Abstract
Integral membrane proteins reside within the bilayer membranes that surround cells and organelles, playing critical roles in movement of molecules across them and the transduction of energy and signals. While their extreme amphipathicity presents technical challenges, biological mass spectrometry has been applied to all aspects of membrane protein chemistry and biology, including analysis of primary, secondary, tertiary and quaternary structure, as well as the dynamics that accompany functional cycles and catalysis.
Keywords: Mass spectrometry, bottom-up proteomics, shotgun proteomics, top-down proteomics, electrospray-ionization, fast-photochemical oxidation of proteins (FPOP), Fourier-transform, ion-cyclotron resonance, human proteome project, lipid
1. Introduction to membrane protein mass spectrometry and proteomics
Proteomics aims to measure all the protein components of a cell with sufficient detail to accurately follow cellular function1. Membranes are critical components of all cells where they mediate the interaction of the contents of a cell or an organelle with its environment. Up to one third of the mass of biological lipid bilayer membranes is made up of integral membrane proteins (IMPs) that reside within the bilayer and peripheral proteins that bind to its surface. IMPs constitute at least one quarter of the proteome2, yet account for a larger proportion of current drug targets, reflecting their critical roles in controlling the flux of molecules, energy and information across membranes3. Mass spectrometry based proteomics technologies have generally lagged with respect to coverage of the membrane proteome due to their bias toward soluble hydrophilic peptides that are easily recovered during sample processing and chromatography, and that ionize and dissociate well during mass spectrometry. IMPs are highly amphipathic with substantial hydrophobic regions and sometimes limited soluble domains such that they yield relatively few, friendly, ‘proteotypic’ peptides for convenient protein identification. Analytical technologies that better target and improve coverage of IMPs and their transmembrane domains are under development and exciting advances have been made in the last few years.
Biological mass spectrometry continues to mature and new developments in hardware and electronics, as well as software, are driving instrument performance to previously unimagined extremes of speed, sensitivity and resolution. Such advances are practically worthless without careful sample preparation, and progress in the analysis of IMPs relies critically on this aspect of the workflow for both top-down approaches where proteins are analyzed intact, and bottom-up or shotgun approaches that digest the proteome into a complex peptide mixture. In this review article, important partnerships of technology and sample preparation will be emphasized as they yield new insights into the structure and function of the bilayer proteome.
There are two main structural classes of IMPs whose primary sequences must be folded to match the surrounding bilayer properties (Figure 1). The most common motif is the transmembrane alpha-helix that is often multiplexed to generate the polyhelix bundle such as that shown for bacteriorhodopsin, an archael protein that uses its bound chromophore to absorb light energy and drive vectorial proton translocation. Since the alpha-helix is constructed from a continuous sequence, it is typical that 20–30 apolar amino-acid residues span the membrane in a single transmembrane helix (TMH), generating a hydrophobic stretch of primary structure. The other, less common motif is the transmembrane beta-barrel, as illustrated with the outer mitochondrial membrane voltage-dependent anion channel (VDAC). In outer membrane pore structures the beta-barrel alternates hydrophobic side-chains toward the interior of the membrane and more polar side-chains toward the hydrated center of the pore. Disruption of the bilayer using detergents or organic solvents is essential to extract intact IMPs for analysis.
Figure 1.
Structural motifs of integral membrane proteins. Integral membrane proteins constitute a significant component of biological membranes and have domains that span the lipid bilayer. The most common motif is the transmembrane alpha-helix that is often multiplexed generating a polyhelix bundle such as that shown for bacteriorhodopsin (A). Since the alpha-helix is constructed from a continuous sequence, it is typical that 20–30 apolar amino-acid residues span the membrane, generating a hydrophobic stretch of primary structure – the red spirals that dominate the figure. The other common motif is the transmembrane beta-barrel, as illustrated with murine VDAC1 (B). The beta structure alternates hydrophobic side-chains toward the interior of the membrane and more polar side-chains toward the hydrated center of the pore. Disruption of the bilayer using detergents or organic solvents is necessary to extract whole integral membrane proteins for analysis while surface ‘shaving’ proteolysis experiments can be used to release peptides for mass spectrometry. The VDAC1 structure was reproduced with permission, Copyright (2008) National Academy of Sciences, USA.
There are many aspects of the structure/function relationships of IMPs that remain poorly understood. Deviations from the general structural motifs described above, and the problem of folding and insertion into the membrane are important current foci of research4,5. Structural dynamics related to functional cycles is an area where mass spectrometry can make spectacular contributions beyond the static structures seen in crystals (see section 7, for example). Finally, the role of structural lipids in modulation of structure/function, and the related but distinct role of general bilayer properties including fluidity, curvature and lateral pressure present a substantial challenge.
2. Current analytical challenges in membrane protein research
It is important to understand how the structure of IMPs determines their function within and beyond the membrane bilayer, and how small molecules, proteins and other cofactors, and the membrane itself modulates their function. Extending such knowledge into mammalian membrane systems will enable better understanding of the mechanism of action of existing pharmaceutical compounds and empower the conception and development of new ones. The basis of understanding structure and function of proteins is a high-resolution atomic structure, usually obtained by X-ray crystallography or NMR, and while a significant number of IMPs have yielded to these analyses, these structures have predominantly been of abundant proteins from relatively simple membrane systems. The combination of structural determination and other biophysical measurements has enabled a detailed understanding of many of the essential processes associated with membranes6, including the mechanism of selective ion transmission by the Na+-coupled transporters7; signal transduction by the G-protein-coupled receptors8; energy conversion in photosynthetic reaction centers for light-driven water-splitting9; energy transduction from transmembrane electrochemical gradients to chemical energy by the rotary ATP synthases10,11,12; and the mechanisms used by IMPs to perform hydrolytic reactions within the bilayer13.
Even armed with a crystal structure there are still years of work required toward understanding functional dynamics and their modulation by ligands, agonists, antagonists and other modulators. Mass spectrometry is playing a central role in these analyses and as a high-throughput proteomics technology will accelerate understanding of membrane protein structure/function relationships. Design and development of experiments to characterize conformational dynamics related to binding events, and complications due to target heterogeneity, and to accommodate larger mammalian receptors and channels from the molecular weight 50 kD class up to and beyond 100 kD will challenge the field in the coming years. Ultimately we seek the ability to rationally design new IMPs to do our bidding. An essential facet of this endeavor will be computational simulation of IMP structure and function. Molecular dynamics studies are now becoming part of the strategy toward understanding IMP structure/function and recently yielded insights into the role of TMH design and oligomerization14 and movement of cations through voltage sensor domains15.
3. Current status of bottom up membrane proteomics
The deficiencies of standard bottom-up or shotgun proteomics protocols toward integral membrane proteins have been acknowledged for some time and a number of improvements have emerged, including advances in sample preparation techniques as well as refinement of the technology employed for their analysis16. Firstly it was recognized that many membrane proteins were insoluble in the conditions used for tryptic digestion and that proteolysis could further lead to precipitation of hydrophobic domains. Inclusion of organic solvent during digestion provided dramatic enhancement of integral membrane protein recovery with Blonder and colleagues using 60% methanol to raise the percentage of membrane proteins detected in their shotgun analysis to over 30% which is approaching the proportion expected for this class of the proteome17. More recently, the benefits of using perfluoro-octanoic acid were reported18. Due to concerns about loss of specificity and activity, several studies have used detergents as an alternative to organic solvents with some success though it is noted that comparisons have typically favored organic solvents19,20. The added complication of needing to remove detergent from the sample prior to analysis has been largely overcome with development of acid-labile surfactants that are hydrolyzed prior to chromatography21 though there has been discussion of problems of column longevity in relation to the surfactant cleavage products. Masuda and colleagues recently introduced the idea of phase-transfer surfactants and demonstrated that deoxycholate was effective for protein digestion and removable via partition to an immiscible solvent such as ethyl acetate22. Spin filters have been used for effective purification of peptides for proteomics23 and a protocol that combines protein extraction in SDS prior to addition of urea and filtration to fully remove dodecyl sulfate prior to digestion has found general applicability and works well for membrane proteins (FASP24). Analysis of phosphorylation sites on brain proteins yielded 4579 proteins of which 23% were derived from plasma membranes. The sites of modification provided useful topology information for a number of channels and transporters25.
