Background: α-Synuclein aggregates and macroautophagy are associated with neurodegeneration.
Results: Modulation of macroautophagic activity does not affect α-synuclein aggregate levels, although these aggregates cause accumulation of immature autophagosomes.
Conclusion: α-Synuclein aggregates are resistant to degradation and impair autophagy by delaying autophagosome maturation.
Significance: Understanding the impact of α-synuclein aggregates on autophagy may help elucidate therapies for α-synuclein-mediated neurodegeneration.
Keywords: Aggregation, α-Synuclein, Autophagy, Parkinson Disease, Protein Degradation
Abstract
Cytoplasmic α-synuclein (α-syn) aggregates, referred to as Lewy bodies, are pathological hallmarks of a number of neurodegenerative diseases, most notably Parkinson disease. Activation of macroautophagy is suggested to facilitate degradation of certain proteinaceous inclusions, but it is unclear if this pathway is capable of degrading α-syn aggregates. Here, we examined this issue by utilizing cellular models in which intracellular Lewy body-like α-syn inclusions accumulate after internalization of pre-formed α-syn fibrils into α-syn-expressing HEK293 cells or cultured primary neurons. We demonstrate that α-syn inclusions cannot be effectively degraded, even though they co-localize with essential components of both the autophagic and proteasomal protein degradation pathways. The α-syn aggregates persist even after soluble α-syn levels have been substantially reduced, suggesting that once formed, the α-syn inclusions are refractory to clearance. Importantly, we also find that α-syn aggregates impair overall macroautophagy by reducing autophagosome clearance, which may contribute to the increased cell death that is observed in aggregate-bearing cells.
Introduction
Synucleinopathies are a family of neurodegenerative diseases characterized by cytoplasmic aggregates of the protein α-synuclein (α-syn)2 and include Parkinson disease (PD), PD with dementia, dementia with Lewy bodies, and multiple system atrophy (1, 2). In PD, PD with dementia, and dementia with Lewy bodies, α-syn forms highly insoluble fibrillar aggregates called Lewy bodies (LBs) and Lewy neurites, which accumulate in the neuronal cytoplasm or neuritic processes, respectively. These inclusions are rich in phosphorylated α-syn (p-α-syn), are often ubiquitinated (3, 4), and are widely distributed in the central nervous system (CNS), where they are associated with neuron loss (5). Moreover, α-syn mutations and α-syn gene duplication/triplication are linked to familial forms of PD (6–9). Collectively, these findings implicate α-syn aggregates in the onset and progression of disease. Thus, it is important to gain a better understanding of whether α-syn inclusions can be effectively degraded by cells, and whether these aggregates might negatively affect key cellular pathways.
The removal of protein aggregates, as well as misfolded and damaged proteins, occurs via two main degradation pathways, the ubiquitin-proteasome system (UPS) and the autophagy-lysosomal pathway (ALP) (10). Macroautophagy, a prominent constituent of the ALP, is responsible for removal of both soluble and aggregated proteins, as well as damaged organelles, via their sequestration into double-membrane vesicles called autophagosomes, followed by their degradation by catalytic enzymes after autophagosome/lysosome fusion (11, 12). Macroautophagy (hereafter, autophagy) has been implicated in bulk degradation of pathologic aggregates, such as the polyglutamine-expanded huntingtin inclusions observed in Huntington disease (13). ALP is a vital cellular process, and impairment of the ALP can lead to neurodegeneration, as demonstrated in animal models that are deficient in genes coding for autophagy proteins in neuronal tissue (14, 15) and in human neurodegenerative lysosomal storage diseases (16, 17).
ALP as well as UPS have been implicated in the degradation of soluble, nonaggregated α-syn (18–21). However, it is not clear if autophagy plays a role in the clearance of LB-like α-syn aggregates, as a heretofore lack of in vitro models that faithfully recapitulate α-syn aggregation has made it difficult to address this question. Unlike many aggregate-prone proteins, simple overexpression of α-syn does not lead readily to the formation of insoluble LB-like inclusions. Therefore, additional manipulations have been used to generate α-syn aggregates in cultured cells, including co-expression of proteins such as synphilin-1 (22) and exposure to proteolytic inhibitors, oxidative stress, or nitrative insult (23–25), which likely directly or indirectly affect protein degradation pathways. However, the α-syn aggregates formed in these various cellular paradigms typically fail to exhibit several important characteristics of LBs, including ubiquitination and the presence of insoluble phosphorylated α-syn. To better model LB-like inclusions in cultured cells, we recently developed models of α-syn aggregation in which the introduction of small amounts of pre-formed α-syn fibrils (Pffs) into α-syn-expressing cells, including primary neurons from wild-type (WT) nontransgenic mice, results in the templated assembly of endogenously expressed α-syn and the formation of insoluble aggregates resembling LBs and Lewy neurites (26, 27). The ability to generate α-syn aggregates efficiently, without using any treatment that directly perturbs protein degradative function, provides model systems to investigate the possible interplay between autophagy and α-syn aggregates.
Here, we have investigated whether Pff-seeded, LB-like α-syn aggregates can be removed by autophagy. We observed that pathologic α-syn inclusions cannot be effectively eliminated and that they cause a partial impairment of autophagosome clearance that could contribute to the decreased viability observed in cells harboring α-syn deposits. Thus, our findings provide novel insights into the effects of α-syn aggregates on cellular metabolism and viability and may have implications regarding potential therapeutic strategies for synucleinopathies.
EXPERIMENTAL PROCEDURES
Mammalian and Primary Neuronal Cell Cultures
HEK293 cells (QBiogene) stably expressing WT or A53T α-syn were generated as described previously (26). Cells were maintained in complete media (DMEM (Invitrogen), supplemented with 10% FBS, penicillin/streptomycin, and l-glutamine), and 500 μg/ml, 100 μg/ml, or no G418 (Invitrogen) was added to the media for WT α-syn, A53T α-syn, and naive cells, respectively. The day before the experiment, HEK293 cells were plated at a density of 60,000–75,000 cells/well on poly-d-lysine (PDL, 0.1 μg/ml in distilled H2O)-coated and swine gelatin (0.1% w/v in distilled H2O)-coated 12-mm coverslips, and 150,000–200,000 cells/well of PDL coated on 12-well plates (Thermo-Fisher Scientific).
HeLa T-Rex (Invitrogen) A53T α-syn cells were generated as per the manufacturer's instructions. Briefly, A53T α-syn cDNA in the pCDNA 5TO vector (Invitrogen) was transfected into HeLa T-Rex cells, and hygromycin B (Thermo-Fisher Scientific)-resistant cells that inducibly express A53T α-syn were screened by light microscopy and IB. HeLa T-Rex A53T α-syn cells were maintained in Tet-Free complete medium (DMEM, supplemented with 10% Tet-Screened FBS penicillin/streptomycin, l-glutamine), including 100 μg/μl G418 and 100 μg/μl hygromycin B. The day before the experiment, HeLa T-Rex cells were plated at a density of 25,000–30,000 cells/well on PDL-coated coverslips, and 50,000–60,000 cells/well on 12-well plates. 1 μg/ml doxycycline (dox) was added at the time of plating to induce α-syn expression.
Primary mouse hippocampal neurons were cultured and maintained as described previously (27). Briefly, hippocampi dissected from E16 to E18 C57BL/6 or CD-1 mouse brains (Charles River, MA) and α-syn KO mice (28) were treated with papain and DNase, and dissociated neurons were plated in plating media (Neurobasal Media (Invitrogen), supplemented with 10% FBS, penicillin/streptomycin, Glutamax, B27) on PDL (0.1 mg/ml, in 0.1 m borate buffer, pH 8.4)-coated coverslips at a density of 60,000–100,000 cells/well or PDL-coated 12-well plates at 400,000–600,000 cells/well. Plating media were changed to neuronal media (Neurobasal Media, supplemented with penicillin/streptomycin, Glutamax, B27) 2–18 h after plating.
Recombinant α-Syn Proteins and Generation of α-Syn Pffs
Recombinant WT and C-terminally truncated α-syn (remaining amino acids, 1–120) with and without a C-terminal Myc tag were expressed in Escherichia coli and were purified as described previously (29). Briefly, E. coli BL21-RIL cells expressing α-syn from the pRK172 expression vector were lysed using a glass mortar and pestle, and cell lysate was cleared through heat denaturation followed by centrifugation, and α-syn was purified from supernatant by gel filtration and subsequent ion exchange chromatography.
α-syn Pffs were generated by incubating purified α-syn (5 mg/ml in PBS) at 37 °C with constant agitation for 5 days. Electron microscopy, thioflavin S, and protein sedimentation assays confirmed the generation of insoluble α-syn amyloid fibrils at the end of incubation (29). Generated Pffs were aliquoted and stored at −80 °C.
Pff Transduction
Unless indicated otherwise, HEK293 or HeLa T-Rex cells expressing A53T α-syn and α-syn Pffs generated from C-terminally Myc-tagged and truncated α-syn (1–120) were used for non-neuronal Pff transductions. By using this combination of cells and Pffs, a 4.7-fold increase in percentage of cells bearing aggregates (78.5 ± 0.7% versus 16.7 ± 3.5%; n = 3, 48 h after transduction) was elicited compared with WT α-syn cells and WT α-syn Pff combinations. Cells were washed with Opti-MEM (Invitrogen), before transduction. For experiments in dox-induced α-syn HeLa cells, cells were rinsed three times with Opti-MEM for 5 min each to completely remove dox. 0.8 μg of sonicated α-syn Pffs in 80 μl of dPBS (Cellgro) was added to a tube of protein transduction reagent Bioporter (Sigma) and incubated for 10 min at room temperature. After incubation, 420 μl of Opti-MEM was added to the reagent tube, and one tube of diluted reagent-Pff complex was used to transduce four wells on a 12-well plate or eight coverslips. After 4 h, Opti-MEM with reagent-Pff complex was removed, and cells were maintained in low serum starvation media (DMEM, 0.25% FBS supplemented with penicillin/streptomycin, l-glutamine) unless indicated otherwise. In neurons, WT α-syn Pffs were diluted in PBS at 100 μg/ml, sonicated 65 times, and diluted in neuronal media. For coverslips 0.5 μg and for 12-well plate wells 5 μg of α-syn Pffs were added directly to culture media at 5 or 10 DIV. Neuronal media of transduced cells were not removed earlier than 3 days after transduction. Where indicated, neurons were starved in starvation medium (DMEM, penicillin/streptomycin) to activate autophagy (30). PBS was used as a negative control in all transductions. In previous studies, we showed that transduction with monomeric α-syn is similar to transduction with PBS and did not lead to the accumulation of insoluble α-syn aggregates or to detectable changes in cellular functions (27).
Drug Treatments
2.5 mg/ml rapamycin in DMSO (Sigma) was stored at −20 °C and was diluted 1:10 in DMSO before each experiment. Lysosome inhibitor mixture components, leupeptin hemisulfate, pepstatin A, and E46d (all from Sigma) were prepared as follows: 20 mm in distilled H2O, 2 mm in DMSO, and 10 mm in DMSO, respectively, and kept at −20 °C, 200 mm chloroquine (Sigma), 100 mm 3-methyladenine (Sigma) solutions were prepared in distilled H2O immediately before each experiment. Working concentrations of each drug are described in the figure legends.
Plasmids and Transfections
The pYFPN1-Htt-Gln-72 plasmid was a generous gift of Dr. Robert Baloh. HEK293 transfections were carried out using FuGENE HD transfection reagent (Promega, Madison, WI) as per the manufacturer's instructions. mRFP-LC3 (Addgene plasmid 21075) (31) was generated by Dr. Tamotsu Yoshimori and deposited to Addgene. Neurons were transfected using Lipofectamine 2000 (Invitrogen) according to manufacturer's instructions. Transfection was done at DIV5, and neurons were transduced at DIV6. Live images were acquired using a Nikon Eclipse TE2000-E microscope equipped with CoolSnap HQ camera (Roper Scientific) and NIS Elements (Nikon Inc., New York). For siRNA transfections, Lipofectamine siRNAMAX (Invitrogen) was used according to the manufacturer's instructions. The sequence of the anti-mouse-α-syn siRNA duplex was 5′-GCAUGAGACUAUGCACCU-3′ and 5′-UAUUUAUAGGUGCAUAGU-3′. Transfection was carried out at DIV6, 1 day after transduction.
