Background: β-Glucosidases hydrolyze cellobiose and are an important step in biomass saccharification.
Results: The structure of JMB19063 is a homodimer, and residues from the partnering monomer form a portion of the active site.
Conclusion: JMB19063 is the first structure in the GH3 family that requires dimerization for catalysis.
Significance: Structural characterization of novel β-glucosidases may provide insights into improving biofuel production.
Keywords: Bacterial Metabolism, Biofuel, Cellulase, Enzyme Kinetics, Structural Biology, β-Glucosidase
Abstract
A recent metagenomic analysis sequenced a switchgrass-adapted compost community to identify enzymes from microorganisms that were specifically adapted to switchgrass under thermophilic conditions. These enzymes are being examined as part of the pretreatment process for the production of “second-generation” biofuels. Among the enzymes discovered was JMB19063, a novel three-domain β-glucosidase that belongs to the GH3 (glycoside hydrolase 3) family. Here, we report the structure of JMB19063 in complex with glucose and the catalytic variant D261N crystallized in the presence of cellopentaose. JMB19063 is first structure of a dimeric member of the GH3 family, and we demonstrate that dimerization is required for catalytic activity. Arg-587 and Phe-598 from the C-terminal domain of the opposing monomer are shown to interact with bound ligands in the D261N structure. Enzyme assays confirmed that these residues are absolutely essential for full catalytic activity.
Introduction
Switchgrass (Panicum virgatum) is one of the leading lignocellulosic feedstock candidates for production of “second-generation” biofuels (1–3), and the development of optimized process technologies for this feedstock has warranted significant attention in the scientific and commercial biofuel sectors. This focus on switchgrass as a feedstock and the desire to pretreat biomass at elevated temperatures has spurred interest in increasing the current repertoire of enzymes used for biofuel production from switchgrass. In a recent metagenomic study, random shotgun sequencing of a switchgrass-adapted compost community was used to identify enzymes from microorganisms that were specifically adapted to switchgrass under thermophilic conditions (4). Sequence analysis identified JMB19063 as a novel β-glucosidase from the GH3 (glycoside hydrolase 3) family. Although the species of origin for JMB19063 is unknown, the sequence most closely resembles proteins from the genera Flavobacterium and Bacteroides, with ∼70–80% identities. In the complex mixture of cellulases required to break down cellulose to glucose, β-glucosidases are the enzymes that hydrolyze cellobiose and other short soluble oligosaccharides to release glucose and are commonly viewed as one of the rate-limiting enzymes (5). The GH3 family represents a collection of >2700 enzymes in the Carbohydrate-Active enZYmes (CAZy) Database, including β-glucosidases (EC 3.2.1.21), xylan 1,4-β-xylosidases (EC 3.2.1.37), and glucan 1,3-β-glucosidases (EC 3.2.1.58). However, there are only eight crystal structures of GH3 enzymes deposited in the Protein Data Bank. Because the majority of β-glucosidases from lignocellulosic biomass microbiomes are members of the GH3 family (4, 6, 7), high-resolution structural characterization of a β-glucosidase from this family may provide insights into alleviating product inhibition and thereby enhancing the overall efficiency of saccharification by increasing the catalytic rate of the bottleneck enzyme.
Here, we present the x-ray crystal structure (resolved to 2.20 Å resolution) and biochemical characterization of the GH3 β-glucosidase JMB19063, discovered from the metagenome sequence of a switchgrass-adapted compost microbial community. It is the first reported structure of a class of dimeric enzymes from the GH3 family, and we demonstrate that dimerization is absolutely required for catalytic activity. JMB19063 contains a C-terminal domain with an Ig-like region that contributes to both substrate binding and dimerization. We have also crystallized the catalytic variant D261N in the presence of cellopentaose to more closely examine the details of substrate binding. The results provide new insight into the GH3 family of enzymes and may enable the identification and optimization of other dimeric β-glucosidases.
EXPERIMENTAL PROCEDURES
Metagenome Sequencing, Assembly, and Analysis
The details of the sequence analysis and identification of the enzyme sequences have been described previously (4). Briefly, metagenomic DNA from a 31-day switchgrass-adapted compost community was shotgun-sequenced using the Roche 454 GS-FLX titanium system by the Department of Energy Joint Genome Institute and assembled using Newbler (version 2). Candidate full-length glycoside hydrolase enzyme sequences were identified by BLASTX hits (E < 1 e−10) matching functionally characterized enzymes in the CAZy Database (8) over at least 90% of their length. Frameshift errors due to 454 homopolymer sequencing errors were identified and manually corrected based on the gapped BLASTX alignment to the closest protein sequences.
Protein Expression and Purification
The codon-optimized β-glucosidase gene without the putative signal peptide sequence was inserted into the pET-DEST42 vector (Invitrogen) and then transformed into the BL21 Star (DE3) Escherichia coli strain (Invitrogen) for protein expression of the β-glucosidase with a C-terminal His6 tag as described previously (4). Overexpression and purification of the recombinant β-glucosidase was performed following a previously reported procedure (9). After purification, the buffer was exchanged with 200 mm NaCl and 25 mm HEPES at pH 7 using an Econo-Pac 10 desalting column (Bio-Rad). The protein solution was passed through a 0.45-μm membrane syringe filter to remove a trace amount of visible precipitants. Purified protein was then concentrated to 10 mg/ml using a Vivaspin 6 centrifugal concentrator (Sartorius Stedim Biotech, Göttingen, Germany) for crystallization. The protein concentrations were determined by the Bradford assay (Bio-Rad). The functional mutants of the β-glucosidase (D261N, R587A, F598A, and R587A/F598A) were created employing the QuikChange site-directed mutagenesis protocol (Stratagene). All recombinant β-glucosidase mutants were expressed and purified by the same methods described above for the wild-type β-glucosidase.