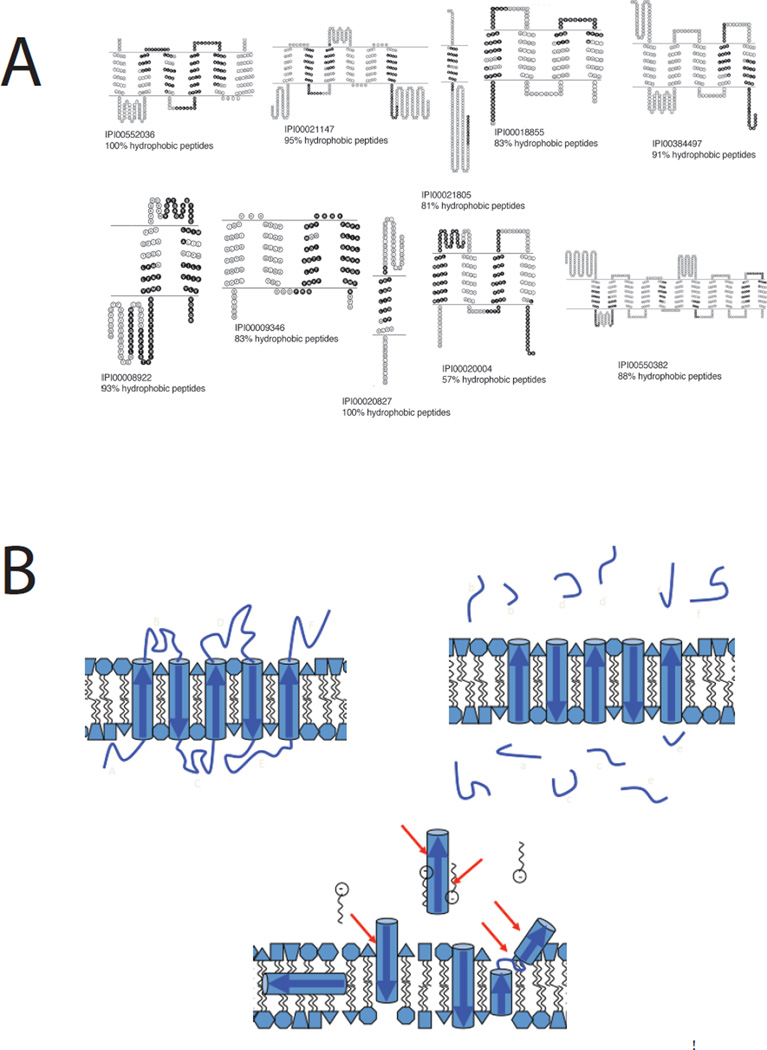
An alternative strategy that has been fruitful involved the use of the alternative digestion enzyme Thermolysin, combined with a high pH that tightens the specificity of hydrolysis and favorably modifies the membrane properties26. This approach has been modified to provide topological information by comparing results obtained for intact membrane vesicles versus broken membranes with both leaflet surfaces exposed27 (Figure 2A). However, the assumption that membrane protein structures do not react to loop proteolysis may not always be valid (Figure 2B). Wu’s group has also made the important observation that recovery of hydrophobic peptides derived from transmembrane domains of IMPs is dramatically enhanced by performing chromatographic separations at 60 °C28. A third strategy that has performed well for coverage of IMPs takes advantage of the fact that they are amenable to analysis by SDS-PAGE. In an experiment nicknamed ‘GeLC-MS’, the whole lane from an SDS-PAGE separation is sliced into small pieces and each analyzed by shotgun technology after ‘in-gel’ trypsin digestion29,30. This approach has been extended toward an exhaustive analysis of the membrane proteome by using the gel separation AND two-dimensional chromatography in a three-dimensional experiment with multiple repeats that the authors acknowledge is a ‘trade-off in heavy instrument time’31. Despite significant progress, sequence coverage of integral membrane proteins remains patchy and there are some transmembrane regions that remain refractory to detection and analysis. Strategies to improve coverage of transmembrane domains include use of CNBr to cleave at Met residues commonly found in bilayer domains thereby making the hydrophobic peptides that are released smaller and more forgiving32. Similarly, others have used mild oxidation with performic acid to oxidize Cys and Met residues in transmembrane domains making them more hydrophilic33. Nevertheless, the only way to be sure that complete coverage is achieved is by working with the intact protein molecule and analyzing it using the top-down approach.
Figure 2.
Peptide recovery in shotgun proteomics. Sophisticated protocols are dramatically improving representation of integral membrane proteins in shot-gun proteomics experiments. A. After shaving soluble loop regions with proteinase K at high pH to release protease-accessible peptides (PAPs) and subsequent cleavage of membrane-embedded polypeptides (MEPs) with Met-directed CNBr both fractions are analyzed by MuDPIT at elevated column temperature. The black shaded polypeptide regions recovered from the MEP fraction resulted in many protein identifications not seen in the PAP fraction (57% of total) and raised the proportion of integral membrane proteins recovered to 38% of the total, in line with the number predicted by informatics approaches. Note that despite landmark advances in proteolysis efficiency and peptide recovery, sequence coverage within the transmembrane domains remains sporadic. Reproduced from reference 27. B. A schematic shows how proteolytic shaving works under idealistic conditions, releasing loop regions for mass spectrometric analysis (upper right). The lower image represents what may happen during a lengthy proteolysis experiment as membrane protein structures react to early cleavage events, emphasizing that interpretation of such experiments should proceed with caution.
4. Post-translational modification of integral membrane proteins
While bottom-up proteomics is closing in on full proteome coverage with respect to identifying the expressed gene products, the comprehensive analysis of post-translational modifications is more challenging. In these experiments the rate of false positive protein identification can be driven close to zero by the requirement that two unique peptides per protein be observed. However, this criterium cannot be applied for localization of modification sites within single peptides. Firm identification based upon a single tandem mass spectrum has benefited greatly from the introduction of hybrid ion-trap instruments with high-resolution Fourier-transform detectors (LTQ FT and Orbitrap) thereby allowing experiments that preserve the fast duty cycle of the ion-trap enabling multiple MSMS experiments during the time it takes for an accurate mass measurement on precursor ions (the ‘high-low’ MSMS strategy). Putative peptides and modified versions of them can be filtered with very narrow precursor ion mass tolerances (0.3 – 5 ppm) lowering occurrence of false positives. Dedicated software filters34 are also used to improve confidence of modified peptide identification and modification site mapping.
There is little doubt that high-resolution mass measurement of product ions will improve the confidence of primary structure assignment and thus localization of post-translational modifications especially during top-down mass spectrometry. High-resolution product-ion measurements (the ‘high-high’ MSMS strategy) in bottom-up proteomics bring improved accuracy to characterization of post-translational modifications at the expense of duty cycle. As the orbitrap analyzer and associated ion-optics and electronics develop and improve, it became possible to compare the performance of optimized instrument methods for ‘high-high’ and ‘high-low’ MSMS strategies35,36. Despite a longer duty cycle, the ‘high-high’ approach gave comparable performance as assessed by detection of phosphopeptides. Work from other groups is providing more evidence to support this observation37,38, and the latest quadrupole-orbitrap mass filter/analyzer39 (Q-Exactive) lacks the low-resolution option of the ion-trap analyzer. Current quadrupole time-of-flight (Q-tof) instruments offer ‘high-high’ MSMS performance but have not yet realized optimal sensitivity. Whether any of the software available for protein identification fully capitalizes on accurate precursor and product ion masses remains an open question40, but serious attempts to address this challenge are being made41.
While not always acknowledged, the importance of careful sample preparation for accurate measurements of post-translational modifications is frequently emphasized by leaders in the field42,43. The problem is the activity of enzymes that can modify post-translational modification status during the period after biological treatment and before analysis; the action of phosphatases on protein phosphorylations, for example. Thus protocols incorporate multiple strategies aimed at rapid inactivation of enzyme activity through denaturation and inhibition. For cells and tissues the most advanced protocols use denaturants such as 8 M urea combined with heat treatment25,44, while a recent protocol that examines the preservation of natural peptide hormones in blood relies upon reduced temperatures, acidification, protease inhibition, isotopic exogenous controls, and dilution45 (RAPID), emphasizing that different systems often require their own dedicated protocols. Examination of post-translational modification status of IMPs using bottom-up methods has the added demand that the proteins remain soluble during trypsin treatment, yet become detergent free for analysis. The FASP protocol, mentioned above, is attractive due to use of high concentrations of SDS that both solubilizes IMPs and effectively denatures enzymes, and then uses 8 M urea to displace bound dodecyl sulfate. When used to study the brain phosphoproteome, 23% of the phosphorylation sites were found on membrane proteins including a large number of ion channels and transporters25.
