Abstract
Products of the myc gene family integrate extracellular signals by modulating a wide range of their targets involved in cellular biogenesis and metabolism; the purpose of this integration is to regulate cell death, proliferation, and differentiation. However, understanding the regulation of myc at the transcription level remains a challenge. We performed rapid amplification of dmyc cDNA ends (5′ RACE) and mapped the transcription start site at P1 promoter, 18 base pairs upstream of the start of the known EST GM01143 and within the 5′ UTR. Our data show that the first TATA box, previously computationally predicted, is utilized to generate dmyc full length mRNA. The largest transcript contains all three exons, generated after the removal of the introns by constitutively regulated splicing events. Further investigation of Downstream Promoter Element (DPE) was achieved by studying lacZ reporter activity; investigation revealed that this element and its upstream cluster of binding sites are required for the dmyc intron 2 activity. These findings may provide valuable tools for further analysis of dmyc cis-elements.
Keywords: dmyc, Drosophila, 5′ RACE, RNA splicing, TATA-box, Downstream Promoter Element (DPE)
Introduction
The spatiotemporal distribution of the right amount of Myc during early stages of development is crucial for all aspects of normal cell growth, proliferation, and differentiation.1–3 In terminally differentiated cells, Myc protein is nearly absent; in adults, myc expression is confined to cell proliferation in tissues and during regenerative processes.4,5 Deregulation of myc possesses high potential towards malignant transformation.6–8myc induces G1-S phase transition by upregulation of cyclin D1, 2, 3/CDK4, 6 as well as cyclin E/CDK2;6,9 repression of cell cycle inhibitors p15, p21, p27; and inactivation of retinoblastoma protein (RB).8 Consequently, tight control of myc expression is required so that it can be activated or repressed rapidly and precisely whenever necessary. Unraveling the structure of the myc promoter provides valuable knowledge for understanding myc biology and its oncogenic behavior.10,11
The evolutionary conservation and structural similarity between human c-myc and fly dmyc makes it possible to study the myc promoter in Drosophila with the sole copy of the gene, dmyc.12–14 In our previous study, we dissected non-coding regions of the dmyc gene and identified separable regulatory units responsible for its patterning during the early stages of Drosophila development.14 We predicted three potential TATA regions for the P1 promoter within the 5′ UTR, namely, GC box1/TATA box1/Inr1, GC box2/TATA box2/Inr2, and GC box3/TATA box3/Inr3. In our current study, 5′ RACE analysis of the dmyc cDNA end at the 5′ UTR region revealed that the P1 promoter initiates transcription from the GC box1/TATA box1/Inr1 region. Transcription initiation from the P1 promoter produces the dmyc full length mRNA. Analysis of the splice junctions revealed that the removal of the introns and joining of the exons is regulated by a constitutive splicing mechanism.15,16 The P-element P{lacW} in the dmyc-lacZ enhancer trap fly line, w67c23P{lacW}dmG0354/FM7c,17–19 was shown to be inserted within the TATA box1 sequences. The insertion of the P-element P{lacW} into the endogenous dmyc promoter20 presumably disrupts transcription, engendering the stock with a lethal copy of the gene. This phenomenon is not unusual for random P-element mutagenesis. We have shown that the dmyc intron 2 full fragment and its truncation (missing the 2 kb upstream sequences) are active during development.14 Here, we have dissected the intron 2 region into its cluster of binding sites and the 3′ end fragment containing the DPE element; we show that the dmyc large intron requires these two elements to express reporter lacZ in the larval brain and discs, in embryos, and in adult female ovaries. Our previous work and the study reported here covering upstream and downstream regulatory regions may serve as a foundation for deciphering of the cis-elements and trans-acting factors that bind to these elements and are likely to be important for dmyc regulation.
Materials and Methods
Generation of dmyc cDNA by rapid amplification of cDNA ends (RACE)
RACE Ready cDNA was prepared using a Smarter™ RACE cDNA Amplification Kit (Cat. No. 634924, Clontech Laboratories, Inc.), and following the manufacturer’s user manual (Protocol No. PT4096- 1, Version No. 011312). We used adult Drosophila total poly(A+) RNA (Clontech Laboratories, Inc.) for synthesis of dmyc cDNA. For the generation of positive control cDNA mouse heart total RNA was used. Total RNA (1 μg) was reverse transcribed into cDNA in a reaction volume of 10 μL using SMART-Scribe reverse transcriptase and SMARTer IIA oligo. Polymerase chain reactions (PCR) of the dmyc and the positive control cDNAs were performed using Advantage® 2 Polymerase Mix and the Universal Primer Mix (UPM) (Cat. No. 639201, Clontech Laboratories, Inc.). RACE PCR reactions were performed with UPM forward primer (recognizes the SMARTer IIA sequence at the 5′ end of the cDNA) and the gene specific primers GSP-C05, and GSPC06 reverse primers (primers 17–19 in Supplemental Table 1). Thermal cycler for PCR 1 (see Results Section) was commenced using the following program for touchdown PCR: 5 cycles of initial denaturation at 94°C for 30 seconds, and annealing at 72°C for 3 minutes; 5 cycles of denaturation at 94°C for 30 seconds, annealing at 70°C for 30 seconds, and extension at 72°C for 3 minutes; and 25 cycles of denaturation at 94°C for 30 seconds, annealing at 68°C for 30 seconds, and extension at 72°C for 3 minutes. For the PCR 2 (see Results Section), the stringency of the reactions in the thermal cycler was increased by raising the annealing temperature in increments of 2 °C.
The amplification products were examined by running on 1.5% agarose gels, and subsequent sequencing at Microsynth AG (Balgach, Switzerland). All primers were synthesized at Microsynth. The sequences of the oligonucleotides used for the synthesis and sequencing of the dmyc cDNA ends are indicated in Supplemental Table 1.
Generation of J8.3, J8.4, and J8.5 LacZ reporter strains
The approximately 1 kb insert for the reporter construct J8.4 is derived from the genomic sequences in pC-RP27 and the 2 kb J8.5 promoter fragment from the genomic sequences in pJ8.14 Each insert was subcloned either into pCaSpeR-NLSlacZ and/or into the site specific pattB-temp del LoxP reporter plasmid. The original vector pUAST-attB was engineered to remove the 5 × UAS-hsp70 and LoxP site sequences, followed by self-ligation to obtain the ready to use pattB-temp del LoxP transforming vector. The promoter in the transgene J8.4 contains multiple conserved sequence block regions, just upstream of the DPE element in the dmyc intron 2. The insert in transgene J8.5 is approximately 2 kb in size; it is derived from the 3′ end of the dmyc large intron. For the creation of J8.4 promoter, pC-RP27 served as the template, the amplified fragment was 2601 bp in size, and the amplifying primers had the names In2E-FNot and In2E-R-Asp (primers 9 and 10 in Supplemental Table 1). For the creation of the J8.5 insert, J8 served as the template, the amplified fragment was 2662 bp in size, and the amplifying primers had the names pDPE-F and pDPE-R (primers 11 and 12 in Supplemental Table 1). Polymerase chain reaction conditions in the thermal cycler were one cycle of initial denaturation at 98 °C for 30 seconds; 30 cycles of denaturation at 98 °C for 10 seconds, annealing at 62 °C for 30 seconds for the J8.4 insert, annealing at 57 °C for 30 seconds for the J8.5 insert, and extension at 72 °C for 2 minutes; and one final extension cycle at 72 °C for 5 minutes, held at 4 °C. The upper primer introduced a NotI site at the 5′ end and the lower primer an Acc65I restriction site at the 3′ end of each amplified product. The polymerase chain reaction fragments were each separately digested with NotI/Acc65I and run on 1.5% agarose gel to isolate the 0.93 kb insert of J8.4 and the 2 kb J8.5 promoter. The isolated fragments were each subcloned separately in 5′ NotI-Acc65I 3′ linearized, dephosphorylated (CIAP, Roche Diagnostics, Indianapolis, IN) pCaSpeR4-NLSlacZ and pattB-temp del LoxP vectors in front of reporter lacZ in order to obtain the transgenes J8.4 and J8.5. The promoter fragment of J8.4 was fused inframe to the promoter of J8.5 in the J8.5 transgene to obtain the construct J8.3. Reporters J8.3 to J8.5 terminate transcription by the SV40 poly (A) signal.
All intermediate plasmid constructs, engineered transforming vectors, and transgenes were sequenced by Microsynth (Balgach, Switzerland). Except for the standard primers that were provided by Microsynth, all other oligonucleotides and sequencing primers were designed using the software tool DNASTAR Lasergene 9.1 (module: Primer Select) and synthesized at Microsynth. All oligonucleotides for polymerase chain reactions and sequencing primers are listed in Supplemental Table 1.
Each reporter transgene was transfected into electrocompetent Max DH10B Escherichia coli cells (Invitrogen). Plasmid DNA for injection was isolated using a Qiagen large construct kit. The amplified inserts were fully sequenced and each plasmid was sequenced at the distal and proximal sites of the dmyc promoter sequences before injection. For the reporter studies based on random P-element insertion, embryos from genotype y[1] w[1118] were used; for the studies with the phage ΦC31 integrase transgenesis system, we used embryos from fly lines carrying different attP attachment sites. Both strains were from Bloomington Stock Center (Bloomington, IN). The attP fly stocks used in this study are listed in Supplemental Table 2.
X-Gal reaction assays and immunoblotting
For each construct, 6 to 10 independent transgenic lines were dissected and X-Gal staining was performed at standardized reaction conditions.21 The incubation temperature for different tissues was as follows: discs 29 °C; embryos 37 °C; and ovaries at room temperature. For each construct, a representative disc, brain, embryo, and ovary was chosen for presentation. dpp-lacZ fly stock (gifted by Dragan Gligorov, Karch’s laboratory) was used as a positive control, while y[1] w[1118] fly stock was used as a negative control.
