Abstract
The γ subunit of the major histocompatibility complex (MHC) class II complex, CD74, is overexpressed in a significant proportion of metastatic breast tumors, but the mechanistic foundation and biologic significance of this phenomenon are not fully understood. Here, we show that when CD74 is overexpressed in human cancer and noncancerous epithelial cells, it interacts and interferes with the function of Scribble, a product of a well-known tumor suppressor gene. Furthermore, using epithelial cell lines expressing CD74 under the control of tetracycline-inducible promoter and quantitative high-resolution mass spectrometry, we demonstrate that, as a result of CD74 overexpression, the phosphorylation pattern of the C-terminal part of Scribble undergoes specific changes. This is accompanied with a translocation of the protein from the sites of cell-to-cell contacts at the plasma membrane to the cytoplasm, which is likely to effectively enhance the motility and invasiveness of the cancer cells.
Introduction
Cancer metastasis is a multistage process that is governed by a complex program of gene expression and signal transduction that ultimately allows the metastasizing cells to invade surrounding stroma, travel through the circulation, and colonize distant sites. This program sequentially switches specific genes on and turns others off, effectively exerting very dramatic changes of the abundance of many hundreds of proteins in the cell. Since the completion of the human genome project, post-genomic approaches based on application of oligonucleotide microarrays and next-generation sequencing are beginning to shed light on the global cancer genomics landscape and its molecular landmarks. In addition, developments in quantitative proteomics now allow us to dig even deeper: to map the posttranslational modifications and protein-protein interactions that are at the core of the regulation of the molecular mechanisms that drive metastasis. In this study, we applied one such quantitative proteomics approach to investigate the role of CD74 in promoting metastasis of triple-negative breast cancer, a particularly malignant type of the disease.
CD74, the γ subunit of the major histocompatibility complex (MHC) class II complex, is frequently overexpressed in malignant tumors of epithelial and mesenchymal origin. The protein has been suggested as a potential target for rationale-based therapies of lymphoma and multiple myeloma and therapeutic agents targeting CD74 or components of its signaling cascade are in advanced stages of clinical development [1–3]. More recently, we and others reported that CD74 overexpression is linked to increased invasion and metastasis of breast tumors, particularly the tumors of the triple-negative phenotype [4,5].
CD74 is a chaperone protein with an important role in innate immunity. It is required for the expression and functions of the MHC class II receptors and, in addition, has been implicated in cytokine and survival signaling [6–8]. However, the mechanistic foundation for the apparent CD74-augmented malignancy of triple-negative breast tumors is not known. To address this, we have engineered human epithelial cells to express CD74 under the control of a highly regulated inducible promoter, which allowed us to study the effect of CD74 overexpression on protein abundance and protein phosphorylation at a system-wide scale. We found that when overexpressed, CD74 affects the phosphorylation state and function of Scribble, a product of the well-known tumor suppressor gene scrib, which is crucial for the proper maintenance of epithelial cell integrity and function and which is frequently deregulated in breast cancer [9]. Scribble function in maintaining polarity was first discovered in Drosophila. Bilder et al. found that scrib mutations cause aberrant cell shape and loss of monolayer organization in epithelia [10] and that scrib acts as a tumor suppressor [11]. In human cells, Scribble is required for E-cadherin-mediated cell-cell adhesion, and when its expression is downregulated, epithelial cells acquire mesenchymal appearance and their migration is augmented [12].
Materials and Methods
Unless indicated otherwise in the text, chemicals and HPLC solvents were purchased from Thermo Fisher (Loughborough, United Kingdom). The highest available grades were used.
Cell Culturing
Breast cancer cell lines MDA-MB-435, MDA-MB-231, MCF7, and ZR-75.1 and human embryonic kidney 293T/s (HEK293T/s) were grown on RPMI 40 Ultraglutamine medium supplemented with 10% fetal calf serum at 37°C and 5% CO2 and split every second day. The cells were grown to about 80% confluency, detached, washed with phosphate-buffered saline (PBS), and stored at -80°C until needed for analysis.
SILAC Labeling
The cell cultures were labeled using the SILAC Labeling Kit from Thermo Fisher following the manufacturer's instructions and as described by Ong et al. [13]. All cell cultures were labeled for at least five doubling times to ensure complete protein labeling.
Transient Transfection
This was performed as previously described using HEK293T and MCF7 cells [5].
Generation of Stably Transfected HEK293s Cells Expressing CD74 under the Tetracycline-Inducible Promoter
To generate these cell lines, we used an HEK293s derivative stably transfected with the TetR construct and expressing the repressor (HEK293s-TetR) [14]. The cells and the TetO plasmid were kind gifts from Dr Phil Reeves from the University of Essex. The CD74 sequence was subcloned into the TetO plasmid using the following primers:
FOR: 5′-GGAATTCGCCACCATGCACAGGAGGAGAAGCAG-3′
REV: 5′-GCGGCCGCTCACATGGGGACTGGGCCCAGATCC-3′
from a construct described previously and using the same cloning strategy as described by Reeves et al. for rhodopsin [14]. The TetOCD74 plasmid was sequenced to validate the sequence and then transfected in to the HEK293s-TetR cells. The transfectants were selected on G418-containing medium for about 3 weeks until G418-resistant TetR/TetO colonies could be isolated. Twelve colonies were isolated initially, and after screening for inducible CD74 expression, five positive colonies were expanded and stored as stock for further experiments.