While bottom-up approaches can detect some peptides that carry more than a single modification, the vast majority of the information concerning number of modifications per protein comes from singly modified peptides. However, it would be naive to consider individual PTMs as independent binary switches, while in many cases a much more sophisticated system logic is achieved through combinations of PTMs in concert with the tertiary/quaternary structure of individual proteins and their binding partners. Thus, bottom-up approaches that reduce the system to peptides render complex behavior invisible. The best example of this to date is the extensive modification of the histone family of proteins46,47. While information encoded in combinatorial sets of PTMs can be preserved by working with intact proteins, highly modified proteins such as histones present incredibly complex mixtures with many isobaric species. Even the most powerful high-resolution mass spectrometers are limited with respect to how well they can fractionate and quantify such species in the gas phase, demanding high-quality separations prior to MS. In practice, this is often achieved using a middle-down approach whereby intact proteins are cleaved into smaller pieces, in the range 4 – 15 kD, that can be chromatographically separated and yield close to full sequence coverage in dissociation experiments. Histones are cleaved with the endopeptidase Glu-C to yield an N-terminal fragment (5 kD) that can be optimally separated by a novel online hydrophilic interaction chromatography (HILIC) strategy identifying hundreds of differentially modified forms of the target polypeptide48. Many groups acknowledge that no single experiment can adequately cope with such complex mixtures and frequently resort to multiple approaches that mix top-down, middle-down and bottom-up experiments49,50. Whether members of the integral membrane proteome exhibit the same combinatorial complexity as that observed for histone modification remains to be determined. A recent study of phosphorylation of the mammalian beta-adrenergic receptor revealed thirteen sites whose phosphorylation is under control of multiple kinases and modulated by bound ligands, resulting in patterns that dictate the conformation, and thus function, of bound arrestin protein cofactors51.
5. Intact mass measurements and top-down high-resolution mass spectrometry of integral membrane proteins
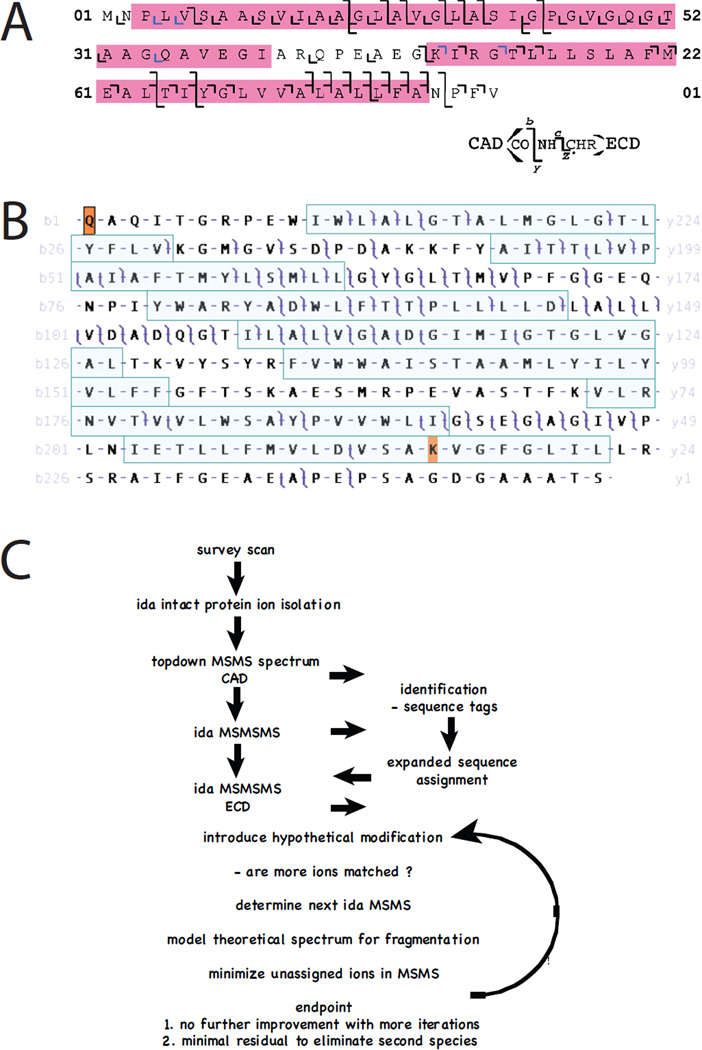
Top-down mass spectrometry seeks to combine an intact protein mass measurement with a dissociation spectrum to define the primary covalent structure in a single experiment. Dissociation of intact proteins after electrospray-ionization was demonstrated by Smith and coworkers52 with subsequent identification of a protein using information from a top-down experiment53. Kelleher and McLafferty presented and named the concept ‘top-down’ in the context of high-resolution Fourier-transform ion cyclotron resonance mass spectrometry, arguing that successful application would require the high mass accuracy afforded by these analyzers54. High-resolution FT-MS was first performed on proteins in the late ‘80s55. Application of top-down mass spectrometry to IMPs required development of chromatographic solvent systems compatible with their solubility and the electrospray ionization process. Both reverse-phase and size-exclusion chromatographies have been described enabling routine electrospray analysis of a wide variety of IMPs ranging from photosynthetic reaction-center polypeptides to G-protein coupled receptors such as rhodopsin56–66 achieving mass accuracy similar to that obtainable for water-soluble proteins − 100 ppm on low resolution quadrupole analyzers. Top-down analyses were applied to smaller IMPs first using low- and then intermediate-resolution Q-tof analyzers allowing charge-state assignment on product ions62,67,68. The mass of the intact seven transmembrane protein bacteriorhodopsin apoprotein was first measured using high-resolution FT-MS achieving mass accuracy within 8 ppm69, though top-down collisionally activated dissociation experiments performed at the time did not yield comparable product ion mass accuracy. Routine 5 ppm mass accuracy on both precursor and product ions was achieved with the introduction of hybrid linear ion trap Fourier-transform ion cyclotron resonance instrumentation and a preliminary top-down experiment on the bacteriorhodopsin holoprotein confirmed all known post-translational modifications including the removal of the C-terminal Asp262 residue70. A detailed top-down analysis of the two transmembrane helix c-subunit of the ATP synthase achieved extensive sequence coverage through the transmembrane domains. Comparison of collisionally activated dissociation (CAD) with electron-capture dissociation (ECD) showed that effective ECD was only achieved when the protein ions were excited using an IR laser prior to ECD71 (activated-ion ECD; aiECD; Figure 3A) achieving more extensive coverage than CAD. By combining CAD product-ion data from six different precursors and the extended m/z range feature of the hybrid instrument, comprehensive coverage of the bacteriorhodopsin holoprotein with full characterization of its post-translational modifications was achieved72 (Figure 3B). This study also demonstarted that mammalian IMPs of the 30 kD porin class such as VDAC are amenable to top-down analysis. Continuing work has demonstrated that 50 kD polytopic chloride channels are perfectly amenable to top-down analysis73 provided that homogenous preparations are available. By ‘peak parking’ an online size-exclusion chromatography experiment we have extended high-resolution data collection to average more than 1000 transients, yielding maximal signal to noise. While sequence coverage in CAD experiments was somewhat limited, the ability to localize photoaffinity-labeling sites was instrumental for interpretation of structure/function data.
Figure 3.
Top-down mass spectrometry of integral membrane proteins. A. Sequence coverage within the transmembrane domains of the c-subunit of chloroplast ATP synthase was improved when activated-ion electron-capture dissociation (aiECD) was compared with collisionally activcated dissociation (CAD). c and z product ions colored blue were assigned manually. Reproduced from reference 71. B. Multiply charged ions generated by electrospray-ionization of intact bacteriorhodopsin were subjected to collisionally activated dissociation (CAD) and product ions analyzed with a high-resolution FT-ICR mass spectrometer. Masses of precursor and product ions were matched at 10 ppm tolerance to the known sequence of the protein with post-translational removal of residues 1 – 13, N-terminal cyclization to form pyrrolidone carboxylic acid, removal of Asp262, and modification of Lys229 with N6-(retinylidene)lysine. Note that many product ions yield overlapping b- and y- ions such that the entire sequence is covered. While some transmembrane helices (shaded) are accessible to CAD, other regions are less so, with their primary structures inferred based upon the genomic translation and the numerous precursor and product ions that span them. Reproduced with permission from Mol. Cell. Proteomics 2010, 9, 791–803, copyright ASBMB. C. In the future data will be interpreted ‘on the fly’ so that experiments are controled in real time to improve coverage across primary sequence and post-translational modifications using MS3 strategies.
High-throughput, top-down proteomics of membrane fractions will require significant developments in separations technologies, possibly involving two dimensional chromatography68, or multi-dimensional mixed electrophoresis/chromatography systems74. Membrane proteins were detected in the latter analysis that identified over 1000 total proteins in ‘discovery’ mode, though it should be noted that classical features of traditional top-down analyses were compromised, resulting in some identifications without intact mass measurements, and a false discovery rate of 5%. Experimental strategies that preserve the high fidelity of the ‘traditional’ top-down approach while enabling high throughput are under development (Figure 3C). The schema presented uses static nanospray at very low flow rate to allow time for transient averaging on the precursor ion, allowing accurate mass of the intact protein to be obtained, and subsequent transient averaging during dissociation experiments with on-the-fly data interpretation to direct choice of product ions for further dissociation analyses (MS3). In this way data is accumulated until sufficient information defines the primary structure including precise localization of post-translational modifications. Technological advances to implement such a strategy include the need to perform multiple ‘fills’ (ion accumulation) prior to MS3 and the development of hardware/software for real-time data interpretation and direction of dissociation experiments.
