Low temperature is one of the major environmental factors restricting plant geographic distribution, growing season, and productivity. Plants combat low temperature by increasing membrane fluidity through the enhancement of membrane lipid unsaturation. This study demonstrates that ADS2 is essential for regulating membrane lipid desaturation in response to low-temperature exposure.
Abstract
Fatty acid desaturation of membrane lipids is a strategy for plants to survive chilling or freezing temperature. To further characterize enzymes involved in this stress response pathway, ACYL-LIPID DESATURASE2 (ADS2; Enzyme Commission 1.14.99) was studied using genetic, cell, and biochemical approaches. ads2 mutant plants appear similar to the wild type under standard growth conditions but display a dwarf and sterile phenotype when grown at 6°C and also show increased sensitivity to freezing temperature. Fatty acid composition analysis demonstrated that ads2 mutant plants at 6°C have reduced levels of 16:1, 16:2, 16:3, and 18:3 and higher levels of 16:0 and 18:0 fatty acids compared with the wild type. Lipid profiling revealed that 34C species of phosphatidylglycerol (PG) and monogalactosyl diacylglycerol (MGDG) content in ads2 mutants were lower and phosphatidic acid, phosphatidylinositol, phosphatidylethanolamine, phosphatidylcholine, lyso-phosphatidylcholine, and phosphatidylserine were higher than the wild type. Subcellular localization of C- and N-terminal enhanced fluorescence fusion proteins indicated that ADS2 localized primarily to the endoplasmic reticulum, although signal was also confirmed in Golgi and plastids. A double mutation with a putative plastid ADS3 paralog exacerbates the growth defects of ads2 mutant plants under low temperature. These observations suggest that ADS2 encodes a 16:0 desaturase of MGDG and PG. We hypothesize that a low temperature–induced shift from the plastid to endoplasmic reticulum pathway for membrane lipid biosynthesis is required for the cold stress response in Arabidopsis thaliana, and ADS2 is essential to adjust the acyl composition of organelle membrane lipid composition in response to cold stress.
INTRODUCTION
Low temperature is one of the major environmental factors that limit the geographical distribution of many plant species. While many tropical and subtropical plants exhibit a marked physiological dysfunction when exposed to chilling temperature (0 to 15°C), most temperate plant species can acquire freezing tolerance by prior exposure to nonfreezing low temperature, a process known as cold acclimation (Guy, 1990). Maximov (1912) first suggested that disruption of the plasma membrane was the primary response of freezing injury. Heber and Santarius (1973) postulated that a hierarchy of injury to various cellular membranes could exist. Cold acclimation may directly alter the stability of the plasma membrane so that it is more tolerant of the consequences of ice formation and cell dehydration induced by freezing temperature (Steponkus et al., 1983). It was speculated that thermotropic phase transition of membrane lipids might play an initiative role in the chilling injury, and much attention has been directed to study the association of low temperature–induced lipid alterations to cold acclimation and chilling tolerance (Lyons, 1973). By measuring the fatty acid or lipid changes in response to cold hardiness, it was suggested that the degree of unsaturation of fatty acids (FAs) was correlated with chilling injury (Lyons et al., 1964; Gerloff et al., 1966; Kuiper, 1970). Comparing the acyl lipid composition between chilling-sensitive and -resistant plants, a positive correlation between the chilling sensitivity of herbaceous plants and the level of saturated and trans-monounsaturated molecular species of phosphatidylglycerol (also termed high-temperature melting phosphatidylglycerol [htm-PG]) was found (Murata, 1982; Murata et al., 1982).
In the past two decades, with advancements in genetic manipulation of enzymes for the biosynthesis and desaturation of FAs as well as the characterization of mutants with defective enzymes, the role of the FA desaturation of membrane lipids in cold tolerance has been reexamined. Wolter et al. (1992) expressed Escherichia coli glycerol-3-phosphate acyltransferase in Arabidopsis thaliana, which resulted in elevated levels of 16:0 in the sn-1 position for all lipid classes without altering the overall proportions of membrane lipids; the transgenic plants showed increased chilling sensitivity. Kodama et al. (1994) introduced an Arabidopsis plastid ω-3 fatty acid desaturase (FAD7) into tobacco (Nicotiana tabacum), resulting in higher levels of trienoic FAs and increased tolerance to low-temperature stress. This experiment suggested that lipid unsaturation or lipid composition is linked to chilling sensitivity.
Characterization of mutants with defective FA desaturases provided additional clues about the correlation between lipid desaturation and cold tolerance. FATTY ACID DESATURASE2 (FAD2) encodes a microsomal oleoyl-phosphatidycholine desaturase, and its mutation resulted in reduced levels of polyunsaturated 18C FAs and an increased proportion of oleate. Extraplastidial membrane lipids, including phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylinositol (PI), showed a marked increase in the content of 18:1 and a concomitant decrease in 18:2 and 18:3, whereas plastid lipids (monogalactosyl diacylglycerol [MGDG], digalactosyl diacylglycerol [DGDG], sulfolipid [SL], and phosphatidylglycerol [PG]) showed a smaller increase in 18:1 (Miquel and Browse, 1992). When Arabidopsis fad2 mutant plants were grown at 6°C, the rosette leaves gradually died, and the plants were inviable. This observation makes it clear that polyunsaturated lipids have an essential role in maintaining cellular function and plant viability at low growth temperature (Miquel et. al., 1993). A mutation in ACYL-LIPID DESATURASE3 (ADS3) (also named FAD5), which encodes a chloroplast 16:0Δ7 desaturase specific to the sn-2 position of MGDG, resulted in elevated amounts of palmitic acid and decreased amounts of unsaturated 16C FAs. When fad5 mutants were grown at 5°C, newly developed leaves were chlorotic (Kunst et al., 1989; Hugly and Somerville, 1992). A similar low-temperature growth defect was also observed with a mutation in FAD6 (also named FADc), which encodes a plastid cis-ω-6 desaturase, specific to 16:1 and 18:1 at the sn-1 and sn-2 of chloroplast glycerolipids. This mutation resulted in reduced levels of 16:3 and 18:3 FAs (Hugly and Somerville, 1992). These observations provided the first direct evidence that plastid lipid polyunsaturation contributes to low-temperature tolerance. However, both the fad5 and fad6 mutants continue to grow and complete their life cycles normally at 6°C (Hugly and Somerville, 1992).
Lipid profiling demonstrated that the fab1 mutant contains a higher amount of htm-PG; however, mutant plants were unaffected by low-temperature treatments (Wu and Browse, 1995). These observations raise questions about the premise that the content of plastid htm-PG is the primary determinant of chilling sensitivity in plants. However, growth of fab1 plants was compromised by a more than 2-week exposure to 2°C due to the disruption of the thylakoid and chloroplast structure (Wu et al., 1997), demonstrating that chloroplast membrane unsaturation of PG is an important aspect for the proper growth and development of plants at low temperature.
Galactoglycerolipids were initially found to be the most abundant lipid species in chloroplasts (Wintermans, 1960) and were assumed to be exclusive to this organelle. Later, Yoshida and Uemura (1986) reported that galactoglycerolipids were also present in extraplastidic membranes. Härtel et al. (2000) demonstrated that phosphate deprivation stimulates extraplastidic galactolipid biosynthesis. Under phosphate limiting growth conditions, galactolipids are also found in the plasma membrane (Andersson et al., 2003), tonoplast (Andersson et al., 2005), and mitochondria (Jouhet et al., 2004). In this study, we demonstrate that a cold-inducible ADS2 gene encodes a 16:0 desaturase for MGDG and PG. The mutation of ADS2 alters leaf membrane lipid composition, and mutant plants show increased sensitivity to chilling and freezing temperature. We demonstrate that ADS2 is an essential component for cold adaption in Arabidopsis.
RESULTS
ads2 Mutant Identification
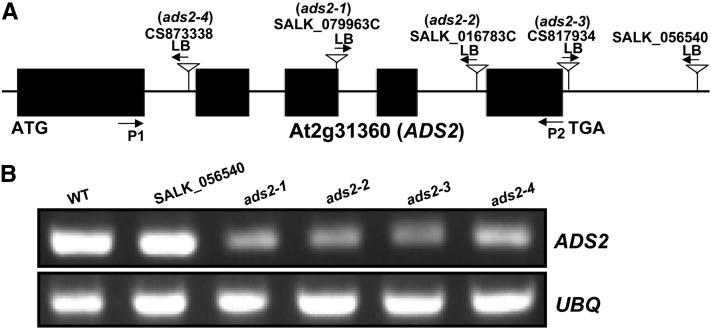
Acyl-lipid desaturases are defined as any desaturases that used acyl-lipid as substrate. Based on this definition, the previously characterized FATTY ACID DESATURASE family (FAD2, FAD3, FAD4, FAD6,, FAD7,, and FAD8) can also be considered as acyl-lipid desaturases, since they all use acyl-lipid as substrate instead of acyl-CoA. Furthermore, nine additional genes in the Arabidopsis genome were cataloged as acyl-lipid desaturases based on sequence similarity with cyanobacterial acyl-lipid desaturases and mammalian acyl-CoA desaturases. These nine members are At1g06080 (ADS1), At1g06090, At1g06100, At1g06120, At1g06350, At1g06360, At2g31360 (ADS2), At3g15850 (ADS3), and At3g15870 (Browse et al., 1985; Kunst et al., 1989; Fukuchi-Mizutani et al., 1998; Mekhedov et al., 2000; Heilmann et al., 2004a, 2004b). ADS1 forms a tightly linked gene cluster with At1g06090, At1g06100, and At1g06120 and another pair of genes (At1g06350 and At1g06360) on chromosome I. ADS3 is linked with At3g15870 on chromosome III (Heilmann et al., 2004a). Phylogenic analysis revealed a close relationship among different members. ADS1 and ADS2 form their own clade (see Supplemental Figure 1A and Supplemental Data Set 1 online), suggesting their common origin through gene duplication during evolutionary history. At the amino acid level, ADS2 shares a higher similarity with ADS1 (75%) compared with other members (see Supplemental Figure 1B online). Sequence alignment indicates that ADS3 and At3g15870 have an extra 64 amino acids at their N termini compared with other members (see Supplemental Figure 1B online), suggesting that duplicated genes could be differentially targeted at the subcellular level, indicative of subfunctionalization and/or neofunctionalization. Previous experiments have established that ADS3 (FAD5) is responsible for the Δ7 desaturation of 16:0 on MGDG at the sn-2 position in plastids (Browse et al., 1985; Kunst et al., 1989; Mekhedov et al., 2000). ADS2 was originally projected to be a Δ9 acyl-lipid desaturase homolog in Arabidopsis based on its sequence similarity with a Δ9 desaturase from rose petals (Rosa hybrida cv Kardinal) (Fukuchi-Mizutani et al., 1995; Fukuchi-Mizutani et al., 1998). Biochemical analysis of yeast and Arabidopsis seeds indicated that ADS2 operates on either galactolipids or phospholipids (such as PC), but the regiospecificity is different depending on the headgroup (Heilmann et al., 2004b). However, assignments of protein function based solely on heterologous expression should be viewed with caution since substrate availability or protein subcellular localization in planta might affect preference and, hence, biological function (Heilmann et al., 2004b). Therefore, we decided to take a genetic approach to understand the biochemical and physiological function of ADS2 in planta. Seven individual T-DNA insertion lines (CS873338, SALK_079963C, SALK_016783C, CS817934, SALK_056540, CS879863, and CS849116) were obtained from The Arabidopsis Information Resource. Using PCR, we could not detect T-DNA insertions in CS879863 and CS849116, although the other five lines were confirmed and homozygous lines were obtained (Figure 1A). RT-PCR was then used to determine ADS2 gene expression. We found that the expression of ADS2 was highly reduced in SALK_079963C, SALK_016783C, CS817934, CS873338, and not affected in SALK_056540 (Figure 1B). Based on this result, for convenience, we renamed SALK_079963C, SALK_016783C, CS817934, and CS873338 as ads2-1, ads2-2, ads2-3, and ads2-4 mutants, respectively.
Figure 1.
ads2 Mutant Identification.
(A) Schematic representation of T-DNA insertion positions in the At2g31360 gene. LB, T-DNA left border.
