Abstract
Protein structure is highly diverse when considering a wide range of protein types, helping to give rise to the multitude of functions that proteins perform. In particular, certain proteins are known to adopt a knotted or slipknotted fold. How such proteins undergo mechanical unfolding was investigated utilizing a combination of single molecule atomic force microscopy (AFM), protein engineering and steered molecular dynamics (SMD) simulations to show the mechanical unfolding mechanism of the slipknotted protein AFV3-109. Our results reveal that the mechancial unfolding of AFV3-109 can proceed via multiple parallel unfolding pathways that all cause the protein slipknot to untie, and the polypeptide chain to completely extend. These distinct unfolding pathways proceed either via a two-state or three-state unfolding process involving the formation of a well-defined, stable intermediate state. SMD simulations predict the same contour length increments for different unfolding pathways as single molecule AFM results, thus provding a plausible molecular mechanism for the mechanical unfolding of AFV3-109. These SMD simulations also reveal that two-state unfolding is initiated from both the N- and C-termini, while three-state unfolding is initiated only from the C-terminus. In both pathways, the protein slipknot was untied during unfolding, and no tightened slipknot conformation observed. Detailed analysis revealed that interactions between key structural elements lock the knotting loop in place, preventing it from shrinking and the formation of a tightened slipknot conformation. Our results demonstrate the bifurcation of the mechancial unfolding pathway of AFV3-109, and point to the generality of a kinetic partitioning mechanism for protein folding/unfolding.
Introduction
Over the last two decades, theoretical and experimental work has significantly advanced our understanding of how proteins fold1-6. Proteins navigate themselves on a high dimensional funnel-shaped energy landscape to achieve robust and fast folding. Most proteins do not possess complex topology, where unfolding most proteins from its N- and C-termini will simply result in a simple linear polypeptide chain7. However, a small proportion of proteins have been observed to exhibit complex topologies involving the formation of knots and slipknots8. Upon being stretched from N- and C-termini, such proteins would form a tightened knot instead of a linear polypeptide chain. Despite their complex topology, these knotted or slipknotted proteins can spontaneously fold into their native conformations in order to carry out their designated biological functions9. Understanding how a linear polypeptide chain is able to overcome the topological difficulty inherent within these proteins to fold into such complex topologies has attracted considerable research interest over the last few years. Both experimental and theoretical work has been used to shed light on the folding mechanism of such proteins9-19; in particular, computational studies have provided certain mechanistic insights into the folding mechanisms of these proteins. For example, the formation of a slipknot conformation was suggested as a key intermediate state to reduce the topological complexity during the spontaneous folding of a knotted protein9,19, revealing the importance of the slipknot conformation in the folding of knotted proteins.
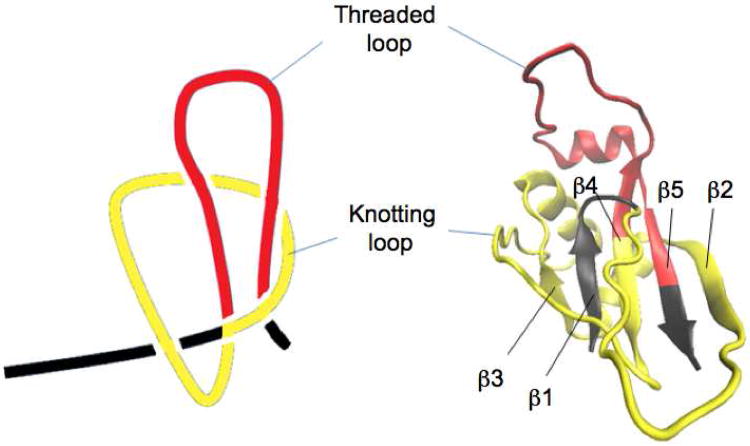
Technically, a slipknot is not a real knot. In a slipknot, the end of the polypeptide chain is threaded back after being threaded through the knotting loop, forming a threaded loop akin to a shoelace (Fig. 1). In this case, pulling on the N- and C-termini of a slipknotted protein will untie the slipknot and lead to a fully extended linear polypeptide chain. Due to its structural importance and its role in the folding of knotted proteins, we combined single molecule force spectroscopy and steered molecular dynamics (SMD) simulations to investigate the unfolding mechanism of the slipknotted protein AFV3-109. AFV3-109 is a protein from Acidianus Filamentous Virus 3 with unknown biological function, although structural analysis of AFV3-109 suggests that it could interact with nucleic acids20. Among the known slipknotted proteins, AFV3-109 has a relatively simple structure. As shown in Fig. 1, the core of AFV3-109 is a β-sheet with five β-strands connected by helices and loops20. A clear slipknot conformation is present in AFV3-109 with a knotting loop (colored in yellow, Fig. 1) near the N- terminus and a threaded loop (colored in red, Fig. 1) near the C- terminus. As the “reverse” process of folding, the unfolding of slipknotted proteins will involve untying the slipknot. Understanding this molecular mechanism of untying a slipknot will help to understand how the protein folded in the first place. Applying a mechanical stretching force to the slipknotted protein, either through experimental single molecule force spectroscopy experiments or SMD simulations, is an ideal approach to investigating this intricate process.
Figure 1.
The three-dimensional structure of AFV3-109 (PDB code: 2J6B). AFV3-109 is an α/β protein. A five stranded β-sheet connected by helices and loops forms the core of AFV3-109. The order of the five strands is β3β1β4β5β2, while β5 is anti-parallel to the others. The knotting loop (colored in yellow) encompasses amino acid residues 8 to 77, including β2, α1, β3, α2 and part of β4. Amino acid residues from 78 to 106, including part of β4, α3 and β5, are threaded through the knotting loop twice and form a threaded loop (colored in red).
Single-molecule force spectroscopy has evolved into a powerful tool to probe the mechanical unfolding and folding dynamics of proteins at the single molecule level 2,21-26. SMD simulations complement single molecular force spectroscopy experiments and provide invaluable insights into describing atomic level molecular events that occur during the mechanical unfolding process, while helping to guide new experimental designs24-30. Both single molecule AFM and simulation have been used to stretch knotted proteins in order to study their mechanical unfolding mechanism. These simulations suggest that the presence of a knot and slipknot in protein structures stabilizes the protein, and that the stretching of slipknotted proteins may involve the formation of a intermediate state comprising a temporarily tightened slipknot during the mechanical unfolding process16. Single molecule AFM experiments showed that the mechanical stretching of phytochrome, a protein with a figure-eight knot, leads to a tightened knot that involves 17 residues15. Here we apply a stretching force to the N- and C-termini of the slipknotted protein AFV3-109 in order to investigate its mechanical unfolding and folding mechanism. Due to its slipknot structure, we believe that AFV3-109 can be unfolded and untied just as a shoelace is untied.