6. Native mass spectrometry of integral membrane protein complexes
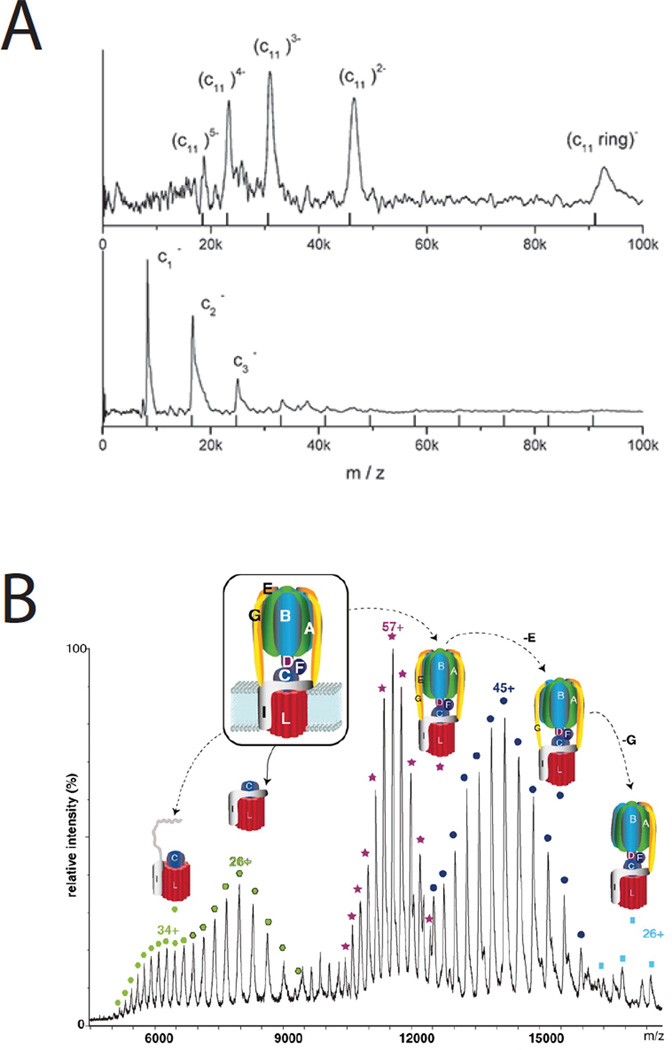
Routine analysis of soluble non-covalent protein complexes has been proceeding in a number of laboratories for some years75–77 while progress with IMP complexes has lagged considerably behind. Fortunately, a number of recent studies have established the feasibility of analyzing quaternary structure of intact membrane protein complexes. In 2004 the bacterial multi-drug resistance transporter EmrE was analyzed by native positive ion ESI-MS78. The analytical challenge is that aqueous solubility of IMP complexes is achieved through coating of the hydrophobic surfaces of the complex with a detergent micelle, so that the resulting ions have a heterogenous mass distribution due to variability in the number of detergent molecules of the micelle. Ilag and coworkers used collisional activation to displace detergent from the micelle enabling native mass spectrometry of the dimeric EmrE protein and demonstration of tetraphenyl phosphonium ligand binding in vacuo. Robinson and coworkers have continued this work and subsequently reported analysis of intact ABC-transporters BtuC2D279 and MacB80, in both cases determining subunit stoichiometry. An alternative strategy for native MS of IMP complexes was used by Brutschy’s group involving a novel methodology called laser induced liquid beam (bead) desorption and ionization (LILBID). Laser excitation tuned to the O-H stretch vibration frequency of water (wavelength = 3 µm) is used to softly ionize complexes from solution phase droplets81,82 for time-of-flight mass spectrometry. Interestingly, LILBID generates stronger negative ion currents than positive currents reflecting the overall charge state of the complex at the neutral pH of the buffered suspension. Increasing laser fluence results in disruption of non-covalent associations so that masses of individual subunits can be determined. LILBID was first used to characterize 2- and 4-subunit cytochrome oxidase and 4-subunit cytochrome bc1 complexes from P. denitrificans83, and to measure ATP synthase c-ring stoichiometry in a number of prokaryotes, providing direct evidence for complexes of 11, 13 and 15 subunits in different species84 (Figure 4A). More recently LILBID was used to analyze mitochondrial inner membrane complex 1 from the yeast Yarrowia85. Under the softest ionization conditions the intact complex measured 980 kDa. After more energetic ionization each of the forty subunits could be assigned by their mass ranging from 6.8 – 75.2 kDa with some subunits requiring modified conditions for detection including both positive and negative ion measurements. The authors also demonstrated the modest sample requirements of LILBID using complex 1 eluted from a blue native gel85. The ability to integrate LILBID into the blue native gel work flow is of great significance to membrane proteomics because of the demonstrated ability of the non-denaturing gel to separate intact integral membrane complexes after mild detergent solubilization of the bilayer. Subunit spectra were somewhat shifted to higher masses suggesting there might be advantages to non-denaturing gel systems that do not include Coomassie Blue dye. The Robinson group has now used positive-ion ESI-MS for measurements of intact ATP synthase and c-ring stoichiometry, emphasizing the importance of specifically bound lipid molecules within the structural complex, and the utility of gas-phase ion mobility to detect subtle conformational changes of functional significance86 (Figure 4B). Mass spectrometers that combine the design features necessary for ionization and desolvation of intact non-covalent complexes with gas-phase ion mobility separations, and the ability to isolate ions and perform top-down dissociation experiments are highly attractive for this work, and will result in a much broader accessibility to the types of experiments described above. Progress in native ESI-MS of IMP complexes has been reviewed87,88.
Figure 4.
Native mass spectrometry of integral membrane protein complexes. There has been long standing interest in the number of c-subunits that make up the ring component of Fo of the ATP synthase. A. The soft ionization technique laser-induced liquid bead ion desorption-MS (LILBID) was used to directly measure c-ring stoichiometry in a number of prokaryotic complexes including Clostridium paradoxum with 11 subunits - the upper panel used minimal laser excitation to retain the intact native complex, while the lower panel uses higher excitation shifting the spectrum toward the monomer. The mass spectra were recorded using a time-of-flight analyzer operated in negative ion mode. Reproduced with permission from reference 83. B. Positive-ion electrospray-ionization was used to measure the m/z of intact ATP-synthase complexes, and sub-complexes where various subunits had been displaced in the gas phase. The complex shown is from Enterococcus hirae and has a measured c-ring stoichiometry of 12. Six molecules of the phospholipid phosphatidylethanolamine were bound to each c-ring. Reproduced from reference 86. Printed with permission from AAAS.
7. Membrane protein dynamics
The overwhelming majority of our understanding of integral membrane protein structure-function relationships has come from high-resolution chrystallographic analyses that provide full description of secondary and tertiary backbone and sidechain configurations6. Details of functional cycles are subsequently modeled onto static structures based on a wide variety of biophysical measurements, and in some cases resolved structures of proteins at different stages of their catalytic cycles. Mass spectrometry can provide numerous structural insights that are highly complementary to crystal structures, and a variety of experiments will play an increasingly significant role in elucidating details of structure-function and, most excitingly, residue-specific dynamics of IMPs in action.
The first strategy to yield structural information at the proteomic scale was the previously mentioned study that used proteinase K treatment at high pH to ‘shave’ surface exposed loops for shotgun proteomics. By working with intact membrane vesicles, and subsequently ruptured vesicles it was possible to correlate specific peptides with orientation in the bilayer26. The disadvantage of a proteolysis experiment is that subsequent cleavage events may arise due to changes in accessibility resulting from changes in conformation in response to earlier cleavage events (Figure 2B). More recently, sites of post-translational modifications provided useful topology information for a number of channels and transporters25, with the advantage that the modifications were made prior to the analysis and thus should be faithful to solvent exposure in vivo.