CD74 Overexpression and Knockdown
The TetR/TetO-CD74 HEK293s cells were induced with 1 µg/ml tetracycline for various periods of time. An untreated TetR/TetO-CD74 control was mock treated for the same periods of time. An additional control with TetR HEK293s cells treated with tetracycline was also included in most of the experiments to identify possible tetracycline-induced phenomena that are independent of CD74.
CD74 knockdown in MDA-435-MB cells was performed using siRNA from Santa Cruz Biotechnology (Santa Cruz, CA) according to the manufacturer's instruction.
Tumor Tissue Acquisition
All tissues were collected under Local Research Ethics Committee (LREC) and National Health Services (NHS) Trust approval as previously described [5,15]. Tumor tissue was placed on ice immediately in the operating theater. The tissue was assessed and cut by the pathologist on ice before being divided up with a proportion of the tissue snap frozen in liquid nitrogen and the remainder fixed in formalin with the shortest possible delay.
Membrane Protein Isolation
Approximately 20 to 50 mg of frozen tissue from a surgery specimen or core biopsy was snap frozen in liquid nitrogen and pulverized in a BioPulverizer stainless steel device (BioSpec Products, Bartlesville, OK). The homogenized sample was placed on ice and mixed with 500 µl of permeabilization solution containing PBS, 0.2% saponin, and protease and phosphatase inhibitors (Roche Diagnostics, Burgess Hill, United Kingdom). For cultured cells, the homogenization step was omitted and the cells were directly permeabilized. The samples were incubated on ice with intermittent mixing for 15 minutes and centrifuged at 14,000 rpm in a refrigerated centrifuge for 45 minutes. Following centrifugation, the supernatant containing the water-soluble proteins was removed and stored frozen until needed. The pellet was washed with 500 µl of PBS containing protease and phosphatase inhibitors and centrifuged as above. The washed permeabilized pellet was extracted with 100 µl of membrane protein extraction buffer containing 1% (vol/vol) IGEPAL in 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, and protease and phosphatase inhibitors as above, resuspended, and incubated on ice for 10 to 15 minutes. Cultured cells were extracted in 1 ml of buffer per 107 cells. The sample was then centrifuged at 4°C and 14,000 rpm for 15 minutes. The supernatant containing the membrane proteins was recovered and either processed immediately as described below or stored at -80°C until needed.
Immunoprecipitation
Membrane protein fractions extracted as above containing about 0.5 mg of total protein (estimated by dye-binding assay) were diluted 1:1 with binding buffer containing 50 mM Tris-HCl (pH 7.4) and 150 mM NaCl and incubated with 6 µg of LN2 antibody for 30 minutes at room temperature to allow antibody binding to the CD74 extracellular domain. To each tube, 60 µl of protein A/G agarose (Santa Cruz Biotechnology) prewashed with 1 ml of binding buffer was then added. The tubes were further incubated for 1 hour at room temperature on a rotator. We found that because of the inclusion of protease and phosphatase inhibitors in the membrane extraction buffer, performing the experiments at room temperature gave better yields and throughput. The agarose beads were washed three times with binding buffers and the precipitated proteins eluted with 30 µl of sodium dodecyl sulfate sample buffer containing 12 mM DTT for 5 minutes at 100°C. The samples were then cooled on ice and alkylated with 60 mM iodoacetamide for 30 minutes in the dark. For Western blot analysis, an aliquot of the sample was separated by denaturing polyacrylamide gel electrophoresis (PAGE) and transferred onto a polyvinylidene fluoride (PVDF) membrane. For immunoprecipitation/mass spectrometry (IP/MS) analysis, the sample was separated and the gel sliced into five size-resolved fractions, digested with trypsin, and analyzed by liquid chromatography/tandem mass spectrometry (LC-MS/MS).
Protein Digestion and Preparation of Samples for Mass Spectrometry
The protein samples were mixed with 2x sodium dodecyl sulfate sample buffer, reduced, alkylated, and subjected to in-gel digestion as previously described [15]. The SILAC-labeled proteins were separated into up to 10 fractions by a semipreparative PAGE before digestion.
Isolation of Phosphopeptides
Aliquots containing about 0.5 mg of total protein were separated by a semipreparative PAGE on single-well minigel. Up to 10 size-resolved fractions were excised and digested as described above. The tryptic peptides were extracted from the gel pieces, and 10% of the extracted peptides were removed for quantitative analysis of protein abundance and dried in a vacuum concentrator. The remaining 90% were also dried and dissolved in 100 µl of 80%acetonitrile containing 2% formic acid and used to isolate phosphorylated peptides. Phosphopeptides were isolated using the Magnetic TiO2 Phosphopeptide Isolation Kit from Thermo Fisher following the manufacturer's instructions.