Our investigations using single molecule AFM clearly show that the mechanical unfolding of AFV3-109 unties the slipknot structure, leading to a fully extended polypeptide chain. The mechanical untying of the slipknot can occur through three distinct mechanical unfolding pathways: one in a simple two-state fashion, and the other two in a three-state fashion that involves the formation of a well-defined intermediate state. SMD simulations of the mechanical unfolding of AFV3-109 reveals a similar bifurcation phenomenon, providing a plausible molecular explanation of the mechanical unfolding process observed in single molecule AFM experiments. These simulations show that the two-state unfolding process initiates simultaneously from the N- and C-termini, while the three-state unfolding process initiates only from the C-terminus. In the three-state unfolding mechanism, the formation of the unfolding intermediate results from the unraveling of the C-terminal threaded β-hairpin loop, while final unfolding corresponds to a complete unraveling of the intermediate state. The agreement between single molecule AFM and SMD simulations results provides a detailed molecular picture of how the slipknotted protein AFV3-109 unfolds; these results contribute towards a clearer understanding of how slipknotted and knotted proteins unfold and fold.
Materials and Methods
Protein Engineering
The protein constructs were built according to recombinant strategies described previously31. The plasmid containing the gene encoding AFV3-109 and restriction sites (5′ BamHI and 3′ KpnI and BglII) was purchased from GeneScript. We used two alternating digestion and ligation steps to construct the gene encoding (GB1)4-AFV3-109-(GB1)4. By digesting the pUC57/AFV3-109 plasmid with the restriction enzymes BamHI and KpnI, we obtained the AFV3-109 insert containing “sticky ends”. The AFV3-109 insert was subsequently ligated by T4 DNA Ligase to the corresponding “sticky ends” of the pQE80L/(GB1)4 vector, which was obtained by digesting the pQE80L/(GB1)4 plasmid with BglII and KpnI. From this ligation, we obtained the pQE80L/(GB1)4-AFV3-109 plasmid, which was digested with BglII and KpnI to get the pQE80L/(GB1)4-AFV3-109 vector. By digesting the pQE80L/(GB1)4 plasmid with BamHI and KpnI, we obtained a (GB1)4 insert, which was ligated to the pQE80L/(GB1)4-AFV3-109 vector in order to construct the final pQE80L/(GB1)4-AFV3-109-(GB1)4 plasmid. To get the pQE80L/Cys-GB1-AFV3-109-Cys plasmid, we digested the pQE80L/Cys-GB1 plasmid with BglII and KpnI to obtain the pQE80L/Cys-GB1 vector, to which the AFV3-109 insert with a cysteine at the C- terminus was ligated.
Proteins were over-expressed in a DH5α strain, purified by using Co2+ affinity chromatography and stored at 4°C in a phosphate-buffered saline (PBS) solution at a protein concentration of ∼1.0 mg/mL. Cysteines at both the N- and C- termini of the heterodimer protein Cys-GB1-AFV3-109-Cys were introduced to react with the intermolecular cross-linker 1,8-Bis-Maleimidotriethyleneglycol, referred as BM(PEO)3. By forming a thioether bond between thiol groups in the heterodimer protein and maleimide groups in BM(PEO)3, polyproteins (GB1-AFV3-109)n were formed32.
Single Molecule Force Spectroscopy
Single-molecule force spectroscopy experiments were carried out on a custom-built AFM31. Before each experiment, we calibrated the spring constant (which ranged from 50 to 80 pN/nm) of each individual AFM cantilever (Si3N4 cantilevers from Vecco) using the equipartition theorem33. In a typical AFM experiment, we deposited ∼1.0 μl of protein solution in PBS (1.0 μg/ml) onto a clean glass coverslip covered by PBS buffer (∼50 μl) and allowed the protein to adsorb onto the substrate for ∼10 min before the force spectroscopy experiment.
SMD Simulations
Both constant velocity and constant force SMD simulations of mechanical unfolding of AFV3-109 were carried out. The coordination file for the AVF3-109 slipknot protein was obtained from the PDB (pdb code: 2J6B) and a protein structure file (psf) created using the modeling software Visual Molecular Dynamics (VMD)34 and the psfgen plug-in. The protein was solvated in an explicit solvent environment. The CHARMM 35 force field topology parameters were used for the protein and water was considered using the TIP3P model36. A total of three protein-water systems were created containing 45589, 82903, 136947 atoms. Solvent boxes with length, width and height dimensions of (100Å, 67Å, 72Å), (240Å, 60Å,60Å) and (405Å, 60Å, 60Å) were utilized. The system was energy minimized and equilibrated, and the resulting coordinates and velocities used as starting point for constant velocity and constant force SMD simulations. The simulations were performed with periodic boundary conditions in the isobaric-isothermic (NPT) ensemble; electrostatic interactions were computed by the PME (Particle Mesh Ewald) method37, non-bonded interactions were treated with a cutoff using a switching function beginning at 10Å and reaching zero at 14Å, and the uniform dielectric constant was set to 1. Pulling velocities used for the constant velocity simulations ranged from 0.0025 Å/ps to 0.5 Å/ps and applied forces in the constant force simulations were 500 pN and 1 nN. The spring constant used in SMD simulations was 7 kcal*mol−1*Å−2 (or 486 pN/Å). The simulation program was NAMD38. Data extraction and analysis were performed in VMD and MATLAB.
Results
The Slipknot in AFV3-109 can be Mechanically Untied
To investigate the mechanical unfolding of AFV3-109 using single molecule AFM, we constructed two different AFV3-109-containing polyprotein chimeras (GB1)4-AFV3-109-(GB1)4 and (GB1-AFV3-109)n. In both polyprotein chimeras, mechanically well characterized GB1 domains are used as fingerprint domains to facilitate the identification of single molecule stretching events and discerning the mechanical unfolding signatures of AFV3-109, as GB1 unfolds at a characteristic force of ∼180 pN with a contour length increment (ΔLC) of 18 nm at a pulling speed of 400 nm/s31.
