Here, we show that the MDO1 gene, which leads to stem cell defects, encodes TEN1, a core component of the CST telomere binding complex. We show that TEN1 is required for telomere integrity, but it also negatively regulates the telomerase enzyme. These findings add an unanticipated layer of complexity to CST function and new insights into how CST promotes telomere stability and stem cell viability.
Abstract
Telomeres protect chromosome ends from being recognized as DNA damage, and they facilitate the complete replication of linear chromosomes. CST [for CTC1(Cdc13)/STN1/TEN1] is a trimeric chromosome end binding complex implicated in both aspects of telomere function. Here, we characterize TEN1 in the flowering plant Arabidopsis thaliana. We report that TEN1 (for telomeric pathways in association with Stn1, which stands for suppressor of cdc thirteen) is encoded by a previously characterized gene, MERISTEM DISORGANIZATION1 (MDO1). A point mutation in MDO1, mdo1-1/ten1-3 (G77E), triggers stem cell differentiation and death as well as a constitutive DNA damage response. We provide biochemical and genetic evidence that ten1-3 is likely to be a null mutation. As with ctc1 and stn1 null mutants, telomere tracts in ten1-3 are shorter and more heterogeneous than the wild type. Mutants also exhibit frequent telomere fusions, increased single-strand telomeric DNA, and telomeric circles. However, unlike stn1 or ctc1 mutants, telomerase enzyme activity is elevated in ten1-3 mutants due to an increase in repeat addition processivity. In addition, TEN1 is detected at a significantly smaller fraction of telomeres than CTC1. These data indicate that TEN1 is critical for telomere stability and also plays an unexpected role in modulating telomerase enzyme activity.
INTRODUCTION
Telomeres are essential for chromosome integrity and consist of tandem arrays of simple G-rich repeats terminating in a single-strand 3′ extension termed the G-overhang. The telomere tract is bound by proteins that protect the terminus from nucleolytic degradation, inappropriate recombination, and activation of a DNA damage response. Telomeres also promote replication of the chromosome terminus through recruitment of telomerase and lagging strand replication machinery. Mammalian telomeres are capped by a six-member protein complex called shelterin (de Lange, 2005), while the telomere ends in budding yeast are bound by the trimeric replication protein A (RPA)-like CST (for Cdc13/Stn1/Ten1) complex (Giraud-Panis et al., 2010).
All three of the yeast CST genes are essential (Garvik et al., 1995; Nugent et al., 1996; Grandin et al., 1997, 2001). CST binds the G-overhang primarily through interactions with Cdc13 (Nugent et al., 1996). Cdc13 plays a central role in coordinating telomeric DNA replication by promoting G-strand synthesis via an interaction with the Est1 component of the telomerase ribonucleoprotein (RNP) and C-strand synthesis through its association with DNA polymerase α/primase (Evans and Lundblad, 1999; Qi and Zakian, 2000; Chandra et al., 2001; Wu and Zakian, 2011). Mutation of any of the CST components triggers nucleolytic degradation of the telomeric C-strand, leading to gross extension of the G-overhang (Garvik et al., 1995; Grandin et al., 1997, 2001).
In budding yeast, Ten1 (for telomeric pathways in association with Stn1) and Stn1 (for suppressor of cdc thirteen) are proposed to regulate telomerase. Telomeres in temperature-sensitive ten1 and stn1 mutants elongate in a telomerase-dependent manner (Grandin et al., 1997, 2001). Furthermore, Stn1 appears to block the binding of Est1 to Cdc13, preventing telomerase action on the telomere (Chandra et al., 2001). Ten1 may act in concert with Stn1 since a Stn1-Ten1 fusion protein rescues the telomere lengthening phenotype of stn1 (Grandin et al., 2001). Ten1 could also regulate telomerase via a separate mechanism as overexpression of TEN1 partially rescues telomere defects in stn1 mutants, hence, its discovery as a partial suppressor of stn1 (Grandin et al., 2001).
The consequences of CST depletion are much less severe in Candida albicans. Null mutations in STN1 or TEN1 are not lethal, and cells do not accumulate single-stranded G-rich telomere DNA (Sun et al., 2009), implying that the essential contributions of CST components are not universally conserved. CST orthologs (CTC1/STN1/TEN1) have been reported in vertebrates and plants (Song et al., 2008; Miyake et al., 2009; Surovtseva et al., 2009; Nakaoka et al., 2012). Although CONSERVED TELOMERE MAINTENANCE COMPONENT1 (CTC1) shares no sequence similarity with Cdc13 from budding yeast, CST complexes appear to function in similar capacities across eukaryotes.
As in yeast, vertebrate and plant CST interact with polymerase (pol) α (Casteel et al., 2009; Price et al., 2010; Huang et al., 2012; Nakaoka et al., 2012). Indeed, mammalian CTC1 and STN1 were first identified as pol α accessory factors (Casteel et al., 2009). Biochemical studies reveal that vertebrate CST stimulates pol α/primase activity on telomeric substrates (Dai et al., 2010; Huang et al., 2012; Nakaoka et al., 2012) and promotes new origin firing at nontelomeric sites (Stewart et al., 2012). CST also interacts with the shelterin components TPP1 in human cells (Wan et al., 2009) and POT1b in mice (Wu et al., 2012). Moreover, recent data suggest that human CST modulates telomerase enzyme activity through primer sequestration (Chen et al., 2012). These findings argue that CST coordinates replication of telomeric C- and G-strands via dynamic interactions with shelterin, pol α, and telomerase.
Despite these biochemical findings, the in vivo function of vertebrate CST remains poorly understood. A conditional CTC1 knockout in mice triggers telomere loss, increased G-overhangs, and ultimately activation of an ATR (Ataxia telangiectasia and Rad3 related)-dependent DNA damage response, primarily in highly proliferating tissues (Gu et al., 2012). Knockdown of CTC1 in HeLa and MCF7 human cell lines leads to similar phenotypes (Surovtseva et al., 2009). More recent studies of CTC1 knockdown in other human cancer lines revealed telomere elongation in one case (Chen et al., 2012) and no significant change in telomere length in another (Wu et al., 2012).
The first reports of CTC1 and STN1 in multicellular eukaryotes came from studies in Arabidopsis thaliana. A null mutation in either CTC1 or STN1 profoundly affects telomere integrity and stem cell proliferation. Although mutant plants are viable, they exhibit dramatic morphological phenotypes, including abnormally small leaves, irregular phyllotaxy, fasciated stems, and reduced fertility (Song et al., 2008; Surovtseva et al., 2009). In addition, telomere tracts are drastically shorter and plants display abundant end-to-end chromosome fusions, enhanced G-overhang signals, and telomeric circles (Song et al., 2008; Surovtseva et al., 2009).
Recently, CST was shown to work in concert with the Ku70/80 heterodimer to promote telomere integrity in Arabidopsis (Kazda et al., 2012). Unlike other model organisms, half of the chromosome ends in Arabidopsis, presumably those replicated by the leading strand machinery, are blunt-ended and protected by Ku. The remaining telomeres, replicated by the lagging strand mechanism, possess a canonical G-overhang bound by CST (Kazda et al., 2012). Plants encode only a subset of the vertebrate shelterin components. This observation coupled with the unusual architecture of plant telomeres suggests that CST evolved a more pivotal role than its vertebrate counterparts in protecting chromosome ends throughout the cell cycle (Nelson and Shippen, 2012).
In this study, we examine the contribution of TEN1 in Arabidopsis. We show that TEN1 is encoded by MERISTEM DISORGANIZATION1 (MDO1), a gene recently discovered by Hashimura and Ueguchi (2011) that is crucial for stem cell viability. A point mutation in MDO1 (mdo1-1/ten1-3) causes severe shoot apical meristem aberrations, including stem cell death or differentiation, developmental defects, and a constitutive DNA damage response. Here, we demonstrate that the defects associated with mdo1-1/ten1-3 result from severe telomere dysfunction. Although most of the mutant phenotypes closely parallel those in plants lacking CTC1 or STN1, an unexpected role for TEN1 in the negative regulation of telomerase repeat addition processivity (RAP) was uncovered. Thus, in conjunction with its essential function in telomere protection/replication as a component of CST, TEN1 plays an additional unanticipated role in modulating telomerase activity.