Enzyme Assays and Kinetic Measurements
The β-glucosidase activity at various temperatures and pH values was measured as described previously (4) using 4-nitrophenyl β-d-glucopyranoside (pNPG).3 The kinetics of pNPG hydrolysis by the β-glucosidase was measured at 50 °C in 50 mm MES buffer (pH 6.5) containing 0.1–10 mm pNPG and 0.5 μg (wild-type) or 5 μg (R587A, F598A, and R587A/F598A) of the recombinant enzyme in 1-ml reaction volume. The hydrolysis reaction was quenched by mixing the reaction mixture with an equal volume of 2% Na2CO3 solution. The amount of hydrolyzed p-nitrophenol was measured by reading the absorbance at 420 nm. Eadie-Hofstee analyses were used to measure the kinetic parameters (Vmax, Km, and kcat) for the wild-type and mutant enzymes.
The hydrolysis of cellobiose, cellotetraose, and maltose by the wild-type β-glucosidase and the three mutants was further qualitatively analyzed by nanostructure-initiator mass spectrometry (10)-based enzyme activity assays (Nimzyme) (11, 12). The principle of Nimzyme has been described previously (11, 12). In brief, hydrolysis reactions were measured at 50 °C in 25 mm MES buffer (pH 6.5). In each case, 0.1 mm per Nimzyme substrate (cellobiose, cellotetraose, or maltose) was incubated with 2 μg (wild-type) or 10 μg (R587A, F598A, and R587A/F598A) of the recombinant enzyme in 25-μl reaction volumes for 15 min. For the hydrolysis of cellotetraose by the wild-type protein, additional time points were added. Reactions were quenched by the addition of an equal volume of methanol. Subsequently, 0.7 μl per sample was spotted onto a nanostructure-initiator mass spectrometry surface and analyzed using an Applied Biosystems 4800 MALDI-TOF/TOF mass analyzer. Enzyme activities were determined as product/substrate ratios. In addition, the enzyme activity on polysaccharides were tested by incubating 35 μg of wild-type JMB19063 with 1 mg of carboxymethylcellulose, glucomannan, arabinoxylan, galactomannose, carboxymethyl curdlan, or lichenan in a 1-ml reaction volume containing 50 mm MES buffer (pH 6.5) under the same conditions used for the pNPG assay. At the end of the incubation, a 3,5-dinitrosalicylic acid assay was performed to detect reducing sugar (33).
Crystallization
Crystallization screening was carried out on a Phoenix robot (Art Robbins Instruments, Sunnyvale, CA) using a sparse matrix screening method (13). Wild-type JMB19063 and the D261N point mutant were crystallized by sitting drop vapor diffusion in drops containing a 2:1 ratio of protein solution and 0.1 m sodium citrate and 30% (w/v) PEG 3350. JMB19063 D261N contained an additional 5 mm cellopentaose in the crystallization buffer. Rod-like crystals were observed within 2 days. For data collection, crystals were flash-frozen in liquid nitrogen directly from the crystallization drop.
X-ray Data Collection and Structure Determination
The x-ray data sets for JMB19063 were collected at the Berkeley Center for Structural Biology on beamlines 8.2.1 and 8.2.2 of the Advanced Light Source at the Lawrence Berkeley National Laboratory. Diffraction data were recorded using ADSC Q315r detectors (Area Detector Systems Corp., San Diego, CA). Processing of image data was performed using the HKL2000 suite of programs (14). Phases were calculated by molecular replacement with the program Phaser (15) using the structure of barley β-d-glucan exohydrolase (Protein Data Bank code 1EX1) (16) as a search model. Automated model building was conducted using RESOLVE (17) from the PHENIX suite of programs (18), resulting in a model that was 85% complete. Manual building using Coot (19) was alternated with reciprocal space refinement using PHENIX (18). Waters were automatically placed using PHENIX and manually added or deleted with Coot according to peak height (3.0σ in the Fo − Fc map) and distance from a potential hydrogen bonding partner (<3.5 Å). TLS refinement (20) using 10 groups, chosen using the TLSMD web server (21), was used in later rounds of refinement. An initial rigid body refinement using PHENIX (18) was performed with the final wild-type model (all waters removed) for JMB19063 D261N. This model was then refined and built in the same manner as the wild-type model. All data collection, phasing, and refinement statistics are summarized in Table 1.
TABLE 1.