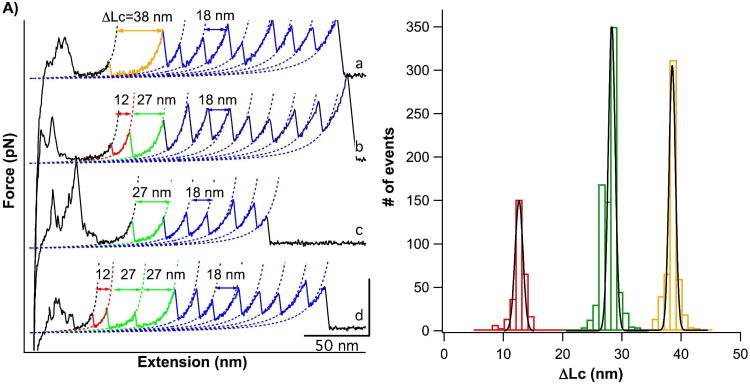
Stretching the polyprotein (GB1)4-AFV3-109-(GB1)4 yielded force-extension curves with a characteristic sawtooth-like pattern, where the force peaks correspond to the mechanical unfolding of individual domains in the polyprotein chain, and the last peak generally corresponds to the stretching and subsequent detachment of the unfolded polypeptide chain. Fig. 2A shows representative curves from the unfolding of the (GB1)4-AFV3-109-(GB1)4 polyprotein. The unfolding of GB1 fingerprint domains (colored in blue) can be readily identified by their unique unfolding signatures: unfolding force of ∼180 pN and ΔLC of ∼18 nm measured by fitting the Worm-like Chain model of polymer elasticity to consecutive unfolding force peaks39,40. As AFV3-109 is flanked by two (GB1)4 at both N- and C-termini, the observation of five or more GB1 unfolding events in a force-extension curve ensures that the unfolding signatures of AFV3-109 are included within the force-extension curve. Therefore, unfolding force peak(s) occurring before GB1 domain unfolding can be attributed to the mechanical unfolding of the AFV3-109 domain within the polyprotein chain.
Figure 2.
The mechanical unfolding of AFV3-109. A) Force-extension curves of (GB1)4-AFV3-109-(GB1)4. Dotted lines are WLC fits to the force-extension curves. The unfolding events of the GB1 fingerprint domains are characterized by a ΔLC of ∼18 nm (colored in blue). The mechanical unfolding of AFV3-109 can proceed through multiple pathways: in curve a), the unfolding of AFV3-109 is an apparent two-state unfolding event with a ΔLC of 38 nm; in curve b), the unfolding of AFV3-109 proceeds in a three-state unfolding pathway involving a stable intermediate state, with a ΔLC(N-I) of 12 nm and a ΔLC(I-U) of 27 nm. In some cases, the N-I unfolding event was not observed, and only the I-U unfolding was observed (curve c). For clarity, the two-state unfolding event is colored in orange, the N-I event is colored in red and the I-U event is colored in green. B) ΔLC histograms for mechanical unfolding events of AFV3-109. Gaussian fits to the experimental data shows a ΔLc of 12.0 ± 1.2nm (n=298), 27.3 ± 1.3nm (n=790), and 37.9 ± 1.1nm (n=521).
In curve a) (Fig. 2A), the unfolding of AFV3-109 occurs in an apparent two-state fashion with a ΔLC of ∼38 nm (colored in orange). As there are 109 residues in AFV3-109, the contour length of the unfolded and fully extended AFV3-109 is 39.2 nm when there is no knot or tightened slipknot. The distance between the N- and C- termini of AFV3-109 in the native state, L0, is 1.2 nm (PDB ID code 2j6b). Therefore, the complete unfolding of AFV3-109, from its slipknotted structure to a fully extended polypeptide chain, would result in a ΔLC of ∼38 nm, which is in excellent agreement with the experimentally measured ΔLC. This result suggests that upon stretching, AFV3-109 can be mechanically unraveled and the slipknot structure is fully untied in a manner akin to how a shoelace is untied.
Untying the Slipknot in Multiple Ways: Bifurcation of the Mechanical Unfolding Pathways of AFV3-109
In addition to two-state unfolding, we also observed that the mechanical unfolding of AFV3-109 can proceed in different pathways. Curve b) (Fig. 2A) shows an example in which the mechanical unfolding of AFV3-109 proceeds in a three-state fashion, with the formation of an intermediate state that gives rise to two unfolding force peaks for one AFV3-109 domain. The first unfolding force peak of the three-state unfolding occurs at ∼ 90 pN with a ΔLC(N→I) of ∼12 nm, corresponding to the transition from the native state to a unfolding intermediate state (N→I) (colored in red). The second step occurs at ∼100 pN with Δ LC(I→U) of ∼27 nm, corresponding to unfolding of the intermediate state (I→U) (colored in green). The sum of the two ΔLC is 39 nm, which is in agreement with the predicted ΔLC of the complete unfolding of AFV3-109. This result again suggests that AFV3-109 has been completely unraveled and the slipknot has been untied. It is of note that the unfolding intermediate state is mechanically stable and unfolds at ∼100 pN.
In addition to two-state and three-state unfolding, we also observed AFV3-109 unfolding events that show a Δ LC(I→U) of only ∼27 nm (curves c and d), suggesting that the unfolding of AFV3-109 in this pathway begins from the same unfolding intermediate state as in the three-state unfolding pathway. It is likely that this unfolding scenario is a special case of the three-state unfolding pathway, in which part of AFV3-109 in the native state is unfolded prior to stretching, or is unfolded at forces that are below our 20 pN detection limit.
The ΔLC histogram of all mechanical AFV3-109 unfolding events displays three well-defined peaks (Fig. 2B). Fitting this data using a Gaussian fit shows three ΔLC of 12.0 ± 1.2nm (n = 298), 27.3 ± 1.3nm (n = 790), and 37.9 ± 1.1nm (n = 521), corresponding to the contour length increments deriving from the N→I, I→U and N→U unfolding steps.
These differences in unfolding behavior clearly indicate that the mechanical unfolding of AFV3-109 can proceed through multiple unfolding pathways, specifically, that such pathways are bifurcated. Since AFV3-109 is completely unfolded in all three pathways, our results suggest that there are multiple ways to untie the slipknot and fully extend AFV3-109. The three distinct unfolding pathways occur roughly at a frequency of 20 (N→I→U):40 (I→U):40 (N→U) (Fig. 2B), revealing the kinetic partitioning that occurs as AFV3-109 is mechanically unfolded.