For Western Blot analysis, ovaries taken from the flies carrying the J8 or J8.5 transgene were treated with Cytobuster™ Protein Extraction Reagent (71009-3, Novagen). The extracts were treated with phreon to remove yolk proteins. Proteins extracted were run on SDS-PAGE denaturing system (20 μg/lane) using PowerEase ® 500 Power Supply (invitrogen), for 40 minutes, at a constant voltage of 200 V. Proteins were then transferred to nitrocellulose membrane using iBlot® Western Detection Kit (Life technologies, IB7410-01). The membranes were processed using a WesternBreeze® Chromogenic Kit-Anti- Rabbit (WB7105, Invitrogen). lacZ protein was visualized with ANTI-BETAGALACTOSIDASE, (Molecular Probes, A11132) and Drosophila actin was detected with Anti-Actin monoclonal antibody (Millipore, MAB150IR).
Reverse transcriptase polymerase chain reaction analysis
Total RNA was isolated from the sample brain, disc, and ovary (Fig. 5K, T, U and Z) using an aMResco phenol-free total RNA purification kit (Code N788 kit) with RNase-free DNase treatment (Promega, Basel, Switzerland) following the manufacturer’s protocol. Total RNA (1 μg) was reverse transcribed into cDNA in a reaction volume of 30 μL using SuperScript™ III reverse transcriptase oligo(dT) 20 primers and reverse transcription reagents from Invitrogen (Carlsbad, CA). Semi-quantitative polymerase chain reactions were performed on the resulting cDNA using a high fidelity Phusion DNA polymerase kit (Finnzymes, Bioconcept, Switzerland). The polymerase chain reaction conditions in the thermal cycler were as follows: one cycle of initial denaturation at 98 °C for 30 seconds; 30 cycles denaturation at 98 °C for 20 seconds, annealing at 57.2 °C for 30 seconds, and extension at 72 °C for 2 minutes; and one cycle final extension at 72 °C for 4 minutes, held at 4 °C. The polymerase chain reactions, 5 μL per lane, were run on 1.5% agarose gel for 100 minutes at a voltage of 120 V.
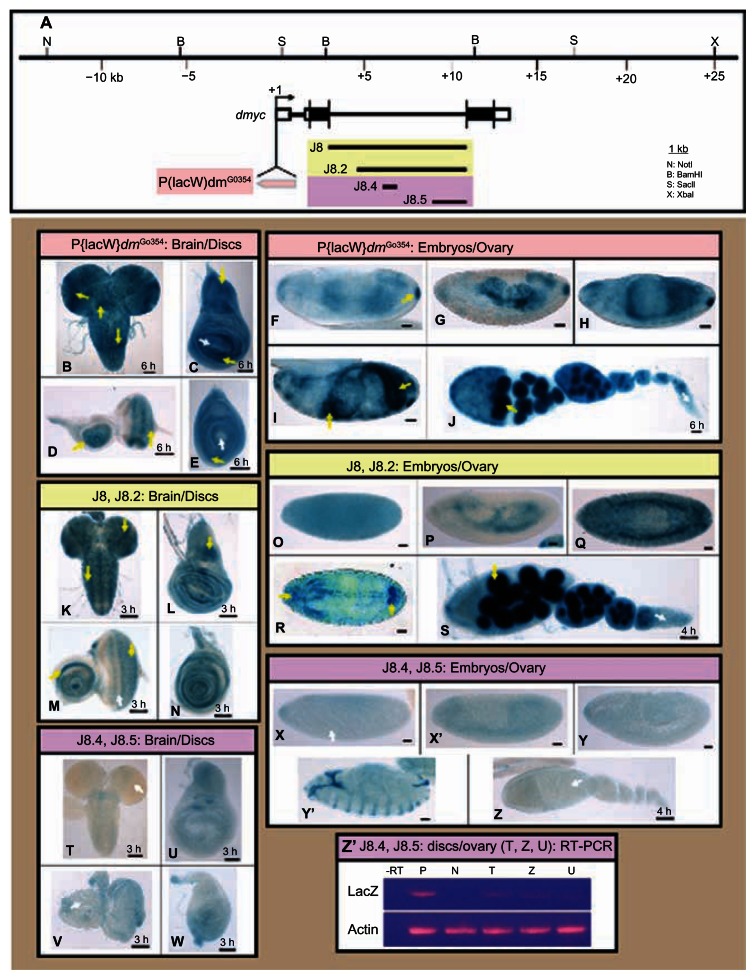
Figure 5.
dmyc downstream promoter element (DPE) and its upstream binding sites cluster are required for the intron 2 activity. (A) The full length intron 2 in the J8 transgene and its truncation, J8.2, the enhancer region in J8.4 and the 3′ truncation J8.5 (containing the DPE element), and their relative location with respect to the dmyc locus and the genomic organization are shown. The insertion site of the wild type dmyc-lacZ P-element in the fly stock w67c23P{lacW}dmG0354/FM7c at the 5′ end of the dmyc gene is depicted. (B–J) In the wild type dmyc-lacZ enhancer trap line, lacZ patterning reflects the pattern reported for the dmyc endogenous mRNA distribution (B, brain; C, wing disc; D, eye disc; E, leg disc). (F–I) In the embryos, lacZ is expressed with high similarities to dmyc mRNA distribution, predominantly in the midgut, hindgut, pharynx, anal pad, and partly in the mesodermal tissue (embryo stages are as follows: F, G, 5–9; H, I, 12–16). The staining in the ovary (J) reflects the dmyc mRNA localization, which has been reported in nurse cells and oocyte, but is weakly expressed at the tip of the germarium. (K–S) The J8 and J8.2 transgenes retain normal patterning of the dmyc in the brain and discs (K–N), throughout different stages of embryogenesis (O–R embryo stages: O, P, 5–9; Q, R, 9–13), and in the ovary (S). (T–Z) J8.4 and the J8.5 transgenes fail to express lacZ in the larval brain and discs (T–W), in the embryos (X–Y′ embryo stages: X, 2–6; X′, Y, 9–12; Y′, 13–16), and in the ovary (Z). (Z′) Reverse transcriptase polymerase chain reaction on J8.4 brain and ovary (shown in T, Z) and J8.5 disc (shown in U) detects no lacZ transcripts beyond background level. Controls used were as follows: -RT, negative control with no transcriptase (brain from K); P, K, positive control; N, y[1] w[1118] leg discs, negative control; actin, Drosophila actin as internal control.
Notes: The yellow arrow indicates lacZ expression and the white arrow indicates lack of lacZ activity. Staining time for discs and ovary is indicated above the scale bar; embryos were stained overnight. Scale bar in (B–Z) indicates 50 μm.
Results
Analysis of the dmyc exon1 cDNA end reveals that the TATA box1 is utilized for the production of the transcript
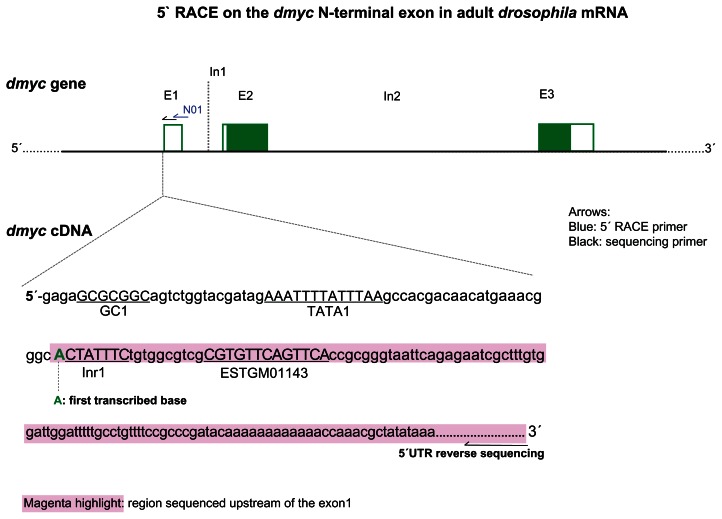
Addition of a poly (A) tail to the 3′ end of an RNA molecule or a mixture of RNA molecules can facilitate studies requiring RNA. One of the positive impacts is to increase the stability of the RNA and enhance its ability for the synthesis of the first strand cDNA.22,23 In addition to poly (A)-tailing, it is well known that the addition of an oligonucleotide to the 5′ end of the mRNA before reverse transcription reaction ensures generation of a full-length cDNA.24 Consequently, the stable and 5′ end extended RNA molecule provides a tool for capturing most of its 5′ end, which helps to identify potential cis-acting regulatory elements and the existence of one or more transcription initiation sites.24 Analysis of the amplified dmyc cDNA (Fig. 1) revealed that transcription of dmyc RNA is most likely to be initiated with an A nucleotide (Fig. 2, Fig. S1). The A residue at the beginning of dmyc cDNA, 24 nucleotides downstream of TATA box1 (Fig. 2), appears to be a potential candidate that precedes the cap structure at the 5′ end of the transcript. Investigation of cap structure in eukaryotic mRNAs has shown that in most RNAs the 7-methyl Guanine is linked through a 5′ to 5′ bond and three phosphate molecules to an A or G residue to be transcribed as first nucleotide.49 The GC box1 (5′-GCGCGGC-3′, SP1 binding site), TATA box1 (5′-AAATTTTATTTAA-3′), and the Initiator 1 element (5′-CTATTTCT-3′) may serve as basal promoter elements in this region (Fig. 2).