RESULTS
Identification of Arabidopsis TEN1
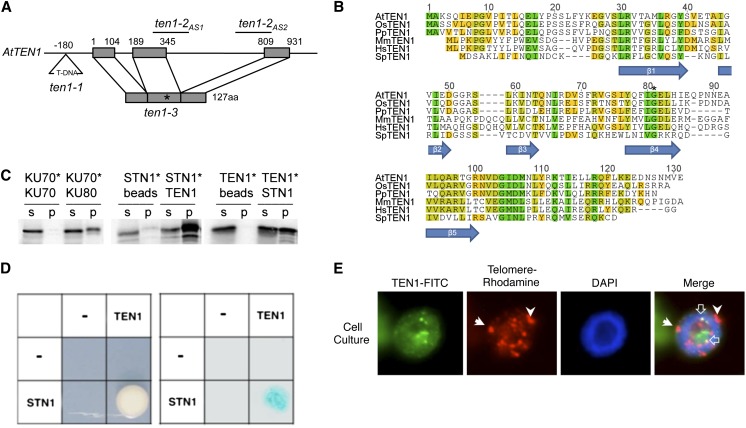
In the second iteration of PSI-BLAST using human TEN1 (Miyake et al., 2009) as a query, we retrieved a single hit: NP_176022.2 (E-value = 2e-07). This gene, At1g56260, corresponds to MDO1 (Hashimura and Ueguchi, 2011) and hereafter is termed TEN1. TEN1 is a single-copy gene with one open reading frame interrupted by two introns (Figure 1A). The open reading frame encodes a 127–amino acid protein with 23% identity/48% similarity to human TEN1. Secondary structure prediction by PSIPRED (McGuffin et al., 2000) revealed a single OB-fold that shares significant similarity TEN1 in other plants as well as in Schizosaccharomyces pombe and humans (Figure 1B). RT-PCR amplified a single TEN1 mRNA species that was expressed widely in Arabidopsis tissues (see Supplemental Figure 1A online).
Figure 1.
Arabidopsis TEN1 Is a Member of CST Complex.
(A) Schematic of TEN1 gene structure. The T-DNA insertion in ten1-1 is illustrated, along with the position of two antisense constructs and the point mutation responsible for the G77E mutation in ten1-3. aa, amino acids.
(B) Alignment of TEN1 proteins from different eukaryotes. At, Arabidopsis thaliana; Os, Oryza sativa (rice); Pt, Populus trichocarpa (poplar); Mm, Mus musculus; Hs, Homo sapiens; Sp, Schizosaccharomyces pombe. The positions of β-strands of the OB-fold are indicated below the alignment. Green, 100% similarity; chartreuse, 80 to 99% similarity; yellow, 60 to 79% similarity; and gray, below 60% similarity.
(C) TEN1 interacts with STN1 in vitro. Results of coimmunoprecipitation performed with recombinant proteins. One protein is [35S]Met labeled (asterisk), and the other is T7 tagged and unlabeled. s, supernatant; p, pellet. Results for the positive (KU70/KU80) and negative (KU70/KU70) controls are shown.
(D) Yeast two-hybrid assay results for STN1 and TEN1. The two proteins fused to GAL4-AD and GAL4-BD were coexpressed and grown on selection plates for His auxotrophy (left) or assayed to detect β-galactosidase activity of positive transformants (right). “−” Indicates empty vector.
(E) Nuclear localization of TEN1 in purified nuclei. TEN1 was detected by anti-TEN1 antibody in hexaploid Arabidopsis suspension cell culture. Telomeres were labeled by FISH using a rhodamine-labeled telomere probe. 4′,6-Diamidino-2-phenylindole (DAPI)–stained nuclei are shown. In the merge, closed white arrows denote subcentromeric stretches of telomeric DNA on chromosome 1. TEN1 colocalization with telomeres is indicated by the open white arrow.
A hallmark of TEN1 is its ability to interact with STN1 (Grandin et al., 2001; Martín et al., 2007; Miyake et al., 2009; Nakaoka et al., 2012). To assay for TEN1–STN1 interaction, recombinant proteins were expressed in rabbit reticulocyte lysate and labeled with [35S]Met. One of the proteins was expressed as T7-tagged fusion. Reciprocal coimmunoprecipitation experiments with T7 antibody showed a direct interaction between STN1 and TEN1 (Figure 1C). Yeast two-hybrid analysis confirmed this association (Figure 1D).
We next asked whether TEN1 colocalizes with telomeres using a polyclonal antibody raised specifically against recombinant TEN1 (see Methods). Immunolocalization was performed with the TEN1 antibody on the nuclei of asynchronously dividing Arabidopsis suspension cell culture. Fluorescence in situ hybridization (FISH) using a rhodamine-labeled telomere probe was used to identify telomeres. TEN1 appeared as punctate spots in the nucleus (Figure 1E). A merged image of revealed that TEN1 colocalized with 13% (15/114) of the telomeres examined (Figure 1E, top). A similar value, 11.1% (34/306), was obtained with Arabidopsis seedlings. These observations were unexpected, as previous experiments with transgenic plants expressing a tagged version of CTC1 found that it associated with ∼50% of the chromosome ends in cycling cells and cells arrested in G1 (Surovtseva et al., 2009). Thus, our data suggest that the association of TEN1 with telomeres may be more transient than that of STN1 or CTC1. Moreover, because some of the punctate spots recognized by the TEN1 antibody do not colocalize with telomeres, TEN1 may have extrachromosomal functions.
To test whether increased TEN1 expression would drive telomere localization, immunolocalization was conducted on seedling nuclei from a genetic complementation line in which TEN1 was expressed from its native promoter in a ten1-3 background (Hashimura and Ueguchi, 2011). In this line, the level of TEN1 mRNA was three to 10-fold higher than in wild-type plants (see Supplemental Figure 1B online). The number of telomeres bound by TEN1 was increased to 35/171 (20.5%) (P value = 0.036), implying that the low frequency of TEN1-telomere association is not due to coincident overlap or inaccessibility of telomere-bound TEN1 to antibody. Instead, the data indicate that TEN1 is associated with a substantially smaller fraction of telomeres than CTC1.
The ten1-3 Mutation Causes Profound Defects in Plant Development and Fertility
To examine the function of TEN1 in vivo, we initially characterized a T-DNA insertion line (ten1-1) that contains a disruption in the 5′ untranslated region of TEN1, 180 bp upstream of the start codon (Figure 1A). Quantitative RT-PCR showed ∼50% reduction in TEN1 mRNA in homozygous ten1-1 mutants (see Supplemental Figure 1C online). In an attempt to achieve a greater TEN1 knockdown, we targeted two regions of TEN1 with antisense RNA (TEN1-2AS1 and TEN1-2AS2) (Figure 1A). Quantitative RT-PCR revealed a wide range of TEN1 mRNA depletion. For example, ten1-2AS1-15 showed an 82% reduction in TEN1 mRNA, while TEN1 mRNA levels were reduced by only 10% in ten1-2AS1-8 (see Supplemental Figure 1C online). Similar results were obtained for ten1-2AS2 knockdown lines. Strikingly, despite the significant reduction of TEN1 mRNA in ten1-2AS1-15, mutant plants were morphologically wild type, indicating that only a small amount of TEN1 is required for wild-type function.
In marked contrast with ten1-1 and ten1-2AS lines, plants harboring the mdo1-1 mutation exhibited severe growth and developmental defects (Hashimura and Ueguchi, 2011) (Figure 2), much like those described for stn1 and ctc1 null mutants (Song et al., 2008; Surovtseva et al., 2009). Hereafter, we term the mdo1-1 mutation ten1-3. Homozygous ten1-3 mutants showed a lack of apical dominance. Many plants had fused stems, smaller leaves, and irregular siliques (Figure 2A, middle). Seeds collected from first generation (G1) ten1-3 mutants displayed remarkably variable germination rates spanning 30 to 90%. Compared with their parents, second-generation (G2) ten1-3 mutants developed even more slowly and the majority had more severe morphological defects, including the complete absence of apical meristems and exceptionally short roots (Figure 2C). As reported earlier (Hashimura and Ueguchi, 2011), overexpression of wild-type TEN1 rescued these morphological phenotypes (Figure 2A, right), confirming that the ten1-3 mutation is responsible for the developmental abnormalities.
Figure 2.
The ten1-3 Mutation Causes Severe Morphological Defects.
(A) First generation (G1) ten1-3 mutants are smaller in stature than the wild type (WT) and harbor smaller leaves, fused stems, and irregular phyllotaxy (middle panel). These phenotypes are rescued by expression of a wild-type copy of TEN1 (right panel).
(B) Second generation (G2) ten1-3 mutants display more severe growth phenotype than G1 mutants and are infertile. Arrowhead denotes aborted siliques.
(C) Two-week-old seedlings of the genotypes indicated were grown on Murashige and Skoog medium without selection. G2 ten1-3 mutants exhibit shoot apical meristem abnormalities and fail to produce true leaves. G2 mutants are shown in a 2× zoom to show abnormal apical meristem.