The Mechanical Unfolding of AFV3-109 Does Not Involve the Formation of a Tightened Slipknot
Coarse grained MD simulations16 on slipknot protein thymidine kinase suggested that during the mechanical unfolding of a slipknot protein, a high stretching force could result in molecular jamming and lead to the formation of a tightened slipknot as an unfolding intermediate state. This possibility raises the question whether the unfolding intermediate state (with a Δ LC(I→U) of 27 nm) observed in our AFM experiments on AFV3-109 could correspond to a tightened slipknot conformation.
If a tightened slipknot could form in the mechanical unfolding of AFV3-109, ΔLC(I→U) of 27 nm would correspond to the length increment due to untying the tightened slipknot, which should be the sum of the length of the polypeptide chain trapped in the tightened slipknot and the size of the tightened slipknot itself. The polypeptide chain trapped in the tightened slipknot would include residues 73 to 106 (β4, α3 and part of β5) and would be ∼12 nm long. Thus, the size of the tightened slipknot would be ∼15 nm (27nm -12 nm). Considering the complexity of this tightened slipknot and tightened figure-eight knot (Fig. S4), the size of a tightened slipknot should be no bigger than a tightened figure-eight knot, which has been measured to be 6. 2 nm using single molecule AFM and SMD simulations15. Therefore, 15 nm is unphysically large for a tightened slipknot conformation. This result strongly suggests that the unfolding intermediate state of AFV3-109 observed in our single molecule AFM experiments does not correspond to the tightened slipknot and the unfolding of AFV3-109 does not involve the formation of a tightened slipknot conformation.
The Fusion of (GB1)4 to the C-Terminus of AFV3-109 Does Not Impede the Folding of AFV3-109
In the slipknot structure of AFV3-109, the threaded loop is at the C-terminus. Since the (GB1)4-AFV3-109-(GB1)4 polyprotein is expressed as a continuous polypeptide chain, it is unknown whether the fusion of (GB1)4 at the C-terminus of AFV3-109 interferes with the intrinsic folding of AFV3-109. To ensure that the fusion of (GB1)4 to the C-terminus of AFV3-109 does not adversely affect the intrinsic folding of AFV3-109, we constructed the polyprotein (GB1-AFV3-109)n based on the polymerization of Cys-GB1-AFV3-109-Cys using thiol-maleimide coupling chemistry (see Materials and Methods)32. In this construct, the C-terminus of AFV3-109 is free, and the folding of AFV3-109 should not be affected as the polyprotein (GB1-AFV3-109)n is formed only after AFV3-109 has folded properly.
Force-extension curves of (GB1-AFV3-109)n are shown in Fig. S1 (Supporting Information). We observed that the unfolding signatures exhibited by AFV3-109 in (GB1-AFV3-109)n are identical to those observed in the original (GB1)4-AFV3-109-(GB1)4 polyprotein; specifically, that multiple unfolding pathways, the same average unfolding force and contour length increment are all demonstrated. In addition, we noticed that in force-extension curves containing more than one unfolding event of AFV3-109-109, an N→I unfolding event is not necessarily followed by an I→U unfolding event. The N→I unfolding of a different AFV3-109 domain can occur between the N→I and I→U events of the preceding AFV3-109 domain, indicating that the mechanical stability of the intermediate state is comparable to that of the native state. This result strongly suggests that the C-terminal fusion protein containing (GB1)4 does not adversely influence the formation of the slipknot structure in AFV3-109, and that the folding of AFV3-109 does not involve complex chain crossing.
Signatures of the Mechanical Unfolding of AFV3-109
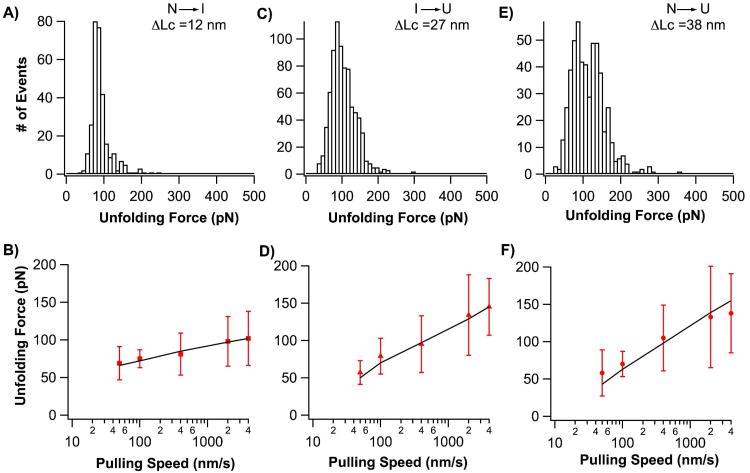
Having identified the mechanical unfolding pathways and mechanical unfolding intermediate state, we can now characterize these unfolding pathways in detail. The average unfolding force is 113 ± 44 pN for N→U (n=521), 90 ± 28 pN for N→I (n=298) and 104 ± 38 pN for I→U (n=790) at a pulling speed of 400 nm/s (Fig. 3). It is evident that the three unfolding steps have similar mechanical resistance. To characterize the underlying energy profiles of these distinct unfolding pathways, we carried out pulling experiments at different pulling velocities (Fig. 3). It is evident that the pulling speed dependence of the unfolding force is similar for N→U and I→U, but that the N→I transition differs significantly, which may be suggestive of the large difference in transition states between in these unfolding steps. We carried out Monte Carlo simulations, to simulate the unfolding behaviors of AFV3-109 in order to estimate the distance to the transition state, Δxu, and the spontaneous unfolding rate constant at zero force, α0, for each individual unfolding step (Fig. 3). These results are shown in Table 1. It is worth noting that Δxu for N→U (0.27 nm) is significantly smaller than that of N→I (0.59 nm), suggesting that the N→U pathway is fundamentally different from that of N→I, eliminating the possibility that the N→U pathway is a special case of the N→I →U pathway, where the intermediate state was missed due to insufficient time resolution.
Figure 3.
