Abstract
Nucleophosmin-anaplastic lymphoma kinase (NPM-ALK) is aberrantly expressed in a subset of T cell lymphoma that commonly affects children and young adults. NPM-ALK possesses significant oncogenic potential that was previously documented using in vitro and in vivo experimental models. The exact mechanisms by which NPM-ALK induces its effects are poorly understood. We have recently demonstrated that NPM-ALK is physically associated with type I insulin-like growth factor receptor (IGF-IR). A positive feedback loop appears to exist between NPM-ALK and IGF-IR through which these two kinases interact to potentiate their effects. We have also found that a single mutation of the Tyr644 or Tyr664 residue of the C terminus of NPM-ALK to phenylalanine decreases significantly, but does not completely abolish, the association between NPM-ALK and IGF-IR. The purpose of this study was to determine whether the dual mutation of Tyr644 and Tyr664 abrogates the association and interactions between NPM-ALK and IGF-IR. We also examined the impact of this dual mutation on the oncogenic potential of NPM-ALK. Our results show that NPM-ALKY644,664F completely lacks association with IGF-IR. Importantly, we found that the dual mutation of Tyr644 and Tyr664 diminishes the oncogenic effects of NPM-ALK, including its ability to induce anchorage-independent colony formation and to sustain cellular transformation, proliferation, and migration. Furthermore, the association between NPM-ALK and IGF-IR through Tyr644 and Tyr664 appears to contribute to maintaining the stability of NPM-ALK protein. Our results provide novel insights into the mechanisms by which NPM-ALK induces its oncogenic effects through interactions with IGF-IR in this aggressive lymphoma.
Introduction
The transmembranous receptor tyrosine kinase anaplastic lymphoma kinase (ALK) is a member of the insulin receptor family [1]. Members of this family display structural similarities, including the YXXXYY motif located within their respective tyrosine kinase domains. The mechanisms of ALK activation are not completely known. Previous studies suggested that some of the heparin-binding growth factors such as midkine and pleiotrophin are capable of binding to and activating ALK [2–4]. The physiological expression of ALK is seen in neural tissues at an early stage of human fetal development [5]. Thereafter, ALK expression is aberrant and is largely limited to malignant neoplasms, including an aggressive type of T cell non-Hodgkin lymphoma known as anaplastic large-cell lymphoma, which is one of the most common hematological neoplasms in children and young adults [6,7]. It is believed that the expression of ALK in this lymphoma is a secondary event resulting from one of several chromosomal aberrations that involve the ALK gene on chromosome 2p23 [8,9]. The most common of these chromosomal aberrations is the t(2;5)(p23;q35) translocation that, in addition to ALK, involves the nucleophosmin (NPM) gene on chromosome 5q35. This translocation leads to the generation of the chimeric oncogenic tyrosine kinase NPM-ALK [8]. The expression of NPM-ALK is detected in approximately 85% of ALK-expressing (ALK+) T cell lymphoma cases [6]. Unlike ALK, NPM-ALK is an entirely intracellular molecule that lacks extracellular and transmembranous domains. Although NPM-ALK lacks a nuclear localization signal, it can translocate to the nucleus by forming heterodimers with wild-type (WT) NPM [10].
There is agreement that NPM-ALK plays a crucial role in the pathogenesis of NPM-ALK+ T cell lymphoma [6,7]. Previous in vitro studies demonstrated that NPM-ALK promotes cellular survival and sustains transformation of rodent fibroblasts [11]. Moreover, in vivo studies in different transgenic mouse models showed that NPM-ALK initiates and maintains lymphomagenesis [12,13]. Regardless of the experimental approach, different transgenic mice models demonstrated that NPM-ALK induces variable types of malignant lymphoma that included T cell, B cell, and plasma cell neoplasms, which is a striking difference from the human NPM-ALK+ lymphoma that develops almost exclusively in T lymphocytes and is clinicopathologically distinct from B cell and plasma cell neoplasms. This important observation implies that, in addition to NPM-ALK, other interacting molecules are most likely required to develop the immunophenotypic and clinicopathologic features that characterize this lymphoma in humans. Previous studies have documented that survival-promoting molecules, such as JAK/STAT, AKT/phosphatidylinositol 3-kinase (PI3K), and mitogen-activated protein (MAP) kinases, are highly expressed and activated in NPM-ALK+ T cell lymphoma, and it is believed that NPM-ALK interacts with and activates through phosphorylation these molecules [14–18]. Nonetheless, our understanding of the mechanisms by which NPM-ALK induces its oncogenic effects is still evolving.
Type I insulin-like growth factor receptor (IGF-IR) tyrosine kinase is another member of the insulin receptor family [19]. It has been reported that IGF-IR is expressed in solid tumors, plasma cell myeloma, and leukemia. It has also been documented that IGF-IR stimulates cellular survival, promotes metastatic dissemination, and initiates therapeutic resistance in these tumors [20–26]. Furthermore, oncogenic molecules such as EWS-FLI-1, epidermal growth factor receptor, platelet-derived growth factor receptor (PDGFR), H-ras, and simian virus 40 large tumor antigen require the contribution of IGF-IR to induce their cellular oncogenic effects [27–31]. We have recently shown that IGF-IR is overexpressed and highly phosphorylated/activated in NPM-ALK+ T cell lymphoma cell lines and primary tumors compared with normal human T lymphocytes that lack the expression of IGF-IR [32]. Importantly, we used laser scanning electron microscopy and immunoprecipitation (IP)/Western blot (WB) and found that NPM-ALK and IGF-IR colocalize and are physically associated, respectively. In addition, our previous results suggested that the two kinases collaborate to sustain their tyrosine phosphorylation and kinase activation levels. A single mutation of the Tyr644 or Tyr664 residues, which are located within the C terminus of NPM-ALK, to phenylalanine decreased significantly the association between IGF-IR and NPM-ALK [32]. In the present study, we tested the impact of a dual mutation of Tyr644 and Tyr664 on the association and interactions between NPM-ALK and IGF-IR, and we also systematically examined the effects of this dual mutation on the oncogenic potential of NPM-ALK. Our data show that the dual mutation of Tyr644 and Tyr664 abrogates completely the association between NPM-ALK and IGF-IR. Importantly, we found novel evidence that the association and functional collaboration between NPM-ALK and IGF-IR maintain high levels of phosphorylation and tyrosine kinase activity of NPM-ALK, which further enhance upregulating the phosphorylation of downstream targets. Taken together, these data support that the interactions between NPM-ALK and IGF-IR sustain the transformation, proliferation, and migration of NPM-ALK+ cells.
Materials and Methods
Cell Lines, Cell Cultures, and Treatments
The previously established NPM-ALK+ T cell lymphoma cell lines Karpas 299, SU-DHL-1, DEL, SUP-M2, and SR-786 (German Collection of Microorganisms and Cell Culture, Braunschweig, Germany) and the NPM-ALK-negative cell line Mac-2A (a gift from Dr George Rassidakis, MD Anderson Cancer Center, Houston, TX) were used [33]. The P6 (mouse BALB/c3T3 fibroblasts overexpressing human IGF-IR) and R- (mouse 3T3-like fibroblasts with targeted ablation of Igflr) cell lines were a generous gift from Dr Renato Baserga (Thomas Jefferson University, Philadelphia, PA) [21,34]. In addition, the 293T and NIH/3T3 cell lines (American Type Culture Collection, Manassas, VA) were used in some experiments. Cell lines were maintained in RPMI 1640 (T cell lymphoma) or Dulbecco's modified Eagle's medium (P6, R-, 293T, NIH/3T3) supplemented with 10% fetal bovine serum (FBS; 15% for Mac-2A; Sigma-Aldrich, St Louis, MO), 2 mM glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C in humidified air with 5% CO2. In some experiments, NPM-ALK+ T cell lymphoma cell lines were cultured in 1% FBS overnight and then treated with 500 ng/ml IGF-I (R&D Systems, Minneapolis, MN).
Picropodophyllin (PPP; Enzo Life Sciences, Farmingdale, NY) or imatinib mesylate (Enzo Life Sciences) was used to treat P6 cells stably transfected with NPM-ALK or NPM-ALKY644,664F. Specific targeting of IGF-IR or phospholipase C-γ (PLC-γ) was achieved using transient transfection of SMARTpool-designed siRNA. The siCONTROL non-targeting siRNA was used as a negative control (Dharmacon, Lafayette, CO). The 293T cell line was transfected using Lipofectamine 2000 (Invitrogen, Grand Island, NY). Electroporation by using the Nucleofector “V” solution was used to transfect all the other cell lines (Amaxa, Atlanta, GA; A-030 program for NPM-ALK+ lymphoma and Mac-2A cell lines and T-030 for P6 and R- cell lines).