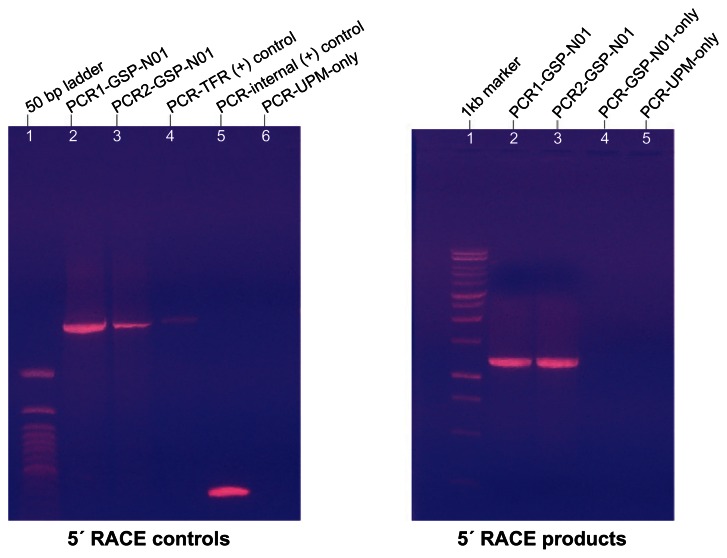
Figure 1.
Extension of the dmyc cDNA at its 5′ end.
Notes: Polymerase chain reaction amplification of the dmyc cDNA 5′ end was performed with the forward primer UPM and the reverse primer GSP-N01; two different settings of annealing and extension temperatures were used (see Materials and Methods). Sequences for the oligonucleotides are listed in Table S1.
Abbreviations: TFR, Transferrin Receptor; UPM, Universal Primer Mix; GSP-N01, Gene Specific Primer N-term.
Figure 2.
5′ RACE analysis of dmyc cDNA at its 5′ site detects TATA box1 as main promoter region. dmyc genomic organization is shown at the top (not drawn to scale).
Notes: Gene specific primer GSP-N01 (blue arrow) was used to amplify the dmyc cDNA 5′ end and the sequencing primer (black arrow) was used to sequence the dmyc cDNA end. (The UPM forward amplification primer and the UPM-sh the forward sequencing primer are not plotted). The 5′ end sequence of the dmyc cDNA is shown at the bottom. The sequenced portion of the cDNA 5′ end is highlighted in magenta and the first transcribed nucleotide is in green. For the sequencing chromatogram, see Figure S1. Sequences for the oligonucleotides are listed in Table S1.
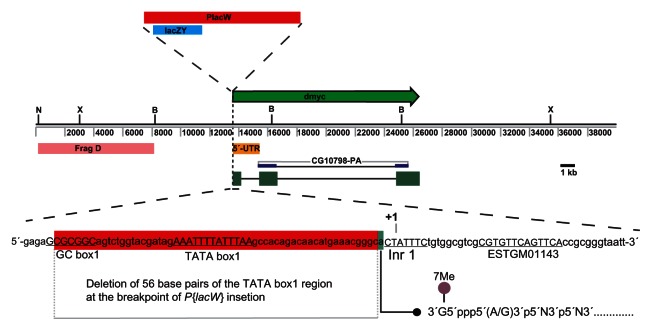
Evidence for the usage of above core promoter elements for the dmyc transcription initiation comes from the fact that the insertion of the P-element P{lacW} in the enhancer trap line w67c23P{lacW} dmG0354 (Fig. 3A) causes the affected allele to be lethal.18,25 The P- element P{lacW} is inserted into the endogenous dmyc promoter20 and presumably disrupts transcription. This phenomenon is not unusual for random P-element mutagenesis. Analysis of the insertion breakpoint revealed that the P-element insertion leads to loss of 56 nucleotides in this region, including TATA box1 and GC box1. These results show that dmyc transcription may be initiated 24 bases downstream of TATA box1, an A residue at this locus being the first base to be transcribed. It further emphasizes that the immediate upstream elements in this region act as basal promoter elements for the assembly of general transcription factors to build a pre-initiation/initiation complex. The exact sequence of each regulatory element and experimental identification of binding factors to these elements remains unresolved.
Figure 3.
Insertion of the P-element P{lacW} at the TATA box1 region of the dmyc gene is lethal.
Notes: Insertion site of the reporter lacZ P-element (10.3 kb in size) at the dmyc locus is shown. Breakpoints of the insertion are as follows: 3D2, X:3267141..3267197, which maps to the region 213 nucleotides upstream of dmyc exon1 start site. Insertion of the P-element P{lacW} causes deletion of 56 base pairs (red highlight), including GC box1 and TATA box1, at the breakpoint of insertion. A residue (green highlight adjacent to the breakpoint) was shown to be the first transcribed base (Fig. 2) that favors capping of the dmyc mRNA.
Identification of splicing signals in the dmyc transcript
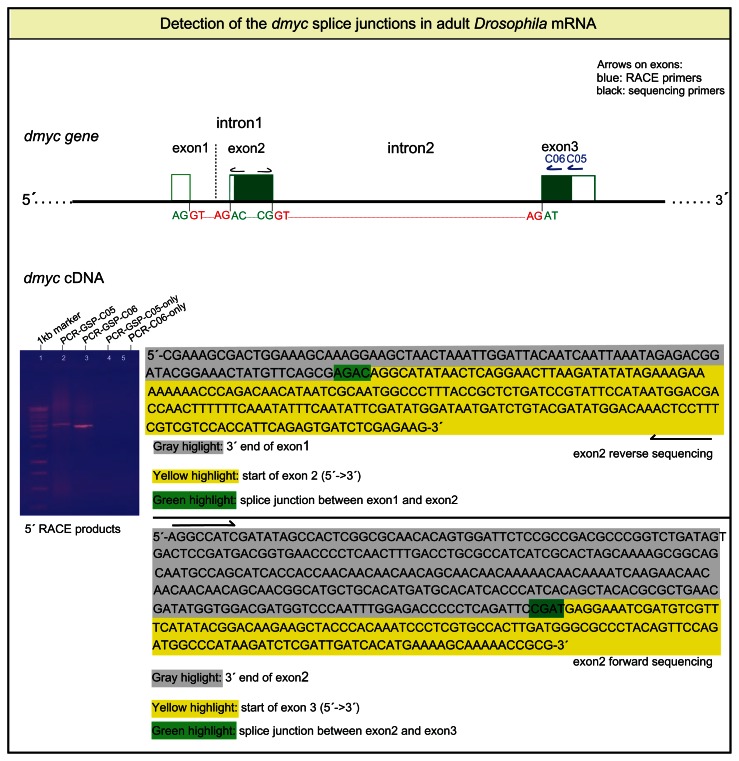
In eukaryotic pre-mRNA molecules, the noncoding intronic sequences are removed through different splice mechanisms and the protein-coding regions are ligated.26,27 The major recognition signals required for splicing involve the cis-regulatory elements 5′ and 3′ splice sites, branchpoint sequence, and the polypyrimidine tract.28,29 Interaction of specific factors and RNAs with these cis-elements stepwise catalyze the formation of the spliceosome that, via two transesterification reactions, removes the intervening sequences and ligates the exons.30,31 We first sought to perform 5′ RACE analysis with two gene specific primers (GSP-C05 and GSP-C06) that hybridize to the C-terminus exon and extend the dmyc mRNA from its 3′ end towards 5′ site (Fig. 4). Sequence analyses of intron ends confirmed that the first two nucleotides are GT, whereas the last two bases are AG (Fig. 4). We hypothesized that like many eukaryotic mRNAs, the GU/AG nucleotide pairs should not appear in mature mRNA. Indeed, after RACE analysis of the different lengths of the dmyc cDNA ends, we did not observe the GU/AG nucleotides in the mRNA; instead we observed the ligation of the 3′ end of each upstream exon to the 5′ end of the following exon in the splice junctions (Fig. 4, Fig. S2). From this result we conclude that the splicing of the dmyc largest transcript is constitutively regulated by ligating each upstream exon to its neighboring downstream exon. Furthermore, we showed that all three exons were detected in the transcript. This observation suggests that the exon1, despite its noncoding characteristic, might contain important translational signals for the synthesis of the dmyc protein. It would be of great interest to characterize putative “weak cis-splicing elements” such as exon splicing enhancers and silencers,32 which may play a role in the dmyc RNA splicing. In adult Drosophila RNA mixture we could not detect a smaller transcript that could emerge through alternative splicing or transcription initiation from a downstream promoter. Investigations for the existence of shorter transcript variants in early developmental stages remain unresolved.
Figure 4.
5′ RACE analysis of the dmyc cDNA ends detects splice junctions of the transcript.
Notes:dmyc genomic organization (not drawn to scale) with the nucleotides at the exon/intron junctions are shown in green and red. Gel picture shows polymerase chain reaction amplification products of the dmyc cDNA with the gene specific reverse primers GSP-C05 and GSP-C06. For all of the amplification reactions, the Universal Primer Mix (UPM, Clontech) was used as forward primer.
dmyc large intron requires Downstream Promoter Element (DPE) and the binding sites cluster for its developmental patterning
In the early stages of animal development, dmyc expression is highly dynamic and is required to ensure both cell growth and size, but also cell proliferation at the right time and at the right place.8,9 Expression pattern of the lacZ reporter in the dmyc-lacZ enhancer trap line w67c23P{lacW}dmG0354/FM7c (Fig. 5A) has been shown to be comparable to the endogenous patterning of dmyc.14,18,19,33 In the third instar larval brain, dmyc is active in the lobes, ventral ganglion, and a limited number of cells in both halves of hemispheres and ventral ganglion (Fig. 5B). In the wing disc, lacZ activity, representing dmyc mRNA, is mainly detected around the wing pouch and in the notum, in the eye imaginal tissue, in proliferative zones anterior and posterior to morphogenetic furrow, around the center of antennal disc, and in the leg disc around the center of the disc (Fig. 5C–E). In the early embryo, maternal transcripts are detectable; later, during germ band extension, dmyc intensifies in the mesoderm, mid-gut, pharynx, and anal pad (Fig. 5F–I). In the ovary, dmyc activity is predominantly detected in the nurse cells and oocyte (Fig. 5J).