Nanoscale LC-MS/MS Analysis
Protein digest analysis by electrospray ionization MS was performed on a hybrid LTQ/Orbitrap Velos instrument (Thermo Fisher) interfaced to a splitless nanoscale HPLC (Ultimate 3000; Dionex, Loughborough, United Kingdom). The peptides were desalted and concentrated online at a flow of 1 µl/min on a 2-cm-long, 0.1-mm i.d. trap column packed with 5-µm C18 particles (Dionex). Following concentration/desalting, the peptides were eluted from the trap column and separated in a 90-minute gradient of 2% to 30% (vol/vol) acetonitrile in 0.1% (vol/vol) formic acid at a flow rate of 0.3 µl/min. The separation column was a 15-cm-long, 0.1-mm i.d. pulled tip packed with 5-µm C18 particles (Nikkyo Technos Co, Tokyo, Japan). The eluting peptides were electrosprayed directly from the packed tip into the LTQ/Orbitrap Velos mass spectrometer by applying 1.75 kV through a liquid junction interface. The LTQ/Orbitrap Velos was operated in the Top 20 data-dependent mode where it first executes two high-resolution scans at a resolution of 30,000 (at 400 m/z) followed by 20 MS/MS scans for the 20 most abundant peptide ions having a charge state > 1. During the high-resolution scans, the Orbitrap analyzer was set to accumulate 106 ions for the maximum of 0.5 second. During MS/MS scans, the LTQ was set to accumulate 5000 precursor ions for the maximum of 0.1 second. The normalized collision energy was set to 30; minimum signal intensity required was set to 500, activation time to 10 milliseconds, and activation Q to 0.250. A dynamic exclusion procedure was implemented to avoid repetitive analysis of abundant peptide ions: After a peptide ion has been analyzed once, its m/z was put in the exclusion list for 30 seconds. The mass calibration was internal by means of lock mass. The ambient ion of 445.12 m/z was used for this purpose throughout all experiments. For targeted detection of Scribble peptides, a mixed mode analysis was used in which a full scan was performed as described above; the next four selected reaction monitoring (SRM) scans were performed to isolate and fragment two Scribble reporter peptides, VSLVGADDLR and VQSPEPPAPER, in their heavy and light SILAC isoforms. Finally, seven data-dependent MS/MS scans were performed to isolate and fragment the seven most abundant peptide ions detected in the high-resolution full scan.
Data Analysis
MS/MS data were analyzed by MaxQuant and the Andromeda search engine as described in [16–18]. The MaxQuant searches were performed using a reverse database to calculate false discovery rate (FDR). Results from the Andromeda engine were filtered at both peptide and protein level. In both cases, the cutoff was at 1% FDR. For SILAC ratios of unmodified peptides and phosphopeptides, the quantitative analysis was performed by MaxQuant and further evaluated by statistical tests using Microsoft Excel.
Western Blot Analysis and Immunohistochemical Analysis of Tumor Tissue
Immunoblot analysis and immunohistochemistry were performed as previously described [15]. For CD74 detection in immunohistochemical (IHC) and for immunoprecipitation, the mouse monoclonal antibody LN2 from Santa Cruz Biotechnology was used. β-Tubulin and Scribble antibodies were also from Santa Cruz Biotechnology. IHC staining was scored using the immunoreactivity score (IRS) scores as implemented for Her2 staining, taking into account staining intensity and the percentage of positive cells: intensity ranges from 1 to 3 for weak, moderate, and strong staining, respectively. Percentage ranges from 1 to 5 for <10%, 10% to 25%, 25% to 50%, 50% to 75%, and >75% staining of the malignant cells; total score = intensity + percentage.
Indirect Immunofluorescence and Colocalization Analysis
Cells to be analyzed were grown in eight-well chamber slides (Lab-Tek; Fisher Scientific, Loughborough, United Kingdom), treated and fixed in 4% buffered paraformaldehyde, and permeabilized in PBS containing 0.5% Triton X-100 for 5 minutes. The cells were blocked in 5% BSA in PBS for 1 hour and stained with mouse anti-Scribble antibody and rabbit anti-CD74 antibody (1:100 dilution) diluted in PBS containing 1%BSA for 1 hour at room temperature (RT). The cells were washed three times in PBS containing 0.1% (vol/vol) Tween 20. The cells were then incubated with secondary antibody solution containing goat anti-mouse Cy5 (Abcam, Cambridge, United Kingdom) and goat anti-rabbit fluorescein isothiocyanate (Abcam), diluted 1:1000 in PBS containing 1% BSA and 0.1% Tween 20 for 45 minutes. The cells were then washed three times in PBS containing 0.1% Tween 20 and analyzed by laser scanning confocal microscopy on a Nikon Eclipse Ti microscope. Colocalized pixels were identified using the ImageJ plugin ColocalizeRGB.
Fluorescence-Activated Cell Sorting (FACS) Analysis
The cells were stained as above and counted in a BD FACS Aria instrument. A secondary antibody-only control was included to estimate nonspecific staining.
Results
In a previous study, we found that CD74 is more abundant in lymph node-metastatic triple-negative tumors compared to nonmetastatic triple-negative breast cancer (TNBC) tumors. This conclusion was based on data obtained by mass spectrometry and pooled tumor protein lysates from six metastatic and six nonmetastatic triple-negative breast tumors [5]. Since this analysis did not discriminate between expression in the malignant cells and other cells in the tumor, we decided to reexamine CD74 expression in another collection of triple-negative breast tumors, this time using immunohistochemical staining instead of mass spectrometry. The results, shown in Figure W1 in the Supplementary information available online, confirmed the previously reported observation that CD74 tends to be more abundant in lymph node-metastatic triple-negative breast tumors. We stained 9 nodepositive tumors and 10 node-negative tumors. Seven of the nodepositive tumors showed strong CD74 staining in the malignant cells, while only 3 of the 10 node-negative tumors showed such staining. This corresponds to a P value of .0409 by the nonparametric Mann-Whitney t test, indicating that aberrant CD74 expression in the malignant cells is associated with increased metastasis. A similar pattern was observed when various established breast cancer cell lines were studied for CD74 expression: The most metastatic line, MDA-435-MD, was the only one to show constitutive CD74 expression. All other tested cell lines did not express the protein normally at detectable level, although some of them could be induced to express small amounts if treated with interferon γ ([4,5]; Supplementary information). To investigate whether CD74 expression contributes to invasion of MDA-435-MB cells, we used siRNA to knock down CD74 and performed wound healing assays. Indeed, when the expression of CD74 was downregulated by siRNA, the wound healing ability of MDA-435-MB cells was diminished (Supplementary information, Figure W2). Thus, tumor specimen analysis and in vitro experiments with cultured cells provided additional evidences for the involvement of CD74 in the invasion and metastasis of breast cancer.