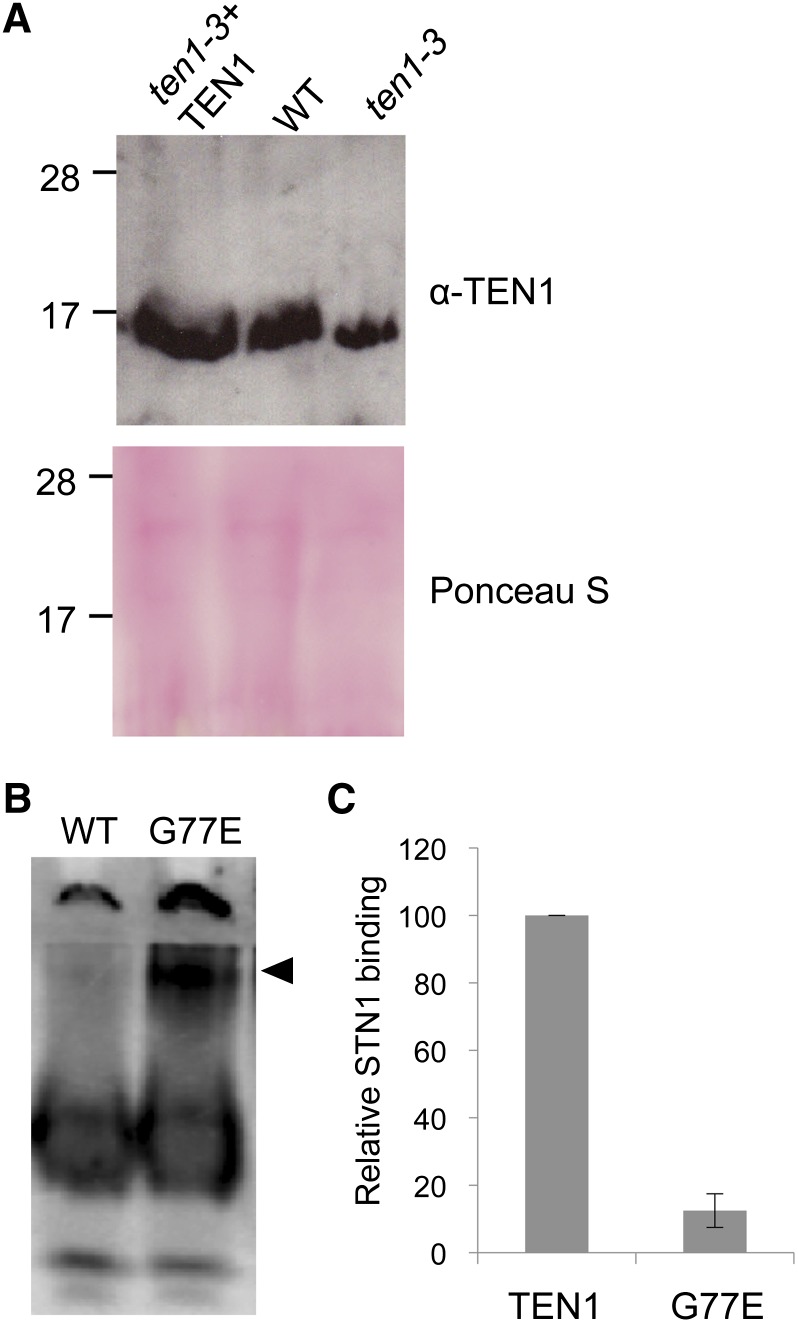
The ten1-3 mutation is caused by a Gly-to-Glu amino acid substitution at position 77 (Figure 1A) (Hashimura and Ueguchi, 2011). As expected, the level of TEN1 mRNA was essentially wild type in ten1-3 plants (see Supplemental Figure 1B online). Protein gel blotting was performed to assess TEN1 protein expression. To confirm the specificity of the TEN1 antibody, FLAG-HA–tagged TEN1 was transiently expressed in tobacco (Nicotiana tabacum) leaves, and the extracted proteins were used for protein gel blot analysis with anti-HA and anti-TEN1 antibodies. A single band corresponding the fusion protein was detected with anti-HA antibody (see Supplemental Figure 2A online). The same size product was detected with the anti-TEN1 antibody, along with another band of 17 kD that may represent partial loss of the tags (see Supplemental Figure 2A online). As expected, a single band corresponding to predicted size of TEN1 (16 kD) was observed in wild-type Arabidopsis (see Supplemental Figure 2A online). We visually see a slight increase in TEN1 abundance in the TEN1 complementation line and decreased TEN1 in ten1-3 plants (Figure 3A). Notably, attempts to express recombinant TEN1G77E in Escherichia coli resulted in significantly lower protein yields than wild-type TEN1, arguing that the mutation reduced TEN1 stability.
Figure 3.
The TEN1G77E Mutant Protein Is Unstable and Does Not Interact with STN1 in Vitro.
(A) Protein gel blot results for the wild type (WT), ten1-3, and the TEN1 complementation line (TEN1-3 + TEN1) are shown. Ponceau S stain loading controls included. Molecular mass size markers in kilodaltons are on the left. The blot was probed with a polyclonal antibody raised against Arabidopsis TEN1.
(B) Native PAGE results for recombinant wild-type TEN1 or TEN1G77E protein expressed in rabbit reticulocyte lysate. Arrow indicates a higher molecular mass polypeptide in the TEN1G77E protein sample.
(C) Quantification of recombinant TEN1 protein binding to STN1. Shown are results of coimmunoprecipitation experiments with recombinant wild-type TEN1 and TEN1G77E. The interaction for wild-type TEN1-STN1 was set to 100%. An example of raw data is shown in Supplemental Figure 2B online.
[See online article for color version of this figure.]
Although recombinant TEN1G77E expressed in rabbit reticulocyte lysate was soluble, a large fraction of the protein migrated much more slowly than wild-type TEN1 on a nondenaturing gel (Figure 3B), consistent with significant structural perturbation. In addition, coimmunoprecipitation experiments showed that the G77E mutation abolished the interaction of TEN1 with STN1 in vitro (Figure 3C; see Supplemental Figure 2B online). Since the Gly-77 residue on TEN1 is not predicted to lie within the STN1 binding interface based on the Stn1-Ten1 crystal structure from S. pombe (Sun et al., 2009), we conclude that the ten1-3 mutation profoundly disrupts TEN1 structure and stability. Thus, ten1-3 mutants can be classified as null or nearly null for TEN1.
TEN1 Is Required for Telomere Length Maintenance
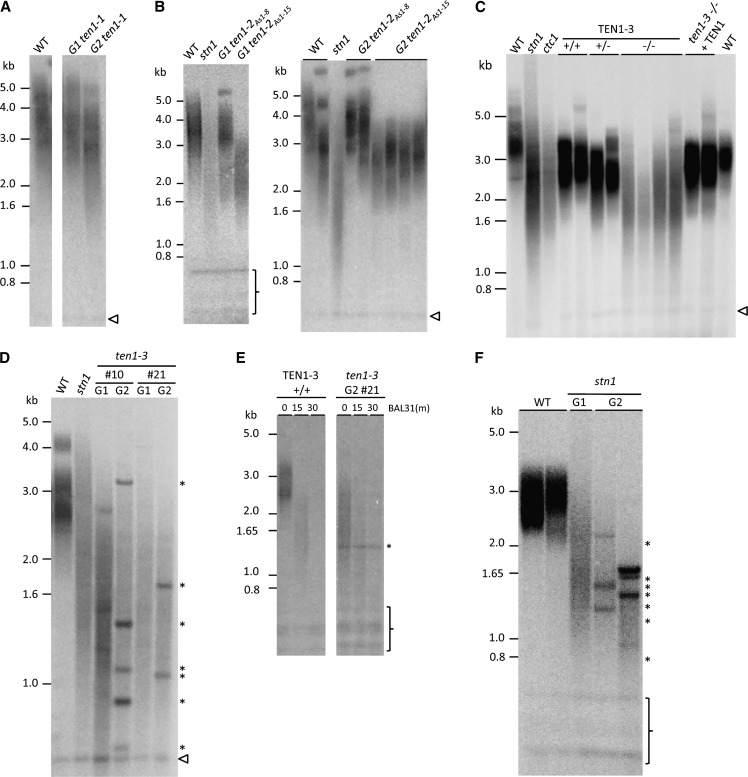
Terminal restriction fragment (TRF) analysis was performed to analyze bulk telomere length in ten1 mutants. Although there was no obvious change in telomere length in ten1-1 or in the ten1-2AS1-8 antisense line relative to the wild type (Figures 4A and 4B), the telomere profile was altered in ten1-2AS1-15 mutants (Figure 4B), where TEN1 mRNA is decreased by 82%. The size range of telomeres contracted and longer telomeres were absent. Stronger evidence that TEN1 is important for telomere length maintenance came from analysis of ten1-3 mutants. Telomere tracts were more heterogeneous, significantly shorter overall than the wild type or any of the ten1-2AS lines, and closely resembled ctc1 and stn1 mutants (Figure 4C). Primer extension telomere repeat assay (PETRA), which measures telomere length on individual chromosome arms, confirmed telomere shortening on all arms tested (see Supplemental Figure 3A online). As expected, telomeres in the TEN1 complementation line were wild type (Figure 4C), verifying that the ten1-3 mutation is responsible for the defects in telomere length maintenance.