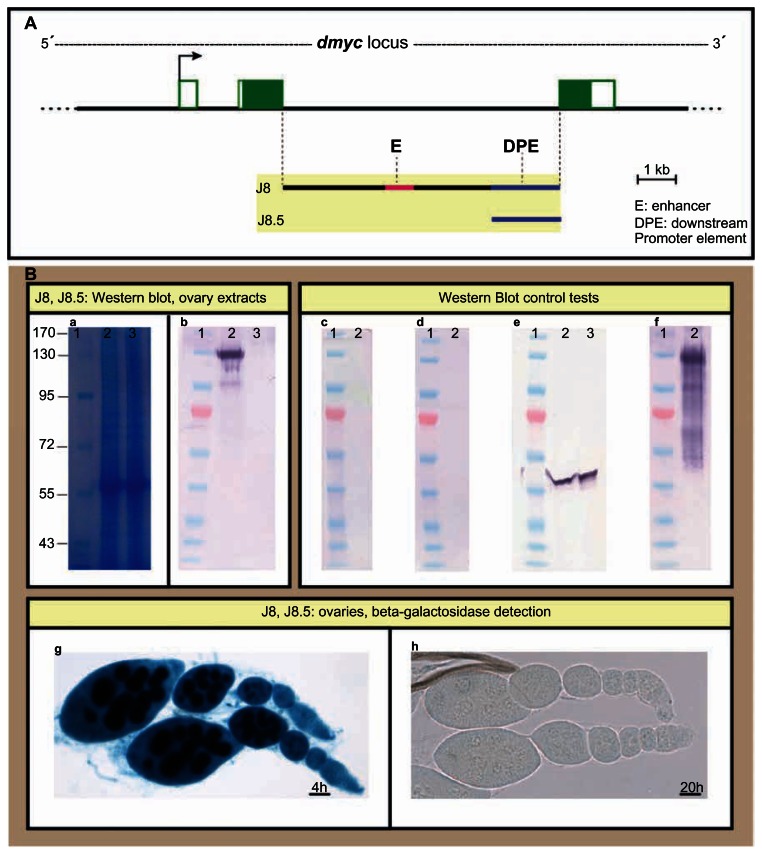
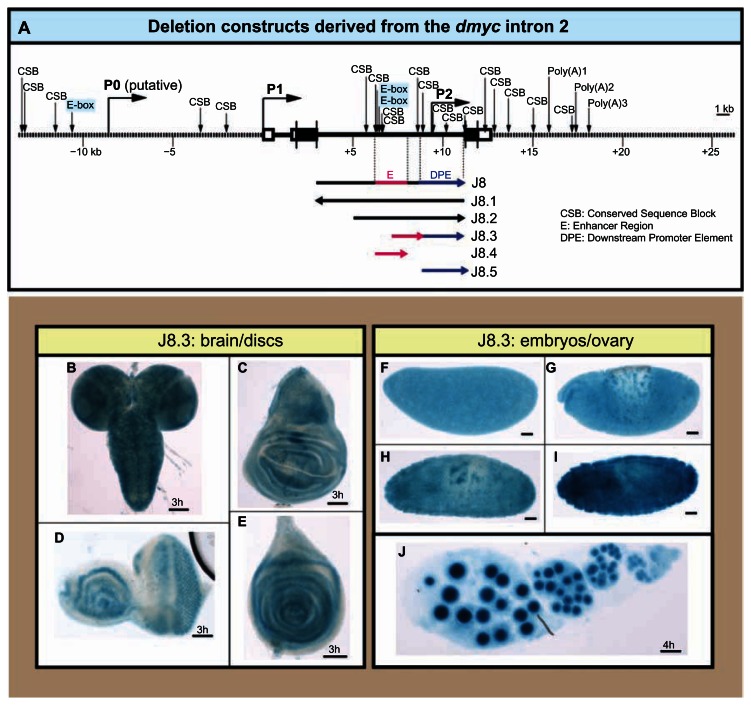
In a previous analysis we have shown that the dmyc large intron can drive expression of a lacZ reporter during the early stages of development,14 as is the case for most developmentally regulated introns in the Drosophila transcriptome.34–36 We have examined the full length large intron and its deletion construct that contains the 6 kb downstream sequences (J8 and J8.2 in Fig. 5A); it was shown that J8 and J8.2 transgenes express lacZ similar to the endogenous dmyc patterning in larval brain and imaginal discs (Fig. 5K–N), in embryos and ovaries (Fig. 5O–S). To gain more insight into the dmyc regulation from this downstream regulatory region, we dissected J8.2 truncation into its cluster of binding sites region and the downstream sequences, including the DPE element (J8.4 and J8.5 transgenes in Fig. 5A), and tested each separately during Drosophila development. In β-gal assays for lacZ expression, under control of each of these elements, no activity was detectable beyond basal levels in larval imaginal tissues, embryos, and adult Drosophila ovaries ((Fig. 5T–Z). Reverse transcription polymerase chain reaction on the discs and ovary shown in Figure 5T, U, and Z did not detect any lacZ transcripts beyond background level in these tissues that were negative for lacZ staining (Fig. 5Z′). We further isolated protein extracts from the ovarian tissues of transgenic animals carrying either the largest construct J8 or the 3′ end deletion, J8.5 transgene (Fig. 6A), and tested them for the existence of lacZ protein (Fig. 6B). As expected, the immunoblotting assay was positive for the full length J8 fragment, but was negative for the J8.5 extracts (Fig. 6B, b). This result is consistent with the β-gal assays for the detection of lacZ protein in the ovaries (Fig. 6B, g, h) that is representative of dmyc mRNA distribution. To confirm these findings, we combined the promoters of the J8.4/J8.5 transgenes in the truncation J8.3 to exclude any other regions within the J8/J8.2 (Fig. 7A). The transgene J8.3 recapitulates the intron 2 activity (Fig. 7B–J), suggesting that the enhancer region with the predicted E-boxes and conserved sequence blocks14 (potential binding sites, Fig. S3) and the downstream promoter element are required for the activity from this region. Investigation of the transcription start site from the regulatory unit in this region remains to be done.
Figure 6.
Detection of lacZ protein by Western Blot analysis of ovary extracts isolated from flies carrying J8 or J8.5 transgenes. (A) The intron 2 full length in the J8 transgene and its truncation, J8.5, and their relative location with respect to the dmyc locus and the genomic organization are shown. (B) Proteins extracted from the ovaries of the female flies carrying J8 or J8.5 transgenes were analyzed by SDS-PAGE and immunoblotting; lacZ protein was detectable in the J8 ovaries (panel b, lane 2), whereas J8.5 extract was negative (panel b, lane 3).
Notes: (a): Gel SafeBlue staining of the protein extracts; 1: protein marker in kDa unit, 2: J8 lysate, 3: J8.5 lysate. (b): The extracts in (a) subjected to immunoblotting with antibody anti-beta Gal. (c): J8 extracts with no primary antibody added, (d): J8 extracts with no secondary antibody added, (e): The extracts in (a) subjected to immunoblotting with antibody anti-Drosophila actin. (f): positive Control: beta-Gal protein. (g): lacZ staining of J8 ovaries and (h): lacZ staining of J8.5 ovaries. The staining time for ovaries is indicated above the scale bar. Scale bar in (g, h) is 50 μm.
Figure 7.
Combined action of the downstream promoter element and its upstream binding sites are required for the intron 2 activity. (A) The 40 kb dmyc locus with the predicted regulatory elements14 and the deletion constructs derived from the intron 2 region are shown. (B–J) Fusion of the enhancer region (J8.4) to the downstream promoter element (J8.5) in J8.3 construct recapitulates the J8/J8.2 transgenes activity in the larval brain and discs (B–E), during embryogenesis (F–I embryo stages: F, 2–5; G, 9–12; H, I, 12–15), and in the ovary (J).
Notes: Staining time for discs and the ovary is indicated above the scale bar; embryos were stained overnight. Scale bar in (B–J) indicates 50 μm.