Data collection and refinement statistics
Statistics for the highest resolution shell are shown in parentheses. r.m.s.d., root mean square deviation.
| WT | D261N | |
|---|---|---|
| Resolution (Å) | 50–2.2 (2.24–2.2) | 50–2.2 (2.24–2.2) |
| Space group | P43212 | P43212 |
| Unit cell dimensions | a = b = 121.7, c = 242.8Å; α = γ = β = 90° | a = b = 121.5, c = 242.9Å; α = γ = β = 90° |
| Total reflections | 914,456 | 990,372 |
| Unique reflections | 88,156 | 93,226 |
| Multiplicity | 10.4 (7.4) | 10.6 (7.3) |
| Completeness (%) | 95.00 (65.30) | 99.50 (94.70) |
| I/σ(I) | 15.44 (1.73) | 20.62 (1.48) |
| Rsym | 0.117 (0.612) | 0.101 (0.649) |
| R-factor | 0.1635 | 0.1895 |
| Rfree | 0.2116 | 0.2244 |
| Number of atoms | 12,343 | 12,031 |
| Protein residues | 1481 | 1487 |
| Water molecules | 843 | 485 |
| r.m.s.d. from ideal geometry | ||
| Bond lengths (Å) | 0.005 | 0.007 |
| Angles | 0.95° | 0.95° |
| Ramachandran plot | ||
| Favored (%) | 97 | 97 |
| Outliers (%) | 0.2 | 0.2 |
| Clashscore | 10.92 | 10.75 |
| Average B-factor | 37.6 | 50.4 |
Creation of Structure-based Sequence Alignment and the Phylogenetic Tree
To build a high-quality sequence alignment in this diverse protein family, we used a combination of structural and sequence information. First, we performed pairwise structural alignments with 3DHIT (22) of six GH3 family structures (chain A of proteins with Protein Data Bank codes 3BMX, 1X38, 2OXN, 2X41, 3F94, and 3ABZ) with the JMB19063 structure. These six structures were selected based on their resolution and to remove redundancy at the 90% sequence identity. For each of these structures, we performed a BLAST search (23) with the GH3 sequences (after removing short sequence fragments) from the CAZy Database (8) to find sequences between 25 and 90% sequence identity to the structure's sequence, and the resulting sequences were aligned with MUSCLE (24). These seven multiple sequence alignments were then combined into one sequence alignment by aligning equivalent positions in the individual sequence alignments using the pairwise structural alignments with the JMB19063 structure. Redundant sequences were filtered out at 90% sequence identity, preferentially keeping sequences with structures, experimental characterization, and longer lengths, in this order of priority.
Gapped positions and their neighbors were trimmed from the above structure-based sequence alignment by removing positions with <60% occupancy and two flanking positions. A tree was built from the resulting trimmed alignment using FastTree 2.1.3 (25) and the tree was rerooted such that the root was the midpoint between the leaves with the farthest evolutionary distance.
RESULTS AND DISCUSSION
Overall Structure of JMB19063
The crystal structure of JMB19063 in complex with β-glucose was solved to 2.20 Å resolution. The structure of the catalytic variant D261N crystallized in the presence of cellopentaose was also determined to 2.20 Å. The electron density map is generally well ordered throughout the entire polypeptide chain, with the only disorder observed in the C-terminal His6 tag and linker region (amino acids 738–775).
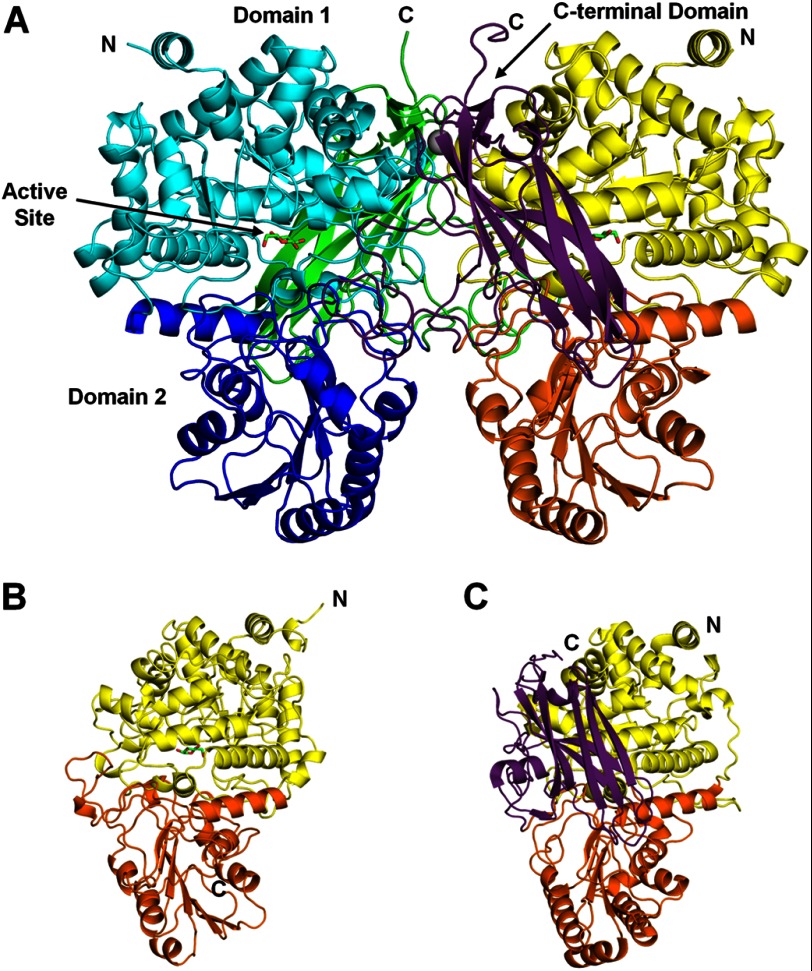
