Abstract
Cockayne syndrome is a rare inherited disorder characterized by accelerated aging, cachectic dwarfism and many other features. Recent work has implicated mitochondrial dysfunction in the pathogenesis of this disease. This is particularly interesting since mitochondrial deficiencies are believed to be important in the aging process. In this review, we will discuss recent findings of mitochondrial pathology in Cockayne syndrome and suggest possible mechanisms for the mitochondrial dysfunction.
Introduction
The accelerated aging disorder Cockayne syndrome (CS) is characterized by progressive brain atrophy, leukodystrophy, cachexia and growth retardation (Koob et al. 2010; Nance and Berry 1992; Natale 2011). CS is caused by mutations in CSB (80% of the cases) and CSA (20%) which both participate in transcription coupled (TC) nucleotide excision DNA repair (NER) (Anindya et al. 2010), transcription (Le et al. 2010) and base excision repair (BER) (Stevnsner et al. 2008). In recent years a potential mitochondrial involvement has been proposed for this disease and CSB was recently reported to be present in the mitochondria (Aamann et al. 2010; Kamenisch et al. 2010). This is particularly interesting because defects in mitochondrial functions are implicated in aging (Balaban et al. 2005). In this review we will discuss the possible mitochondrial pathogenesis in Cockayne syndrome. For thorough information regarding other aspects of this disease we will refer the readers to other sections of this review.
Mitochondria
Mitochondria are small tubular organelles localized in the cytoplasm of almost all mammalian cells. They consist of an outer membrane, inter membrane space, a highly folded inner membrane and a matrix compartment. Mitochondria function as the powerplant of the cell supplying ATP production through oxidative phosphorylation. Interestingly, mitochondria are believed to have evolved via the engulfment of an oxygen consuming prokaryote by an anaerobic eukaryote in the primordial sea 1.5 billion years ago (Gray 2012. This notion is supported by the fact that mitochondria have their own circular genome similar to many bacteria. During the course of evolution the genes have slowly been transferred to the nucleus with only 13 protein encoding genes remaining encoded by a 16.6 kb mitochondrial DNA (mtDNA) molecule. All these genes encode subunits of the electron transport chain, the ATP productive and oxygen consuming mitochondrial super-complex. It therefore follows that all other mitochondrial proteins, including all proteins involved in mtDNA metabolism, are encoded in the nucleus and a nuclear defect can therefore translate into mitochondrial dysfunction.
A substantial amount of research has been invested in understanding the role of mitochondria in aging and age associated diseases. In aging, mitochondrial free radical production is thought to lead to the accumulation of damage to various macromolecules including DNA. Accumulating DNA damage will lead to further deterioration of mitochondrial function, increased free radical production and more DNA damage etc. (Balaban et al. 2005). This mitochondrial theory of aging is supported by several observations: 1) Reactive oxygen species (ROS), the main endogenous damaging agents, are primarily produced in mitochondria. 2) MtDNA lies in close proximity to the mitochondrial inner membrane where ROS is predominantly produced. 3) Mitochondria do not have the same capacity for DNA repair as the nucleus (Gredilla et al. 2010). A number of findings have reinforced this theory, for example oxidative mtDNA damage has been found to accumulate with age in rodent liver (Hudson et al. 1998), increased mtDNA deletions have been found in the brain of elderly people compared to young (Corral-Debrinski et al. 1992) and a mouse model with a mutation in the proof reading domain of the mitochondrial DNA polymerase display mitochondrial mutagenesis and accelerated aging (Trifunovic et al. 2004). Further, overexpression of the antioxidant enzyme catalase targeted to the mitochondria extends lifespan in mice (Schriner et al. 2005). In addition, mitochondria and oxidative stress has been linked to major age related pathologies such as cardiovascular disease, neurodegeneration, diabetes etc. (Lin and Beal 2006; Lowell and Shulman 2005; Mercer et al. 2010).
CS is a highly complex neurological disorder with variation in severity and large differences in age of onset (Koob et al. 2010; Nance and Berry 1992; Natale 2011). Strikingly, most signs and symptoms seen in CS patients are also found in mitochondrial diseases (Table 1). Indeed, like CS mitochondrial diseases often display neurological involvement, a complex phenotype and large variations in age of onset (Haas et al. 2007; Schapira 2006). Corroborating the idea of a mitochondrial dysfunction in CS we recently showed marked alterations in the metabolism of CSBm/m mice as well as in a number of CSB deficient cell lines (Scheibye-Knudsen et al. 2012). These changes corresponded to a rather unusual phenotype with an increase in mitochondrial content, membrane potential and ROS production. In addition we found increased oxygen consumption rates in a number of CSB deficient cell lines (Scheibye-Knudsen et al. 2012). As we shall discuss in greater detail below these alterations seemed to stem from an inability to degrade damaged mitochondria through autophagy; a process called mitophagy. Interestingly, another recent paper also investigated the mitochondrial phenotype of CS cells (Pascucci et al. 2012). Here the authors showed mitochondrial membrane depolarization using the dye JC-1. The dye fluoresce red in normal mitochondrial and green upon membrane depolarization. Using this approach the authors find an increased population of cells undergoing mitochondrial depolarization in good agreement with previous data suggesting increased apoptosis in CS cells (Laposa et al. 2007; McKay et al. 2001). The authors also found increased ROS production in agreement with our findings. Further they investigated the oxygen consumption rate of the cells after culturing them in media where glucose has been exchanged for galactose. The rational for this is that the cells will become more dependent on oxidative phosphorylation if they are unable to use glycolysis (Gohil et al. 2010). Under these circumstances glutamine is added to the media and utilized by the cells as a source of alpha-ketoglutarate that is then metabolized in the citric acid cycle. Cells will switch more towards oxidative phosphorylation if the cells are not able to metabolize galactose, since galactolysis will yield the same amount of ATP as glucose if the Leloir pathway is active. This pathway is predominantly active in the liver, however, a compensatory activation of this pathway can occur in non-liver cells grown in galactose containing media (Christopher et al. 1977). A good way to examine if cells are able to metabolize galactose is using the Seahorse XF analyzer (Gohil et al. 2010). This instrument will report the extracellular acidification rate as a measurement of glycolysis simultaneously with the measurement of oxygen consumption rates. Pascucci et al. did use the Seahorse analyzer however they did not show the extracellular acidification rates.. Interestingly, at least one of their cell lines was able to metabolize galactose as was evident from the increase in oxygen consumption after addition of 2-deoxyglucose. Based on this it is difficult to interpret the oxygen consumption results from their paper. In addition, the authors interestingly observed increased lactate production in CSA and CSB deficient cells in good agreement with our results (Scheibye-Knudsen et al. 2012). This is important since lactic acidosis is a hallmark feature of mitochondrial diseases and this metabolite has been found to accumulate in the brain of CS patients (Koob et al. 2010). We should underscore that much of the work investigating mitochondrial involvement in CS has been focused on the mutations in the CSB complementation group. Thus much work is needed to investigate whether the same alterations are seen with CSA deficiencies.
Table 1. Striking similarities in the phenotype of mitochondrial diseases and Cockayne syndrome.
This is a list of signs and symptoms seen in Cockayne syndrome and whether that particular trait is also seen in mitochondrial diseases. The traits are ordered with the most prevalent first.
