Background: The sphingosine 1-phosphate (S1P) system may contribute to lung fibrosis.
Results: S1P receptor (S1PR) agonists with different receptor subtype selectivity profiles varied in their potential to induce fibrotic responses in human lung fibroblasts.
Conclusion: S1P2R and S1P3R signaling contributes to fibrotic responses in lung fibroblasts.
Significance: Improving S1P1R modulator selectivity may lead to an improved safety profile of compounds for autoimmune therapy.
Keywords: Collagen, Fibroblast, Fibrosis, Myofibroblast, Sphingosine-1-phosphate, Transforming Growth Factor Beta (TGFbeta), Impedance, S1P Receptor Agonists
Abstract
Synthetic sphingosine 1-phosphate receptor 1 modulators constitute a new class of drugs for the treatment of autoimmune diseases. Sphingosine 1-phosphate (S1P) signaling, however, is also involved in the development of fibrosis. Using normal human lung fibroblasts, we investigated the induction of fibrotic responses by the S1P receptor (S1PR) agonists S1P, FTY720-P, ponesimod, and SEW2871 and compared them with the responses induced by the known fibrotic mediator TGF-β1. In contrast to TGF-β1, S1PR agonists did not induce expression of the myofibroblast marker α-smooth muscle actin. However, TGF-β1, S1P, and FTY720-P caused robust stimulation of extracellular matrix (ECM) synthesis and increased pro-fibrotic marker gene expression including connective tissue growth factor. Ponesimod showed limited and SEW2871 showed no pro-fibrotic potential in these readouts. Analysis of pro-fibrotic signaling pathways showed that in contrast to TGF-β1, S1PR agonists did not activate Smad2/3 signaling but rather activated PI3K/Akt and ERK1/2 signaling to induce ECM synthesis. The strong induction of ECM synthesis by the nonselective agonists S1P and FTY720-P was due to the stimulation of S1P2 and S1P3 receptors, whereas the weaker induction of ECM synthesis at high concentrations of ponesimod was due to a low potency activation of S1P3 receptors. Finally, in normal human lung fibroblast-derived myofibroblasts that were generated by TGF-β1 pretreatment, S1P and FTY720-P were effective stimulators of ECM synthesis, whereas ponesimod was inactive, because of the down-regulation of S1P3R expression in myofibroblasts. These data demonstrate that S1PR agonists are pro-fibrotic via S1P2R and S1P3R stimulation using Smad-independent pathways.
Introduction
Sphingosine 1-phosphate (S1P)2 is a bioactive sphingolipid that is involved in a variety of physiological processes such as cell growth, survival, cytoskeletal organization, migration, and lymphocyte trafficking (1). S1P levels, which are high in blood and much lower in tissues (2), are controlled by coordinated activities of two sphingosine kinases (SphK1 and SphK2) and several degrading enzymes (3). S1P binds with low nanomolar affinity to five related G protein-coupled receptors, named S1P receptors (S1P1R, S1P2R, S1P3R, S1P4R, and S1P5R). The S1P1R, S1P2R, and S1P3R subtypes are widely expressed within the human body, whereas S1P4R and S1P5R tissue expression is much more restricted (3).
Synthetic S1P1R modulators constitute a new class of drugs for the treatment of T-cell-mediated autoimmune diseases, and fingolimod (FTY720/Gilenya; Novartis) is the first nonselective S1P1R modulator approved for the treatment of relapsing-remitting multiple sclerosis (2). Fingolimod is a pro-drug that is phosphorylated within the body to its active form (S)-fingolimod-P (FTY720-P), which shares structural homology to the natural ligand S1P. To current knowledge FTY720-P ameliorates multiple sclerosis symptoms by acting as an S1P1R desensitizing and internalizing agonist on T-cells and neural cells. Importantly, S1P1R internalization leads to a depletion of circulating lymphocytes, because S1P1R signaling is required to allow lymphocytes to egress from lymph nodes (2).
In addition to its prominent role in lymphocyte trafficking, there is increasing evidence that S1P and S1PR signaling plays a role in pro-fibrotic responses in various tissues and isolated cells. Fibrosis is a process that can be triggered by chronic tissue damage because of toxic substances, viral infection, inflammation, or mechanical stress and eventually leads to changes in organ architecture and to organ failure. Fibrosis is often described as disordered wound healing that is characterized by accumulation of myofibroblasts with increased proliferative capacity and excessive extracellular matrix (ECM) deposition (4). S1P, S1PR, and SphK1 were shown to be up-regulated in serum, bronchoalveolar lavage, and lung tissue of patients with idiopathic pulmonary fibrosis (5), human liver fibrosis (6), experimental mouse models of bleomycin-induced lung fibrosis (7), and cholestasis-induced liver fibrosis (8). Moreover, chronic fingolimod treatment of rats and monkeys caused fibrotic lung changes, including increased lung weight associated with smooth muscle hypertrophy, hyperdistension of the alveoli, and increased collagen deposition. In addition, insufficient or lack of pulmonary collapse at necroscopy, indicative of excessive collagen presence, was observed in rats, dogs, and monkeys (9). In vitro, S1P and FTY720-P displayed pro-fibrotic activities, such as induction of the myofibroblast marker α-smooth muscle actin (αSMA) and increases in collagen synthesis (10–12).
Based on the described lung findings after prolonged fingolimod treatment (9), we employed normal human lung fibroblasts (NHLF) to study S1PR agonist-mediated induction of fibrotic responses, i.e., myofibroblast transformation, fibrotic gene expression, and excessive production of ECM. We investigated responses that were induced by the nonselective S1PR agonists S1P and FTY720-P (3, 13, 14), the highly selective S1P1R agonist SEW2871 (15), and the selective S1P1R agonist ponesimod (16, 17), which is currently under clinical investigation for treatment of multiple sclerosis and psoriasis, and we compared them with responses induced by the well described pro-fibrotic mediator TGF-β1 (18). Here, we show that TGF-β1 and the two nonselective S1PR agonists, S1P and FTY720-P, were highly pro-fibrotic, whereas ponesimod and SEW2871 were less active or inactive. In contrast to TGF-β1, the S1PR agonists did not induce Smad2/3 phosphorylation but rather activated PI3K/Akt and ERK1/2 signaling to induce ECM synthesis. Furthermore, we found that S1PR agonist-induced ECM synthesis was mediated by S1P2R and S1P3R signaling, explaining why the intensity of the response to nonselective S1PR agonists was stronger than to selective S1P1R agonists, such as ponesimod or SEW2871. Improving S1P1R modulator selectivity may thus lead to an improved safety profile of compounds targeted for autoimmune therapy.
EXPERIMENTAL PROCEDURES
Materials
The following reagents were used: S1P (Enzo Life Sciences, Lausen, Switzerland); TGF-β1, PDGF-bb, and SB431542 (Sigma-Aldrich); W146 (Avanti Polar Lipids, Inc., Alabaster, AL); JTE-013, PD0325901, and SEW2871 (Tocris Bioscience, Abingdon, UK); and PI-103 and FTY720-P (Cayman Chemical, Tallinn, Estonia). TY-52156 (19) and ponesimod were synthesized by Actelion Pharmaceuticals Ltd. (Allschwil, Switzerland). The following antibodies were used: mouse anti-α smooth muscle actin (Sigma-Aldrich); rabbit (rb) anti-phospho-Smad2, rb anti-Smad2, mouse anti-phospho-ERK1/2, rb anti-Phospho-S6 ribosomal protein (Ser-473), rb anti-S6 ribosomal protein, and rb anti-α/β-tubulin (Cell Signaling Technology, Boston, MA); rb anti-phospho-Smad3, rb anti-Smad3, and rb anti-ERK1/2 (Millipore, Zug, Switzerland); goat anti-collagen I and goat anti-collagen III (SouthernBiotech, Birmingham, AL); goat anti-fibronectin and donkey anti-goat IgG-HRP (Santa Cruz Biotechnology, Heidelberg, Germany); goat anti-mouse Alexa 480 (Invitrogen); and sheep anti-mouse HRP and donkey anti-rb-HRP (GE Healthcare).
Cell Culture
NHLF (donor 1: female 11 years; donor 2: female 19 years; Lonza, Verviers, Belgium) were cultivated in fibroblast growth medium 2 (Lonza) following the manufacturer's instructions and were used at passages 5–7 for experiments. CHO-K1 cells with stable expression of human S1P2R were grown in Ham's F-12, 10% FBS, 100 units/ml penicillin, 1 μg/ml streptomycin in presence of 1 mg/ml geneticin (all Invitrogen). NHLF were transformed to myofibroblasts by incubation with 1 ng/ml TGF-β1 for 72 h in fibroblast growth medium 2, before experiments were performed in the presence of 1 ng/ml TGF-β1. For experiments, NHLF and myofibroblasts were starved in fibroblast basal medium (Lonza) with 0.1% FFA-BSA (Calbiochem, Darmstadt, Germany) for 24 h before stimulation with compounds at various concentrations for the indicated times. When kinase inhibitors (concentrations as indicated) or S1PR antagonists were used, the cells were preincubated with inhibitors or antagonists for 1 h. S1P1R antagonist W146 was used at 1 μm, S1P2R antagonist JTE-013 was used at 0.2 μm, and S1P3R antagonist TY-52156 was used at 1.25 μm. Controls and all samples were supplemented with equal concentrations of Me2SO or methanol (vehicles).
siRNA Transfection
NHLF (104 cells/well) were transfected in 96-well tissue culture plates (for qPCR) or on E-plates (Roche Applied Science, Rotkreuz, Switzerland, for impedance measurements) with Lipofectamine RNAiMAX (Invitrogen) following the manufacturer's protocol. The following siRNAs were used: HA044100078 and HA044100082 for S1P2R with the respective negative control SIC001 at 33 nm (Sigma-Aldrich) and s4454 and s4455 for S1P3R with respective Silencer® select negative control No. 1 siRNA (4390843) at 11 nm (Ambion, Austin, TX). Knockdown of S1P2,3R mRNA expression was controlled by qPCR. Knockdown of receptor function was analyzed after 48 h using impedance measurements and compared with siRNA transfection with the respective negative control siRNA.