Antibodies
Antibodies were obtained from Cell Signaling Technology (Boston, MA): pIGF-IR (Tyr1131, Catalog No. 3021), pIGF-IR (Tyr1135/1136, 3024), pSTAT3 (Tyr705, 9131) pERK (Thr202/Try201, 9101), pTyr-100 (9411), PLC-γ (2822), AKT (9272), pAKT (Ser473, 4051), Hsp70 (4872), Hsp90 (4874), CHIP (2080), pALK (Tyr1586 equivalent to Tyr646 in pNPM-ALK, 3343), and PDGFRβ (3169); Calbiochem (Darmstadt, Germany): STAT3 (569388); Dako (Carpinteria, CA): ALK (M7195); Santa Cruz Biotechnology (Santa Cruz, CA): IGF-IRβ (sc-9038), ERK (sc-94), and pPDGFRβ (Tyr1021, sc-12909-R); Roche (South San Francisco, CA): Myc (11667149001); Abcam (Cambridge, MA): pPLC-γY1235 (ab73993); and Sigma-Aldrich: β-actin (A-2228).
In Situ Proximity Ligation Assay
We used in situ proximity ligation assay (PLA) technique to compare the physical association between IGF-IR and NPM-ALK versus IGF-IR and NPM-ALKY644,664F [35]. R- cells were transfected with empty vector (EV), WT NPM-ALK, or NPM-ALKY644,664F using Lipofectamine 2000 for 48 hours. Cytospin slides were prepared using 10 x 105 cells and fixed with cold methanol for 5 minutes. Slides were then washed with phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde for 20 minutes, permeabilized using 0.1% Triton X-100 in PBS for 20 minutes, and subsequently stained as recommended by the manufacturer (Olink Bioscience, Uppsala, Sweden). Briefly, slides were blocked with the blocking solution for 30 minutes at 37°C. The anti-ALK (354300, mouse; Invitrogen) and anti-IGF-IR (sc-9038, rabbit; Santa Cruz Biotechnology) antibodies were added simultaneously, each at a 1:25 concentration, and the slides were incubated overnight at 4°C. The slides were then washed, and the anti-mouse and anti-rabbit PLA probes were added for 1 hour at 37°C. To ligate the two PLA probes together, the ligation mixture was added for 30 minutes at 37°C. The amplification-polymerase mixture was added for 100 minutes at 37°C in the dark. Finally, slides were washed and mounted using a mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI).
Expression Plasmids
Full-length NPM-ALK (amino acids 1–680) in pCDNA3.1(+) was mutated to generate the NPM-ALKY644F and NPM-ALKY664F constructs by using the QuikChange II XL site-directed mutagenesis kit (200521-5; Stratagene, Santa Clara, CA) according to the manufacturer's instructions [32]. The mutants IGF-IRY950F and IGF-IRK1003A were generated using a similar approach (primer sequence is shown in Table 1). To prepare IGF-IR (1–1245) where the C terminus of IGF-IR was deleted, IGF-IRΔ1245-F-EcoR1 (5′-TAATGAATCAAGGGAATTTCGTCGAAT-3′) and IGF-IRΔ1245-R-Not1 (5′-TATATGCGGCCGCCCGGSSGCCAGGCTCCAT-3′) were used to amplify portions of IGF-IR, which were subsequently digested with EcoRJ/Not1, subcloned into pCDNA3.1(+).pCMV-myc-3B vector (Stratagene), and reconstructed into pCMV-3x-myc vector by using the QuikChange II XL site-directed mutagenesis kit (200521-5; Stratagene). Then, IGF-IR and the different IGF-IR mutants were subcloned from pCDNA3.1(+) (Invitrogen) into the EcoRI and ApaI sites of pCMV-3x-myc vector. Mutant DNA was sequenced using the ABI 3730xl Genetic Analyzer (Applied Biosystems, Carlsbad, CA).
Table 1.
Primers Used in the Study.
| NPM-ALKY644F: F, 5′-CCCTTGTGGGAATGTCAATTTCGGCTACCAGC-3′; R, 5′-AATTGACATTCCCACAAGGGAAGTGACGTA-3′ |
| NPM-ALKY664F: F,5′-TGCCCTGGAGCATGGTCATTTCGAGGATAC CAT-3′; R, 5′-AATGACCAGCTCCAGGGGCAGTAGCGGCTT-3′ |
| IGF-IRY950F: F, 5′-TGCCTCTGTGAACCCGGAGTTCTTCAGCGCTG-3′; R, 5′-AACTCCGGGTTCACAGAGGCATACAGCACTC-3′ |
| IGF-IRK1003A: F, 5′-AGAGTGGCCATTGCAACAGTGAACGAGGCC-3′; R, 5′-GGCCTCGTTCACTGTTGCAATGGCCACTCT-3′ |
Establishment of the P6 and R- Stable Cell Lines That Permanently Express NPM-ALK or NPM-ALKY644,664F
The lentiviral-based expression vector pLove (Addgene, Cambridge, MA), the pENTR 3C vector (Invitrogen), and the third-generation lentivirus packaging vectors pMDLg/pRRE, pRSV-Rev, and pMD2.G (Addgene) were used to create the stable P6 and R- cell lines. First, we reconstructed the pENTR 3C vector into p3C-green fluorescent protein (GFP) using the QuikChange II XL site-directed mutagenesis kit. Then, NPM-ALK and NPM-ALKY644,664F mutant cDNA were subcloned into pENTR 3C-GFP vector and then into pLove expression vector using the Gateway Cloning System (Life Technologies, Grand Island, NY). For lentiviral production, pLove expression vector was cotransfected with third-generation lentivirus packaging vectors into 293T cells using Lipofectamine (Invitrogen). Briefly, 293T cells (1 x 106 cells) were split into 25.0-cm2 flasks 24 hours before transfection, and then the cells were transfected with 2.5 µg of pLove vector together with 1.875 µg of the packaging vectors pMDLg and pRSV-Rev and 0.625 µg of the envelope vector pMD2.G. After 5 hours of incubation at 37°C, fresh culture medium was added for 48 hours. The lentivirus-containing medium was collected and spun down at 1500 rpm for 5 minutes to pellet cellular debris, and the supernatant was filtered through 0.45-µm pores. P6 and R- cells were infected with fresh lentivirus containing medium supplemented with 8 µg/ml Polybrene (Millipore, Billerica, MA) to enhance transfection efficiency. Forty-eight to 72 hours after infection, P6 and R- cells were stably infected with viral particles. Stable clones were selected by GFP cell sorting through the Becton Dickinson FACSVantage SE Turbo cell sorter (BD, Franklin Lakes, NJ).
WB and IP
Cell lysates were obtained using standard techniques [the lysis buffer contained 25 mM Hepes (pH 7.7), 400 mM NaCl, 1.5 mM MgCl2, 2 mM EDTA, 0.5% Triton X-100, 0.1 mM PMSF, 3 mM DTT, phosphatase inhibitor cocktail (20 mM β-GP, 1 mM Na3VO4; Roche), and protease inhibitor cocktail (10 µg/ml leupeptin, 2 µg/ml pepstatin, 50 µg/ml antipain, 1x benzamidine, 2 µg/ml aprotinin, 20 µg/ml chymostatin; Roche)]. For IP, lysates were incubated with primary antibody overnight at 4°C. Agarose beads conjugated with A/G were then added and incubated for 2 hours at 4°C. The immunocomplexes were spun, washed three times with cold PBS and once with lysis buffer, and then subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). For WB, 50 to 80 µg of total protein was electrophoresed on 8% to 12% SDS-PAGE. The proteins were transferred to nitrocellulose membranes and probed with specific primary antibodies and then with the appropriate HRP-conjugated secondary antibodies (GE Healthcare, Piscataway, NJ). Proteins were detected using a chemiluminescence-based kit (GE Healthcare).
Glutathione S-Transferase Pull-Down Experiments
Glutathione S-transferase (GST) fusion constructs were expressed in BL21 Escherichia coli cells, and crude bacterial lysates were prepared by sonication in 50 mM Tris-HCl (pH 7.4), 1.5 mM EDTA, 1 mM DTT, 10% (vol/vol) glycerol, and 0.4 M NaCl in the presence of the protease inhibitor mixture. The in vitro transcription and translation experiments were performed using rabbit reticulocyte lysate (TNT Systems; Promega, Madison, WI) and FluroTectTM Greenlys tRNA labeled according to the manufacturer's recommendation. In GST pulldown assays, 10 µg of the appropriate GST fusion protein was mixed with 30 to 50 µl of the in vitro transcribed/translated products and incubated in binding buffer (75 mM NaCl, 50 mM Hepes, pH 7.9) at room temperature for 60 minutes in the presence of the protease inhibitor mixture. The binding reaction mixture was then added to 30 µl of glutathione-sepharose beads and mixed at 4°C for 2 hours. The beads were washed three times with binding buffer, resuspended in 30 µl of 2x SDS-PAGE loading buffer, and resolved on 12% gels. Then, proteins were detected using a chemiluminescence-based kit (Promega).