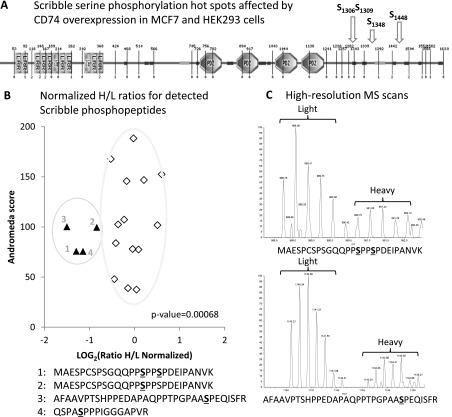
To identify likely targets of CD74 in this context, we decided to follow a quantitative approach based on stable isotope labeling, in which total protein abundance and protein phosphorylation were quantified simultaneously in cells engineered to express the protein in a highly regulated way. This allowed the identification of phosphorylation hotspots: protein phosphorylation sites that are significantly affected by CD74 overexpression while the corresponding total protein abundances are not. This combined genome-scale proteomic screen pinpointed the tumor suppressor protein Scribble as a likely target of CD74 in breast cancer. While the total amount of Scribble, as measured by quantitative high-resolution mass spectrometry, did not change dramatically after 24 hours of CD74 expression, three specific phosphopeptides decreased significantly in the cells overexpressing CD74. Figures 1 and W3 illustrate the results obtained using HEK293s cells expressing CD74 under the control of a tetracycline-inducible promoter. Similar results were obtained in transiently transfected MCF7 cells. Three of the phosphorylation sites on two of the phosphopeptides conform well to the canonical mitogen-activated protein kinase (MAPK) phosphorylation consensus, P-X-S/T-P, and have been detected previously in large-scale phosphoproteomics studies [19,20]. The third peptide is also phosphorylated on a proline-directed site and could be either MAPK or cyclin-directed kinase target.
Figure 1.
CD74 overexpression affects the pattern of posttranslational modifications of the tumor suppressor protein Scribble. (A) Domain map of Scribble based on a cartoon from the SMART database showing the four phosphorylation hotspots in the C-terminal part of the protein affected by CD74 overexpression in HEK293 and MCF7 cells. The three sites were identified in quantitative proteomics/phosphoproteomics screens using transiently transfected MCF7 and HEK293 cells and stably transfected HEK293s cells expressing CD74 under the control of tetracycline-inducible promoter. (B) Scatterplot of Scribble phosphopeptides detected by high-resolution LC-MS/MS analysis. The normalized heavy-to-light ratios are plotted against the Andromeda score. Two clusters of data points are apparent: a set of phosphopeptides that center around H/L ratio of 1, which are not affected by CD74 overexpression, and another cluster that contains the phosphopeptides that are strongly decreased in the CD74-overexpressing cells. The P value was calculated by the Student's t test using the normalized H/L ratios of the two clusters indicated by gray borders. (C) Zoom-in of the isotope envelops of two phosphopeptide ions showing large difference between the intensities of the light peptide ions and the heavy counterparts deriving from CD74-overexpressing cells. The phosphopeptide sequences are indicated under the spectra.
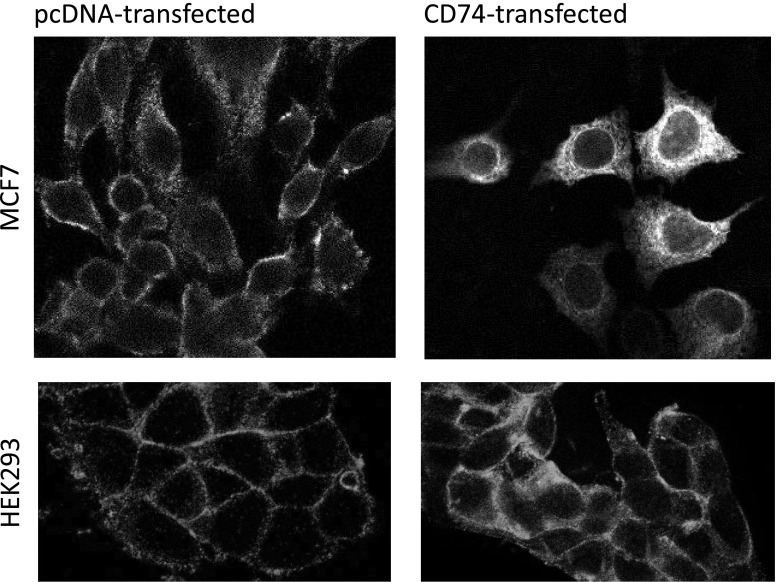
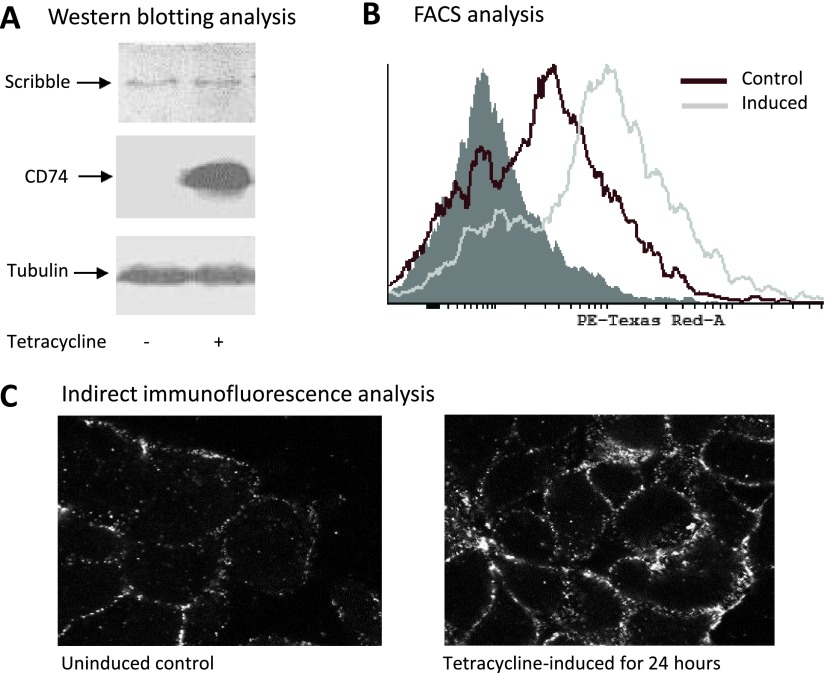
Following the identification of Scribble as likely target of CD74, we examined its intracellular localization in control HEK293T and MCF7 cells and in cells transiently transfected with a construct encoding full-length CD74. Strikingly, in the transfected cells, Scribble appeared to change its typical localization at the adherence junctions and translocate to the cytoplasm. This is shown in Figure 2. In the stably transfected HEK293s cells, the protein did not change localization as dramatically after 24 hours of induction, but its immunoreactivity increased and the CD74-expressing cells consistently showed significantly higher fluorescence in confocal images (Figure 3C). This increased immunoreactivity was reproduced in FACS experiments (Figure 3B) but not in Western blot analyses (Figure 3A).