Lysosomal Function Assays
Magic Red Cathepsin MR-(RR)2 (Immunochemistry Technologies) was used to monitor cathepsin-B activity in HEK293 cells, according to the manufacturer's instructions. Briefly, 48 h after transduction, cells were incubated with the Magic Red reagent for 15 min and washed in imaging buffer (10 mm HEPES, 136 mm NaCl, 2.5 mm KCl, 2 mm CaCl2, 1.3 mm MgCl2, 10 mm glucose), and the fluorescent signal generated by proteolytic activity was observed using a fluorescence microscope. For EGFR degradation assay, 48 h after transduction, HEK293 cells were incubated with 200 ng/ml murine EGF (PeproTech) or BSA for 30 and 120 min. As a positive control, a set of EGF-treated wells were preincubated with 100 μm chloroquine for 1 h. At the end of the incubations, cells were harvested in 2% SDS lysis buffer for immunoblotting.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde in PBS followed by permeabilization with 0.1% Triton X-100. For LC3 and Lamp1 staining, digitonin (100 μg/ml) used for permeabilization. For 20 S proteasome staining, cells were permeabilized during fixation with 1% Triton X-100 to reduce nuclear staining. After blocking with bovine serum albumin (BSA, 3% w/v in PBS), cells were incubated in primary antibodies (supplemental Table S1) followed by Alexa Fluor-conjugated secondary antibodies (Invitrogen). Cells on coverslips were incubated with DAPI and mounted on glass slides in Fluoromount-G (SouthernBiotech, AL). For neurons, 4% w/v sucrose was included in the fixation solution. Images were acquired using an Olympus BX 51 microscope equipped with a digital camera DP71 and DP manager (Olympus), except for Figs. 7A and 9A where images were acquired using a Nikon Eclipse TE2000-E microscope equipped with CoolSnap HQ camera (Roper Scientific) and NIS Elements (Nikon Inc.).
FIGURE 7.
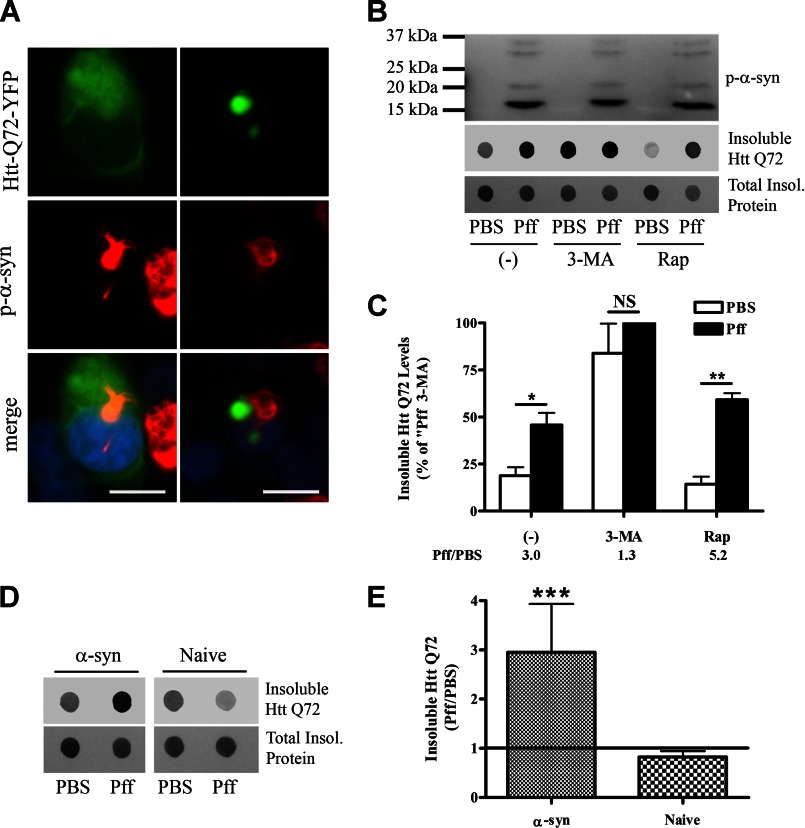
Autophagy function is reduced in α-syn aggregate-bearing cells. A–C, 48 h after PBS or Pff transduction, HEK293 α-syn cells were transfected with YFP-tagged Htt Gln-72. 20 h after transfection, the cells were treated with 10 mm 3-MA, 0.2 μm Rap, or vehicle (−) for 24 h. A, diffuse, soluble (left) or aggregated (right) Htt Gln-72 (green) did not co-localize with p-α-syn (red) aggregates. B, representative IBs for p-α-syn and GFP (recognizes YFP tag of Htt) as well as Ponceau S total protein staining. The Htt Gln-72 within Triton X-100-insoluble cellular fractions was visualized by dot IBs. C, quantification of dot blots (n = 4). Ratio of insoluble Htt Gln-72 levels in Pff-td and PBS-td cells for each treatment condition is indicated below the graph. Pff transduction caused increased insoluble Htt Gln-72 protein levels. 3-MA treatment abolished the difference between Htt Gln-72 levels of PBS-td and Pff-td cells, suggesting that this difference in untreated cells resulted from impaired autophagy. D and E, 48 h after transduction, HEK293 α-syn or naive HEK293 cells were transfected with YFP-tagged Htt Gln-72 and 48 h after transfection were harvested for IB. D, representative IBs for GFP (insoluble Htt Gln-72) and Ponceau S (total protein staining). The Htt Gln-72 within Triton X-100-insoluble cellular fractions was visualized by dot IBs. E, quantification of dot blots (n = 4), shown as ratio of insoluble Htt Gln-72 levels of Pff-td and PBS-td cells. The significant increase in insoluble Htt Gln-72 levels observed in Pff-td α-syn HEK293 cells was absent in naive HEK293 cells. Error bars, ±S.E. Statistical significance was determined by Student's paired t test (B and E) and one-way ANOVA (D) with Tukey's post hoc analysis. NS, *, **, and *** indicate p > 0.05, p < 0.05, p < 0.01, and p < 0.001, respectively. Scale bars, 20 μm.
FIGURE 9.
Lysosome morphology, but not catalytic function, is affected in Pff-td cells. A, Pff-td cells have altered lysosomal morphology. 24 h after Pff transduction, cells were treated for 24 h with a lysosomal protease inhibitor mixture (Lysosome Inh.) or vehicle (−). The IF images show examples of giant (>2.5 μm in diameter) Lamp1-positive (green) vesicles in Pff-td cells (arrowheads). Quantification of giant Lamp1 vesicles, shown as number of vesicles per 100 nuclei (blue), is shown on the right. Pff-td cells harbored significantly more giant lysosomes than PBS-td cells. Lysosomal inhibitor treatment eliminated this difference. Co-localization between p-α-syn (red) and Lamp1 (green) was not observed in the absence or presence of inhibitor treatment. B, lysosome function was not affected in Pff-td cells. The fluorogenic lysosomal substrate, Magic Red MR-(RR)2, is cleaved by the lysosomal protease cathepsin-B (Cath-B) to yield a fluorescent product. Both normal lysosomes and the giant lysosomes in Pff-td cells converted the fluorogenic substrate, suggesting that lysosomal function was unaffected by the α-syn aggregates. Treatment with lysosomal inhibitors (top right) diminished lysosomal fluorescence, confirming dependence of Magic Red MR-(RR)2 cleavage on proper lysosome function. C, lysosomal degradation of EGFR is unaffected in Pff-td cells. A representative IB (n ≥3) is shown, which reveals similar EGF-induced EGFR degradation by lysosomes in PBS-td and Pff-td cells 30 and 120 min after addition of EGF. EGFR was not substantially degraded in PBS-td or Pff-td cells in the absence of EGF (EGF −). Cq treatment inhibited EGFR degradation, confirming that the reduction of EGFR after EGF treatment was dependent on lysosome function. Statistical significance was determined with Student's t test. NS, *** indicate p > 0.05, and p < 0.001, respectively. Scale bars, 20 μm.
Immunoblotting
For sequential extractions, cells were scraped into 1% Triton X-100 in Tris-buffered saline (TBS) (50 mm Tris, 150 mm NaCl, pH 7.4) and a protease/phosphatase inhibitor mixture at 4 °C. Lysates were sonicated and centrifuged at 100,000 × g for 30 min. The pellet was washed and suspended in 2% SDS in TBS. For total protein extractions, cells were scraped and sonicated directly in 2% SDS in TBS. Protein concentrations were estimated by BCA assay (Thermo-Fisher Scientific), and equal amounts of protein were loaded onto gels for each sample unless indicated otherwise. For sequential extractions, protein concentrations of Triton X-100 fractions were used to calculate the volume of lysate to be loaded. For a given sample, Triton X-100 and SDS fractions were loaded at a volume ratio of 1:2 to better visualize the less abundant SDS-soluble proteins. This difference in loading amounts was considered when Triton X-100 and SDS fractions were compared in the quantification panels. Samples were run on 5–20% gradient SDS-polyacrylamide gels, and proteins were transferred to nitrocellulose or PVDF membranes for IB. 5% fat-free milk in TBS was used for blocking, except for p-α-syn IB, where 7.5% BSA in TBS was used. For dot blot assays, samples were diluted 1:10 in TBS and loaded to Bio-Dot SF Microfiltration Apparatus (Bio-Rad). Proteins captured by nitrocellulose membrane were visualized by IB. The primary antibodies used in this study are listed in supplemental Table 1. Immunoreactivity was detected via chemiluminescence by use of HRP-conjugated secondary antibodies (Jackson ImmunoResearch), Western Lightning ECL substrate (PerkinElmer Life Sciences), and LAS-3000 imager (Fuji, Tokyo, Japan) or by use of IRDye secondary antibodies and ODY-2816 Imager (Li-Cor Biosciences).
Immunoelectron Microscopy
Immuno-EM was conducted as described previously (27). Briefly, 14 days after Pff treatment, neurons were fixed in periodate/lysine/paraformaldehyde and permeabilized with 0.05% saponin in PBS with 2% fish gelatin and 0.05% thimerosal followed by incubation in p-α-syn antibody 81A. Neurons were then incubated with goat anti-mouse IgG coupled to Nanogold (Nanoprobes, Yaphank, NY). The Nanogold-labeled neurons were postfixed and gold-toned with 0.05% gold chloride.
Toxicity Assays
CytoTox 96® nonradioactive cytotoxicity assay (Promega) was used to monitor LDH release as an indicator of cytotoxicity, and the CellTiter 96® AQueous One Solution cell proliferation assay (Promega) was used to monitor methanethiosulfonate tetrazolium reduction as an estimate of cell viability, according to the manufacturer's instructions. HEK293 cells were treated for 48 h with rapamycin, 48 h after transduction. Complete media indicate that transduced cells were incubated in nutrient-rich media, starting 4 h after transduction for 92 h. For neurons, cells were starved in DMEM or treated with rapamycin 7 days after transduction for 7 days. For viability assays, HEK293 cells and neurons were incubated with MTS tetrazolium substrate for 1 and 4 h, respectively. Results are shown as the ratios of Pff- and PBS-td cells that received the same treatment.
Quantifications, Image Processing, and Statistics
Co-localization of p-α-syn aggregates and inclusions of Pdp-related proteins was quantified by manual counting using ImageJ (National Institutes of Health, Bethesda). Inclusion was defined as a cytoplasmic region with clearly increased IF signal compared with surrounding cytoplasmic areas of the same cell. IF pictures from 3 or 4 independent experiments were used for quantification, and a minimum of 100 p-α-syn aggregates from four random fields (∼30 cells per field) were counted for each condition. IB signals were quantified by MultiGauge version 2.3 (Fujifilm) or Image Studio version 2.0 (Li-Cor Biosciences). Acquired images were processed with Adobe Photoshop CS2 (Adobe Systems Inc.) to facilitate visualization where needed. The adjustments were applied equally across the entire image and to controls. All experiments were repeated a minimum of three times. The number following “±” sign indicates S.E. Statistical analyses used for each experiment were indicated in the figure legends. The supplemental Table S2 summarized data used for statistical analyses. All analyses were carried out using Graphpad Prism 4 (Graphpad Software Inc.).