Tyrosine Kinase Activity
The tyrosine kinase activity of IGF-IR was determined after using NPM-ALK or NPM-ALKY644,664F as a direct substrate by using the ADP-Glu Kit (Promega). Briefly, IP was used to pull down IGF-IR protein after transfection of IGF-IR-myc into 293T cells. Coimmunoprecipitated IGF-IR was incubated with in vitro translated NPM-ALK or NPM-ALKY644,664F in a buffer containing 150 µM ultra adenosine triphosphate (ATP), 25 mM Tris-HCl (pH 7.5), 2 mM DTT, 0.1 mM Na3VO4, and 10 mM MgCl2 for 1 hour at 37°C. The reaction was stopped with the ADP-Glu reagent. Thereafter, the luciferase intensity, by which the IGF-IR kinase activity was quantified, was measured using the ADP-Glu Luciferase Assay System (Promega).
The tyrosine kinase activity of NPM-ALK and its different mutants was measured using a commercially available kit (Takara Bio, Otsu, Shiga, Japan). Cell lysates were prepared and the specific antibody was used for IP. Agarose beads conjugated with protein A/G were added. Kinase reactions were initiated by adding ATP-2Na into immobilized wells coated with peptide substrate. Next, HRP-conjugated anti-phosphotyrosine (PY20) antibody was added to each well and developed by the addition of HRP and 3,3′,5,5′-tetramethylbenzidine. The reaction was stopped with 1N H2SO4, and absorbance was measured at 450 nm in a microplate reader (MRX II; Dynex Technologies, Chantilly, VA). The ATP-free reaction was used as a negative control.
Confocal Laser Scanning Electron Microscopy
The 293T cells were placed into the cell culture chambers, washed with PBS, and subjected to fixation/permeabilization in 4% paraformaldehyde for 20 minutes, followed by 20 minutes in 0.1% Triton X-100 at room temperature. After blocking with 10% FBS in PBS for 45 minutes, cells were incubated with a mixture of polyclonal rabbit anti-ALK antibody (Cell Signaling; diluted 1:25 in PBS) and mouse anti-IGF-IRβ antibody (Invitrogen; diluted 1:100 in PBS) overnight at 4°C and stained for 90 minutes with a mixture of secondary antibodies, including Alexa Fluor 594 (Catalog No. A21203)-conjugated donkey anti-mouse IgG (1:300) and Alexa Fluor 488 (A21206)-conjugated donkey anti-rabbit IgG (1:800; Invitrogen). Images were visualized by confocal laser scanning microscopy (LSM510; Carl Zeiss, Thornwood, NY).
Colony Formation Assay
Colony formation was detected using a cell transformation detection kit (Chemicon, Billerica, MA). Harvested cells were resuspended in 0.4% agar in culture medium at a density of 1.25 x 103 cells/0.25 ml/plate in a 24-well plate and seeded into solidified 0.8% agar in culture medium. Plates were incubated for 3 to 4 weeks at 37°C in 5% CO2. Fresh culture medium was added one to two times per week. Colonies were stained and photographed using the FluorChem 8800 Imaging System (Alpha Innotech, Santa Clara, CA).
Cell Migration Assay
Cell migration was studied using 24-well Transwell plates with polycarbonate membranes (Corning Costar, Cambridge, MA). Briefly, P6 or R- cells permanently transfected with EV, NPM-ALK, or NPM-ALKY644,664F were loaded onto the upper compartment. Simultaneously, culture medium with 10% FBS was loaded onto the lower compartment. Plates were incubated for 48 hours at 37°C, and cells migrating through the membrane into the lower chamber were counted using light microscopy (Coulter, Miami, FL).
MTT Assay
Cell proliferation was evaluated by using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay and a commercially available ELISA-based kit (Promega). Cells were seeded into 96-well plates, a total of 100 µl of an MTT solution was added to each well, and the cells were then incubated at 37°C and 5% CO2 for 2 hours. Thereafter, cell proliferation was assessed by measuring the absorbance at 540 nm.
Protein Degradation Assay
Cells were transfected with NPM-ALK plasmids, and NPM-ALK protein degradation was measured after treatment with cycloheximide (CHX, c4859; Sigma-Aldrich; 100 µg/ml).
Statistical Analysis
Statistical analysis was performed using a two-tailed Student's t test or analysis of variance for repeated measures and Bonferroni post-hoc test. A P value of <.05 was considered statistically significant.
Results
The Physical Association between NPM-ALK and IGF-IR Is Tyrosine Phosphorylation Dependent
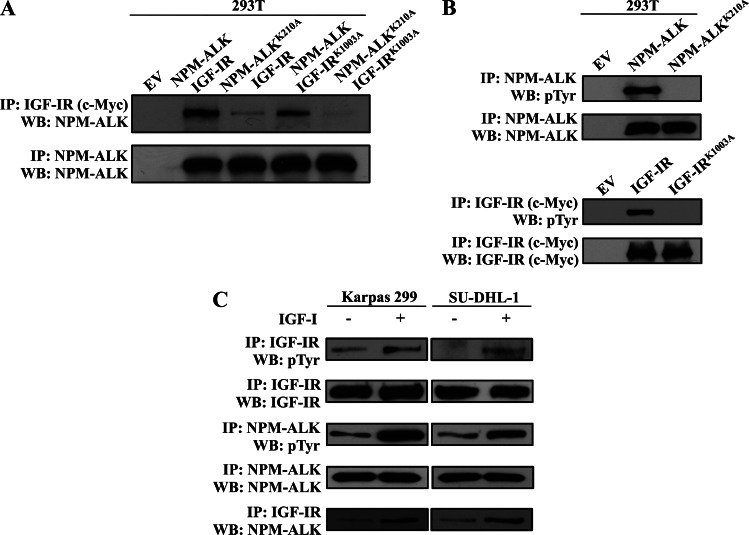
We previously used laser scanning electron microscopy and IP/WB experimental approaches and detected cellular colocalization and physical association between NPM-ALK and IGF-IR, respectively [32]. In this study, we obtained consistent results using a different experimental approach, i.e., GST pull-down of recombinant NPM-ALK and in vitro translated IGF-IR protein (Figure W1). To examine whether adequate total tyrosine phosphorylation of NPM-ALK and IGF-IR is a prerequisite for their physical association, we performed transfection experiments in 293T cells that showed that the association between NPM-ALK and IGF-IR was moderately decreased when the IGF-IRK1003A mutant was used and was markedly decreased when the NPM-ALKK210A mutant was used (Figure 1A). In further support of the necessity of adequate tyrosine phosphorylation, the association between NPM-ALKK210A and IGF-IRK1003A was almost completely abrogated (Figure 1A). Figure 1B confirms the lack of tyrosine phosphorylation in the NPM-ALKK210A and IGF-IRK1003A mutants. Moreover, when Karpas 299 and SUDHL-1 cell lines cultured in 1% FBS overnight were stimulated with IGF-I, the total tyrosine phosphorylation of endogenously expressed IGF-IR and NPM-ALK was markedly upregulated (Figure 1C). Importantly, this up-regulation was associated with a notable increase in the association between NPM-ALK and IGF-IR.
Figure 1.
Tyrosine phosphorylation is a prerequisite for the physical association between NPM-ALK and IGF-IR. (A) Transfection studies in 293T cells followed by IP using anti-c-Myc (the label for transfected IGF-IR) and WB using anti-ALK antibody show that WT NPM-ALK and WT IGF-IR are physically associated. Whereas a moderate decrease in the association between WT NPM-ALK and IGF-IRK1003A was noted, a large decrease in the association between WT IGF-IR and NPM-ALKK210A was detected. Importantly, the association between NPM-ALKK210A and IGF-IRK1003A was almost completely abrogated. Control experiment where anti-ALK antibody was used for IP and WB is shown. (B) Total tyrosine phosphorylation of the NPM-ALKK210A and IGF-IRK1003A mutants was dramatically reduced compared with tyrosine phosphorylation of WT NPM-ALK and IGF-IR, respectively. Control experiments where anti-ALK or anti-c-Myc antibodies were used for IP and WB are shown. (C) IGF-I increased the total tyrosine phosphorylation of endogenous IGF-IR and NPM-ALK in serum-deprived Karpas 299 and SU-DHL-1 cell lines. The increase in tyrosine-phosphorylated IGF-IR and NPM-ALK was associated with a dramatic enhancement of their association.