Figure 2.
Scribble is mislocalized in CD74-overexpressing cells. Laser scanning confocal images of control and transfected MCF7 and HEK293 cells stained with mouse monoclonal anti-Scribble primary antibodies and anti-mouse fluorescein isothiocyanate conjugate. Representative focal planes taken from the midsection of the cells are shown.
Figure 3.
Scribble expression in stably transfected HEK293s cells expressing CD74 under the control of tetracycline-inducible promoter. (A) Western blot analysis showing the same Scribble abundance in the fully denatured and unfolded samples extracted from control and tetracycline-induced cells. (B) The same cell cultures as in A show increased Scribble staining in the CD74-overexpresing cells in FACS assays despite the fact that the same anti-Scribble primary antibody is used. The filled-in gray histogram is from secondary antibody-only control. (C) The same cell cultures as in A and B show increased Scribble staining by confocal microscopy. The same primary antibody is used.
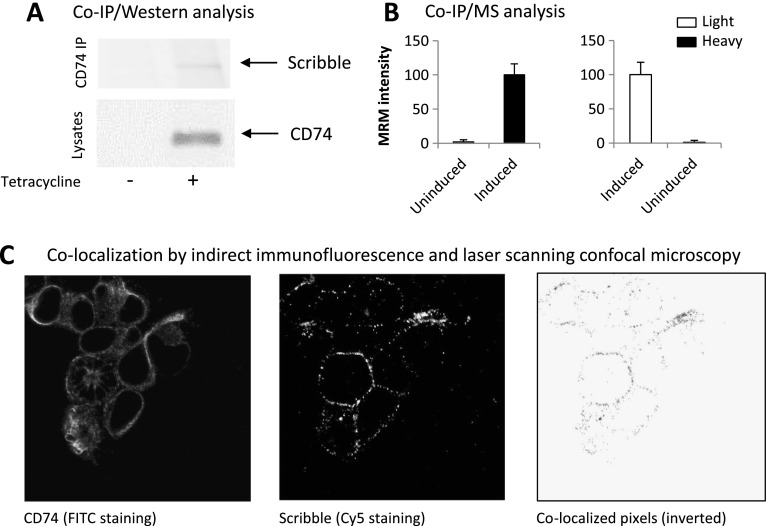
To shed more light on the underlying mechanisms, we then asked whether CD74 and Scribble interact and/or colocalize in the stably transfected cells. Binding of CD74 to a particular part of Scribble might, for example, prevent the interaction of Scribble with a third protein and effectively ensure that the epitope recognized by the anti-Scribble antibody is exposed and detectable in immunofluorescence and FACS experiments. Indeed, in both co-IP/Western blot and co-IP/MS experiments, CD74 and Scribble copurified as shown in Figure 4. We first performed co-IP/Western blot experiments, in which CD74 was immunoprecipitated using an antibody recognizing its extracellular part. The immunoprecipitated proteins were probed with anti-Scribble antibody and a band with the same electrophoretic mobility as Scribble was detected only in the co-IP from cells overexpressing CD74 but not from control uninduced cells (Figure 4A). To rule out any possibility that this result might have been due to a nonspecific binding or antibody cross-reactivity, we performed independent co-IP/MS experiments. These were again designed around the quantitative SILAC technology; its implementation for protein interaction analysis is described by Blagoev et al. [21] and took advantage of the high sensitivity and mass accuracy of the hybrid LTQ/Orbitrap technology. To ensure quantitative precision and rule out false-positive results, the co-IP/MS experiments were performed twice independently and in the following manner: In the first experiment, we labeled one cell culture with heavy arginine and lysine and had another cell culture grown on light amino acids. We then induced CD74 expression for 24 hours in the heavy-labeled culture but left the light-labeled culture uninduced. Then, CD74 was immunoprecipitated from both cell cultures under identical conditions and using equal total protein amounts. The proteins immunoprecipitated from “heavy” and “light” cultures were mixed, separated by denaturing PAGE, digested with trypsin, and analyzed by high-resolution LC-MS/MS. To increase the sensitivity of detection, we performed a mixed mode data-dependent/targeted analysis. This allowed a very precise quantitative assessment of the ability of CD74 to coimmunoprecipitate Scribble. In the alternative labeling experiment, this procedure was repeated but in reverse: The light-labeled culture was induced with tetracycline, while the heavy-labeled sample was left uninduced. The results from these experiments are shown in Figure 4B. In both assays, Scribble reporter peptides were only detected in the CD74 IP from the induced samples but not from the uninduced samples.