This is the first structure described for a dimeric GH3 enzyme, and the enzyme forms a dimer of identical monomers, each composed of three distinct domains (Fig. 1A). Domain 1 (amino acids 1–349) forms an (α/β)8-TIM barrel. Domain 2 (amino acids 350–571) is an α/β-sandwich comprising a six-stranded β-sheet sandwiched between three α-helices on either side. The majority of all structurally described enzymes from the GH3 family contain this two-domain architecture, corresponding to the Glyco_hydro_3 (Pfam PF00933) and Glyco_hydro_3_C (Pfam PF01915) domain families. The first of these structures solved was the barley exohydrolase (HvExoI) (Fig. 1B) (16). JMB19063 contains an additional domain at the C terminus (amino acids 572–737) that consists of a 50-amino acid linker and an Ig-like β-sandwich fold region. The Ig-like region is most similar to the family of CARDB domains (Pfam PF07705), which are bacterial cell adhesion domains. A structural Ca2+ ion is observed in the Ig-like regions of both monomers. It is coordinated by Asp-664, the backbone carbonyl of Val-666, and four water molecules. The calcium is positioned similarly to Ca2+ ions in the structurally related cellulose-binding domains from the cellulosomal scaffoldin subunit of Clostridium cellulolyticum (Protein Data Bank code 1G43) (26) and the Cel9V glycoside hydrolase from Clostridium thermocellum (code 2WNX) (27). In JMB19063, the Ca2+ ion helps position a loop, consisting of residues 664–674, that is at the dimer interface and makes five hydrogen bonds with the other monomer. Another three-domain β-glucosidase from the GH3 family has recently been described in Thermotoga neapolitana (TnBgl3B) (28). The majority of the TnBgl3B structure superposes well with JMB19063 (Fig. 1C), and TnBgl3B also has a third Ig-like domain at the C terminus. In the structure of TnBgl3B and all other previously described structures from the GH3 family, the active site is at the interface of domains 1 and 2, and residues from both domains play a role in substrate binding. The C-terminal domain of TnBgl3B does not contact the active site and is described as having no known function (28). In our structure of JMB19063, the C-terminal domain from the opposing monomer composes a portion of the enzyme active site, and residues from this domain play key roles in substrate binding (Fig. 2).
FIGURE 1.
A, JMB19063 is a dimer of identical monomers (cyan, blue, and green or yellow, orange, and purple). The enzyme is composed of three domains. The N-terminal domain (cyan/yellow) is an (α/β)8-TIM barrel that contains the catalytic Asp and Glu residues. Domain 2 (blue/orange) is an α/β-sandwich that makes up a portion of the active site. The C-terminal domain (green/purple) is composed of a 50-residue linker and a region with an Ig-like fold, which shows structural homology to the CARDB family of domains, which are cell wall-binding domains. B, the structure of the two-domain HvExoI protein (Protein Data Bank code 1EX1) is highly similar to domain 1 (yellow) and domain 2 (orange) of JMB19063. C, TnBgl3B (code 2X40) also contains a third Ig-like domain (purple) in addition to domain 1 (yellow) and domain 2 (orange).
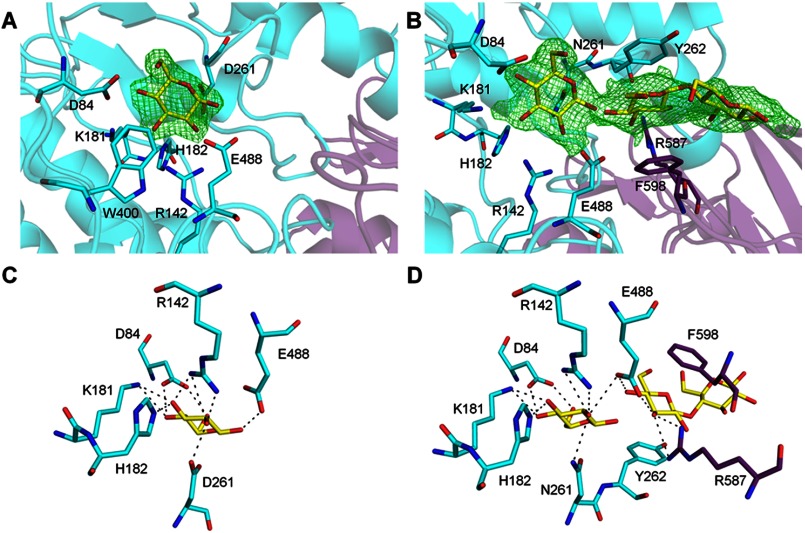
FIGURE 2.
Detailed view of the active site. Amino acids from the partnering monomer are shown in purple. A, electron density for the glucose bound to wild-type JMB19063 is shown in green wire mesh. The Fo − Fc omit map is contoured to 2.8σ. B, electron density for the glucose and cellobiose moiety bound to JMB19063 D261N is shown in green wire mesh. The Fo − Fc omit map is contoured to 2.8σ. C, stick representation of glucose (yellow) bound to wild-type JMB19063. D, stick representation of glucose and cellobiose (yellow) bound to JMB19063 D261N.