[3H]Proline Incorporation Assay
NHLF or myofibroblasts were stimulated in 96-well plates (104 cells/well) for 24 h as described in the presence of 7.4 kBq/well l-[2,3-3H]proline (PerkinElmer Life Sciences). The cells were lysed in 0.15 m NaOH for 30 min on ice and proteins were TCA-precipitated (20%) on ice for 30 min. The proteins were transferred to GF/C filter plates (PerkinElmer Life Sciences) with a cell harvester (Filtermate; Packard). Microscint 20 (PerkinElmer Life Sciences) was added to dried plates, and radioactive proline incorporation was determined by liquid scintillation counting using a TopCount (PerkinElmer Life Sciences). The data were normalized to base line (defined by preincubation with vehicle, S1PR antagonist or inhibitors without S1PR agonist stimulation) and expressed as percentages of increase over control.
Indirect Immunofluorescence
The cells were fixed in 3% paraformaldehyde, incubated in PBS (Mg2+/Ca2+), 10% FBS, 0.1% saponine (Sigma-Aldrich) for 45 min before mouse anti-α smooth muscle actin antibody incubation for 1 h followed by goat anti-mouse Alexa 480 and Hoechst (Invitrogen) incubation for 30 min. Images were acquired under identical conditions with an Axiovert 200M fluorescence microscope (Carl Zeiss, Feldbach, Switzerland) and Openlab software (Improvision; PerkinElmer Life Sciences).
Western Blotting
After stimulation for the indicated time points, the cells were lysed in radioimmune precipitation assay buffer supplemented with 1× MiniComplete Protease Stop and PhosStop (Roche Applied Sciences; for (P-)Smad2/3, (P-)ERK1/2), or 1× cell lysis buffer (Cell Signaling Technology; for (P-)S6 ribosomal protein) followed by sonication. For detection of ECM proteins, the cells were lysed by addition of boiling radioimmune precipitation assay buffer supplemented with 6 m urea (Sigma-Aldrich) and 1× MiniComplete Protease Stop followed by scraping and sonication. The samples were subjected to SDS-PAGE and immunoblotting according to the manufacturers' protocols. Detection was performed with Western Lightning ECL (PerkinElmer Life Sciences), and digital images were taken with LAS-4000 (FUJIFILM).
Quantitative PCR
The fibrotic marker gene analysis was performed according to the TaqMan Fast Cells-to-Ct protocol (Ambion) and run on an ABI 7500 machine using TaqMan assays (Applied Biosystems, Foster City, CA). The results were calculated with the ΔΔCt method and expressed as ratios of treated NHLF versus controls. For analysis of basal S1PR expression, qPCR experiments were performed as previously described (20). TaqMan assays used for mRNA detection are listed in supplemental Table S1.
Impedance Measurements
CHO cells were seeded at 40,000 cells/well into gelatin-coated E-plates (Roche Applied Science). NHLF were seeded at 10,000 cells/well into E-plates. Both cell types were subjected to continued impedance sampling over the whole experimental period (xCELLigence system; Roche Applied Science). After overnight growth, the medium was exchanged with starvation medium for 1 h (CHO) or 24 h (NHLF) and stimulation was performed as described followed by continued impedance sampling. For data analysis, impedance raw traces were normalized at the time point of agonist addition, and the base line response (vehicle-treated cells) was subtracted. The EC50 values in CHO-S1P2 cells were calculated using the proprietary software IC50 witch and an assigned fixed minimum at 5000 nm FTY720-P. The geometric mean of three independent experiments was calculated.
Statistics
For statistical analysis, the unpaired two-tailed Student's t test or one-way analysis of variance with Dunnett's post hoc test was performed. When the p value was < 0.05, the results were considered significant (GraphPad 5 Software, San Diego, CA).
RESULTS
TGF-β1 and S1PR-mediated Pro-fibrotic Responses Show Commonalities and Differences
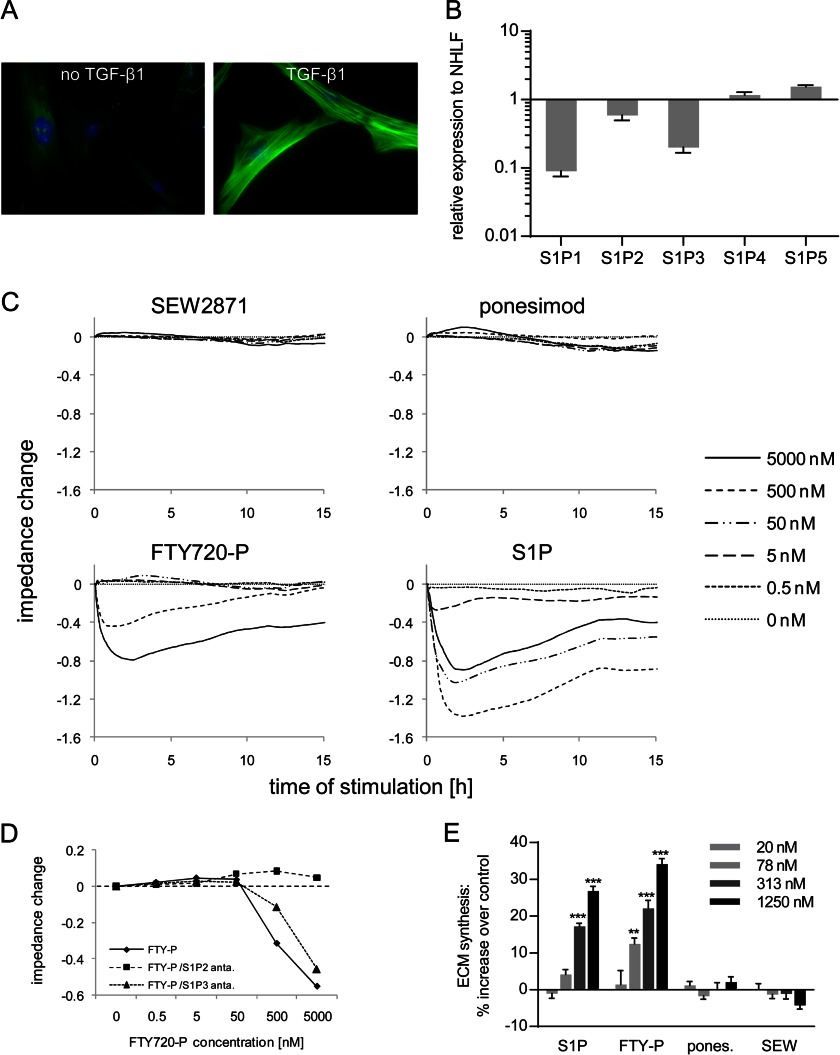
Differentiation of resident fibroblasts into collagen-secreting, αSMA-expressing myofibroblasts is an established hallmark of fibrosis, and TGF-β1 is a known inducer of this differentiation process (18). To study whether S1P and a selection of synthetic S1PR agonists were also able to induce cell differentiation, NHLF were stimulated with S1P, FTY720-P, ponesimod, or SEW2871 (5 μm) for 72 h, and αSMA expression was analyzed by indirect immunofluorescence (Fig. 1A). In vehicle-treated control cells, αSMA was not detectable. Stimulation with 5 ng/ml TGF-β1 induced pronounced, stress fiber-like αSMA expression. In contrast, none of the S1PR agonists induced αSMA expression, suggesting that neither the natural ligand S1P nor synthetic S1PR agonists were able to transform NHLF into myofibroblasts under these conditions.
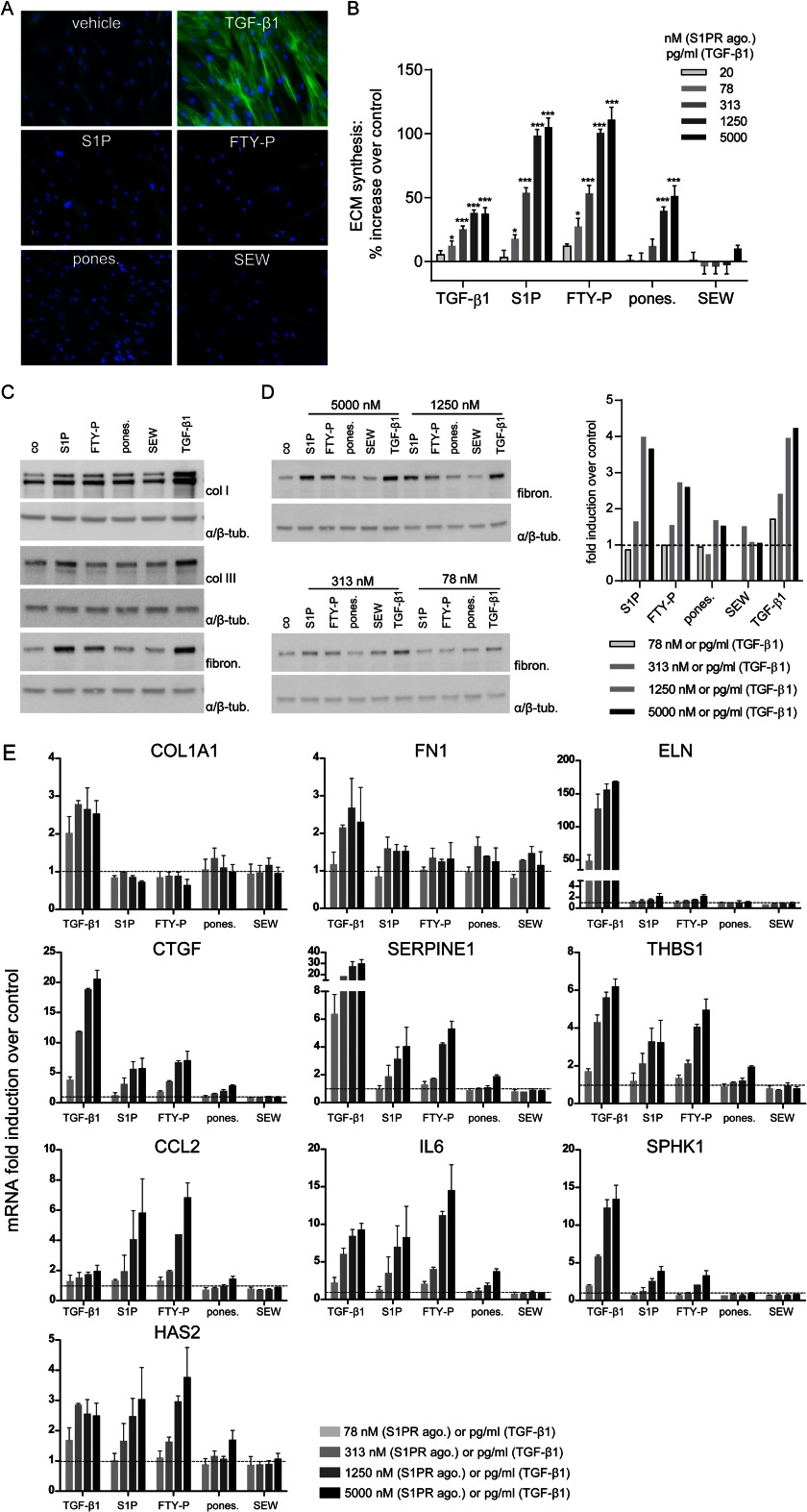
FIGURE 1.