Figure 4.
Scribble and CD74 copurify from membrane protein isolates and colocalize in stably transfected HEK293s cells. (A) Co-IP/Western blot analysis. Scribble is detected only in the CD74 IP from cells induced with tetracycline to express CD74. (B) Co-IP/MS. Themultiple reaction monitoring (MRM) signal for the reporter peptides is normalized to 100. The values are mean of three replicate LC-MS/MS analyses ± SD. This experimentwas performed twice. First, CD74 expression was induced in the heavy SILAC-labeled cells. Then, the protein was overexpressed in the light SILAC-labeled cells. In both experiments, MaxQuant detected Scribble peptides only in the CD74 IP from tetracycline-induced cells. (C) Colocalization of Scribble and CD74 in HEK293s cells overexprerssing CD74.
Thus, CD74 and Scribble copurify, which suggests that the two proteins might interact physically, either directly or through a third protein. If this is the case and the interaction takes place in vivo, then subpopulations of CD74 and Scribble molecules should also be seen at the same sites in the cells. We therefore asked whether the two proteins colocalize and performed double-labeling immunofluorescence experiments to test this hypothesis. The results, illustrated in Figure 4C, show that, indeed, this is the case and portions of the pools of the two proteins clearly colocalize in the CD74-overexpressing cells.
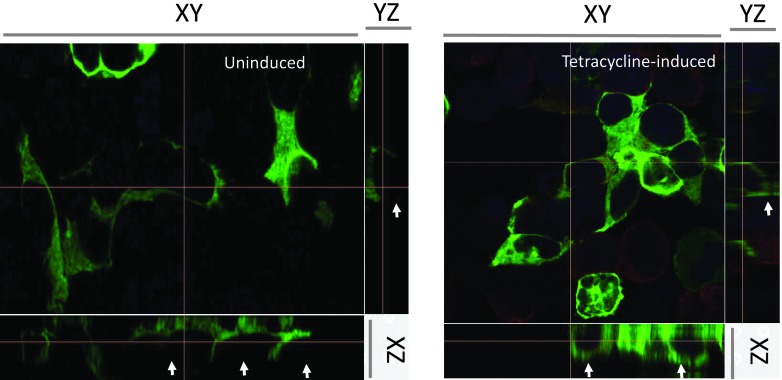
In another independent line of investigations, we transfected TetO-CD74 HEK293 cells with a green fluorescent protein (GFP)-tagged Scribble construct and asked whether GFP-Scribble would change localization when the expression of CD74 is turned on by tetracycline. The results from these experiments are illustrated in Figure 5. The GFP-Scribble reporter localized to the basolateral parts of the membrane in the uninduced cells and was never seen on the apical side of the cell. In contrast, when CD74 was overexpressed for 24 hours, GFP-Scribble clearly appeared at the apical side of the cells. This was also seen in indirect immunofluorescence experiments, in which we stained endogenous Scribble with a monoclonal antibody as in the experiments presented in Figure 4 and examined its localization by confocal microscopy (data not shown).
Figure 5.
Overexpression of CD74 induces translocation of a subpopulation of Scribble molecules from the basolateral side to the apical side of the cells. GFP-Scribble was transiently transfected in TetO-CD74 HEK293s cells and the expression of CD74 was induced with tetracycline. After 24 hours of induction, the cells were fixed and stained with anti-CD74 antibody and analyzed by laser scanning confocal microscopy. The image is presented as “Slice View,” in which the three orthogonal planes XY (the midsection of the stack), YZ, and XZ are formed by slicing the stack as indicated by the crossed orange lines. The white arrows point to the apical side of the cells where GFP-Scribble can be seen only in the cells overexpressing CD74.
Discussion
In recent tumor proteomics study focusing on triple-negative breast cancer, we identified CD74 as frequently overexpressed in the lymph node-metastatic tumors [5]. Here, we show that when CD74 is overexpressed in cultured epithelial and cancer cells, the phosphorylation state of several sites in the C-terminal part of the tumor suppressor protein Scribble changes significantly. The corresponding phosphopeptides were detected and quantified by high-resolution mass spectrometry using SILAC-labeled cells. Altogether, we mapped 19 different Scribble phosphorylation sites and also quantified a large number of unmodified Scribble peptides. This enabled statistical analysis that pinpointed four phosphorylation sites as hotspots—phosphorylations that change significantly in response to CD74 overexpression. Three of these sites conform to the canonical MAPK consensus, which is intriguing as CD74 was previously implicated in an MAPK signaling: As a cellular receptor for the cytokine macrophage inhibitory factor (MIF), CD74 was shown to interact and activate the RAF/mitogen-activated protein kinase kinase (MEK)/extracellular signal-regulated kinase (ERK) cascade [6].