RESULTS
Insoluble α-Syn Aggregates Interact with Protein Degradation Machinery
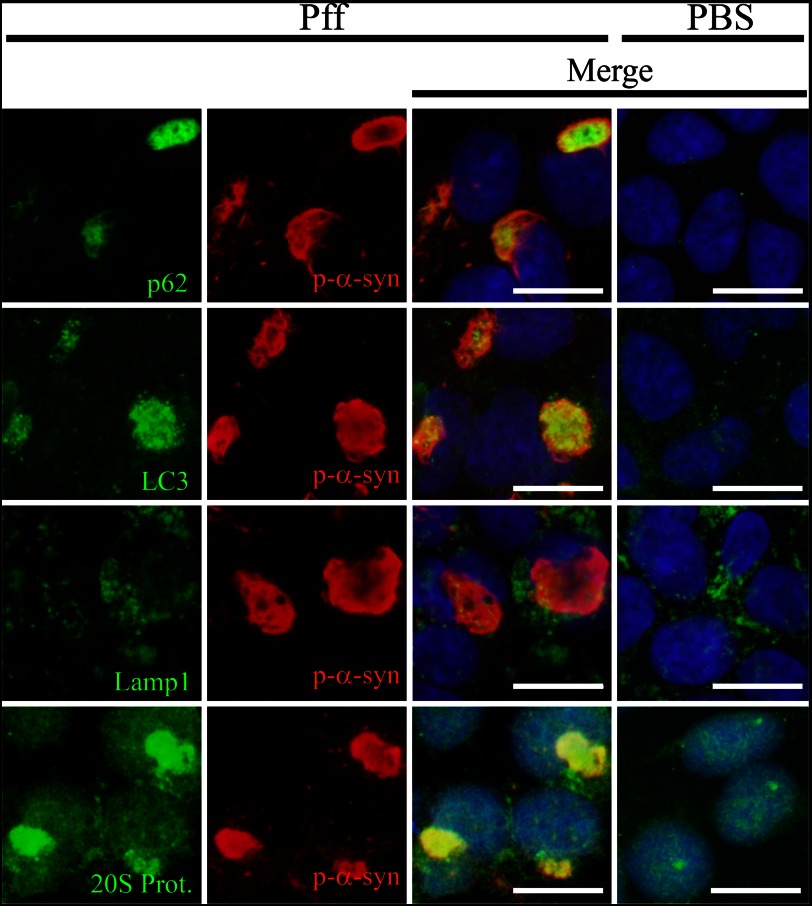
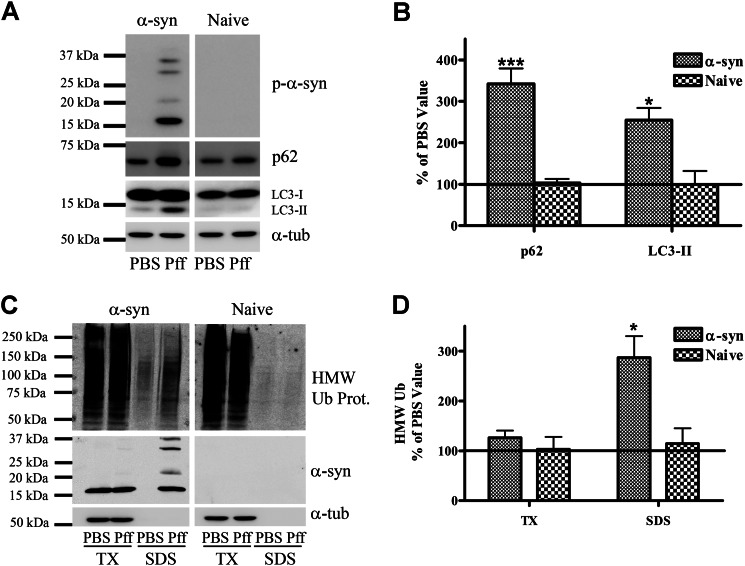
The HEK293 model of LB-like α-syn inclusions, in which α-syn-expressing cells are transduced with α-syn Pffs to induce aggregate formation (26), provides a cellular system to evaluate whether α-syn aggregates undergo proteolytic degradation. Utilizing this model, we first assessed by IF the localizations of several proteins involved in the ALP (Fig. 1). These include the following: p62, an adaptor protein that recruits autophagy machinery to protein aggregates (32); LC3, a protein specifically localized to autophagosome membranes when in its lipidated form (LC3-II) (33); and Lamp1, a marker of lysosomes and autophagolysosomes (34). In control PBS-transduced (PBS-Td) cells without α-syn aggregates, p62 and LC3 staining was faint and punctate and spread across the cytoplasm (Fig. 1). In contrast, in Pff-transduced (Pff-td) cells bearing α-syn aggregates, p62 and LC3 staining was largely redistributed (Fig. 1), such that the majority of the p-α-syn aggregates showed co-localization with accumulations of these proteins (97.4 ± 1.3% for p62 and 77.8 ± 5.5% for LC3, n = 3). Autophagy substrates should progressively localize to autophagolysosomes, which are the product of autophagosome-lysosome fusion and where substrates are normally degraded by lysosomal proteases (35). In contrast with p62 and LC3, the lysosome-autophagolysosome marker Lamp1 did not show appreciable co-localization with α-syn aggregates (10.2 ± 6.0%, n = 4; Fig. 1). These data suggest that the α-syn inclusions are associated with the early autophagy machinery but may not be delivered to autophagolysosomes.
FIGURE 1.
Co-localization of protein degradation pathway components with α-syn aggregates. IF images of HEK293 α-syn cells 24 h after Pff transduction, stained with p-α-syn antibody (red), DAPI (blue), or antibodies to p62, LC3, Lamp1, and 20 S proteasome (green). Dispersed and punctate cytoplasmic staining of p62 and LC3 was observed in PBS-td cells. In Pff-td cells, p-α-syn aggregates showed strong co-localization with p62 and LC3. Similarly, there was diffuse staining of the 20 S proteasome in the PBS-td cells, whereas Pff-td cells had cytoplasmic 20 S proteasome staining that largely co-localized with α-syn aggregates. In contrast, Lamp1-positive vesicles were observed in both PBS-td and Pff-td cells, and these did not typically co-localize with p-α-syn aggregates. Scale bars, 20 μm.
In addition to co-localization with p62 and LC3, the α-syn aggregates in Pff-td cells were also observed to be associated with the 20 S proteasome (Fig. 1). It was previously demonstrated that most α-syn inclusions in the HEK293 model are ubiquitinated and that they co-localize with the molecular chaperone Hsp70 (26). These findings suggest that proteasome components, as well as those from the ALP, are recruited to the α-syn aggregates, likely in an attempt to degrade the inclusions. Alternatively, these proteasome and autophagy constituents may interact aberrantly with aggregates, an event that could affect their function.
Insoluble α-Syn Aggregates Are Not Degraded by Autophagy and Their Clearance Is Severely Impaired
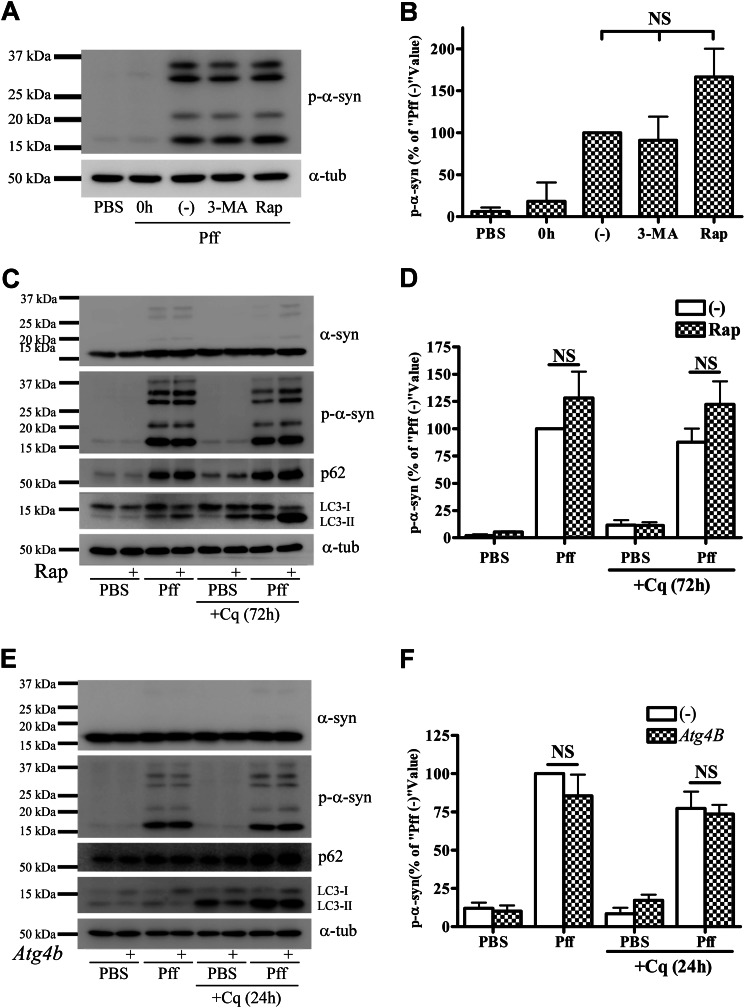
To elucidate if α-syn aggregates are degraded by autophagy, we evaluated whether autophagy inhibition or activation affected the levels of p-α-syn. HEK293 cells stably expressing α-syn (HEK293 α-syn) were treated for 24 h with either an inhibitor of autophagosome formation, 3-methyladenine (3-MA) (33, 36), or an activator of autophagy, rapamycin (Rap). Drug treatment was initiated 4 h after PBS or Pff transduction, a time when only minimal α-syn aggregation had occurred. Immunoblotting revealed that neither 3-MA nor Rap caused a significant change in p-α-syn levels, although rapamycin treatment trended toward an increase of p-α-syn (Fig. 2, A and B). Examination of parallel cultures 4 h after drug addition, a time that allows for the detection of cell-associated Myc-tagged Pffs, revealed that neither drug affected cellular Pff levels (data not shown). The effectiveness of the drug treatments was seen in an LC3 immunoblot, with the expected decrease and increase of LC3-II formation after 3-MA and Rap treatment, respectively (data not shown). Likewise, there were corresponding drug-induced changes in the abundance of LC3-positive puncta as visualized by IF (data not shown). We also noted that there was a significant decrease in the amount of LC3 that was recruited to the aggregates when the cells were treated with 3-MA (data not shown), confirming that the α-syn inclusions normally associate with autophagosome-conjugated LC3-II and not with cytoplasmic forms of LC3 that are increased after 3-MA treatment.
FIGURE 2.
Autophagy does not degrade α-syn aggregates. A and B, after 4 h of Pff transduction, HEK293 α-syn cells were treated with the autophagy inhibitor 3-MA (10 mm) or the autophagy activator Rap (0.2 μm) in starvation media for 24 h. Cells were subsequently extracted with SDS for IB. A, representative IB of p-α-syn and α-tubulin (α-tub), and B, quantification of IBs (n = 3). “0h” refers to cells that were harvested prior to drug treatment, and “(−)” refers to vehicle-treated cells. Minimal α-syn pathology was present when drug treatment was initiated, as shown in the 0h lane. Neither 3-MA nor rapamycin caused a significant change in p-α-syn levels. C and D, extended rapamycin treatment in a nutrient-rich condition does not induce clearance of aggregates. 4 h after Pff transduction, HEK293 α-syn cells were maintained in nutrient-rich media for 20 h, followed by rapamycin treatment for 72 h in the absence or presence of 15 μm Cq in nutrient-rich conditions. C, IB of p-α-syn, p62, LC3, and the loading control, α-tubulin. D, quantification of p-α-syn levels in IBs from three independent experiments, with data normalized to the mean p-α-syn level from vehicle-treated Pff-td cells. Significant activation of autophagy by rapamycin was evident because LC3-II accumulated in Cq and rapamycin co-treated cells, but this activation did not reduce aggregated α-syn levels. E and F, genetic inhibition of autophagy does not cause accumulation of p-α-syn aggregates. HEK293 α-syn-stable cells were transfected with the dominant negative autophagy inhibitor, mutant ATG4BC74A-strawberry, or with empty vector (−). Cells were transduced with Pff for 24 h, treated with 30 μm Cq for an additional 24 h, and then harvested 72 h after transfection. E, IB of p-α-syn, p62, LC3, and the loading control, α-tubulin. F, quantification of p-α-syn levels from three independent experiments, with data normalized to the mean p-α-syn level from empty vector-transfected Pff-td cells. ATG4B mutant (ATG4BC74A) prevents lipidation of LC3 to LC3-II, thereby inhibiting autophagosome closure and autophagy (38) resulting in a significant decrease in total and LC3-I-normalized LC3-II levels in ATG4B-transfected cells (2.34 ± 0.30 versus 0.63 ± 0.05 in Pff-td mock and ATG4B-transfected cells, respectively, p < 0.01) without changing aggregated p-α-syn levels. Error bars, ± S.E. One-way ANOVA with Tukey's post-hoc analysis (B) or Student's paired t test (D and F) was used to test for statistically significant differences. NS indicates p > 0.05.
Although Rap treatment of the HEK293 α-syn cells resulted in an enhancement of autophagy, Rap-induced autophagy activation may have been attenuated because of prior activation of autophagy caused by serum deprivation during transduction (37). To eliminate this possibility, HEK293 α-syn cells were treated with Rap for 72 h under nutrient-rich conditions, and Rap did not significantly affect p-α-syn levels in the Pff-td cells, although the observed increase of LC3-II confirmed activation of autophagy (Fig. 2, C and D). Similarly, p-α-syn levels were not affected by treatment with the lysosome inhibitor, chloroquine (Cq) (Fig. 2, C and D), even though this drug clearly affected LC3-II clearance. Importantly, inhibition of autophagy by expression of a dominant negative ATG4B mutant (38) did not lead to an elevation of p-α-syn (Fig. 2, E and F), although this mutant protein greatly reduced the conversion of LC3-I to LC3-II. These data thus confirm that seeded α-syn aggregates in HEK293 cells are not appreciably degraded by autophagy.