Mitochondrial deficiencies in Cockayne syndrome
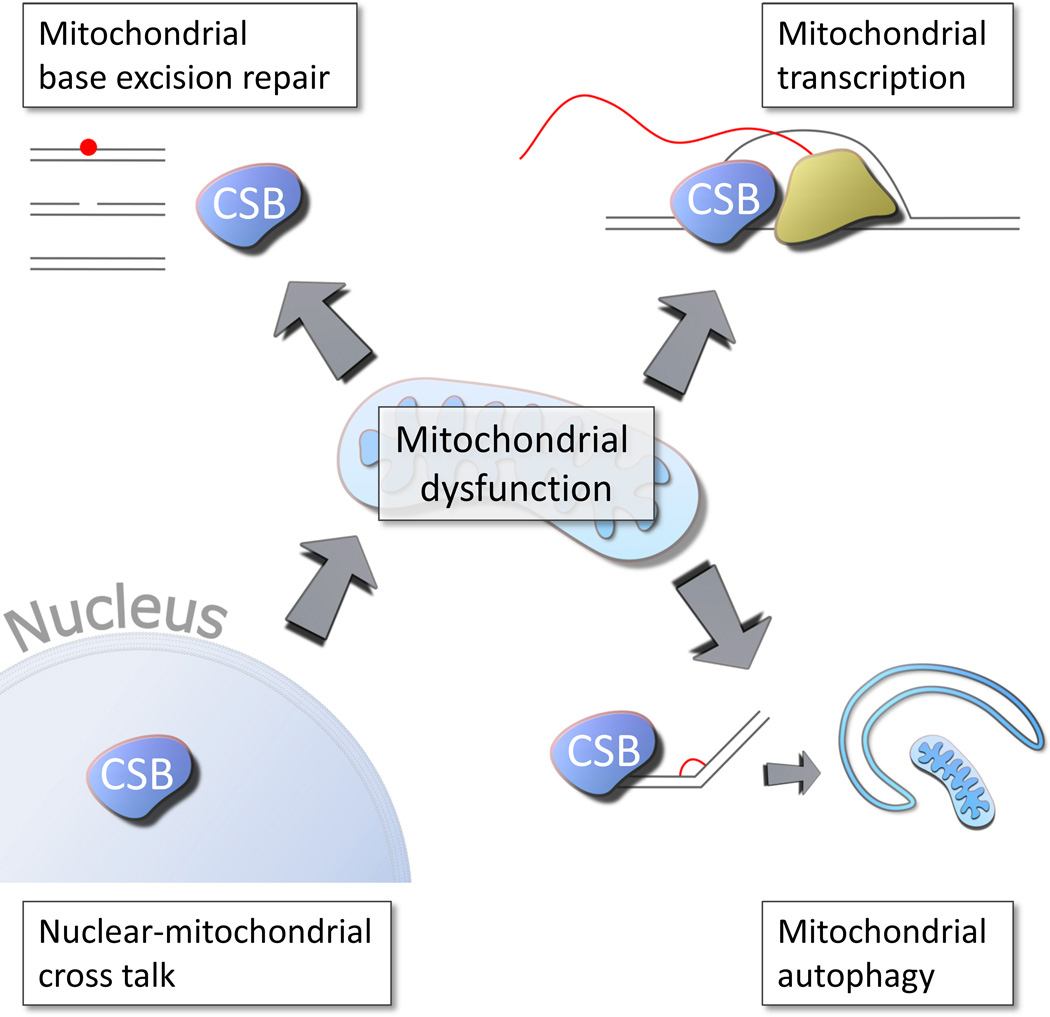
The observation that CS patients have a much more severe phenotype than individuals with mutations in other genes involved in NER, such as the xeroderma pigmentosum (XP) genes, led to the suggestion that CS proteins may be involved in other pathways than NER (Hanawalt 1994). In particular, it remains an unsolved mystery why mutations in XP complementation group A (XPA), a necessary gene in both global genomic (GG) and TC-NER, lead to a milder neurological phenotype than mutations in CSA or CSB. Interestingly, a mitochondrial NER deficiency cannot explain the XPA phenotype since NER is not present in mitochondria (Clayton et al. 1974). The severe neurological deficiency could therefore be secondary to the canonical NER deficiency (Figure 1). Although we would postulate that the difference in phenotype may stem from a mitochondrial dysfunction in CS other pathways may of course contribute. These could include transcriptional deficiencies, deficiencies in base excision repair, and other. We shall discuss these possibilities in greater detail below.
Figure 1. Possible pathways to mitochondrial dysfunction in Cockayne syndrome.
Defects in base excision DNA repair, the primary repair route for oxidative lesions, have been found in Cockayne syndrome (See figure 2). In addition mitochondrial transcription was shown recently to be decreased in Cockayne syndrome (See figure 3). Further defects in mitochondrial autophagy, mitophagy, has been found in Cockayne syndrome (See figure 4). Finally the mitochondrial dysfunction could stem from a nuclear defect (See figure 5).
Cockayne syndrome and base excision repair
In the last decade we and others have shown that CSB is intimately involved in base excision repair (BER) through direct or indirect interactions with the enzymes OGG1, NEIL1, APE1 and PARP1 (Figure 2) (Muftuoglu et al. 2009; Thorslund et al. 2005; Tuo et al. 2002; Wong et al. 2007). BER is critical for the repair of oxidative single base lesions and deficiencies could perhaps drive the neurological phenotype in CS since some BER deficiencies are associated with neurodegeneration (Jeppesen et al. 2011). Further, since BER is the main DNA repair pathway in mitochondria a defect here could lead to a mitochondrial phenotype. Supporting this idea recent data from our and another lab showed the presence of CSB in mitochondria (Aamann et al. 2010; Kamenisch et al. 2010). In these papers an increase in mitochondrial DNA deletions and mutations were found in Csb deficient mice, adipose tissue and human cells, respectively. Interestingly, mtDNA deletions have also been reported in Neil1 deficient mice (Vartanian et al. 2006). Decreased mitochondrial Neil1 activity in Csb deficient mice could therefore possibly explain the increased deletions found in adipocytes in the Csbm/m mouse. Alternatively this effect could be mediated in a similar fashion through Ogg1. Like Neil1, Ogg1 knockout in lower organisms leads to increased mtDNA deletions (Singh et al. 2001). The mitochondrial phenotype could therefore possibly be explained by a mitochondrial DNA repair deficiency since all BER related interacting partners of Csb are present in mitochondria. Indeed, decreased Ogg1 incision activity has been found in mitochondrial extracts isolated from CSB mutant mice as well as in cells deficient in CSB although no difference in oxidative mitochondrial DNA damage was found using a gene specific repair assay and FPG incision (Stevnsner et al. 2002). The finding of decreased Ogg1 activity was, however, not dependent upon the catalytic activity of CSB since the mitochondrial DNA repair deficiency could be rescued in cells expressing catalytic dead CSB (Stevnsner et al. 2002). Supporting these observations it was later found that CSB deficient cells did accumulate more damage (Osenbroch et al. 2009). Although these results indicate a connection between BER and CSB, other evidence have suggested this may not be the case. For example, Csbm/m/Ogg1−/− double transgenic mice do not accumulate more oxidative mitochondrial DNA damage than WT as measured using an assay where supercoiled mtDNA was relaxed after incision by the oxidative DNA damage specific endonuclease FPG (Trapp et al. 2007). This view has recently been corroborated using long range qPCR showing no increased mitochondrial DNA damage in CSB deficient cells compared to controls (Berquist et al. 2012). This result was supported by investigating mitochondrial 8-oxo-G levels by HPLC-ED and finding no accumulation in various CSB deficient cell lines (Pascucci et al. 2012). Additionally, using mass spectrometry, no accumulation of 8-oxo-G was found in mtDNA isolated from various tissues from Csbm/m mice, however, nuclear accumulation of 8-oxo-G was present (Muftuoglu et al. 2009). Interestingly, in this study accumulation of the oxidized DNA lesion, FapyA, was found in mitochondria. This particular lesion is a ring opened adenine that is a prime NEIL1 substrate. These findings indicate that CSB may not exert its mitochondrial effect through OGG1 but rather through NEIL1. While a mitochondrial BER deficiency may contribute to the CS phenotype, it is still an open question as to whether it plays a significant role in the neurodegenerative phenotype seen in CS. For further insight into the potential role of oxidative damage in CS we recommend perusing the article from Dr. Bernd Epe in this issue of Mechanisms of Ageing and Development.
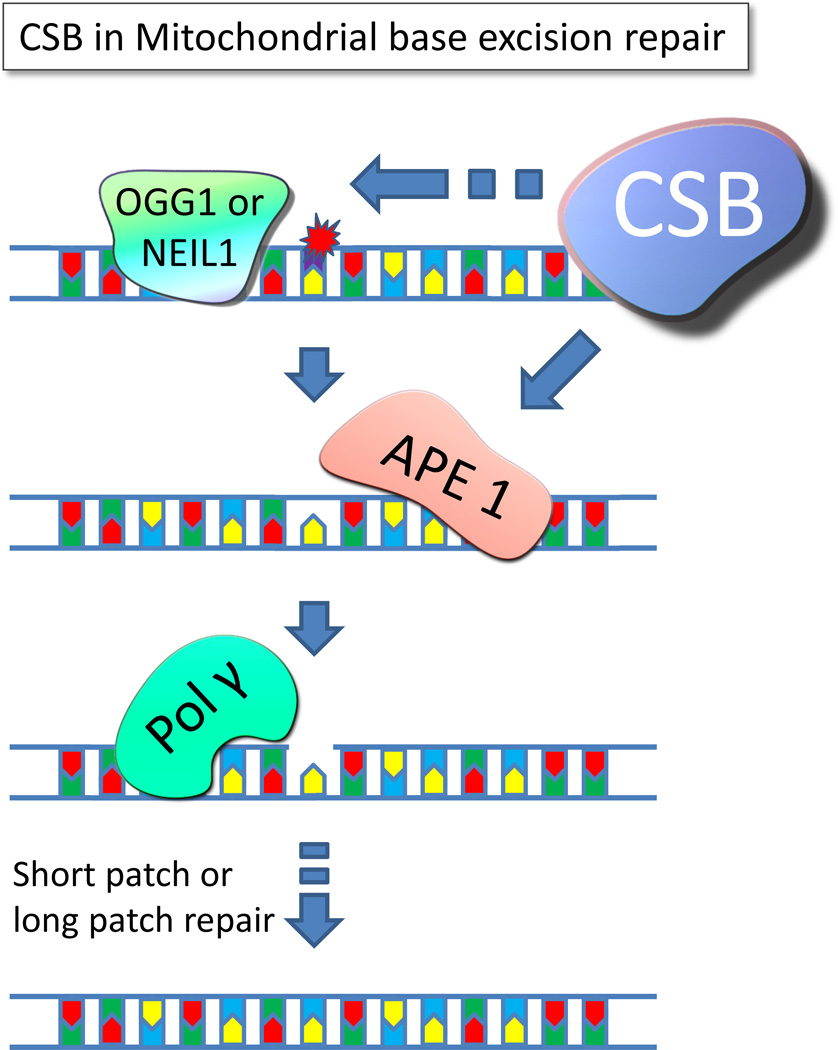
Figure 2. Cockayne syndrome protein B regulates base excision repair.