(B) ADS2 expression was highly reduced in SALK_079963c, SALK_016783c, CS817934, and CS873338, and was not affected in SALK_056540. RT-PCR was repeated three times to detect gene expression, with similar results. UBQ was used as loading control, primer pairs (P1 and P2) as indicated in (A) (arrows) were used to amplify ADS2 cDNA. WT, the wild type.
ads2 Mutant Plants Display Reduced Chilling Tolerance
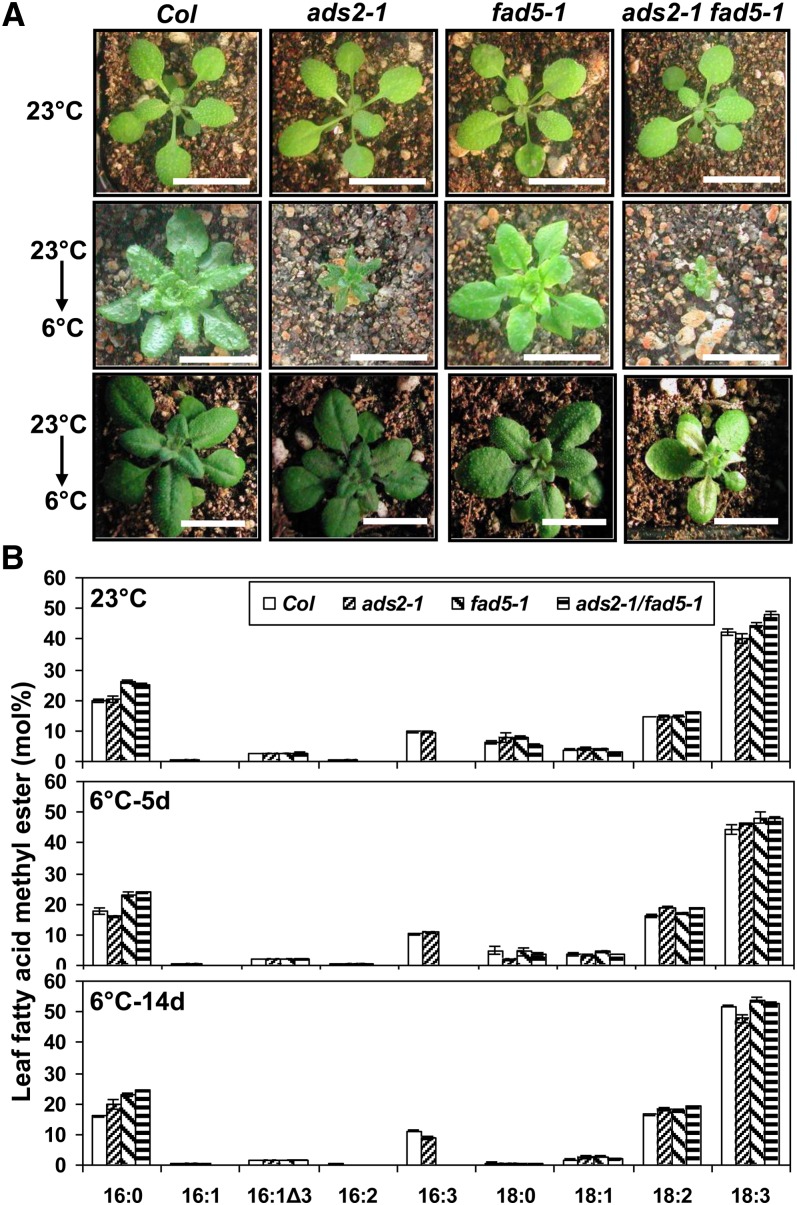
ADS2 expression is quickly induced when plants are shifted from 22 to 10°C (Fukuchi-Mizutani et al., 1998; Maruyama, et al., 2004), suggesting ADS2 is involved in cold acclimation or chilling tolerance. To test this hypothesis, wild-type, ads2-1, ads2-2, ads2-3, and ads2-4 mutants were grown at 23 and 6°C. At 23°C, all four ads2 mutant lines grew normally and there were no visible growth defects compared with the wild type (Figures 2A to 2E). At 6°C, wild-type Arabidopsis plants were not injured by cold exposure and continued to grow and develop during prolonged incubation, although the growth rate was significantly reduced relative to at 23°C. However, the growth rate of ads2 mutants was significantly lower than the wild type at 6°C (Figures 2F to 2J). The retarded growth phenotype became severe with prolonged exposure to low temperature, especially after reproductive transition (Figures 2K to 2O). ads2 mutants began bolting at a similar time to that of the wild type, but all four ads2 mutants were sterile, while the wild type produced abundant, viable seeds at 6°C (Figure 3A). In the wild type, anther opening follows flower opening such that pollen grains are not prematurely released (Figure 3B). In ads2 mutant plants, the anther failed to open after flowering (Figure 3C), which might be one of the factors contributing to the sterile phenotype. When flowering ads2 mutants were moved from 6°C to room temperature, the phenotype (dwarf and sterility) was partially suppressed (Figure 3D). The ads2 mutant plants eventually produced viable seeds several weeks after the temperature shift, indicating that the low growth temperature is one of the main factors responsible for these developmental defects.
Figure 2.
ads2 Mutant Plants Show Increased Chilling Sensitivity.
(A) to (E) The wild type and mutants were grown at 23°C for 18 d under long-day conditions (bars = 2 cm). Col, Columbia.
(F) to (J) Plants were first grown at 23°C under long-day conditions for 10 d and were then moved to a 6°C cold chamber and grown for an additional 82 d under continuous white light (bars = 1.8 cm).
(K) to (O) Plants were first grown at 23°C under long-day conditions for 10 d and then were moved to 6°C under continuous white light conditions for 6 months (bars = 3 cm).
[See online article for color version of this figure.]
Figure 3.
ads2 Mutant Plants Are Sterile under Chilling Condition.
(A) Wild-type (WT) and ads2-2 mutant plants grown at 6°C for 6 months.
(B) and (C) Wild-type and ads2-2 mutant flowers (bars = 0.5 mm).
(D) ads2-2 mutant plants grown at 6°C until flowering and then moved to 23°C under continuous white light (bar = 3 cm).
[See online article for color version of this figure.]
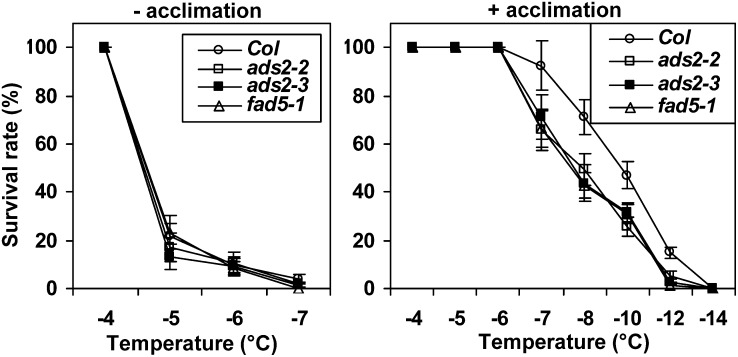
ads2 Mutants Display Reduced Freezing Tolerance
Since ads2 mutant plants had reduced cold tolerance, we then tested their sensitivity to freezing temperature using a previously established protocol (Xin and Browse, 1998). Wild-type, ads2, and fad5-1 mutant plants were grown side by side for 10 d at 23°C, and then one group of plants was moved to 6°C and grown an additional 5 d before being exposed to a gradient of freezing temperatures. Another group of plants was directly used for freezing treatment without cold acclimation. The nonacclimated plants showed a tolerance to a narrow range of freezing temperatures (−4 to −7°C); ∼80% of plants were killed when temperature was shifted from −4 to −5°C. Without acclimation, the temperature for 50% survival is essentially the same for wild-type, ads2, and fad5-1 mutant plants (Figure 4, left). After acclimation, wild-type and mutant plants can survive through a much lower freezing temperature (Figure 4, right). ads2 and fad5-1 mutant plants had a lower survival rate than the wild type between −7 and −12°C, and the temperatures that resulted in 50% survival were −10.0, −8.0, −7.9, and −7.8°C for the wild type, ads2-2, ads2-3, and fad5-1, respectively (Figure 4, right). Since cold-acclimated ads2 and fad5-1 mutant plants can tolerate lower freezing temperatures compared with respective nonacclimated control plants, this observation suggests that ads2 mutant plants still retain the capacity to acclimate to low, nonfreezing temperatures, although not as efficiently as the wild type.
Figure 4.
ads2 Mutant Plants Display Reduced Freezing Tolerance.
Percentage of survival of nonacclimated and acclimated plants after freezing at different temperatures. The data are means ± se for triplicates. Col, Columbia.
Low Temperature–Induced FA Desaturation Was Disrupted in ads2 Mutant Plants
To understand the biochemical basis for reduced cold and freezing tolerance of ads2 mutants, leaf FA composition was analyzed from two independent knockdown lines (ads2-2 and ads2-3). When grown at 23°C, the two ads2 mutants did not have significant differences in FA composition compared with the wild type. By contrast, fad5-1 mutant plants had significantly lower levels of 16:1, 16:2, and 16:3 and a higher level of 16:0 than the wild type (Table 1), in agreement with ADS3 encoding a palmitoyl-Δ7-desaturase (Kunst et al., 1989; Hugly and Somerville, 1992). When grown at 6°C, 16:2 and 16:3 levels were 26 and 27-35% lower than wild-type levels under the same conditions, respectively, in the ads2-2 and ads2-3 mutant plants, while 16:0 content was 26 to 30% greater than wild-type levels in the same two independent ads2 mutant lines (Table 1). These results indicate that ADS2 also encodes a palmitic acid desaturase.
Table 1. FAME Analysis of ads2 Mutant Plants Grown under Different Temperatures.
| Col |
ads2-2 |
ads2-3 |
fad5-1 |
|||||
|---|---|---|---|---|---|---|---|---|
| FAME | 23°C | 6°C | 23°C | 6°C | 23°C | 6°C | 23°C | 6°C |
| 16:0 | 15.5 ± 0.15 | 17.0 ± 0.57 | 16.0 ± 0.2 | 21.5 ± 1.0* | 16.0 ± 0.37 | 22.2 ± 1.01* | 22.0 ± 0.20* | 23.1 ± 0.15* |
| 16:1 | 0.76 ± 0.03 | 0.28 ± 0.02 | 0.69 ± 0.01 | 0.23 ± 0.01* | 0.67 ± 0.03 | 0.29 ± 0.02 | 0.22 ± 0.02* | 0.24 ± 0.01 |
| t16:1 | 3.23 ± 0.06 | 1.18 ± 0.03 | 3.15 ± 0.03 | 1.24 ± 0.12 | 3.16 ± 0.16 | 1.26 ± 0.19 | 3.17 ± 0.03 | 1.27 ± 0.08 |
| 16:2 | 0.71 ± 0.06 | 0.50 ± 0.04 | 0.74 ± 0.06 | 0.37 ± 0.03* | 0.79 ± 0.04 | 0.37 ± 0.09* | 0.05 ± 0.03* | 0.12 ± 0.01* |
| 16:3 | 11.3 ± 0.27 | 11.7 ± 0.33 | 10.8 ± 0.05 | 8.48 ± 0.39* | 11.1 ± 0.18 | 7.60 ± 0.86* | 0.61 ± 0.03* | 1.16 ± 0.39* |
| 18:0 | 6.73 ± 0.24 | 3.57 ± 0.45 | 5.89 ± 0.36 | 6.22 ± 0.83* | 5.93 ± 0.56 | 6.80 ± 0.92* | 5.26 ± 0.44* | 4.66 ± 0.73 |
| 18:1 | 4.58 ± 0.14 | 2.23 ± 0.27 | 4.36 ± 0.09 | 4.68 ± 0.69* | 4.32 ± 0.17 | 4.18 ± 0.75* | 4.04 ± 0.17 | 3.03 ± 0.19 |
| 18:2 | 17.6 ± 0.11 | 11.7 ± 0.09 | 17.8 ± 0.21 | 11.46 ± 0.53 | 17.9 ± 0.17 | 10.90 ± 0.97 | 17.40 ± 0.16 | 12.8 ± 0.26* |
| 18:3 | 39.6 ± 0.40 | 51.8 ± 0.83 | 40.57 ± 0.42 | 46.00 ± 1.25* | 40.12 ± 0.90 | 46.56 ± 1.76* | 47.27 ± 0.89* | 53.51 ± 0.74 |
Wild-type, ads2, and fad5-1 mutant plants were grown in a growth chamber at 23°C for 25 d, after which aerial leaf tissues were harvested for FAME analysis. For cold-grown Arabidopsis, plants were grown in a growth chamber at 23°C for 2 weeks and then moved to a 6°C cold room for an additional 37 d before aerial leaves were harvested for FAME analysis. Data are expressed as average ± se of mol % (n = 4). Student’s t test was performed to calculate the P value. *P < 0.05. Col, Columbia.
