIF3 cDNAs into pGADT7 and pGBKT7 vectors. All constructs were transformed into yeast strain AH109 by the lithium acetate method, and yeast cells were grown on minimal medium/-Leu-Trp according to the manufacturer’s instructions. Transformed colonies were plated onto minimal medium/-Leu/Trp-His/x-α-gal containing 3-amino-1,2,4-triazole to test for possible interactions.
GST Pull-Down Assays
The GST pull-down assay was performed as described previously with some modifications (Yang et al., 2008; Yu et al., 2011). GST and GST-HDA15 recombinant proteins were incubated with 30 mL of GST resin in a binding buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.25% Triton X-100, and 35 mM β-mercaptoethanol) for 2 h at 4°C, the binding reaction was washed three times with the binding buffer and then the PIF3-His recombinant protein was added and incubated for an additional 2 h at 4°C. After extensive washing (at least five times), the pulled-down proteins were eluted by boiling, separated by 10% SDS-PAGE, and detected by immunoblotting using an anti-His antibody.
Semi–in Vivo Pull-Down Assays
PIF3-His recombinant protein (0.25 µg) was added to 1 mL of pull-down buffer with 50 μL of Nickel-nitrilotriacetic acid (Ni-NTA) agarose, and the mixtures were preincubated at 4°C for 1 h. One milligram of soluble plant total proteins extracted from Col or 35S:HDA15-GFP seedlings grown in the dark for 2 d was added into the mixtures and incubated for 3 h. After washing three times, the pull down proteins were detected by immunoblotting using an anti-GFP antibody.
BiFC Assays
Full-length cDNA fragments of HDA15 and PIF3 were subcloned into the pCR8/GW/TOPO vectors and then recombined into the YN (pEarleyGate201-YN) and YC (pEarleyGate202-YC) vectors (Lu et al., 2010). The constructed vectors were transformed into Arabidopsis protoplasts by polyethylene glycol for transient expression (Yoo et al., 2007). Transfected cells were imaged using a TCS SP5 confocal spectral microscope imaging system (Leica). To detect the interaction in tobacco, leaves of 2- to 4-week-old tobacco plants (Nicotiana benthamiana) were infiltrated with Agrobacterium tumefaciens strains (GV3101) containing HDA15 and PIF3 BiFC construct pairs. Epidermal cell layers were examined 3 to 4 d after infiltration using the YFP filter.
Coimmunoprecipitation Assays
Total proteins were extracted from 2-d-etiolated Col, hda15, and PIF3-MYC seedlings or seedlings exposed to RL (10 µmol m−2 s−1) for 10 min, 30 min, and 2 h in an extraction buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM MgCl2, 1 mM DTT, 20% glycerol, and 1% CA-630) containing protease inhibitor cocktail (Roche). Protein extracts were incubated with rabbit polyclonal anti-HDA15 antibody overnight at 4°C and then protein G Mag Sepharose beads (GE Healthcare) were added. After incubation for 2 h at 4°C, the beads were centrifuged and washed four times with a wash buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM MgCl2, 1 mM DTT, 10% glycerol, and 1% CA-630). The immunoprecipitated proteins were detected by SDS-PAGE gel with an anti-myc antibody. Endogenous HDA15 protein was detected with a mouse polyclonal anti-HDA15 antibody after immunoprecipitation with anti-HDA15 Rb. Input PIF3-MYC protein was detected with an anti-myc antibody.
HDAC Activity Assays
HDA15-GFP was purified according to the manual for the Magnetic GFP-Trap (ChromoTek). Soluble total proteins extracted from Col, 35S:GFP, or 35S:HDA15-GFP seedlings grown in the dark for 2 d were mixed with 25 μL magnetic particles in immunoprecipitation buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, and 0. 5% Nonidet P-40) and incubated overnight at 4°C. After washing three times with cold buffer [0.2 M (NH4)HCO3], the proteins were harvested for further analysis.