CSB has been shown to increase the incision activity of OGG1 and NEIL1; two glycosylases responsible for the initial step of removing oxidized lesions such as 8-oxo-G from the ribose backbone. This creates an abasic site that is recognized and removed by APE1 yielding a gab in the DNA strand. APE1 has been shown to physically and functionally interact with CSB. The resulting gap in the DNA is then further processed by short patch or long patch repair involving proteins such as DNA polymerase gamma, DNA ligase III, Fen1 and others.
Cockayne syndrome and mitochondrial transcription
A hallmark feature of cells deficient in CSA or CSB is a delayed resumption of transcription after UV irradiation (Mayne and Lehmann 1982). A defect in transcription coupled repair is the conventional explanation for this phenomenon. As an alternative theory this deficiency could be due to general loss of transcription as has been suggested (Compe and Egly 2012). This explanation is supported by the findings that CSB and a number of other NER factors are recruited to promoters of genes and are associated with downstream RNA polymerase II transcription as well (Le et al. 2010). Notably, CSB was not required for initial binding of the transcription complex, however, CSB was necessary for association of other NER factors during elongation. This fits well with previous findings showing a role for CSB in RNA elongation by RNA polymerase II (Selby and Sancar 1997). Interestingly, CSB also stimulates RNA polymerase I and III activity (Bradsher et al. 2002; Yu et al. 2000; Yuan et al. 2007). These polymerases are, among other things, responsible for transcribing rDNA that consists of highly repetitive sequences. rDNA genes therefore have a high propensity for forming secondary structures leading to stalling of RNA polymerases. In this context CSB may help the polymerase elongate through these highly structured regions (Yu et al. 2000). The function of CSB in relation to transcription may therefore be very similar to its role in TC-NER. In transcription CSB could resolve stalled polymerases at secondary structures while in DNA repair CSB is known to resolve polymerases at stalling lesions such as cylopyrimidine dimers, 6–4 photoproducts and bulky adducts (Fousteri et al. 2006; Laine and Egly 2006). Although, the role of CSB in nuclear transcription seem well documented the possible downstream pathogenesis is less clear and begs the question of how a transcriptional defect could lead to neurodegeneration. For a thorough insight into the transcriptional role of CSA and CSB we will recommend reading the sections by Drs. Jean-Marc Egly and Bruce McKay in this special issue of Mechanisms of Ageing and Development. The role of CSB in regards to the mitochondrial RNA polymerase (mtRNA pol) was until recently unknown. MtRNA pol is, contrary to the other polymerases, a single subunit polymerase related to the bacteriophage T7 RNA polymerase (Arnold et al. 2012). Following our and others finding that CSB was present in mitochondria, we recently showed strong stimulation of the elongation activity of mtRNA pol in vitro by CSB (Berquist et al. 2012). Additionally, we found decreased transcription from the heavy strand promotor in CSB deficient CS1AN cells compared to isogenic cells expressing WT CSB. The exact mechanism for how CSB would stimulate elongation is still not known. Based on the enzymatic activity of CSB two possible mechanisms could be proposed. First, the DNA annealing activity could possibly aid in transcription after the strands have been separated by the polymerase. Second, and perhaps more likely, CSB is known to be able to push other DNA binding proteins off the strand (Berquist and Wilson, III 2009). CSB could therefore facilitate transcription by removing proteins bound to the strand in front of the transcription machinery (Figure 3). In either case a defect in mitochondrial transcription via loss of CSB could possibly explain the mitochondrial phenotype seen in CS. The role of CSA in this process remains to be determined.
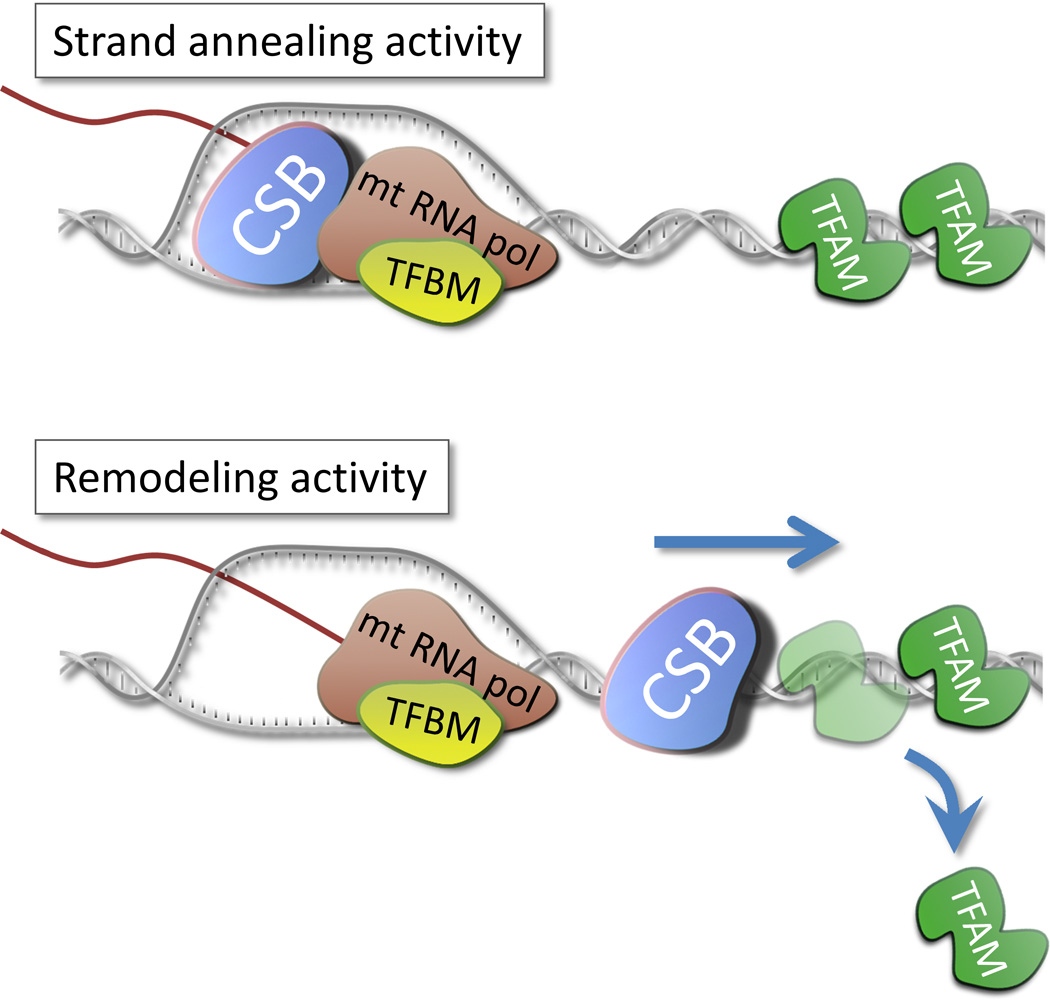
Figure 3. Possible roles for CSB in mitochondrial transcription.
CSB was recently shown to stimulate the RNA elongation activity of the mitochondrial polymerase. In this regard CSB could act either as a DNA annealing enzyme closing the transcription bubble after the RNA polymerase. Alternatively, CSB has been shown to be able to displace TFAM from the DNA strand. In this regard CSB could act to remove bound proteins from the DNA strand in front of the polymerase thereby facilitating transcription.
Cockayne syndrome and mitophagy
As mentioned, we recently found increased mitochondrial content, increased membrane potential and increased free radical production in CSB deficient cells (Scheibye-Knudsen et al. 2012). In addition, increased FCCP uncoupled respiration (spare respiratory capacity) and increased oxygen consumption rates were found in CSB deficient cells possibly due to increased ATP consumption. These changes did not appear to be related to increased mitochondrial biogenesis since no consistent changes in the canonical mitochondrial transcription factors, PGC-1alpha, TFAM and ERR alpha, were found in CSB deficient cells. Since the amount of mitochondria present in the cells must be determined by biogenesis as well as degradation, we investigated autophagy, which is the primary degradation route for mitochondria. Macro autophagy is the process by which degradation of large protein aggregates and whole organelles are removed from the cells (Laplante and Sabatini 2012). Mitophagy is the process by which mitochondria are selectively degraded. A number of proteins are involved in this pathway which is thought to be initiated either by the accumulation of NIX or PINK1 at the outer mitochondrial membrane. NIX was initially found to be upregulated in reticulocytes during maturation to erythrocytes and this upregulation was responsible for the programmed removal of mitochondria from red blood cells (Schweers et al. 2007). Now NIX has also been implicated in the role of selectively removing damaged mitochondria although more work needs to be done to fully establish this relationship (Ding et al. 2010). On the other hand the role of PINK1 in selective mitophagy seems well established. Here PINK1 is constitutively imported into the mitochondria and degraded when the membrane potential is high. Upon depolarization PINK1 accumulates at the outer membrane where it recruits Parkin. Parkin is an E3-ubiquitin ligase that ubiquitinates outer membrane proteins. This mitochondrial ubiquitination leads to the recruitment of the scaffolding protein P62 that will facilitate the association and engulfment of the mitochondria with an autophagosome coated by the linkage protein LC-3B isoform II. Fusion of the autophagosome with a lysosome will conclude the autophagic process through degradation of the content of the autophagosome. Interestingly, in the case of CS we found decreased colocalization of LC3, P62 and ubiquitin with the mitochondria in response to stress perhaps explaining the mitochondrial phenotype we observed (Scheibye-Knudsen et al. 2012). We were also able to reverse this phenotype by treating the CSB deficient cells with the autophagy stimulating drug rapamycin. This FDA approved drug has previously been shown to lead to lifespan extension through inhibition of the mTOR pathway and activation of autophagy (Harrison et al. 2009). In addition, rapamycin is thought to be neuroprotective and decrease glial activation and could thereby secondarily attenuate some neurological features of CS (Bove et al. 2011; Dello et al. 2012). Recently, a similar mitochondrial phenotype was reported in ataxia-telangiectasia mutated (ATM) deficient cells (Valentin-Vega et al. 2012). Increased oxygen consumption rate and decreased mitophagy were found corresponding well with our study. In addition the authors isolated mitochondria and found ATM present in this organelle. We therefore propose that CSB and ATM act as part of a mitochondrial DNA damage response pathway which promotes mitophagy in response to irreparable mitochondrial DNA damage (Figure 4).