Even though the leaf FA profile of ads2 mutant plants shares many similarities with fad5-1 mutant plants (e.g., reduced 16:2 and 16:3 and increased 16:0) when plants were grown at 6°C, differences emerged regarding 18C FAs. Under 6°C, the 18:0 content in ads2 mutant plants increased 74 to 90% and 18:1 content increased 87 to 110%, while 18:3 content decreased 10 to 11% relative to the wild type. The changes in these 18C FAs were less dramatic in fad5-1 mutant plants relative to the wild type under 6°C (Table 1). The varying patterns regarding 18C FAs between ads2 and ads3 mutants suggests that ADS2 is also involved in desaturation of 18C FAs.
Lipidomic Analysis Identifies Two Potential Lipid Substrates for ADS2 Desaturation
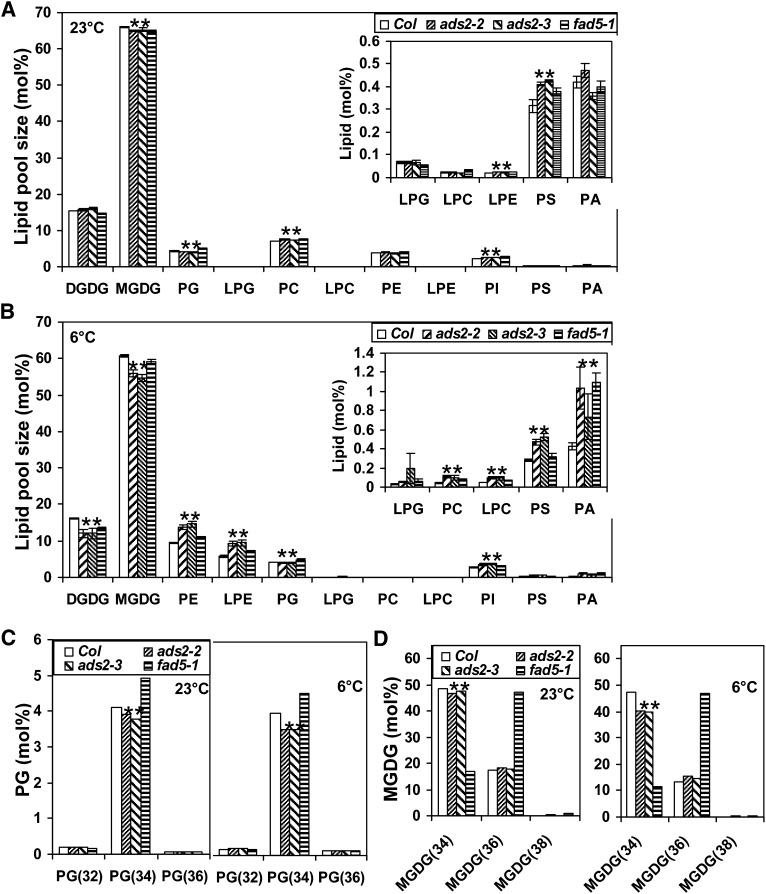
Fatty acid methyl ester (FAME) analysis demonstrated that ADS2 desaturates palmitic acid, and possibly stearic acid (Table 1), but did not identify which lipid species were the substrates for ADS2 desaturation. Thus, lipidomic analysis was conducted to identify the lipid species that were specifically reduced by ADS2 mutations. Wild-type, ads2, and fad5-1 plants were grown at 23 or 6°C respectively, and leaf lipids were isolated for lipidomic analysis (see Supplemental Table 1 online). At 23°C, although the FA composition in ads2 mutant and wild-type plants was similar (Table 1), several lipid species showed small but statistically significant changes. For example, total PI, lysophosphatidylethanolamine (LPE), PC, and phosphatidylserine (PS) content was significantly higher, and total PG and MGDG content was lower in ads2 mutant plants than in the wild type (Figure 5A; see Supplemental Table 1 online). At 23°C, there was no difference regarding the content of total phosphatidic acid (PA), PE, lysophosphatidylcholine (LPC), and DGDG between the ads2 mutant plants and the wild type; however, upon exposure to low temperature, ads2 mutant plants showed more dramatic changes in their leaf lipid profile, with total PA, PI, PE, LPE, PC, LPC, and PS content being significantly higher, while total PG, MGDG and DGDG decreased substantially compared with the wild type (Figure 5B; see Supplemental Data Set 2 online).
Figure 5.
Lipidomic Analysis Reveal Membrane Lipid Change Pattern in ads2 Mutant Plants.
(A) and (B) The relative total content (mol %) of different lipid species when plants were grown at 23 or 6°C. Col, Columbia.
(C) The size of the PG subpool (mol %) when plants were grown at 23 or 6°C.
(D) The size of the MGDG subpool (mol %) when plants were grown at 23 or 6°C.
Wild-type, ads2-2, ads2-3, and fad5-1 mutant plants were grown in a growth chamber at 23°C for 25 d, and aerial leaf tissues were harvest for lipid extraction. For cold-grown Arabidopsis, plants were grown in a growth chamber at 23°C for 2 weeks and then moved to a 6°C cold room for an additional 37 d before aerial leaves were harvested for lipid extraction. Data are expressed as average ± se of mol % (n = 4), and statistically significant differences between wild-type and ads2 mutant plants are labeled with asterisks.
Next, we examined which lipid species subpools contribute to the reduction of total PG at lower growth temperature. In Arabidopsis leaf, total PG is mainly composed of four subpools: 16\16 PG [PG(32)], 16\18 PG and 18\16 PG [PG(34)], and 18\18 PG [PG(36)]. At 23°C, total leaf PG contained 94% PG(34), 4.4% PG(32), and 1.6% PG(36) (mol %) in wild-type plants (see Supplemental Data Set 2 online). The reduction of total PG in ads2 mutant plants exclusively contributed to the decrease in PG(34) pools, including 34:1, 34:2, 34:3, and 34:4, (Figure 5C, left; see Supplemental Data Set 2 online); this was also the case with plants grown at 6°C (Figure 5C, right). We also observed that the reduction in the PG(34) pool in ads2 mutant plants compared with the wild type was greater at 6°C than 23°C. These data suggest that PG(34:0) is a substrate for ADS2 desaturation. Unlike ads2 mutant plants, fad5-1 mutant plants have increased content of total PG primarily due to increased levels of PG(34:3 and 34:4) (Figure 5C; see Supplemental Data Set 2 online), suggesting that ADS3 does not desaturate PG(34).
We then examined which lipid species subpools contribute to the reduction of the total MGDG pool size. In wild-type Arabidopsis leaves, the total MGDG pool is primarily composed of three different lipids; MGDG(34) and MGDG(36) account for 74 and 26% (mol %), respectively, of the total MGDG pool, whereas MGDG(38) is only a minor species (see Supplemental Data Set 2 online). The content of MGDG(34) in two ads2 mutant lines was always lower than the wild type control regardless of growth temperature and contributed exclusively to the reduced content of the total MGDG pool (Figures 5A, 5B, and 5D; see Supplemental Data Set 2 online). We then checked which MGDG(34) molecular species was significantly reduced. In Arabidopsis leaves, MGDG(34) has different unsaturation states, including MGDG(34:1), MGDG(34:2), MGDG(34:3), MGDG(34:4), MGDG(34:5), and MGDG(34:6). At 23°C, MGDG(34:5) and MGDG(34:6) were significantly reduced in ads2-2, and MGDG(34:2), MGDG(34:3), MGDG(34:4), and MGDG(34:5) were significantly lower in ads2-3 mutant plants compared with the wild type (see Supplemental Data Set 2 online). At 6°C, MGDG(34:2), MGDG(34:4), MGDG(34:5), and MGDG(34:6) content in both ads2 mutant plants was significantly lower than that of the wild type (see Supplemental Data Set 2 online). These data support the notion that ADS2 also desaturates MGDG(34:1) to MGDG(34:2) in planta.
The MGDG(34) content in the fad5-1 mutant was reduced a great degree compared with ads2 mutant plants, coinciding with concomitantly higher levels of MGDG(36) (Figure 5D). The data suggest that ADS3 desaturates palmitic MGDG in the plastid, since galactolipids are found predominantly in the chloroplast. Our observation also supports the notion that suppressing the prokaryotic pathway leads to enhanced MGDG biosynthesis through the endoplasmic reticulum (ER) pathway (Browse et al., 1985; Kunst et al., 1989; Mekhedov et al., 2000). The fact that MGDG(34:1) and MGDG(34:3) increased with a concomitant reduction in MGDG(34:2) and MGDG(34:4, 34:5, and 34:6) in fad5-1 mutant plants (see Supplemental Data Set 2 online) suggests that ADS3 converts MGDG(34:1) to MGDG(34:2) as well as MGDG(34:3) to MGDG(34:4).
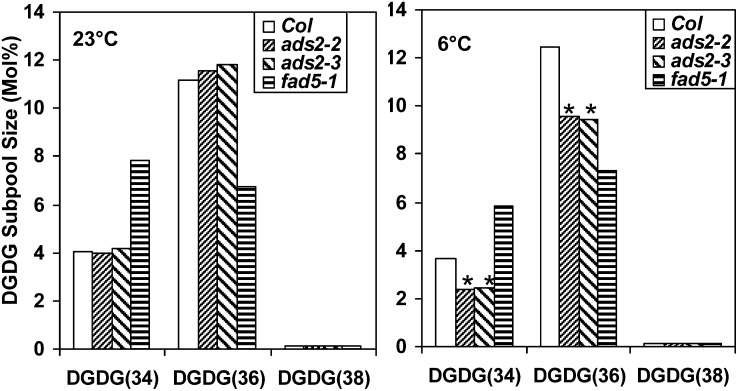
Since total DGDG content in ads2 mutant plants is significantly lower than the wild type when plants were grown at 6°C, we examined the changes in the subpool size of DGDG. In wild-type Arabidopsis leaves, the total DGDG pool is primarily composed of three different lipids; DGDG(34) and DGDG(36) account for 26.3 and 72.8% (mol %), respectively, of the total DGDG pool, whereas DGDG(38) accounts for <1% (mol %) (see Supplemental Data Set 2 online). At 23°C, the DGDG(34) and DGDG(36) subpool size in two ads2 mutant plants showed small changes (Figure 6, left), but the statistical significance cannot be confirmed for the total DGDG pool (Figure 5A); at 6°C, the DGDG(34) and DGDG(36) subpool size in two ads2 mutant lines displayed a significant reduction compared with the wild type (Figure 6, right). The reduction in the DGDG(34) subpool mainly results from the reduced content in DGDG(34:3), DGDG(34:5), and DGDG(34:6); similarly, the reduced content in DGDG(36:5) and DGDG(36:6) contributes to most of the reduction in the DGDG(36) subpool in ads2 mutant plants (see Supplemental Data Set 2 online). Since MGDG serves as precursor for DGDG biosynthesis, the reduced MGDG(34) content in ads2 mutant plants could result in a reduction of DGDG(34), but the mechanisms leading to the DGDG(36) reduction in ads2 mutants remain unclear, since MGDG(36) was not significantly affected (Figure 5D, right).
Figure 6.
The DGDG Subpool Size Changed at Different Temperatures.
Comparison of the size of leaf DGDG subpool among wild-type, ads2,, and fad5-1 mutant plants at 23°C (left) and 6°C (right). Plant growth conditions were the same as described in Figure 5. Statistically significant differences between wild-type and ads2 mutant plants are labeled with asterisks. Col, Columbia.