Furthermore, Scribble localization changed and apparent immunoreactivity increased in cells overexpressing CD74 (Figure 2). Conceivably, there could be at least three explanations for this increased immunoreactivity: 1) Scribble protein abundance might be upregulated in response to CD74 overexpression; 2) the epitope recognized by the anti-Scribble antibody could overlap with and be masked by the MAPK-dependent phosphorylations discussed above, which would mean that overexpression of CD74, by decreasing the phosphorylation on these sites, would increase the immunoreactivity of Scribble; 3) the epitope is masked by a protein that binds to it in the control cells, but this interaction is disrupted in CD74-overexpresing cells.
The first model is immediately disproved because the SILAC analysis showed no significant change in total Scribble after 24 hours of tetracycline treatment. A Western blot analysis confirmed this (Figure 3A), although in cells induced for longer periods of time Scribble abundance did decrease moderately (Figure W4). Thus, CD74 overexpression increased the immunoreactivity of native Scribble but did not affect it in Western blots where the protein is fully unfolded and denatured. The Western blot results also disprove the second model: If increased Scribble immunoreactivity in the CD74-overexpressing cells was due to a hypophosphorylated epitope, it is likely that it would have shown up in the Western blot results as well. Thus, the only valid model is explanation 3, which assumes, for specific changes in the network of protein-protein interactions of Scribble, its immediate molecular microenvironment. This is consistent with the fact that Scribble appeared to change localization when CD74 was overexpressed (Figures 2 and 5).
Furthermore, when overexpressed, CD74 was able to coimmunoprecipitate Scribble and was seen to colocalize with a subset of Scribble molecules (Figure 4).
Taken together, our results show that when overexpressed, CD74 engages in a functional interaction with Scribble, which initially affects not only the total abundance of Scribble but also the pattern of its posttranslational modifications in the C-terminal part of the protein. This, by as yet to be elucidated mechanisms, causes the tumor suppressor protein to shift its localization from the basolateral membrane and the sites of cell-to-cell contacts to the cytoplasm and the apical membrane. Apparently, this also leads to a down-regulation of Scribble in the long run as the protein abundance decreased after 48 hours of induced CD74 expression (see Figure W4 available online). Thus, when CD74 is overexpressed for a prolonged period of time, it appears to cause an overall decrease in Scribble abundance. This is consistent with data from a large-scale LC-MS/MS analysis of a collection of breast tumors we carried out recently and will describe in a separate publication. In this data set, summarized in Figure W5 available online, CD74 and Scribble showed a clear negative correlation: The tumors with the highest CD74 abundance showed lowest Scribble abundance and vice versa. These data further corroborate the link between CD74 overexpression and Scribble deregulation established in our experiments with cell-based models and transient and stable inducible overexpression of CD74.
Scribble is a potential tumor suppressor and its deregulation and abnormal localization in breast tumors has been documented already [9]. The protein plays a crucial role in the maintenance of epithelial polarity [10,22,23]. It is known to interact with the mitogen-activated protein kinases of the ERK family, apparently downregulating their activation and ability to migrate to the nucleus [24,25]. As reported in the study of Nagasaka et al., in normal epithelial cells, Scribble is localized at the basolateral membrane and this is required for its ability to inhibit G1-to-S transition [26].
Here, we show that overexpression of CD74, a frequently observed phenomenon in breast cancer, is a direct cause for deregulation of Scribble. This finding can explain why triple-negative breast tumors overexpressing CD74 tend to be more aggressive and with a heightened metastatic propensity. It also suggests that the pathway involved in this mechanism could be a good target for developing rationale-based therapies and companion diagnostics for the treatment of triple-negative breast cancer.
Supplementary Material
Acknowledgments
We are grateful to all members of the Breast Unit, Broomfield Hospital (Broomfield, Chelmsford) and to Debbie Eden and the Histopathology Department, Broomfield Hospital. We thank Phil Reeves for providing the TetR-HEK293s cells and the TetO plasmid and Ian Macara for kindly providing the GFP-Scribble construct.
Footnotes
This research was supported by National Institutes of Health grant 1RO3CA150131 to M.V.M., by a grant from the Greendale Fund to M.V.M., and by a Computational Proteomics Analysis System installation and customization award from Canary Foundation to M.V.M. The authors declare no competing financial interests in relation to the described work.
This article refers to supplementary materials, which are designated by Figures W1 to W5 and are available online at www.neoplasia.com.