Signatures of the mechanical unfolding of the slipknot protein AFV3-109. A, C and E) Unfolding force histograms for the unfolding step N→I (A), I→U (B) and N→U (C). B, D and F) Pulling speed dependence of the unfolding force for each individual unfolding step. Solid lines correspond to the Monte Carlo simulation results using the following α0 and Δxu: (a) N→I unfolding force; (b) I→U unfolding force; (c) two-state unfolding force. The red bars (right) represent the standard deviation of unfolding forces.
Table 1. Kinetic parameters characterizing the mechanical unfolding of AFV3-109.
| ΔLc (nm) | α0 (s−1) | Δxu (nm) | |
|---|---|---|---|
|
| |||
| N→U pathway | 38 | 1.8 | 0.24 |
|
| |||
| N→I→U pathway | |||
| N→I | 12 | 0.011 | 0.59 |
| I→U | 27 | 0.72 | 0.27 |
|
| |||
| N…I→U pathway | |||
| N→I | N/A | N/A | N/A |
| I→U | 27 | 0.72 | 0.27 |
SMD Simulations Uncover the Molecular Mechanism Behind Bifurcation of Mechanical Unfolding Pathways of AFV3-109
To understand the molecular mechanism underlying the bifurcation of mechanical unfolding pathways and the nature of the mechanical unfolding intermediate state of AFV3-109, we carried out all-atom SMD simulations to simulate the mechanical unfolding of AFV3-109. In total, 23 simulations were carried out with a total duration of 0.387 μs.
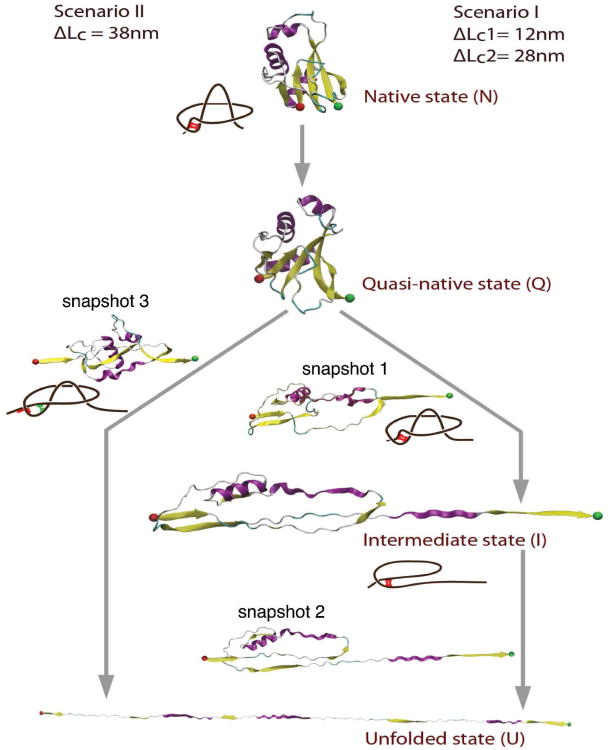
The first event that occurs when steering force is applied is the unzipping of antiparallel strands β4β5, where the five-strand-β sheet is separated into two parts consisting of β3β1β4 strands and β2β5 strands respectively, forming a quasi-native state (Fig. 4). The extension gain from unzipping is approximately 30Å (where the unzipping of β4β5 is completed when a total N-to-C extension of 45Å is reached). Upon continued application of stretching force, the unfolding of AFV3-109 exhibits two distinct types of pathways for all unfolding trajectories: one with an unfolding intermediate state and one without. These results are summarized in Fig. 4 and a description of these pathways is included below.
Figure 4.
SMD simulations reveal two distinct mechanical unfolding pathways of AFV3-109. Scenario I corresponds to unfolding pathways involving an unfolding intermediate state, and scenario II corresponds to the two-state unfolding pathway. Along each step of the unfolding process, snapshots of AFV3-109 are shown (as indicated by snapshots 1, 2 and 3) to indicate the structural changes of AFV3-109 during the unfolding process. In both scenarios, the slipknot structure was untied and no tightened slipknot conformation observed. Interactions (specific or non-specific) between key structural elements are responsible for preventing the formation of the “tightened knot” structure. Such interactions, which are indicated in the schematic drawing of AFV3-109 beside the snapshots, prevent the shrinking of the knotting loop and help untie the slipknot. Red bars indicate specific interactions between strands β1 and β4, while green bars indicate the non-native interactions formed after the sliding of strands β1 and β4.
In the first pathway, unfolding initiates from the C-terminus. The applied steering force leads to unzipping of antiparallel strands β2β5, as well as the sliding of α3 through the loop formed by β2 and α1 (snapshot 1 in Fig. 4). This causes the β5α3β4 arm to become fully extended, fully exposing the β1β4 parallel strands to steering force. During this process, the threaded loop is pulled out of the knotting loop, leading to the untying of the slipknot in AFV3-109. The resultant state is mechanically resistant, and serves as a well-defined intermediate state (shown as I in Fig. 4).
Further stretching AFV3-109 leads to the unraveling of this intermediate state. The major barrier during this process corresponds to the mechanical unraveling of the β1β4 (snapshot 2 in Fig. 4); following the breakage of the β1β4 barrier, there is no mechanical barrier and the protein stretches to a fully extended state.
Detailed analysis of trajectories following this type of pathway reveals that the intermediate state I always occurs at around 13 nm. However, the initial peak occurs at a much lower frequency, and in some trajectories this step does not lead to an observable unfolding force peak (Fig. S2 and Fig. S3 in Supporting Information). Comparing Fig. S3 with the contour length figure of the AFM data, we can conclude the two peaks at 3 nm and 13 nm are corresponding to the N→I transition and I→U transitions, respectively.
Additionally, there are typically more peaks than accounted for by the initial and intermediate peaks (Fig. S3). Fig. S3 shows a typical example a trajectory with three extra peaks at 45 Å, 55 Å, and 70 Å. To further study these peaks, we collected peaks at all trajectories (inset of Fig. S3). We found that these extra peaks are mostly caused by friction that occurs when pulling the threaded loop out of the knotting loop; friction originates from non-specific interactions, which are different in different trajectories and thus occurs at different locations during different unfolding events. In contrast, the initial unfolding force peak (N→I) and intermediate peak (I→U) were due to specific interactions.
In contrast to this, such additional peaks were not observed within AFM experiments. This is likely due to the pulling speed at which SMD simulations were performed (0.0025 Å/ps to 0.5 Å/ps), which is at least six orders of magnitude faster than the pulling speed used in the AFM experiments; the decreased speed at which AFM experiments are conducted could eliminate or reduce the friction that causes additional peaks.