As the 20 S proteasome subunit was found associated with p-α-syn aggregates (Fig. 1), we also queried whether inhibition of proteasome function would affect the levels of α-syn aggregates. Addition of the proteasome inhibitor clasto-lactacystin-β-lactone did not increase Triton X-100-insoluble p-α-syn or α-syn in the Pff-td cells, although clasto-lactacystin-β-lactone treatment led to a significant increase in high molecular weight ubiquitinated proteins (HMW Ub; data not shown), thereby demonstrating inhibition of proteasomal degradation. These results suggest that α-syn aggregates are not cleared effectively by either autophagic or proteasomal pathways.
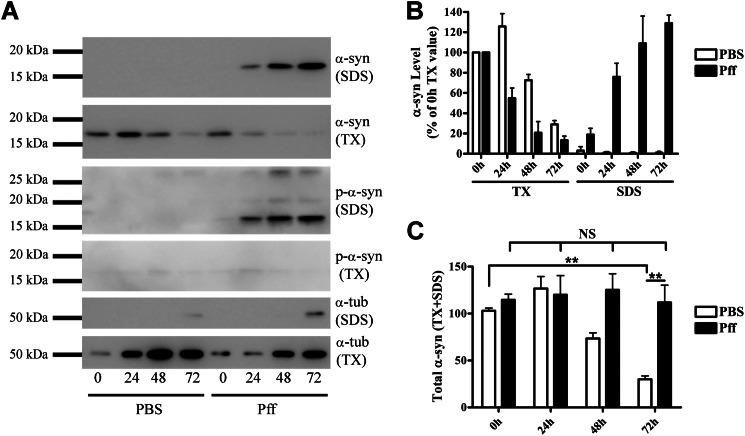
To further confirm this conclusion, another α-syn expression model was utilized in which α-syn was under the control of a dox-inducible promoter. After growth for 16 h in dox-containing medium to induce α-syn expression, the cells were transduced with α-syn Pffs and subsequently grown for varying lengths of time in dox-free medium to suppress α-syn synthesis. The cells underwent sequential extractions, and samples were analyzed by immunoblotting, using a C-terminal α-syn antibody that recognizes endogenous α-syn and not the C-terminal truncated 1–120myc α-syn Pffs. In PBS-td cells, Triton X-100-soluble α-syn was degraded, with the amount of α-syn remaining 72 h after dox removal being 29.9 ± 5.1% of the amount observed immediately (0 h) after transduction (Fig. 3, A and B). In Pff-td cells, Triton X-100-soluble α-syn decreased at a faster rate over time, concomitant with a progressive increase in Triton X-100-insoluble/SDS-soluble α-syn levels (Fig. 3, A and B). However, no net degradation of total α-syn was detected in the Pff-td cells 72 h after dox removal, although there was a substantial decrease of total α-syn in the PBS-treated cells (Fig. 2, B and C). Taken together, these data indicate that α-syn aggregates are not effectively degraded by any cellular mechanism in the time frame of this study, including but not limited to the UPS and ALP.
FIGURE 3.
Clearance of aggregated α-syn is severely impaired. HeLa T-Rex cells that express α-syn under the control of a dox-inducible promoter were induced with dox for 16 h. After removal of dox, cells were transduced for 4 h and then incubated in dox-free starvation media for 0, 24, 48, or 72 h until they were extracted in Triton X-100, followed by SDS. A, representative IB for endogenous α-syn, p-α-syn, and α-tubulin (α-tub). Equal volumes of cell extract were loaded. B, quantification of IBs (n = 4). The α-syn levels in Triton X-100 fractions at 0 h after transduction were used to normalize α-syn levels at other time points. C, graph showing total (Triton X-100 + SDS) α-syn levels (n = 4). In PBS-td cells, α-syn was rapidly cleared from cells, whereas in Pff-td cells, no net α-syn reduction was observed. Statistical significance was determined with Student's t test. NS and ** indicate p > 0.05 and p < 0.01, respectively.
Typical Autophagy Substrates Accumulate in α-Syn Aggregate-bearing Cells
As noted previously, LC3-II and p62 are autophagy substrates whose levels are routinely used to monitor autophagic function (39–41). Interestingly, both of these proteins accumulated in Pff-td HEK293 α-syn cells but not in Pff-td-naive HEK293 cells that have negligible α-syn expression (Fig. 4C) (42) and therefore do not form α-syn aggregates (Fig. 4, A and B). Additionally, the levels of Triton X-100-insoluble HMW-Ub proteins, which are typically degraded by autophagy (43, 44), were significantly increased in Pff-td HEK293 α-syn cells in the absence or presence of proteasome inhibition (data not shown) but not in Pff-td naive HEK293 cells (Fig. 4, C and D). These data suggest an impairment of autophagy in aggregate-bearing cells.
FIGURE 4.
Typical autophagy substrates accumulate in α-syn aggregate-bearing cells. A and B, 48 h after Pff transduction, α-syn and naive HEK293 cells were harvested, and proteins were extracted in SDS. A, representative IB for p-α-syn, p62, LC3, and the loading control, α-tubulin (α-tub). B, quantification of IBs (n ≥3), with protein levels in Pff-td cells expressed as a percentage of the corresponding PBS control level. C and D, 48 h after transduction, α-syn and naive HEK293 cells were extracted in Triton X-100 (TX) and then SDS. C, representative IB for HMW ubiquitin (Ub), α-syn, with α-tubulin as a loading control. D, quantification of IBs (n ≥3), with Pff-td cell levels expressed as a percentage of the corresponding PBS-td cell levels. Insoluble Ub protein levels did not increase in Pff-td naive HEK293 cells. Error bars, ±S.E. Student's paired t test was used to test for statistically significant differences. * indicates p < 0.05 and *** indicates p < 0.001, respectively.
α-Syn Aggregates Also Interact with Autophagy and Proteasome Components in Neurons but Are Resistant to Degradation
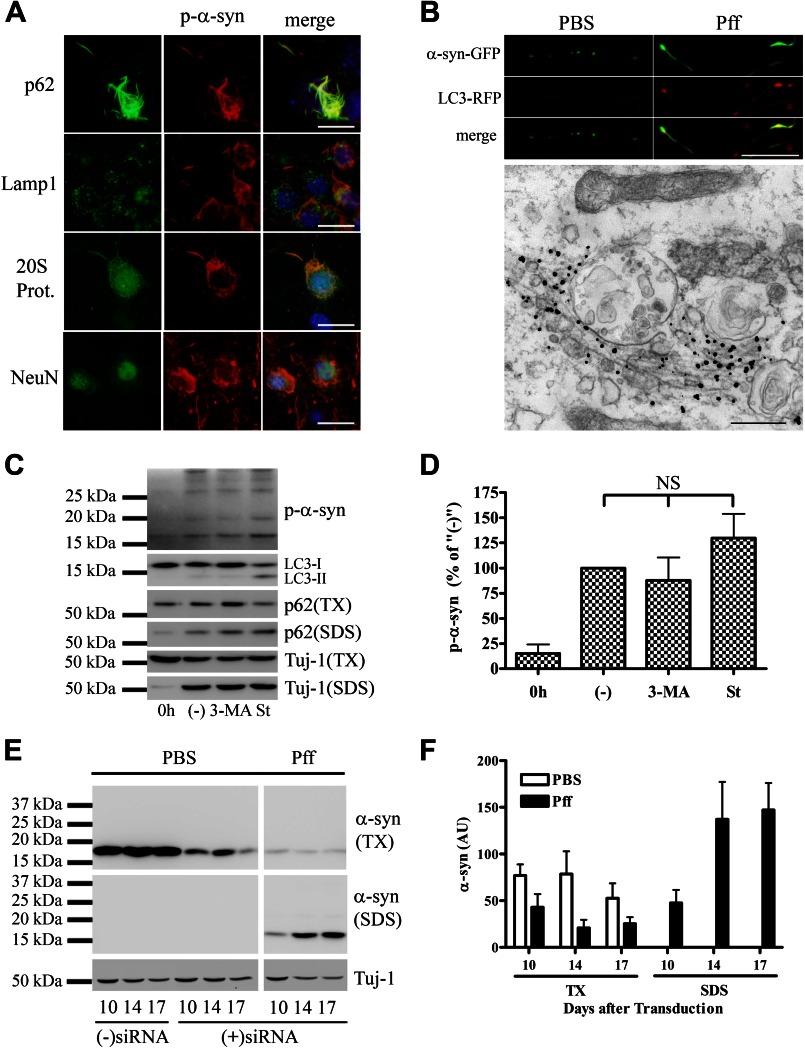
To determine whether α-syn aggregates in primary wild-type neurons are also resistant to degradation, we utilized a recently developed neuronal model of α-syn pathology where endogenous mouse α-syn (m-α-syn) readily aggregates to form LB-like α-syn inclusions when transduced with α-syn Pffs (27). The α-syn aggregates in neurons, like those in HEK293 α-syn cells, co-localized with ubiquitin (27), p62, and the 20 S proteasome but not with Lamp1 (Fig. 5A). Live imaging of LC3-mRuby-transfected neurons revealed that LC3-positive vesicles co-localized with α-syn aggregates and accumulated in Pff-td neuronal processes, and immuno-EM with p-α-syn antibodies demonstrated that these aggregates can be found in close association with vesicle-like structures, but not inside these vesicles (Fig. 5B). These data suggest that α-syn inclusions associate with autophagy components in neurons, but they are not efficiently delivered to autophagolysosomes for degradation.
FIGURE 5.
α-Syn aggregates interact with protein degradation pathway components but cannot be effectively degraded in neurons. At 5 DIV, primary hippocampal neurons were transduced with human WT α-syn Pffs as described (27). At 19 DIV, cells were either fixed for IF or harvested for IB analysis. A, IF using p62, Lamp1, 20 S proteasome, neuronal nuclei (NeuN), and p-α-syn antibodies, demonstrating that neuronal α-syn aggregates co-localized with p62 and 20 S proteasome but not with Lamp1. B, top, fluorescence images of neurons transfected with LC3-mRuby and α-syn-GFP, showing accumulation of LC3 puncta that co-localized with p-α-syn aggregates in Pff-td neuronal processes. Bottom, immuno-EM, showing nano-gold-labeled p-α-syn aggregates at ×50,000 magnification, with proximal autophagic vesicle-like structures. Fibrillar nano-gold-labeled aggregates were typically not localized inside these structures despite close association. C and D, at DIV10, neurons were transduced with WT α-syn Pffs, and at DIV13 they were starved to activate autophagy or treated with 10 mm 3-MA to inhibit autophagy. Pff-td cells in one well were harvested at DIV13 to monitor pretreatment levels of p-α-syn (denoted as “0h”). The remaining cells were harvested at DIV15. C, representative IB for p-α-syn, LC3, p62, and neuron-specific βIII-tubulin (Tuj-1). TX, Triton X-100. D, quantification of IBs (n = 3), where “(−)” designates vehicle-treated cells. Minimal α-syn pathology was present when drug treatment was initiated, as shown in the 0h lane. 3-MA treatment increased soluble p62 levels, whereas starvation reduced it. However, neither treatment caused a significant change in p-α-syn levels. E and F, at DIV5, neurons were transduced with PBS or WT α-syn Pffs, and after 24 h, they were transfected with anti-α-syn siRNA or vehicle. Cells were harvested 10, 14, and 17 days after transduction. (−) siRNA denotes vehicle-transfected PBS-td cells harvested 10 days after transduction. E, representative IB for m-α-syn and Tuj-1. F, quantification of IBs from three independent experiments. Reduction of α-syn expression neither stopped aggregate growth nor caused clearance of aggregates. Error bars, ±S.E. One-way ANOVA with Tukey's post-hoc analysis was used to test for statistically significant differences. NS indicates p > 0.05. Scale bars, 20 μm (A), 20 μm and 500 nm (B).
To further confirm that neuronal α-syn aggregates, like those in HEK293 α-syn cells, are not effectively cleared by autophagy, Pffs were introduced into neurons that had been cultured in vitro for 10 days (DIV10). At DIV13, before significant amounts of α-syn aggregates were generated (27), the Pff-td neurons were treated for 48 h with 3-MA to inhibit autophagy or with starvation media to activate autophagy (30). As expected, 3-MA caused accumulation of the autophagy substrate p62 in the Triton X-100-soluble fraction, whereas starvation slightly reduced its levels (Fig. 5C). However, neither treatment significantly affected p-α-syn levels (Fig. 5, C and D), suggesting that α-syn inclusions in neurons are not significantly degraded by autophagy during the time frame of these studies.
The clearance of α-syn aggregates in neurons was more closely examined by reducing α-syn expression in PBS or Pff-td WT neurons through transfection of an anti-α-syn siRNA. Endogenous m-α-syn levels were monitored by immunoblotting with a species-specific antibody that does not recognize added human α-syn Pffs (27). Despite a significant reduction in m-α-syn expression after siRNA treatment (i.e. decrease by 77 ± 5%; compare −siRNA and +siRNA at DIV10 in Fig. 5E), insoluble endogenous m-α-syn progressively accumulated in the siRNA-treated Pff-td neurons during the course of the experiment (Fig. 5, E and F). Thus, even a low level of α-syn expression was enough to sustain the growth of α-syn aggregates, suggesting that α-syn aggregates are not cleared from the neurons at a significant rate.