Dual Mutation of Tyr644 and Tyr664 in NPM-ALK Abrogates Its Physical Association with IGF-IR
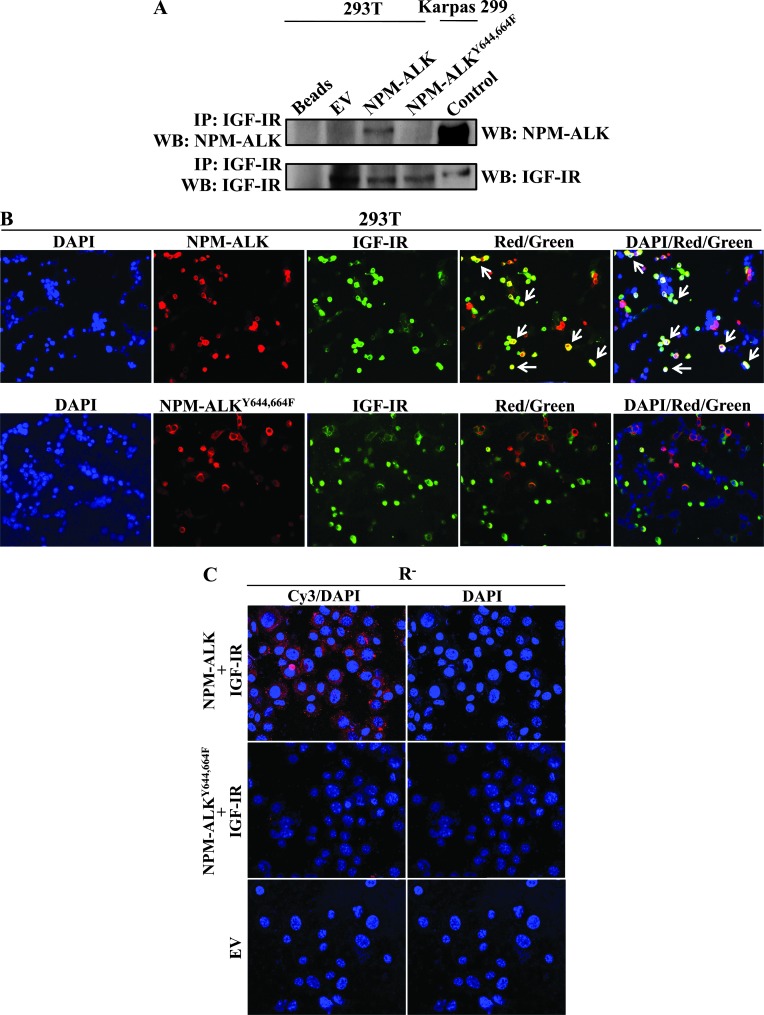
We previously showed that an individual mutation of the Tyr644 or Tyr664 residue of NPM-ALK diminishes its association with IGF-IR [32]. Herein, transfection experiments in 293T cells showed a complete lack of association between IGF-IR and NPM-ALKY644,664F (Figure 2A). In addition, confocal laser scanning electron microscopy showed the same cellular colocalization for NPM-ALK and IGF-IR but not for NPM-ALKY644,664F and IGF-IR (Figure 2B). Our results were further supported by the in situ PLA, which provided another evidence that NPM-ALK and IGF-IR are physically associated and that this association is abolished when Tyr644 and Tyr664 are simultaneously mutated to phenylalanine (Figure 2C). Our experiments also demonstrated that Tyr950 of IGF-IR is the site where it physically associates with NPM-ALK (Figure W2).
Figure 2.
Tyr644 and Tyr664 of NPM-ALK are the sites of its association with IGF-IR. (A) IP, using anti-IGF-IR antibody, and WB, using anti-ALK antibody, were performed in 293T cells transfected with WT NPM-ALK or NPM-ALKY644,664F. The association between WT NPM-ALK and IGF-IR was abrogated when Tyr644 and Tyr664 were simultaneously mutated to phenylalanine. Control experiment where IP and WB were performed using anti-IGF-IR antibody is shown. In addition, control WB showing the expression of NPM-ALK and IGF-IR was concurrently performed on cell lysates from Karpas 299 cells. In addition, a negative control where only beads were used is depicted. (B) Laser scanning electron microscopy shows the expression of transfected WT NPM-ALK or NPM-ALKY644,664F (red) and IGF-IR (green) proteins in 293T cells. Bright yellow signals (white arrows) were detected in cells transfected with WT NPM-ALK and IGF-IR (red/green and DAPI/red/green overlaps). Conversely, these signals were not detected in cells transfected with NPM-ALKY644,664F and IGF-IR. (C) In situ PLA performed in R- cells shows that transfected WT NPM-ALK associates with transfected IGF-IR. The association between the two proteins is shown as speckled red signals confined to the cytoplasm and absent in the nucleus. The red signals were not seen in R- cells simultaneously transfected with NPM-ALKY644,664F and IGF-IR, indicating lack of association of the two proteins. Control experiment where the cells were transfected with EV is shown.
NPM-ALKY644,664F Possesses Significantly Less Potential to Interact with IGF-IR
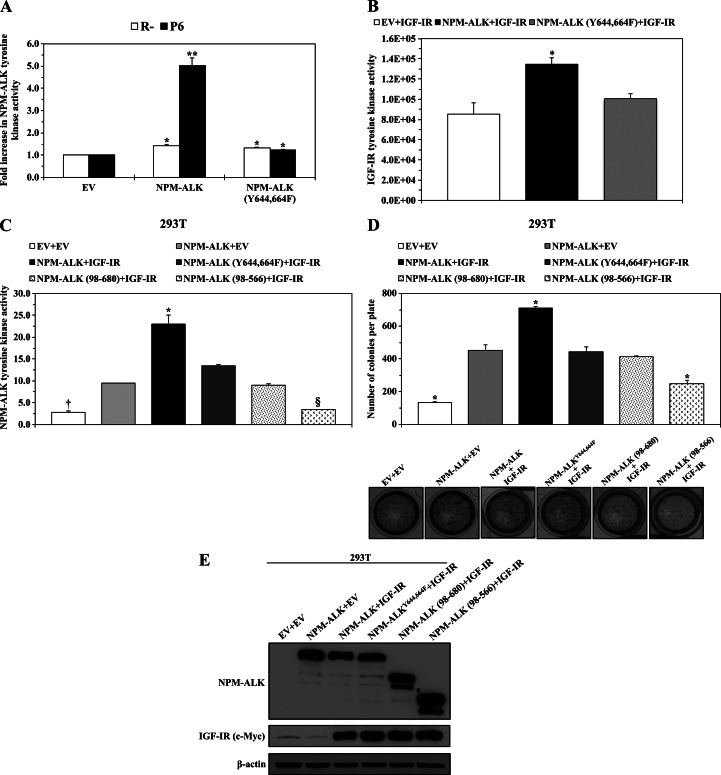
To further investigate the effects of dual mutation of Tyr644 and Tyr664 on the interactions between NPM-ALK and IGF-IR, we performed transfection experiments in R- and P6 cell lines that lack or express, respectively, the IGF-IR protein (Figure 3A). Compared with EV, transient transfection of NPM-ALK or NPM-ALKY644,664F in R- or P6 cells was associated with a modest, albeit statistically significant, increase in NPM-ALK tyrosine kinase activity (Figure 3A). On the contrary, transfection of NPM-ALK into P6 cells was associated with a dramatic enhancement of NPM-ALK tyrosine kinase activity (Figure 3A).
Figure 3.
NPM-ALKY644,664F demonstrates significantly decreased interactions with IGF-IR. (A) Transfection of WT NPM-ALK or NPM-ALKY644,664F in R- cells that lack expression of IGF-IR was associated with a relatively small but statistically significant increase in NPM-ALK tyrosine kinase activity. Similarly, there was a slight increase in tyrosine kinase activity when NPM-ALKY644,664F was transfected into P6 cells that express IGF-IR. Notably, NPM-ALK tyrosine kinase activity was much more pronounced when WT NPM-ALK was transfected into P6 cells. The results are shown as means ± SE of three different experiments (*P < .05, **P < .01). (B) Measuring IGF-IR tyrosine kinase activity by using in vitro translated WT NPM-ALK or NPM-ALKY644,664F as a direct substrate of IGF-IR shows that IGF-IR tyrosine kinase activity significantly increased compared to the baseline level only when WT NPM-ALK was used as a substrate. IGF-IR tyrosine kinase activity is depicted as luciferase activity (in relative light unit). The results are shown as means ± SE of three experiments (*P < .05 compared with other experimental conditions). (C) Transfection of WT NPM-ALK or different functionally defective constructs with IGF-IR in 293T cells showed that WT NPM-ALK possesses significantly greater tyrosine kinase activity when transfected with IGF-IR compared to WT NPM-ALK transfected with EV or compared to NPM-ALKY644,664F, NPM-ALK (98–680), or NPM-ALK (98–566) transfected with IGF-IR. The tyrosine kinase activity of NPM-ALKY644,664F transfected with IGF-IR was similar to WT NPM-ALK transfected with EV and higher than that of the mutants NPM-ALK (98–680) and NPM-ALK (98–566) transfected with IGF-IR. The results are shown as means ± SE of three experiments (*P < .01 versus all other experimental conditions, †P < .01 versus all except NPM-ALK (98–566) + IGF-IR, §P < .01 versus all except EV + EV). (D) Transfection of WT NPM-ALK and IGF-IR into 293T cells was associated with a significantly higher anchorage-independent colony formation potential when compared with all other conditions. Transfection of EV and WT NPM-ALK or of IGF-IR and NPM-ALKY644,664F or NPM-ALK (98–680) resulted in a significantly higher number of colonies than transfection of EV only or of IGF-IR and NPM-ALK (98–566). The results are shown as means ± SE of three experiments (*P < .01 versus all other experimental conditions). (E) The expression levels of IGF-IR and the different NPM-ALK constructs in 293T cells after transfection. β-Actin demonstrates equal loading of the different proteins.