In the second type of unfolding pathway, unfolding is initiated from both the N-and C-termini, where it seems that there is only one major unfolding barrier to unfolding. From the native state, the 5-strand β sheet breaks into β2β5 and β3β1β4 beta sheets (Fig. 4). This is immediately followed by breakage of parallel strands β3β1β4. Continued application of steering force leads to the unzipping of anti parallel strands β2β5 (snapshot 3 in Fig. 4, where β2 and β5 are shown in the process of breaking). Following the continuous application of steering force, the protein unfolds to full extension. The second pathway corresponds to two-state unfolding behaviour.
Unfolding pathways predicted by SMD simulations yield contour length increments ΔLc that are identical to those measured by single molecule AFM experiments, suggesting that the unfolding pathways observed in SMD simulations likely correspond to the ones observed in single molecule AFM experiments. Thus, SMD simulations provide a plausible mechanistic description of the mechanical barrier crossing by AFV3-109 as it is steered along its mechanical unfolding pathways.
For the N→I transition, the major barrier corresponds to the breakage of interface between strands β4 and β5 (snapshot 1 in Fig. 4), allowing for an extension of 120Å up to the β1β4 barrier. During this process, the threaded loop unravels and the slipknot structure is effectively untied, leading to a mechanically stable intermediate state. The subsequent breakage of the β1β4 β strands results in an additional 280 Å extension, corresponding to the I→U transition. For the N→U pathway, the major barrier is the unzipping of β4β5 and the concurrent breakage of β1β4.
In both pathways, it seems that the unzipping of β4β5 is a crucial event in the mechanical unfolding of AFV3-109. After this event, the unfolding pathways bifurcate into two macroscopically and microscopically distinct pathways. It is important to note that in both pathways, the threaded loop is pulled out of the knotting loop, and the tightened knot intermediate, which was predicted for a different slipknot protein thymidine kinase in coarse grain MD simulations16, is not observed. This result is in good agreement with our single molecule AFM results, and thus SMD simulations provide a complimentary and plausible molecular mechanism for the mechanical unfolding of AFV3-109.
Discussion
A Kinetic Partitioning Mechanism for the Mechanical Unfolding of AFV3-109
A kinetic partitioning mechanism has been predicted as a general mechanism that governs protein folding and unfolding dynamics5,6,43,44. Kinetic traps on the folding/unfolding energy landscape lead to the formation of intermediate states and the bifurcation of folding and unfolding pathways. A growning number of proteins have shown clear evidence that kinetic partitioning mechanism defining their folding/unfolding dynamics45-48. Here, we combine single molecule AFM and SMD simulations to investigate the unfolding mechanism of AFV3-109 in detail. Our results revealed that AFV3-109 unfolds via three distinct unfolding pathways. If the unfolding is initiated at the C-terminus of AFV3-109, a stretching force pulls the threaded loop out of the knotting loop and an intermediate state forms, where β1 and β4 constitute a shear topology that provides mechanical stability; if the unfolding is initiated from both termini, the potential intermediate state is avoided and the protein unfolds in a two-state fashion. In the latter case, the threaded loop must be pulled out of the knotting loop before the threaded loop is tightened. This requires that the unraveling of β4β5 and β2β5 to be faster than the unraveling of β1 and β4. These results demonstrate that multiple parallel pathways are available as AFV3-109 unfolds and unties the slipknot under mechanical force, providing a clear example of how the kinetic partitioning mechanism determines protein folding/unfolding mechanisms in general. Since the folding of a slipknotted protein from a completely unfolded polypeptide chain involves complex chain movement, it is likely that more kinetic traps will form along the folding pathway. Thus, kinetic partitioning may be more pronounced in the folding pathway.
SMD simulations provides an accurate description of the molecular events leading to the unfolding of AFV3-109, revealing that the interactions between β strands β4 and β5 are critical for the mechanical integrity of AFV3-109. It is of note that though the disruption of β4β5 is obligatory for AFV3-109 to unfold, the two pathways bifurcate from this point on, leading to very different means of untying the slipknot. That these two unfolding pathways are fundamentally different is shown by the large difference in mechanical unfolding distance between the two pathways (0.59 nm for N-I-U versus 0.29 nm for N-U). It seems that the relative stability of the β1β4 sheet may serve as a “gear box” to steer the mechanical unfolding into one pathway or the other; however, the detailed mechanism in which this “gear box” operates remains to be elucidated.
Although SMD simulations provide a plausible mechanism for the mechanical unfolding of AFV3-109, it is important to note that the pulling speed used in SMD simulations (∼0.25 m/s) is orders of magnitude faster than that used in single molecule AFM experiments (∼400 nms). Such differences could partially account for the differences between AFM experiments and SMD simulations. For example, the friction seen in SMD simulations is largely absent in AFM results; the relative population of different pathways is quite different between AFM experiments and SMD simulations: in AFM experiments, a large fraction (40%) of AFV3-109 was observed to unfold in a two-state manner, while only a small fraction (∼10%) of SMD trajectories correspond to two state unfolding.
Comparing all-Atom SMD Simulations to Coarse Grain MD Simultaions
The mechancial unfolding of the slipknotted protein thymidine kinase has been previously studied using coarse grain MD simulations16. These simulations suggested two different and general mechanical unfolding pathways depending on the relative shrinking rates of the two loops, which would vary depending on the stretching force. Under low stretching forces, the threaded loop shrinks faster than the knotting loop and the slipknot loosens. Under high stretching force, the knotting loop shrinks faster and the stretching force temporarily tightens the slipknot. As a result, a tightened slipknot is formed as the intermediate state in this unfolding pathway16. However, in our all-atom SMD simulations on AFV3-109, we did not observe such a “tightened knot” intermediate state.
Instead, we observed that the slipknot loosens and an intermediate formed in which the slipknot has been untied and the protein is in a deformed state, where a shearing geometry provides the necessary mechanical resistance (β strands 1 and 4). Moreover, in both single molecule AFM experiments and SMD simulations, we observed that the pulling velocity did not affect unfolding pathways of AFV3-109 in two vastly different pulling speed regimes (50 nm/s to 4000 nm/s for AFM experiments and 0.0025 Å/ps to 0.5 Å/ps for SMD simulations), as the relative ratio between the two pathways remains roughly the same at different pulling velocities. However, it remains worth investigating whether pathway switching can occur in the relevant pulling speed regime predicted by coarse grained MD simulations.49 Coarse grained MD and SMD simulations on two different model proteins revealed that different slipknot proteins can unravel following different pathways, suggesting that detailed interactions in different slipknot proteins play important roles in determining their specific unfolding pathways and kinetics.