Many other studies targeting single proteins illustrate the power of mass spectrometry to impact studies of membrane protein function. Carbodiimide reagents that target protonated carboxylates were used to study the relationship between Asp/Glu side chains of a sugar-proton symporter protein and sugar substrate binding, leading to the conclusion that a hydrogen bond was formed between the Glu269 carboxylate side chain and the C3-OH of the substrate galactopyranosyl ring during the membrane transport cycle89. A subsequent high-resolution crystal structure of the twelve-TMH lactose permease showed this structural constraint to be correct to within 1 Å90. Bifunctional crosslinking reagents have been used to measure distance constraints across membrane protein structures, such as the seven transmembrane helix G-protein coupled receptor rhodopsin91, and to identify a cell surface receptor for extracellular vitamin A-retinol-binding protein92.
Chemical modification of a protein can lead to changes in conformation, potentially complicating structural interpretations. In order to complete the labeling reaction before the protein has a chance to respond, a highly reactive reagent must be introduced quickly. In practice this is achieved using single-shot bursts of hydroxyl radicals produced either by synchrotron X-ray degradation of water, or by UV laser degradation of hydrogen peroxide. Hydroxyl radicals oxidize nearly all protein amino-acid residues, with a range of kinetics, resulting in a permanent record of accessibility to the aqueous environment that can be easily read by bottom-up mass spectrometry. Chance and coworkers used synchrotron-generated hydroxyl radical to study the relationship between bound water molecules and the activation process in rhodopsin93,94. Fast photochemical oxidation of proteins95 (FPOP) has been applied to membrane proteins with the unexpected and unexplained result that modifications were mostly confined to the sulfur containing residues methionine and cysteine, limiting the spatial resolution usefully achieved96,97.
Use of backbone amide hydrogen-deuterium exchange (HDX) provides a complementary approach to covalent modification that emphasizes not only accessibility but also structural dynamics. The major advantage of this technique is that it allows the native structure to be analyzed, while the disadvantage lies in the labile nature of the modification, demanding rapid analysis of samples under conditions that limit back exchange. HDX was used to study model TMH peptides in bilayers98 and was first applied to IMPs to study the response of microsomal glutathione transferase 1 (MGST1) to glutathione (GSH) binding99. MGST1 is trimeric and each each subunit has a Cysteine residue (Cys49) in the N-terminal cytosolic domain that plays a central role in the protein’s function as a sensor of oxidative and chemical stress. Binding of a single GSH per trimer induces a cooperative change in conformation across the whole trimeric structure. GSH binding resulted in site-specific changes in HDX rates, accelerating or retarding exchange. Introduction of phosphatidylcholine also resulted in site-specific changes in HDX emphasizing the sensitivity of the structure to the surrounding lipid environment. IMP structure and function should not be considered independent of the surrounding bilayer environment. Individual lipids can play direct structural roles as components of protein-lipid complexes, while the bilayer itself can influence function through general features such as composition, curvature and lateral pressure100,101.
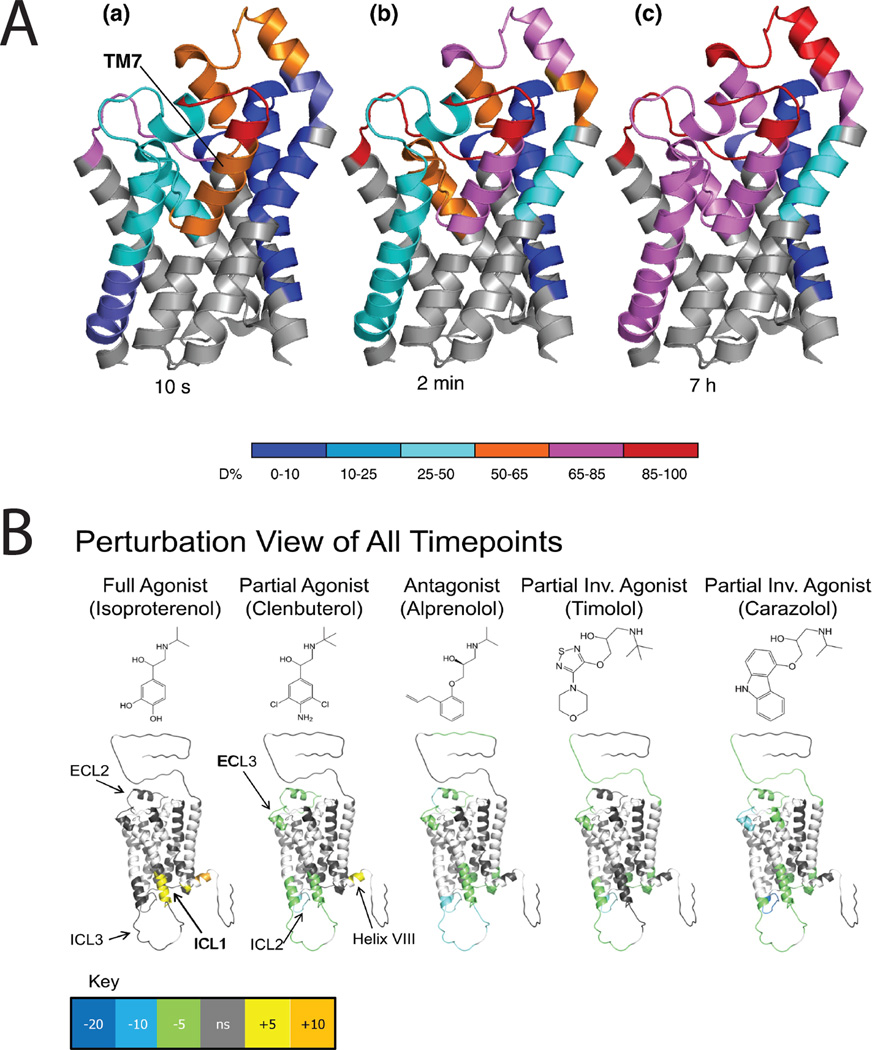
A significant structural insight to emerge from HDX analysis of IMPs is that transmembrane alpha-helices are not always rigid bodies. This was first observed for the lactose permease of E. coli when global HDX was monitored by Fourier-transform infra-red spectroscop102 (FTIR) revealing fast exchange, with 80–90 % HDX within 10–20 minutes, for the intact protein. These results have been reproduced by others, and extended, using site-specific HDX monitored by mass spectrometry, to one helix of the glycerol facilitor96 (Figure 5A). Thus regions of some polyhelix IMPs exist in a molten state where their backbone hydrogen bonding is unstable allowing rapid HDX. It is likely that such dynamic behavior is critical to their function, such that substrate binding is not stable, but induces conformational change as part of a rapid catalytic cycle, in the examples above, facilitating transmembrane transport.
Figure 5.
Hydrogen-deuterium exchange (HDX) provides insights into dynamics of integral membrane proteins. A. Transmembrane helix 7 of the glycerol facilitator (GF) is highly dynamic. Deuteration behavior of GF depicted by color-coding individual protein segments according to their deuteration levels. (a) t = 10 s. (b) t = 2 min. (c) t = 7 h. Note that exchange requires breakage of alpha-helix backbone amide H-bonds. Reprinted from J. Mol. Biol. 416(3). Pan Y, Piyadasa H, O'Neil JD, Konermann L. Conformational dynamics of a membrane transport protein probed by H/D exchange and covalent labeling: the glycerol facilitator. 400–413. Copyright 2012, with permission from Elsevier. B. HDX data overlaid onto a modified crystal structure for the beta-adrenergic receptor (β2AR). The HDX data are shown using color gradients where the differences in the average percentage deuterium uptake across all time points between the apo- and ligand-bound receptor for peptides in a given region of the receptor sequence are assigned a color as shown in the key, and then overlaid onto the structure. Here, white indicates regions of the receptor that were not resolved in the HDX experiment, and gray indicates no significant change. Ligand structures are indicated above the protein structures. Reproduced from Structure 19(10), West GM, Chien EY, Katritch V, Gatchalian J, Chalmers MJ, Stevens RC, Griffin PR. Ligand-dependent perturbation of the conformational ensemble for the GPCR β2 adrenergic receptor revealed by HDX. 1424-32. Copyright 2011, with permission from Elsevier.
G-protein coupled receptors are a class of IMPs that modulate transmembrane signaling, their importance highlighted by the recent award of the 2012 Nobel Prize in Chemistry to Lefkowitz and Kobilka. Structural dynamics of the beta-adrenergic receptor has been investigated using HDX mass spectrometry by two groups lead by Virgil Woods and Patrick Griffin103–106. Site-specific changes in the dynamics of the receptor toward agonists, antagonists and inverse agonists are now being investigated, despite challenges to sequence coverage arising from N-terminal glycosylation and C-terminal fatty acylation. As shown in Figure 5B, each different ligand introduces subtle site-specific changes in dynamics that may relate to the mode of action of these molecules in vivo. These experiments were performed on isolated protein suspended in a detergent micelle so it is not clear how location in a native bilayer membrane might affect the observed behavior. In this respect, the development of the bilayer nanodisc is attractive107. In principal similar to the bicelle108, the nanodisc uses an amphipathic protein instead of detergent to surround and solubilize a small circular lipid bilayer disc with just tens of lipid molecules providing a potentially native environment for IMPs. Nanodiscs have been used for HDX of the IMP gamma-Glutamyl carboxylase109 and have been analyzed by native MS though not yet with an embedded IMP110. Amphipathic polymers are also being investigated in this respect111.