Figure 4.
TEN1 Is Important for Telomere Length Regulation and Genome Maintenance.
TRF analysis of ten1 mutants. Blots were hybridized with a radiolabeled G-rich telomeric probe.
(A) Results for first (G1) and second (G2) generation ten1-1 are shown relative to the wild type (WT).
(B) Telomere length in first (left) and second (right) generations of two antisense knockdown lines of TEN1. For comparison, results are shown with first generation stn1-1 mutants.
(C) TRF analysis of ten1-3 mutants. Results for offspring of ten1-3 heterozygous plants are analyzed.
(D) Parent-progeny analysis for two different ten1-3 mutants.
(E) BAL31 time course of DNA with the wild type and a G2 ten1-3 mutant.
(F) Telomere profile of G1 and G2 stn1-1 mutants. Asterisks indicated abnormally sharp TRF bands. Interstitial telomeric DNA repeats are denoted by the bracket or an arrowhead.
Unlike tert mutants, which suffer progressive telomere shortening in subsequent plant generations (Riha et al., 2001), bulk telomeres did not shorten further in second generation (G2) ten1-3 mutants (Figure 4D). However, TRF analysis revealed a new profile of products, consisting of heterogeneous telomere tracts punctuated by multiple discrete bands (Figure 4D). G2 ten1-3 DNA was digested with BAL31 exonuclease prior to TRF analysis to determine whether the bands correspond to terminal DNA sequences (Figure 4E; see Supplemental Figure 3B online). Although bulk telomeric DNA was completely degraded within 30 min, the sharp bands were insensitive to exonuclease treatment. Because this banding profile was not observed in G1 ctc1 or G1 stn1 mutants (Song et al., 2008; Surovtseva et al., 2009), we asked whether it represented a general response to the prolonged absence of CST. TRF analysis performed with DNA from G2 stn1 mutants also revealed sharp bands (Figure 4F). We hypothesize that these products reflect gross rearrangements of telomeric DNA, resulting from chromosome end deprotection and multiple rounds of the breakage-fusion-bridge cycle (see below).
TEN1 Promotes Telomere Integrity
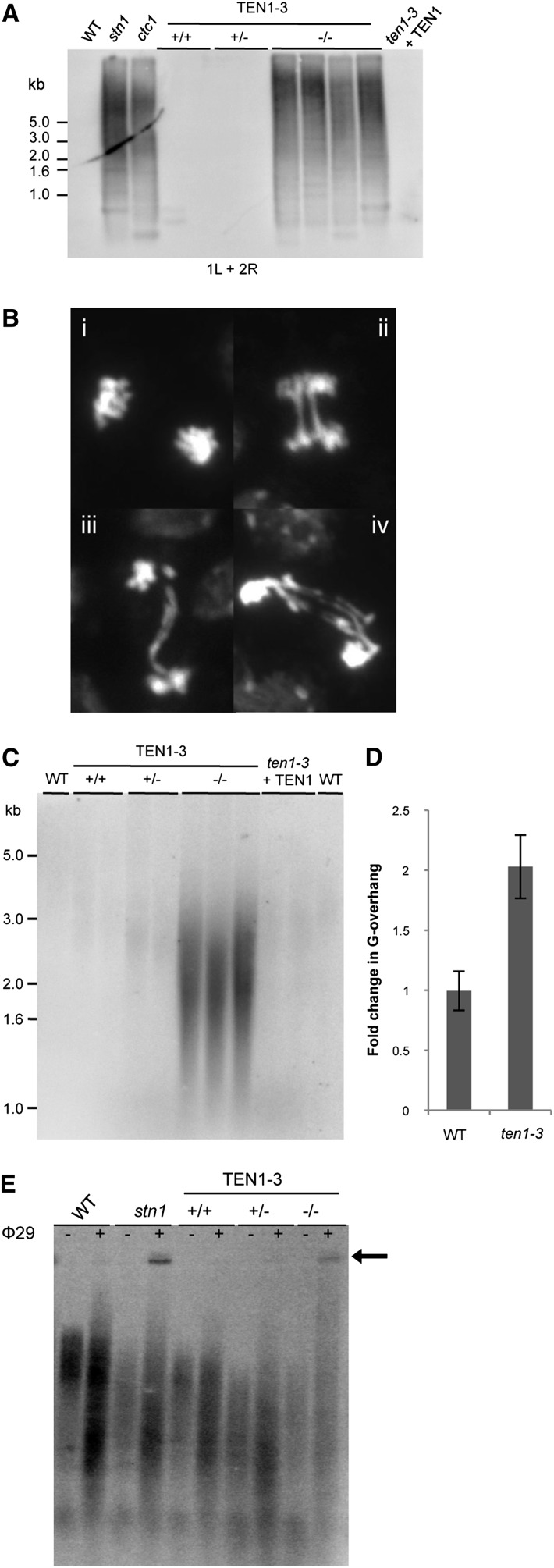
Telomere fusion PCR (TF-PCR) was employed to ask if TEN1 is needed to prevent end-to-end chromosome fusions. As for ctc1 and stn1 mutants, abundant TF-PCR products were generated in reactions with ten1-3, but not wild-type DNA samples (Figure 5A; see Supplemental Figure 4 online). Cloning and sequence analysis showed evidence of extensive nucleolytic resection prior to chromosome end-joining (see Supplemental Table 1 online). To obtain a quantitative estimate of telomere fusion events, mitotic chromosomes were examined for evidence of anaphase bridges (Figure 5B). Thirty to fifty percent of anaphases examined in ten1-3 mutants contained bridged chromosomes, with several involving multiple chromosomes. Notably, this value is significantly higher than number of bridges found in ctc1 or stn1 mutants (18 to 30%).
Figure 5.
TEN1 Prevents End-to-End Chromosome Fusions and Promotes Proper Telomere Architecture.
(A) Telomere fusion PCR products obtained from the wild type (WT) and ten1-3 mutants are shown. Primer pair used to amplify specific subtelomeric regions are indicated.
(B) Cytology of mitotic chromosomes in the wild type (i) and ten1-3 mutants (ii to iv) are shown. DAPI–stained chromosome spreads were prepared from pistils.
(C) In-gel hybridization analysis of DNA isolated from the wild type and ten1-3 mutants using a C-strand telomeric probe under native conditions.
(D) Quantification of the G-overhang signal for ten1-3 mutants. A DNA gel blot of interstitial telomere DNA or ethidium bromide staining of DNA was used as a DNA loading control for quantification of G-overhang signal. Data represent seven individual biological replicates of ten1-3. Error bars indicate sd for biological replicates.
(E) TCA was performed with the wild type, ten1-3 +/− offspring, and stn1-1 DNA in the presence (+) or absence (−) of phi (ϕ) 29 polymerase. Arrow indicates extrachromosomal telomere circles.
In-gel hybridization was performed to determine if TEN1 modulates G-overhang architecture. Compared with wild-type siblings, ten1-3 mutants displayed a twofold increase in G-overhang signal, similar to stn1 and ctc1 mutants (Figures 5C and 5D) (Song et al., 2008; Surovtseva et al., 2009). Exonuclease controls confirmed that the hybridization signal was derived from terminal single-stranded telomeric DNA (see Supplemental Figure 5A online). As expected, the enhanced G-overhang signal was absent in the TEN1 complementation line (Figure 5C).
One other hallmark of telomere instability is an increase in the frequency of telomere recombination. The telomere circle assay (TCA) (Zellinger et al., 2007) was used to monitor telomere recombination events. As in stn1 and ctc1 mutants, ten1-3 plants displayed increased production of extrachromosomal telomere circles (Figure 5E). We conclude that TEN1, like the other components of CST, promotes telomere integrity by maintaining telomere length and proper architecture of the chromosome terminus.