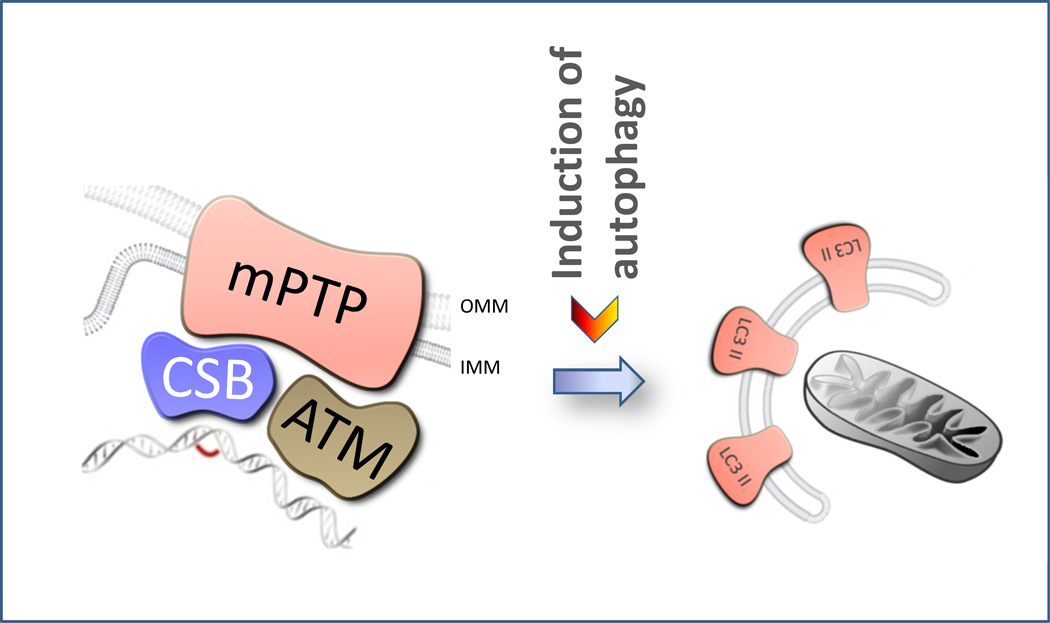
Figure 4. A possible role for CSB in mitophagy.
CSB deficient cells were recently shown to be defective in mitophagy. In this regard CSB could act as a mitochondrial DNA damage sensor either directly or through currently unknown proteins inducing mitophagy through the mitochondrial permeability transition pore (mPTP) when an irreparable lesion is encountered. Ataxia telangiectasia mutated (ATM) protein could function in a similar fashion. OMM: outer mitochondrial membrane. IMM: Inner mitochondrial membrane.
Nuclear-mitochondrial crosstalk in CS
A caveat to the above hypotheses come from the observation that membrane potential, oxygen consumption rate and FCCP uncoupled respiration are all increased in CSB deficient cells (Scheibye-Knudsen et al. 2012). This is unusual for a primary mitochondrial defect where disruption of the electron transport chain would normally lead to decreased oxygen consumption and decreased membrane potential. Another possibility is that the mitochondrial phenotype could be a secondary compensatory response to a nuclear DNA repair or transcriptional defect (Figure 5). In the following we will present to possible scenarios that could explain the cellular and organismal features seen in CS. We wish to underscore that much of this is still hypothetical and need further evaluation.
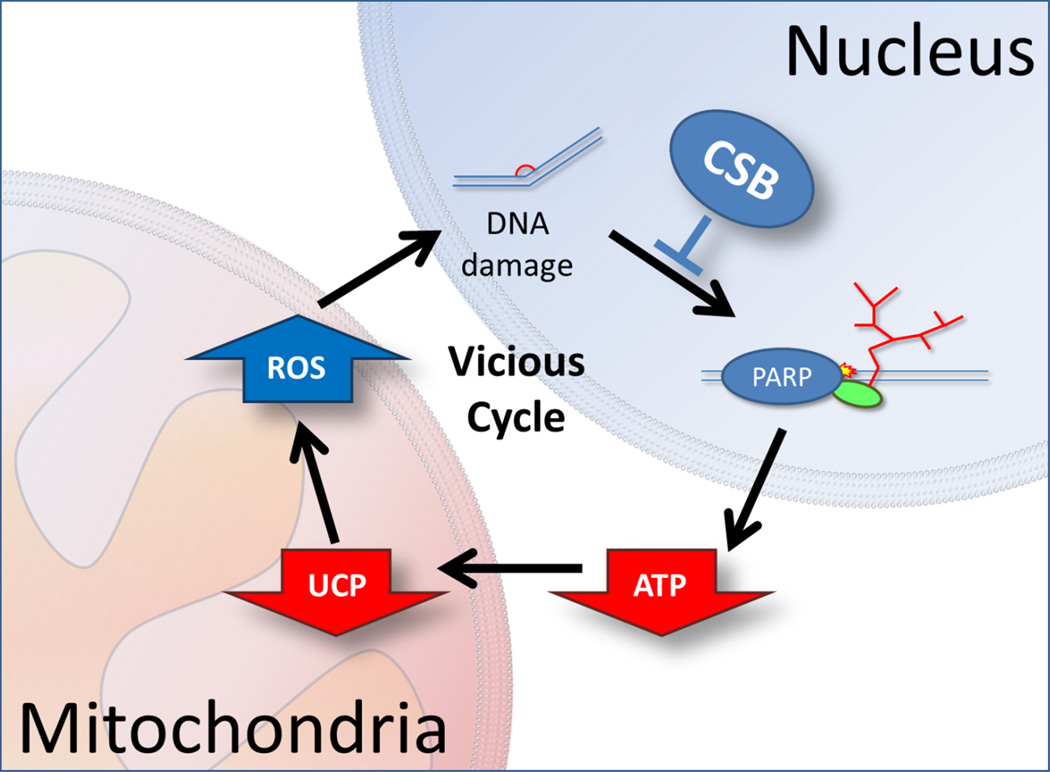
Figure 5. Mitochondrial dysfunction secondary to a nuclear deficiency.
Due to the unusual nature of the mitochondrial dysfunction in CSB with increased oxygen consumption, increased membrane potential and increased ROS production the phenotype could stem from a compensatory response to a nuclear defect. Here CSB acts through the canonical transcription coupled nucleotide DNA repair. A defect leads to increased PARP1 activation. Poly-ADP-ribosylation is an energetically demanding process leading to increased ATP consumption and thereby decreased ATP levels. To increase energy output from the mitochondria the cells decrease the amount of uncoupling proteins. This leads to increased membrane potential and consequently increased ROS formation that can further damage DNA leading to a vicious cycle.
In the case of a CS-related DNA repair deficiency one could propose that decreased DNA repair would lead to increased PARP1 and perhaps ATM activation because of accumulated DNA damage. Activation of these pathways will lead to decreased NAD+ levels and increased ATP consumption. Increased ATP consumption could explain the cachectic phenotype in the CS patients as well as the increased metabolism in Csbm/m mice, and increased ATP consumption was indeed found in CSB deficient cells (Scheibye-Knudsen et al. 2012). To compensate for this increase in ATP consumption cells could downregulate mitochondrial uncoupling proteins leading to a more efficient ATP production. Decreasing levels of uncoupling proteins could explain the increased membrane potential and relative increase in FCCP uncoupled respiration seen in these cells. ROS production is also regulated by membrane potential with increasing membrane potential leading to increased ROS. Furthermore, as we discussed in the previous paragraph, mitophagy is intimately regulated by the membrane potential where an increase in membrane potential will lead to a decrease in mitophagy. Thus, CS cells may be carrying around mitochondria that otherwise should have been degraded and in fact, more damaged mitochondria are seen in CS cells (Scheibye-Knudsen et al. 2012). Interestingly, increased ROS production is known to augment mitochondrial biogenesis through activation of PGC-1alpha. A downstream effect of increased PGC-1alpha activity is increased expression of uncoupling proteins. Under normal circumstances increased ROS activates a negative feedback loop that increased UCPs thereby lowering membrane potential. Interestingly, PGC-1alpha is not consistently activated in CSB deficient cells indicating that other so far unknown transcription factors may play a role (Scheibye-Knudsen et al. 2012).