We also generated in vitro translated NPM-ALK and NPM-ALKY644,664F proteins and compared their effects as direct substrates on IGF-IR tyrosine kinase activity. In vitro incubation of IGF-IR and NPM-ALK was associated with a significantly higher level of IGF-IR tyrosine kinase activity compared with the incubation of IGF-IR and NPM-ALKY644,664F (Figure 3B).
Next, we further analyzed the interactions between IGF-IR and NPM-ALKY644,664F compared with other NPM-ALK mutants that are known to possess inherent functional defects. In this regard, we used the NPM-ALK (98–680) mutant that lacks NPM and the ability to form homodimers and the NPM-ALK (98–566) mutant that, in addition to lacking NPM, also lacks the C terminus that binds with downstream targets. Our results showed that cotransfection of NPM-ALK with IGF-IR into 293T cells induced much higher tyrosine activity of NPM-ALK than EV cotransfected with NPM-ALK. Notably, NPM-ALKY644,664F demonstrated significantly reduced tyrosine activity, compared with NPM-ALK, when cotransfected with IGF-IR (Figure 3C). Nonetheless, the tyrosine activity of NPM-ALKY644,664F was still higher than the tyrosine activity of the NPM-ALK (98–680) and NPM-ALK (98–566) mutants cotransfected with IGF-IR.
In addition, cotransfection of NPM-ALK and IGF-IR into 293T cells, compared with cotransfection of NPM-ALK with EV or any of the NPM-ALK mutants with IGF-IR, significantly increased the anchorage-independent colony formation potential of the cells grown for 3 to 4 weeks (Figure 3D). Cotransfection of NPM-ALKY644,664F with IGF-IR was associated with a colony formation potential similar to that of cotransfection of NPM-ALK with EV or to cotransfection of NPM-ALK (98–680) with IGF-IR but significantly higher than that of NPM-ALK (98–566) cotransfected with IGF-IR (Figure 3D). Levels of expression of the constructs transfected into 293T cells are shown in Figure 3E.
Dual Mutation of Tyr644 and Tyr664 Diminishes the Oncogenic Potential of NPM-ALK
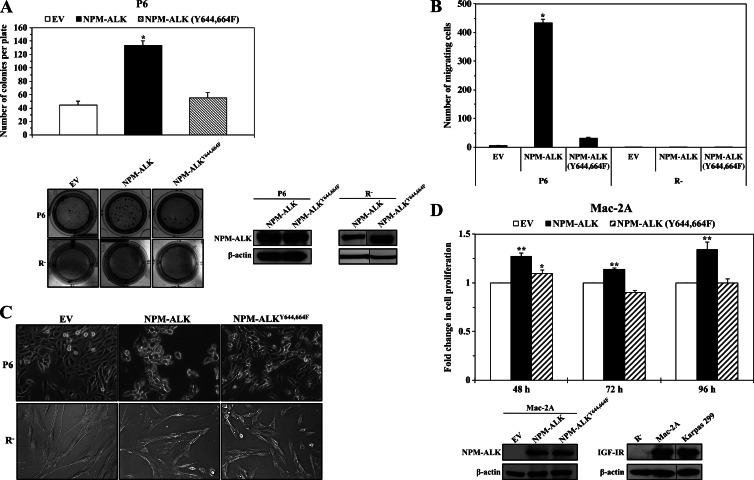
Next, we studied the oncogenic potential of the NPM-ALKY644,664F mutant. We used P6 and R- cells that express or lack IGF-IR, respectively, to generate clones that stably express NPM-ALK or NPM-ALKY644,664F. There was a notable increase in the anchorage-independent colony formation potential of P6 cells permanently transfected with NPM-ALK, but P6 cells transfected with NPM-ALKY644,664F demonstrated a colony formation potential similar to that of P6 cells transfected with EV (Figure 4A). Importantly, similar to R- cells stably transfected with EV or NPM-ALKY644,664F, R- cells stably transfected with NPM-ALK failed to develop anchorage-independent colonies (Figure 4A). Levels of expression of NPM-ALK and NPM-ALKY644,664F in P6 and R- cells are shown in Figure 4A. Furthermore, whereas NPM-ALK significantly increased the migration of P6 cells, NPM-ALKY644,664F failed to induce a similar effect (Figure 4B). However, stable transfection of NPM-ALK failed to stimulate the migration of R- cells, and its effect was similar to EV and NPM-ALKY644,664F (Figure 4B). We also noted differences in the morphologic growth pattern of P6 cells that were permanently transfected with NPM-ALK versus those transfected with NPM-ALKY644,664F (Figure 4C). NPM-ALK induced morphologic changes consistent with cellular transformation, which included three-dimensional clumping, with irregular and polyhedral cellular morphology. In contrast, NPM-ALKY644,664F induced monolayer growth pattern and spindle-shape cellular morphology that were similar to control cells transfected with EV. Similar to its effects on colony formation and cellular migration, NPM-ALK could not induce noticeable changes in the monolayer growth pattern and spindle shape of R- cells. The effects of stably transfected NPM-ALK on R- cells were similar to the effects of EV and NPM-ALKY644,664F (Figure 4C). We also used an ALK-negative T cell lymphoma cell line, Mac-2A, that express endogenous IGF-IR to further examine the contribution of Tyr644 and Tyr664 to the oncogenic potential of NPM-ALK. Transfection of NPM-ALK significantly increased the proliferation of Mac-2A cells, but this effect was not present when NPM-ALKY644,664F was transfected instead (Figure 4D).
Figure 4.
Oncogenic defects of NPM-ALKY644,664F. (A) Stable expression of WT NPM-ALK in P6 cells that express considerable levels of IGF-IR was associated with a remarkable increase in the anchorage-independent colony formation potential of these cells. In contrast, the potential of P6 cells permanently transfected with NPM-ALKY644,664F to form colonies was comparable to that of P6 cells transfected with EV (upper panel). The results are shown as means ± SE of three consistent experiments (*P < .0001 versus EV and NPM-ALKY644,664F). Examples of the P6 cell colonies grown in soft agar are shown (left lower panel). In contrast, R- cells stably transfected with NPM-ALK did not demonstrate evidence of anchorage-independent colony formation and were similar to the R- cells stably transfected with EV or NPM-ALKY644,664F (lower left panel). The expression levels of WT NPM-ALK, NPM-ALKY644,664F, and β-actin in P6 and R- cells are illustrated. A vertical line has been inserted to indicate repositioned gel lanes. Transfection of EV did not induce protein expression (data not shown). (B) P6 cells permanently transfected with WT NPM-ALK demonstrate remarkably increased migration potential compared to P6 cells transfected with EV or NPM-ALKY644,664F. Notably, R- cells stably transfected with NPM-ALK or NPM-ALKY644,664F failed to undergo cellular migration. The results are shown as means ± SE of three experiments (*P < .0001 versus EV and NPM-ALKY644,664F in P6 cells). (C) P6 cells permanently transfected with WT NPM-ALK demonstrated morphologic features consistent with transformation including three-dimensional clumping and polyhedral cellular morphology. Conversely, P6 cells transfected with NPM-ALKY644,664F maintained the spindle-shape morphology and the monolayer growth pattern characteristic of cells transfected with EV (upper panel). R- cells transfected with NP-ALK did not show evidence of cellular transformation and were similar to R- cells transfected with EV or with NPM-ALKY644,664F (lower panel). (D) Transient transfection of WT NPM-ALK, but not NPM-ALKY644,664F, induced a significant increase in the proliferation of the ALK-negative T cell lymphoma cell line Mac-2A. The expression levels of WT NPM-ALK and NPM-ALKY644,664F in Mac-2A cells are shown in the left lower panel. WB shown in the right lower panel demonstrated that Mac-2A cells express endogenous IGF-IR. Karpas 299 and R- cells were used as positive and negative controls, respectively, for the expression of IGF-IR. β-Actin demonstrates equal loading of the different proteins. A vertical line has been inserted to indicate repositioned gel lanes. The results represent the means ± SE of three consistent experiments (*P < .05 versus EV, **P < .01 versus EV and NPM-ALKY644,664F).