ADS2 Localizes to the Endoplasmic Reticulum, Golgi, and Chloroplast
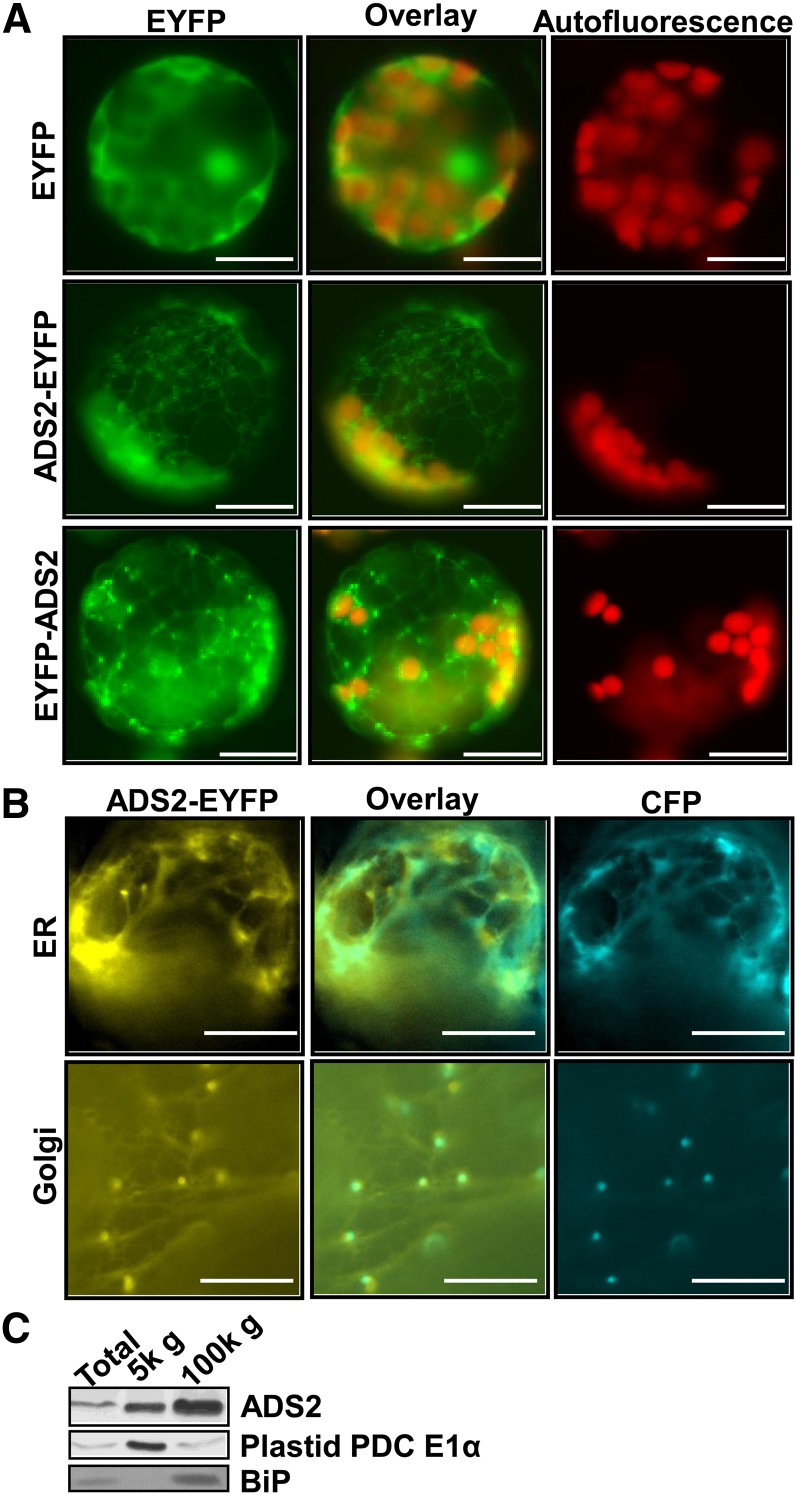
A comparison of eight subcellular prediction algorithms produced mixed results for ADS2. While SubLoc and WoLFPSORT predicted cytoplasmic localization, iPSORT, MitoPred, MitoProt2, MultiLoc, PeroxP, Predotar, and TargetP algorithms did not offer a conclusive answer. To experimentally determine the subcellular localization of ADS2, enhanced yellow fluorescence tag protein (EYFP) was fused in frame with ADS2 at its C terminus, and the fused protein was stably expressed in Arabidopsis. Protoplasts were prepared from stable transgenic plants, and subcellular localization was observed by fluorescence microscopy. The EYFP vector control signal was detected in the cytoplasm (Figure 7A, top panel). When EYFP was fused to the C terminus of ADS2, the EYFP signal appeared primarily in reticulate structures (Figure 7A, middle panel). Closer examination revealed that punctate spots were within the reticular network (Figure 7A, middle panel). A relatively weak EYFP signal also colocalized with chloroplasts as evidenced from the change from red (autofluorescence) to orange in the color overlay (Figure 7A, middle panel). These observations suggested ADS2 may have multiple subcellular destinations. To investigate if the distribution pattern of ADS2 was specific for the C-terminal fusion, we then examined other transgenic plants in which EYFP was fused in frame with ADS2 at the N terminus. The EYFP signal was also present in a visible reticulate pattern, and this focal position also revealed details of the punctate spots, which were mainly located at the reticular node (Figure 7A, bottom panel). Again, a relatively weak EYFP signal was also detected in chloroplasts (Figure 7A, bottom panel). These observations demonstrated that the EYFP signal pattern is similar regardless of N- or C-terminal fusions. However, the punctate spots were brighter when EYFP was fused at the N terminus of ADS2 than at the C terminus (Figure 7A). However, this could be due to the higher expression level of ADS2 in the N-terminal fusion transgenic plants that we chose for protoplast preparation.
Figure 7.
ADS2 Localizes to the Endoplasmic Reticulum, Golgi, and Plastid.
(A) Fluorescence micrographs of stably transformed Arabidopsis cell expressing EYFP vector, ADS2-EYFP, and EYFP-ADS2 chimeric proteins. Top panel, plant cells transformed with binary vector only as control; middle panel, plant cells transformed with ADS2-EYFP chimeric protein; bottom panel, plant cells transformed with EYFP-ADS2 chimeric protein. Left column, EYFP; middle column, overlay of autofluorescence with EYFP signal; right column, chlorophyll autofluorescence. Bars =10 µm.
(B) Transient coexpression of ADS2-EYFP protein with ER marker (CD3-953) or Golgi marker (CD3-961) in tobacco epidermal cells. EYFP was fused to the ADS2 C terminus, and the ER and Golgi markers were both fused with CFP. Bars =10 µm.
(C) ads1 fad5-1 double mutant leaves were subfractionated and assayed by immunoblotting. 5k g = 5000 RCF. Antibodies to the plastid PDC E1α subunit were used to show plastid enrichment, and antibodies to BiP were used to show ER enrichment.
To confirm the subcellular compartment of the punctate spots, ADS2-EYFP was transiently coexpressed with ER, Golgi, or plastid organelle markers in tobacco leaves. Coexpression of ADS2-EYFP and ER marker CD3-953 revealed partial overlapping signal within the reticular ER networks (Figure 7B, top panel). Coexpression of ADS2-EYFP with Golgi marker (CD3-961) revealed that the punctate EYFP spots colocalize with the Golgi cyan fluorescent protein (CFP) marker (Figure 7B, bottom panel).
When coexpressing ADS2-EYFP with chloroplast marker CD3-993, a weak signal was observed for both EYFP and CFP channels when adjusting the focal plane to the palisade mesophyll cell layer to detect plastids (see Supplemental Figure 2 online). Although the fluorescent signals overlapped, the low intensity necessitated an orthogonal approach to confirm plastid localization. Subcellular fractionation of leaf homogenates followed by immunoblotting confirmed ADS2 presence within the 5000× RCF (5000g) plastid fraction at a comparable level to plastid PDC E1α (Figure 7C). A comparison with the ER marker, luminal binding protein (BIP), also confirmed ADS2 is enriched within the microsomal membrane fraction, confirming dual localization within both plastids and microsomes.
Mutation of ADS3 Enhances Growth Defects of ads2 Mutant Plants under Low Temperature
A mutation in ADS3 has been reported to affect chloroplast biogenesis and plant growth under low temperature (Hugly and Somerville, 1992). To test a possible genetic interaction between ADS2 and ADS3, ads2-1 fad5-1 double mutants were constructed, and their growth performance was evaluated at both 23 and 6°C. When grown at 23°C, there was essentially no difference among wild-type, ads2-1, fad5-1, and ads2-1 fad5-1 double mutant plants (Figure 8, top panel). However, when grown at 6°C, reduced stature phenotypes were observed for ads2-1, fad5-1,, and ads2-1 fad5-1 double mutants depending on the developmental stage for cold treatment and the duration of low-temperature exposure. When 9-d-old seedlings were used for cold treatment and kept in the cold for a long period (81 d), ads2-1 mutant plants showed much smaller stature than the wild type and fad5-1 mutants and the leaves remained dark green. By contrast, the growth rate of fad5-1 was only slightly affected; the leaves that developed during cold exposure were chlorotic (Figure 8A, middle panel). The ads2-1 fad5-1double mutant plants were smaller than ads2-1 single mutant plants and the leaves were also chlorotic, like fad5-1 (Figure 8A, middle panel).
Figure 8.
Mutation in ADS3 Enhances Growth Defects of ads2 Mutant Plants under Low Temperature.
(A) Wild-type, ads2-1, fad5-1, and ads2-1 fad5-1 double mutant plants were grown at 23 or 6°C. Top panel, plants were grown at 23°C for 18 d; middle panel, plants were first grown at 23°C under long-day conditions for 9 d and were then moved into a 6°C chamber under continuous light for an additional 81 d; bottom panel, plants were first grown at 23°C under long-day conditions for 21 d and were then moved into a 6°C chamber under continuous light for an additional 14 d (bars = 1.2 cm). Col, Columbia.
(B) FAME analysis of plants before and after cold treatment. Top, FA composition of plants grown at 23°C for 3 weeks; middle, FA composition of plants after exposure to cold for 5 d; bottom, FA composition of plants after exposure to cold for 14 d.
[See online article for color version of this figure.]
If plants grew at 23°C for a long period (21 d) and were then subjected to cold treatment (5 d of growth at 6°C), there was no apparent difference among wild-type, single mutant, or double mutant plants. However, 14 d later at 6°C, ads2-1 fad5-1 double mutant plants were smaller than the parentals, and the chlorotic leaf phenotype was accentuated, with part of the leaf area being bleached (Figure 8A, bottom panel). These observations suggest that ADS2 and ADS3 have additive effects on chilling tolerance, since a mutation in ADS3 enhanced ads2 growth defects under low temperature.
To give a quantitative overview of the mutant phenotype and its association with FA unsaturation, leaf FA composition in wild-type, ads2-1, fad5-1, and ads2-1 fad5-1 double mutant plants was measured immediately before cold treatment and after cold exposure for 5 and 14 d. When plants were grown at 23°C for 21 d, the leaf FA composition in ads2-1 fad5-1 double mutants was similar to the fad5-1 single mutant, including higher content of 16:0 and much lower content of 16:2 and 16:3. The 18:2 and 18:3 content in the double mutant was higher than in the wild type and fad5-1 (Figure 8B, top panel). After cold treatment at 6°C for 5 d, leaf FA composition in ads2-1 fad5-1 double mutants was still similar to that of fad5-1 (Figure 8B, middle panel). Although the 16:1 content in the ads2-1 single mutant was slightly lower than the wild type, 16:2, 16:3, and 16:0 content was not significantly altered compared with the wild type (Figure 8B, middle panel). After 14 d of cold treatment, higher 16:0 content and lower 16:2 and 16:3 content in ads2-1 was observed (Figure 8B, bottom panel). ads2-1 and fad5-1 showed lower and higher 18:3 contents than the wild type, respectively. Accordingly, 18:3 content in ads2-1 fad5-1 double mutant plants was similar to the wild type (Figure 8B, bottom). Both ads2-1 and fad5-1 had a higher 16:0 content than the wild type, and the 16:0 content in the double mutant was even higher than in the parental lines (Figure 8B, bottom). Thus, we concluded that ADS2 and ADS3 have additive effects on 16:0 and 18:3 accumulation.