In principle, AFV3-109 could form a tightened slipknot under a stretching force. The absence of a tightened slipknot intermediate in our experiments suggests that forming a tightened slipknot may not be energetically favorable. The formation of a tightened knot requires the rupture of interactions between β-strands 1 and 4, 1 and 3 as well as interactions between β-strands 2 and 5. In contrast, unknotting would require the rupture of fewer interactions: only those between β-strands 2 and 5 as well as 4 and 5. Intuitively, unknotting should be favorable. In order to understand why the “tightened knot” did not form in AFV3-109 more thoroughly, we carried out a detailed analysis of our SMD trajectories. We found that in both pathways, interactions (specific or nonspecific) between key structural elements prevent the formation of the “tightened knot” structure. In scenario I (Fig. 4), specific hydrogen bonds between β1 and β4 strands are at the two ends of the knotting loop and form a lock to prevent the loop from shrinking (highlighted in red in snapshot 1 in Fig. 4). Thus, no jamming would appear in this case and the β1- β4 ruptures after the threaded loop is pulled out. In scenario II, although the β1-β4 breaks earlier after the sliding of the two strands, a non-native (weaker) patch forms which again prevents the knotting loop from shrinking (highlighted in green in snapshot 3 in Fig. 4). These insights reflect the importance of detailed interactions within protein structure in determining the unfolding pathways and unfolding mechanism of knotted or slipknotted proteins.
Implication for the Folding Mechanism of Slipknot and Knot Proteins
Our results on the unfolding of AFV3-109 has interesting implications concerning the folding mechanism of slipknotted and knotted proteins. Coarse grained MD simulations on the folding of knotted proteins suggested two different mechanisms for the formation of knotted structures during the folding of knotted proteins: where the slipknot forms as an intermediate state or through plug movement of the threaded loop9. In the N-I-U pathway of AFV3-109, the formation of the intermediate state involves the unthreading of the threaded loop in the knotting loop. The reverse of this step would be the formation of the slipknot structure. Our preliminary refolding experiments by single molecule AFM showed that the unfolded AFV3-109 can refold back to its native state when it is relaxed to zero force (Fig. S5), suggesting that the folding of AFV3-109 is a spontaneous process. In addition, AFV3-109 can refold into the unfolded intermediate state prior to successfully refolding into the native state (Fig. S5), suggesting that threading the threaded loop in the knotting loop may represent a major barrier for AFV3-109 folding. In this sense, the folding of AFV3-109 can serve as a model system to study the mechanism of slipknot formation in knotted proteins.
Furthermore, our preliminary results show that the folding process of AFV3-109 is slow and shows large variation from molecule to molecule. For some “efficient” cases, around 10% AFV3-109 could refold back to its native in 15 seconds. For most cases, AFV3-109 does not fold within our instrument observation time window (∼30 s). How nature solves this knotting problem to make slipknot and knot proteins fold efficiently remains an open question. Jackson and coworkers recently discovered that chaperones can accelerate the folding of knotted proteins50, raising an interesting question whether chaperones can also facilitate the folding of slipknot proteins in a similar fashion. Future experiments will be needed to examine the molecular mechanism of slipknot formation and whether chaperones can speed up the folding of slipknot proteins.
Supplementary Material
Acknowledgments
This work is supported by the Natural Sciences and Engineering Research Council of Canada, Canada Research Chairs Program and Canada Foundation of Innovation and is also supported by the National Science Foundation through Teragrid resources provided by TACC-Lonestar (TG-MCB090005) and T32HL007692 from the National Heart, Lung, and Blood Institute of U.S.A.
Abbreviations
- AFM
single molecule atomic force microscopy
- SMD
steered molecular dynamics
Footnotes
Supporting Information. Force-extension curves of (GB1-AFV3-109)n; A typical unfolding trajectory without a clear initial unfolding force peak; Non-specific interactions can lead to extra unfolding force peaks in SMD simulations; Comparison between the tightened slipknot of AFV3-109 and tightened figure-eight knot in Phytochrome; Unfolded AFV3-109 can refold back to its native state as well as unfolding intermediate state prior to its complete folding. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Onuchic JN, Luthey-Schulten Z, Wolynes PG. Annu Rev Phys Chem. 1997;48:545. doi: 10.1146/annurev.physchem.48.1.545. [DOI] [PubMed] [Google Scholar]
- 2.Borgia A, Williams PM, Clarke J. Annu Rev Biochem. 2008;77:101. doi: 10.1146/annurev.biochem.77.060706.093102. [DOI] [PubMed] [Google Scholar]
- 3.Fersht AR, Daggett V. Cell. 2002;108:573. doi: 10.1016/s0092-8674(02)00620-7. [DOI] [PubMed] [Google Scholar]
- 4.Wolynes PG. Q Rev Biophys. 2005;38:405. doi: 10.1017/S0033583505004075. [DOI] [PubMed] [Google Scholar]
- 5.Thirumalai D, Klimov DK, Woodson SA. Theoretical Chemistry Accounts. 1997;96:14. [Google Scholar]
- 6.Thirumalai D, O'Brien EP, Morrison G, Hyeon C. Annu Rev Biophys. 2010;39:159. doi: 10.1146/annurev-biophys-051309-103835. [DOI] [PubMed] [Google Scholar]
- 7.Mansfield ML. Nat Struct Biol. 1994;1:213. doi: 10.1038/nsb0494-213. [DOI] [PubMed] [Google Scholar]
- 8.Mallam AL. Febs Journal. 2009;276:365. doi: 10.1111/j.1742-4658.2008.06801.x. [DOI] [PubMed] [Google Scholar]