Dimerization
The buried solvent-accessible surface of the dimer interface as calculated by the protein-protein interaction interface server (PISA) (29) is 3286 Å2 and is composed of 71 hydrogen bonds and 19 salt bridges. The majority of the interactions occur between the C-terminal linker region and domains 1 and 2 of the other monomer. One notable exception to this is the Ca2+-binding loop (positions 664–674) in the Ig-like region, which forms five hydrogen bonds with domain 1. The results from structural analysis were confirmed by size-exclusion chromatography, indicating that the enzyme is indeed homodimeric in nature. Only one other structure of a multimeric GH3 enzyme has been described. A β-glucosidase from Kluyveromyces marxianus (KmBglI) forms a tetramer (30). The crystal structure of KmBglI suggests that tetramer formation is not essential for catalysis because the subunit interfaces are far from the active site. Because the active site contribution of the C-terminal domain comes from residues of the opposing monomer, dimerization is essential for the activity of JMB19063. However, it appears that in the GH3 family, neither the role of a third domain in substrate binding nor the necessity of multimerization for catalytic activity has heretofore been described. Recently, the structure of a GH3 enzyme from Synechococcus sp. PCC 7002 was deposited in the Protein Data Bank (code 3SQL). Although it is currently unpublished, the structure suggests that, like JMB19063, it is dimeric. The enzyme is two-domain and therefore does not have the same dimer interface as JMB19063. However, the interface is close to the active site, and thus, it is possible that residues from the partnering monomer may play a role in substrate binding.
Active Site
In the structure of wild-type JMB19063, although it was not specifically added to crystallization buffer, well resolved density for a β-glucose is observed at the active site cleft formed by the interface of domains 1 and 2 in both monomers, and the glucose molecule occupies the −1 subsite (Fig. 2A). Asp-261 in domain 1 and Glu-488 in domain 2 are positioned similarly to the catalytic residues (Asp-285 and Glu-491) in HvExoI, suggesting that Asp-261 and Glu-488 act as the catalytic nucleophile and acid/base, respectively, in JMB19063. The glucose is tightly bound by a network of hydrogen bonds with Asp-84, Arg-142, Lys-181, His-182, Asp-261, and Glu-488 and is further stabilized by hydrophobic interaction with Trp-430 (Fig. 2C). The source of the glucose molecule is most likely from protein expression, as there is some glucose contamination in yeast extract (31) that is carried through the purification process.
The catalytic mutant JMB19063 D261N was crystallized in the presence of cellopentaose. However, in both subunits, interpretable density was observed only for three of the five sugars in the −1, +1, and +2 subsites (Fig. 2B). Despite being catalytically inactive, it was not possible to model an intact cellotriose moiety into the electron density. It was necessary to introduce a break between the sugars in the −1 and +1 subsites to correctly fit the density. Therefore, a β-glucose was modeled into the −1 subsite, and a cellobiose moiety was modeled into the +1 and +2 subsites. A small residual activity coupled with long crystallographic time scales is the probable cause of the broken bond between the sugars in the −1 and +1 subsites. Some weak electron density exists beyond the +2 subsite; however, it was not clear enough to justify modeling in either of the remaining two sugars. This disorder is likely due to the lack of any interactions between the protein and sugar units beyond the +1 subsite. The glucose in the −1 subsite almost exactly overlaps with the wild-type JMB19063 glucose. Additionally, all active site residues, including Asn-261, are positioned nearly identically to their wild-type counterparts. Therefore, it is likely that the ligands are bound to the D261N variant in the same manner as we would expect from the wild-type enzyme. The sugar in the +1 subsite is stabilized by hydrogen bonds to Glu-488 and hydrophobic stacking with Tyr-262 (Fig. 2D). Additional hydrogen bonding and hydrophobic stacking are provided by Arg-587 and Phe-598, respectively, from the C-terminal domain of the opposing monomer. These interactions are spatially conserved in HvExoI at Arg-291 and Trp-434, which are both in domain 1.
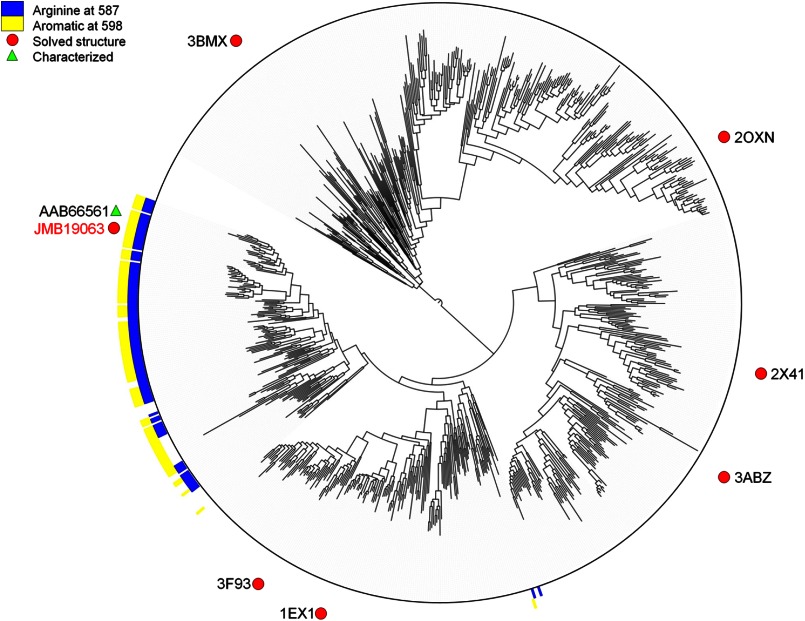
Because Arg-587 and Phe-598 from the C-terminal domain of the opposing monomer contribute to substrate binding, it is necessary for the enzyme to be in a dimeric state to have full catalytic activity. Therefore, we hypothesized that any GH3 enzyme that has similar amino acids conserved at those sites is a functional dimer. To investigate this, we built a phylogenetic tree using the existing GH3 crystal structures to provide a map of the relationships in the family (Fig. 3; for a high-resolution version, including accession numbers, see supplemental Fig. S2). A large clade containing JMB19063 was found to have a conserved arginine at the position corresponding to Arg-587 and an aromatic residue at a position analogous to Phe-598. Of the enzymes in this clade, only a β-glucosidase from Flavobacterium meningosepticum has been biochemically characterized (32). Like JMB19063, this enzyme was found to be dimeric.