DISCUSSION
ADS2 Is an Essential Component for Cold Tolerance in Arabidopsis
Genetic studies demonstrated that the ability of plants to acclimate to cold conditions is a quantitative trait involving the action of many genes with small additive effects (Thomashow, 1990). In support of this, several hundred cold-responsive genes have been identified by transcriptome analysis (Vogel et al., 2005; Oono et al., 2006; Robinson and Parkin, 2008). However, only a small number of these cold-responsive genes have been investigated. In this study, we demonstrate that ads2 mutant plants grow normally at room temperature and show much slower growth than the wild type when grown at a low temperature (Figure 2). In addition, pollen development was aborted, producing sterile ads2 mutant plants under cold conditions (Figure 3). Usually, when cold-sensitive plants are exposed to low temperature, germination and growth are reduced. Pollen maturation is especially sensitive to chilling temperature, resulting in reproductive failure (Satake and Koike, 1983). Arabidopsis (Columbia ecotype) is regarded as a cold-resistant plant as it can continuously grow and complete its life cycle at low temperatures (Figure 2). The stunted growth and sterile phenotypes of ads2 mutant plants under low-growth temperature suggest that mutations in ADS2 convert Arabidopsis from cold resistant to cold sensitive, indicating it plays an essential role in cold fitness.
MGDG Is the Substrate for ADS2 Desaturation in Chloroplasts
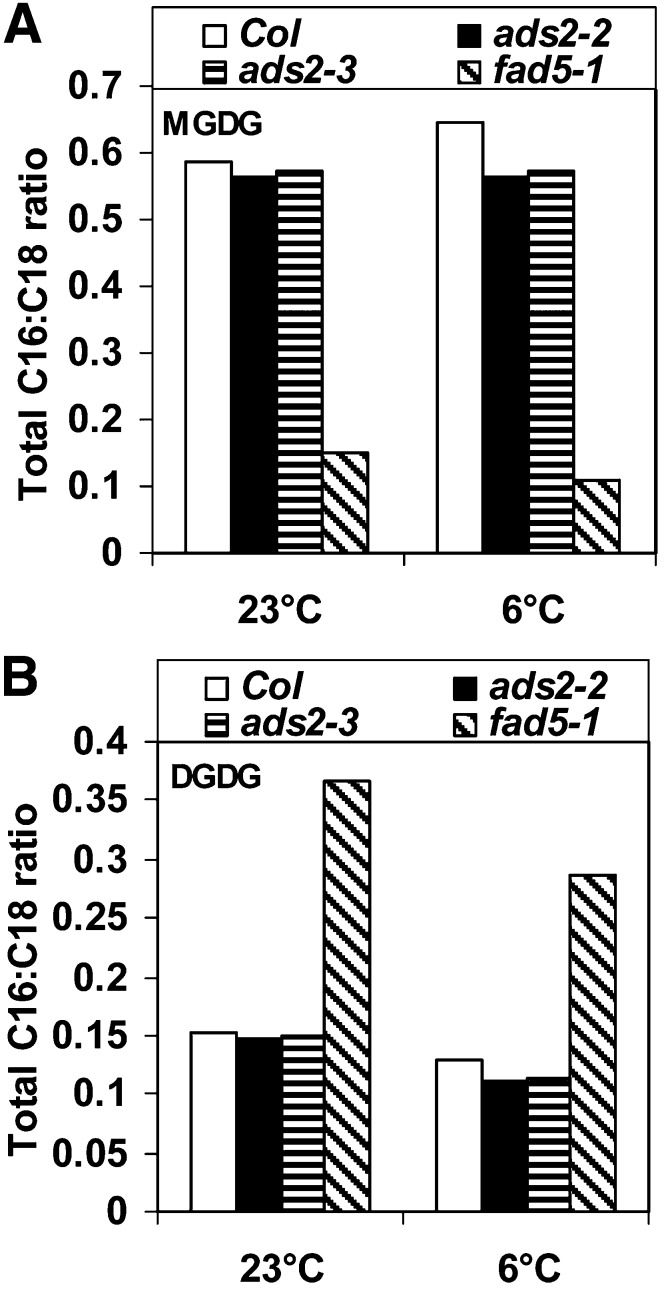
Galactolipids MGDG and DGDG are primarily located in thylakoids as well as the inner and outer envelope membranes of the chloroplast (Browse and Somerville, 1991), although a small amount of galactolipids is also present in plant mitochondria (Schwertner and Biale, 1973; Edman and Ericson, 1987). In Arabidopsis, the plastidic pathway for lipid synthesis takes place in the inner envelope membrane of the chloroplast and produces lipids with 16C FAs in the sn-2 position of the glycerol backbone, whereas the ER pathway for lipid synthesis gives rise to phospholipids with 18C FAs in this position. Extraplastidic galactolipids are synthesized at the outer envelope membrane of chloroplasts by type-B MGDG synthase (MGD2/3) and DGD2 using the precursors derived from the ER pathway, and these galactolipids are then transported and integrated into extraplastidic membranes through an as yet undiscovered mechanism (Awai et al., 2001; Kobayashi et al., 2009a, 2009b). However, questions remain regarding desaturation of galactolipids. In this study, we provide several lines of evidence that ADS2 is a 16:0Δ7 desaturase of MGDG. First, under a 6°C growth temperature, the leaf FA content in two ads2 mutant plants was 27 to 35% lower in 16:3 and 26 to 30% higher in 16:0 content than the wild type. A similar trend was also observed in fad5-1 mutant plants, which lack ADS3, a plastid 16:0Δ7 desaturase of MGDG at the sn-2 position (Table 1), suggesting that ADS2 possesses similar activity. Second, lipid profiling reveals a significant reduction of total MGDG as well as the MGDG(34) subpool in both ads2 mutant lines compared with the wild type (Figure 5; see Supplemental Data Set 2 online), suggesting that MGDG(34) is a candidate substrate for ADS2 desaturation. Third, the ratio of total 16C to 18C FAs in MGDG is lower in the two ads2 mutant lines compared with the wild type, especially when plants are grown under cold conditions (Figure 9A). An even larger decrease was also observed in fad5-1 mutant lines relative to the wild type regardless of whether the plants were grown at 23 or 6°C (Figure 9A). Lastly, some ADS2 protein is located in plastids. This is unexpected since it does not contain an apparent plastid targeting peptide (Figure 7; see Supplemental Figure 1 online). However, a noncanonical pathway for plastid targeting has previously been reported. α-Carbonic anhydrase has been demonstrated to be transported to plastids through the secretory pathway en route (Villarejo et al., 2005). However, it remains unclear if ADS2 adopts a similar mechanism for plastid targeting.
Figure 9.
FA Composition of Galactolipids in Wild-Type, ads2, and fad5-1 Mutant Lines.
(A) Ratio of total 16C to 18C FAs in MGDG.
(B) Ratio of total 16C to 18C FAs in DGDG.
The calculation is based on each MGDG(34) or DGDG(34) containing one 16C and one 18C FA and each MGDG(36) or DGDG(36) containing two 18C FAs. Col, Columbia.
ADS2 Also Desaturates PG in the ER
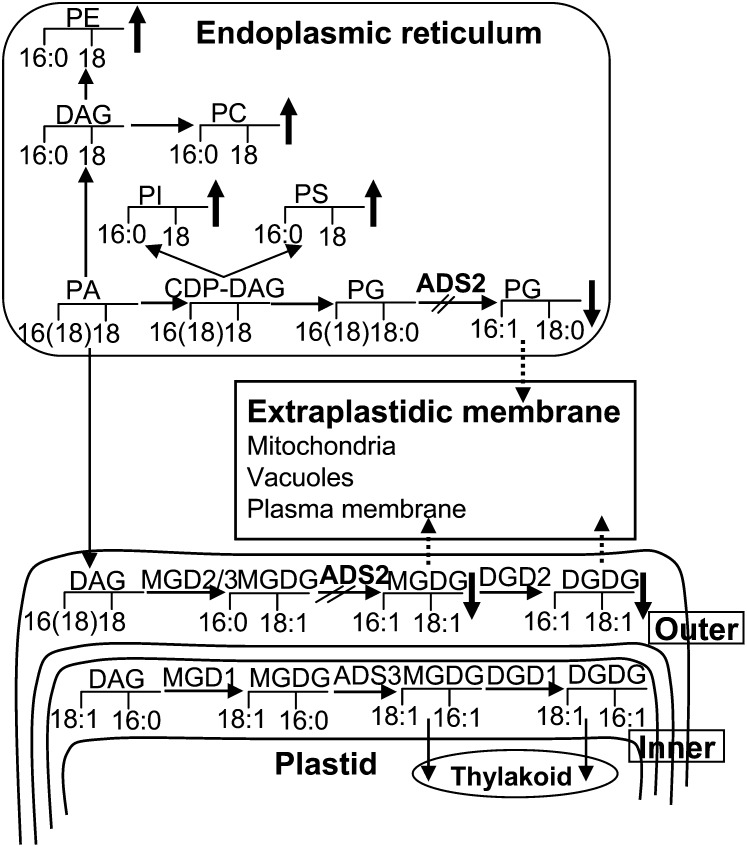
In higher plants, PG is synthesized in three cellular compartments: plastids, ER, and mitochondria. Radioisotopic tracer analyses established that the PG synthesized through the ER versus plastid pathway is roughly at a 1:5 mole ratio, respectively, when plants were grown at 22°C (Browse et al., 1986). PG(34) is the major component and accounts for more than 90% of the total PG in Arabidopsis, followed by minor levels of PG(32) (16/16 PG) and PG(36) (18/18 PG) (see Supplemental Dataset 2 online). These subpools of PG have different origins; PG(32) is synthesized by the plastid pathway, while PG(36) is produced by the ER pathway. Although the plastid and ER both synthesize PG(34), their products have distinct molecular signatures and metabolic fates. PG(34) synthesized in the plastid has palmitic acid esterified at the sn-2 position and can be desaturated by FAD4 at the Δ3 position, introducing an unusual trans–double bond (18/t16:1 PG) (Gao et al., 2009). The PG(34) synthesized by the plastid pathway is thought to be exclusively targeted to plastid membranes. By contrast, the PG(34) synthesized through the ER pathway has palmitic acid esterified at the sn-1 position of the glycerol backbone of PG (16/18 PG). These PGs presumably could integrate into multiple subcellular membranes. Based on the following evidence, we suggest that ADS2 can desaturate 16:0 at the sn-1 position of PG: First, the two ads2 mutant plants both showed significantly lower content of total PG than the wild type, which was exclusive to PG(34) (Figure 5). By contrast, fad5-1 mutant plants had a higher content of total PG than the wild type, contributed mostly by the rising content of PG(34) (Figure 5; see Supplemental Table 1 online), indicating that ADS3 cannot desaturate PG(34) in plastids. Second, other glycerolipids, including LPE, PC, PI, and PS, which are exclusively produced through the ER pathway and regarded as markers for extraplastidic membranes, are higher in the two ads2 mutant plants when grown at 23°C (Figure 5; see Supplemental Table 1 online). An even greater increase in these lipid species was observed when plants were grown at 6°C. Assuming that ADS2 desaturates both PG(34) and MGDG(34), its mutation should reduce flux into these two lipid species. Indeed, decreased PG(34) and MGDG(34) was observed in these two mutant lines (Figure 5; see Supplemental Table 1 online). On the other hand, biosynthesis of these extraplastidic lipids is interconnected and shares several common intermediates, such as PA(34) and DAG(34). Reduced flux into PG(34) and MGDG(34) in the ads2 mutant plants could generate a feedback effect and push more intermediates into other branch pathways, which would explain why other extraplastidic lipids, including PA, PI, PE, LPE, PC, LPC, and PS, were correspondingly higher (Figure 10; see Supplemental Table 1 online).
Figure 10.
Schematic Representation of the Roles Played by ADS2 in MGDG and PG Biosynthesis in Arabidopsis Leaves.