Discussion
Tight regulation of c-myc transcription is essential for its appropriate function during animal development37 as deregulation of c-myc is associated with malignant transformation in humans.7,38,39myc is linked to major processes in cell physiological behavior such as proliferation, growth, differentiation, apoptosis, and metabolism.9,40,41 In turn, the involved pathways signal back to fine tune the controlled expression of myc.40,42 A large body of evidence suggests that one mechanism for c-myc promoter regulation involves the presence of multiple transcription initiation sites in the noncoding regulatory region of the gene.10 We suggested that the sole Drosophila homolog of the c-myc gene, dmyc, initiates its transcription from multiple start sites.14 In the study reported here, we have investigated the initiation site of the P1 promoter in the 5′ UTR region, the splice signals of the transcript, and have dissected the intronic promoter to gain more insight into the dmyc regulation that is responsible for its patterned expression in development. First, using a gene specific primer that hybridizes to dmyc exon1, we performed 5′ RACE experiments to synthesize the 5′ portion of the dmyc cDNA that subsequently served as template to amplify the fragment. Our results indicate that only one initiation site is detectable in this region, and the core promoter consists of GC box1, TATA box1, and Inr1. This observation confirms our former computational prediction of the above regulatory elements in the 5′ UTR.14 Examination of a dmyc haplo-lethal fly stock (w67c23P{lacW}dmG0354/FM7c; Bloomington Stock Number 11981) showed that the insertion point of the P- element P{lacW}20 into the P1 promoter is downstream of the TATA box1 elements and it has broken away 56 bp of the TATA region, including the two elements GC box1 and the TATA box1. The insertion of the P- element and loss of TATA sequences presumably disrupts transcription since these elements are crucial for the assembly of pre-initiation complex (PIC)43,44 and subsequent recruitment of RNA Pol II to the PIC. Consequently, the affected copy of the dmyc gene has become nonfunctional. Furthermore, the finding is consistent with the fact that most eukaryotic promoters consist of a TFIIB Recognition Element (BRE, immediately upstream of TATA box), a TATA box, and an initiator element that is located 25 to 30 bp downstream of the TATA box.45 It has been shown that the largest and second-largest subunits of RNAP II interact with approximately 50 bp DNA sequences in the promoter region, including sequences upstream and downstream of the transcription start.46 The number and the relative distance of the core promoter elements appear to be appropriate to confer dmyc—as immediate early gene—with strength for the action of its promoter. This work provides a starting point for investigating the exact sequences of each element and the assembly of general transcription machinery.
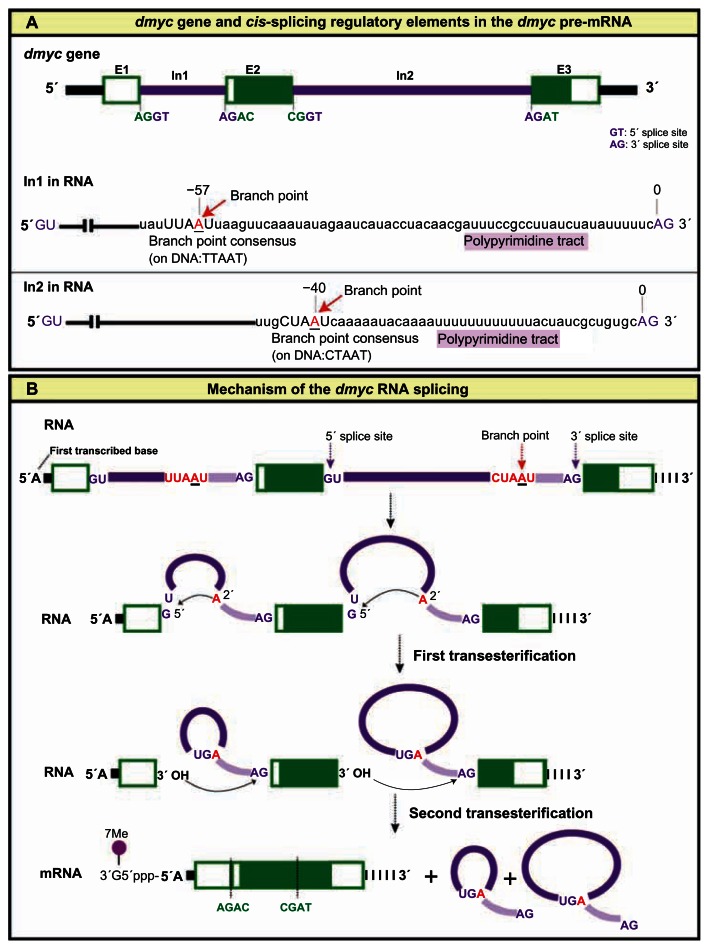
In higher eukaryotes, the removal of introns from pre-mRNA molecules is mediated through a complex process that involves many factors. The splice cis-elements participate in two trans-esterification reactions performed by a complex structure known as spliceosome; these reactions have the effect of removing the introns and joining the exons together.29,47,48 Indeed, analysis of the dmyc full length cDNA for major splice signals in the transcript revealed that, like virtually all eukaryotic introns, dmyc introns include 5′ and 3′ end splice signals, a branch point consensus sequence, and a polypyrimidne tract located adjacent to the AG sequence within each intron (Fig. 8A). The branchpoint consensus sequence in Drosophila resembles 5′ CTAAT 3′ and occasionally 5′ TTAAT 3′; in both cases, branch formation occurs at the underlined A, the branchpoint.28 The consensus sequence is present in intron 2, whereas intron 1 contains the less conserved sequence (Fig. 8A). Sequencing of the amplified cDNA ends showed that the dmyc introns are spliced out at the 5′-GU/3′-AG sites by a constitutively regulated splicing mechanism, which includes the accomplishment of two trans- esterification reactions. Consequently, exon1 is ligated to exon2, and exon2 to exon3, so that the full length mRNA contains all three exons. Indeed, the dmyc protein is 717 amino acids, while c-myc is about 439 amino acids; both proteins contain domains that are required for myc function.13 In our current study, we have shown the splice mechanism for the full length dmyc transcript by considering main splice cis-elements. However, we cannot rule out the possibility that alternative splicing during early stages of development may produce shorter transcripts. The knowledge obtained here can serve as a base for studying the detailed mechanism of splicing, including the identification of splice trans-factors and putative weak splice cis-factors such as exon splice enhancers and exon splice silencers.
Figure 8.
Major splice cis-elements and mechanism of the dmyc RNA splicing. (A) dmyc genomic organization (not drawn to scale) with the nucleotides at the 5′ splice donor and 3′ splice acceptor sites (GT/AG), are shown on top. Splicing cis-elements of the dmyc RNA are shown below and include the following: 5′ and 3′ splice signals (GU/AG), branchpoint consensus sequence, and a polypyrimidine tract located adjacent to the AG sequence at the 3′ end of each intron. (B) dmyc RNA splicing is regulated constitutively, following the strict GU/AG splicing rules and includes the ligation of each upstream exon to its adjacent downstream exon. The A residue (branchpoint) within the branchpoint consensus sequence participates in the formation of the ester bond between the 5′ phosphate of the intron and the 2′ oxygen of the branchpoint A residue.
Notes: The second transesterification reaction between the free 3′ oxygen of the upstream exon and the 5′ phosphate of the downstream exon initiates the joining of the exons together and the excision of the intron as a lariat. The exon/exon splice junctions in the mRNA molecule are indicated at the bottom.
We had previously indicated that the full length intron 2 and its truncation, missing the 2 kb upstream sequences, are still active in dmyc patterning. However, the region of multiple conserved sequence blocks and E-boxes, or the 2 kb sequences at the 3′ end of the intron 2 which contains the downstream promoter, each separately abrogates the intronic activity in virtually all the examined tissues. In light of this finding, analysis of RNA extracts taken from Drosophila ovary or embryonic tissues for the identification of a shorter form of dmyc transcript, will be of great interest. The knowledge gained here, combined with further molecular genetic approaches, will serve for further understanding of the tight regulation of dmyc at transcriptional level.
Acknowledgements
We thank F. Karch, R. Maeda and D. Bopp, for their critical comments and discussions, and O. Georgiev, for recommending the Clontech Race Kit. We thank L. M. Quinn and T. Brody for the comments on the revision of the manuscript. The fly strains were received from Bloomington Stock Center.
Footnotes
Authors Contributions
Conceived and designed the experiments: JK. Analyzed the data: JK, CM. Wrote first draft of the manuscript: JK. Agree with manuscript results and conclusions: JK, CM. Made critical revisions and approved final version: JK, CM. Both authors reviewed and approved the final manuscript.
Competing Interests
Author(s) disclose no potential conflicts of interest.
Disclosures and Ethics
As a requirement of publication the authors have provided signed confirmation of their compliance with ethical and legal obligations including but not limited to compliance with ICMJE authorship and competing interests guidelines, that the article is neither under consideration for publication nor published elsewhere, of their compliance with legal and ethical guidelines concerning human and animal research participants (if applicable), and that permission has been obtained for reproduction of any copyrighted material. This article was subject to blind, independent, expert peer review. The reviewers reported no competing interests.
Funding
This work was partially supported by Lica Pharmaceuticals A/S, Copenhagen, Denmark.
Supplementary materials
Sanger sequencing of the PCR products derived from rapid amplification of the dmyc cDNA at its 5′ end. Notes: Chromatogram on the top: Sequencing of the amplification product with the GSP-N01, hybridizing to the 5′ end of exon1 (see Fig. 2). The forward sequencing primer is UPM-sh. Except an A residue upstream of the Inr1, no further exact base pair matching to the dmyc genomic template strand could be sequenced (red highlight on the chromatogram). The extended region with mismatch is due to the ligation of an oligonucleotide to the 5′ end of the RACE cDNA to ensure capturing of the transcript’s start site. Chromatogram at the bottom: The GSP-N01 amplification product was sequenced with the reverse sequencing primers Exn1-R2 and Exn1-R3 to obtain the readout of the sequences downstream of the known ESTGM01143. The PCR products were sequenced at Microsynth AG, Balgach, Switzerland.
5′ RACE analysis of the dmyc cDNA ends determines splice junctions of the largest transcript.
Notes: Chromatograms show sanger sequencing results after analysis of the 5′ RACE products that were amplified with the gene specific primers GSP-C05 and GSP-C06 (see Fig. 4). GSP-C05 and GSP-C06 hybridize to the dmyc exon3 and extend the full length cDNA from its 3′ towards the 5′ end. The PCR products were sequenced with the reverse primer Exn2-R2 and the forward primer Exn2-F1 (see Fig. 4). The PCR products were sequenced at Microsynth AG, Balgach, Switzerland. Splice junctions between exon1/exon2 and exon2/exon3 are indicated in green.