FIGURE 3.
Phylogenetic tree built from alignment of GH3 family members with representative structures using FastTree 2.1.3 (25).
Enzyme Kinetics
JMB19063 hydrolyzed a variety of substrates with β-1,4 linkages up to cellotetraose, including cellotriose, cellobiose, pNPG, and 4-nitrophenyl β-d-cellobioside. The enzyme was unable to hydrolyze 4-nitrophenyl β-d-xylopyranoside and a range of polysaccharide substrates that we tested, including carboxymethylcellulose, glucomannan, lichenan, galactomannose, and carboxymethyl curdlan. The Michaelis-Menten parameters of the β-glucosidase activity were determined using pNPG as the substrate at pH and temperature optima (Fig. 4). JMB19063 hydrolyzed pNPG with a kcat of 150 s−1 and a Km of 0.79 mm (Table 2). The crystal structure of the cellopentaose-bound D261N mutant revealed that, as a consequence of dimerization, Arg-587 and Phe-598 interact with the bound cellopentaose in the catalytic pocket of the opposing subunit. Therefore, we hypothesized that these non-catalytic residues may influence enzyme activity by complementing the catalytic pocket of the other subunit as a result of dimerization. To determine whether Arg-587 and Phe-598 influence enzyme activity, each of these residues was mutated to Ala, and Michaelis-Menten parameters were measured using pNPG as the substrate. The results show that both the R587A and the F598A mutations substantially decreased the catalytic efficiency (kcat/Km) by decreasing kcat and increasing Km (Table 2); the kcat values for R587A and F598A were 37 and 0.87% of that of the wild-type enzyme, respectively, and the Km values for R587A and F598A were 2.2- and 4.9-fold over that of the wild-type enzyme, respectively. The double mutant (R587A/F598A) had no detectable activity on pNPG, so the Michaelis-Menten parameters were not measured. Size-exclusion column chromatograms of the purified mutant enzymes overlapped the chromatogram of the wild-type enzyme at the 161-kDa region (supplemental Fig. S1), indicating that the differences in kinetic parameters are direct results of the mutations. Therefore, these results strongly suggest that Arg-587 and Phe-598 are essential for formation of the JMB19063 catalytic pocket with the dimerized subunit and are important in the enzyme activity of the β-glucosidase.
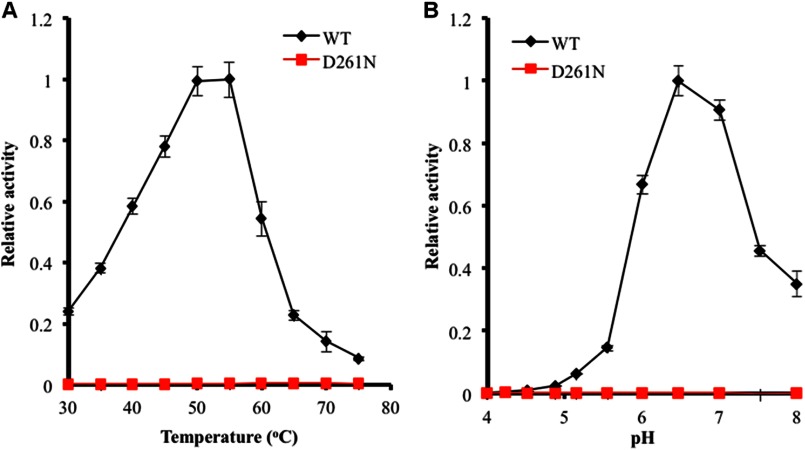
FIGURE 4.
pH and temperature activity profiles of JMB19063 measured by pNPG assay. The enzyme activity of JMB19063 on pNPG at various pH values was measured at 50 °C (A), and the temperature profile of JMB19063 was measured at pH 6.5 (B).
TABLE 2.
Enzymatic analysis of JMB19063
The Michaelis-Menten parameters were determined using pNPG as the substrate as described under “Experimental Procedures.” Reactions were incubated for 30 min at enzyme concentrations of 0.5 μg (wild-type) and 5 μg (R587A, F598A, and R587A/F598A) of the recombinant enzyme in a 1-ml reaction volume. Data represent the means ± S.E. of four independent experiments for each enzyme. The R587A/F598A mutant did not have any detectable activity.