The glycerol backbones with the typical carbon length of FAs in the sn-1 and sn-2 positions are indicated to illustrate the main molecular species derived from each pathway. The head groups are always at the glycerol sn-3 position. The MGD1-DGD1 pathway represents the plastid pathway and contributes to the bulk of galactolipid synthesis at the inner envelope membranes of chloroplasts. These galactolipids are assumed to transport and integrate into the thylakoid. The alternative MGD2/3-DGD2 pathway (extraplastidic pathway) synthesizes galactolipid at the outer envelope membranes using precursor derived from the ER pathway, and these galactolipids are assumed to be transported out and integrated into extraplastidic membrane through unresolved mechanisms. ER-derived PG was assumed to integrate into multiple subcellular membranes. Reactions that are blocked in ads2 mutants are indicated. The arrow illustrates how the ads2 mutation affects carbon flux into different pathways of lipid biosynthesis. The upward-pointing arrow represents higher lipid content, and the downward-pointing arrow represents lower lipid content in ads2 mutant plants compared with the wild type. Dashed arrows represent reactions that have not been experimentally confirmed yet. DAG, diacylglycerol; CDP, cytidine-5′-diphosphate.
It is unlikely that ADS2 encodes a palmitic-CoA Δ7-desaturase, since we would expect that PI(34:2, 34:3), PE(34:1, 34:2, 34:3, 34:4), and PS(34:1, 34:2, 34:3, 34:4) should display reduced content similar to PG(34). The fact that only PG(34:2, 34:3, 34:4) was reduced supports the notion that ADS2 converts PG(34:0)(16:0/18:0-PG) to PG(34:1) (16:1/18:0-PG) in the ER.
Other Biochemical Activities of ADS2 in Planta
Although lipidomic analyses identified MGDG and PG as potential substrates for ADS2 desaturation, it remains possible that there are additional substrates for ADS2 desaturation in planta since the lipid extraction protocol and the detection method was aimed to identify prominent lipid species. Recently, Smith et al. (2013) reported that ADS2 can introduce a double bond at the n-9 position of very-long-chain fatty acids (VLCFAs) 24:0-CoA and 26:0-CoA based upon evidence of seed FA compositional changes and leaf lipid compositional alternations in ads2 mutant plants. Our lipidomic data also revealed differences in lipid species containing 24:1(n-9). Both PE(42:2) and PS(42:2) content in two ads2 mutant lines were significantly higher than the wild type, and PE(42:4) and PS(42:4) content was significantly lower than the wild type regardless of growth temperature (see Supplemental Data Set 2 online). However, VLCFAs, including 24C- and 26C-FAs, are minor components of Arabidopsis leaves. If we assume that ADS2 is only involved in 24:1(n-9) and 26:1(n-9) biosynthesis, the changes in these VLCFAs would not explain the dramatic alteration to 16C- and 18C-FAs in ads2 mutants at 6°C (Table 1). Rather, these data suggest that ADS2 can desaturate FAs with different chain lengths (e.g., 16:0, 18:0, 24:0, and 26:0) but with a stringent carbon position requirement (n-9) relative to the methyl end of the FA. FAME analyses indicated that 18:0 content in ads2 mutants was higher than the wild type with a concomitant decrease in 18:3 content when plants were grown at 6°C, suggesting that ADS2 can also convert 18:0 to 18:1Δ9. Inconsistent with this notion, 18:1 content in two ads2 mutant lines was slightly higher than the wild type when grown at 6°C. However, considering that 18:1 is present at low levels relative to 18:2 and 18:3, any reduced flux from 18:1 to 18:2 or 18:2 to 18:3 collaterally associated with the ADS2 cold stress phenotype could lead to an apparent increase in 18:1 content. Therefore, the mixed results from FAME analyses do not categorically exclude the possibility that ADS2 is also involved in 18:1Δ9 biosynthesis (Table 1).
The evidence obtained from genetic and physiological tests on ads2 mutants clearly demonstrated that ADS2 is involved in cold and freezing tolerance in planta (Figures 2 to 4). Parallel biochemical comparisons between wild-type and ads2 mutant plants also demonstrated that ADS2 plays a significant role in adjusting membrane lipid unsaturation and composition in response to low-temperature stress (Table 1, Figures 5 to 10). The identification of multiple substrates for ADS2 (Smith et al., 2013) further supports this notion. This raises questions as to which lipid species play prominent roles in the low-temperature stress response and how membrane lipid unsaturation and compositional alteration affects membrane properties, which may be important for maintaining proper membrane function under low temperature.
Dual subcellular targeting of proteins and multifunctional enzymes is a widespread phenomenon in diverse biological systems, and enzyme activity could potentially be affected by different metabolic contexts. The observation that ADS2 functions in both the ER and plastids is surprising in view of the marked differences in lipid composition, type of electron donors present, redox state, and pH between these two subcellular compartments. FAD5 remained active when it was redirected to the cytoplasm from the plastid, suggesting that partnering with native electron donors is not essential for its activity (Heilmann et al., 2004a). This might also be the case for ADS2. It is unclear if temperature affects ADS2 substrate preference, subcellular localization, or activity in the ER and plastids, and this needs to be determined to fully understand the functionality of ADS2 in planta.
Contribution of the Prokaryotic and Extraplastidic Pathway to the Biosynthesis of PG and DGDG under Different Temperatures
For plants grown at 6°C, the total PG(32) mol % pool size [PG(32:0) + PG(32:1)] in the wild type, ads2-2, and ads2-3 was 0.15, 0.21, and 0.20% (Figure 5C, right; see Supplemental Data Set 2 online), respectively, so the PG(32) pool is ∼40% higher in two ads2 mutant lines compared with the wild type. This increase in the PG(32) pool size was not seen when plants were grown at 23°C (Figure 5C, left; see Supplemental Table 1 online). These observations suggested that the ADS2 mutation resulted in enhanced PG(32) synthesis in the plastid only when plants were exposed to low temperature.
The total PG(36) pool [= PG(36:1) + PG(36:2) + PG(36:3) + PG(36:4) + PG(36:5) + PG (36:6)] generally showed little difference between wild-type and two ads2 mutant lines regardless of whether plants were grown at 23 or 6°C (Figure 5C; see Supplemental Data Set 2 online), suggesting that the ADS2 mutation did not affect PG(36) biosynthesis. The total PG(34) pool [= (PG(34:0) + PG(34:1) + PG(34:2) + PG(34:3) + PG(34:4)] is ∼10% lower in two ads2 mutant lines compared with the wild type at 6°C; by contrast, at 23°C, the total PG(34) pool in two ads2 mutant lines is only ∼5 to 7% lower than the wild type (Figure 5C; see Supplemental Data Set 2 online). The reduction in total PG(34) content in ads2 mutant plants is likely contributed by the ER pathway, since the 16:1Δ3 FAs in ads2 mutant plants was not reduced, which was present exclusively in the PG(34) synthesized by the plastid pathway (Table 1). These data indicate that a significant portion of PG(34) is synthesized through an ADS2-dependent pathway when plants are exposed to low temperature; blocking PG(34) synthesis in the ER resulted in the enhancement of PG(32) synthesis in the plastid.
When plants were grown at 23°C, leaf total DGDG content in ads2 mutant plants and its subpool DGDG(34), DGDG(36), and DGDG(38) was not significantly affected compared with the wild type (Figures 5A and 6, left; see Supplemental Data Set 2 online); however, at 6°C, leaf total DGDG content in ads2 mutant plants and its subpool DGDG(34) and DGDG(36) was significantly lower than the wild type (Figures 5B and 6, right; see Supplemental Data Set 2 online). Since DGDG(34) can be synthesized by both the plastidic and extraplastidic pathway, we chose DGDG(36) as an example since its precursor is exclusively derived from the ER pathway and synthesized in the outer envelope membrane of plastids. The significant reduction in DGDG(36) in ads2 mutant plants at 6°C suggested that wild-type plants rely more heavily on the ER-derived precursor for DGDG biosynthesis when grown at 6°C than at 23°C. Interestingly, phosphate depletion has also been demonstrated to stimulate DGDG biosynthesis through an ER-derived precursor (Härtel et al., 2000; Kobayashi et al., 2009a), suggesting that lipid remodeling might be a common response to diverse stress conditions and is mediated by differential biosynthesis through multiple lipid synthesis pathways.
Association of FA Desaturation with Cold and Freezing Tolerance
It is generally agreed that membrane lipid unsaturation is associated with cold and freezing tolerance. However, it remains controversial as to which lipid species is most relevant, partly due to the fact that knockout or overexpression of any desaturase often results in comprehensive modification of the lipidome (Somerville, 1995; Penfield, 2008). Mutations for FAD2 and FAD5 desaturases impair plant growth under low temperature, and fad2 mutant plants show a much more severe phenotype than the fad5 mutant under low temperature growth (Kunst et al., 1989; Hugly and Somerville, 1992; Miquel et al., 1993). The FAD2 mutation abolishes 18:1 desaturation in the ER pathway, thus reducing desaturation of ER-derived MGDG and DGDG in the plastid and desaturation of mitochondrial membrane lipids, including PC, PE, DPG, PI, and PG (Caiveau et al., 2001). In this study, we found that the ADS2 mutation resulted in higher 16:0 and 18:0 and lower 16:3 and 18:3 content under 6°C growth conditions, indicating that ads2 mutant plants lose their ability to increase FA desaturation in response to low temperature exposure (Table 1). Since FAD2, ADS2, and FAD5 are involved in lipid desaturation in ER, extraplastid, and plastid pathways, respectively, their mutation logically results in different degrees of growth defects under low temperature (fad2>ads2>fad5). This suggests that membrane lipid composition or desaturation has important roles for cold tolerance in multiple cellular compartments.
Although ads2 mutant plants showed reduced tolerance to both chilling and freezing temperature (Figures 2 and 4), the cold exposure time required for visible phenotypic differentiation is variable. The retarded growth phenotype of ads2 mutant plants at 6°C took more than a 2-week-long exposure (Figure 7A), which is closely associated with changes in 16C- and 18C-FA desaturation (Figure 7B). By contrast, a differential freezing response in ads2 mutant plants than the wild type was observed after 5 d of cold acclimation (Figure 4), a time point that ads2 mutant plants did not show dramatic alternation on its FA desaturation level compared with the wild type (Figure 7B) . These observations suggest that plant cold tolerance is more closely associated with membrane FA desaturation than freezing tolerance. Although several lipid species in ads2 mutant plants showed small, but statistically significant changes compared with the wild type when plants were grown at 23°C (Figure 5A), these differences seem insufficient, since freezing responses between wild-type and ads2 mutant plants are essentially the same for unacclimated plants (Figure 4, left). Since reduced freezing tolerance in ads2 mutant plants developed only after cold acclimation, this suggests that the ADS2 mutation resulted in additional changes beyond FA desaturation during cold acclimation. Alternatively, an unidentified ADS2-derived signaling pathway is required to activate some aspects of cold acclimation, which may be important for the development of freezing tolerance. Smith et al. (2013) reported that some sphingolipid species are significantly altered in ads2 mutant plants, and its unsaturation has been shown to contribute to cold tolerance in Arabidopsis (Chen et al., 2012), raising the possibility that sphingolipids might play a role in the reduced capacity of cold acclimation in ads2 mutant plants. In addition, since PG is the substrate for cardiolipin biosynthesis in mitochondria, reduced PG content in ads2 mutant plants could result in reduced biosynthesis of cardiolipin, which has also been demonstrated to be essential for optimal mitochondrial function and growth under temperature stress (Jiang et al., 1999; Zhong et al., 2004). Characterization of ads2 mutant plants suggests that ADS2 is essential to adjust the acyl unsaturation and composition of organelle membrane lipids in response to cold stress.
METHODS
Plant Materials
T-DNA insertion lines and fad5-1 were ordered from the ABRC. Seeds were sown in a 1:1 mixture of water-saturated vermiculite and peat moss–enriched soil and grown in a growth chamber under long-day conditions (14-h-light/10-h-dark cycle, 23°C day/20°C night, 50% humidity, and light intensity of 200 µmol m−2 s−1] or continuous white light (40 µmol m−2 s−1).