Finally, it should be noted that while exquisite detail can be afforded through HDX or FPOP strategies, both have demanding experimental constraints and yield complex datasets that require extensive processing. Many questions can be answered with simpler experiments using site-specific reagents. For example, carboxyl group footprinting was used to map phosphorylation-induced conformational changes and the dimerization interface of a membrane bound tyrosine kinase112, and to resolve the question as to the orientation of the FMO photosynthetic antenna protein of C. tepidum113.
8. Conclusions
Huge advances have been made in the analysis of integral membrane proteins by mass spectrometry, and the membrane proteome can be fully covered provided appropriate protocols are used. The coming years will see dramatic advances in native mass spectrometry of integral membrane protein complexes, and widespread use of hydrogen-deuterium exchange and fast photochemical oxidation strategies for routine study of membrane protein dynamics through their catalytic cycles. Of paramount importance will be acknowledgement that these studies should be relevant to the behavior of the protein within the bilayer, with appropriate understanding of the role of closely bound structural lipid molecules and the more general influence of the bilayer environment in vivo.
ACKNOWLEDGMENTS
This article is dedicated to Professor James Barber FRS. Anonymous reviewers as well as Bill Cramer and Kym Faull are thanked for helpful suggestions on the manuscript. Support from NIH for instrument purchase (S10 RR023045) and research projects (GM088499) is gratefully acknowledged. The image of bacteriorhodopsin (pdbid=1py6) was modeled with the program (PPM 1.0; http://opm.phar.umich.edu) and reproduced with the permission of Andrei L. Lomize.
Biography
The author is a Professor in the NPI-Semel Institute of the David Geffen School of Medicine and directs proteomics research within the Pasarow Mass Spectrometry Laboratory. He applies biological mass spectrometry to study structure/function relationships of integral membrane proteins, protein misfolding and neurodegenerative diseases, and other systems biomedicine projects involving peptides, lipids and proteins.
REFERENCES
- 1.Whitelegge JP. Proc. Natl. Acad. Sci. U.S.A. 2002;99(18):11564–11566. doi: 10.1073/pnas.192449199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fagerberg L, Jonasson K, von Heijne G, Uhlén M, Berglund L. Proteomics. 2010;10(6):1141–1149. doi: 10.1002/pmic.200900258. [DOI] [PubMed] [Google Scholar]
- 3.Weinglass AB, Whitelegge JP, Kaback HR. Curr. Opin. Drug Discov. Devel. 2004;7(5):589–599. [PubMed] [Google Scholar]
- 4.von Heijne G. Annu. Rev. Biochem. 2011;80:157–160. doi: 10.1146/annurev-biochem-111910-091345. [DOI] [PubMed] [Google Scholar]
- 5.von Heijne G. Biochem. Soc. Trans. 2011;39(3):747–750. doi: 10.1042/BST0390747. [DOI] [PubMed] [Google Scholar]
- 6.White SH. Nature. 2009;459(7245):344–346. doi: 10.1038/nature08142. [DOI] [PubMed] [Google Scholar]
- 7.Krishnamurthy H, Piscitelli CL, Gouaux E. Nature. 2009;459(7245):347–355. doi: 10.1038/nature08143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenbaum DM, Rasmussen SG, Kobilka BK. Nature. 2009;459(7245):356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barber J. Cold Spring Harb. Symp. Quant. Biol. 2012 [Google Scholar]
- 10.Junge W, Sielaff H, Engelbrecht S. Nature. 2009;459(7245):364–370. doi: 10.1038/nature08145. [DOI] [PubMed] [Google Scholar]
- 11.Boyer PD. Mol. Cell. 2001;8(2):246–247. doi: 10.1016/s1097-2765(01)00315-x. [DOI] [PubMed] [Google Scholar]
- 12.Watt IN, Montgomery MG, Runswick MJ, Leslie AG, Walker JE. Proc. Natl. Acad. Sci. U. S. A. 2010;107(39):16823–16827. doi: 10.1073/pnas.1011099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erez E, Fass D, Bibi E. Nature. 2009;459(7245):371–378. doi: 10.1038/nature08146. [DOI] [PubMed] [Google Scholar]
- 14.Hsin J, LaPointe LM, Kazy A, Chipot C, Senes A, Schulten K. J. Am. Chem. Soc. 2011;133(35):14071–14081. doi: 10.1021/ja204869h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khalili-Araghi F, Tajkhorshid E, Roux B, Schulten K. Biophys. J. 2012;102(2):258–267. doi: 10.1016/j.bpj.2011.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilmore JM, Washburn MP. J. Proteomics. 2010;73(11):2078–2091. doi: 10.1016/j.jprot.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 17.Blonder J, Goshe MB. [Google Scholar]
- 18.Moore RJ, Pasa-Tolic L, Masselon CD, Lipton MS, Smith RD. J. Proteome Res. 2002;1(4):351–360. doi: 10.1021/pr0255248. [DOI] [PubMed] [Google Scholar]
- 19.Kadiyala CS, Tomechko SE, Miyagi M. PLoS One. 2010;5(12):e15332. doi: 10.1371/journal.pone.0015332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitra SK, Gantt JA, Ruby JF, Clouse SD, Goshe MB. J. Proteome Res. 2007;6(5):1933–1950. doi: 10.1021/pr060525b. [DOI] [PubMed] [Google Scholar]
- 21.Zhang N, Chen R, Young N, Wishart D, Winter P, Weiner JH, Li L. Proteomics. 2007;7(4):484–493. doi: 10.1002/pmic.200600518. [DOI] [PubMed] [Google Scholar]
- 22.Yu YQ, Gilar M, Lee PJ, Bouvier ES, Gebler JC. Anal. Chem. 2003;75(21):6023–6028. doi: 10.1021/ac0346196. [DOI] [PubMed] [Google Scholar]
- 23.Masuda T, Tomita M, Ishihama Y. J. Proteome Res. 2008;7(2):731–740. doi: 10.1021/pr700658q. [DOI] [PubMed] [Google Scholar]
- 24.Manza LL, Stamer SL, Ham AJ, Codreanu SG, Liebler DC. Proteomics. 2005;5(7):1742–1745. doi: 10.1002/pmic.200401063. [DOI] [PubMed] [Google Scholar]
- 25.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Nat. Methods. 2009;6(5):359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 26.Wiśniewski JR, Nagaraj N, Zougman A, Gnad F, Mann M. J. Proteome Res. 2010;9(6):3280–3289. doi: 10.1021/pr1002214. [DOI] [PubMed] [Google Scholar]
- 27.Wu CC, MacCoss MJ, Howell KE, Yates JR., 3rd Nat. Biotechnol. 2003;21(5):532–538. doi: 10.1038/nbt819. [DOI] [PubMed] [Google Scholar]
- 28.Blackler AR, Speers AE, Ladinsky MS, Wu CC. J. Proteome Res. 2008;7(7):3028–3034. doi: 10.1021/pr700795f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Speers AE, Blackler AR, Wu CC. Anal. Chem. 2007;79(12):4613–4620. doi: 10.1021/ac0700225. [DOI] [PubMed] [Google Scholar]
- 30.Taylor SW, Fahy E, Zhang B, Glenn GM, Warnock DE, Willy S, Murphy AN, Gaucher SP, Capaldi RA, Gibson BW, Ghos SS. Nat. Biotechnol. 2003;21(3):281–286. doi: 10.1038/nbt793. [DOI] [PubMed] [Google Scholar]
- 31.Lundby A, Olsen JV. Methods Mol. Biol. 2011;753:143–155. doi: 10.1007/978-1-61779-148-2_10. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Yu J, Wang Y, Griffin NM, Long F, Shore S, Oh P, Schnitzer JE. Mol. Cell. Proteomics. 2009;8(6):1219–1235. doi: 10.1074/mcp.M800215-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Washburn MP, Wolters D, Yates JR., 3rd Nat. Biotechnol. 2001;19(3):242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 34.Mitra SK, Goshe MB. Methods Mol. Biol. 2009;528:311–326. doi: 10.1007/978-1-60327-310-7_22. [DOI] [PubMed] [Google Scholar]