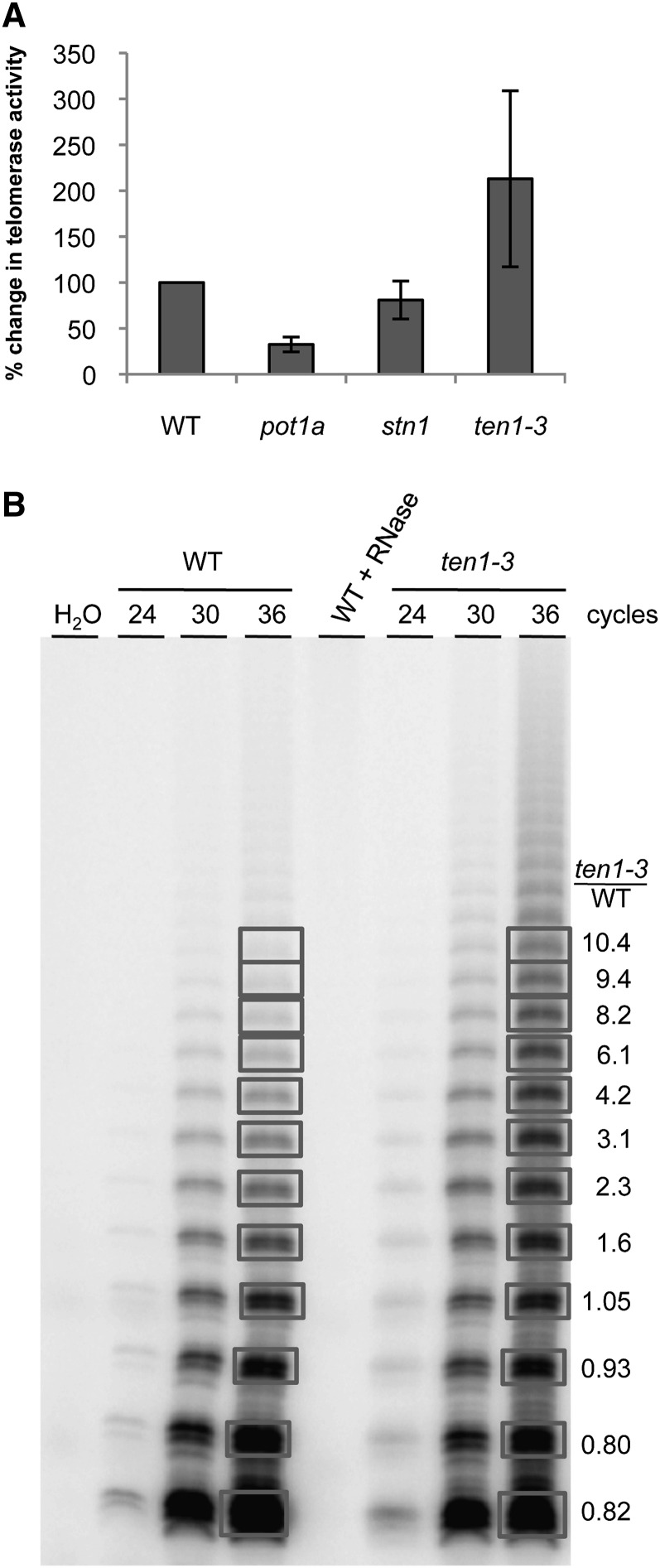
TEN1 Negatively Regulates Telomerase RAP
Plants lacking STN1 or CTC1 exhibit no change in telomerase activity levels (Song et al., 2008; Surovtseva et al., 2009). Therefore, we were surprised to find that telomerase activity was elevated in ten1-3 mutants (Figure 6A). Quantitative telomere repeat amplification protocol (Q-TRAP) revealed, on average, a twofold increase in enzyme activity in ten1-3 mutants compared with wild-type siblings (P value = 0.0013). The alteration in enzyme activity was somewhat variable with some plants showing only slightly increased activity, while others showed a threefold increase (see Supplemental Figure 6A online). When products of the TRAP reaction were resolved by denaturing PAGE, it was evident that ten1-3 mutants generated substantially more of the longer telomere repeat arrays than the wild type (Figure 6B). Importantly, the ratio of shorter products generated with ten1-3 versus the wild type was slightly less than 1:1 but increased to more than 10:1 for longer products. This skewed ratio indicates that the ten1-3 extracts exhibit qualitatively different telomerase activity. Specifically, the findings indicate that telomere RAP was increased in the absence of TEN1.
Figure 6.
TEN1 Is a Negative Regulator of Telomerase Activity.
(A) Telomerase activity in flowers measured by Q-TRAP. Data are normalized to the wild type (WT); each data point represents two or three biological replicates, with three technical replicates. Error bars represent the sd between biological replicates.
(B) TRAP products from the wild type and ten1-3 mutants at 24, 30, and 36 cycles of PCR resolved by PAGE. Quantification (right) represents the signal for the corresponding bands of ten1-3 divided by the wild type for the 36-cycle PCR reaction.
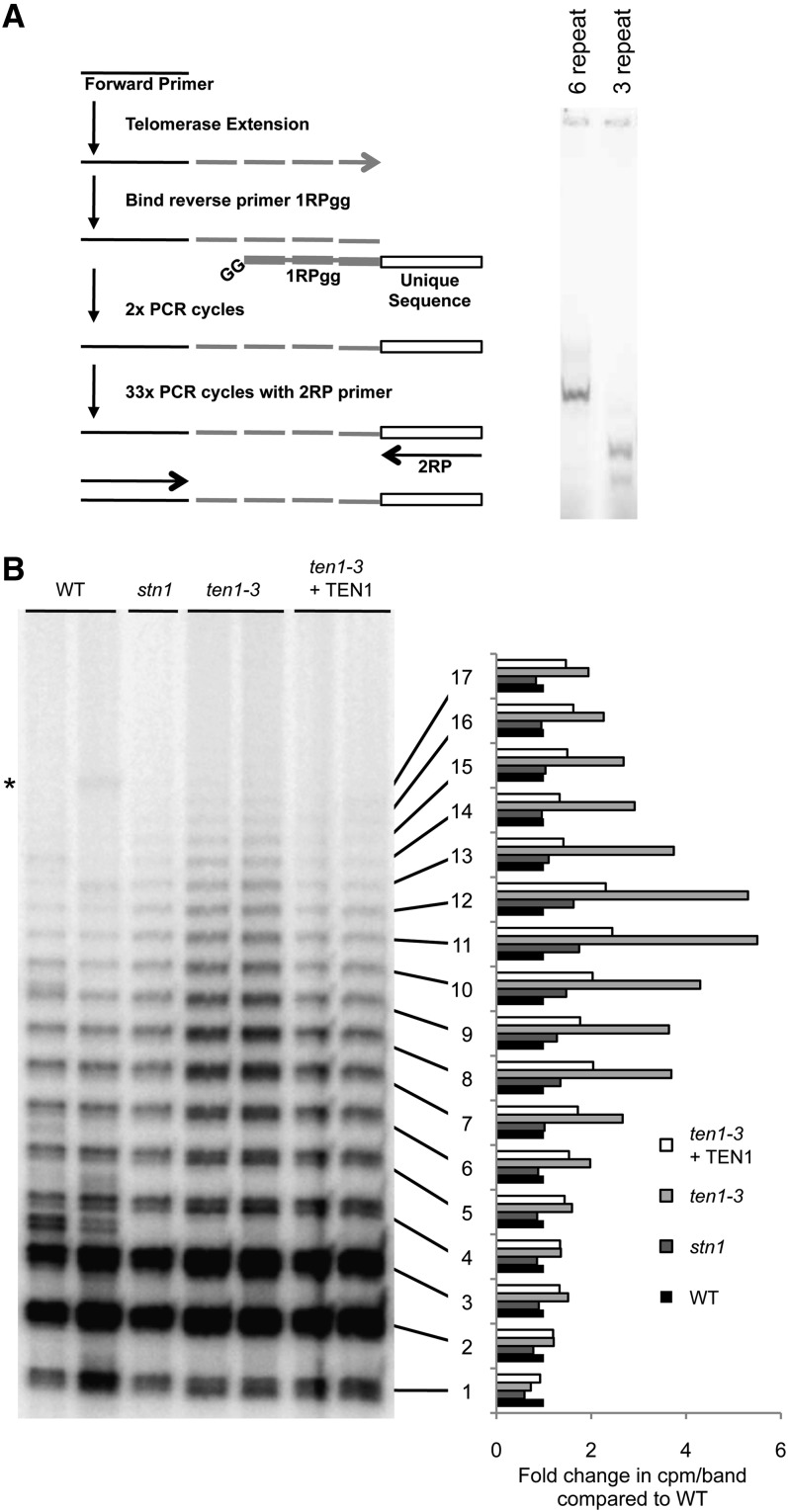
A direct, non-PCR-based telomerase activity assay is not available for Arabidopsis. Therefore, we modified the processivity TRAP (pTRAP) assay devised for human telomerase (Szatmari and Aradi, 2001) to assess RAP in extracts from ten1-3 mutants. In this assay, telomerase extension products are tagged with a unique sequence that is used for amplification in conventional PCR (Figure 7A). Control reactions with synthetic oligonucleotide substrates bearing either three or six telomere repeats yielded the expected products (Figure 7A). Examination of pTRAP products showed that the abundance of short products was essentially the same for wild-type, ten1-3, and stn1-1 mutants. However, the longer products were strongly overrepresented in reactions with ten1-3 (Figure 7B). Although the TEN1 complementation line had a slightly elevated processivity, the increase was not statistically significant and the product profile more closely resembled that of the wild type than the mutant (Figure 7B). We conclude that TEN1 negatively regulates telomerase enzyme activity by decreasing RAP.