Biochemical Consequences of the Dual Mutation of Tyr644 and Tyr664 in NPM-ALK
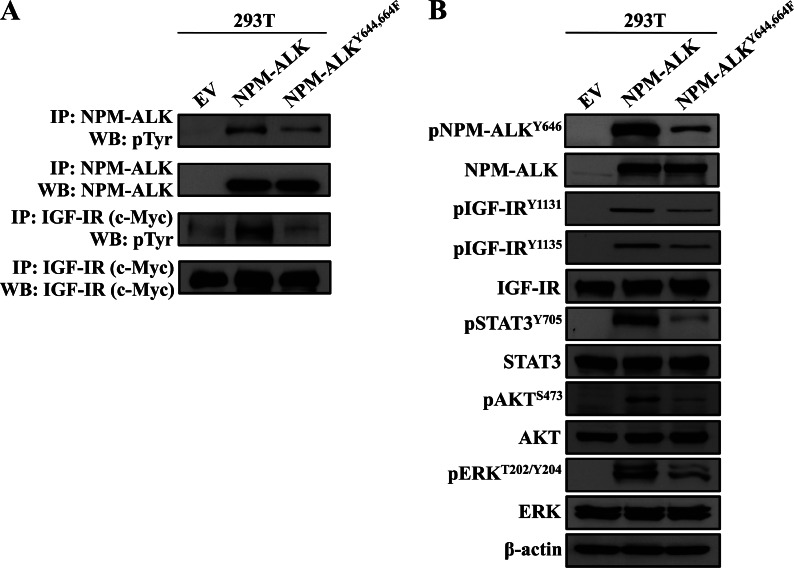
To understand the functional defects of lack of association and interactions between NPM-ALK and IGF-IR, we examined the biochemical changes induced by the NPM-ALKY644,664F mutant. Transfection of IGF-IR with NPM-ALK or NPM-ALKY644,664F was performed in 293T cells. Total tyrosine phosphorylation of NPM-ALKY644,664F was markedly diminished compared with total phosphorylation of NPM-ALK (Figure 5A). Notably, transfection of NPM-ALKY644,664F was also associated with a dramatic decrease in total tyrosine phosphorylation of IGF-IR (Figure 5A). In addition, the phosphorylation of Tyr646, which is also located within the C terminus of NPM-ALK, was diminished when NPM-ALKY644,664F was transfected instead of NPM-ALK (Figure 5B). Another important observation was that the phosphorylation of NPM-ALK/IGF-IR common downstream targets, including STAT3, AKT, and ERK, was remarkably decreased when NPM-ALKY644,664F was transfected instead of NPM-ALK (Figure 5B).
Figure 5.
Biochemical alterations resulting from dual mutation of Tyr644 and Tyr664 in NPM-ALK. (A) Cotransfection of IGF-IR and WT NPM-ALK in 293T cells shows that the two kinases possess high levels of total tyrosine phosphorylation, whereas cotransfection of IGF-IR and NPM-ALKY644,664F shows that not only NPM-ALKY644,664F but also IGF-IR demonstrate much less total tyrosine phosphorylation levels. The expressions of IGF-IR and NPM-ALK constructs in 293T cells are shown. (B) Transfection of NPM-ALKY644,664F was associated with a pronounced decrease in pNPM-ALKY646, pIGF-IRY1131/Y1135, pSTAT3Y705, pAKTS473, and pERKT202/Y204 levels. β-Actin supports equal protein loading.
Dual Mutation of Tyr644 and Tyr664 Decreases NPM-ALK Protein Stability
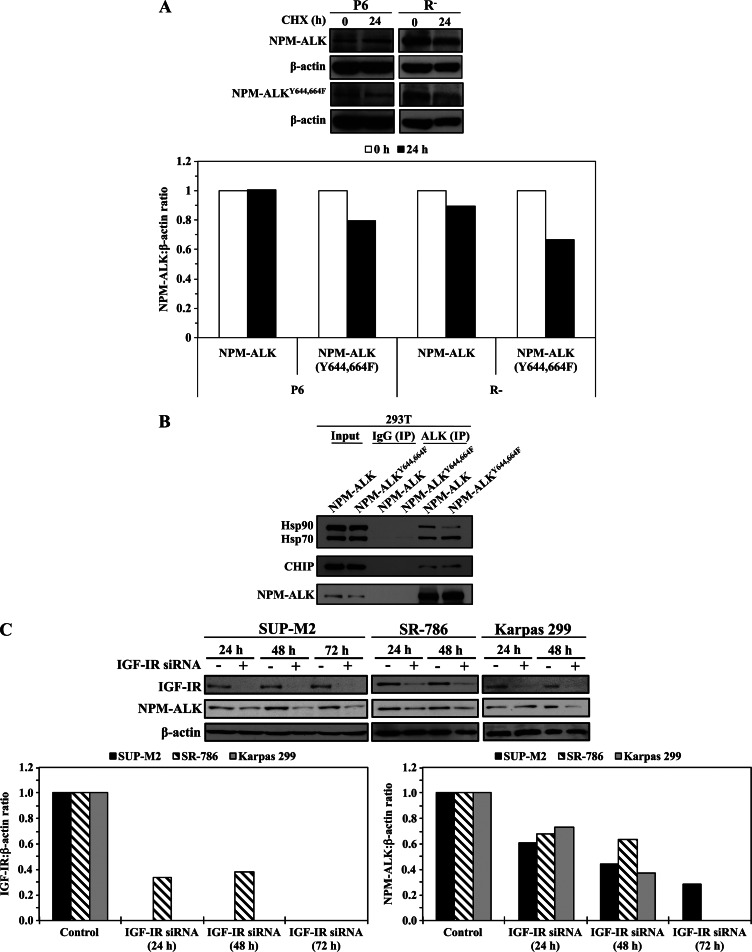
It was previously demonstrated that the expression of NPM-ALK protein can be regulated by the proteasome degradation pathway [36,37]. A screening experiment using MG132 to inhibit proteasome degradation in R- cells transfected with NPM-ALKY644,664F also showed that this mutant is most likely regulated by the protein degradation pathway (Figure W3). Thereafter, the effects of the dual mutation of Tyr644 and Tyr664 that causes lack of binding with IGF-IR on the stability of NPM-ALK protein were studied. P6 and R- cells were transfected with NPM-ALK or NPM-ALKY644,664F and then treated with CHX to inhibit new protein synthesis. There was no change in NPM-ALK protein levels at 24 hours after treatment of P6 cells (Figure 6A, upper panel). In contrast, NPM-ALKY644,664F was less stable in P6 cells, and its levels decreased at 24 hours after treatment. Importantly, these findings were more prominent when R- cells were used (Figure 6A, upper panel). Densitometry of the WB bands and extrapolation of the NPM-ALK-to-β-actin ratio showed that, compared to no change in P6 cells, NPM-ALK decreased to approximately 89%of its baseline level in R- cells (Figure 6A, lower panel). In addition, NPM-ALKY644,664F decreased to 79% and 66% of its baseline levels in P6 and R- cells, respectively (Figure 6A, lower panel). To further analyze these observations, we examined the association between NPM-ALK and NPM-ALKY644,664F with proteins known to regulate the stability of NPM-ALK protein [36,38]. Compared with NPM-ALK, NPM-ALKY644,664F maintained the ability to associate with Hsp70 and CHIP and demonstrated a remarkably reduced ability to associate with Hsp90 (Figure 6B). Thereafter, we set to directly examine whether the association between NPM-ALK and IGF-IR, independently from Hsp90, contributes to maintaining the stability of NPM-ALK protein. Specific down-regulation of IGF-IR by siRNA was associated with a marked decrease in NPM-ALK protein levels in SUP-M2, SR-786, and Karpas 299 cell lines (Figure 6C).
Figure 6.
NPM-ALKY644,664F is a less stable protein than WT NPM-ALK. (A) Although treatment of P6 cells that express IGF-IR with CHX did not induce a notable decrease in transiently transfected WT NPM-ALK levels at 24 hours, CHX decreased NPM-ALKY644,664F levels in these cells (upper panel). These effects were more pronounced when the R- cells that lack IGF-IR were treated with CHX. Densitometry of the WB bands was performed and the NPM-ALK-to-β-actin ratio is shown (lower panel). Treatment of P6 cells by CHX induced no changes in WT NPM-ALK and modestly decreased its levels in R- cells to 89% of the baseline levels. The effect of CHX on NPM-ALKY644,664F was more pronounced, as it decreased its level to 79% and 66% of the baseline levels in P6 and R- cells, respectively. (B) Transfection studies in 293T cells showed that WT NPM-ALK associates with Hsp70, Hsp90, and CHIP proteins. Whereas NPM-ALKY644,664F maintained the association with Hsp70 and CHIP, its association with Hsp90 was markedly decreased compared with WT NPM-ALK. (C) Specific downregulation of IGF-IR by siRNA was associated with a remarkable decrease in NPM-ALK protein levels in three different NPM-ALK+ T cell lymphoma cell lines including SUP-M2, SR-786, and Karpas 299 (upper panel). Densitometric analysis of the ratios between IGF-IR and β-actin and NPM-ALK and β-actin bands are shown (lower panel).