HDAC assays were performed using the HDAC activity colorimetric assay kit (BioVision) following the manufacturer's instructions. Briefly, 4 µg of purified proteins was diluted to 85 μL in each well of the 96-well plate, and 10 μL 10× HDAC assay buffer and 5 μL colorimetric substrate [10 mM Ac-Lys(Ac)-pNA] were added to each well and incubated at 37°C for 1 h. The reaction was stopped by adding 10 μL Lys developer and incubating the plate at 37°C for 30 min. The HDAC activity was then measured by spectrophotometer at 405 nm. HeLa nuclear extracts (4 µg) were used as the positive control. The HDAC inhibitor TSA was also used to demonstrate the specificity of deacetylation activities.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome initiative or GenBank/EMBL databases under the following accession numbers: PIF3 (At1g09530), HDA15 (AT3G18520), PIF4 (At2g43010), PIF5 (At3g59060), HDA19 (At4g38130), ACTIN2 (AT3G18780), UBQ10 (At4g05320), CHLD (AT1G08520), GUN5 (AT5G13630), LHCB2.2 (AT2G05070), LHCB4.2 (AT3G08940), LHCB6 (AT1G15820), PSBQ (AT4G05180), PSAE1 (AT4G28750), PSAK (AT1G30380), and LHB1B1 (AT2G34430).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. HDA15 Specifically Interacts with PIF3 in Yeast.
Supplemental Figure 2. HDA15 Interacts with PIF3 in Tobacco Epidermal Cells in Dark.
Supplemental Figure 3. HDA15 Is Constitutively Localized in the Nucleus under Dark or Light Conditions.
Supplemental Figure 4. Expression Profiles of Genes Involved in the Chlorophyll Biosynthetic Pathway in hda15-1.
Supplemental Figure 5. Microarray Analysis of HDA15-Regulated Genes in 2-d-Old Etiolated Seedlings.
Supplemental Figure 6. Nucleosome Occupancy Analysis of the Chlorophyll Biosynthetic and Photosynthetic Genes in Etiolated Col, hda15, pif3, and hda15 pif3 Seedlings.
Supplemental Figure 7. Identification and Analysis of 35S:HDA15-MYC Transgenic Plants.
Supplemental Figure 8. Detection and Validation of HDA15 Target Genes.
Supplemental Figure 9. Phenotypic Analysis of HDA15 RNAi Seedlings (CS30922 and CS30923) Grown in RL and FR for 4 d.
Supplemental Figure 10. HDA15 and HDA19 Antagonistically Regulate Hypocotyl Elongation.
Supplemental Table 1. List of the Genes That Are Induced in 2-d-Old Etiolated hda15-1 (in This Study) and pifq (Leivar et al., 2009) Seedlings Relative to the Wild Type.
Supplemental Table 2. List of Primers Used in This Study.
Supplemental Data Set 1A. Genes Upregulated in hda15-1 in Darkness.
Supplemental Data Set 1B. Genes Downregulated in hda15-1 in Darkness.
Supplementary Material
Acknowledgments
We thank Giltsu Choi (Korea Advanced Institute of Science and Technology) for providing 35S:PIF3-MYC seeds and Peter Quail (University of California, Berkeley) for PIF3:YFP-PIF3 seeds. We also thank Technology Commons, College of Life Science, National Taiwan University for the convenient use of the Bio-Rad real-time PCR system and the confocal spectral microscope imaging system. This study was funded by grants from the National Science Council of Taiwan (99-2321-B-002-027-MY3, 101-2311-B-002-012-MY3, and 101-2923-B-002-005-MY3), National Taiwan University (101R892005), Academia Sinica (AS-1102-TP-B05), National Basic Research Program of China (973 Program 2012CB910900), and the National Natural Science Foundation of China (31128001).
AUTHOR CONTRIBUTIONS
X.L. and K.W. conceived this project and designed all research. X.L., C.C., K.W, L.Y., M.Z., M.L., R.T., S.Y., and G.T. performed the research. X.L., Y.C., H.H., and K.W. analyzed data. X.L. and K.W. wrote the article.