Figure 7.
TEN1 Decreases Telomerase Repeat Addition Processivity.
(A) Left: Schematic of processivity TRAP (Szatmari and Aradi, 2001). (1) Telomerase extends a forward primer substrate. (2) and (3) Binding of reverse primer 1RPgg, which contains a unique sequence tag on the 5′ end and terminates in two 3′ noncomplementary G nucleotides that precisely position the primer at the terminus of the extension product. Two cycles of PCR are used to tag the telomerase product with the unique sequence tag. (4) Thirty-three cycles of PCR using the forward primer and 2RP, a reverse primer complementary to the unique sequence tag. Right: Results for control reactions with oligonucleotides containing three (PT3) or six (PT6) telomere repeats (see Supplemental Table 2 online) subjected to steps 2 to 4 of processivity TRAP.
(B) Results of processivity TRAP for floral extracts from wild-type (WT), stn1, and ten1-3 mutants. Left: Telomerase extension products displayed by PAGE. Asterisk denotes nonspecific PCR amplification products. Right: Quantification of processivity TRAP. Signal was quantified for the individual bands indicated. The average signal was calculated for each genotype (except stn1) and that average was compared with the average for the wild type.
DISCUSSION
Telomere integrity is critical for genome stability and the long-term proliferative capacity of stem cell pools. The MDO1/TEN1 gene was originally identified in a forward genetic screen for defects in meristem maintenance (Hashimura and Ueguchi, 2011). Here, we demonstrate that the molecular basis for stem cell failure is telomere dysfunction. Plants lacking TEN1 harbor short, highly heterogeneous telomere tracts with aberrant G-overhangs that are subjected to inappropriate recombination including massive end-to-end chromosome fusions. These phenotypes are strikingly similar to those of ctc1 and stn1 null mutants (Song et al., 2008; Surovtseva et al., 2009), and together with biochemical data showing that TEN1 physically interacts with STN1, our findings argue that TEN1 is a key component of the Arabidopsis CST complex required for genome integrity.
A critical role for CST in cell proliferation in humans is highlighted by a spate of genetic studies demonstrating that compound heterozygous mutations in CTC1 underlie the stem cell disorders Coats plus and Dyskeratosis congenita (Anderson et al., 2012; Keller et al., 2012; Mangino et al., 2012; Polvi et al., 2012). Interestingly, only a subset of patients with CTC1 mutations exhibit telomere shortening (Walne et al., 2013), consistent with the prevailing model that mammalian CTC1 is a multifunctional protein that contributes to different facets of DNA metabolism. We found that only a small amount of TEN1 is sufficient in Arabidopsis, suggesting that conflicting reports pertaining to CST deficiency in mammalian cell lines (Surovtseva et al., 2009; Chen et al., 2012; Gu et al., 2012; Wu et al., 2012) could reflect different levels of depletion. Studies to elucidate how CST contributes to human disease are in their infancy, and thus far no mutations in TEN1 or STN1 have been reported. Relative to CTC1, these genes are much smaller targets for mutation, and given the essential role of STN1 and TEN1 in plants (Song et al., 2008; Hashimura and Ueguchi, 2011; this study), disease-related mutations in their human counterparts may ultimately be recovered.
Several lines of evidence indicate that TEN1 does not always function in concert with CTC1 and STN1. Purification of pol α accessory factor from human cells revealed the presence of CTC1 and STN1, but not TEN1 (Casteel et al., 2009). In addition, we found that TEN1 associates with a smaller fraction of Arabidopsis telomeres than CTC1. The current model for telomere protection in Arabidopsis proposes that CST is bound to half of the chromosome ends (Kazda et al., 2012). Consistent with this model, CTC1 colocalizes with ∼50% of the telomeres (Surovtseva et al., 2009). By contrast, we found only 13 to 20% of telomeres were bound by TEN1. These results are particularly striking, given that the same relatively low fraction of mammalian telomeres are bound by CST (13 to 20%) (Miyake et al., 2009). Unlike Arabidopsis CST, mammalian CST plays no significant role in chromosome end protection. Therefore, an intriguing possibility is that Arabidopsis TEN1, unlike CTC1 and perhaps STN1, promotes telomere integrity through transient interactions with the chromosome terminus.
The most compelling argument that TEN1 makes an unanticipated contribution outside the context of the trimeric CST complex comes from the unexpected observation that telomerase enzyme activity is elevated in plants lacking TEN1, but not CTC1 or STN1 (Song et al., 2008; Surovtseva et al., 2009; this study). In both conventional TRAP and pTRAP assays, significantly longer telomere repeat arrays were generated in ten1-3 reactions than with stn1 or wild-type extracts, indicating that TEN1 negatively regulates telomerase activity by modulating RAP. Studies in other model systems have uncovered telomerase-associated OB-fold proteins that stimulate RAP. These include p82 (Teb1), a stable component of the Tetrahymena thermophila telomerase RNP (Min and Collins, 2009), and the mammalian shelterin components TPP1/POT1 (de Lange, 2009). TPP1 stimulates the DNA binding ability of POT1 and the heterodimer then provides a bridge linking telomerase to the telomere (Wang et al., 2007; Xin et al., 2007). We were unable to detect telomerase activity in TEN1 immunoprecipitates. This result may reflect the transient nature of the TEN1–telomerase interaction, or the association of TEN1 with telomerase may result in nonprocessive elongation (e.g., the addition of less than one full telomere repeat). Such products would not be readily detected by TRAP.
It is noteworthy that studies in budding yeast (Grandin et al., 2001) and more recently in human cancer cells (Chen et al., 2012) suggest that CST contributes to the negative regulation of telomerase at chromosome ends. Although we did not observe telomere elongation in ten1-3 mutants, enhanced telomerase action at chromosome ends would be masked by profound telomere deprotection in this background. Our data are thus consistent with yeast and mammalian CST studies and go a step further by directly implicating TEN1 in telomerase regulation.
METHODS
Plant Materials and Plasmids
The ten1-1 mutant of Arabidopsis thaliana was obtained from the ABRC. Plants were genotyped using TEN1-1 F and TEN1-1 R (see Supplemental Table 2 online). The ten1-2AS lines were created using the Gateway vector pB7WG2 with a 35S promoter (Karimi et al., 2002); two separate constructs were created, targeting two separate regions of TEN1 (Figure 1A; see Supplemental Table 2 online). Antisense constructs were introduced using Agrobacterium tumefaciens–mediated transformation (Zhang et al., 2006). Transformed plants were selected on Murashige and Skoog + Basta plates. The ten1-3 mutant and the complementation line were obtained from the Ueguchi lab at the Bioscience and Biotechnology Center (Nagoya University, Nagoya, Japan). Plants were genotyped as previously described (Hashimura and Ueguchi, 2011). Plants were grown in the conditions described (Surovtseva et al., 2007).
Cytology, Immunofluorescence, and FISH
To examine anaphase bridge formations, flower pistils were prepared and analyzed as described (Riha et al., 2001; Song et al., 2008). Immunolocalization and FISH were performed on Arabidopsis suspension cells, MM2d (Menges and Murray, 2002), that were grown under continuous darkness at 130 rpm and a temperature of 25°C. Nuclei were extracted from 1-week-old cells (Song et al., 2008). TEN1 was detected with rabbit anti-TEN1 (1:200) antibody, and the signal was amplified using fluorescein isothiocyanate donkey anti-rabbit antibody (1:200; Jackson ImmunoResearch). FISH was performed as described (Armstrong et al., 2009).
Antibody Preparation, Protein Extraction, and Protein Gel Blot Analysis
Escherichia coli (200 μg) expressed TEN1 and adjuvant were mixed and injected into rabbits. Blood was collected from the central ear artery, and clotted blood was clarified by centrifugation at 2500 rpm for 20 min. Protein A purification was performed to purify anti-TEN1 antibody. To determine a suitable dilution of the antibody, immunoblotting was conducted with serial dilutions of antigen and stored at −20°C.