| Parameter | WT | R587A | F598A |
|---|---|---|---|
| Km (mm) | 0.79 ± 0.1 | 1.7 ± 0.41 | 3.9 ± 0.19 |
| kcat (s−1) | 150 ± 12 | 56 ± 6.5 | 1.3 ± 0.28 |
| kcat/Km | 198 ± 39 | 37 ± 15 | 0.33 ± 0.053 |
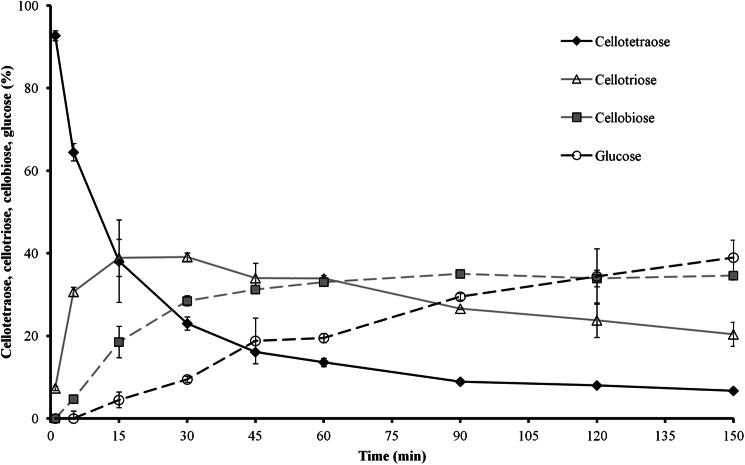
Additionally, the hydrolysis of cellobiose, cellotetraose, and maltose was qualitatively analyzed using the nanostructure-initiator mass spectrometry (10)-based Nimzyme assay (11, 12). Whereas the wild-type protein hydrolyzed >90% of cellobiose under our prescribed assay conditions, R587A converted only ∼5% and F598A only ∼10% of cellobiose (Table 3). Considering that five times more mutant than wild-type protein was used for these assays, the catalytic activity of R587A was only ∼1.0% and that of F598A was only ∼2.3% of that of the wild-type enzyme. The double mutant (R587A/F598A) had no detectable activity. Furthermore, the wild-type β-glucosidase was able to utilize cellotetraose, whereas none of the mutant proteins showed this activity (Table 3). Monitoring the hydrolysis of cellotetraose by the wild-type protein over time (Fig. 5) clearly showed the stepwise degradation from cellotetraose via cellotriose and cellobiose down to glucose. None of the tested enzyme variants was able to cleave the α-1,4 linkages in maltose (Table 3). Together, these data also suggest the involvement of Arg-587 and Phe-598 in the formation of the catalytic pocket.
TABLE 3.
Conversion of cellobiose, cellotetraose, xylobiose, lactose, and maltose determined in a Nimzyme assay
Cellobiose and cellotetraose were the only substrates that were hydrolyzed by JMB19063 among the substrates that we tested by Nimzyme assay. Each reaction was carried out in a 25-μl reaction volume containing 25 mm MES buffer (pH 6.5), 0.1 mm cellobiose or cellotetraose, and 2 μg (WT) or 10 μg (R587A or F598A) of recombinant enzyme at 50 °C for 15 min. All activities were corrected by the values of the negative control samples without enzyme. Data represents the means ± S.E. of three independent experiments.
| WT | R587A | F598A | |
|---|---|---|---|
| Hydrolysis of cellobiose (%) | 92.4 ± 2.4 | 4.8 ± 0.3 | 10.4 ± 1.2 |
| Hydrolysis of cellotetraose (%) | 61.9 ± 5.8 | No activity | No activity |
FIGURE 5.
Time course of stepwise conversion of cellotetraose by the wild-type protein to cellotriose, cellobiose, and glucose monitored in a Nimzyme assay. All activities were corrected by the values of negative control samples without enzyme. Error bars represent S.E. of three independent experiments.
Conclusions
We have presented the crystal structures of JMB19063 and a D261N catalytic variant of JMB19063. The structure of JMB19063, corroborated by size-exclusion chromatography, revealed that the enzyme is homodimeric. Each monomer is composed of three distinct domains. In addition to the two N-terminal domains, the C-terminal domain from the partnering subunit forms a portion of the active site cavity. Arg-587 and Phe-598 from this domain were shown to interact with bound ligands in the D261N catalytic variant structure. Enzyme assays confirmed that these residues are absolutely essential for full catalytic activity. Phylogenetic analysis showed that JMB19063 may be the first structure from a large subclass of enzymes in the GH3 family that has a unique and previously undescribed mode of substrate binding that requires dimerization.
Supplementary Material

This article contains supplemental Figs. S1 and S2.
- pNPG
- 4-nitrophenyl β-d-glucopyranoside.