Chilling and Freezing Tolerance Assays
For the chilling tolerance assay, wild-type and mutant seeds were first germinated in potting soil at 23°C under long-day conditions and then moved to a 6°C growth chamber under continuous light conditions (40 µmol m−2 s−1).
The freezing tolerance assay was performed using a modified method from Xin and Browse (1998). Sterilized Arabidopsis thaliana seeds (60 to 120 seeds per line) were sown on Petri dishes containing 0.43% Murashige and Skoog salt, 0.05% MES, 0.011% vitamin B5 mix, and 0.8% agar and were stratified at 6°C in a dark room for 2 d. Seeds were germinated at 25°C under 40 µmol quanta m−2 s−1 continuous white light for 10 d, and then plants were divided into two groups: One group of plants was transferred to a 6°C growth chamber and grown under continuous white light for an additional 5 d for cold acclimation, while another group of plants was used for freezing treatment without prior cold acclimation. Petri dishes with plants were placed on trays filled with ice chips and were moved to a dark growth chamber (Percival) with a temperature setting of −1°C for at least 24 h to equilibrate the temperature evenly. A thermocoupler (Omega) was used to monitor medium temperature. The chamber was cooled at 4°C per h, and dishes of plants were removed at the desired temperature (−4 to −14°C), and kept in a 6°C cold room in the dark for 18 h and then returned to the original growth conditions. Viable plants were counted 2 d later.
Lipidomic and FAME Analysis
Lipid extraction and quantitative profiling were performed according to Devaiah et al. (2006). Unfractionated lipid extracts in chloroform containing appropriate amounts of internal standards were introduced by continuous infusion into the electrospray ionization source of a triple quadrupole mass spectrometer (Applied Biosystems API 4000) with an autosampler (LC Mini PAL; CTC Analytics). The internal standards used for quantification include the following: lysoPG 14:0, lysoPG 18:0, di 14:0 PG, di Phy (20:0)PG, lysoPE 14:0, lysoPE 18:0, di 12:0 PE, di 23:0 PE, lysoPC 13:0, lysoPC 19:0, di 12:0 PC, di 24:1 PC, di14:0 PA, di Phy (20:0)PA, di 14:0 PS, di Phy PS, PI 16:0-18:0, PI di 18:0, DGDG 34:0, DGDG 36:0, MGDG 34:0, and MGDG 36:0. The analyses of MGDG and DGDG were performed in the same solvent mixture as that used for the phospholipid analysis and with different scan parameters for galactolipids. Neutral loss scans in positive ion mode were used to detect the [M + NH4]+ ions of MGDG (NL179.06) and DGDG (NL 341.11). Collision energies, using nitrogen as the collision gas (gas pressure set at 2 arbitrary units) were 21 V for MGDG and 24 V for DGDG. Declustering potentials were 90 V, entrance potentials 10 V, and exit potentials 23 V for both MGDG and DGDG. Data processing was performed in a Lipidome DB Data Calculation Environment (http://lipidome.bcf.ku.edu:9000/Lipidomics) with normalization to internal standards (Zhou et al., 2011).
For FAME analysis, lipids were hydrolyzed and trimethylsilylated. Hydrolysis was performed with 0.5 mL of 1.25 M HCl in methanol and 0.8 mL chloroform at 50°C for 5 h. Following hydrolysis, solvents and HCl were evaporated under nitrogen. Samples were then resuspended in 80 μL pyridine and 20 μL N-methyl-N-TMS-trifluoroacetamide and 1% (w/v) chlorotrimethylsilane and incubated for 1 h at 50°C to produce FAMEs. FAME mixtures were analyzed using a Hewlett Packard 6890 gas chromatograph with a flame ionization detector and 7683 series injector (Agilent). Samples (1 µL) were injected with a split ratio of 30:1 and resolved on a 30 m × 0.25 mm × 0.25-µm DB-23 column (Agilent). Injection temperature was 150°C, the interface was set to 250°C, and the ion source was adjusted to 200°C. The helium flow rate was 1 mL min−1. After a 5-min solvent delay at 150°C, the oven temperature was increased at 2°C min−1 to 200°C and then held at 200°C for 5 min. Data processing was performed using Chemstation software (Agilent).
Subcellular Localization Study
ADS2 cDNA was PCR amplified from a cDNA library by a pair of gene-specific primers; BamHI and NotI restriction enzyme cutting sites were added to forward and reverse primers, respectively. The primer sequences are 5′-CGGGATCCATGTCGGTGACATCAACGGTG-3′ and 5′-ATAAGAATGCGGCCGCACGAACTATAGCCATACGACG-3′.
The PCR products were digested by BamHI and NotI and then inserted into entry vectors PE6c and PE6n, such that an EYFP fluorescence tag was in frame with ADS2 at the C terminus and N terminus, respectively (Dubin et al., 2008). The cassette was then moved into binary vector pSITE-0B by Gateway LR clonase (Chakrabarty et al., 2007). The constructs were transformed into wild-type Arabidopsis by the Agrobacterium tumefaciens–mediated floral dip method (Clough and Bent, 1998). T1 seeds were grown on Murashige and Skoog medium containing 50 µg/mL kanamycin, and the viable green seedlings were further examined under a Leica MZFL III stereomicroscope equipped with epifluorescence excitation and Chroma HiQ ET YFP filters for the detection of EYFP signal. Lines with positive EYFP signal were transferred into soil. Protoplasts were isolated from the leaves of ADS2 transgenic plants by following the method developed by Yoo et al. (2007). Protoplasts were observed by fluorescence microscopy (Olympus BX61), and the EYFP signal was captured using a fluorescein isothiocyanate filter with an excitation wavelength of between 470 and 490 nm and emission wavelength of between 500 and 540 nm; the chloroplast autofluorescence signal was captured using a Cy5 filter with an excitation wavelength of between 600 and 700 nm and emission wavelength of between 650 and 700 nm. The automatic filter switch, sequential image capture, and overlay were controlled by MicroSuit Basic Edition software.
For the colocalization assay, ER (CD3-953 and CD3-954), Golgi (CD3-961 and CD3-962), and plastid (CD3-993 and CD3-994) CFP fusion binary plasmids were ordered from the ABRC (Nelson et al., 2007). The ADS2-EYFP C-terminal fusion construct was mixed with organelle markers individually and cotransformed into tobacco (Nicotiana tabacum) leaves. Transformation was performed by following the procedure described by Sparkes et al. (2006). An Olympus IX70 microscope was controlled by MetaMorph software for image capture (version 6.3 v6; Molecular Devices), and Chroma filters 49001 and 49003 were used for the CFP and EYFP channels, respectively.
For subcellular fractionation, ads1 fad5-1 double mutant leaves were harvested and homogenized, and subcellular protein fractions were prepared as described by Bao et al. (2003). Protein from each fraction was resolved by SDS-PAGE and transferred to polyvinylidene difluoride membranes for antibody probing. The ADS1 peptide that we used to produce anti-ADS1 antibody is 80% identical to the ADS2 sequence, and initial experiments demonstrated that polyclonal ADS1 peptide antibody cross-reacts with ADS2 (Chen et al., 2009) since the ads1 mutant is a complete knockout (see Supplemental Figure 3A online), but a proper size band is still detected with ADS1 antibody (see Supplemental Figure 3B online). Rabbit polyclonal antibody to plastid PDC E1α was used to assay plastid enrichment, and a mouse monoclonal antibody to the ER protein BiP was used to determine ER enrichment (Stressgen). The dilution factors for ADS1, PDC E1α, and BiP antibody are 1000, 2000, and 2000, respectively. Anti-rabbit or anti-mouse IgG-peroxidase antibodies were diluted at 30,000× (Sigma-Aldrich).
Double Mutant Construction
To construct the double mutant of ads2 and fad5-1, the ads2-1 mutant was crossed with fad5-1 (CS206). F1 seeds were germinated and self-pollinated, and the resultant F2 seeds were grown on soil. Individual plants were first genotyped by PCR to identify homozygous mutants for ads2-1, and then one leaf was excised for FAME analysis by gas chromatography (Li et al., 2006). Plants with extremely low levels of 16:1Δ7, 16:2, and 16:3 FAs were regarded as ads2-1 fad5-1 double mutant plants.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At1g06080 (ADS1), At1g06090, At1g06100, At1g06120, At1g06350, At1g06360, At2g31360 (ADS2), At3g15850 (ADS3), and At3g15870. Accession numbers for T-DNA lines are SALK_079963C (ads2-1), SALK_016783C (ads2-2), CS817934 (ads2-3), and CS873338 (ads2-4).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phylogenic Analysis and Sequence Alignment of Nine Acyl-Lipid Desaturase Genes in Arabidopsis.
Supplemental Figure 2. ADS2 Protein Partially Colocalize with Chloroplast Marker.
Supplemental Figure 3. ADS1 Peptide Antibody Cross Reacts with ADS2 Protein.
Supplemental Data Set 1. Sequences Used to Generate the Phylogeny Presented in Supplemental Figure 1A.
Supplemental Data Set 2. Lipidomic Profiling in Wild-Type ads2-2, ads2-3, and fad5-1 Mutant Plants.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Stock Center for providing T-DNA insertion lines and organelle marker constructs, the Kansas Lipidomics Research Center for lipid profiling, and Carlotta Woods, Kirby Swatek, and Rashaun Wilson for editing the article. Financial support was provided by National Science Foundation Plant Genome Research Program Grant DBI-0332418.
AUTHOR CONTRIBUTIONS
J.J.T. contributed to experimental design, data analysis, and article writing. M.C. contributed to experimental design, experiment execution, data analysis, and article writing.