- 35.Beausoleil SA, Villén J, Gerber SA, Rush J, Gygi SP. Nat. Biotechnol. 2006;24(10):1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 36.Nagaraj N, D'Souza RC, Cox J, Olsen JV, Mann M. J. Proteome Res. 2010;9(12):6786–6794. doi: 10.1021/pr100637q. [DOI] [PubMed] [Google Scholar]
- 37.Nagaraj N, D'Souza RC, Cox J, Olsen JV, Mann MJ. Proteome Res. 2012;11(6):3506–3508. [Google Scholar]
- 38.Frese CK, Altelaar AF, Hennrich ML, Nolting D, Zeller M, Griep-Raming J, Heck AJ, Mohammed S. J. Proteome Res. 2011;10(5):2377–2388. doi: 10.1021/pr1011729. [DOI] [PubMed] [Google Scholar]
- 39.Phanstiel DH, Brumbaugh J, Wenger CD, Tian S, Probasco MD, Bailey DJ, Swaney DL, Tervo MA, Bolin JM, Ruotti V, Stewart R, Thomson JA, Coon JJ. Nat. Methods. 2011;8(10):821–827. doi: 10.1038/nmeth.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michalski A, Damoc E, Hauschild JP, Lange O, Wieghaus A, Makarov A, Nagaraj N, Cox J, Mann M, Horning S. Mol. Cell. Proteomics. 2011;10(9):M111.011015. doi: 10.1074/mcp.M111.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. J. Proteome Res. 2011;10(4):1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 42.Baliban RC, Dimaggio PA, Plazas-Mayorca MD, Garcia BA, Floudas CA. J. Proteome Res. 2012;11(9):4615–4629. doi: 10.1021/pr300418j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao L, Yu K, Banh C, Nguyen V, Ritz A, Raphael BJ, Kawakami Y, Kawakami T, Salomon AR. J. Immunol. 2007;179(9):5864–5876. doi: 10.4049/jimmunol.179.9.5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tobe BT, Hou J, Crain AM, Singec I, Snyder EY, Brill LM. Stem Cell Rev. 2012;8(1):16–31. doi: 10.1007/s12015-011-9317-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Svensson M, Boren M, Sköld K, Fälth M, Sjögren B, Andersson M, Svenningsson P, Andren PE. J. Proteome Res. 2009;8(2):974–981. doi: 10.1021/pr8006446. [DOI] [PubMed] [Google Scholar]
- 46.Stengel A, Keire D, Goebel M, Evilevitch L, Wiggins B, Tache Y, Reeve JR., Jr Endocrinology. 2009;150(11):5113–5118. doi: 10.1210/en.2009-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strahl BD, Allis CD. Nature. 2000;403(6765):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 48.Gardner KE, Allis CD, Strahl BD. J. Mol. Biol. 2011;409(1):36–46. doi: 10.1016/j.jmb.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Young NL, DiMaggio PA, Plazas-Mayorca MD, Baliban RC, Floudas CA, Garcia BA. Mol. Cell. Proteomics. 2009;8(10):2266–2284. doi: 10.1074/mcp.M900238-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bonet-Costa C, Vilaseca M, Diema C, Vujatovic O, Vaquero A, Omeñac N, Castejón L, Bernués J, Giralt E, Azorín F. J. Proteomics. 2012;75(13):4124–4138. doi: 10.1016/j.jprot.2012.05.034. [DOI] [PubMed] [Google Scholar]
- 51.Wu S, Lourette NM, Tolić N, Zhao R, Robinson EW, Tolmachev AV, Smith RD, Pasa-Tolić L. J. Proteome Res. 2009;8(3):1347–1357. doi: 10.1021/pr800720d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nobles KN, Xiao K, Ahn S, Shukla AK, Lam CM, Rajagopal S, Strachan RT, Huang TY, Bressler EA, Hara MR, Shenoy SK, Gygi SP, Lefkowitz RJ. Sci. Signal. 2011;4(185):ra51. doi: 10.1126/scisignal.2001707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loo JA, Edmonds CG, Smith RD. Anal. Chem. 1991;63(21):2488–2499. doi: 10.1021/ac00021a018. [DOI] [PubMed] [Google Scholar]
- 54.Mørtz E, O'Connor PB, Roepstorff P, Kelleher NL, Wood TD, McLafferty FW, Mann M. Proc. Natl. Acad. Sci. U. S. A. 1996;93(16):8264–8267. doi: 10.1073/pnas.93.16.8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kelleher NL, Lin HY, Valaskovic GA, Aaserud DJ, Fridriksson EK, McLafferty FW. J. Am. Chem. Soc. 1999;121(4):806–812. [Google Scholar]
- 56.Henry KD, Williams ER, Wang BH, McLafferty FW, Shabanowitz J, Hunt DF. Proc. Natl. Acad. Sci. U. S. A. 1989;86(23):9075–9078. doi: 10.1073/pnas.86.23.9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schindler PA, Van Dorsselaer A, Falick AM. Anal. Biochem. 1993;213(2):256–263. doi: 10.1006/abio.1993.1418. [DOI] [PubMed] [Google Scholar]
- 58.Fearnley IM, Walker JE. Biochem. Soc. Trans. 1996;24(3):912–917. doi: 10.1042/bst0240912. [DOI] [PubMed] [Google Scholar]
- 59.Hufnagel P, Schweiger U, Eckerskorn C, Oesterhelt D. Anal. Biochem. 1996;243(1):46–54. doi: 10.1006/abio.1996.0480. [DOI] [PubMed] [Google Scholar]
- 60.Sharma J, Panico M, Shipton CA, Nilsson F, Morris HR, Barber J. J. Biol. Chem. 1997;272(52):33158–33166. doi: 10.1074/jbc.272.52.33158. [DOI] [PubMed] [Google Scholar]
- 61.Whitelegge JP, Gundersen C, Faull KF. Protein Sci. 1998;7(6):1423–1430. doi: 10.1002/pro.5560070619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Whitelegge JP, le Coutre J, Lee JC, Engel CK, Privé GG, Faull KF, Kaback HR. Proc. Natl. Acad. Sci. U. S. A. 1999;96(19):10695–10698. doi: 10.1073/pnas.96.19.10695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Whitelegge JP, Aguilera R, Zhang H, Taylor R, Cramer WA. Mol. Cell. Proteomics. 2002;1(10):816–827. doi: 10.1074/mcp.m200045-mcp200. [DOI] [PubMed] [Google Scholar]
- 64.le Coutre J, Whitelegge JP, Gross A, Turk E, Wright EM, Kaback HR, Faull KF. Biochemistry. 2000;39(15):4237–4242. doi: 10.1021/bi000150m. [DOI] [PubMed] [Google Scholar]
- 65.Gómez SM, Nishio JN, Faull KF, Whitelegge JP. Mol. Cell. Proteomics. 2002;1(1):45–59. doi: 10.1074/mcp.m100007-mcp200. [DOI] [PubMed] [Google Scholar]
- 66.Gómez SM, Bil' KY, Aguilera R, Nishio JN, Faull KF, Whitelegge JP. Mol. Cell. Proteomics. 2003;2(10):1068–1085. doi: 10.1074/mcp.M300062-MCP200. [DOI] [PubMed] [Google Scholar]
- 67.Carroll J, Fearnley IM, Walker JE. Proc. Natl. Acad. Sci. U. S. A. 2006;103(44):16170–16175. doi: 10.1073/pnas.0607719103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Whitelegge JP. Plant Physiol. Biochem. 2004;42(12):919–927. doi: 10.1016/j.plaphy.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 69.Whitelegge JP. Trends Anal. Chem. 2005;24(7):576–582. [Google Scholar]
- 70.Whitelegge JP. Photosyn. Res. 2003;78(3):265–277. doi: 10.1023/B:PRES.0000006828.65688.0d. [DOI] [PubMed] [Google Scholar]
- 71.Whitelegge JP, Halgand F, Souda P, Zabrouskov V. Expert Rev. Proteomics. 2006;3(6):585–596. doi: 10.1586/14789450.3.6.585. [DOI] [PubMed] [Google Scholar]
- 72.Zabrouskov V, Whitelegge JP. J. Proteome Res. 2007;6(6):2205–2210. doi: 10.1021/pr0607031. [DOI] [PubMed] [Google Scholar]
- 73.Ryan CM, Souda P, Bassilian S, Ujwal R, Zhang J, Abramson J, Ping P, Durazo A, Bowie JU, Hasan S, Baniulis D, Cramer WA, Faull KF, Whitelegge JP. Mol. Cell. Proteomics. 2010;9(5):791–803. doi: 10.1074/mcp.M900516-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Howery AE, Elvington S, Abraham SJ, Choi KH, Philips S, Ryan CM, Sanford RL, Simpson-Dworschak S, Almqvist J, Tran K, Chew TA, Zachariae U, Andersen OS, Whitelegge JP, Matulef K, Du Bois J, Maduke M. Chem. Biol. 2012;19(11):1460–1470. doi: 10.1016/j.chembiol.2012.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tran JC, Zamdborg L, Ahlf DR, Lee JE, Catherman AD, Durbin KR, Tipton JD, Vellaichamy A, Kellie JF, Li M, Wu C, Sweet SM, Early BP, Siuti N, LeDuc RD, Compton PD, Thomas PM, Kelleher NL. Nature. 2011;480(7376):254–258. doi: 10.1038/nature10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Loo JA. Mass Spectrom. Rev. 1997;16(1):1–23. doi: 10.1002/(SICI)1098-2787(1997)16:1<1::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 77.Miranker AD. Curr. Opin. Struct. Biol. 2000;10(5):601–606. doi: 10.1016/s0959-440x(00)00137-8. [DOI] [PubMed] [Google Scholar]