Another possible hypothesis could link the transcriptional defect observed in CS with the mitochondrial phenotype. When RNA polymerases hit a secondary DNA structures, such as a stem-loop, stalling of the polymerase will occur. Here CSB is able to facilitate the resolution of the structure and continuation of elongation. Without CSB the polymerase will stall and may fall off the strand. Another polymerase will hit the stalling structure and fall off etc. These futile transcription cycles will lead to higher energy expenditures because of the ATP needed for RNA synthesis. Futile transcription cycles by RNA pol II have indeed been found although a link to energy metabolism needs further examination (Thiebaut et al. 2008). Then the aforementioned downstream compensation of downregulation of uncoupling proteins, increased membrane potential and increased ROS may still occur.
Concluding remark
Dysfunctional mitochondrial may very well contribute to the spectrum of problems patients with CS confront although the pathogenesis is contested at the moment. We sought in this review to give a broad introduction to what we think are the most likely scenarios behind the potential role of mitochondria in CS. At the moment, we feel all options are open and the phenotype could quite possibly stem from any combination of defects. However, some encouraging news have come out of this research with the discovery that rapamycin may reverse some of the mitochondrial features seen in CS. We will hopefully soon have more answers.
Highlights.
Increasing evidence suggests a role of mitochondria in the pathogenesis of Cockayne syndrome grp B.
Altered mitochondrial base excision repair, transcription or autophagy may play a role in the mitochondrial dysfunction.
Nuclear-mitochondrial cross-talk defects could also play a role in the mitochondrial phenotype in Cockayne syndrome.
Acknowledgement
This research was supported entirely by the Intramural Research Program of the NIH, National Institute on Aging.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Aamann MD, Sorensen MM, Hvitby C, Berquist BR, Muftuoglu M, Tian J, de Souza-Pinto NC, Scheibye-Knudsen M, Wilson DM, III, Stevnsner T, Bohr VA. Cockayne syndrome group B protein promotes mitochondrial DNA stability by supporting the DNA repair association with the mitochondrial membrane. FASEB J. 2010;24:2334–2346. doi: 10.1096/fj.09-147991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angle B, Burton BK. Risk of sudden death and acute life-threatening events in patients with glutaric acidemia type II. Mol.Genet.Metab. 2008;93:36–39. doi: 10.1016/j.ymgme.2007.09.015. [DOI] [PubMed] [Google Scholar]
- Anindya R, Mari PO, Kristensen U, Kool H, Giglia-Mari G, Mullenders LH, Fousteri M, Vermeulen W, Egly JM, Svejstrup JQ. A ubiquitin-binding domain in Cockayne syndrome B required for transcription-coupled nucleotide excision repair. Mol.Cell. 2010;38:637–648. doi: 10.1016/j.molcel.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T, Souri M, Ushikubo S, Kamijo T, Yamaguchi S, Kelley RI, Rhead WJ, Uetake K, Tanaka K, Hashimoto T. Purification of human very-long-chain acyl-coenzyme A dehydrogenase and characterization of its deficiency in seven patients. J.Clin.Invest. 1995;95:2465–2473. doi: 10.1172/JCI117947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold JJ, Smidansky ED, Moustafa IM, Cameron CE. Human mitochondrial RNA polymerase: structure-function, mechanism and inhibition. Biochim.Biophys.Acta. 2012;1819:948–960. doi: 10.1016/j.bbagrm.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Baloh RH, Salavaggione E, Milbrandt J, Pestronk A. Familial parkinsonism and ophthalmoplegia from a mutation in the mitochondrial DNA helicase twinkle. Arch.Neurol. 2007;64:998–1000. doi: 10.1001/archneur.64.7.998. [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Weinberger MJ, Kobori JA, Rinaldo P, Burlina AB. Mitochondrial short-chain L-3-hydroxyacyl-coenzyme A dehydrogenase deficiency: a new defect of fatty acid oxidation. Pediatr.Res. 1996;39:185–188. doi: 10.1203/00006450-199601000-00031. [DOI] [PubMed] [Google Scholar]
- Berquist BR, Canugovi C, Sykora P, Wilson DM, III, Bohr VA. Human Cockayne syndrome B protein reciprocally communicates with mitochondrial proteins and promotes transcriptional elongation. Nucleic Acids Res. 2012;40:8392–8405. doi: 10.1093/nar/gks565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berquist BR, Wilson DM., III Nucleic acid binding activity of human Cockayne syndrome B protein and identification of Ca(2+) as a novel metal cofactor. J.Mol.Biol. 2009;391:820–832. doi: 10.1016/j.jmb.2009.06.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdon A, Minai L, Serre V, Jais JP, Sarzi E, Aubert S, Chretien D, de LP, Paquis-Flucklinger V, Arakawa H, Nakamura Y, Munnich A, Rotig A. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat.Genet. 2007;39:776–780. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- Bove J, Martinez-Vicente M, Vila M. Fighting neurodegeneration with rapamycin: mechanistic insights. Nat.Rev.Neurosci. 2011;12:437–452. doi: 10.1038/nrn3068. [DOI] [PubMed] [Google Scholar]
- Bradsher J, Auriol J, Proietti de SL, Iben S, Vonesch JL, Grummt I, Egly JM. CSB is a component of RNA pol I transcription. Mol.Cell. 2002;10:819–829. doi: 10.1016/s1097-2765(02)00678-0. [DOI] [PubMed] [Google Scholar]
- Camm JH, Carpenter JV. Dental treatment of a patient with Friedreich's ataxia. Spec.Care Dentist. 1987;7:117–119. doi: 10.1111/j.1754-4505.1987.tb00619.x. [DOI] [PubMed] [Google Scholar]
- Carrozzo R, Dionisi-Vici C, Steuerwald U, Lucioli S, Deodato F, Di GS, Bertini E, Franke B, Kluijtmans LA, Meschini MC, Rizzo C, Piemonte F, Rodenburg R, Santer R, Santorelli FM, van RA, Vermunt-de KD, Morava E, Wevers RA. SUCLA2 mutations are associated with mild methylmalonic aciduria, Leigh-like encephalomyopathy, dystonia and deafness. Brain. 2007;130:862–874. doi: 10.1093/brain/awl389. [DOI] [PubMed] [Google Scholar]
- Christopher CW, Colby WW, Ullrey D, Kalckar HM. Comparative studies of glucose-fed and glucose-starved hamster cell cultures: responses in galactose metabolism. J.Cell Physiol. 1977;90:387–405. doi: 10.1002/jcp.1040900303. [DOI] [PubMed] [Google Scholar]
- Clayton DA, Doda JN, Friedberg EC. The absence of a pyrimidine dimer repair mechanism in mammalian mitochondria. Proc.Natl.Acad.Sci.U.S.A. 1974;71:2777–2781. doi: 10.1073/pnas.71.7.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compe E, Egly JM. TFIIH: when transcription met DNA repair. Nat.Rev.Mol.Cell Biol. 2012;13:343–354. doi: 10.1038/nrm3350. [DOI] [PubMed] [Google Scholar]
- Corral-Debrinski M, Horton T, Lott MT, Shoffner JM, Beal MF, Wallace DC. Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nat.Genet. 1992;2:324–329. doi: 10.1038/ng1292-324. [DOI] [PubMed] [Google Scholar]
- de Vries MC, Rodenburg RJ, Morava E, van Kaauwen EP, ter LH, Mullaart RA, Snoeck IN, van Hasselt PM, Harding P, van den Heuvel LP, Smeitink JA. Multiple oxidative phosphorylation deficiencies in severe childhood multi-system disorders due to polymerase gamma (POLG1) mutations. Eur.J.Pediatr. 2007;166:229–234. doi: 10.1007/s00431-006-0234-9. [DOI] [PubMed] [Google Scholar]
- Dello RC, Lisi L, Feinstein DL, Navarra P. mTOR kinase, a key player in the regulation of glial functions: Relevance for the therapy of multiple sclerosis. Glia. 2012 doi: 10.1002/glia.22433. [DOI] [PubMed] [Google Scholar]