Finally, Pff-td WT neurons, but not Pff-td neurons from α-syn knock-out (KO) mice, showed increased p62, LC3-II, and Triton X-100-insoluble HMW-Ub protein levels (Fig. 6, A–D), demonstrating that accumulation of these proteins are not caused simply by the addition of Pffs, but rather by the endogenous m-α-syn inclusions that are seeded by Pffs. Moreover, these data suggest that α-syn aggregates impair autophagy in neurons, as was observed in HEK293 cells.
FIGURE 6.
Canonical autophagy substrates accumulate in neurons with α-syn accumulations. Primary hippocampal neurons from WT or α-syn knock-out (KO) mouse embryos were cultured in nutrient-rich neuronal media. At DIV5, neurons were transduced with human α-syn Pffs or PBS, and neurons were harvested for IB at DIV19. A and C, representative IB for p-α-syn, m-α-syn, Ub, p62, LC3 and α-tubulin (α-tub). B and D, effect of Pff transduction on levels of immunoblotted proteins in WT and α-syn KO neurons (n ≥3 for B and n = 5 for D). Protein levels are shown as a percentage of the values of PBS-td cells. Triton X-100 (TX)-insoluble HMW Ub proteins, p62, LC3-II, and m-α-syn levels were significantly increased in Pff-treated neurons, suggesting that the removal of these proteins was impaired. No change was observed in Pff-td α-syn KO neurons. Error bars, ±S.E. Student's paired t test was used to test for statistically significant differences. No asterisk, *, and ** indicate p > 0.05, p < 0.05, and p < 0.01, respectively.
Autophagy Function Is Impaired in α-Syn Aggregate-bearing Cells
To further substantiate that α-syn aggregates impair autophagy function, we examined the degradation of the established autophagy substrate, YFP-tagged huntingtin exon 1 fragment, with 72 glutamine repeats (Htt Gln-72), which forms highly insoluble aggregates that are degraded by autophagy (13, 45). Htt Gln-72 and p-α-syn did not associate with each other when examined by IF in Pff-td cells (Fig. 7A). However, insoluble Htt Gln-72 levels were greatly increased in Pff-td cells compared with PBS-td cells (Fig. 7, B and C). The difference in insoluble Htt Gln-72 levels between the PBS-td and Pff-td cells was largely negated when autophagy was inhibited with 3-MA, and it was enhanced when autophagy was further activated with Rap (Fig. 7, B and C). This indicates that the increase in Htt Gln-72 levels in Pff-td cells is due to impairment of autophagic function that cannot be overcome by enhancement of autophagosome formation. That the amount of insoluble Htt Gln-72 was increased in Pff-td cells upon 3-MA addition reveals there was not a complete cessation of autophagy-mediated degradation of Htt Gln-72 in the α-syn aggregate-bearing cells in the absence of 3-MA. Although there was a modest increase of Htt Gln-72 clearance in the Rap-treated PBS-td cells, the Rap effect was likely muted in both the PBS-td and Pff-td cells because autophagy was already activated due to maintenance under starvation conditions after transduction. As expected, p-α-syn levels remained essentially unaffected by the 3-MA and Rap (Fig. 7B). Likewise, Pff transduction did not increase Htt Gln-72 levels in naive HEK293 cells that do not form α-syn aggregates (Fig. 7, D and E), demonstrating that α-syn Pffs do not by themselves impair autophagy, nor do they seed Htt Gln-72 aggregates.
Together, these and previous data indicate that autophagy function is compromised in α-syn aggregate-bearing cells, as evidenced by the accumulation of canonical autophagy substrates and the partial inhibition of Htt Gln-72 degradation.
Autophagosome Clearance Is Impaired in α-Syn Aggregate-bearing Cells
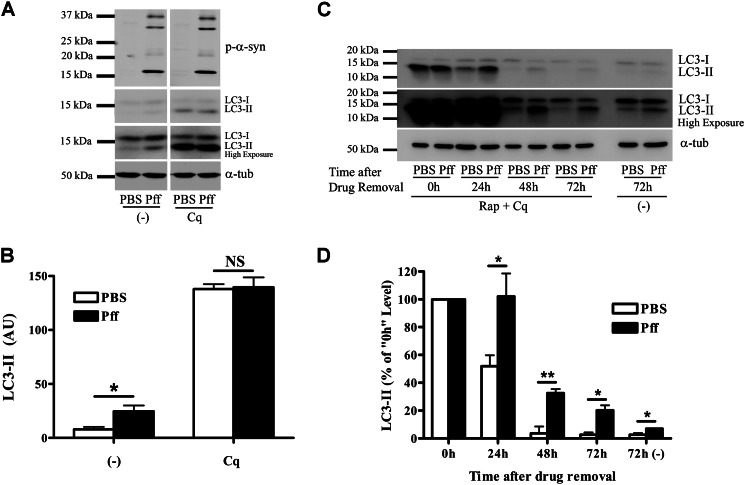
As the autophagy pathway consists of multiple steps, including autophagosome formation, maturation, and subsequent clearance of autophagosomes, we investigated whether α-syn aggregates specifically interfere with any of these steps. Our data suggest that autophagosome formation is not prevented, as demonstrated by the large number of LC3-II-positive autophagosomes that are associated with α-syn aggregates and the increased LC3-II levels observed in Pff-td cells (Figs. 1, 2, C and E, and 4, A and B). However, LC3-II levels could also be elevated as the result of an elevation in autophagosome synthesis, rather than or in addition to an inhibition of autophagosome clearance. To distinguish between these possibilities, we utilized a, LC3-II turnover assay (41). PBS-td and Pff-td HEK293 α-syn cells were treated for 4 h with a high concentration of chloroquine (100 μm) to inhibit all lysosomal degradations. This treatment abolished the differences in LC3-II levels between PBS-td and Pff-td cells (Fig. 8, A and B), and such a result is typically observed when there is an impairment of autophagosome clearance (39, 41). This was further confirmed when PBS-td and Pff-td cells were simultaneously treated with Rap and Cq for 4 h to induce accumulation of a large pool of autophagic vesicles. After removal of both drugs, LC3-II levels rapidly decreased in the PBS-td cells, so more than half of the LC3-II was cleared in 24 h and nearly all LC3-II was degraded by 48–72 h (Fig. 8, C and D). Remarkably, there was no net clearance of LC3-II in Pff-td cells after 24 h, and LC3-II levels remained significantly higher than the corresponding level observed in PBS-td cells at each later time point (Fig. 8, C and D). It should be noted that the residual effects of Cq likely caused a transient slowing in the elimination of LC3-II in both PBS and Pff-td cells (46). Nonetheless, these data suggest that α-syn aggregates do not inhibit formation of autophagosomes but rather impair autophagosome clearance.
FIGURE 8.
Autophagosome clearance is impaired in α-syn aggregate-bearing cells. A and B, HEK293 α-syn cells were incubated in nutrient-rich media for 4 h after Pff transduction, and the cells were treated with 100 μm of the lysosome inhibitor Cq 48 h after transduction. After 4 h of incubation with Cq, the cells were lysed in SDS buffer. A, representative IB for p-α-syn, LC3, and the loading control, α-tubulin (α-tub). B, quantification of IBs (n = 4). Pff-td and PBS-td cells showed comparable LC3-II levels upon complete inhibition of autophagosome clearance by Cq, suggesting that autophagosome clearance is impaired. C and D, HEK293 α-syn cells were incubated in nutrient-rich media 4 h after Pff transduction, and cells were treated with 0.2 μm rapamycin and 100 μm Cq 48 h after transduction. This “pulse” generated a large pool of autophagosomes in both Pff-td and PBS-td cells. After 4 h, the drugs were removed, and cells were harvested 0, 24, 48, and 72 h later. As a control, a set of cells was treated only with vehicle and were harvested 72 h after treatment. C, representative IB for LC3 and α-tubulin. D, quantification of IBs (n ≥3). Control cells rapidly cleared accumulated autophagosomes, whereas this clearance was significantly inhibited in Pff-td cells. Error bars, ±S.E. B and D, Student's t test was used to test for statistically significant differences. NS, *, and ** indicate p > 0.05, p < 0.05, and p < 0.01, respectively.
Lysosome Function Appears Normal in α-Syn Aggregate-bearing Cells
The absence of Lamp1 co-localization with α-syn aggregates (Fig. 1) suggests that the resistance of α-syn deposits to degradation and the impairment of autophagosome clearance may be caused by a defect in autophagosome maturation, rather than by an impairment of lysosomal degradation. Nevertheless, lysosomal defects could contribute to decreased autophagosome clearance by inhibiting lysosome-autophagosome fusion (47). To investigate whether α-syn aggregates affect Lamp1-positive lysosomes, we conducted IF analyses in PBS-td and Pff-td HEK293 cells. Although lysosomes appeared primarily as abundant fine puncta in PBS-td cells, they were less abundant and enlarged in Pff-td cells. The frequency of giant lysosomes (>2.5 μm in diameter), which were rarely observed in PBS-td cells, was significantly increased in Pff-td cells (Fig. 9A). No such change in lysosome morphology was observed in Pff-td naive HEK293 cells (data not shown), demonstrating that formation of giant lysosomes was caused by the formation of LB-like α-syn aggregates, rather than simply Pff treatment. Giant lysosomes could also be readily observed in PBS-td cells after treatment with lysosomal protease inhibitors (Fig. 9, A and B). Notably, lysosomal inhibition did not result in the accumulation of p-α-syn aggregates in giant lysosomes within Pff-td cells or increased Lamp1/p-α-syn co-localization (7.0 ± 2.6% with inhibitor versus 10.2 ± 6.0% without inhibitor, n = 3). This suggests that the lysosomal dilation observed in Pff-td cells was not due to the sequestration and subsequent degradation of α-syn aggregates in lysosomal compartments (Fig. 9A).
Lysosomal enlargement may indicate possible lysosomal dysfunction, which could lead to impaired autophagosome clearance. Therefore, we asked if the giant lysosomes in Pff-td cells are functional. The lysosomal processing of a fluorogenic cathepsin-B substrate, Magic Red-RR(2), as well as the lysosomal degradation of the epidermal growth factor (EGF) receptor after addition of EGF were assessed in Pff-td cells. Notably, lysosome function was maintained in these cells, as both processing of the Magic Red substrate (Fig. 9B) and degradation of the EGFR (Fig. 9C) were observed. The integrity of lysosomes in Pff-td cells was also supported by experiments that demonstrated that the enlarged lysosomes had a normal pH and that the lysosomal protease, cathepsin D, could be properly processed from its procathepsin D precursor (data not shown).