- 78.Sobott F, Robinson CV. Curr. Opin. Struct. Biol. 2002;12(6):729–734. doi: 10.1016/s0959-440x(02)00400-1. [DOI] [PubMed] [Google Scholar]
- 79.Ilag LL, Ubarretxena-Belandia I, Tate CG, Robinson CV. J. Am. Chem. Soc. 2004;126(44):14362–14363. doi: 10.1021/ja0450307. [DOI] [PubMed] [Google Scholar]
- 80.Barrera NP, Di Bartolo N, Booth PJ, Robinson CV. Science. 2008;321(5886):243–246. doi: 10.1126/science.1159292. [DOI] [PubMed] [Google Scholar]
- 81.Lin HT, Bavro VN, Barrera NP, Frankish HM, Velamakanni S, van Veen HW, Robinson CV, Borges-Walmsley MI, Walmsley AR. J. Biol. Chem. 2009;284(2):1145–1154. doi: 10.1074/jbc.M806964200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kleinekofort W, Pfenninger A, Plomer T, Christian G, Brutschy B. Int. J. Mass Spectrom. Ion Proc. 1996;156(3):195–202. [Google Scholar]
- 83.Morgner N, Kleinschroth T, Barth HD, Ludwig B, Brutschy B. J. Am. Soc. Mass Spectrom. 2007;18(8):1429–1438. doi: 10.1016/j.jasms.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 84.Meier T, Morgner N, Matthies D, Pogoryelov D, Keis S, Cook GM, Dimroth P, Brutschy B. Mol. Microbiol. 2007;65(5):1181–1192. doi: 10.1111/j.1365-2958.2007.05857.x. [DOI] [PubMed] [Google Scholar]
- 85.Morgner N, Zickermann V, Kerscher S, Wittig I, Abdrakhmanova A, Barth HD, Brutschy B, Brandt U. Biochim. Biophys. Acta. 2008;1777(10):1384–1391. doi: 10.1016/j.bbabio.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 86.Sokolova L, Wittig I, Barth HD, Schägger H, Brutschy B, Brandt U. Proteomics. 2010;10(7):1401–1407. doi: 10.1002/pmic.200900756. [DOI] [PubMed] [Google Scholar]
- 87.Zhou M, Morgner N, Barrera NP, Politis A, Isaacson SC, Matak-Vinković D, Murata T, Bernal RA, Stock D, Robinson CV. Science. 2011;334(6054):380–385. doi: 10.1126/science.1210148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Barrera NP, Robinson CV. Annu. Rev. Biochem. 2011;80:247–271. doi: 10.1146/annurev-biochem-062309-093307. [DOI] [PubMed] [Google Scholar]
- 89.Laganowsky A, Reading E, Hopper JTS, Robinson CV. Nat. Protocols. 2012 doi: 10.1038/nprot.2013.024. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Weinglass A, Whitelegge JP, Hu Y, Verner G, Faull KF, Kaback HR. EMBO J. 2003;22(7):1467–1477. doi: 10.1093/emboj/cdg145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Science. 2003;301(5633):610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 92.Jacobsen RB, Sale KL, Ayson MJ, Novak P, Hong J, Lane P, Wood NL, Kruppa GH, Young MM, Schoeniger JS. Protein Sci. 2006;15(6):1303–1317. doi: 10.1110/ps.052040406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kawaguchi R, Yu J, Honda J, Hu J, Whitelegge JP, Ping P, Wiita P, Bok D, Sun H. Science. 2007;315(5813):820–825. doi: 10.1126/science.1136244. [DOI] [PubMed] [Google Scholar]
- 94.Angel TE, Gupta S, Jastrzebska B, Palczewski K, Chance MR. Proc. Natl. Acad. Sci. U. S. A. 2009;106(34):14367–14372. doi: 10.1073/pnas.0901074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Orban T, Jastrzebska B, Gupta S, Wang B, Miyagi M, Chance MR, Palczewski K. Structure. 2012;20(5):826–840. doi: 10.1016/j.str.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hambly DM, Gross ML. J. Am. Soc. Mass Spectrom. 2005;16(12):2057–2063. doi: 10.1016/j.jasms.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 97.Pan Y, Piyadasa H, O'Neil JD, Konermann L. J. Mol. Biol. 2012;416(3):400–413. doi: 10.1016/j.jmb.2011.12.052. [DOI] [PubMed] [Google Scholar]
- 98.Pan Y, Ruan X, Valvano MA, Konermann L. J. Am. Soc. Mass Spectrom. 2012;23(5):889–988. doi: 10.1007/s13361-012-0342-x. [DOI] [PubMed] [Google Scholar]
- 99.Demmers JA, Haverkamp J, Heck AJ, Koeppe RE, 2nd, Killian JA. Proc. Natl. Acad. Sci. U. S. A. 2000;97(7):3189–3194. doi: 10.1073/pnas.050444797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Busenlehner LS, Codreanu SG, Holm PJ, Bhakat P, Hebert H, Morgenstern R, Armstrong RN. Biochemistry. 2004;43(35):11145–11152. doi: 10.1021/bi048716k. [DOI] [PubMed] [Google Scholar]
- 101.Whitelegge J. Science. 2011;334(6054):320–321. doi: 10.1126/science.1214084. [DOI] [PubMed] [Google Scholar]
- 102.Barrera NP, Zhou M, Robinson CV. Trends Cell Biol. 2013;23(1):1–8. doi: 10.1016/j.tcb.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 103.le Coutre J, Kaback HR, Patel CK, Heginbotham L, Miller C. Proc. Natl. Acad. Sci. U. S. A. 1998;95(11):6114–6117. doi: 10.1073/pnas.95.11.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang X, Chien EY, Chalmers MJ, Pascal BD, Gatchalian J, Stevens RC, Griffin PR. Anal. Chem. 2010;82(3):1100–1108. doi: 10.1021/ac902484p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chung KY, Rasmussen SG, Liu T, Li S, DeVree BT, Chae PS, Calinski D, Kobilka BK, Woods VL, Jr, Sunahara RK. Nature. 2011;477(7366):611–615. doi: 10.1038/nature10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.West GM, Chien EY, Katritch V, Gatchalian J, Chalmers MJ, Stevens RC, Griffin PR. Structure. 2011;19(10):1424–1432. doi: 10.1016/j.str.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Westfield GH, Rasmussen SG, Su M, Dutta S, DeVree BT, Chung KY, Calinski D, Velez-Ruiz G, Oleskie AN, Pardon E, Chae PS, Liu T, Li S, Woods VL, Jr, Steyaert J, Kobilka BK, Sunahara RK, Skiniotis G. Proc. Natl. Acad. Sci. U. S. A. 2011;108(38):16086–16091. doi: 10.1073/pnas.1113645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. J. Am. Chem. Soc. 2004;126(11):3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 109.Ujwal R, Bowie JU. Methods. 2011;55(4):337–341. doi: 10.1016/j.ymeth.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hebling CM, Morgan CR, Stafford DW, Jorgenson JW, Rand KD, Engen JR. Anal. Chem. 2010;82(13):5415–5419. doi: 10.1021/ac100962c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Marty MT, Zhang H, Cui W, Blankenship RE, Gross ML, Sligar SG. Anal Chem. 2012;84(21):8957–8960. doi: 10.1021/ac302663f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Leney AC, McMorran LM, Radford SE, Ashcroft AE. Anal. Chem. 2012;84(22):9841–9847. doi: 10.1021/ac302223s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang H, Shen W, Rempel D, Monsey J, Vidavsky I, Gross ML, Bose R. Mol. Cell. Proteomics. 2011;10(6):1–16. doi: 10.1074/mcp.M110.005678. M110.005678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wen J, Zhang H, Gross ML, Blankenship RE. Proc. Natl. Acad. Sci. U. S. A. 2009;106(15):6134–6139. doi: 10.1073/pnas.0901691106. [DOI] [PMC free article] [PubMed] [Google Scholar]