- Di FA, Ronchi D, Lodi T, Fassone E, Tigano M, Lamperti C, Corti S, Bordoni A, Fortunato F, Nizzardo M, Napoli L, Donadoni C, Salani S, Saladino F, Moggio M, Bresolin N, Ferrero I, Comi GP. The mitochondrial disulfide relay system protein GFER is mutated in autosomal-recessive myopathy with cataract and combined respiratory-chain deficiency. Am.J.Hum.Genet. 2009;84:594–604. doi: 10.1016/j.ajhg.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding WX, Ni HM, Li M, Liao Y, Chen X, Stolz DB, Dorn GW, Yin XM. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J.Biol.Chem. 2010;285:27879–27890. doi: 10.1074/jbc.M110.119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diomedi-Camassei F, Di GS, Santorelli FM, Caridi G, Piemonte F, Montini G, Ghiggeri GM, Murer L, Barisoni L, Pastore A, Muda AO, Valente ML, Bertini E, Emma F. COQ2 nephropathy: a newly described inherited mitochondriopathy with primary renal involvement. J.Am.Soc.Nephrol. 2007;18:2773–2780. doi: 10.1681/ASN.2006080833. [DOI] [PubMed] [Google Scholar]
- Durr A, Cossee M, Agid Y, Campuzano V, Mignard C, Penet C, Mandel JL, Brice A, Koenig M. Clinical and genetic abnormalities in patients with Friedreich's ataxia. N.Engl.J.Med. 1996;335:1169–1175. doi: 10.1056/NEJM199610173351601. [DOI] [PubMed] [Google Scholar]
- Elpeleg O, Miller C, Hershkovitz E, Bitner-Glindzicz M, Bondi-Rubinstein G, Rahman S, Pagnamenta A, Eshhar S, Saada A. Deficiency of the ADP-forming succinyl-CoA synthase activity is associated with encephalomyopathy and mitochondrial DNA depletion. Am.J.Hum.Genet. 2005;76:1081–1086. doi: 10.1086/430843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fousteri M, Vermeulen W, van Zeeland AA, Mullenders LH. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol.Cell. 2006;23:471–482. doi: 10.1016/j.molcel.2006.06.029. [DOI] [PubMed] [Google Scholar]
- Fratter C, Gorman GS, Stewart JD, Buddles M, Smith C, Evans J, Seller A, Poulton J, Roberts M, Hanna MG, Rahman S, Omer SE, Klopstock T, Schoser B, Kornblum C, Czermin B, Lecky B, Blakely EL, Craig K, Chinnery PF, Turnbull DM, Horvath R, Taylor RW. The clinical, histochemical, and molecular spectrum of PEO1 (Twinkle)-linked adPEO. Neurology. 2010;74:1619–1626. doi: 10.1212/WNL.0b013e3181df099f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freisinger P, Futterer N, Lankes E, Gempel K, Berger TM, Spalinger J, Hoerbe A, Schwantes C, Lindner M, Santer R, Burdelski M, Schaefer H, Setzer B, Walker UA, Horvath R. Hepatocerebral mitochondrial DNA depletion syndrome caused by deoxyguanosine kinase (DGUOK) mutations. Arch.Neurol. 2006;63:1129–1134. doi: 10.1001/archneur.63.8.1129. [DOI] [PubMed] [Google Scholar]
- Gelfand JM, Duncan JL, Racine CA, Gillum LA, Chin CT, Zhang Y, Zhang Q, Wong LJ, Roorda A, Green AJ. Heterogeneous patterns of tissue injury in NARP syndrome. J.Neurol. 2011;258:440–448. doi: 10.1007/s00415-010-5775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohil VM, Sheth SA, Nilsson R, Wojtovich AP, Lee JH, Perocchi F, Chen W, Clish CB, Ayata C, Brookes PS, Mootha VK. Nutrient-sensitized screening for drugs that shift energy metabolism from mitochondrial respiration to glycolysis. Nat.Biotechnol. 2010;28:249–255. doi: 10.1038/nbt.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz A, Isohanni P, Pihko H, Paetau A, Herva R, Saarenpaa-Heikkila O, Valanne L, Marjavaara S, Suomalainen A. Thymidine kinase 2 defects can cause multi-tissue mtDNA depletion syndrome. Brain. 2008;131:2841–2850. doi: 10.1093/brain/awn236. [DOI] [PubMed] [Google Scholar]
- Gray MW. Mitochondrial evolution. Cold Spring Harb.Perspect.Biol. 2012;4:a011403. doi: 10.1101/cshperspect.a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gredilla R, Bohr VA, Stevnsner T. Mitochondrial DNA repair and association with aging--an update. Exp.Gerontol. 2010;45:478–488. doi: 10.1016/j.exger.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillausseau PJ, Massin P, Dubois-LaForgue D, Timsit J, Virally M, Gin H, Bertin E, Blickle JF, Bouhanick B, Cahen J, Caillat-Zucman S, Charpentier G, Chedin P, Derrien C, Ducluzeau PH, Grimaldi A, Guerci B, Kaloustian E, Murat A, Olivier F, Paques M, Paquis-Flucklinger V, Porokhov B, Samuel-Lajeunesse J, Vialettes B. Maternally inherited diabetes and deafness: a multicenter study. Ann.Intern.Med. 2001;134:721–728. doi: 10.7326/0003-4819-134-9_part_1-200105010-00008. [DOI] [PubMed] [Google Scholar]
- Haas RH, Parikh S, Falk MJ, Saneto RP, Wolf NI, Darin N, Cohen BH. Mitochondrial disease: a practical approach for primary care physicians. Pediatrics. 2007;120:1326–1333. doi: 10.1542/peds.2007-0391. [DOI] [PubMed] [Google Scholar]
- Hammans SR, Sweeney MG, Hanna MG, Brockington M, Morgan-Hughes JA, Harding AE. The mitochondrial DNA transfer RNALeu(UUR) A-->G(3243). A clinical and genetic study. Brain. 1995;118(Pt 3):721–734. doi: 10.1093/brain/118.3.721. [DOI] [PubMed] [Google Scholar]
- Hanawalt PC. Transcription-coupled repair and human disease. Science. 1994;266:1957–1958. doi: 10.1126/science.7801121. [DOI] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, Pahor M, Javors MA, Fernandez E, Miller RA. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt IJ, Harding AE, Petty RK, Morgan-Hughes JA. A new mitochondrial disease associated with mitochondrial DNA heteroplasmy. Am.J.Hum.Genet. 1990;46:428–433. [PMC free article] [PubMed] [Google Scholar]
- Hudson EK, Hogue BA, Souza-Pinto NC, Croteau DL, Anson RM, Bohr VA, Hansford RG. Age-associated change in mitochondrial DNA damage. Free Radical Research. 1998;29:573–579. doi: 10.1080/10715769800300611. [DOI] [PubMed] [Google Scholar]
- Huoponen K. Leber hereditary optic neuropathy: clinical and molecular genetic findings. Neurogenetics. 2001;3:119–125. doi: 10.1007/s100480100115. [DOI] [PubMed] [Google Scholar]
- Iafolla AK, Thompson RJ, Jr, Roe CR. Medium-chain acyl-coenzyme A dehydrogenase deficiency: clinical course in 120 affected children. J.Pediatr. 1994;124:409–415. doi: 10.1016/s0022-3476(94)70363-9. [DOI] [PubMed] [Google Scholar]
- Ito M, Kobashi H, Naito E, Saijo T, Takeda E, Huq AH, Kuroda Y. Decrease of pyruvate dehydrogenase phosphatase activity in patients with congenital lactic acidemia. Clin.Chim.Acta. 1992;209:1–7. doi: 10.1016/0009-8981(92)90327-m. [DOI] [PubMed] [Google Scholar]
- Jeppesen DK, Bohr VA, Stevnsner T. DNA repair deficiency in neurodegeneration. Prog.Neurobiol. 2011;94:166–200. doi: 10.1016/j.pneurobio.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamenisch Y, Fousteri M, Knoch J, von Thaler AK, Fehrenbacher B, Kato H, Becker T, Dolle ME, Kuiper R, Majora M, Schaller M, van der Horst GT, van SH, Rocken M, Rapaport D, Krutmann J, Mullenders LH, Berneburg M. Proteins of nucleotide and base excision repair pathways interact in mitochondria to protect from loss of subcutaneous fat, a hallmark of aging. J.Exp.Med. 2010;207:379–390. doi: 10.1084/jem.20091834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karadimas CL, Vu TH, Holve SA, Chronopoulou P, Quinzii C, Johnsen SD, Kurth J, Eggers E, Palenzuela L, Tanji K, Bonilla E, De Vivo DC, DiMauro S, Hirano M. Navajo neurohepatopathy is caused by a mutation in the MPV17 gene. Am.J.Hum.Genet. 2006;79:544–548. doi: 10.1086/506913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob M, Laugel V, Durand M, Fothergill H, Dalloz C, Sauvanaud F, Dollfus H, Namer IJ, Dietemann JL. Neuroimaging in Cockayne syndrome. AJNR Am.J.Neuroradiol. 2010;31:1623–1630. doi: 10.3174/ajnr.A2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskinen T, Santavuori P, Sainio K, Lappi M, Kallio AK, Pihko H. Infantile onset spinocerebellar ataxia with sensory neuropathy: a new inherited disease. J.Neurol.Sci. 1994;121:50–56. doi: 10.1016/0022-510x(94)90156-2. [DOI] [PubMed] [Google Scholar]