Analysis of fibrotic responses in NHLF subjected to S1PR agonists and TGF-β1: myofibroblast transformation, ECM synthesis, and regulation of pro-fibrotic genes. A, NHLF were stimulated with TGF-β1 (5 ng/ml) or S1PR agonists (5 μm) for 72 h, and immunofluorescent staining was performed. Green, α-smooth muscle actin; blue, nuclei. B, NHLF were stimulated with TGF-β1 (20–5000 pg/ml) and S1PR agonists (20–5000 nm), and ECM synthesis was measured after 24 h with the [3H]proline incorporation assay. The data represent the means ± S.E. of three independent experiments. *, p < 0.05; ***, p < 0.001, one-way analysis of variance, Dunnett's post test. C, NHLF were stimulated with S1PR agonists (5000 nm) or TGF-β1 (5 ng/ml) for 24 h, proteins were isolated, and the expression of collagen I (col I), collagen III (col III), and fibronectin (fibron.) was analyzed by Western blotting. Equal protein amounts were confirmed by α/β-tubulin (α/β-tub.) analysis. D, NHLF were stimulated with S1PR agonists (78–5000 nm) or TGF-β1 (78–5000 pg/ml) for 24 h, proteins were isolated, and the expression of fibronectin was analyzed by Western blotting (left panel). The fibronectin expression was quantified and normalized against α/β-tubulin and expressed as fold induction over control (dashed line, right panel). The data in C and D show representative experiments (n = 3). E, qPCR of pro-fibrotic genes after stimulation of NHLF with TGF-β1 (78–5000 pg/ml) or S1PR agonists (78–5000 nm) for 8 h. Normalization was performed using B2M, HPRT1, 18 S, and PPIA, which were selected by GENORM application (43). The data are the means ± S.E. of two independent experiments. Dashed lines show the control level of gene expression.
Next, we assessed ECM synthesis using the [3H]proline incorporation assay as readout. This assay represents a highly sensitive quantitative method for assessing neosynthesis of proline-rich extracellular matrix proteins such as collagens and fibronectin (21). To this end, NHLF were treated with different concentrations of TGF-β1, S1P, FTY720-P, ponesimod, or SEW2871 or with vehicle, and [3H]proline incorporation was measured after 24 h (Fig. 1B). TGF-β1 enhanced ECM synthesis in a concentration-dependent manner starting with a lowest effective concentration (LEC) of 78 pg/ml, and a plateau was reached at the maximal effective concentration (MEC) of 1250 pg/ml, at which TGF-β1 induced a 38% increase in ECM synthesis compared with control. S1P and FTY720-P also enhanced ECM synthesis in a concentration-dependent manner (LEC, 78 nm; MEC, 5000 nm) and induced an even higher maximal response than TGF-β1. ECM levels increased to 105 and 111% above control for S1P and FTY720-P, respectively. Ponesimod enhanced ECM synthesis with a 16-fold lower potency (LEC, 1250 nm) and showed a lower maximal ECM increase (51% above control at MEC) than either S1P or FTY720-P. SEW2871 was inactive. To elucidate the composition of the up-regulated ECM, we stimulated NHLF with S1PR agonists (5 μm) and TGF-β1 (5 ng/ml) for 24 h and performed Western blotting for collagen I, collagen III, and fibronectin (Fig. 1C). An increase in collagen I expression was detectable in cells treated with TGF-β1 and to a lower extent in cells treated with S1P, FTY720-P, and ponesimod, but not with SEW2871. Collagen III expression was not detectably up-regulated by S1PR agonists and only slightly increased by TGF-β1. In contrast, fibronectin expression was considerably increased by TGF-β1, S1P, and FTY720-P treatment, moderately increased by ponesimod, and not changed by SEW2871 treatment. Because the amplitude of the fibronectin response was suitable for quantification, we performed concentration-response experiments using the fibronectin readout. Strikingly, fibronectin expression showed the same regulation pattern after stimulation with concentration series of TGF-β1 and S1PR agonists as seen before with [3H]proline incorporation (Fig. 1D).
In fibroblasts, fibrotic mediators such as TGF-β1 can induce the gene expression of other pro-fibrotic factors, which stimulate the fibrotic process even further, and together they increase the expression of fibrosis response genes, such as extracellular matrix components, leading to increased ECM deposition and organ restructuring. We therefore analyzed the mRNA expression levels of a selection of known fibrosis-related genes in NHLF after stimulation for 8 h with a dilution series of TGF-β1, S1PR agonists, or vehicle using qPCR (Fig. 1E). The expression of genes that encode for the ECM proteins collagen 1a (COL1A1), fibronectin (FN1), or elastin (ELN) were strongly increased by TGF-β1, whereas S1PR agonists induced no COL1A1 and only weak expression of FN1 and ELN. The expression of several pro-fibrotic genes such as CTGF, SERPINE1, thrombospondin 1 (THBS1), or SphK1 was up-regulated by TGF-β1, S1P, and FTY720-P, whereas ponesimod induced no response (SphK1) or only low responses (CTGF, SERPINE1, and thrombospondin 1). CCL2 (chemokine (C-C motif) ligand 2), IL6, and hyaluronan synthase 2 (HAS2) were strongly and concentration-dependently induced by S1P and FTY720-P and to a lower extent by TGF-β1. Ponesimod induced clearly lower responses of these genes. SEW2871 induced none of the genes.
To exclude a potential donor-specific bias in these results, ECM synthesis and the expression of the fibrosis-related gene set were studied in NHLF from a second donor. Essentially, these cells showed the same responses (supplemental Fig. S1).
In summary, TGF-β1 and S1PR agonists showed commonalities and differences in inducing pro-fibrotic responses. Although only TGF-β1 induced myofibroblast transformation, ECM synthesis and a set of fibrotic target genes were robustly induced by TGF-β1, S1P, and FTY720-P. Ponesimod was less active, and SEW2871 was inactive in all of these readouts.
PI3K/Akt/mTOR and ERK1/2 Signaling, but Not TGFβRI/Smad2/3 Signaling, Is Involved in S1PR Agonist-induced ECM Synthesis
TGF-β1 and S1PR agonists showed differences in their pro-fibrotic profiles, which might be caused by the initiation of different pro-fibrotic signaling pathways. To this end, we focused on signaling pathways known to be activated by TGF-β1 or S1P (3, 22–26). Smad2/3 phosphorylation is a canonical event in the pro-fibrotic signaling of TGF-β1, and therefore we analyzed Smad2/3 phosphorylation after 30 min of stimulation with S1PR agonists (5 μm) or TGF-β1 (5 ng/ml) using Western blotting. Only TGF-β1 induced phosphorylation of Smad2 and Smad3, whereas S1PR agonists did not cause any detectable phosphorylation (Fig. 2A). To fully exclude an indirect involvement of the TGF-β pathway in ECM synthesis by S1PR agonists via the induction of TGF-β1 secretion, the ALK inhibitor SB431542 was employed in the [3H]proline incorporation assay to block TGFβR1 signaling. The inhibitor did not reduce ECM synthesis induced by S1P, FTY720-P, ponesimod, or PDGF-bb (Fig. 2B), excluding an indirect role of this pathway in ECM induction by S1PR agonists. As expected, TGF-β1-induced ECM synthesis was reduced to control levels in presence of the ALK inhibitor.
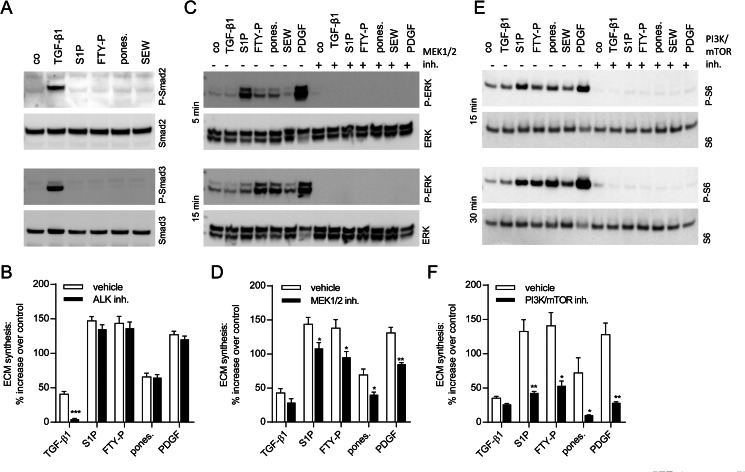
FIGURE 2.