Glossary
- FA
fatty acid
- htm-PG
high-temperature melting phosphatidylglycerol
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PI
phosphatidylinositol
- MGDG
monogalactosyl diacylglycerol
- DGDG
digalactosyl diacylglycerol
- PG
phosphatidylglycerol
- FAME
fatty acid methyl ester
- LPE
lysophosphatidylethanolamine
- PS
phosphatidylserine
- LPC
lysophosphatidylcholine
- PA
phosphatidic acid
- ER
endoplasmic reticulum
- EYFP
enhanced yellow fluorescence tag protein
- CFP
cyan fluorescent protein
- VLCFA
very-long-chain fatty acid
References
- Andersson M.X., Larsson K.E., Tjellström H., Liljenberg C., Sandelius A.S. (2005). Phosphate-limited oat. The plasma membrane and the tonoplast as major targets for phospholipid-to-glycolipid replacement and stimulation of phospholipases in the plasma membrane. J. Biol. Chem. 280: 27578–27586 [DOI] [PubMed] [Google Scholar]
- Andersson M.X., Stridh M.H., Larsson K.E., Liljenberg C., Sandelius A.S. (2003). Phosphate-deficient oat replaces a major portion of the plasma membrane phospholipids with the galactolipid digalactosyldiacylglycerol. FEBS Lett. 537: 128–132 [DOI] [PubMed] [Google Scholar]
- Awai K., Maréchal E., Block M.A., Brun D., Masuda T., Shimada H., Takamiya K., Ohta H., Joyard J. (2001). Two types of MGDG synthase genes, found widely in both 16:3 and 18:3 plants, differentially mediate galactolipid syntheses in photosynthetic and nonphotosynthetic tissues in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 98: 10960–10965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X., Thelen J.J., Bonaventure G., Ohlrogge J.B. (2003). Characterization of cyclopropane fatty-acid synthase from Sterculia foetida. J. Biol. Chem. 278: 12846–12853 [DOI] [PubMed] [Google Scholar]
- Browse J., McCourt P., Somerville C.R. (1985). A mutant of Arabidopsis lacking a chloroplast-specific lipid. Science 227: 763–765 [DOI] [PubMed] [Google Scholar]
- Browse J., Somerville C. (1991). Glycerolipid synthesis: Biochemistry and regulation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42: 467–506 [Google Scholar]
- Browse J., Warwick N., Somerville C.R., Slack C.R. (1986). Fluxes through the prokaryotic and eukaryotic pathways of lipid synthesis in the ‘16:3’ plant Arabidopsis thaliana. Biochem. J. 235: 25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiveau O., Fortune D., Cantrel C., Zachowski A., Moreau F. (2001). Consequences of ω -6-oleate desaturase deficiency on lipid dynamics and functional properties of mitochondrial membranes of Arabidopsis thaliana. J. Biol. Chem. 276: 5788–5794 [DOI] [PubMed] [Google Scholar]
- Chakrabarty R., Banerjee R., Chung S.M., Farman M., Citovsky V., Hogenhout S.A., Tzfira T., Goodin M. (2007). PSITE vectors for stable integration or transient expression of autofluorescent protein fusions in plants: Probing Nicotiana benthamiana-virus interactions. Mol. Plant Microbe Interact. 20: 740–750 [DOI] [PubMed] [Google Scholar]
- Chen M., Markham J.E., Cahoon E.B. (2012). Sphingolipid Δ8 unsaturation is important for glucosylceramide biosynthesis and low-temperature performance in Arabidopsis. Plant J. 69: 769–781 [DOI] [PubMed] [Google Scholar]
- Chen M.J., Mooney B.P., Hajduch M., Joshi T., Zhou M.Y., Xu D., Thelen J.J. (2009). System analysis of an Arabidopsis mutant altered in de novo fatty acid synthesis reveals diverse changes in seed composition and metabolism. Plant Physiol. 150: 27–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Devaiah S.P., Roth M.R., Baughman E., Li M., Tamura P., Jeannotte R., Welti R., Wang X. (2006). Quantitative profiling of polar glycerolipid species from organs of wild-type Arabidopsis and a phospholipase Dalpha1 knockout mutant. Phytochemistry 67: 1907–1924 [DOI] [PubMed] [Google Scholar]
- Dubin M.J., Bowler C., Benvenuto G. (2008). A modified Gateway cloning strategy for overexpression tagged proteins in plants. Plant Methods 4: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edman K., Ericson I. (1987). Phospholipid and fatty acid composition in mitochondria from spinach (Spinacia oleracea) leaves and petioles. A comparative study. Biochem. J. 243: 575–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi-Mizutani M., Savin K., Cornish E., Tanaka Y., Ashikari T., Kusumi T., Murata N. (1995). Senescence-induced expression of a homologue of delta 9 desaturase in rose petals. Plant Mol. Biol. 29: 627–635 [DOI] [PubMed] [Google Scholar]
- Fukuchi-Mizutani M., Tasaka Y., Tanaka Y., Ashikari T., Kusumi T., Murata N. (1998). Characterization of delta 9 acyl-lipid desaturase homologues from Arabidopsis thaliana. Plant Cell Physiol. 39: 247–253 [DOI] [PubMed] [Google Scholar]
- Gao J., Ajjawi I., Manoli A., Sawin A., Xu C., Froehlich J.E., Last R.L., Benning C. (2009). FATTY ACID DESATURASE4 of Arabidopsis encodes a protein distinct from characterized fatty acid desaturases. Plant J. 60: 832–839 [DOI] [PubMed] [Google Scholar]
- Gerloff E.D., Richardson T., Stahmann M.A. (1966). Changes in fatty acids of alfalfa roots during cold hardening. Plant Physiol. 41: 1280–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy C.L. (1990). Cold acclimation and freezing stress tolerance: Role of protein metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 41: 187–223 [Google Scholar]
- Härtel H., Dörmann P., Benning C. (2000). DGD1-independent biosynthesis of extraplastidic galactolipids after phosphate deprivation in Arabidopsis. Proc. Natl. Acad. Sci. USA 97: 10649–10654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heber, U., and Santarius, K.A. (1973). Cell death by cold and heat and resistance to extreme temperatures. Mechanisms of hardening and dehardening. In Temperature and Life, H. Precht, J. Christophersen, H. Henseel, and W. Larcher, eds (Berlin: Springer-Verlag), pp. 232–263. [Google Scholar]
- Heilmann I., Mekhedov S., King B., Browse J., Shanklin J. (2004a). Identification of the Arabidopsis palmitoyl-monogalactosyldiacylglycerol delta7-desaturase gene FAD5, and effects of plastidial retargeting of Arabidopsis desaturases on the fad5 mutant phenotype. Plant Physiol. 136: 4237–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann I., Pidkowich M.S., Girke T., Shanklin J. (2004b). Switching desaturase enzyme specificity by alternate subcellular targeting. Proc. Natl. Acad. Sci. USA 101: 10266–10271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugly S., Somerville C. (1992). A role for membrane lipid polyunsaturation in chloroplast biogenesis at low temperature. Plant Physiol. 99: 197–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F., Gu Z., Granger J.M., Greenberg M.L. (1999). Cardiolipin synthase expression is essential for growth at elevated temperature and is regulated by factors affecting mitochondrial development. Mol. Microbiol. 31: 373–379 [DOI] [PubMed] [Google Scholar]
- Jouhet J., Maréchal E., Baldan B., Bligny R., Joyard J., Block M.A. (2004). Phosphate deprivation induces transfer of DGDG galactolipid from chloroplast to mitochondria. J. Cell Biol. 167: 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Awai K., Nakamura M., Nagatani A., Masuda T., Ohta H. (2009a). Type-B monogalactosyldiacylglycerol synthases are involved in phosphate starvation-induced lipid remodeling, and are crucial for low-phosphate adaptation. Plant J. 57: 322–331 [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Nakamura Y., Ohta H. (2009b). Type A and type B monogalactosyldiacylglycerol synthases are spatially and functionally separated in the plastids of higher plants. Plant Physiol. Biochem. 47: 518–525 [DOI] [PubMed] [Google Scholar]
- Kodama H., Hamada T., Horiguchi G., Nishimura M., Iba K. (1994). Genetic enhancement of cold tolerance by expression of a gene for chloroplast ω-3 fatty acid desaturase in transgenic tobacco. Plant Physiol. 105: 601–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper P.J.C. (1970). Lipids in alfalfa leaves in relation to cold hardiness. Plant Physiol. 45: 684–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst L., Browse J., Somerville C. (1989). A mutant of Arabidopsis deficient in desaturation of palmitic acid in leaf lipids. Plant Physiol. 90: 943–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.H., Beisson F., Pollard M., Ohlrogge J. (2006). Oil content of Arabidopsis seeds: The influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67: 904–915 [DOI] [PubMed] [Google Scholar]
- Lyons J.M. (1973). Chilling injury in plants. Annu. Rev. Plant Physiol. 24: 445–466 [Google Scholar]
- Lyons J.M., Wheaton T.A., Pratt H.K. (1964). Relationship between the physical nature of mitochondrial membranes and chilling sensitivity in plants. Plant Physiol. 39: 262–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K., Sakuma Y., Kasuga M., Ito Y., Seki M., Goda H., Shimada Y., Yoshida S., Shinozaki K., Yamaguchi-Shinozaki K. (2004). Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J. 38: 982–993 [DOI] [PubMed] [Google Scholar]
- Maximov N.A. (1912). Chemische Schutzmittel der Pflanzen gegan Erfrieren. Ber. Dtsch. Bot. Ges. 30: 52–56 [Google Scholar]
- Mekhedov S., de Ilárduya O.M., Ohlrogge J. (2000). Toward a functional catalog of the plant genome. A survey of genes for lipid biosynthesis. Plant Physiol. 122: 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel M., Browse J. (1992). Arabidopsis mutants deficient in polyunsaturated fatty acid synthesis. Biochemical and genetic characterization of a plant oleoyl-phosphatidylcholine desaturase. J. Biol. Chem. 267: 1502–1509 [PubMed] [Google Scholar]
- Miquel M., James D., Jr, Dooner H., Browse J. (1993). Arabidopsis requires polyunsaturated lipids for low-temperature survival. Proc. Natl. Acad. Sci. USA 90: 6208–6212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata N. (1982). Molecular species composition of phosphatidylglycerols from chilling-sensitive and chilling-resistant plants. Plant Cell Physiol. 24: 81–86 [Google Scholar]
- Murata N., Sato N., Takahashi N., Hamazaki Y. (1982). Compositions and positional distributions of fatty acids in phospholipids from leaves of chilling sensitive and chilling-resistant plants. Plant Cell Physiol. 23: 1071–1079 [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Oono Y., Seki M., Satou M., Iida K., Akiyama K., Sakurai T., Fujita M., Yamaguchi-Shinozaki K., Shinozaki K. (2006). Monitoring expression profiles of Arabidopsis genes during cold acclimation and deacclimation using DNA microarrays. Funct. Integr. Genomics 6: 212–234 [DOI] [PubMed] [Google Scholar]
- Penfield S. (2008). Temperature perception and signal transduction in plants. New Phytol. 179: 615–628 [DOI] [PubMed] [Google Scholar]
- Robinson S.J., Parkin I.A. (2008). Differential SAGE analysis in Arabidopsis uncovers increased transcriptome complexity in response to low temperature. BMC Genomics 9: 434–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satake, T., and Koike, S. (1983). Sterility caused by cooling treatment at the flowering stage in rice plants. Jpn. J. Crop Sci. 52: 207–213.
- Schwertner H.A., Biale J.B. (1973). Lipid composition of plant mitochondria and of chloroplasts. J. Lipid Res. 14: 235–242 [PubMed] [Google Scholar]
- Smith M.A., Dauk M., Ramadan H., Yang H., Seamons L.E., Haslam R.P., Beaudoin F., Ramirez-Erosa I., Forseille L. (2013). Involvement of Arabidopsis ACYL-COENZYME A DESATURASE-LIKE2 (At2g31360) in the biosynthesis of the very-long-chain monounsaturated fatty acid components of membrane lipids. Plant Physiol. 161: 81–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C. (1995). Direct tests of the role of membrane lipid composition in low-temperature-induced photoinhibition and chilling sensitivity in plants and cyanobacteria. Proc. Natl. Acad. Sci. USA 92: 6215–6218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I.A., Runions J., Kearns A., Hawes C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1: 2019–2025 [DOI] [PubMed] [Google Scholar]
- Steponkus P.L., Dowgert M.F., Gordon-Kamm W.J. (1983). Destabilization of the plasma membrane of isolated plant protoplasts during a freeze-thaw cycle: The influence of cold acclimation. Cryobiology 20: 448–465 [DOI] [PubMed] [Google Scholar]
- Thomashow M.F. (1990). Molecular genetics of cold acclimation in higher plants. Adv. Genet. 28: 99–131 [Google Scholar]
- Villarejo A., et al. (2005). Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat. Cell Biol. 7: 1224–1231 [DOI] [PubMed] [Google Scholar]
- Vogel J.T., Zarka D.G., Van Buskirk H.A., Fowler S.G., Thomashow M.F. (2005). Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 41: 195–211 [DOI] [PubMed] [Google Scholar]
- Wintermans J.F.G.M. (1960). Concentrations of phosphatides and glycolipids in leaves and chloroplasts. Biochim. Biophys. Acta 44: 49–54 [DOI] [PubMed] [Google Scholar]
- Wolter F.P., Schmidt R., Heinz E. (1992). Chilling sensitivity of Arabidopsis thaliana with genetically engineered membrane lipids. EMBO J. 11: 4685–4692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Browse J. (1995). Elevated levels of high-melting-point phosphatidylglycerols do not induce chilling sensitivity in an Arabidopsis mutant. Plant Cell 7: 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Lightner J., Warwick N., Browse J. (1997). Low-temperature damage and subsequent recovery of fab1 mutant Arabidopsis exposed to 2°C. Plant Physiol. 113: 347–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Z., Browse J. (1998). Eskimo1 mutants of Arabidopsis are constitutively freezing-tolerant. Proc. Natl. Acad. Sci. USA 95: 7799–7804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yoshida S., Uemura M. (1986). Lipid composition of plasma membranes and tonoplasts isolated from etiolated seedlings of mung bean (Vigna radiata L.). Plant Physiol. 82: 807–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q., Gohil V.M., Ma L., Greenberg M.L. (2004). Absence of cardiolipin results in temperature sensitivity, respiratory defects, and mitochondrial DNA instability independent of pet56. J. Biol. Chem. 279: 32294–32300 [DOI] [PubMed] [Google Scholar]
- Zhou Z., Marepally S.R., Nune D.S., Pallakollu P., Ragan G., Roth M.R., Wang L., Lushington G.H., Visvanathan M., Welti R. (2011). LipidomeDB data calculation environment: Online processing of direct-infusion mass spectral data for lipid profiles. Lipids 46: 879–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.