Glossary
- HAT
histone acetytransferase
- HDAC
histone deacetylase
- GST
glutathione S-transferase
- BiFC
bimolecular fluorescence complementation
- YFP
yellow fluorescent protein
- RL
red light
- GFP
green fluorescent protein
- TSA
trichostatin A
- RNAi
RNA interference
- Col
Columbia
- qRT-PCR
quantitative RT-PCR
- RNAPII
RNA polymerase II
- FR
far-red
References
- Alinsug M.V., Chen F.F., Luo M., Tai R., Jiang L., Wu K. (2012). Subcellular localization of class II HDAs in Arabidopsis thaliana: Nucleocytoplasmic shuttling of HDA15 is driven by light. PLoS ONE 7: e30846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sady B., Ni W., Kircher S., Schäfer E., Quail P.H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23: 439–446 [DOI] [PubMed] [Google Scholar]
- Arguello-Astorga G., Herrera-Estrella L. (1998). Evolution of light-regulated plant promoters. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 525–555 [DOI] [PubMed] [Google Scholar]
- Ballesteros M.L., Bolle C., Lois L.M., Moore J.M., Vielle-Calzada J.P., Grossniklaus U., Chua N.H. (2001). LAF1, a MYB transcription activator for phytochrome A signaling. Genes Dev. 15: 2613–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer D., Viczián A., Kircher S., Nobis T., Nitschke R., Kunkel T., Panigrahi K.C., Adám E., Fejes E., Schäfer E., Nagy F. (2004). Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamed M., Bertrand C., Servet C., Zhou D.X. (2006). Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell 18: 2893–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamed M., et al. (2008). Genome-scale Arabidopsis promoter array identifies targets of the histone acetyltransferase GCN5. Plant J. 56: 493–504 [DOI] [PubMed] [Google Scholar]
- Berger S.L. (2007). The complex language of chromatin regulation during transcription. Nature 447: 407–412 [DOI] [PubMed] [Google Scholar]
- Bertrand C., Benhamed M., Li Y.F., Ayadi M., Lemonnier G., Renou J.P., Delarue M., Zhou D.X. (2005). Arabidopsis HAF2 gene encoding TATA-binding protein (TBP)-associated factor TAF1, is required to integrate light signals to regulate gene expression and growth. J. Biol. Chem. 280: 1465–1473 [DOI] [PubMed] [Google Scholar]
- Charron J.B., He H., Elling A.A., Deng X.W. (2009). Dynamic landscapes of four histone modifications during deetiolation in Arabidopsis. Plant Cell 21: 3732–3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheminant S., Wild M., Bouvier F., Pelletier S., Renou J.P., Erhardt M., Hayes S., Terry M.J., Genschik P., Achard P. (2011). DELLAs regulate chlorophyll and carotenoid biosynthesis to prevent photooxidative damage during seedling deetiolation in Arabidopsis. Plant Cell 23: 1849–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua Y.L., Brown A.P., Gray J.C. (2001). Targeted histone acetylation and altered nuclease accessibility over short regions of the pea plastocyanin gene. Plant Cell 13: 599–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua Y.L., Watson L.A., Gray J.C. (2003). The transcriptional enhancer of the pea plastocyanin gene associates with the nuclear matrix and regulates gene expression through histone acetylation. Plant Cell 15: 1468–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Eberharter A., Becker P.B. (2002). Histone acetylation: A switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 3: 224–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C., Chory J. (1997). Light control of plant development. Annu. Rev. Cell Dev. Biol. 13: 203–229 [DOI] [PubMed] [Google Scholar]
- Gendrel A.V., Lippman Z., Martienssen R., Colot V. (2005). Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2: 213–218 [DOI] [PubMed] [Google Scholar]
- Guo L., Zhou J., Elling A.A., Charron J.B., Deng X.W. (2008). Histone modifications and expression of light-regulated genes in Arabidopsis are cooperatively influenced by changing light conditions. Plant Physiol. 147: 2070–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke C.S., Gohda K., Osterlund M.T., Oyama T., Okada K., Deng X.W. (2000). HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J. 19: 4997–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques R., Jang I.C., Chua N.H. (2009). Regulated proteolysis in light-related signaling pathways. Curr. Opin. Plant Biol. 12: 49–56 [DOI] [PubMed] [Google Scholar]
- Hill K., Wang H., Perry S.E. (2008). A transcriptional repression motif in the MADS factor AGL15 is involved in recruitment of histone deacetylase complex components. Plant J. 53: 172–185 [DOI] [PubMed] [Google Scholar]