Analysis of TGF-β1- and S1PR agonist-induced signaling pathways and their involvement in ECM synthesis. Activation of Smad2/3, ERK1/2, and PI3K/Akt signaling was studied by Western blotting (A, C, and E), and pathway involvement in ECM synthesis was measured with the [3H]proline incorporation assay (B, D, and F). A, NHLF were stimulated for 30 min with TGF-β1 (5 ng/ml) or S1PR agonists (5 μm), proteins were isolated, and Smad2 and Smad3 phosphorylation compared with total Smad2 and Smad3 was analyzed. B, NHLF were preincubated with vehicle or the ALK inhibitor SB431542 (0.625 μm) for 1 h before stimulation with TGF-β1 (5 ng/ml), S1PR agonists (5 μm), or PDGF-bb (50 ng/ml) for 24 h. C, NHLF were preincubated with vehicle or the MEK1/2 inhibitor PD0325901 (1 μm) for 1 h followed by stimulation for 5 and 15 min with TGF-β1 (5 ng/ml), S1PR agonists (5 μm), or PDGF-bb (50 ng/ml). Proteins were isolated, and ERK1/2 phosphorylation compared with total ERK1/2 was analyzed. D, NHLF were preincubated with vehicle or the MEK1/2 inhibitor (1 μm) for 1 h before stimulation with TGF-β1, S1PR agonists, or PDGF-bb for 24 h with concentrations as under B. E, NHLF were preincubated with vehicle or PI3K/mTOR inhibitor PI-103 (0.5 μm) for 1 h before stimulation with TGF-β1, S1PR agonists, or PDGF-bb for 15 and 30 min with concentrations as under C. Proteins were isolated, and S6 ribosomal protein phosphorylation compared with total S6 ribosomal protein was analyzed. F, NHLF were preincubated with vehicle or PI3K/mTOR inhibitor (0.5 μm) for 1 h before stimulation with TGF-β1, S1PR agonists, or PDGF-bb for 24 h with concentrations as under B. The data in A, C, and E show representative experiments (n = 2). The data in B, D, and F represent the means ± S.E. of three or four independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001 (t test, vehicle versus inhibitor treatment).
Next, we analyzed induction of ERK1/2 phosphorylation by the various stimulants. S1P, FTY720-P, and ponesimod (5000 nm) led to increased ERK1/2 phosphorylation 5 and 15 min after stimulation (Fig. 2C), which was completely inhibited by the MEK1/2 inhibitor PD0325901. SEW2871 showed weak ERK1/2 phosphorylation after 15 min. In contrast, TGF-β1 did not lead to ERK1/2 phosphorylation. As expected, PDGF-bb induced ERK1/2 phosphorylation, which was also blocked by the MEK1/2 inhibitor (Fig. 2C). We then performed the [3H]proline incorporation assay in the presence of the MEK1/2 inhibitor to determine the contribution of ERK1/2 signaling to S1PR agonist-induced ECM synthesis. We observed a small but significant reduction in ECM synthesis induced by S1P, FTY720-P, ponesimod, and PDGF-bb, but not by TGF-β1 (Fig. 2D), suggesting a minor contribution of ERK signaling in S1PR agonist-induced ECM synthesis.
We then studied activation of the PI3K/Akt pathway by analyzing phosphorylation of S6 ribosomal protein, which represents a downstream target of Akt. As shown in Fig. 2E, S1P, FTY720-P, ponesimod (5 μm), and, as expected, PDGF-bb (50 ng/ml) induced S6 ribosomal protein phosphorylation after 15 and 30 min. SEW2871 showed weak phosphorylation after 30 min. Importantly, S6 ribosomal protein phosphorylation was completely inhibited by the PI3K/mTOR inhibitor PI-103, demonstrating that the PI3K/Akt signaling pathway was activated. In contrast to the S1PR agonists and PDGF-bb, TGF-β1 did not cause S6 ribosomal protein phosphorylation. To analyze the contribution of PI3K/Akt signaling to S1PR agonist-induced ECM synthesis, [3H]proline incorporation assays were performed in the presence of the PI3K/mTOR inhibitor. As shown in Fig. 2F, S1P-, FTY720-P-, ponesimod-, and PDGF-bb-induced, but not TGF-β1-induced, ECM synthesis was significantly attenuated in the presence of the inhibitor.
Thus, in NHLF, S1PR agonists and TGF-β1 utilized different pathways to induce ECM synthesis. The S1PR agonists did not induce Smad2/3 phosphorylation but activated ERK1/2 and PI3K/Akt pathways with PI3K/Akt signaling being central for ECM synthesis.
S1P Receptor Subtype Selectivity Accounts for Different Levels of Maximal ECM Synthesis
The S1PR agonists S1P, FTY720-P, and ponesimod displayed different potencies and maximal efficacies in activating ECM synthesis and inducing pro-fibrotic gene expression, whereas SEW2871 was inactive in these assays (Fig. 1). These agonists vary in their selectivity towards S1PR subtypes, with S1P and FTY720-P being the least selective (3, 13, 14), ponesimod displaying improved S1P1R selectivity (16, 17), and SEW2871 being highly selective for S1P1R (15).
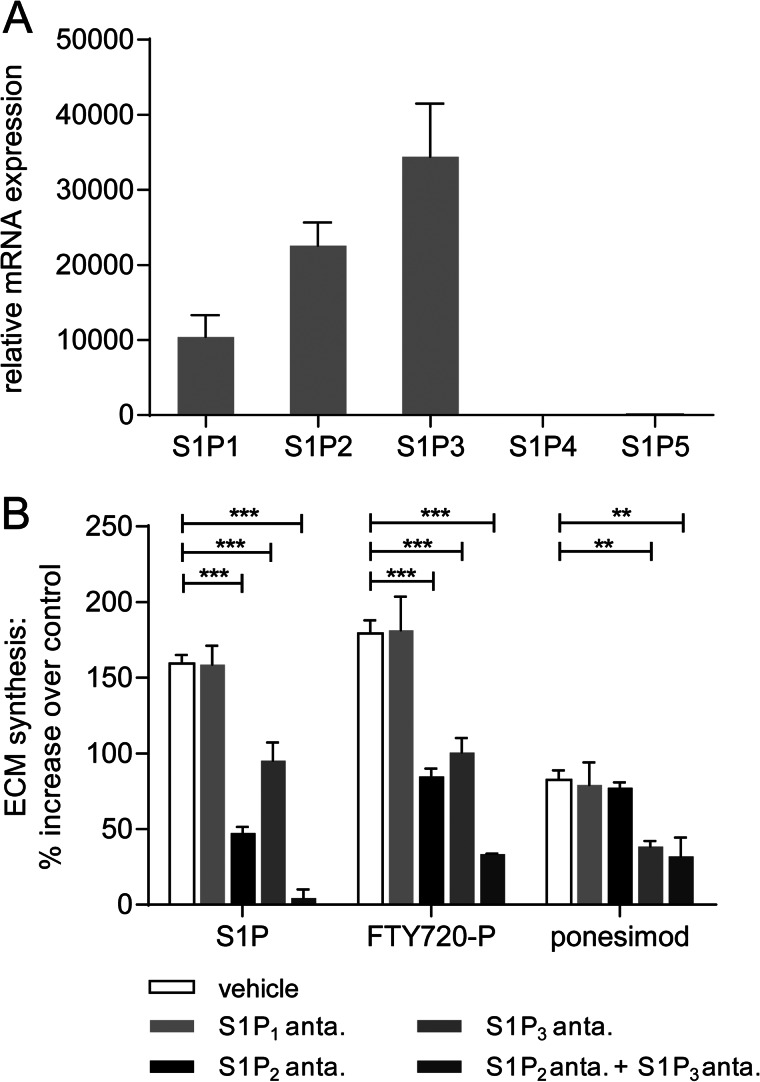
We therefore investigated whether the different receptor selectivity profiles would account for the observed differences in increasing ECM synthesis. First, the S1PR subtype mRNA expression in NHLF was analyzed, and Fig. 3A illustrates that S1P1R, S1P2R, and S1P3R mRNAs were expressed in these cells, whereas S1P4R and S1P5R mRNAs were not detected. We then investigated the contribution of S1P1,2,3R subtype signaling and performed [3H]proline incorporation assays in the presence of specific S1PR antagonists (Fig. 3B). The S1PR agonists S1P, FTY720-P, and ponesimod were tested at MEC (5 μm). The presence of the S1P1R antagonist W146 did not affect the induction of ECM synthesis, which confirmed that S1P1R signaling was not involved in the stimulation of ECM synthesis as already suggested by the inactivity of SEW2871 in this assay. In contrast, preincubation with the S1P2R antagonist JTE-013 significantly attenuated the S1P- and FTY720-P-induced ECM synthesis (Fig. 3B). Ponesimod-induced ECM synthesis, however, was not changed by this antagonist. These results suggested a role for S1P2R signaling in ECM induction by S1P and, surprisingly, FTY720-P, but not by ponesimod. Preincubation with the S1P3R antagonist TY-52156 reduced the induction of ECM synthesis by S1P, FTY720-P, and ponesimod, suggesting a role for S1P3R activation in ECM synthesis induction for all three compounds. To analyze whether S1P2R and S1P3R contributed in an additive way, we performed the [3H]proline incorporation assay in the presence of both S1P2R and S1P3R antagonists. The S1P- and FTY720-P-induced ECM increases were even further attenuated, whereas the combination of both antagonists did not result in any further reduction of the ponesimod-induced ECM synthesis, when compared with using an S1P3R antagonist alone.
FIGURE 3.
Analysis of S1PR subtype contribution to S1PR agonist-induced ECM synthesis. A, relative S1PR mRNA expression in NHLF was analyzed by qPCR. The data show the means ± S.E. of three independent experiments. For normalization, 18 S, HPRT1, and PPIA were used. B, NHLF were preincubated with vehicle, S1P1R, S1P2R, or S1P3R antagonist or a combination of S1P2R and S1P3R antagonists for 1 h before stimulation with S1PR agonists (5 μm) for 24 h. ECM synthesis was then measured with the [3H]proline incorporation assay. The data represent the means ± S.E. of three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001 (t test, vehicle versus antagonist treatment).