EvoDifference analysis of the top strand D. melanogaster dmyc intron 2 region.
Notes: Each block of uppercase bases represents a cluster of conserved sequences. Uppercase black nucleotides represent bases in the D. melanogaster dm EvoP intron 2 reference sequence that are conserved in all or all but one of the other 5 orthologous DNAs, D. sechellia, D. yakuba, D. erecta, D. willistoni or D. virilis. The conserved bHLH binding sites and E-boxes are in white text with black highlight, as is the repeat motif ATGTTGCCA and the nearby sub-repeat TGTTGC. The cluster CGCGTGGGAAAA, a dead-ringer for the HLHm-3-2 enhancer, is in white text with dark blue highlight. The consensus binding site GTGGGAA for Su(H) (Suppressor of Hairy) is in the cluster. The DPE in the intron 2 region is lower case bold and underlined. The 5′ and the 3′ end of the transgene J8.4 is with yellow highlighted and the 5′ and the 3′ end of the transgene J8.5 is with light blue highlighted. Sequence orientation is 5′ → 3′ (see also Kharazmi et al. 2011, GRSB).
Positive and negative controls for lacZ staining. For each lacZ staining of transgenes negative and positive control staining was performed. As negative control different tissues were taken from the “y w” and “attp-flies” listed in Table 2. dpp-lacZ flies served as source for positive control. Depicted are negative controls (A–M) and positive controls (N–T), (A, B, N, brain; C–H, O–Q, discs; I–L, R, S, embryos; and M, T, ovaries).
Notes: Staining times for the discs and ovaries are indicated above the scale bars. Embryo staining took place overnight. Scale bar in (A–T) indicates 50 μm.
Table S1.
Analytical primer pairs.
| No. | Name | Primer Sequence (5′→3′) | bp | Usage |
|---|---|---|---|---|
| 1 | BAC-F | CTT CGC GTC CAA CAG ATG | 18 | pC-RP27 sequencing |
| 2 | BAC-R | GCG CCA AAG CAA GAG GGA ATG | 21 | |
| 3 | pZpA-F | CCA AAA AAG GGT TTC ATT AAC TTG TAC ACA TAC | 33 | pLacZ-SV40 p(A) sequencing |
| 4 | pZpA-R | TTG GGG ATT TTA ATA GCG GGC CCT GTG TGT | 30 | |
| 5 | SV40 | GTT CAG GGG GAG GTG TGG G | 19 | SV40 P(A) sequencing |
| 6 | pZR | CGG GCC TCT TCG CTA TTA CG | 20 | pJ8 reverse sequencing |
| 7 | pcaF | GAC GGC GAT ATT TCT GTG GAC | 21 | pCaSpeR4 and |
| 8 | pcaR | CCT TAG CAT GTC CGT GGG GTT TGA | 24 | pJ8.4 sequencing |
| 9 | In2E-F-Not |
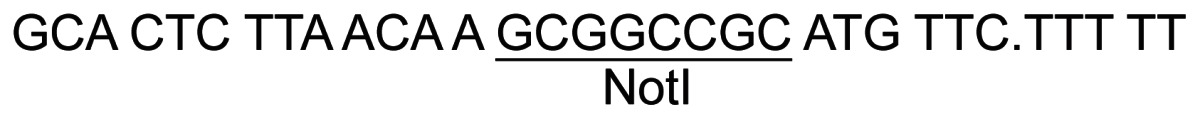
|
32 34 |
PCR – pJ8.4 |
| 10 | In2E-R-Asp |
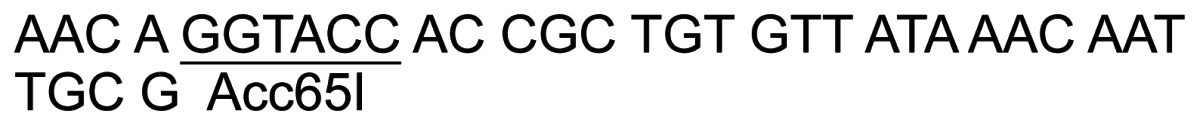
|
||
| 11 | pDPE-F |
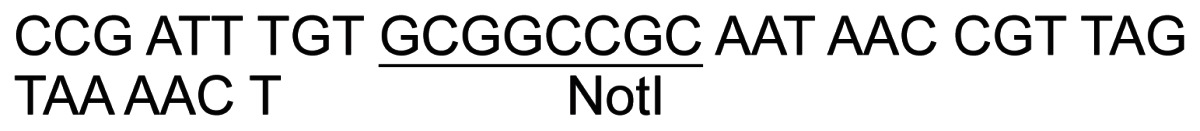
|
36 | PCR – pJ8.5 |
| 12 | pDPE-R | ATG CGC TCA GGT CAA ATT CAG ACG | 24 | |
| 13 | Intr2-R 01 | CCT CAT CTG CAC AGC GAT AGT A | 22 | pJ8.5 sequencing |
| 14 | Intr2-R 02 | TCA TCT GCA CAG CGA TAG TAA | 21 | |
| 15 | Intr2-R 03 | CTG CAC AGC GAT AGT AAA | 18 | |
| 16 | Intr2-R 04 | GGG GAA GGG GGA GGT GGA GAA GA | 23 | |
| 17 | GSP-N01 | GCC GGG CGG TAT TAA ATG GAC CTC GTC CTG | 30 | 5′ RACE cDNA synthesis |
| 18 | GSP-C05 | CTA GCC CTA CGC CGC CGC TTT AAG CCG ATA GTA TGA GAC | 39 | 5′ RACE cDNA synthesis |
| 19 | GSP-C06 | CAC TGC GCC GCT GCC AAT TAC GAT CCA TG | 29 | |
| 20 | UPM-sh | CTA ATA CGA CTC ACT ATA GGG C | 22 | 5′ UTR RACE cDNA sequencing |
| 21 | Exn1-R2 | AAA CAG GCA AAA ATC CAA TCC ACA | 24 | 5′ UTR RACE cDNA sequencing |
| 22 | Exn1-R3 | GGT TAT TTC TGG GAT TTA GCA CTT | 24 | |
| 23 | Exn2-F | ACC GGC CAG CAG CAG TCC A | 19 | 5′ RACE cDNA sequencing |
| 24 | Exn2-R1 | GAA TGG TGG ACG ACG AAA GGA GTT | 24 | |
| 25 | Exn2-R2 | CTC TGG CTT CAT ATC CTC CTC TAA | 24 |
Notes: The oligonucleotides were designed with Bioinformatics tool Laser Gene 9.1 module Primer Select and synthesized at Microsynth, Switzerland. As indicated the primer sets were used for sequencing and/or amplification of dmyc non-coding regions, all 5′ RACE experiments, and for sequencing of cloning vectors and injection plasmids.
Abbreviations: bp, base pairs; E, exon; F, forward; PCR, polymerase chain reaction; R, reverse; RACE, rapid amplification of cDNA ends; Exn, exon; Intr, intron; DPE, downstream promoter element; UPM-sh, Universal Primer Short.
Table S2.
List of the Fly Stocks used in the study.
| No. | Bloomington Stock # | Name | Genotype with FlyBase links |
|---|---|---|---|
| 1 | 6598 | y w | y1 w1118 |
| 2 | 23648 | attp-86F | P{hsp70-flp}1, y1 w*; M{3xP3-RFP.attP}ZH-86Fb; M{vas-int.B}ZH-102D |
| 3 | 24480 | attp-2A | y1 M{3xP3-RFP.attP}ZH-2A w*; M{vas-int.Dm}ZH-102D |
| 4 | 24485 | attp-68E | y1 M{vas-int.Dm}ZH-2A w*; M{3xP3-RFP.attP’}ZH-68E |
| 5 | 8622 | attp-8622 | y1 w67c23; P{CaryP}attP2 |
| 6 | 25709 | attp-25709 | y1 v1 P{nos-phiC31int.NLS}X; P{CaryP}attP40 |
| 7 | 25710 | attp-25710 | y1 sc1 v1 P{nos-phiC31int.NLS}X; P{CaryP}attP2 |
| 8 | 24872 | attp-24872 | y1 M{vas-int.Dm}ZH-2A w*; PBac{y+-attP-3B}VK00037 |
| 9 | 11981 | dmyc-lacZ | w67c23 P{lacW}dmG0354/FM7c |
| 10 | 2475 | double balancer | w*; T(2;3)apXa, apXa/CyO; TM3, Sb1 |
| 11 | 11108 | blue balancer | Cyo, P{lArB}A66.2F2/b1 Adh* cn* l(2)**; ry506 |
| 12 | 8412 | dpp-lacZ | y1 w1118; P{dpp-lacZ.Exel.2}3 |
Notes: For random P-element mediated transgene introduction embryos were taken from the y[1] w[1118] flies (Bloomington Stock number, 6598), and for phi-C31 transgenesis embryos taken from different attp lines were used. The fly stock “blue balancer” that expresses lacZ in embryos and ovaries, was used as a source for positive control in the embryos and ovaries staining. The dpp-lacZ line was used as positive control for the imaginal discs staining.