To analyze the expression of TEN1, protein was extracted from wild-type and mutant seedlings using CelLytic P protein extraction buffer (Sigma-Aldrich). Forty-five micrograms of each protein was used for SDS-PAGE, followed by protein gel blotting. The polyvinylidene difluoride (PVDF) membrane was blocked in 6% (w/v) nonfat dry milk in 1× TBST (50mM Tris, 150mM NaCl, 0.1% Tween-20) buffer for 2 h at room temperature. The membrane was incubated with anti-TEN1 antibody (1:7500) in 6% (w/v) nonfat dry milk in 1× TBST buffer for 2 h at room temperature. Anti-rabbit-horseradish peroxidase secondary antibody (1:6667 of 0.4 mg/mL in 50% glycerol; Jackson ImmunoResearch) was incubated in the same conditions using an ECL prime protein gel blotting detection kit (GE Healthcare).
Transient expression of Flag-HA-TEN1 was performed as described (Zhu et al., 2011) with the following modifications. Leaves were collected 20 h after agroinfiltration, ground in liquid nitrogen, and resuspended in 1× SDS loading dye (1 mL/g of tissue). Twenty-five microliters of Nicotiana benthamiana and 45 μg of Arabidopsis total protein were run on an SDS-PAGE gel. Protein gel blotting with anti-HA was performed with anti-HA antibody (1:3000; Sigma-Aldrich) and anti-mouse-horseradish peroxidase secondary antibody (1:3000; GE Healthcare). The membrane was stripped and probed with anti-TEN1 antibody.
Protein Interaction Assays
STN1, TEN1, Ku70, and Ku80 cDNA were cloned into pET28a (T7-tag fusion) and pCITE4a vectors (Novagen). Proteins were expressed in rabbit reticulocyte lysate (Promega) according to the manufacturer’s instructions with [35S]Met (Perkin-Elmer) to label the protein expressed from pCITE4a, and in some cases pET28a. Coimmunoprecipitation was performed as described (Karamysheva et al., 2004). Quantification was performed by calculating the ratio of TEN1:STN1 signal and comparing with the wild-type interaction set to 100%. For yeast two-hybrid assays, GAL4-AD or GAL4-BD constructs of TEN1 and STN1 cDNA were transformed and expressed in yeast strain PJ69-4A. To eliminate false positives, the yeast two-hybrid assay was conducted under stringent media conditions consisting of synthetic dropout/-Leu/-Trp/-His/-Ade selection medium with 50 mM 3-aminotriazole. To confirm positive interactions, we switched inserts from the GAL4-AD vector to the GAL4-BD vector and repeated the assay.
RNA Analysis
Total RNA was extracted from plant tissue using a plant RNA extraction kit (Omega). Reverse transcription was performed with cDNA Supermix (Quanta) per the manufacturer’s instructions. TEN1 mRNA levels were measured by quantitative RT-PCR with primers TEN1 Q RT-PCR 1F and TEN1 Q RT-PCR 1R (see Supplemental Table 2 online) using Sso Fast Eva Green Supermix (Bio-Rad) in accordance with manufacturer’s specifications. mRNA levels were normalized to GAPDH and TIP41L mRNA levels in corresponding samples.
TRF, PETRA, and TF-PCR
DNA from individual plants was extracted as described (Cocciolone and Cone, 1993). TRF analysis was performed with 50 μg of DNA as previously described (Fitzgerald et al., 1999). For Bal31 assay, 200 μg of DNA was incubated with 65 units of Bal31 (New England Biolabs) in 1× Bal31 reaction buffer. Equal amounts of sample were taken out at 15- or 30-min intervals for 60 or 90 min. Phenol:choloroform extraction was performed followed by isopropanol precipitation. Resuspended DNA was digested with Tru1I following the TRF protocol. PETRA and TF-PCR were performed as described (Heacock et al., 2004).
In-Gel Hybridization and Telomere Circle Amplification
In-gel hybridization was performed as described with the following modifications (Heacock et al., 2007). Exonuclease treatment was performed with T4 DNA polymerase (New England Biolabs) at 37°C for 10 min. Prior to drying the gel, the lower portion containing the interstitial telomere repeats was removed and a DNA gel blot was performed using a [32P] 5′ end labeled (T3AG3)4 oligonucleotide probe. The relative amount of single-strand G-overhang was calculated by quantifying the hybridization signal obtained from the native gel and then normalizing this value with the loading control of either interstitial telomere signal from the DNA gel blot or ethidium bromide staining. The value for the wild type was set to one. TCA was performed as previously described (Zellinger et al., 2007).
Telomerase Activity Assays
Protein for TRAP assays was extracted from flowers and reactions were conducted as described (Fitzgerald et al., 1996). Q-TRAP was performed as discussed (Kannan et al., 2008). pTRAP protocol was adapted from Szatmari and Aradi (2001). Telomerase extension was performed with reaction mix containing 1× Go Taq MasterMix, colorless (Promega), 50 ng protein extract, 0.1 μM TRAP forward (see Supplemental Table 2 online), and ∼5 μCi [α-32P]dGTP (Perkin-Elmer). After telomerase extension, 0.04 μM 1RPgg (see Supplemental Table 2 online) was added to the reaction followed by two PCR cycles (95°C for 30 s, 60°C for 1 min, and 72°C for 45 s). After addition of 0.1 μM 2RP (see Supplemental Table 2 online), 33 PCR cycles were conducted (95°C for 30 s, 64°C for 30 s, and 72°C for 1 min). Products were ethanol precipitated and resolved on 6% denaturing PAGE, followed by autoradiography.
Accession Numbers
Sequence data from this article can be found in The Arabidopsis Information Resource data library under the following accession numbers: MDO1/TEN1 (AT1G56260) and STN1 (AT1G07130).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. TEN1 mRNA Expression Levels Monitored by Quantitative Real-Time PCR.
Supplemental Figure 2. G77E Mutation Alters TEN1 Expression in Vivo and Ability to Bind STN1 in Vitro.
Supplemental Figure 3. Telomere Shortening and Length Deregulation in ten1-3 Mutants.
Supplemental Figure 4. Telomere Fusion PCR ten1 Mutants.
Supplemental Figure 5. Increased G-Overhang Signals in ten1-3 Mutants Are Exonuclease Sensitive and Represent Terminal Single-Strand DNA.
Supplemental Figure 6. Analysis of Telomerase Enzyme Activity in ten1-3 Mutants.
Supplemental Table 1. Sequence Analysis of ten1 Mutant Telomere Fusion PCR Products.
Supplemental Table 2. Primers.
Supplementary Material
Acknowledgments
We thank Chiharu Ueguchi for providing mdo1 seeds. We also thank Kara Boltz for insightful comments on the article and Andrew Nelson for assistance with microscopy and data analysis. This work was supported by a grant from the National Institutes of Health (R01-GM065383) to D.E.S.
AUTHOR CONTRIBUTIONS
K.A.L. and D.E.S. wrote the article. K.A.L., X.S., J.R.L., and D.E.S designed the research. K.A.L, J.R.L., X.S., and K.B.R. performed research and analyzed data.