- Kurt B, Jaeken J, Van HJ, Lagae L, Lofgren A, Everman DB, Jayakar P, Naini A, Wierenga KJ, Van GG, Copeland WC, DiMauro S. A novel POLG gene mutation in 4 children with Alpers-like hepatocerebral syndromes. Arch.Neurol. 2010;67:239–244. doi: 10.1001/archneurol.2009.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine JP, Egly JM. Initiation of DNA repair mediated by a stalled RNA polymerase IIO. EMBO J. 2006;25:387–397. doi: 10.1038/sj.emboj.7600933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laposa RR, Huang EJ, Cleaver JE. Increased apoptosis, p53 up-regulation, and cerebellar neuronal degeneration in repair-deficient Cockayne syndrome mice. Proc.Natl.Acad.Sci.U.S.A. 2007;104:1389–1394. doi: 10.1073/pnas.0610619104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le MN, Mota-Fernandes D, Velez-Cruz R, Iltis I, Biard D, Egly JM. NER factors are recruited to active promoters and facilitate chromatin modification for transcription in the absence of exogenous genotoxic attack. Mol.Cell. 2010;38:54–66. doi: 10.1016/j.molcel.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Lee HF, Lee HJ, Chi CS, Tsai CR, Chang TK, Wang CJ. The neurological evolution of Pearson syndrome: case report and literature review. Eur.J.Paediatr.Neurol. 2007;11:208–214. doi: 10.1016/j.ejpn.2006.12.008. [DOI] [PubMed] [Google Scholar]
- Lee HF, Tsai CR, Chi CS, Lee HJ, Chen CC. Leigh syndrome: clinical and neuroimaging follow-up. Pediatr.Neurol. 2009;40:88–93. doi: 10.1016/j.pediatrneurol.2008.09.020. [DOI] [PubMed] [Google Scholar]
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- Lodi R, Tonon C, Valentino ML, Manners D, Testa C, Malucelli E, La MC, Barboni P, Carbonelli M, Schimpf S, Wissinger B, Zeviani M, Baruzzi A, Liguori R, Barbiroli B, Carelli V. Defective mitochondrial adenosine triphosphate production in skeletal muscle from patients with dominant optic atrophy due to OPA1 mutations. Arch.Neurol. 2011;68:67–73. doi: 10.1001/archneurol.2010.228. [DOI] [PubMed] [Google Scholar]
- Lonnqvist T, Paetau A, Valanne L, Pihko H. Recessive twinkle mutations cause severe epileptic encephalopathy. Brain. 2009;132:1553–1562. doi: 10.1093/brain/awp045. [DOI] [PubMed] [Google Scholar]
- Lorenzoni PJ, Scola RH, Kay CS, Arndt RC, Freund AA, Bruck I, Santos ML, Werneck LC. MELAS: clinical features, muscle biopsy and molecular genetics. Arq Neuropsiquiatr. 2009;67:668–676. doi: 10.1590/s0004-282x2009000400018. [DOI] [PubMed] [Google Scholar]
- Lorenzoni PJ, Scola RH, Kay CS, Arndt RC, Silvado CE, Werneck LC. MERRF: Clinical features, muscle biopsy and molecular genetics in Brazilian patients. Mitochondrion. 2011;11:528–532. doi: 10.1016/j.mito.2011.01.003. [DOI] [PubMed] [Google Scholar]
- Lowell BB, Shulman GI. Mitochondrial dysfunction and type 2 diabetes. Science. 2005;307:384–387. doi: 10.1126/science.1104343. [DOI] [PubMed] [Google Scholar]
- Maj MC, MacKay N, Levandovskiy V, Addis J, Baumgartner ER, Baumgartner MR, Robinson BH, Cameron JM. Pyruvate dehydrogenase phosphatase deficiency: identification of the first mutation in two brothers and restoration of activity by protein complementation. J.Clin.Endocrinol.Metab. 2005;90:4101–4107. doi: 10.1210/jc.2005-0123. [DOI] [PubMed] [Google Scholar]
- Mancuso M, Filosto M, Bellan M, Liguori R, Montagna P, Baruzzi A, DiMauro S, Carelli V. POLG mutations causing ophthalmoplegia, sensorimotor polyneuropathy, ataxia, and deafness. Neurology. 2004;62:316–318. doi: 10.1212/wnl.62.2.316. [DOI] [PubMed] [Google Scholar]
- Massin P, Dubois-LaForgue D, Meas T, Laloi-Michelin M, Gin H, Bauduceau B, Bellanne-Chantelot C, Bertin E, Blickle JF, Bouhanick B, Cahen-Varsaux J, Casanova S, Charpentier G, Chedin P, Dupuy O, Grimaldi A, Guerci B, Kaloustian E, Lecleire-Collet A, Lorenzini F, Murat A, Narbonne H, Olivier F, Paquis-Flucklinger V, Virally M, Vincenot M, Vialettes B, Timsit J, Guillausseau PJ. Retinal and renal complications in patients with a mutation of mitochondrial DNA at position 3,243 (maternally inherited diabetes and deafness). A case-control study. Diabetologia. 2008;51:1664–1670. doi: 10.1007/s00125-008-1073-1. [DOI] [PubMed] [Google Scholar]
- Mayne LV, Lehmann AR. Failure of RNA synthesis to recover after UV irradiation: an early defect in cells from individuals with Cockayne's syndrome and xeroderma pigmentosum. Cancer Res. 1982;42:1473–1478. [PubMed] [Google Scholar]
- McKay BC, Becerril C, Ljungman M. P53 plays a protective role against UV- and cisplatin-induced apoptosis in transcription-coupled repair proficient fibroblasts. Oncogene. 2001;20:6805–6808. doi: 10.1038/sj.onc.1204901. [DOI] [PubMed] [Google Scholar]
- Mercer JR, Cheng KK, Figg N, Gorenne I, Mahmoudi M, Griffin J, Vidal-Puig A, Logan A, Murphy MP, Bennett M. DNA damage links mitochondrial dysfunction to atherosclerosis and the metabolic syndrome. Circ Res. 2010;107:1021–1031. doi: 10.1161/CIRCRESAHA.110.218966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morava E, Sengers R, ter LH, Van Den Heuvel L, Janssen A, Trijbels F, Cruysberg H, Boelen C, Smeitink J. Congenital hypertrophic cardiomyopathy, cataract, mitochondrial myopathy and defective oxidative phosphorylation in two siblings with Sengers-like syndrome. Eur.J.Pediatr. 2004;163:467–471. doi: 10.1007/s00431-004-1465-2. [DOI] [PubMed] [Google Scholar]
- Muftuoglu M, de Souza-Pinto NC, Dogan A, Aamann M, Stevnsner T, Rybanska I, Kirkali G, Dizdaroglu M, Bohr VA. Cockayne syndrome group B protein stimulates repair of formamidopyrimidines by NEIL1 DNA glycosylase. J.Biol.Chem. 2009;284:9270–9279. doi: 10.1074/jbc.M807006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance MA, Berry SA. Cockayne syndrome: review of 140 cases. Am.J.Med.Genet. 1992;42:68–84. doi: 10.1002/ajmg.1320420115. [DOI] [PubMed] [Google Scholar]
- Natale V. A comprehensive description of the severity groups in Cockayne syndrome. Am.J.Med.Genet.A. 2011;155A:1081–1095. doi: 10.1002/ajmg.a.33933. [DOI] [PubMed] [Google Scholar]
- Nikoskelainen E, Wanne O, Dahl M. Pre-excitation syndrome and Leber's hereditary optic neuroretinopathy. Lancet. 1985;1:696. doi: 10.1016/s0140-6736(85)91354-6. [DOI] [PubMed] [Google Scholar]
- Nishino I, Spinazzola A, Papadimitriou A, Hammans S, Steiner I, Hahn CD, Connolly AM, Verloes A, Guimaraes J, Maillard I, Hamano H, Donati MA, Semrad CE, Russell JA, Andreu AL, Hadjigeorgiou GM, Vu TH, Tadesse S, Nygaard TG, Nonaka I, Hirano I, Bonilla E, Rowland LP, DiMauro S, Hirano M. Mitochondrial neurogastrointestinal encephalomyopathy: an autosomal recessive disorder due to thymidine phosphorylase mutations. Ann.Neurol. 2000;47:792–800. [PubMed] [Google Scholar]
- Osenbroch PO, Auk-Emblem P, Halsne R, Strand J, Forstrom RJ, van dPI, Eide L. Accumulation of mitochondrial DNA damage and bioenergetic dysfunction in CSB defective cells. FEBS J. 2009;276:2811–2821. doi: 10.1111/j.1742-4658.2009.07004.x. [DOI] [PubMed] [Google Scholar]
- Ostergaard E, Schwartz M, Batbayli M, Christensen E, Hjalmarson O, Kollberg G, Holme E. A novel missense mutation in SUCLG1 associated with mitochondrial DNA depletion, encephalomyopathic form, with methylmalonic aciduria. Eur.J.Pediatr. 2010;169:201–205. doi: 10.1007/s00431-009-1007-z. [DOI] [PubMed] [Google Scholar]
- Pascucci B, Lemma T, Iorio E, Giovannini S, Vaz B, Iavarone I, Calcagnile A, Narciso L, Degan P, Podo F, Roginskya V, Janjic BM, Van HB, Stefanini M, Dogliotti E, D'Errico M. An altered redox balance mediates the hypersensitivity of Cockayne syndrome primary fibroblasts to oxidative stress. Aging Cell. 2012;11:520–529. doi: 10.1111/j.1474-9726.2012.00815.x. [DOI] [PubMed] [Google Scholar]
- Rahman S, Blok RB, Dahl HH, Danks DM, Kirby DM, Chow CW, Christodoulou J, Thorburn DR. Leigh syndrome: clinical features and biochemical and DNA abnormalities. Ann.Neurol. 1996;39:343–351. doi: 10.1002/ana.410390311. [DOI] [PubMed] [Google Scholar]
- Riley LG, Cooper S, Hickey P, Rudinger-Thirion J, McKenzie M, Compton A, Lim SC, Thorburn D, Ryan MT, Giege R, Bahlo M, Christodoulou J. Mutation of the mitochondrial tyrosyl-tRNA synthetase gene, YARS2, causes myopathy, lactic acidosis, and sideroblastic anemia--MLASA syndrome. Am.J.Hum.Genet. 2010;87:52–59. doi: 10.1016/j.ajhg.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosing HS, Hopkins LC, Wallace DC, Epstein CM, Weidenheim K. Maternally inherited mitochondrial myopathy and myoclonic epilepsy. Ann.Neurol. 1985;17:228–237. doi: 10.1002/ana.410170303. [DOI] [PubMed] [Google Scholar]