References
- 1.King MW, Roberts JM, Eisenman RN. Expression of the c-myc proto-oncogene during development of Xenopus laevis. Mol Cell Biol. 1986;6:4499–508. doi: 10.1128/mcb.6.12.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nishikura K. Expression of c-myc proto-oncogene during the early development of Xenopus laevis. Oncogene Res. 1987;1:179–91. [PubMed] [Google Scholar]
- 3.Gallant P, Shiio Y, Cheng PF, Parkhurst SM, Eisenman RN. Myc and Max homologs in Drosophila. Science. 1996;274:1523–7. doi: 10.1126/science.274.5292.1523. [DOI] [PubMed] [Google Scholar]
- 4.Chin L, et al. Contrasting roles for Myc and Mad proteins in cellular growth and differentiation. Proc Natl Acad Sci U S A. 1995;92:8488–92. doi: 10.1073/pnas.92.18.8488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sodir NM, Evan GI. Nursing some sense out of Myc. J Biol. 2009;8:77. doi: 10.1186/jbiol181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–99. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 7.Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–45. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 8.Gomez-Casares MT, et al. MYC antagonizes the differentiation induced by imatinib in chronic myeloid leukemia cells through downregulation of p27(KIP1) Oncogene. 2012 doi: 10.1038/onc.2012.246. [DOI] [PubMed] [Google Scholar]
- 9.Duman-Scheel M, Johnston LA, Du W. Repression of dMyc expression by Wingless promotes Rbf-induced G1 arrest in the presumptive Drosophila wing margin. Proc Natl Acad Sci U S A. 2004;101:3857–62. doi: 10.1073/pnas.0400526101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wierstra I, Alves J. The c-myc promoter: still MysterY and challenge. Adv Cancer Res. 2008;99:113–333. doi: 10.1016/S0065-230X(07)99004-1. [DOI] [PubMed] [Google Scholar]
- 11.Yang W, et al. PKM2 Phosphorylates Histone H3 and Promotes Gene Transcription and Tumorigenesis. Cell. 2012;150:685–96. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trumpp A, et al. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature. 2001;414:768–73. doi: 10.1038/414768a. [DOI] [PubMed] [Google Scholar]
- 13.Benassayag C, et al. Human c-Myc isoforms differentially regulate cell growth and apoptosis in Drosophila melanogaster. Mol Cell Biol. 2005;25:9897–909. doi: 10.1128/MCB.25.22.9897-9909.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kharazmi J, Moshfegh C, Brody T. Identification of cis-Regulatory Elements in the dmyc Gene of Drosophila Melanogaster. Gene Regulation and Systems Biology. 2011;6:15. doi: 10.4137/GRSB.S8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–13. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoue K, Ohno M, Shimura Y. Aspects of splice site selection in constitutive and alternative pre-mRNA splicing. Gene Expr. 1995;4:177–82. [PMC free article] [PubMed] [Google Scholar]
- 17.Cranna N, Quinn L. Impact of steroid hormone signals on Drosophila cell cycle during development. Cell Div. 2009;4:3. doi: 10.1186/1747-1028-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitchell NC, et al. Hfp inhibits Drosophila myc transcription and cell growth in a TFIIH/Hay-dependent manner. Development. 2010;137:2875–84. doi: 10.1242/dev.049585. [DOI] [PubMed] [Google Scholar]
- 19.Cranna NJ, Mitchell NC, Hannan RD, Quinn LM. Hfp the Drosophila homolog of the mammalian c-myc transcriptional-repressor and tumour suppressor FIR, inhibits dmyc transcription and cell growth. Fly (Austin) 2011;5:129–33. doi: 10.4161/fly.5.2.14482. [DOI] [PubMed] [Google Scholar]
- 20.Bourbon HM, et al. A P-insertion screen identifying novel X-linked essential genes in Drosophila. Mech Dev. 2002;110:71–83. doi: 10.1016/s0925-4773(01)00566-4. [DOI] [PubMed] [Google Scholar]
- 21.Song H, Hasson P, Paroush Z, Courey AJ. Groucho oligomerization is required for repression in vivo. Mol Cell Biol. 2004;24:4341–50. doi: 10.1128/MCB.24.10.4341-4350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drummond DR, McCrae MA, Colman A. Stability and movement of mRNAs and their encoded proteins in Xenopus oocytes. J Cell Biol. 1985;100:1148–56. doi: 10.1083/jcb.100.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krug MS, Berger SL. First-strand cDNA synthesis primed with oligo(dT) Methods Enzymol. 1987;152:316–25. doi: 10.1016/0076-6879(87)52036-5. [DOI] [PubMed] [Google Scholar]
- 24.Zhu YY, Machleder EM, Chenchik A, Li R, Siebert PD. Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. Biotechniques. 2001;30:892–7. doi: 10.2144/01304pf02. [DOI] [PubMed] [Google Scholar]
- 25.McQuilton P, St Pierre SE, Thurmond J. FlyBase 101—the basics of navigating FlyBase. Nucleic Acids Res. 2012;40:D706–14. doi: 10.1093/nar/gkr1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dogan RI, Getoor L, Wilbur WJ, Mount SM. Features generated for computational splice-site prediction correspond to functional elements. BMC Bioinformatics. 2007;8:410. doi: 10.1186/1471-2105-8-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qi L, Haurwitz RE, Shao W, Doudna JA, Arkin AP. RNA processing enables predictable programming of gene expression. Nat Biotechnol. 2012 doi: 10.1038/nbt.2355. [DOI] [PubMed] [Google Scholar]
- 28.Mount SM, et al. Splicing signals in Drosophila: intron size, information content, and consensus sequences. Nucleic Acids Res. 1992;20:4255–62. doi: 10.1093/nar/20.16.4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao K, Masuda A, Matsuura T, Ohno K. Human branch point consensus sequence is yUnAy. Nucleic Acids Res. 2008;36:2257–67. doi: 10.1093/nar/gkn073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2006;3 doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wachtel C, Manley JL. Splicing of mRNA precursors: the role of RNAs and proteins in catalysis. Mol Biosyst. 2009;5:311–6. doi: 10.1039/b820828j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pozzoli U, Sironi M. Silencers regulate both constitutive and alternative splicing events in mammals. Cell Mol Life Sci. 2005;62:1579–604. doi: 10.1007/s00018-005-5030-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siddall NA, Lin JI, Hime GR, Quinn LM. Myc—what we have learned from flies. Curr Drug Targets. 2009;10:590–601. doi: 10.2174/138945009788680400. [DOI] [PubMed] [Google Scholar]
- 34.Daines B, et al. The Drosophila melanogaster transcriptome by paired-end RNA sequencing. Genome Res. 2011;21:315–24. doi: 10.1101/gr.107854.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy S, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–97. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gasch A, Hinz U, Renkawitz-Pohl R. Intron and upstream sequences regulate expression of the Drosophila beta 3-tubulin gene in the visceral and somatic musculature, respectively. Proc Natl Acad Sci U S A. 1989;86:3215–8. doi: 10.1073/pnas.86.9.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J, Levens D. Making myc. Curr Top Microbiol Immunol. 2006;302:1–32. doi: 10.1007/3-540-32952-8_1. [DOI] [PubMed] [Google Scholar]
- 38.Lutz W, Leon J, Eilers M. Contributions of Myc to tumorigenesis. Biochim Biophys Acta. 2002;1602:61–71. doi: 10.1016/s0304-419x(02)00036-7. [DOI] [PubMed] [Google Scholar]
- 39.Soucek L, Evan G. Myc-Is this the oncogene from Hell? Cancer Cell. 2002;1:406–8. doi: 10.1016/s1535-6108(02)00077-6. [DOI] [PubMed] [Google Scholar]
- 40.Johnston LA, Prober DA, Edgar BA, Eisenman RN, Gallant P. Drosophila myc regulates cellular growth during development. Cell. 1999;98:779–90. doi: 10.1016/s0092-8674(00)81512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010;10:301–9. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- 42.Herranz H, Perez L, Martin FA, Milan MA. Wingless and Notch double-repression mechanism regulates G1-S transition in the Drosophila wing. EMBO J. 2008;27:1633–45. doi: 10.1038/emboj.2008.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kornberg RD. The molecular basis of eukaryotic transcription. Proc Natl Acad Sci U S A. 2007;104:12955–61. doi: 10.1073/pnas.0704138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee TI, Young RA. Transcription of eukaryotic protein-coding genes. Annu Rev Genet. 2000;34:77–137. doi: 10.1146/annurev.genet.34.1.77. [DOI] [PubMed] [Google Scholar]
- 45.Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu Rev Biochem. 2003;72:449–79. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- 46.Kim TK, et al. Trajectory of DNA in the RNA polymerase II transcription preinitiation complex. Proc Natl Acad Sci U S A. 1997;94:12268–73. doi: 10.1073/pnas.94.23.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith HC, Harris SG, Zillmann M, Berget SM. Evidence that a nuclear matrix protein participates in premessenger RNA splicing. Exp Cell Res. 1989;182:521–33. doi: 10.1016/0014-4827(89)90255-3. [DOI] [PubMed] [Google Scholar]
- 48.Green MR. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559–99. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- 49.Latchman DS. Gene Control. 1st ed. New York, NY: Garland Science, Taylor and Francis Group LLC; 2010. p. 176. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sanger sequencing of the PCR products derived from rapid amplification of the dmyc cDNA at its 5′ end. Notes: Chromatogram on the top: Sequencing of the amplification product with the GSP-N01, hybridizing to the 5′ end of exon1 (see Fig. 2). The forward sequencing primer is UPM-sh. Except an A residue upstream of the Inr1, no further exact base pair matching to the dmyc genomic template strand could be sequenced (red highlight on the chromatogram). The extended region with mismatch is due to the ligation of an oligonucleotide to the 5′ end of the RACE cDNA to ensure capturing of the transcript’s start site. Chromatogram at the bottom: The GSP-N01 amplification product was sequenced with the reverse sequencing primers Exn1-R2 and Exn1-R3 to obtain the readout of the sequences downstream of the known ESTGM01143. The PCR products were sequenced at Microsynth AG, Balgach, Switzerland.
5′ RACE analysis of the dmyc cDNA ends determines splice junctions of the largest transcript.