Effects of Specific Down-Regulation of PLC-γ on the Proliferation of NPM-ALK+ T Cell Lymphoma Cells
We have identified Tyr664 as a site that partially associates with IGF-IR. Because previous studies have shown that Tyr664 of NPM-ALK is the site of its association with PLC-γ, and also suggested that this association is required by NPM-ALK to induce mitogenic effects [39], we set to test the direct effect of the down-regulation of PLC-γ on NPM-ALK+ T cell lymphoma cells. Specific down-regulation of PLC-γ by siRNA (Figure W4A, upper panel) neither decreased the proliferation of Karpas 299 and DEL cells (Figure W4A, lower panel) nor reduced the phosphorylation of STAT3, AKT, and ERK in these cells (Figure W4B).
Expression Levels of IGF-IR and pIGF-IR in the Mouse Fibroblast NIH/3T3 Cells
Earlier studies have commonly used rodent fibroblasts to examine the oncogenic effects of exogenously expressed NPM-ALK. However, the similarities between the biologic backgrounds in rodent fibroblasts and the lymphoma cells are questionable. Because we have observed that NPM-ALK+ T cell lymphoma cell lines and primary human tumors overexpress IGF-IR and pIGF-IR, we were curious to analyze the endogenous levels of expression of these proteins in the mouse fibroblast NIH/3T3 cells. WB demonstrated that NIH/3T3 cells express low levels of IGF-IR protein and lack the expression of pIGF-IR (Figure W5).
Effect of Selective Inhibition of IGF-IR and PDGFRβ on NPM-ALK and NPM-ALKY644,664F-Expressing P6 Cells
A recent study demonstrated that PDGFR supports the survival of NPM-ALK+ T cell lymphoma [40]. WB analysis showed that PDGFR and pPDGFRY1021 are expressed in P6 cells stably transfected with NPM-ALK or NPM-ALKY644,664F (Figure W6). Thereafter, we set to test the effects of targeting IGF-IR by using PPP versus targeting PDGFR by using imatinib mesylate on the proliferation of P6 cells. Compared to PDGFR inhibition, inhibition of IGF-IR induced a slightly more pronounced decrease in the proliferation of the two types of P6 cells. In addition, combining PPP and imatinib mesylate induced effects almost similar to the effects of PPP alone (Figure W6).
Discussion
We have previously used laser scanning electron microscopy and IP/WB and found that NPM-ALK and IGF-IR are physically associated [32]. Herein, we further demonstrated the physical association between NPM-ALK and IGF-IR by using two other experimental approaches: in situ PLA and GST pull-down of recombinant NPM-ALK and in vitro translated IGF-IR protein. Both approaches consistently supported the association between the two proteins. Furthermore, we identified the Tyr950 residue of IGF-IR as the major site where its association with NPM-ALK occurs. It has been previously shown that IGF-IR uses Tyr950 for its physical association with the downstream modulators IRS-1 and Shc [41–43]. Notably, IRS-1 and Shc also associate with NPM-ALK [11].
Thereafter, we aimed at performing a systematic analysis of the association between NPM-ALK and IGF-IR and defining the functional implications of this association. In this regard, we first examined whether adequate tyrosine phosphorylation of NPM-ALK and IGF-IR is a prerequisite for their association. It has been previously documented that NPM-ALK and IGF-IR with functional kinase domains demonstrate high levels of total tyrosine phosphorylation [10,44,45]. It has also been shown that the total tyrosine phosphorylation and the functional integrity of the kinase domains are largely dependent on single lysine residues located within the ATP-binding domains, namely Lys210 in NPM-ALK and Lys1003 in IGF-IR. Mutations of these lysine residues to other amino acids such as arginine or alanine resulted in defected mutants with markedly reduced total tyrosine phosphorylation [32,46,47]. Cotransfection of IGF-IRK1003A and NPM-ALK in 293T cells modestly decreased their association. However, the association between NPM-ALKK210A and IGF-IR was significantly diminished. More importantly, when NPM-ALKK210A and IGF-IRK1003A were cotransfected, the association between NPM-ALK and IGF-IR was completely abrogated. In another approach, we used NPM-ALK+ T cell lymphoma cell lines to examine the effect of the phosphorylation levels of endogenously expressed NPM-ALK and IGF-IR on their physical association. Stimulation of serum-deprived Karpas 299 and SU-DHL-1 cells with IGF-I increased the total tyrosine phosphorylation of IGF-IR and NPM-ALK, which remarkably enhanced their association. Collectively, these results underscore that adequate total tyrosine phosphorylation is necessary for the association between NPM-ALK and IGF-IR. Similar to these findings, it was previously shown that adequate levels of total tyrosine phosphorylation are required for NPM-ALK and IGF-IR to physically associate with other signaling partners [48,49].
Our previous studies using segment deletion mutants suggested that the C terminus of NPM-ALK is the site of its association with IGF-IR [32]. Indeed, a mutation of Tyr644 or Tyr664 but not Tyr567 or Tyr646 that are all located within the C terminus of NPM-ALK to phenylalanine significantly decreased the association of NPM-ALK with IGF-IR [32]. To further examine the contribution of the Tyr644 and Tyr664 residues of NPM-ALK to its association with IGF-IR, we exploited three different strategies—IP/WB, laser scanning electron microscopy, and in situ PLA—which all failed to show an association between IGF-IR and NPM-ALKY644,664F. It is important to mention that these experiments were performed in two different types of cells: 293T cells, where the association with endogenously expressed IGF-IR was examined, and R- cells, where the association with forcibly expressed exogenous IGF-IR was analyzed.
Next, the functional interplay between IGF-IR and NPM-ALK or NPM-ALKY644,664F was studied in P6 cells that overexpress IGF-IR and compared with that in R- cells with targeted ablation of IGF-IR [21,34]. A relatively modest, yet statistically significant, increase in NPM-ALK tyrosine kinase activity was observed when NPM-ALK was transfected into R- cells and also when NPM-ALKY644,664F was transfected into P6 or R- cells. Like NPM-ALK, NPM-ALKY644,664F contains the first 117 amino acids of the N terminus of NPM, which have been implicated in the homodimerization and subsequent autoactivation of NPM-ALK [10]. Therefore, the increase in NPM-ALK tyrosine kinase activity most likely resulted from the inherent ability of NPM-ALK or NPM-ALKY644,664F to dimerize. Nonetheless, NPM-ALK tyrosine kinase activity was dramatically increased when it was transfected into P6 cells. In further support of functional collaborations between NPM-ALK and IGF-IR, the tyrosine kinase activity of IGF-IR significantly increased when in vitro translated NPM-ALK protein, but not NPM-ALKY644,664F, was used as a direct substrate of IGF-IR. These data suggest that the presence of IGF-IR and its association with NPM-ALK can reciprocally enhance the basal levels of their tyrosine kinase activities.
Next, we compared the interactions between IGF-IR and NPM-ALK or NPM-ALKY644,664F with its interactions with other NPM-ALK mutants that harbor specific functional defects. Cotransfection of IGF-IR and NPM-ALKY644,664F in 293T cells did not affect the basal levels of NPM-ALK tyrosine kinase activity as it induced effects similar to cotransfection of EV with NPM-ALK. As with the findings discussed above, these results could be explained by the ability of NPM-ALK or NPM-ALKY644,664F to form homodimers that possess basal levels of autoactivated tyrosine kinase. Compared with cotransfection of IGF-IR and NPM-ALK, which was associated with dramatic enhancement of NPM-ALK tyrosine kinase activity, transfection of IGF-IR with the functionally defective mutant NPM-ALK (98–680), which lacks NPM and the ability to form dimers, was associated with lessened NPM-ALK tyrosine kinase activity. Finally, transfection of IGF-IR with the most defective mutant, NPM-ALK (98–566), was associated with the smallest level of NPM-ALK tyrosine kinase activity. NPM-ALK (98–566) lacks NPM and the distal 114 amino acid residues of the C terminus that contain the sites where NPM-ALK physically associates with Shc, Grb2, PLC-γ, and IGF-IR, and possibly other unidentified molecules [11,32,39,50].
In addition to its effects on NPM-ALK tyrosine kinase activity, the collaborative interplay between NPM-ALK and IGF-IR also enhances the cellular transformation potential of NPM-ALK because cotransfection of IGF-IR and NPM-ALK significantly increased the anchorage-independent colony formation of 293T cells. In contrast, transfection of NPM-ALK alone or IGF-IR with NPM-ALKY644,664F or with NPM-ALK (98–680) induced lesser effects. As expected, cotransfecting IGF-IR and NPM-ALK (98–566) was associated with the least number of 293T cell colonies.