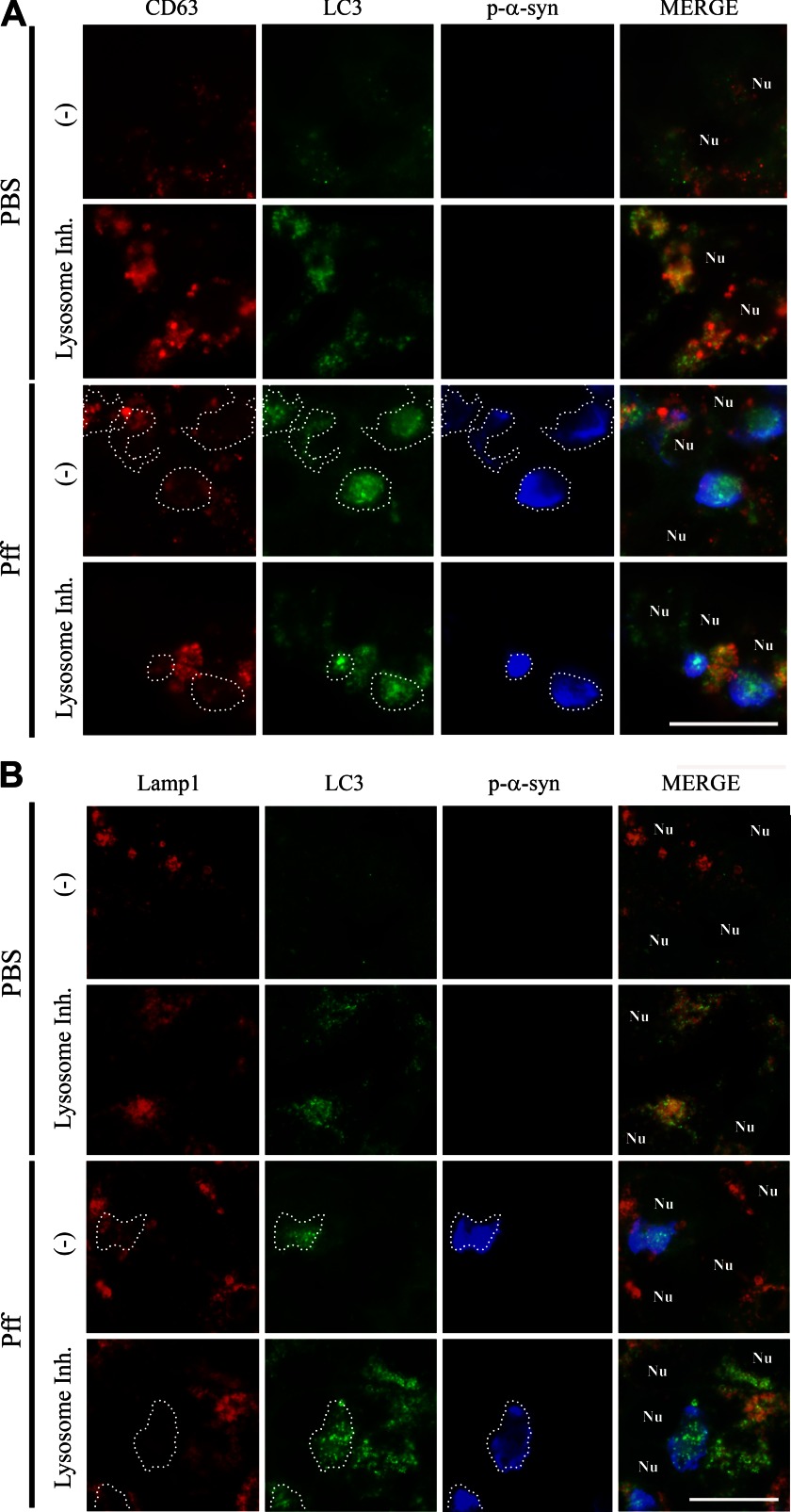
As lysosomal function in Pff-td cells appeared to be normal, we asked if autophagy impairment in α-syn aggregate-bearing cells might result from a defect in autophagosome maturation. This could be due to reduced autophagosome/lysosome fusion or to a reduced number of fully matured autophagosomes that are available for lysosomal fusion. A step in autophagosome maturation that precedes lysosomal fusion is amphisome formation, during which autophagosomes fuse with multivesicular bodies (MVBs) (48). Utilizing the MVB marker CD63 (49), the localizations of CD63, LC3, Lamp1, and p-α-syn were compared with PBS-td and Pff-td cells. PBS-td cells showed light and diffuse staining for LC3 and CD63, with some co-localization of these markers (Fig. 10A). However, inhibition of lysosome function caused accumulation of LC3 vesicles that largely co-localized with both CD63 and Lamp1, demonstrating that these vesicles matured to amphisomes and autophagolysosomes before their clearance was blocked by the lysosomal inhibitors (Fig. 10, A and B). LC3-positive vesicles accumulated and co-localized with p-α-syn in Pff-td cells in the absence of lysosomal inhibition, but there was little overlap of LC3 with CD63 (Fig. 10A) or Lamp1 (Fig. 10B) at the sites of p-α-syn aggregates. As with PBS-td cells, inhibition of lysosomal function in Pff-td cells resulted in an increase of CD63 and Lamp1 co-localization with LC3, with the exception that CD63 and Lamp1 staining was generally not observed in LC3-positive p-α-syn aggregates (Fig. 10, A and B). Although there was minimal localization of CD63 with p-α-syn aggregates in Pff-td cells, either in the absence or presence of lysosomal inhibition (11.8 ± 2.6% and 6.7 ± 4.1%, respectively), the observed CD63-containing MVBs were not associated with α-syn aggregates. However, they appeared to be enlarged, as was seen with Lamp1-positive vesicles (Figs. 9A and 10A). These data support the interpretation that the LC3-positive autophagosomes that associate with α-syn aggregates do not mature to amphisomes. Moreover, the enlarged LC3/CD63-positive amphisomes that do form in Pff-td cells and that are not associated with α-syn aggregates may have altered properties that could provide an explanation for the partial impairment of autophagic function observed in the cells harboring α-syn inclusions.
FIGURE 10.
Autophagosomes associated with p-α-syn aggregates are impaired in maturation. 24 h after PBS or Pff transduction, HEK293 α-syn cells were treated for another 24 h with a lysosomal inhibitor mixture (Lysosome Inh.) or vehicle (−) before they were fixed for IF. A, IF images showing CD63 (red), LC3 (green), p-α-syn (blue, white dotted outline), and nuclei (denoted as “Nu”). There were few LC3 and CD63 puncta in PBS-td cells, and these did not co-localize in the absence of lysosomal inhibitor. However, LC3 and CD63 co-localization was observed in the PBS-td cells when the clearance of autophagic vesicles was prevented by lysosomal inhibitor treatment. In Pff-td cells, LC3 vesicles that accumulate with p-α-syn aggregates were not generally co-localized with CD63 vesicles. Inhibition of lysosomes resulted in some overlap of CD63 and LC3, but CD63 did not localize to LC3 vesicles associated with p-α-syn. Although they did not co-localize with the p-α-syn aggregates, CD63 vesicles in Pff-td cells appeared to be larger and more variable in size compared with those in PBS-td cells. B, IF images showing Lamp1 (red), LC3 (green), p-α-syn (blue, white dotted outline), and nuclei (denoted as “Nu”). Lamp1 co-localized with LC3 in PBS-td cells treated with lysosomal inhibitor and in cells without α-syn aggregates in Pff-td cells after addition of lysosome inhibitor. However, Lamp1 was largely excluded from LC3 vesicles that overlapped with α-syn aggregates in Pff-td cells in the absence or presence of lysosome inhibitor. n ≥3 for all experiments. Scale bars, 20 μm.
Autophagy Activation Enhances Cellular Toxicity in α-Syn Aggregate-bearing Cells
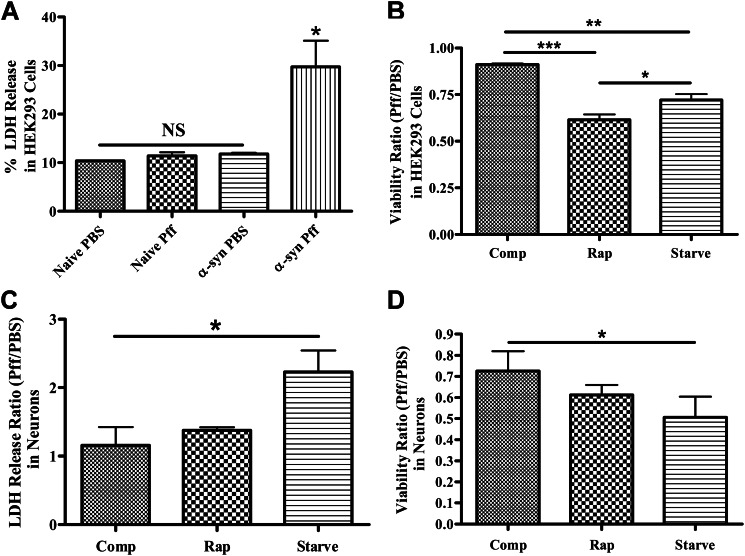
Previous studies have shown that activation of autophagy may attenuate cytotoxicity of aggregate-prone proteins that are degraded by autophagy (30, 50). However, our data suggest that the resistance of α-syn aggregates to autophagy is not the result of a defect in autophagy initiation but rather a problem of autophagosome maturation. Thus, a strategy of autophagy activation may not be productive in cells containing α-syn inclusions, and in fact the cell death that is observed in both aggregate-bearing HEK293 cells (Fig. 11A) and neurons (27) is exacerbated upon activation of autophagy with rapamycin or serum starvation (Fig. 11, B–D). Although it is possible that the increase in toxicity observed with these treatments results from cellular mechanisms other than autophagy, these data are consistent with the notion that increased autophagosome generation can be detrimental in cells when autophagosome clearance is impaired (51–53).
FIGURE 11.
Treatments that activate autophagy increase the toxicity of α-syn aggregates. A, lactate dehydrogenase release assay was performed to monitor cell death in naive HEK293 cells or HEK293 cells that express α-syn. Only Pff-tdHEK293 α-syn cells (α-syn Pff) showed significantly increased LDH release (n = 2). B, PBS- or Pff-tdHEK293 α-syn cells were maintained in nutrient-rich complete media (comp), treated with rapamycin or maintained in starvation media after transduction, until a tetrazolium-based viability assay was carried out. Both starvation and rapamycin treatment significantly reduced viability in transduced cells (n = 3). C and D, Pff- or PBS-td WT neurons were starved or treated with rapamycin, and the LDH release assay (n = 3) (C) and the viability assay (n = 2) (D) were performed. Starvation significantly increased cell death and decreased viability in neurons, and rapamycin treatment trended toward the same pattern. (Error bars, ±S.E. A and B, one-way ANOVA with Tukey's post hoc analysis used to test for statistically significant differences. C and D, Student's paired t test was used to test for statistically significant differences. NS, *, ** and *** indicate p > 0.05, p < 0.05, p < 0.01, and p < 0.001, respectively.)
DISCUSSION
Utilizing recently developed cellular models of Pff-seeded α-syn aggregation, we have demonstrated that LB-like inclusions are resistant to degradation despite their interaction with ALP and UPS components. These aggregates persist even when soluble α-syn levels are substantially reduced, indicating that once seeding occurs, pathologic α-syn aggregates are refractory to clearance from the cells. Interestingly, overall autophagic clearance is impaired in the α-syn aggregate-bearing cells, which may be a factor contributing to the observed reduction in cellular viability. Importantly, alterations of autophagy were observed in both non-neuronal cells and in primary cultured neurons harboring α-syn aggregates. Indeed, the neuronal Pff transduction model of α-syn aggregation is unique in that α-syn aggregates can be formed with very high efficiency in the absence of α-syn overexpression or toxic insults that may perturb autophagy or proteasome function. Moreover, our cellular models of α-syn seeding with recruitment of endogenous α-syn to yield LB-like aggregates may replicate key aspects of synucleinopathies, as several recent lines of evidence suggest that the spreading of amyloid protein pathology, such as seen with α-syn in PD, may result from the neuronal release and uptake of extracellular α-syn seeds (54–58).
Previous studies revealed that LBs in PD co-localize with UPS and ALP components such as ubiquitin, 20 S proteasome, p62, and LC3 (3, 59–61). However, it was unclear whether these associations resulted in degradation of LBs. Although LB-like inclusions co-localized with UPS and ALP components in our cell model, such interactions did not lead to degradation of α-syn aggregates. A similar recruitment of ALP components has also been observed with other aggregated proteins that are resistant to autophagic degradation (62). The failure to degrade α-syn aggregates cannot be explained by a complete inhibition of autophagy, as Htt Gln-72 appeared to be degraded in aggregate-bearing cells, albeit at a significantly reduced rate, suggesting that α-syn aggregates are selectively resistant to degradation. As with a number of other protein aggregates subject to autophagy, Htt Gln-72 aggregates occur as “aggresomes,” which are believed to be actively generated by cells to sequester misfolded proteins within the cytoplasm (63–65). However, Pff-seeded α-syn aggregates do not show robust co-localization with aggresomal markers such as vimentin (26, 66), suggesting that they are a distinct type of inclusion that is resistant to degradation.
The demonstration that cells harboring aggregated α-syn have partial impairment of autophagy complements prior studies that showed that soluble α-syn species may reduce autophagic activity (19, 67, 68). The α-syn inclusions appear to impair autophagy by reducing autophagosome clearance. This disruption of autophagy may contribute to the cellular toxicity seen in the aggregate-bearing cells, and we observed further cell death upon activation of autophagy. Autophagosome clearance defects have been previously suggested in neurodegenerative diseases. For instance, the abnormal accumulation of autophagosomes in Alzheimer disease brain tissue is thought to be caused by impaired autophagosome clearance (52), and LC3-II levels are elevated in PD brain tissues with abundant LB pathology (60). An impairment of autophagosome clearance is likely to be detrimental, as removal of both unwanted and damaged proteins and defective organelles such as mitochondria (69, 70) is vital for cells. Therefore, impairment of autophagy may lead to cell death through several different mechanisms, including oxidative stress, which is strongly implicated in PD pathogenesis (71, 72). Indeed, several proteins that are implicated in PD pathogenesis, including ATP13A2, LRRK2 and Pink-1, may also affect autophagy function (73–75), suggesting that autophagic dysfunction may be a primary cause of cell death in PD.