Notes: Chromatograms show sanger sequencing results after analysis of the 5′ RACE products that were amplified with the gene specific primers GSP-C05 and GSP-C06 (see Fig. 4). GSP-C05 and GSP-C06 hybridize to the dmyc exon3 and extend the full length cDNA from its 3′ towards the 5′ end. The PCR products were sequenced with the reverse primer Exn2-R2 and the forward primer Exn2-F1 (see Fig. 4). The PCR products were sequenced at Microsynth AG, Balgach, Switzerland. Splice junctions between exon1/exon2 and exon2/exon3 are indicated in green.
EvoDifference analysis of the top strand D. melanogaster dmyc intron 2 region.
Notes: Each block of uppercase bases represents a cluster of conserved sequences. Uppercase black nucleotides represent bases in the D. melanogaster dm EvoP intron 2 reference sequence that are conserved in all or all but one of the other 5 orthologous DNAs, D. sechellia, D. yakuba, D. erecta, D. willistoni or D. virilis. The conserved bHLH binding sites and E-boxes are in white text with black highlight, as is the repeat motif ATGTTGCCA and the nearby sub-repeat TGTTGC. The cluster CGCGTGGGAAAA, a dead-ringer for the HLHm-3-2 enhancer, is in white text with dark blue highlight. The consensus binding site GTGGGAA for Su(H) (Suppressor of Hairy) is in the cluster. The DPE in the intron 2 region is lower case bold and underlined. The 5′ and the 3′ end of the transgene J8.4 is with yellow highlighted and the 5′ and the 3′ end of the transgene J8.5 is with light blue highlighted. Sequence orientation is 5′ → 3′ (see also Kharazmi et al. 2011, GRSB).
Positive and negative controls for lacZ staining. For each lacZ staining of transgenes negative and positive control staining was performed. As negative control different tissues were taken from the “y w” and “attp-flies” listed in Table 2. dpp-lacZ flies served as source for positive control. Depicted are negative controls (A–M) and positive controls (N–T), (A, B, N, brain; C–H, O–Q, discs; I–L, R, S, embryos; and M, T, ovaries).
Notes: Staining times for the discs and ovaries are indicated above the scale bars. Embryo staining took place overnight. Scale bar in (A–T) indicates 50 μm.
Table S1.
Analytical primer pairs.
| No. | Name | Primer Sequence (5′→3′) | bp | Usage |
|---|---|---|---|---|
| 1 | BAC-F | CTT CGC GTC CAA CAG ATG | 18 | pC-RP27 sequencing |
| 2 | BAC-R | GCG CCA AAG CAA GAG GGA ATG | 21 | |
| 3 | pZpA-F | CCA AAA AAG GGT TTC ATT AAC TTG TAC ACA TAC | 33 | pLacZ-SV40 p(A) sequencing |
| 4 | pZpA-R | TTG GGG ATT TTA ATA GCG GGC CCT GTG TGT | 30 | |
| 5 | SV40 | GTT CAG GGG GAG GTG TGG G | 19 | SV40 P(A) sequencing |
| 6 | pZR | CGG GCC TCT TCG CTA TTA CG | 20 | pJ8 reverse sequencing |
| 7 | pcaF | GAC GGC GAT ATT TCT GTG GAC | 21 | pCaSpeR4 and |
| 8 | pcaR | CCT TAG CAT GTC CGT GGG GTT TGA | 24 | pJ8.4 sequencing |
| 9 | In2E-F-Not |
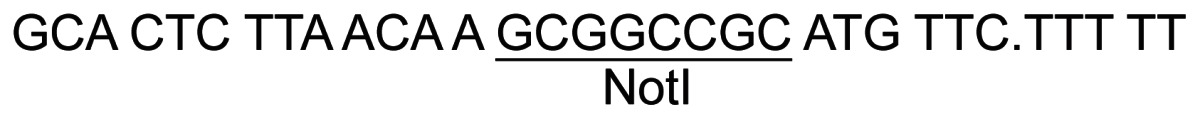
|
32 34 |
PCR – pJ8.4 |
| 10 | In2E-R-Asp |
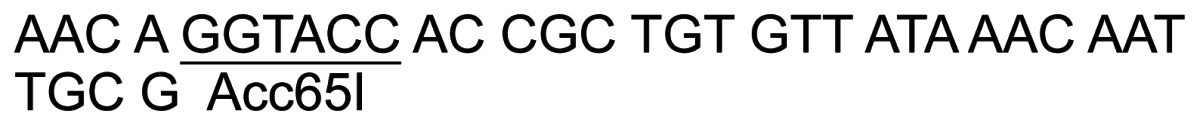
|
||
| 11 | pDPE-F |
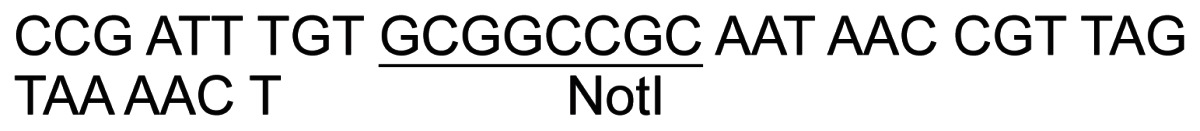
|
36 | PCR – pJ8.5 |
| 12 | pDPE-R | ATG CGC TCA GGT CAA ATT CAG ACG | 24 | |
| 13 | Intr2-R 01 | CCT CAT CTG CAC AGC GAT AGT A | 22 | pJ8.5 sequencing |
| 14 | Intr2-R 02 | TCA TCT GCA CAG CGA TAG TAA | 21 | |
| 15 | Intr2-R 03 | CTG CAC AGC GAT AGT AAA | 18 | |
| 16 | Intr2-R 04 | GGG GAA GGG GGA GGT GGA GAA GA | 23 | |
| 17 | GSP-N01 | GCC GGG CGG TAT TAA ATG GAC CTC GTC CTG | 30 | 5′ RACE cDNA synthesis |
| 18 | GSP-C05 | CTA GCC CTA CGC CGC CGC TTT AAG CCG ATA GTA TGA GAC | 39 | 5′ RACE cDNA synthesis |
| 19 | GSP-C06 | CAC TGC GCC GCT GCC AAT TAC GAT CCA TG | 29 | |
| 20 | UPM-sh | CTA ATA CGA CTC ACT ATA GGG C | 22 | 5′ UTR RACE cDNA sequencing |
| 21 | Exn1-R2 | AAA CAG GCA AAA ATC CAA TCC ACA | 24 | 5′ UTR RACE cDNA sequencing |
| 22 | Exn1-R3 | GGT TAT TTC TGG GAT TTA GCA CTT | 24 | |
| 23 | Exn2-F | ACC GGC CAG CAG CAG TCC A | 19 | 5′ RACE cDNA sequencing |
| 24 | Exn2-R1 | GAA TGG TGG ACG ACG AAA GGA GTT | 24 | |
| 25 | Exn2-R2 | CTC TGG CTT CAT ATC CTC CTC TAA | 24 |
Notes: The oligonucleotides were designed with Bioinformatics tool Laser Gene 9.1 module Primer Select and synthesized at Microsynth, Switzerland. As indicated the primer sets were used for sequencing and/or amplification of dmyc non-coding regions, all 5′ RACE experiments, and for sequencing of cloning vectors and injection plasmids.
Abbreviations: bp, base pairs; E, exon; F, forward; PCR, polymerase chain reaction; R, reverse; RACE, rapid amplification of cDNA ends; Exn, exon; Intr, intron; DPE, downstream promoter element; UPM-sh, Universal Primer Short.
Table S2.
List of the Fly Stocks used in the study.
| No. | Bloomington Stock # | Name | Genotype with FlyBase links |
|---|---|---|---|
| 1 | 6598 | y w | y1 w1118 |
| 2 | 23648 | attp-86F | P{hsp70-flp}1, y1 w*; M{3xP3-RFP.attP}ZH-86Fb; M{vas-int.B}ZH-102D |
| 3 | 24480 | attp-2A | y1 M{3xP3-RFP.attP}ZH-2A w*; M{vas-int.Dm}ZH-102D |
| 4 | 24485 | attp-68E | y1 M{vas-int.Dm}ZH-2A w*; M{3xP3-RFP.attP’}ZH-68E |
| 5 | 8622 | attp-8622 | y1 w67c23; P{CaryP}attP2 |
| 6 | 25709 | attp-25709 | y1 v1 P{nos-phiC31int.NLS}X; P{CaryP}attP40 |
| 7 | 25710 | attp-25710 | y1 sc1 v1 P{nos-phiC31int.NLS}X; P{CaryP}attP2 |
| 8 | 24872 | attp-24872 | y1 M{vas-int.Dm}ZH-2A w*; PBac{y+-attP-3B}VK00037 |
| 9 | 11981 | dmyc-lacZ | w67c23 P{lacW}dmG0354/FM7c |
| 10 | 2475 | double balancer | w*; T(2;3)apXa, apXa/CyO; TM3, Sb1 |
| 11 | 11108 | blue balancer | Cyo, P{lArB}A66.2F2/b1 Adh* cn* l(2)**; ry506 |
| 12 | 8412 | dpp-lacZ | y1 w1118; P{dpp-lacZ.Exel.2}3 |
Notes: For random P-element mediated transgene introduction embryos were taken from the y[1] w[1118] flies (Bloomington Stock number, 6598), and for phi-C31 transgenesis embryos taken from different attp lines were used. The fly stock “blue balancer” that expresses lacZ in embryos and ovaries, was used as a source for positive control in the embryos and ovaries staining. The dpp-lacZ line was used as positive control for the imaginal discs staining.