- 9.Sulkowska JI, Sulkowski P, Onuchic J. Proceedings of the National Academy of Sciences of the United States of America; 2009; p. 3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mallam AL, Jackson SE. Journal of Molecular Biology. 2005;346:1409. doi: 10.1016/j.jmb.2004.12.055. [DOI] [PubMed] [Google Scholar]
- 11.King NP, Yeates EO, Yeates TO. Journal of Molecular Biology. 2007;373:153. doi: 10.1016/j.jmb.2007.07.042. [DOI] [PubMed] [Google Scholar]
- 12.Wallin S, Zeldovich KB, Shakhnovich EI. Journal of Molecular Biology. 2007;368:884. doi: 10.1016/j.jmb.2007.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeates T, Norcross TS, King NP. Current Opinion in Chemical Biology. 2007;11:595. doi: 10.1016/j.cbpa.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mallam AL, Morris ER, Jackson SE. Proceedings of the National Academy of Sciences of the United States of America; 2008; p. 18740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bornschlogl T, Anstrom DM, Mey E, Dzubiella J, Rief M, Forest KT. Biophysical Journal. 2009;96:1508. doi: 10.1016/j.bpj.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sulkowska JI, Sulkowski P, Onuchic JN. Physical Review Letters. 2009;103 doi: 10.1103/PhysRevLett.103.268103. [DOI] [PubMed] [Google Scholar]
- 17.Sulkowska JI, Sulkowski P, Szymczak P, Cieplak M. Journal of the American Chemical Society. 2010;132:13954. doi: 10.1021/ja102441z. [DOI] [PubMed] [Google Scholar]
- 18.King NP, Jacobitz AW, Sawaya MR, Goldschmidt L, Yeates TO. Proceedings of the National Academy of Sciences of the United States of America; 2010; p. 20732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noel JK, Sulkowska JI, Onuchic JN. Proceedings of the National Academy of Sciences of the United States of America; 2010; p. 15403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keller J, Leulliot N, Cambillau C, Campanacci V, Porciero S, Prangishvili D, Forterre P, Cortez D, Quevillon-Cheruel S, Tilbeurgh H. Virology Journal. 2007;4:10. doi: 10.1186/1743-422X-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carrion-Vazquez M, Oberhauser AF, Fisher TE, Marszalek PE, Li H, Fernandez JM. Prog Biophys Mol Biol. 2000;74:63. doi: 10.1016/s0079-6107(00)00017-1. [DOI] [PubMed] [Google Scholar]
- 22.Puchner EM, Gaub HE. Curr Opin Struct Biol. 2009;19:605. doi: 10.1016/j.sbi.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 23.Brockwell DJ. Curr Nanosci. 2007;3:3. [Google Scholar]
- 24.Krammer A, Lu H, Isralewitz B, Schulten K, Vogel V. Proc Natl Acad Sci U S A. 1999;96:1351. doi: 10.1073/pnas.96.4.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu H, Isralewitz B, Krammer A, Vogel V, Schulten K. Biophys J. 1998;75:662. doi: 10.1016/S0006-3495(98)77556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu H, Schulten K. Proteins. 1999;35:453. [PubMed] [Google Scholar]
- 27.Sharma D, Feng G, Khor D, Genchev GZ, Lu H, Li H. Biophys J. 2008;95:3935. doi: 10.1529/biophysj.108.134072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sotomayor M, Schulten K. Science. 2007;316:1144. doi: 10.1126/science.1137591. [DOI] [PubMed] [Google Scholar]
- 29.Paci E, Karplus M. Proc Natl Acad Sci U S A. 2000;97:6521. doi: 10.1073/pnas.100124597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Genchev GZ, Kallberg M, Gursoy G, Mittal A, Dubey L, Perisic O, Feng G, Langlois R, Lu H. Cell Biochem Biophys. 2009;55:141. doi: 10.1007/s12013-009-9064-5. [DOI] [PubMed] [Google Scholar]
- 31.Cao Y, Li HB. Nature Materials. 2007;6:109. doi: 10.1038/nmat1825. [DOI] [PubMed] [Google Scholar]
- 32.Zheng P, Cao Y, Li H. Langmuir. 2011;27:5713. doi: 10.1021/la200915d. [DOI] [PubMed] [Google Scholar]
- 33.Florin EL, Rief M, Lehmann H, Ludwig M, Dornmair C, Moy VT, Gaub HE. Biosensors & Bioelectronics. 1995;10:895. [Google Scholar]
- 34.Humphrey W, Dalke A, Schulten K. J Mol Graph. 1996;14:33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 35.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. J Phys Chem B. 1998;102:3586. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 36.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Journal of Chemical Physics. 1983;79:926. [Google Scholar]
- 37.Darden T, York D, Pedersen L. Journal of Chemical Physics. 1993;98:10089. [Google Scholar]
- 38.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. J Comput Chem. 2005;26:1781. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marko JF, Siggia ED. Macromolecules. 1995;28:8759. [Google Scholar]
- 40.Cao Y, Lam C, Wang MJ, Li HB. Angewandte Chemie-International Edition. 2006;45:642. doi: 10.1002/anie.200502623. [DOI] [PubMed] [Google Scholar]
- 41.Carrion-Vazquez M, Oberhauser AF, Fowler SB, Marszalek PE, Broedel SE, Clarke J, Fernandez JM. Proceedings of the National Academy of Sciences of the United States of America; 1999; p. 3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rief M, Fernandez JM, Gaub HE. Physical Review Letters. 1998;81:4764. [Google Scholar]
- 43.Guo ZY, Thirumalai D. Biopolymers. 1995;36:83. [Google Scholar]
- 44.Thirumalai D, Hyeon C. Biochemistry. 2005;44:4957. doi: 10.1021/bi047314+. [DOI] [PubMed] [Google Scholar]
- 45.Kiefhaber T. Proc Natl Acad Sci U S A. 1995;92:9029. doi: 10.1073/pnas.92.20.9029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wright CF, Lindorff-Larsen K, Randles LG, Clarke J. Nat Struct Biol. 2003;10:658. doi: 10.1038/nsb947. [DOI] [PubMed] [Google Scholar]
- 47.Mickler M, Dima RI, Dietz H, Hyeon C, Thirumalai D, Rief M. Proc Natl Acad Sci U S A. 2007;104:20268. doi: 10.1073/pnas.0705458104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng Q, Li H. Proc Natl Acad Sci U S A. 2008;105:1885. doi: 10.1073/pnas.0706775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hyeon C, Dima RI, Thirumalai D. Structure. 2006;14:1633. doi: 10.1016/j.str.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 50.Mallam AL, Jackson SE. Nat Chem Biol. 2012;8:147. doi: 10.1038/nchembio.742. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.