Glossary
- FISH
fluorescence in situ hybridization
- TRF
terminal restriction fragment
- PETRA
primer extension telomere repeat assay
- TF-PCR
telomere fusion PCR
- TCA
telomere circle assay
- Q-TRAP
quantitative telomere repeat amplification protocol
- RAP
repeat addition processivity
- pTRAP
processivity telomere repeat amplification protocol
References
- Anderson B.H., et al. (2012). Mutations in CTC1, encoding conserved telomere maintenance component 1, cause Coats plus. Nat. Genet. 44: 338–342 [DOI] [PubMed] [Google Scholar]
- Armstrong S.J., Sanchez-Moran E., Franklin F.C. (2009). Cytological analysis of Arabidopsis thaliana meiotic chromosomes. Methods Mol. Biol. 558: 131–145 [DOI] [PubMed] [Google Scholar]
- Casteel D.E., Zhuang S., Zeng Y., Perrino F.W., Boss G.R., Goulian M., Pilz R.B. (2009). A DNA polymerase-alphamiddle dotprimase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J. Biol. Chem. 284: 5807–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra A., Hughes T.R., Nugent C.I., Lundblad V. (2001). Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 15: 404–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.Y., Redon S., Lingner J. (2012). The human CST complex is a terminator of telomerase activity. Nature 488: 540–544 [DOI] [PubMed] [Google Scholar]
- Cocciolone S.M., Cone K.C. (1993). Pl-Bh, an anthocyanin regulatory gene of maize that leads to variegated pigmentation. Genetics 135: 575–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X., Huang C., Bhusari A., Sampathi S., Schubert K., Chai W. (2010). Molecular steps of G-overhang generation at human telomeres and its function in chromosome end protection. EMBO J. 29: 2788–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. (2005). Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev. 19: 2100–2110 [DOI] [PubMed] [Google Scholar]
- de Lange T. (2009). How telomeres solve the end-protection problem. Science 326: 948–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans S.K., Lundblad V. (1999). Est1 and Cdc13 as comediators of telomerase access. Science 286: 117–120 [DOI] [PubMed] [Google Scholar]
- Fitzgerald M.S., McKnight T.D., Shippen D.E. (1996). Characterization and developmental patterns of telomerase expression in plants. Proc. Natl. Acad. Sci. USA 93: 14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald M.S., Riha K., Gao F., Ren S., McKnight T.D., Shippen D.E. (1999). Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc. Natl. Acad. Sci. USA 96: 14813–14818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvik B., Carson M., Hartwell L. (1995). Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 15: 6128–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud-Panis M.J., Teixeira M.T., Géli V., Gilson E. (2010). CST meets shelterin to keep telomeres in check. Mol. Cell 39: 665–676 [DOI] [PubMed] [Google Scholar]
- Grandin N., Damon C., Charbonneau M. (2001). Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 20: 1173–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandin N., Reed S.I., Charbonneau M. (1997). Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 11: 512–527 [DOI] [PubMed] [Google Scholar]
- Gu P., Min J.N., Wang Y., Huang C., Peng T., Chai W., Chang S. (2012). CTC1 deletion results in defective telomere replication, leading to catastrophic telomere loss and stem cell exhaustion. EMBO J. 31: 2309–2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimura Y., Ueguchi C. (2011). The Arabidopsis MERISTEM DISORGANIZATION 1 gene is required for the maintenance of stem cells through the reduction of DNA damage. Plant J. 68: 657–669 [DOI] [PubMed] [Google Scholar]
- Heacock M., Spangler E., Riha K., Puizina J., Shippen D.E. (2004). Molecular analysis of telomere fusions in Arabidopsis: Multiple pathways for chromosome end-joining. EMBO J. 23: 2304–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heacock M.L., Idol R.A., Friesner J.D., Britt A.B., Shippen D.E. (2007). Telomere dynamics and fusion of critically shortened telomeres in plants lacking DNA ligase IV. Nucleic Acids Res. 35: 6490–6500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Dai X., Chai W. (2012). Human Stn1 protects telomere integrity by promoting efficient lagging-strand synthesis at telomeres and mediating C-strand fill-in. Cell Res. 22: 1681–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan K., Nelson A.D., Shippen D.E. (2008). Dyskerin is a component of the Arabidopsis telomerase RNP required for telomere maintenance. Mol. Cell. Biol. 28: 2332–2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamysheva Z.N., Surovtseva Y.V., Vespa L., Shakirov E.V., Shippen D.E. (2004). A C-terminal Myb extension domain defines a novel family of double-strand telomeric DNA-binding proteins in Arabidopsis. J. Biol. Chem. 279: 47799–47807 [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kazda A., Zellinger B., Rössler M., Derboven E., Kusenda B., Riha K. (2012). Chromosome end protection by blunt-ended telomeres. Genes Dev. 26: 1703–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R.B., Gagne K.E., Usmani G.N., Asdourian G.K., Williams D.A., Hofmann I., Agarwal S. (2012). CTC1 Mutations in a patient with dyskeratosis congenita. Pediatr. Blood Cancer 59: 311–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangino M., et al. (2012). Genome-wide meta-analysis points to CTC1 and ZNF676 as genes regulating telomere homeostasis in humans. Hum. Mol. Genet. 21: 5385–5394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín V., Du L.L., Rozenzhak S., Russell P. (2007). Protection of telomeres by a conserved Stn1-Ten1 complex. Proc. Natl. Acad. Sci. USA 104: 14038–14043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin L.J., Bryson K., Jones D.T. (2000). The PSIPRED protein structure prediction server. Bioinformatics 16: 404–405 [DOI] [PubMed] [Google Scholar]
- Menges M., Murray J.A. (2002). Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J. 30: 203–212 [DOI] [PubMed] [Google Scholar]
- Min B., Collins K. (2009). An RPA-related sequence-specific DNA-binding subunit of telomerase holoenzyme is required for elongation processivity and telomere maintenance. Mol. Cell 36: 609–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake Y., Nakamura M., Nabetani A., Shimamura S., Tamura M., Yonehara S., Saito M., Ishikawa F. (2009). RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol. Cell 36: 193–206 [DOI] [PubMed] [Google Scholar]
- Nakaoka H., Nishiyama A., Saito M., Ishikawa F. (2012). Xenopus laevis Ctc1-Stn1-Ten1 (xCST) protein complex is involved in priming DNA synthesis on single-stranded DNA template in Xenopus egg extract. J. Biol. Chem. 287: 619–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson A.D., Shippen D.E. (2012). Blunt-ended telomeres: An alternative ending to the replication and end protection stories. Genes Dev. 26: 1648–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent C.I., Hughes T.R., Lue N.F., Lundblad V. (1996). Cdc13p: A single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274: 249–252 [DOI] [PubMed] [Google Scholar]
- Polvi A., Linnankivi T., Kivelä T., Herva R., Keating J.P., Mäkitie O., Pareyson D., Vainionpää L., Lahtinen J., Hovatta I., Pihko H., Lehesjoki A.E. (2012). Mutations in CTC1, encoding the CTS telomere maintenance complex component 1, cause cerebroretinal microangiopathy with calcifications and cysts. Am. J. Hum. Genet. 90: 540–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price C.M., Boltz K.A., Chaiken M.F., Stewart J.A., Beilstein M.A., Shippen D.E. (2010). Evolution of CST function in telomere maintenance. Cell Cycle 9: 3157–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H., Zakian V.A. (2000). The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev. 14: 1777–1788 [PMC free article] [PubMed] [Google Scholar]
- Riha K., McKnight T.D., Griffing L.R., Shippen D.E. (2001). Living with genome instability: Plant responses to telomere dysfunction. Science 291: 1797–1800 [DOI] [PubMed] [Google Scholar]
- Song X., Leehy K., Warrington R.T., Lamb J.C., Surovtseva Y.V., Shippen D.E. (2008). STN1 protects chromosome ends in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 105: 19815–19820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart J.A., Wang F., Chaiken M.F., Kasbek C., Chastain P.D., II, Wright W.E., Price C.M. (2012). Human CST promotes telomere duplex replication and general replication restart after fork stalling. EMBO J. 31: 3537–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Yu E.Y., Yang Y., Confer L.A., Sun S.H., Wan K., Lue N.F., Lei M. (2009). Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres. Genes Dev. 23: 2900–2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtseva Y.V., Churikov D., Boltz K.A., Song X., Lamb J.C., Warrington R., Leehy K., Heacock M., Price C.M., Shippen D.E. (2009). Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol. Cell 36: 207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtseva Y.V., Shakirov E.V., Vespa L., Osbun N., Song X., Shippen D.E. (2007). Arabidopsis POT1 associates with the telomerase RNP and is required for telomere maintenance. EMBO J. 26: 3653–3661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatmari I., Aradi J. (2001). Telomeric repeat amplification, without shortening or lengthening of the telomerase products: A method to analyze the processivity of telomerase enzyme. Nucleic Acids Res. 29: E3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walne A., Bhagat T., Kirwan M., Gitaux C., Desguerre I., Leonard N., Nogales E., Vulliamy T., Dokal I. (2013). Mutations in the telomere capping complex in bone marrow failure and related syndromes. Haematologica 98: 334–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan M., Qin J., Songyang Z., Liu D. (2009). OB fold-containing protein 1 (OBFC1), a human homolog of yeast Stn1, associates with TPP1 and is implicated in telomere length regulation. J. Biol. Chem. 284: 26725–26731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Podell E.R., Zaug A.J., Yang Y., Baciu P., Cech T.R., Lei M. (2007). The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature 445: 506–510 [DOI] [PubMed] [Google Scholar]
- Wu P., Takai H., de Lange T. (2012). Telomeric 3′ overhangs derive from resection by Exo1 and Apollo and fill-in by POT1b-associated CST. Cell 150: 39–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Zakian V.A. (2011). The telomeric Cdc13 protein interacts directly with the telomerase subunit Est1 to bring it to telomeric DNA ends in vitro. Proc. Natl. Acad. Sci. USA 108: 20362–20369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin H., Liu D., Wan M., Safari A., Kim H., Sun W., O’Connor M.S., Songyang Z. (2007). TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature 445: 559–562 [DOI] [PubMed] [Google Scholar]
- Zellinger B., Akimcheva S., Puizina J., Schirato M., Riha K. (2007). Ku suppresses formation of telomeric circles and alternative telomere lengthening in Arabidopsis. Mol. Cell 27: 163–169 [DOI] [PubMed] [Google Scholar]
- Zhang X., Henriques R., Lin S.S., Niu Q.W., Chua N.H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1: 641–646 [DOI] [PubMed] [Google Scholar]
- Zhu H., Hu F., Wang R., Zhou X., Sze S.H., Liou L.W., Barefoot A., Dickman M., Zhang X. (2011). Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell 145: 242–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.