The LC3-positive autophagic vesicles that accumulate with α-syn aggregates were not decorated with markers of amphisomes and autophagolysosomes, indicating that autophagosome maturation is likely impaired prior to amphisome formation. The resistance of α-syn aggregates to degradation may be caused by an inability of aggregate-associated autophagosomes to migrate within the cell and/or an inability of MVBs to fuse with the autophagosomes. Alternatively, it is possible that α-syn aggregates interfere with the ability of autophagosomes to mature, such that the LC3-positive structures that accumulate in aggregate-bearing cells are incomplete or abnormal. In addition to the α-syn inclusions being refractory to autophagic degradation, it appears that there is a more general impairment of autophagy in aggregate-containing cells. Interestingly, the Pff-td cells show an enlargement of both CD63- and Lamp1-positive vesicular bodies, although lysosome function seems to be unaltered. These changes may suggest that the α-syn aggregates have an effect on vesicle-related cellular processes, including autophagy. Indeed, α-syn aggregates perturb endosomal trafficking in yeast by interacting with Rab proteins (76, 77).
After completion of our studies, a published study (78) demonstrated clearance of α-syn Pffs after their introduction into naive HEK293 cells. In this model, p-α-syn immunoreactive structures were found in a small population of cells shortly after addition of Pffs, and these largely disappeared within 24 h. These data would seem to contradict our observations of α-syn aggregates being resistant to clearance. However, the p-α-syn structures observed in this recent study likely consisted of the introduced exogenous Pffs, with little to no recruitment of endogenous α-syn due to the very low expression of α-syn in naive HEK293 cells. Thus, although these new data may provide insights into the clearance of internalized α-syn Pff seeds, this model differs substantially from that employed in our studies and may not recapitulate key features of human synucleinopathies, where LBs are formed from endogenous α-syn that misfolds within the cytoplasm. In this regard, our findings of impaired autophagy after the formation of LB-like inclusions composed of endogenous α-syn in both neurons and non-neuronal cells may provide a more accurate representation of the biology of LBs and their potential role in neurodegeneration.
In conclusion, we have demonstrated that seeded α-syn aggregates are resistant to degradation and impair autophagosome clearance. These findings have bearing on therapeutic strategies in PD and related synucleinopathies, as prior studies have indicated that autophagy activation via rapamycin treatment may attenuate disease phenotype in certain in vivo models of PD (79). Although the models used in these studies may provide important insights, they typically rely on oxidative insults or proteasome inhibitors to achieve the desired phenotype, and these treatments could increase the cellular dependence on autophagic removal of damaged mitochondria and unwanted proteins (80, 81). In addition, neurons in these models lack the insoluble, LB-like α-syn aggregates that are observed in our cellular models and in synucleinopathies. Our studies suggest that activation of autophagy may not be effective once α-syn aggregates form, which is likely to occur before clinical symptoms manifest. On the contrary, autophagy activation may be harmful to α-syn aggregate-bearing cells because of the impaired autophagosome clearance. However, it is possible that alternative therapeutic approaches, such as restoration of normal autophagic flux (82, 83), may reduce the ALP impairment caused by the α-syn aggregates and be effective in enhancing the survival of affected cells. Further studies will determine whether these approaches can reduce neurodegeneration in PD and related synucleinopathies.
Supplementary Material
Acknowledgments
We are grateful to D. M. Riddle, V. Kehm, and A. Stieber for their technical assistance. We thank J. Q. Trojanowski, T. Cohen, K. Luk, and J. Guo for their critical reading of the manuscript. We also thank R. Baloh (Cedars-Sinai Medical Center) for generously providing pYFPN1-Htt-Gln-72 plasmid.
This work was supported, in whole or in part, by National Institutes of Health Grant NS053488. This work was also supported by the J. P. B. Foundation and the Jeff and Anne Keefer Fund.
This article was selected as a Paper of the Week.

This article contains supplemental Tables 1 and 2 and additional references.
- α-syn
- α-synuclein
- 3-MA
- 3-methyladenine
- ALP
- autophagy-lysosome pathway
- Cq
- chloroquine
- EGFR
- epidermal growth factor receptor
- HMW
- high molecular weight
- IF
- immunofluorescence
- LB
- Lewy body
- PD
- Parkinson disease
- Pff
- pre-formed α-synuclein fibrils
- Rap
- rapamycin
- Ub
- ubiquitin
- UPS
- ubiquitin proteasome system
- p-α-syn
- phospho-α-synuclein
- IB
- immunoblot
- PDL
- poly-d-lysine
- ANOVA
- analysis of variance
- DIV
- days in vitro
- dox
- doxycyline
- m-α-syn
- mouse-specific endogenous α-syn
- Pff-td
- Pff-transduced
- MVB
- multivesicular body.
REFERENCES
- 1. Spillantini M. G. (1999) Parkinson's disease, dementia with Lewy bodies, and multiple system atrophy are α-synucleinopathies. Parkinsonism Relat. Disord. 5, 157–162 [DOI] [PubMed] [Google Scholar]
- 2. Galvin J. E., Lee V. M., Trojanowski J. Q. (2001) Synucleinopathies: clinical and pathological implications. Arch. Neurol. 58, 186–190 [DOI] [PubMed] [Google Scholar]
- 3. Kuzuhara S., Mori H., Izumiyama N., Yoshimura M., Ihara Y. (1988) Lewy bodies are ubiquitinated. A light and electron microscopic immunocytochemical study. Acta Neuropathol. 75, 345–353 [DOI] [PubMed] [Google Scholar]
- 4. Fujiwara H., Hasegawa M., Dohmae N., Kawashima A., Masliah E., Goldberg M. S., Shen J., Takio K., Iwatsubo T. (2002) α-Synuclein is phosphorylated in synucleinopathy lesions. Nat. Cell Biol. 4, 160–164 [DOI] [PubMed] [Google Scholar]
- 5. Wakabayashi K., Tanji K., Mori F., Takahashi H. (2007) The Lewy body in Parkinson's disease: molecules implicated in the formation and degradation of α-synuclein aggregates. Neuropathology 27, 494–506 [DOI] [PubMed] [Google Scholar]
- 6. Polymeropoulos M. H., Lavedan C., Leroy E., Ide S. E., Dehejia A., Dutra A., Pike B., Root H., Rubenstein J., Boyer R., Stenroos E. S., Chandrasekharappa S., Athanassiadou A., Papapetropoulos T., Johnson W. G., Lazzarini A. M., Duvoisin R. C., Di Iorio G., Golbe L. I., Nussbaum R. L. (1997) Mutation in the α-synuclein gene identified in families with Parkinson's disease. Science 276, 2045–2047 [DOI] [PubMed] [Google Scholar]
- 7. Krüger R., Kuhn W., Müller T., Woitalla D., Graeber M., Kösel S., Przuntek H., Epplen J. T., Schöls L., Riess O. (1998) Ala30Pro mutation in the gene encoding α-synuclein in Parkinson's disease. Nat. Genet. 18, 106–108 [DOI] [PubMed] [Google Scholar]
- 8. Singleton A. B., Farrer M., Johnson J., Singleton A., Hague S., Kachergus J., Hulihan M., Peuralinna T., Dutra A., Nussbaum R., Lincoln S., Crawley A., Hanson M., Maraganore D., Adler C., Cookson M. R., Muenter M., Baptista M., Miller D., Blancato J., Hardy J., Gwinn-Hardy K. (2003) α-Synuclein locus triplication causes Parkinson's disease. Science 302, 841. [DOI] [PubMed] [Google Scholar]
- 9. Zarranz J. J., Alegre J., Gómez-Esteban J. C., Lezcano E., Ros R., Ampuero I., Vidal L., Hoenicka J., Rodriguez O., Atarés B., Llorens V., Gomez Tortosa E., del Ser T., Muñoz D. G., de Yebenes J. G. (2004) The new mutation, E46K, of α-synuclein causes Parkinson and Lewy body dementia. Ann. Neurol. 55, 164–173 [DOI] [PubMed] [Google Scholar]
- 10. Rubinsztein D. C. (2006) The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 443, 780–786 [DOI] [PubMed] [Google Scholar]
- 11. Seglen P. O., Berg T. O., Blankson H., Fengsrud M., Holen I., Strømhaug P. E. (1996) Structural aspects of autophagy. Adv. Exp. Med. Biol. 389, 103–111 [DOI] [PubMed] [Google Scholar]
- 12. Mortimore G. E., Miotto G., Venerando R., Kadowaki M. (1996) Autophagy. Subcell. Biochem. 27, 93–135 [DOI] [PubMed] [Google Scholar]
- 13. Ravikumar B., Duden R., Rubinsztein D. C. (2002) Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet. 11, 1107–1117 [DOI] [PubMed] [Google Scholar]
- 14. Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Mizushima N. (2006) Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889 [DOI] [PubMed] [Google Scholar]
- 15. Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Tanaka K. (2006) Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441, 880–884 [DOI] [PubMed] [Google Scholar]
- 16. Futerman A. H., van Meer G. (2004) The cell biology of lysosomal storage disorders. Nat. Rev. Mol. Cell Biol. 5, 554–565 [DOI] [PubMed] [Google Scholar]
- 17. Nixon R. A., Yang D. S., Lee J. H. (2008) Neurodegenerative lysosomal disorders: a continuum from development to late age. Autophagy 4, 590–599 [DOI] [PubMed] [Google Scholar]
- 18. Webb J. L., Ravikumar B., Atkins J., Skepper J. N., Rubinsztein D. C. (2003) α-Synuclein is degraded by both autophagy and the proteasome. J. Biol. Chem. 278, 25009–25013 [DOI] [PubMed] [Google Scholar]
- 19. Cuervo A. M., Stefanis L., Fredenburg R., Lansbury P. T., Sulzer D. (2004) Impaired degradation of mutant α-synuclein by chaperone-mediated autophagy. Science 305, 1292–1295 [DOI] [PubMed] [Google Scholar]
- 20. Ebrahimi-Fakhari D., Cantuti-Castelvetri I., Fan Z., Rockenstein E., Masliah E., Hyman B. T., McLean P. J., Unni V. K. (2011) Distinct roles in vivo for the ubiquitin-proteasome system and the autophagy-lysosomal pathway in the degradation of α-synuclein. J. Neurosci. 31, 14508–14520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Emmanouilidou E., Melachroinou K., Roumeliotis T., Garbis S. D., Ntzouni M., Margaritis L. H., Stefanis L., Vekrellis K. (2010) Cell-produced α-synuclein is secreted in a calcium-dependent manner by exosomes and impacts neuronal survival. J. Neurosci. 30, 6838–6851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Engelender S., Kaminsky Z., Guo X., Sharp A. H., Amaravi R. K., Kleiderlein J. J., Margolis R. L., Troncoso J. C., Lanahan A. A., Worley P. F., Dawson V. L., Dawson T. M., Ross C. A. (1999) Synphilin-1 associates with α-synuclein and promotes the formation of cytosolic inclusions. Nat. Genet. 22, 110–114 [DOI] [PubMed] [Google Scholar]
- 23. McLean P. J., Kawamata H., Hyman B. T. (2001) α-Synuclein-enhanced green fluorescent protein fusion proteins form proteasome-sensitive inclusions in primary neurons. Neuroscience 104, 901–912 [DOI] [PubMed] [Google Scholar]
- 24. Paxinou E., Chen Q., Weisse M., Giasson B. I., Norris E. H., Rueter S. M., Trojanowski J. Q., Lee V. M., Ischiropoulos H. (2001) Induction of α-synuclein aggregation by intracellular nitrative insult. J. Neurosci. 21, 8053–8061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee H. J., Shin S. Y., Choi C., Lee Y. H., Lee S. J. (2002) Formation and removal of α-synuclein aggregates in cells exposed to mitochondrial inhibitors. J. Biol. Chem. 277, 5411–5417 [DOI] [PubMed] [Google Scholar]
- 26. Luk K. C., Song C., O'Brien P., Stieber A., Branch J. R., Brunden K. R., Trojanowski J. Q., Lee V. M. (2009) Exogenous α-synuclein fibrils seed the formation of Lewy body-like intracellular inclusions in cultured cells. Proc. Natl. Acad. Sci. U.S.A. 106, 20051–20056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Volpicelli-Daley L. A., Luk K. C., Patel T. P., Tanik S. A., Riddle D. M., Stieber A., Meaney D. F., Trojanowski J. Q., Lee V. M. (2011) Exogenous α-synuclein fibrils induce Lewy body pathology leading to synaptic dysfunction and neuron death. Neuron 72, 57–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Abeliovich A., Schmitz Y., Fariñas I., Choi-Lundberg D., Ho W. H., Castillo P. E., Shinsky N., Verdugo J. M., Armanini M., Ryan A., Hynes M., Phillips H., Sulzer D., Rosenthal A. (2000) Mice lacking α-synuclein display functional deficits in the nigrostriatal dopamine system. Neuron 25, 239–252 [DOI] [PubMed] [Google Scholar]