Taken together, we could attribute the pronounced ECM response of S1P and FTY720-P to the activation of both S1P2R and S1P3R, which acted in an additive fashion. The less robust and less potent response to ponesimod was due to an activation of S1P3R and the lack of S1P2R agonism.
FTY720-P, but Not Ponesimod and SEW2871, Induces Signaling in Recombinant CHO-S1P2 Cells
FTY720-P is often described as a nonselective S1P1,3,4,5R agonist that does not activate the S1P2R (2, 13). However, several publications have described FTY720-P as an S1P2 R agonist of moderate potency in recombinant systems (14, 27, 28).
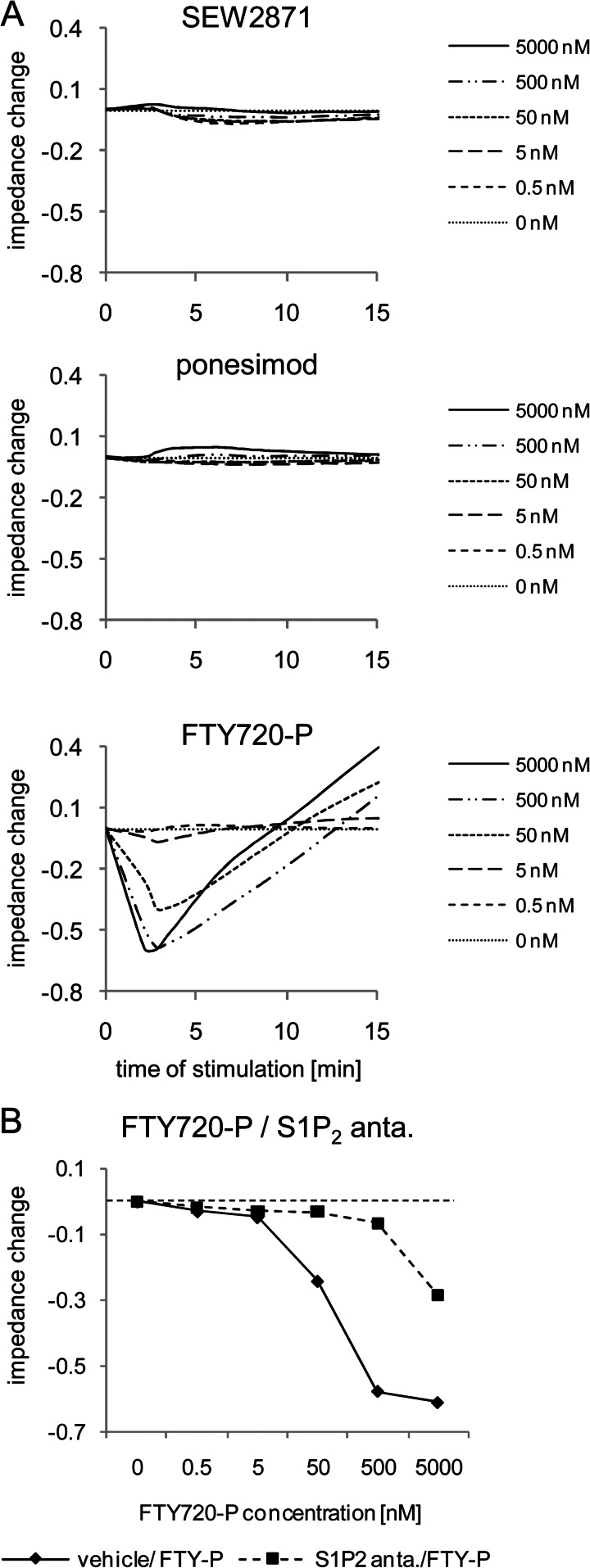
We therefore employed signaling assays using the label-free impedance technique to confirm our data on S1P2R activation by FTY720-P. This technique allows for noninvasive analysis of receptor activation within cell monolayers, and it is a well established technology to analyze integrated responses of G protein-coupled receptor signaling (29, 30). CHO-K1 cells with stable expression of the human S1P2R (CHO-S1P2) were stimulated with different concentrations of FTY720-P, ponesimod, or SEW2871, and impedance responses were monitored for up to 15 min (Fig. 4A). SEW2871 and ponesimod did not induce any change in impedance up to 5000 nm. In contrast, FTY720-P induced a rapid, concentration-dependent and saturable decrease of impedance with a maximal decrease reached after 2.5 min. Impedance values at 2.5 min were used to generate concentration-response curves, and an EC50 of 33 nm (18-69 nm) for FTY720-P (n = 3, geometric mean (range of values)) was determined. In the presence of the S1P2R antagonist JTE-013, the FTY720-P-induced impedance response was strongly inhibited, as shown by the rightward shift of the FTY720-P concentration-response curve (Fig. 4B). In the CHO-K1 pcDNA3.1 control cell line, all agonists were inactive (data not shown). Thus, using impedance measurements, we show that FTY720-P acted as an S1P2R agonist, whereas ponesimod and SEW2871 were inactive at this receptor.
FIGURE 4.
Analysis of S1P2R signaling in recombinant CHO-S1P2 cells using impedance assays. A, CHO-S1P2 cells were stimulated with SEW2871, ponesimod, or FTY720-P (0.5–5000 nm), and impedance responses were followed for 15 min. B, cells were preincubated with or without S1P2R antagonist (anta.) and then stimulated with dilution series of FTY720-P. Impedance responses were monitored over 15 min. Concentration-response curves of FTY720-P in the absence or presence of S1P2R antagonist were then generated with impedance values at 2.5 min. The data show a representative experiment (n = 2).
S1PR Agonists Activate Different S1PR Subtypes in NHLF
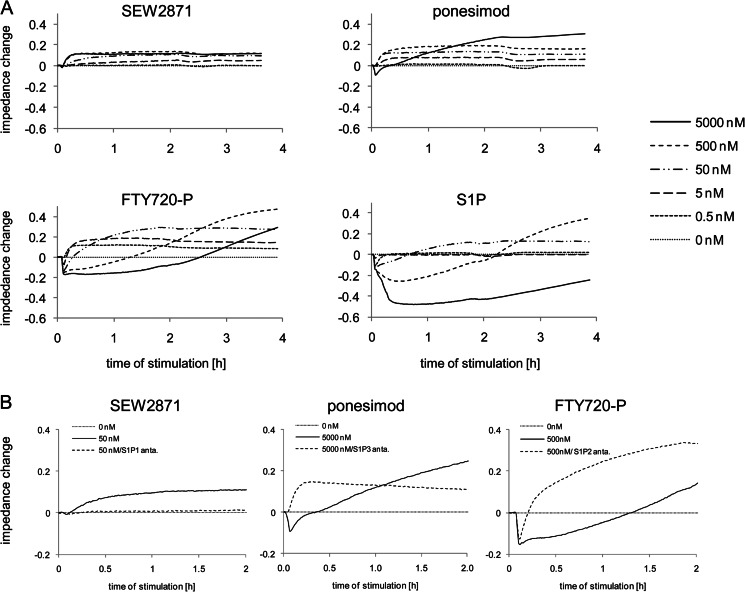
We then investigated the S1PR agonist-induced signaling in NHLF, which expressed S1P1R, S1P2R, and S1P3R mRNA (Fig. 3A), using impedance measurements. NHLF were stimulated with different concentrations of SEW2871, ponesimod, FTY720-P, or S1P, and impedance changes versus base line were followed for up to 4 h (Fig. 5A). The highly selective S1P1R agonist SEW2871 induced a rapid, concentration-dependent and saturable impedance increase reaching a plateau within 15 min of agonist addition. Ponesimod induced a similar increase in impedance at concentrations between 0.5 and 500 nm. At 5000 nm (solid line) a biphasic response was observed that showed a rapid impedance decrease with a maximal reduction at 3 min, followed by a gradual increase over base line reaching a plateau after ∼2 h. FTY720-P induced a signaling profile with up to three different response phases depending on the concentration. At the lowest concentration of 0.5 nm, FTY720-P induced a response that was identical to the responses to SEW2871 and ponesimod. At 5 and 50 nm, FTY720-P showed a biphasic response reminiscent of the ponesimod response at 5000 nm. At 500 and 5000 nm, FTY720-P induced a triphasic response, in which the rapid decrease in impedance at 3 min was followed by a prolonged decrease below base line for up to 2.5 h after agonist addition. Thereafter, impedance increased over base line. S1P induced a signaling profile similar to that of FTY720-P, showing a triphasic response at the higher concentrations.
FIGURE 5.
Analysis of S1PR agonist-induced signaling in NHLF using impedance assays. A, NHLF were stimulated with S1PR agonists (0.5–5000 nm), and impedance responses were monitored over 4 h. B, NHLF were preincubated with or without S1P1R, S1P2R, or S1P3R antagonists for 1 h and then stimulated with SEW2871 (50 nm), ponesimod (5000 nm), or FTY720-P (500 nm). Signaling was monitored over 2 h to reveal S1PR subtype signaling.
To link the individual responses in impedance to S1PR subtype activation, the impedance assays were performed in the presence of S1PR subtype-specific antagonists. As seen in Fig. 5B, preincubation with the S1P1R antagonist W146 reversed the SEW2871-induced impedance (50 nm) to base line, demonstrating that the observed effects were S1P1R-mediated. The ponesimod-induced increase of impedance that was observed at concentrations between 0.5 and 500 nm was blocked by preincubation with the S1P1R antagonist, which confirmed selective S1P1R activation at these concentrations (supplemental Fig. S2A). The rapid decrease of impedance only observed at the highest tested concentration of ponesimod (5000 nm) was due to S1P3R activation, because preincubation with the S1P3R antagonist TY-52156 converted the biphasic signal to a monophasic signal that showed the characteristics of a pure S1P1R response (Fig. 5B). The FTY720-P induced prolonged decrease of impedance observed at 500 and 5000 nm was blocked by preincubation with the S1P2R antagonist JTE-013, confirming S1P2R activation by FTY720-P also in NHLF. Only the rapid impedance decrease (S1P3 response), which was followed by an increase (S1P1 response), remained (Fig. 5B). Supplemental Fig. S2 shows detailed data that corroborate these interpretations.