- Rotig A, Mollet J, Rio M, Munnich A. Infantile and pediatric quinone deficiency diseases. Mitochondrion. 2007;7(Suppl):S112–S121. doi: 10.1016/j.mito.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Schapira AH. Mitochondrial disease. Lancet. 2006;368:70–82. doi: 10.1016/S0140-6736(06)68970-8. [DOI] [PubMed] [Google Scholar]
- Scheibye-Knudsen M, Ramamoorthy M, Sykora P, Maynard S, Lin PC, Minor RK, Wilson DM, III, Cooper M, Spencer R, de CR, Croteau DL, Bohr VA. Cockayne syndrome group B protein prevents the accumulation of damaged mitochondria by promoting mitochondrial autophagy. J.Exp.Med. 2012;209:855–869. doi: 10.1084/jem.20111721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schriner SE, Linford NJ, Martin GM, Treuting P, Ogburn CE, Emond M, Coskun PE, Ladiges W, Wolf N, Van RH, Wallace DC, Rabinovitch PS. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science. 2005;308:1909–1911. doi: 10.1126/science.1106653. [DOI] [PubMed] [Google Scholar]
- Schulte C, Synofzik M, Gasser T, Schols L. Ataxia with ophthalmoplegia or sensory neuropathy is frequently caused by POLG mutations. Neurology. 2009;73:898–900. doi: 10.1212/WNL.0b013e3181b78488. [DOI] [PubMed] [Google Scholar]
- Schweers RL, Zhang J, Randall MS, Loyd MR, Li W, Dorsey FC, Kundu M, Opferman JT, Cleveland JL, Miller JL, Ney PA. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc.Natl.Acad.Sci.U.S.A. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby CP, Sancar A. Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc.Natl.Acad.Sci.U.S.A. 1997;94:11205–11209. doi: 10.1073/pnas.94.21.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KK, Sigala B, Sikder HA, Schwimmer C. Inactivation of Saccharomyces cerevisiae OGG1 DNA repair gene leads to an increased frequency of mitochondrial mutants. Nucleic Acids Res. 2001;29:1381–1388. doi: 10.1093/nar/29.6.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinazzola A, Santer R, Akman OH, Tsiakas K, Schaefer H, Ding X, Karadimas CL, Shanske S, Ganesh J, Di MS, Zeviani M. Hepatocerebral form of mitochondrial DNA depletion syndrome: novel MPV17 mutations. Arch.Neurol. 2008;65:1108–1113. doi: 10.1001/archneur.65.8.1108. [DOI] [PubMed] [Google Scholar]
- Stevnsner T, Muftuoglu M, Aamann MD, Bohr VA. The role of Cockayne Syndrome group B (CSB) protein in base excision repair and aging. Mech.Ageing Dev. 2008;129:441–448. doi: 10.1016/j.mad.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevnsner T, Nyaga S, de Souza-Pinto NC, van der Horst GT, Gorgels TG, Hogue BA, Thorslund T, Bohr VA. Mitochondrial repair of 8-oxoguanine is deficient in Cockayne syndrome group B. Oncogene. 2002;21:8675–8682. doi: 10.1038/sj.onc.1205994. [DOI] [PubMed] [Google Scholar]
- Taanman JW, Kateeb I, Muntau AC, Jaksch M, Cohen N, Mandel H. A novel mutation in the deoxyguanosine kinase gene causing depletion of mitochondrial DNA. Ann.Neurol. 2002;52:237–239. doi: 10.1002/ana.10247. [DOI] [PubMed] [Google Scholar]
- Tein I, Elpeleg O, Ben-Zeev B, Korman SH, Lossos A, Lev D, Lerman-Sagie T, Leshinsky-Silver E, Vockley J, Berry GT, Lamhonwah AM, Matern D, Roe CR, Gregersen N. Short-chain acyl-CoA dehydrogenase gene mutation (c.319C>T) presents with clinical heterogeneity and is candidate founder mutation in individuals of Ashkenazi Jewish origin. Mol.Genet.Metab. 2008;93:179–189. doi: 10.1016/j.ymgme.2007.09.021. [DOI] [PubMed] [Google Scholar]
- Thiebaut M, Colin J, Neil H, Jacquier A, Seraphin B, Lacroute F, Libri D. Futile cycle of transcription initiation and termination modulates the response to nucleotide shortage in S. cerevisiae. Mol.Cell. 2008;31:671–682. doi: 10.1016/j.molcel.2008.08.010. [DOI] [PubMed] [Google Scholar]
- Thorslund T, von KC, Harrigan JA, Indig FE, Christiansen M, Stevnsner T, Bohr VA. Cooperation of the Cockayne syndrome group B protein and poly(ADP-ribose) polymerase 1 in the response to oxidative stress. Mol.Cell Biol. 2005;25:7625–7636. doi: 10.1128/MCB.25.17.7625-7636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapp C, McCullough AK, Epe B. The basal levels of 8-oxoG and other oxidative modifications in intact mitochondrial DNA are low even in repair-deficient (Ogg1(-/-)/Csb(-/-)) mice. Mutat.Res. 2007;625:155–163. doi: 10.1016/j.mrfmmm.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly Y, Gidlof S, Oldfors A, Wibom R, Tornell J, Jacobs HT, Larsson NG. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- Tuo J, Chen C, Zeng X, Christiansen M, Bohr VA. Functional crosstalk between hOgg1 and the helicase domain of Cockayne syndrome group B protein. DNA Repair (Amst) 2002;1:913–927. doi: 10.1016/s1568-7864(02)00116-7. [DOI] [PubMed] [Google Scholar]
- Tyni T, Palotie A, Viinikka L, Valanne L, Salo MK, von DU, Jackson S, Wanders R, Venizelos N, Pihko H. Long-chain 3-hydroxyacyl-coenzyme A dehydrogenase deficiency with the G1528C mutation: clinical presentation of thirteen patients. J.Pediatr. 1997;130:67–76. doi: 10.1016/s0022-3476(97)70312-3. [DOI] [PubMed] [Google Scholar]
- Valentin-Vega YA, Maclean KH, Tait-Mulder J, Milasta S, Steeves M, Dorsey FC, Cleveland JL, Green DR, Kastan MB. Mitochondrial dysfunction in ataxia-telangiectasia. Blood. 2012;119:1490–1500. doi: 10.1182/blood-2011-08-373639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartanian V, Lowell B, Minko IG, Wood TG, Ceci JD, George S, Ballinger SW, Corless CL, McCullough AK, Lloyd RS. The metabolic syndrome resulting from a knockout of the NEIL1 DNA glycosylase. Proc.Natl.Acad.Sci.U.S.A. 2006;103:1864–1869. doi: 10.1073/pnas.0507444103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterthun S, Ferrari G, He L, Taylor RW, Zeviani M, Turnbull DM, Engelsen BA, Moen G, Bindoff LA. Autosomal recessive mitochondrial ataxic syndrome due to mitochondrial polymerase gamma mutations. Neurology. 2005;64:1204–1208. doi: 10.1212/01.WNL.0000156516.77696.5A. [DOI] [PubMed] [Google Scholar]
- Wong HK, Muftuoglu M, Beck G, Imam SZ, Bohr VA, Wilson DM., III Cockayne syndrome B protein stimulates apurinic endonuclease 1 activity and protects against agents that introduce base excision repair intermediates. Nucleic Acids Res. 2007;35:4103–4113. doi: 10.1093/nar/gkm404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A, Fan HY, Liao D, Bailey AD, Weiner AM. Activation of p53 or loss of the Cockayne syndrome group B repair protein causes metaphase fragility of human U1, U2, and 5S genes. Mol.Cell. 2000;5:801–810. doi: 10.1016/s1097-2765(00)80320-2. [DOI] [PubMed] [Google Scholar]
- Yu-Wai-Man P, Griffiths PG, Gorman GS, Lourenco CM, Wright AF, Auer-Grumbach M, Toscano A, Musumeci O, Valentino ML, Caporali L, Lamperti C, Tallaksen CM, Duffey P, Miller J, Whittaker RG, Baker MR, Jackson MJ, Clarke MP, Dhillon B, Czermin B, Stewart JD, Hudson G, Reynier P, Bonneau D, Marques W, Jr, Lenaers G, McFarland R, Taylor RW, Turnbull DM, Votruba M, Zeviani M, Carelli V, Bindoff LA, Horvath R, Amati-Bonneau P, Chinnery PF. Multi-system neurological disease is common in patients with OPA1 mutations. Brain. 2010;133:771–786. doi: 10.1093/brain/awq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Feng W, Imhof A, Grummt I, Zhou Y. Activation of RNA polymerase I transcription by cockayne syndrome group B protein and histone methyltransferase G9a. Mol.Cell. 2007;27:585–595. doi: 10.1016/j.molcel.2007.06.021. [DOI] [PubMed] [Google Scholar]
- Zelnik N, Axelrod FB, Leshinsky E, Griebel ML, Kolodny EH. Mitochondrial encephalomyopathies presenting with features of autonomic and visceral dysfunction. Pediatr.Neurol. 1996;14:251–254. doi: 10.1016/0887-8994(96)00046-x. [DOI] [PubMed] [Google Scholar]