- 29. Giasson B. I., Murray I. V., Trojanowski J. Q., Lee V. M. (2001) A hydrophobic stretch of 12 amino acid residues in the middle of α-synuclein is essential for filament assembly. J. Biol. Chem. 276, 2380–2386 [DOI] [PubMed] [Google Scholar]
- 30. Young J. E., Martinez R. A., La Spada A. R. (2009) Nutrient deprivation induces neuronal autophagy and implicates reduced insulin signaling in neuroprotective autophagy activation. J. Biol. Chem. 284, 2363–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kimura S., Noda T., Yoshimori T. (2007) Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3, 452–460 [DOI] [PubMed] [Google Scholar]
- 32. Bjørkøy G., Lamark T., Johansen T. (2006) p62/SQSTM1: a missing link between protein aggregates and the autophagy machinery. Autophagy 2, 138–139 [DOI] [PubMed] [Google Scholar]
- 33. Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fukuda M. (1991) Lysosomal membrane glycoproteins. Structure, biosynthesis, and intracellular trafficking. J. Biol. Chem. 266, 21327–21330 [PubMed] [Google Scholar]
- 35. Xie Z., Klionsky D. J. (2007) Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9, 1102–1109 [DOI] [PubMed] [Google Scholar]
- 36. Seglen P. O., Bohley P. (1992) Autophagy and other vacuolar protein degradation mechanisms. Experientia 48, 158–172 [DOI] [PubMed] [Google Scholar]
- 37. Mortimore G. E., Pösö A. R. (1987) Intracellular protein catabolism and its control during nutrient deprivation and supply. Annu. Rev. Nutr. 7, 539–564 [DOI] [PubMed] [Google Scholar]
- 38. Fujita N., Hayashi-Nishino M., Fukumoto H., Omori H., Yamamoto A., Noda T., Yoshimori T. (2008) An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol. Biol. Cell 19, 4651–4659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rubinsztein D. C., Cuervo A. M., Ravikumar B., Sarkar S., Korolchuk V., Kaushik S., Klionsky D. J. (2009) In search of an “autophagomometer”. Autophagy 5, 585–589 [DOI] [PubMed] [Google Scholar]
- 40. Klionsky D. J., Cuervo A. M., Seglen P. O. (2007) Methods for monitoring autophagy from yeast to human. Autophagy 3, 181–206 [DOI] [PubMed] [Google Scholar]
- 41. Mizushima N., Yoshimori T., Levine B. (2010) Methods in mammalian autophagy research. Cell 140, 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zagranichnaya T. K., Wu X., Danos A. M., Villereal M. L. (2005) Gene expression profiles in HEK-293 cells with low or high store-operated calcium entry: can regulatory as well as regulated genes be identified? Physiol. Genomics 21, 14–33 [DOI] [PubMed] [Google Scholar]
- 43. Bartlett B. J., Isakson P., Lewerenz J., Sanchez H., Kotzebue R. W., Cumming R. C., Harris G. L., Nezis I. P., Schubert D. R., Simonsen A., Finley K. D. (2011) p62, Ref(2)P, and ubiquitinated proteins are conserved markers of neuronal aging, aggregate formation and progressive autophagic defects. Autophagy 7, 572–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145 [DOI] [PubMed] [Google Scholar]
- 45. Pollitt S. K., Pallos J., Shao J., Desai U. A., Ma A. A., Thompson L. M., Marsh J. L., Diamond M. I. (2003) A rapid cellular FRET assay of polyglutamine aggregation identifies a novel inhibitor. Neuron 40, 685–694 [DOI] [PubMed] [Google Scholar]
- 46. Zetser A., Levy-Adam F., Kaplan V., Gingis-Velitski S., Bashenko Y., Schubert S., Flugelman M. Y., Vlodavsky I., Ilan N. (2004) Processing and activation of latent heparanase occurs in lysosomes. J. Cell Sci. 117, 2249–2258 [DOI] [PubMed] [Google Scholar]
- 47. Kawai A., Uchiyama H., Takano S., Nakamura N., Ohkuma S. (2007) Autophagosome-lysosome fusion depends on the pH in acidic compartments in CHO cells. Autophagy 3, 154–157 [DOI] [PubMed] [Google Scholar]
- 48. Eskelinen E. L. (2005) Maturation of autophagic vacuoles in mammalian cells. Autophagy 1, 1–10 [DOI] [PubMed] [Google Scholar]
- 49. Escola J. M., Kleijmeer M. J., Stoorvogel W., Griffith J. M., Yoshie O., Geuze H. J. (1998) Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 273, 20121–20127 [DOI] [PubMed] [Google Scholar]
- 50. Sarkar S., Ravikumar B., Floto R. A., Rubinsztein D. C. (2009) Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death Differ. 16, 46–56 [DOI] [PubMed] [Google Scholar]
- 51. Wong E., Cuervo A. M. (2010) Autophagy gone awry in neurodegenerative diseases. Nat. Neurosci. 13, 805–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Boland B., Kumar A., Lee S., Platt F. M., Wegiel J., Yu W. H., Nixon R. A. (2008) Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer's disease. J. Neurosci. 28, 6926–6937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang X., Li L., Chen S., Yang D., Wang Y., Zhang X., Wang Z., Le W. (2011) Rapamycin treatment augments motor neuron degeneration in SOD1(G93A) mouse model of amyotrophic lateral sclerosis. Autophagy 7, 412–425 [DOI] [PubMed] [Google Scholar]
- 54. Angot E., Steiner J. A., Hansen C., Li J. Y., Brundin P. (2010) Are synucleinopathies prion-like disorders? Lancet Neurol. 9, 1128–1138 [DOI] [PubMed] [Google Scholar]
- 55. Brundin P., Melki R., Kopito R. (2010) Prion-like transmission of protein aggregates in neurodegenerative diseases. Nat. Rev. Mol. Cell Biol. 11, 301–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Soto C. (2012) In vivo spreading of tau pathology. Neuron 73, 621–623 [DOI] [PubMed] [Google Scholar]
- 57. Luk K. C., Kehm V. M., Zhang B., O'Brien P., Trojanowski J. Q., Lee V. M. (2012) Intracerebral inoculation of pathological α-synuclein initiates a rapidly progressive neurodegenerative α-synucleinopathy in mice. J. Exp. Med. 209, 975–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Luk K. C., Kehm V., Carroll J., Zhang B., O'Brien P., Trojanowski J. Q., Lee V. M. (2012) Pathological α-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science 338, 949–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zatloukal K., Stumptner C., Fuchsbichler A., Heid H., Schnoelzer M., Kenner L., Kleinert R., Prinz M., Aguzzi A., Denk H. (2002) p62 Is a common component of cytoplasmic inclusions in protein aggregation diseases. Am. J. Pathol. 160, 255–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Higashi S., Moore D. J., Minegishi M., Kasanuki K., Fujishiro H., Kabuta T., Togo T., Katsuse O., Uchikado H., Furukawa Y., Hino H., Kosaka K., Sato K., Arai H., Wada K., Iseki E. (2011) Localization of MAP1-LC3 in vulnerable neurons and Lewy bodies in brains of patients with dementia with Lewy bodies. J. Neuropathol. Exp. Neurol. 70, 264–280 [DOI] [PubMed] [Google Scholar]
- 61. Ii K., Ito H., Tanaka K., Hirano A. (1997) Immunocytochemical co-localization of the proteasome in ubiquitinated structures in neurodegenerative diseases and the elderly. J. Neuropathol. Exp. Neurol. 56, 125–131 [DOI] [PubMed] [Google Scholar]
- 62. Wong E. S., Tan J. M., Soong W. E., Hussein K., Nukina N., Dawson V. L., Dawson T. M., Cuervo A. M., Lim K. L. (2008) Autophagy-mediated clearance of aggresomes is not a universal phenomenon. Hum. Mol. Genet. 17, 2570–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kopito R. R. (2000) Aggresomes, inclusion bodies, and protein aggregation. Trends Cell Biol. 10, 524–530 [DOI] [PubMed] [Google Scholar]
- 64. Markossian K. A., Kurganov B. I. (2004) Protein folding, misfolding, and aggregation. Formation of inclusion bodies and aggresomes. Biochemistry 69, 971–984 [DOI] [PubMed] [Google Scholar]
- 65. Kaganovich D., Kopito R., Frydman J. (2008) Misfolded proteins partition between two distinct quality control compartments. Nature 454, 1088–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kawaguchi Y., Kovacs J. J., McLaurin A., Vance J. M., Ito A., Yao T. P. (2003) The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 115, 727–738 [DOI] [PubMed] [Google Scholar]
- 67. Winslow A. R., Chen C. W., Corrochano S., Acevedo-Arozena A., Gordon D. E., Peden A. A., Lichtenberg M., Menzies F. M., Ravikumar B., Imarisio S., Brown S., O'Kane C. J., Rubinsztein D. C. (2010) α-Synuclein impairs macroautophagy: implications for Parkinson's disease. J. Cell Biol. 190, 1023–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Martinez-Vicente M., Talloczy Z., Kaushik S., Massey A. C., Mazzulli J., Mosharov E. V., Hodara R., Fredenburg R., Wu D. C., Follenzi A., Dauer W., Przedborski S., Ischiropoulos H., Lansbury P. T., Sulzer D., Cuervo A. M. (2008) Dopamine-modified α-synuclein blocks chaperone-mediated autophagy. J. Clin. Invest. 118, 777–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Elmore S. P., Qian T., Grissom S. F., Lemasters J. J. (2001) The mitochondrial permeability transition initiates autophagy in rat hepatocytes. FASEB J. 15, 2286–2287 [DOI] [PubMed] [Google Scholar]
- 70. Novak I., Kirkin V., McEwan D. G., Zhang J., Wild P., Rozenknop A., Rogov V., Löhr F., Popovic D., Occhipinti A., Reichert A. S., Terzic J., Dötsch V., Ney P. A., Dikic I. (2010) Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 11, 45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Jenner P., Olanow C. W. (1996) Oxidative stress and the pathogenesis of Parkinson's disease. Neurology 47, S161–S170 [DOI] [PubMed] [Google Scholar]
- 72. Zhang Y., Dawson V. L., Dawson T. M. (2000) Oxidative stress and genetics in the pathogenesis of Parkinson's disease. Neurobiol. Dis. 7, 240–250 [DOI] [PubMed] [Google Scholar]
- 73. Michiorri S., Gelmetti V., Giarda E., Lombardi F., Romano F., Marongiu R., Nerini-Molteni S., Sale P., Vago R., Arena G., Torosantucci L., Cassina L., Russo M. A., Dallapiccola B., Valente E. M., Casari G. (2010) The Parkinson-associated protein PINK1 interacts with Beclin1 and promotes autophagy. Cell Death Differ. 17, 962–974 [DOI] [PubMed] [Google Scholar]
- 74. Gómez-Suaga P., Luzón-Toro B., Churamani D., Zhang L., Bloor-Young D., Patel S., Woodman P. G., Churchill G. C., Hilfiker S. (2012) Leucine-rich repeat kinase 2 regulates autophagy through a calcium-dependent pathway involving NAADP. Hum. Mol. Genet. 21, 511–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Usenovic M., Tresse E., Mazzulli J. R., Taylor J. P., Krainc D. (2012) Deficiency of ATP13A2 leads to lysosomal dysfunction, α-synuclein accumulation, and neurotoxicity. J. Neurosci. 32, 4240–4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Soper J. H., Kehm V., Burd C. G., Bankaitis V. A., Lee V. M. (2011) Aggregation of α-synuclein in S. cerevisiae is associated with defects in endosomal trafficking and phospholipid biosynthesis. J. Mol. Neurosci. 43, 391–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gitler A. D., Bevis B. J., Shorter J., Strathearn K. E., Hamamichi S., Su L. J., Caldwell K. A., Caldwell G. A., Rochet J. C., McCaffery J. M., Barlowe C., Lindquist S. (2008) The Parkinson's disease protein α-synuclein disrupts cellular Rab homeostasis. Proc. Natl. Acad. Sci. U.S.A. 105, 145–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Watanabe Y., Tatebe H., Taguchi K., Endo Y., Tokuda T., Mizuno T., Nakagawa M., Tanaka M. (2012) p62/SQSTM1-dependent autophagy of Lewy body-like α-synuclein inclusions. PloS one 7, e52868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bové J., Martínez-Vicente M., Vila M. (2011) Fighting neurodegeneration with rapamycin: mechanistic insights. Nat. Rev. Neurosci. 12, 437–452 [DOI] [PubMed] [Google Scholar]
- 80. Ogata M., Hino S., Saito A., Morikawa K., Kondo S., Kanemoto S., Murakami T., Taniguchi M., Tanii I., Yoshinaga K., Shiosaka S., Hammarback J. A., Urano F., Imaizumi K. (2006) Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol. Cell. Biol. 26, 9220–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lee J., Giordano S., Zhang J. (2012) Autophagy, mitochondria, and oxidative stress: cross-talk and redox signalling. Biochem. J. 441, 523–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lee J. A., Gao F. B. (2009) Inhibition of autophagy induction delays neuronal cell loss caused by dysfunctional ESCRT-III in frontotemporal dementia. J. Neurosci. 29, 8506–8511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Li L., Wang X., Fei X., Xia L., Qin Z., Liang Z. (2011) Parkinson's disease involves autophagy and abnormal distribution of cathepsin L. Neurosci. Lett. 489, 62–67 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.