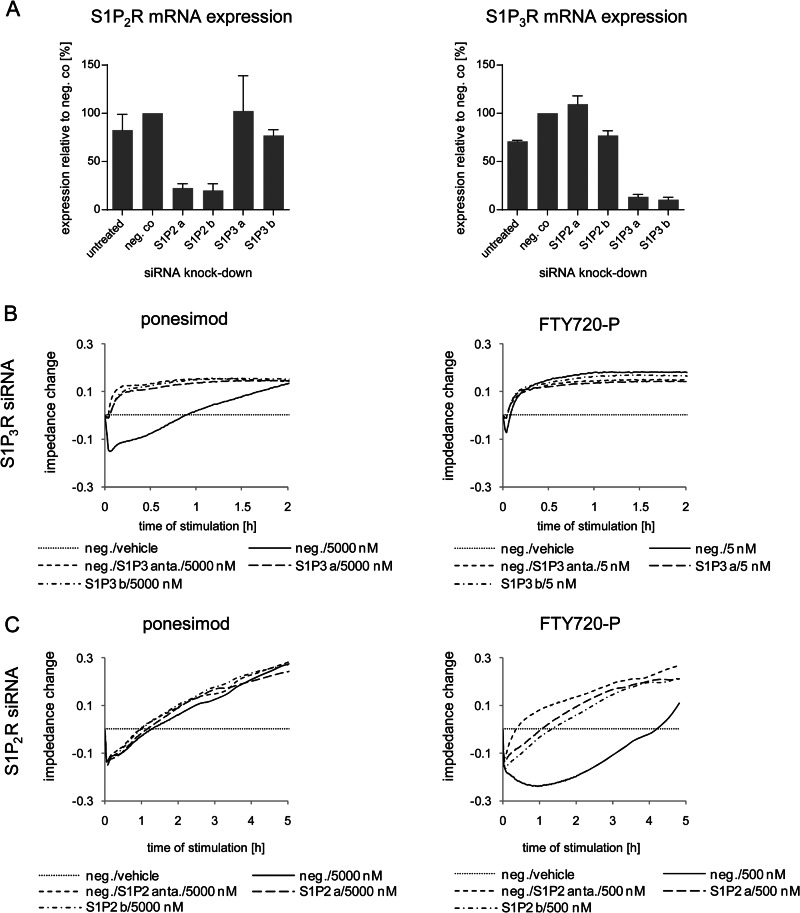
To confirm these conclusions on receptor subtype activation in NHLF, we used the siRNA technology as an independent approach to inactivate S1P receptor subtypes. To this end, we performed similar impedance experiments as described above using siRNA against S1P2R and S1P3R. These receptors are activated by S1P and FTY720-P and had been identified to play an important role in ECM induction. Forty-eight hours after siRNA transfection ∼80% of the S1P2R and ∼90% of the S1P3R mRNAs were down-regulated by S1P2R and S1P3R siRNAs, respectively, compared with NHLF transfected with negative control siRNA (Fig. 6A). At this time point, NHLF were subjected to FTY720-P (5 or 500 nm) or ponesimod (5000 nm), and impedance changes were recorded. As shown in Fig. 6B, knockdown of S1P3R by two independent siRNAs abolished the first response phase and thus converted the biphasic ponesimod-induced response (5000 nm) into a monophasic response. This confirmed the result using the S1P3R antagonist. Comparable results were obtained after stimulation with 5 nm FTY720-P (Fig. 6B). Knockdown of S1P2R blocked the prolonged decrease of impedance observed after treatment with FTY720-P (500 nm) similarly as pretreatment with the S1P2R antagonist (Fig. 6C). Ponesimod-induced responses were affected neither by the S1P2R antagonist nor by S1P2R knockdown (Fig. 6C). Thus, highly comparable results were obtained with selective S1PR antagonists and S1P2R and S1P3R mRNA knockdown, confirming the specificity of the pharmacological receptor antagonists and confirming the previously deduced S1P2R and S1P3R activation by FTY720-P.
FIGURE 6.
Analysis of S1P2,3R subtype signaling in NHLF using siRNA knockdown and impedance assays. A, relative mRNA expression changes of S1P2R and S1P3R in NHLF 48 h after transfection with negative control (neg. co) siRNA, S1P2R siRNA a, S1P2R siRNA b, S1P3R siRNA a, or S1P3R siRNA b compared with nontransfected NHLF (untreated). Expression of NHLF transfected with negative control siRNA was set to 100%. The data represent the means ± S.E. of two independent experiments. B and C, NHLF were transfected with negative control siRNA (neg.; B and C), two S1P3R siRNAs (S1P3 a and S1P3 b; B), or two S1P2R siRNAs (S1P2 a and S1P2 b; C). After 48 h, NHLF transfected with negative control siRNAs were preincubated with or without S1P2R (C) or S1P3R (B) antagonists for 1 h. Then all samples were stimulated with vehicle, ponesimod (5000 nm; B and C), or FTY720-P (5 nm, B; 500 nm, C). Signaling was monitored over 2–5 h to confirm S1P2R and S1P3R subtype signaling deciphered with pharmacological S1PR antagonists. The data show representative experiments (n = 3).
In summary, the four tested S1PR agonists activated S1PR subtypes to different extent in NHLF. Although all agonists activated S1P1R, ponesimod additionally displayed minimal activity on S1P3R, but only S1P and FTY720-P efficiently activated S1P2R and S1P3R.
In Myofibroblasts, S1P and FTY720-P Effectively Induce ECM Synthesis, whereas Ponesimod or SEW2871 Are Inactive
Myofibroblasts represent an important cell type in physiological and pathophysiological processes (31). To analyze S1PR-induced responses in myofibroblasts, we incubated NHLF with TGF-β1 for 72 h and verified myofibroblast differentiation using αSMA immunofluorescent staining (Fig. 7A). Analysis of the S1PR expression levels of such differentiated myofibroblasts (Fig. 7B) revealed that S1P1R and S1P3R mRNAs were 11- and 5-fold down-regulated compared with nontransformed NHLF, whereas S1P2R mRNA was less affected (1.7-fold down-regulated).
FIGURE 7.
Analysis of S1PR mRNA expression, signaling, and ECM induction in NHLF-derived myofibroblasts. A, NHLF were treated with or without 1 ng/ml TGF-β1 for 72 h, reseeded, and starved for 24 h before immunofluorescent staining was performed. Green, α-smooth muscle actin; blue, nuclei. B, expression changes of S1PR mRNA in myofibroblasts compared with nontransformed NHLF. The data show the means ± S.E. of three independent experiments. C, myofibroblasts were stimulated with SEW2871, ponesimod, FTY720-P, or S1P (0.5–5000 nm), and impedance responses were monitored for 15 h. D, concentration response curves for FTY720-P in the absence or presence of S1P2R or S1P3R antagonists generated from the impedance values at 90 min. The data in C and D show representative experiments (n = 2–3). E, myofibroblasts were stimulated with S1PR agonists (20–1250 nm), and ECM synthesis was measured after 24 h with the [3H]proline incorporation assay. The data represent the means ± S.E. of three independent experiments. *, p < 0.05; ***, p < 0.001, one-way analysis of variance, Dunnett's post test.
Next, we performed impedance assays using myofibroblasts and stimulated them with different concentrations of SEW2871, ponesimod, FTY720-P, and S1P. SEW2871 and ponesimod did not induce any impedance change, indicating the lack of S1P1R and S1P3R functionality in these cells (Fig. 7C). In contrast, FTY720-P induced a long lasting (>15 h) decrease in impedance (LEC, 500 nm), whereas the lower tested concentrations of FTY720-P were inactive. The S1P-induced impedance was comparable to that of FTY720-P, although the potency was higher (LEC, 5 nm). To decipher the S1PR subtype activation by FTY720-P, we performed concentration-response experiments in absence or presence of S1P2R or S1P3R antagonists and generated concentration-response curves at 90 min, the time of maximal signal induction (Fig. 7D). The FTY720-P-induced response was essentially unaffected by the S1P3R antagonist but was completely blocked by the S1P2R antagonist, demonstrating that the observed response to FTY720-P was S1P2R-mediated. These data suggest that in myofibroblasts mainly S1P2R was activated, whereas S1P1R and S1P3R signaling was essentially absent. Finally, we tested whether the changes in the receptor expression status and activation in myofibroblasts also translated in an altered ECM response. As seen in Fig. 7E, SEW2871 and ponesimod did not stimulate any increase in ECM synthesis in myofibroblasts, whereas S1P (LEC, 313 nm) and FTY720-P (LEC, 78 nm) induced a concentration-dependent and significant increase in ECM synthesis. ECM levels were increased 27 and 34% over control at MEC (1250 nm) for S1P and FTY720-P, respectively (Fig. 7E). In summary, the reduced S1P3R expression of myofibroblasts caused a loss of pro-fibrotic activity for ponesimod at all tested concentrations, whereas S1P and FTY720-P remained potent inducers of ECM synthesis via S1P2R activation.
DISCUSSION
The S1P system is known to be involved in many different physiological processes, and more recently, the potential role of S1P signaling in fibrosis was described. In this study, we characterized the pro-fibrotic effects of S1PR signaling in normal human lung fibroblasts and demonstrated that the nonselective S1PR agonists S1P and FTY720-P were robust inducers of ECM synthesis and fibrotic gene expression in NHLF, whereas the compounds with higher S1P1R selectivity, ponesimod and SEW2871, were less active or fully inactive. Interestingly, TGF-β1 but not S1PR agonists induced fibroblast to myofibroblast differentiation. These different response patterns led us to investigate the molecular pathways leading to enhanced ECM synthesis in more detail. Our investigations revealed that TGF-β1 and S1PR agonists use different pathways to stimulate ECM synthesis in NHLF. TGF-β1 signaled via the TGFβR1/Smad pathway, whereas S1PR agonists induce ECM synthesis through PI3K/Akt- and ERK1/2-dependent and Smad2/3-independent pathways.