References
- 1.Burton JD, Ely S, Reddy PK, Stein R, Gold DV, Cardillo TM, Goldenberg DM. CD74 is expressed by multiple myeloma and is a promising target for therapy. Clin Cancer Res. 2004;10:6606–6611. doi: 10.1158/1078-0432.CCR-04-0182. [DOI] [PubMed] [Google Scholar]
- 2.Gold DV, Stein R, Burton J, Goldenberg DM. Enhanced expression of CD74 in gastrointestinal cancers and benign tissues. Int J Clin Exp Pathol. 2010;4:1–12. [PMC free article] [PubMed] [Google Scholar]
- 3.Sapra P, Stein R, Pickett J, Qu Z, Govindan SV, Cardillo TM, Hansen HJ, Horak ID, Griffiths GL, Goldenberg DM. Anti-CD74 antibody-doxorubicin conjugate, IMMU-110, in a human multiple myeloma xenograft and in monkeys. Clin Cancer Res. 2005;11:5257–5264. doi: 10.1158/1078-0432.CCR-05-0204. [DOI] [PubMed] [Google Scholar]
- 4.Leth-Larsen R, Lund RR, Ditzel HJ. Plasma membrane proteomics and its application in clinical cancer biomarker discovery. Mol Cell Proteomics. 2010;9:1369–1382. doi: 10.1074/mcp.R900006-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greenwood C, Metodieva G, Al-Janabi K, Lausen B, Alldridge L, Leng L, Bucala R, Fernandez N, Metodiev MV. Stat1 and CD74 overexpression is co-dependent and linked to increased invasion and lymph node metastasis in triple-negative breast cancer. J Proteomics. 2012;75:3031–3040. doi: 10.1016/j.jprot.2011.11.033. [DOI] [PubMed] [Google Scholar]
- 6.Leng L, Metz CN, Fang Y, Xu J, Donnelly S, Baugh J, Delohery T, Chen Y, Mitchell RA, Bucala R. MIF signal transduction initiated by binding to CD74. J Exp Med. 2003;197:1467–1476. doi: 10.1084/jem.20030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lue H, Kapurniotu A, Fingerle-Rowson G, Roger T, Leng L, Thiele M, Calandra T, Bucala R, Bernhagen J. Rapid and transient activation of the ERK MAPK signalling pathway by macrophage migration inhibitory factor (MIF) and dependence on JAB1/CSN5 and Src kinase activity. Cell Signal. 2006;18:688–703. doi: 10.1016/j.cellsig.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 8.Shi X, Leng L, Wang T, Wang W, Du X, Li J, McDonald C, Chen Z, Murphy JW, Lolis E, et al. CD44 is the signaling component of the macrophage migration inhibitory factor-CD74 receptor complex. Immunity. 2006;25:595–606. doi: 10.1016/j.immuni.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhan L, Rosenberg A, Bergami KC, Yu M, Xuan Z, Jaffe AB, Allred C, Muthuswamy SK. Deregulation of Scribble promotes mammary tumori-genesis and reveals a role for cell polarity in carcinoma. Cell. 2008;135:865–878. doi: 10.1016/j.cell.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bilder D, Perrimon N. Localization of apical epithelial determinants by the basolateral PDZ protein Scribble. Nature. 2000;403:676–680. doi: 10.1038/35001108. [DOI] [PubMed] [Google Scholar]
- 11.Bilder D, Li M, Perrimon N. Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science. 2000;289:113–116. doi: 10.1126/science.289.5476.113. [DOI] [PubMed] [Google Scholar]
- 12.Qin Y, Capaldo C, Gumbiner BM, Macara IG. The mammalian Scribble polarity protein regulates epithelial cell adhesion and migration through E-cadherin. J Cell Biol. 2005;171:1061–1071. doi: 10.1083/jcb.200506094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 14.Reeves PJ, Thurmond RL, Khorana HG. Structure and function in rhodopsin: high level expression of a synthetic bovine opsin gene and its mutants in stable mammalian cell lines. Proc Natl Acad Sci USA. 1996;93:11487–11492. doi: 10.1073/pnas.93.21.11487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alldridge L, Metodieva G, Greenwood C, Al-Janabi K, Thwaites L, Sauven P, Metodiev M. Proteome profiling of breast tumors by gel electrophoresis and nanoscale electrospray ionization mass spectrometry. J Proteome Res. 2008;7:1458–1469. doi: 10.1021/pr7007829. [DOI] [PubMed] [Google Scholar]
- 16.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 17.Luber CA, Cox J, Lauterbach H, Fancke B, Selbach M, Tschopp J, Akira S, Wiegand M, Hochrein H, O'Keeffe M, et al. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity. 2010;32:279–289. doi: 10.1016/j.immuni.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 18.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 19.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 20.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blagoev B, Kratchmarova I, Ong SE, Nielsen M, Foster LJ, Mann M. A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat Biotechnol. 2003;21:315–318. doi: 10.1038/nbt790. [DOI] [PubMed] [Google Scholar]
- 22.Bilder D. Epithelial polarity and proliferation control: links from the Drosophila neoplastic tumor suppressors. Genes Dev. 2004;18:1909–1925. doi: 10.1101/gad.1211604. [DOI] [PubMed] [Google Scholar]
- 23.Bilder D, Schober M, Perrimon N. Integrated activity of PDZ protein complexes regulates epithelial polarity. Nat Cell Biol. 2003;5:53–58. doi: 10.1038/ncb897. [DOI] [PubMed] [Google Scholar]
- 24.Nagasaka K, Massimi P, Pim D, Subbaiah VK, Kranjec C, Nakagawa S, Yano T, Taketani Y, Banks L. The mechanism and implications of hScrib regulation of ERK. Small GTPases. 2010;1:108–112. doi: 10.4161/sgtp.1.2.13649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagasaka K, Pim D, Massimi P, Thomas M, Tomaic V, Subbaiah VK, Kranjec C, Nakagawa S, Yano T, Taketani Y, et al. The cell polarity regulator hScrib controls ERK activation through a KIM site-dependent interaction. Oncogene. 2010;29:5311–5321. doi: 10.1038/onc.2010.265. [DOI] [PubMed] [Google Scholar]
- 26.Nagasaka K, Nakagawa S, Yano T, Takizawa S, Matsumoto Y, Tsuruga T, Nakagawa K, Minaguchi T, Oda K, Hiraike-Wada O, et al. Human homolog of Drosophila tumor suppressor Scribble negatively regulates cell-cycle progression from G1 to S phase by localizing at the basolateral membrane in epithelial cells. Cancer Sci. 2006;97:1217–1225. doi: 10.1111/j.1349-7006.2006.00315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplementary Reference
- 1.Luber CA, Cox J, Lauterbach H, Fancke B, Selbach M, Tschopp J, Akira S, Wiegand M, Hochrein H, O'Keeffe M, et al. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity. 2010;32:279–289. doi: 10.1016/j.immuni.2010.01.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.