- Holm M., Ma L.G., Qu L.J., Deng X.W. (2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 16: 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E., Al-Sady B., Hudson M., Kim C., Apel K., Quail P.H. (2004). Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305: 1937–1941 [DOI] [PubMed] [Google Scholar]
- Huq E., Quail P.H. (2002). PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 21: 2441–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H.W., Liu M.J., Chen I.C., Huang C.H., Chao L.Y., Hsieh H.L. (2010). A glutathione S-transferase regulated by light and hormones participates in the modulation of Arabidopsis seedling development. Plant Physiol. 154: 1646–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Lau O.S., Deng X.W. (2007). Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Jiao Y., Ma L., Strickland E., Deng X.W. (2005). Conservation and divergence of light-regulated genome expression patterns during seedling development in rice and Arabidopsis. Plant Cell 17: 3239–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., De Meyer B., Hilson P. (2005). Modular cloning in plant cells. Trends Plant Sci. 10: 103–105 [DOI] [PubMed] [Google Scholar]
- Khanna R., Shen Y., Marion C.M., Tsuchisaka A., Theologis A., Schäfer E., Quail P.H. (2007). The basic helix-loop-helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell 19: 3915–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Yi H., Choi G., Shin B., Song P.S., Choi G. (2003). Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15: 2399–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.C., Lai Z., Fan B., Chen Z. (2008). Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 20: 2357–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W., Benhamed M., Servet C., Latrasse D., Zhang W., Delarue M., Zhou D.X. (2009). Histone acetyltransferase GCN5 interferes with the miRNA pathway in Arabidopsis. Cell Res. 19: 899–909 [DOI] [PubMed] [Google Scholar]
- Kobayashi, K., Baba, S., Obayashi, T., Sato, M., Toyooka, K., Keranen, M., Aro, E.M., Fukaki, H., Ohta, H., Sugimoto, K., and Masuda, T. (2012). Regulation of root greening by light and auxin/cytokinin signaling in Arabidopsis Plant Cell 24: 1081–1095. [DOI] [PMC free article] [PubMed]
- Laherty C.D., Yang W.M., Sun J.M., Davie J.R., Seto E., Eisenman R.N. (1997). Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell 89: 349–356 [DOI] [PubMed] [Google Scholar]
- Lee J., He K., Stolc V., Lee H., Figueroa P., Gao Y., Tongprasit W., Zhao H., Lee I., Deng X.W. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Tepperman J.M., Monte E., Calderon R.H., Liu T.L., Quail P.H. (2009). Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21: 3535–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Yu C.W., Duan J., Luo M., Wang K., Tian G., Cui Y., Wu K. (2012). HDA6 directly interacts with DNA methyltransferase MET1 and maintains transposable element silencing in Arabidopsis. Plant Physiol. 158: 119–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Tang X., Tian G., Wang F., Liu K., Nguyen V., Kohalmi S.E., Keller W.A., Tsang E.W., Harada J.J., Rothstein S.J., Cui Y. (2010). Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J. 61: 259–270 [DOI] [PubMed] [Google Scholar]
- Luo M., Yu C.W., Chen F.F., Zhao L., Tian G., Liu X., Cui Y., Yang J.Y., Wu K. (2012). Histone deacetylase HDA6 is functionally associated with AS1 in repression of KNOX genes in Arabidopsis. PLoS Genet. 8: e1003114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Li J., Qu L., Hager J., Chen Z., Zhao H., Deng X.W. (2001). Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13: 2589–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein R., Roth S.Y. (2001). Histone acetyltransferases: Function, structure, and catalysis. Curr. Opin. Genet. Dev. 11: 155–161 [DOI] [PubMed] [Google Scholar]
- Martínez-García J.F., Huq E., Quail P.H. (2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288: 859–863 [DOI] [PubMed] [Google Scholar]
- Mochizuki N., Brusslan J.A., Larkin R., Nagatani A., Chory J. (2001). Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl. Acad. Sci. USA 98: 2053–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte E., Tepperman J.M., Al-Sady B., Kaczorowski K.A., Alonso J.M., Ecker J.R., Li X., Zhang Y., Quail P.H. (2004). The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc. Natl. Acad. Sci. USA 101: 16091–16098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M., Tepperman J.M., Quail P.H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667 [DOI] [PubMed] [Google Scholar]