Although Smad2 and Smad3 phosphorylation and αSMA up-regulation are well established pro-fibrotic effects of TGF-β1, only a few groups have examined the signaling effects of S1P or FTY720-P in fibroblasts in molecular detail. Gellings Lowe et al. (10) and Urata et al. (12) showed weak induction of αSMA, but both studies lack comparison with the known mediator TGF-β1. Interestingly, Keller et al. (11) found comparable Smad3-dependent induction of αSMA by TGF-β1, S1P, and FTY720-P in human dermal foreskin fibroblasts. However, induction of αSMA by TGF-β1 was surprisingly low in their study. We show that, in NHLF, TGF-β1 is a very strong inducer of αSMA expression as well as Smad phosphorylation, identifying this cell type as a sensitive system to study TGF-β pathway activation. In direct comparison with TGF-β1 we found that S1PR agonists did not induce myofibroblast transformation or Smad phosphorylation, despite showing robust activation of ECM synthesis and pro-fibrotic gene transcription. Instead of activating pro-fibrotic Smad signaling in NHLF, S1PR agonists activated ERK1/2 and PI3K/Akt pathways, both of which have been previously described to be activated by S1PR agonists in rat renal mesangial cells (25, 26). Moreover, we established a causal link between the activation of these pathways and ECM synthesis using pathway-specific inhibitors and demonstrated that the PI3K/Akt/mTOR pathway is predominantly responsible in the induction of ECM synthesis by S1PR agonists. In contrast, TGF-β1-induced ECM synthesis was independent of these pathways.
We also analyzed gene expression changes in NHLF in response to TGF-β1 and S1PR agonist incubation and focused on genes that were previously described to be involved in tissue fibrosis (32–40). We thus uncovered additional commonalities and differences of the pro-fibrotic activity of TGF-β1 and S1PR agonists. The gene set that contained genes encoding ECM components, i.e., collagen Ia1, fibronectin, and elastin, were only regulated by TGF-β1. In contrast, other fibrotic genes, such as CTGF, SERPINE1, thrombospondin 1, CCL2, IL6, SphK1, or hyaluronan synthase 2 were up-regulated by TGF-β1, S1P, and FTY720-P and to a lower extent by ponesimod. Induction of CTGF especially is of significance because it is known to be one of the major mediators of fibrosis (33, 41, 42). SphK1 is discussed as a link between TGF-β1 and S1P signaling, allowing cross-talk between both pathways (7).
Another important finding was the different intensity of the fibrotic response that was induced by the different S1PR agonists and their link to receptor subtype activation: S1P and FTY720-P activated S1P2R and S1P3R and were robust inducers of ECM synthesis, whereas ponesimod activated S1P3R only at high concentrations and was inactive on S1P2R, leading to a less effective and less potent induction of ECM synthesis. SEW2871, a highly selective S1P1R agonist, was inactive in all pro-fibrotic readouts despite the activation of S1P1R signaling as shown by impedance assays. These data strongly suggest that S1P1R are not involved in pro-fibrotic responses in NHLF, whereas S1P2R and S1P3R activation contribute to pro-fibrotic processes in an additive fashion. The extent of the individual receptor contribution to the final fibrotic response depends on receptor expression levels of the target cell as exemplified in NHLF-derived myofibroblasts, in which pro-fibrotic responses were entirely driven by S1P2R after down-regulation of S1P3R. Taken together, our data suggest that, depending on the differentiation status of fibroblasts or the expression ratio of S1P2R/S1P3R, the pro-fibrotic effects of S1PR agonists may vary. In agreement with these findings, several reports in the literature underline the importance of S1P2R and S1P3R for fibrosis. FTY720-P-induced myofibroblast transformation of dermal foreskin fibroblasts was shown to be dependent on S1P3R activation (11), whereas the S1P-induced αSMA and collagen expression were S1P2R-dependent in mouse cardiac fibroblasts (10). Gil et al. (23) proposed activation of S1P1R/S1P3R in S1P-directed chemotaxis in dermal foreskin fibroblasts. Finally, involvement of S1P/S1P3R signaling in vivo was suggested in several animal models (8, 39).
We were surprised to see the S1P2R dependence of FTY720-P-induced fibrotic responses, because most publications describe this compound as an S1P1,3,4,5R agonist lacking S1P2R agonism (13). However, several reports raised the possibility of biased S1P2R agonism of FTY720-P, suggesting that G protein-coupled responses measured by classical GTPγS assays did not fully capture S1P2R activation (14, 27, 28). To further study S1P2R activation by FTY720-P, we thus employed recombinant huS1P2R-expressing CHO cells or NHLF and used label-free impedance technology, which measures integrated cell responses in real time without using any label. Impedance is therefore an unbiased technology that allows the detection of GPCR agonism irrespective of its type of G protein coupling. We clearly showed that FTY720-P was an S1P2R agonist in CHO-S1P2 cells, whereas ponesimod and SEW2871 were inactive. Furthermore, by using selective S1PR antagonists and S1P2,3R siRNA, we were able to assign the individual phases of the responses to the individual receptor subtypes in NHLF and showed activation of S1P1R, S1P2R, and S1P3R by S1P and FTY720-P, activation of S1P1R and S1P3R by ponesimod, and activation of S1P1R by SEW2871. Impedance therefore allowed us to confirm our findings on receptor subtype contributions to ECM induction, showing increased ECM synthesis upon activation of S1P2R and S1P3R by S1P and FTY720-P in NHLF. The less pronounced and less potent response to ponesimod was due to the low potency activation of S1P3R and the lack of S1P2R agonism.
In conclusion, we analyzed S1PR biology in human lung fibroblasts and investigated their pro-fibrotic potential. We showed that in NHLF, S1P2R and S1P3R activation and signaling via the PI3K/Akt pathway but not the TGFβR1/Smad pathway results in pro-fibrotic responses such as increased ECM synthesis and fibrotic gene expression. Therefore, avoiding S1P2R and S1P3R activity, as achieved for ponesimod and SEW2871, reduces the pro-fibrotic potential of this promising new class of immunomodulators.
Supplementary Material
Acknowledgments
We thank Patrick Brossard for careful review of the manuscript and Axel Klenk for help with statistics.

This article contains supplemental Table S1 and Figs. S1 and S2.
- S1P
- sphingosine 1-phosphate
- ALK
- activin receptor-like kinase
- αSMA
- α-smooth muscle actin
- CTGF
- connective tissue growth factor
- ECM
- extracellular matrix
- LEC
- lowest effective concentration
- MEC
- maximal effective concentration
- mTOR
- mammalian target of rapamycin
- NHLF
- normal human lung fibroblasts
- rb
- rabbit
- SERPINE1
- serpine peptidase inhibitor 1
- SphK
- sphingosine kinase
- S1PR
- sphingosine 1-phosphate receptor
- TGFβR1
- TGF-β receptor 1
- qPCR
- quantitative PCR.