REFERENCES
- 1. Bouton J. H. (2007) Molecular breeding of switchgrass for use as a biofuel crop. Curr. Opin. Genet. Dev. 17, 553–558 [DOI] [PubMed] [Google Scholar]
- 2. Rubin E. M. (2008) Genomics of cellulosic biofuels. Nature 454, 841–845 [DOI] [PubMed] [Google Scholar]
- 3. Simmons B. A., Loque D., Blanch H. W. (2008) Next-generation biomass feedstocks for biofuel production. Genome. Biol. 9, 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Allgaier M., Reddy A., Park J. I., Ivanova N., D'haeseleer P., Lowry S., Sapra R., Hazen T. C., Simmons B. A., VanderGheynst J. S., Hugenholtz P. (2010) Targeted discovery of glycoside hydrolases from a switchgrass-adapted compost community. PLoS ONE 5, e8812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shewale J. G. (1982) β-Glucosidase: its role in cellulase synthesis and hydrolysis of cellulose. Int. J. Biochem. 14, 435–443 [DOI] [PubMed] [Google Scholar]
- 6. Warnecke F., Luginbühl P., Ivanova N., Ghassemian M., Richardson T. H., Stege J. T., Cayouette M., McHardy A. C., Djordjevic G., Aboushadi N., Sorek R., Tringe S. G., Podar M., Martin H. G., Kunin V., Dalevi D., Madejska J., Kirton E., Platt D., Szeto E., Salamov A., Barry K., Mikhailova N., Kyrpides N. C., Matson E. G., Ottesen E. A., Zhang X., Hernández M., Murillo C., Acosta L. G., Rigoutsos I., Tamayo G., Green B. D., Chang C., Rubin E. M., Mathur E. J., Robertson D. E., Hugenholtz P., Leadbetter J. R. (2007) Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450, 560–565 [DOI] [PubMed] [Google Scholar]
- 7. Brulc J. M., Antonopoulos D. A., Miller M. E., Wilson M. K., Yannarell A. C., Dinsdale E. A., Edwards R. E., Frank E. D., Emerson J. B., Wacklin P., Coutinho P. M., Henrissat B., Nelson K. E., White B. A. (2009) Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc. Natl. Acad. Sci. U.S.A. 106, 1948–1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) The Carbohydrate-Active enZYmes Database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Park J. I., Kent M. S., Datta S., Holmes B. M., Huang Z., Simmons B. A., Sale K. L., Sapra R. (2011) Enzymatic hydrolysis of cellulose by the cellobiohydrolase domain of CelB from the hyperthermophilic bacterium Caldicellulosiruptor saccharolyticus. Bioresour. Technol. 102, 5988–5994 [DOI] [PubMed] [Google Scholar]
- 10. Northen T. R., Yanes O., Northen M. T., Marrinucci D., Uritboonthai W., Apon J., Golledge S. L., Nordström A., Siuzdak G. (2007) Clathrate nanostructures for mass spectrometry. Nature 449, 1033–1036 [DOI] [PubMed] [Google Scholar]
- 11. Reindl W., Deng K., Gladden J. M., Cheng G., Wong A., Singer S. W., Singh S., Lee J. C., Yao C. H., Hazen T. C., Singh A. K., Simmons B. A., Adams P. D., Northen T. R. (2011) Colloid-based multiplexed screening for plant biomass-degrading glycoside hydrolase activities in microbial communities. Energy Environ. Sci. 4, 2884–2893 [Google Scholar]
- 12. Northen T. R., Lee J. C., Hoang L., Raymond J., Hwang D. R., Yannone S. M., Wong C. H., Siuzdak G. (2008) A nanostructure-initiator mass spectrometry-based enzyme activity assay. Proc. Natl. Acad. Sci. U.S.A. 105, 3678–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jancarik J., Kim S. H. (1991) Sparse matrix sampling: a screening method for crystallization of proteins. J. Appl. Crystallogr. 24, 409–411 [Google Scholar]
- 14. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Method Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 15. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Varghese J. N., Hrmova M., Fincher G. B. (1999) Three-dimensional structure of a barley β-d-glucan exohydrolase, a family 3 glycosyl hydrolase. Structure 7, 179–190 [DOI] [PubMed] [Google Scholar]
- 17. Terwilliger T. C. (2003) Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr. D Biol. Crystallogr. 59, 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 20. Painter J., Merritt E. A. (2006) Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 62, 439–450 [DOI] [PubMed] [Google Scholar]
- 21. Painter J., Merritt E. A. (2006) TLSMD web server for the generation of multi-group TLS models. J. Appl. Crystallogr. 39, 109–111 [Google Scholar]
- 22. Plewczyński D., Paś J., von Grotthuss M., Rychlewski L. (2002) 3D-Hit: fast structural comparison of proteins. Appl. Bioinformatics 1, 223–225 [PubMed] [Google Scholar]
- 23. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 24. Edgar R. C. (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Price M. N., Dehal P. S., Arkin A. P. (2010) FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shimon L. J., Pagès S., Belaich A., Belaich J. P., Bayer E. A., Lamed R., Shoham Y., Frolow F. (2000) Structure of a family IIIa scaffoldin CBD from the cellulosome of Clostridium cellulolyticum at 2.2 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 56, 1560–1568 [DOI] [PubMed] [Google Scholar]
- 27. Petkun S., Jindou S., Shimon L. J., Rosenheck S., Bayer E. A., Lamed R., Frolow F. (2010) Structure of a family 3b′ carbohydrate-binding module from the Cel9V glycoside hydrolase from Clostridium thermocellum: structural diversity and implications for carbohydrate binding. Acta Crystallogr. D Biol. Crystallogr. 66, 33–43 [DOI] [PubMed] [Google Scholar]
- 28. Pozzo T., Pasten J. L., Karlsson E. N., Logan D. T. (2010) Structural and functional analyses of β-glucosidase 3B from Thermotoga neapolitana: a thermostable three-domain representative of glycoside hydrolase 3. J. Mol. Biol. 397, 724–739 [DOI] [PubMed] [Google Scholar]
- 29. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 30. Yoshida E., Hidaka M., Fushinobu S., Koyanagi T., Minami H., Tamaki H., Kitaoka M., Katayama T., Kumagai H. (2010) Role of a PA14 domain in determining substrate specificity of a glycoside hydrolase family 3 β-glucosidase from Kluyveromyces marxianus. Biochem. J. 431, 39–49 [DOI] [PubMed] [Google Scholar]
- 31. Baran R., Bowen B. P., Northen T. R. (2011) Untargeted metabolic footprinting reveals a surprising breadth of metabolite uptake and release by Synechococcus sp. PCC 7002. Mol. Biosyst. 7, 3200–3206 [DOI] [PubMed] [Google Scholar]
- 32. Li Y.-K., Lee J.-A. (1999) Cloning and expression of β-glucosidase from Flavobacterium meningosepticum: a new member of family B β-glucosidase. Enzyme Microb. Technol. 24, 144–150 [Google Scholar]
- 33. Miller G. L. (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31, 426–428 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.