- Oyama T., Shimura Y., Okada K. (1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11: 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R., Müller A., Napoli C.A., Selinger D.A., Pikaard C.S., Richards E.J., Bender J., Mount D.W., Jorgensen R.A. (2002). Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 30: 5036–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E., Kim J., Lee Y., Shin J., Oh E., Chung W.I., Liu J.R., Choi G. (2004). Degradation of phytochrome interacting factor 3 in phytochrome-mediated light signaling. Plant Cell Physiol. 45: 968–975 [DOI] [PubMed] [Google Scholar]
- Reyes J.C., Hennig L., Gruissem W. (2002). Chromatin-remodeling and memory factors. New regulators of plant development. Plant Physiol. 130: 1090–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V., Shen Y., Saijo Y., Liu Y., Gusmaroli G., Dinesh-Kumar S.P., Deng X.W. (2005). An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J. 41: 767–778 [DOI] [PubMed] [Google Scholar]
- Schaffer R., Ramsay N., Samach A., Corden S., Putterill J., Carré I.A., Coupland G. (1998). The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219–1229 [DOI] [PubMed] [Google Scholar]
- Sentandreu M., Martín G., González-Schain N., Leivar P., Soy J., Tepperman J.M., Quail P.H., Monte E. (2011). Functional profiling identifies genes involved in organ-specific branches of the PIF3 regulatory network in Arabidopsis. Plant Cell 23: 3974–3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Moon J., Huq E. (2005). PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J. 44: 1023–1035 [DOI] [PubMed] [Google Scholar]
- Shin J., Kim K., Kang H., Zulfugarov I.S., Bae G., Lee C.H., Lee D., Choi G. (2009). Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 106: 7660–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Park E., Choi G. (2007). PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J. 49: 981–994 [DOI] [PubMed] [Google Scholar]
- Song C.P., Agarwal M., Ohta M., Guo Y., Halfter U., Wang P., Zhu J.K. (2005). Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17: 2384–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soy J., Leivar P., González-Schain N., Sentandreu M., Prat S., Quail P.H., Monte E. (2012). Phytochrome-imposed oscillations in PIF3 protein abundance regulate hypocotyl growth under diurnal light/dark conditions in Arabidopsis. Plant J. 71: 390–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson P.G., Fankhauser C., Terry M.J. (2009). PIF3 is a repressor of chloroplast development. Proc. Natl. Acad. Sci. USA 106: 7654–7659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R., Tanaka A. (2007). Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 58: 321–346 [DOI] [PubMed] [Google Scholar]
- Tepperman J.M., Zhu T., Chang H.S., Wang X., Quail P.H. (2001). Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc. Natl. Acad. Sci. USA 98: 9437–9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessadori F., et al. (2009). Phytochrome B and histone deacetylase 6 control light-induced chromatin compaction in Arabidopsis thaliana. PLoS Genet. 5: e1000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Rodríguez-Concepción M. (2010). Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 107: 11626–11631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Y., Tobin E.M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217 [DOI] [PubMed] [Google Scholar]
- Wu K., Zhang L., Zhou C., Yu C.W., Chaikam V. (2008). HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J. Exp. Bot. 59: 225–234 [DOI] [PubMed] [Google Scholar]
- Yang J.Y., Iwasaki M., Machida C., Machida Y., Zhou X., Chua N.H. (2008). betaC1, the pathogenicity factor of TYLCCNV, interacts with AS1 to alter leaf development and suppress selective jasmonic acid responses. Genes Dev. 22: 2564–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.J., Grégoire S. (2005). Class II histone deacetylases: From sequence to function, regulation, and clinical implication. Mol. Cell. Biol. 25: 2873–2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yu C.W., Liu X., Luo M., Chen C., Lin X., Tian G., Lu Q., Cui Y., Wu K. (2011). HISTONE DEACETYLASE6 interacts with FLOWERING LOCUS D and regulates flowering in Arabidopsis. Plant Physiol. 156: 173–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Zhao M., Shi T., Shi H., An F., Zhao Q., Guo H. (2009). EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc. Natl. Acad. Sci. USA 106: 21431–21436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., et al. (2013). HISTONE DEACETYLASE19 interacts with HSL1 and participates in the repression of seed maturation genes in Arabidopsis seedlings. Plant Cell 25: 134–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.