REFERENCES
- 1. Strub G. M., Maceyka M., Hait N. C., Milstien S., Spiegel S. (2010) Extracellular and intracellular actions of sphingosine-1-phosphate. Adv. Exp. Med. Biol. 688, 141–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brinkmann V., Billich A., Baumruker T., Heining P., Schmouder R., Francis G., Aradhye S., Burtin P. (2010) Fingolimod (FTY720). Discovery and development of an oral drug to treat multiple sclerosis. Nat. Rev. Drug Discov. 9, 883–897 [DOI] [PubMed] [Google Scholar]
- 3. Brinkmann V. (2007) Sphingosine 1-phosphate receptors in health and disease. Mechanistic insights from gene deletion studies and reverse pharmacology. Pharmacol. Ther. 115, 84–105 [DOI] [PubMed] [Google Scholar]
- 4. Hardie W. D., Glasser S. W., Hagood J. S. (2009) Emerging concepts in the pathogenesis of lung fibrosis. Am. J. Pathol. 175, 3–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Milara J., Navarro R., Juan G., Peiró T., Serrano A., Ramón M., Morcillo E., Cortijo J. (2012) Sphingosine-1-phosphate is increased in patients with idiopathic pulmonary fibrosis and mediates epithelial to mesenchymal transition. Thorax 67, 147–156 [DOI] [PubMed] [Google Scholar]
- 6. Li C., Zheng S., You H., Liu X., Lin M., Yang L., Li L. (2011) Sphingosine 1-phosphate (S1P)/S1P receptors are involved in human liver fibrosis by action on hepatic myofibroblasts motility. J. Hepatol. 54, 1205–1213 [DOI] [PubMed] [Google Scholar]
- 7. Kono Y., Nishiuma T., Nishimura Y., Kotani Y., Okada T., Nakamura S., Yokoyama M. (2007) Sphingosine kinase 1 regulates differentiation of human and mouse lung fibroblasts mediated by TGF-β1. Am. J. Respir. Cell Mol. Biol. 37, 395–404 [DOI] [PubMed] [Google Scholar]
- 8. Li C., Jiang X., Yang L., Liu X., Yue S., Li L. (2009) Involvement of sphingosine 1-phosphate (SIP)/S1P3 signaling in cholestasis-induced liver fibrosis. Am. J. Pathol. 175, 1464–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.NDA 02257-FDA Approved Labeling Text for Gilenya (fingolimod) capsules. September 21, 2010. (www.accessdata.fda.gov/drugsatfda_ docs/label/2010/022527s000lbl.pdf)
- 10. Gellings Lowe N., Swaney J. S., Moreno K. M., Sabbadini R. A. (2009) Sphingosine-1-phosphate and sphingosine kinase are critical for transforming growth factor-β-stimulated collagen production by cardiac fibroblasts. Cardiovasc. Res. 82, 303–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Keller C. D., Rivera Gil P., Tölle M., van der Giet M., Chun J., Radeke H. H., Schäfer-Korting M., Kleuser B. (2007) Immunomodulator FTY720 induces myofibroblast differentiation via the lysophospholipid receptor S1P3 and Smad3 signaling. Am. J. Pathol. 170, 281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Urata Y., Nishimura Y., Hirase T., Yokoyama M. (2005) Sphingosine 1-phosphate induces α-smooth muscle actin expression in lung fibroblasts via Rho-kinase. Kobe J. Med. Sci. 51, 17–27 [PubMed] [Google Scholar]
- 13. Brinkmann V., Davis M. D., Heise C. E., Albert R., Cottens S., Hof R., Bruns C., Prieschl E., Baumruker T., Hiestand P., Foster C. A., Zollinger M., Lynch K. R. (2002) The immune modulator FTY720 targets sphingosine 1-phosphate receptors. J. Biol. Chem. 277, 21453–21457 [DOI] [PubMed] [Google Scholar]
- 14. Wetter J. A., Revankar C., Hanson B. J. (2009) Utilization of the Tango β-arrestin recruitment technology for cell-based EDG receptor assay development and interrogation. J. Biomol. Screen 14, 1134–1141 [DOI] [PubMed] [Google Scholar]
- 15. Sanna M. G., Liao J., Jo E., Alfonso C., Ahn M. Y., Peterson M. S., Webb B., Lefebvre S., Chun J., Gray N., Rosen H. (2004) Sphingosine 1-phosphate (S1P) receptor subtypes S1P1 and S1P3, respectively, regulate lymphocyte recirculation and heart rate. J. Biol. Chem. 279, 13839–13848 [DOI] [PubMed] [Google Scholar]
- 16. Bolli M. H., Abele S., Binkert C., Bravo R., Buchmann S., Bur D., Gatfield J., Hess P., Kohl C., Mangold C., Mathys B., Menyhart K., Müller C., Nayler O., Scherz M., Schmidt G., Sippel V., Steiner B., Strasser D., Treiber A., Weller T. (2010) 2-Imino-thiazolidin-4-one derivatives as potent, orally active S1P1 receptor agonists. J. Med. Chem. 53, 4198–4211 [DOI] [PubMed] [Google Scholar]
- 17. Piali L., Froidevaux S., Hess P., Nayler O., Bolli M. H., Schlosser E., Kohl C., Steiner B., Clozel M. (2011) The selective sphingosine 1-phosphate receptor 1 agonist ponesimod protects against lymphocyte-mediated tissue inflammation. J. Pharmacol. Exp. Ther. 337, 547–556 [DOI] [PubMed] [Google Scholar]
- 18. Border W. A., Noble N. A. (1994) Transforming growth factor β in tissue fibrosis. N. Engl. J. Med. 331, 1286–1292 [DOI] [PubMed] [Google Scholar]
- 19. Murakami A., Takasugi H., Ohnuma S., Koide Y., Sakurai A., Takeda S., Hasegawa T., Sasamori J., Konno T., Hayashi K., Watanabe Y., Mori K., Sato Y., Takahashi A., Mochizuki N., Takakura N. (2010) Sphingosine 1-phosphate (S1P) regulates vascular contraction via S1P3 receptor. Investigation based on a new S1P3 receptor antagonist. Mol. Pharmacol. 77, 704–713 [DOI] [PubMed] [Google Scholar]
- 20. Morrison K., Ernst R., Hess P., Studer R., Clozel M. (2010) Selexipag. A selective prostacyclin receptor agonist that does not affect rat gastric function. J. Pharmacol. Exp. Ther. 335, 249–255 [DOI] [PubMed] [Google Scholar]
- 21. Gore-Hyer E., Pannu J., Smith E. A., Grotendorst G., Trojanowska M. (2003) Selective stimulation of collagen synthesis in the presence of costimulatory insulin signaling by connective tissue growth factor in scleroderma fibroblasts. Arthritis Rheum. 48, 798–806 [DOI] [PubMed] [Google Scholar]
- 22. Biernacka A., Dobaczewski M., Frangogiannis N. G. (2011) TGF-β signaling in fibrosis. Growth Factors 29, 196–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gil P. R., Japtok L., Kleuser B. (2010) Sphingosine 1-phosphate mediates chemotaxis of human primary fibroblasts via the S1P-receptor subtypes S1P1 and S1P3 and Smad-signalling. Cytoskeleton 67, 773–783 [DOI] [PubMed] [Google Scholar]
- 24. Sauer B., Vogler R., von Wenckstern H., Fujii M., Anzano M. B., Glick A. B., Schäfer-Korting M., Roberts A. B., Kleuser B. (2004) Involvement of Smad signaling in sphingosine 1-phosphate-mediated biological responses of keratinocytes. J. Biol. Chem. 279, 38471–38479 [DOI] [PubMed] [Google Scholar]
- 25. Xin C., Ren S., Eberhardt W., Pfeilschifter J., Huwiler A. (2006) The immunomodulator FTY720 and its phosphorylated derivative activate the Smad signalling cascade and upregulate connective tissue growth factor and collagen type IV expression in renal mesangial cells. Br. J. Pharmacol. 147, 164–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xin C., Ren S., Kleuser B., Shabahang S., Eberhardt W., Radeke H., Schäfer-Korting M., Pfeilschifter J., Huwiler A. (2004) Sphingosine 1-phosphate cross-activates the Smad signaling cascade and mimics transforming growth factor-β-induced cell responses. J. Biol. Chem. 279, 35255–35262 [DOI] [PubMed] [Google Scholar]
- 27. Gräler M. H., Goetzl E. J. (2004) The immunosuppressant FTY720 down-regulates sphingosine 1-phosphate G-protein-coupled receptors. FASEB J. 18, 551–553 [DOI] [PubMed] [Google Scholar]
- 28. Mandala S., Hajdu R., Bergstrom J., Quackenbush E., Xie J., Milligan J., Thornton R., Shei G. J., Card D., Keohane C., Rosenbach M., Hale J., Lynch C. L., Rupprecht K., Parsons W., Rosen H. (2002) Alteration of lymphocyte trafficking by sphingosine-1-phosphate receptor agonists. Science 296, 346–349 [DOI] [PubMed] [Google Scholar]
- 29. Nayler O., Birker-Robaczewska M., Gatfield J. (eds.) (2010) Integration of Label-free Detection Methods in GPCR Drug Discovery, Wiley, Hoboken [Google Scholar]
- 30. Yu N., Atienza J. M., Bernard J., Blanc S., Zhu J., Wang X., Xu X., Abassi Y. A. (2006) Real-time monitoring of morphological changes in living cells by electronic cell sensor arrays. An approach to study G protein-coupled receptors. Anal. Chem. 78, 35–43 [DOI] [PubMed] [Google Scholar]
- 31. Hinz B., Phan S. H., Thannickal V. J., Prunotto M., Desmoulière A., Varga J., De Wever O., Mareel M., Gabbiani G. (2012) Recent developments in myofibroblast biology. Paradigms for connective tissue remodeling. Am. J. Pathol. 180, 1340–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ghosh A. K., Vaughan D. E. (2012) PAI-1 in tissue fibrosis. J. Cell Physiol. 227, 493–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Leask A., Parapuram S. K., Shi-Wen X., Abraham D. J. (2009) Connective tissue growth factor (CTGF, CCN2) gene regulation. A potent clinical bio-marker of fibroproliferative disease? J. Cell Commun. Signal 3, 89–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li Y., Jiang D., Liang J., Meltzer E. B., Gray A., Miura R., Wogensen L., Yamaguchi Y., Noble P. W. (2011) Severe lung fibrosis requires an invasive fibroblast phenotype regulated by hyaluronan and CD44. J. Exp. Med. 208, 1459–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Meléndez G. C., McLarty J. L., Levick S. P., Du Y., Janicki J. S., Brower G. L. (2010) Interleukin 6 mediates myocardial fibrosis, concentric hypertrophy, and diastolic dysfunction in rats. Hypertension 56, 225–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pierce R. A., Mariani T. J., Senior R. M. (1995) Elastin in lung development and disease. CIBA Found. Symp. 192, 199–214 [DOI] [PubMed] [Google Scholar]
- 37. Prasse A., Müller-Quernheim J. (2009) Non-invasive biomarkers in pulmonary fibrosis. Respirology 14, 788–795 [DOI] [PubMed] [Google Scholar]
- 38. Sweetwyne M. T., Murphy-Ullrich J. E. (2012) Thrombospondin1 in tissue repair and fibrosis. TGF-β-dependent and independent mechanisms. Matrix Biol. 31, 178–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Takuwa N., Ohkura S., Takashima S., Ohtani K., Okamoto Y., Tanaka T., Hirano K., Usui S., Wang F., Du W., Yoshioka K., Banno Y., Sasaki M., Ichi I., Okamura M., Sugimoto N., Mizugishi K., Nakanuma Y., Ishii I., Takamura M., Kaneko S., Kojo S., Satouchi K., Mitumori K., Chun J., Takuwa Y. (2010) S1P3-mediated cardiac fibrosis in sphingosine kinase 1 transgenic mice involves reactive oxygen species. Cardiovasc. Res. 85, 484–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wynn T. A. (2011) Integrating mechanisms of pulmonary fibrosis. J. Exp. Med. 208, 1339–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ponticos M., Holmes A. M., Shi-wen X., Leoni P., Khan K., Rajkumar V. S., Hoyles R. K., Bou-Gharios G., Black C. M., Denton C. P., Abraham D. J., Leask A., Lindahl G. E. (2009) Pivotal role of connective tissue growth factor in lung fibrosis. MAPK-dependent transcriptional activation of type I collagen. Arthritis Rheum. 60, 2142–2155 [DOI] [PubMed] [Google Scholar]
- 42. Sonnylal S., Shi-Wen X., Leoni P., Naff K., Van Pelt C. S., Nakamura H., Leask A., Abraham D., Bou-Gharios G., de Crombrugghe B. (2010) Selective expression of connective tissue growth factor in fibroblasts in vivo promotes systemic tissue fibrosis. Arthritis Rheum. 62, 1523–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, RESEARCH0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.