When the biologic characteristics of the different clones of stably transfected P6 cells were assessed, the potential of P6 cells that expressed NPM-ALKY644,664F to form anchorage-independent colonies in soft agar or to migrate was much less pronounced than that of P6 cells that expressed NPM-ALK. Moreover, only P6 cells that expressed NPM-ALK demonstrated morphologic features consistent with cellular transformation. In a striking difference, NPM-ALK failed to induce anchorage-independent colony formation, migration, or cellular transformation when stably expressed in R- cells that lack IGF-IR expression.
In addition to the mouse fibroblast P6 cells, we also compared the effects of NPM-ALK and NPM-ALKY644,664F using the NPM-ALK-negative T cell anaplastic large-cell lymphoma cell line Mac-2A, which expresses endogenous IGF-IR. Transient transfection of NPM-ALK increased the proliferation of Mac-2A cells, but NPM-ALKY644,664F failed to induce similar effects. Collectively, these results indicate that Tyr644 and Tyr664 are required for NPM-ALK to associate and collaborate with IGF-IR, and such collaboration potentiates the survival-promoting and cellular transformation potential of NPM-ALK.
WT NPM is a ubiquitously expressed RNA-binding phospho-protein [51]. Its physiological functions include the shuttling of ribonucleoproteins between the nucleolus and cytoplasm. The first 117 amino acids of NPM comprise the proximal portion of the N terminus of NPM-ALK, but they lack the nuclear localization signal of NPM. Nevertheless, NPM-ALK forms dimers with WT NPM, and the NPM/NPM-ALK heterodimer can translocate to the nucleus. In addition, earlier studies have demonstrated that NPM-ALK can also form homodimers through the NPM domain, which subsequently induces autoactivation of the ALK tyrosine kinase domain. This process is believed to be a major mechanism by which NPM-ALK induces its deregulated signaling [10]. Whether constitutive activation through homodimerization is the sole mechanism that leads to NPM-ALK phosphorylation/activation and whether constitutively active NPM-ALK can induce cellular transformation independently from other interacting molecules remain the subjects of debate [49,52].
The conclusions of some of the earlier studies could have been affected by their experimental conditions. Notably, the majority of these studies were performed using exogenous NPM-ALK that was forcibly expressed in rodent fibroblasts, e.g., mouse NIH/3T3 or rat-1 fibroblasts. It is possible that the phenotypic backgrounds of these cells are not ideal for drawing definitive conclusions related to the biologic characteristics of NPM-ALK. For example, in contrast to the NPM-ALK+ T cell lymphoma cell lines and primary tumor cells that express high levels of IGF-IR and pIGF-IR [32], we found that NIH/3T3 cells express low levels of IGF-IR and lack the expression of pIGF-IR. It is still possible, however, that NPM-ALK induced cellular transformation of rodent fibroblasts such as NIH/3T3 cells through reciprocal interaction/activation of endogenously expressed IGF-IR, albeit at low levels, in these cells. Nonetheless, when we used R- cells that lack the expression of IGF-IR, NPM-ALK failed to induce cellular transformation. In the work presented herein, a plethora of variable cell lines, including mouse fibroblasts with different levels of expression of IGF-IR, human embryonic kidney cells, and NPM-ALK-negative and NPM-ALK+ T cell lymphoma cell lines, were used with consistent results.
The relevant observation that modulations of FBS levels through supplementation, withdrawal, or deprivation can affect the growth pattern of NPM-ALK+ cells in culture argues against the idea that the survival of these cells is contingent upon homodimerized and constitutively activated NPM-ALK independent from interactions with other extracellular or intracellular molecules that might also affect the phosphorylation and tyrosine kinase activation levels of NPM-ALK. In line with this concept, we previously found that specific down-regulation of NPM-ALK by ALK siRNA was associated with decreased levels of pSTAT3 and pAKT in NPM-ALK+ T cell lymphoma cells [32]. However, the decrease in pSTAT3 and pAKT was largely recovered when the cells were simultaneously treated with ALK siRNA and IGF-I, which is known to be present in FBS [53]. These results suggest that important survival proteins could be activated through alternative pathways that are complementary to NPM-ALK. In the current study, we additionally found that extracellular stimulation of serum-deprived NPM-ALK+ cells by IGF-I not only enhances total tyrosine phosphorylation of IGF-IR but also increases total tyrosine phosphorylation of NPM-ALK. Because NPM-ALK lacks extracellular and transmembranous domains, the effects of IGF-I on NPM-ALK were most likely mediated through stimulation of IGF-IR by IGF-I. These observations strongly suggest that extracellular mediators could interact with intermediary molecules that subsequently affect the basal levels of the phosphorylation and constitutive tyrosine kinase activation of NPM-ALK.
In further support of the contribution of other molecules to the activation and oncogenic effects of NPM-ALK, recent studies demonstrated that constitutively activated NPM-ALK was only capable of promoting cellular immortalization and that interactions with H-ras were required for NPM-ALK to induce cellular transformation [52]. In another study, the Src kinase pp60c-src was found to be physically associated with NPM-ALK. Importantly, pp60c-src appeared to use NPM-ALK as a substrate and to significantly increase its tyrosine phosphorylation levels [49]. In a similar model, Src kinases were found to induce phosphorylation of Bcr-Abl and to modulate its cellular transformation activity [54]. In addition, Src kinases induced phosphorylation of IGF-IR at tyrosine residues known to undergo autophosphorylation [55].
To understand the basis underlying the biologic defects resulting from lack of association and interactions between NPM-ALK and IGF-IR, we analyzed the biochemical consequences of dual mutation of NPM-ALK's Tyr644 and Tyr664 to phenylalanine. This dual mutation was associated with a marked decrease in the total tyrosine phosphorylation of NPM-ALK and IGF-IR, providing additional evidence that the effective association and interplay between these two kinases can sustain ample levels of total tyrosine phosphorylation. In further support of this notion, transfection of NPM-ALKY644,664F into 293T cells was associated with reduced phosphorylation of Tyr646, one of the four tyrosine residues located within the C terminus of NPM-ALK. Furthermore, transfection of NPM-ALKY644,664F was associated with decreased phosphorylation of IGF-IR at Tyr1131 and Tyr1135, which comprise the first and second tyrosine residues, respectively, of the YXXXYY motif of the IGF-IR kinase domain. These findings are quite significant, considering that Tyr1131 and Tyr1135 are used by IGF-IR to induce cellular transformation [56,57]. Transfection of NPM-ALKY644,664F was also associated with significant down-regulation of pSTAT3Y750, pAKTS473, and pERKT202/Y204. These oncogenic proteins contribute significantly to the signaling of both NPM-ALK and IGF-IR [14–18,58–62].
Because the treatment of R- cells transfected with NPM-ALKY644,664F with MG132 induced a marked increase in the levels of this double mutant, we postulated that NPM-ALKY644,664F is regulated posttranslationally by the proteasome degradation system, which is also a proposed mechanism for the regulation of NPM-ALK expression [36,37]. Using CHX to abort new protein formation, we noticed no changes in the stability of NPM-ALK protein transfected into P6 cells. Conversely, NPM-ALK demonstrated lesser stability when transfected into R- cells. These results suggest that the association between IGF-IR and NPM-ALK may lead to stabilization of NPM-ALK protein. Indeed, the stability of NPM-ALKY644,664F was notably decreased in P6 cells compared with the stability of NPM-ALK in the same cells. Because the stability of NPM-ALKY644,664F was lower than that of NPM-ALK in R- cells, we assumed that the association between NPM-ALK and IGF-IR through Tyr644 and Tyr664 may also involve other proteins that sustain the stability of NPM-ALK. Previous studies have shown that the proteasome degradation pathway proteins, including Hsp90, Hsp70, and CHIP, associate with and regulate the stabilization/degradation of NPM-ALK [36,38]. These studies demonstrated that the association of NPM-ALK with the chaperon Hsp90 enhances the stability of NPM-ALK protein. In contrast, the association with Hsp70 facilitates its degradation, and this process is accelerated by the CHIP ligase [36]. In this study, we found that NPM-ALKY644,664F retained the ability to associate with Hsp70 and CHIP, but its ability to associate with Hsp90 was greatly diminished, which could explain, at least partially, the decreased stability of this mutant compared with NPM-ALK. Thereafter, we questioned whether, in addition to Hsp90, IGF-IR also supports the stability of NPM-ALK protein. Specific targeting of IGF-IR by siRNA was associated with a remarkable down-regulation of NPM-ALK protein levels in NPM-ALK+ T cell lymphoma cells. These novel data suggest that the presence of Hsp90 and IGF-IR in the same protein complex with NPM-ALK contributes to maintaining the stability of NPM-ALK.
The NPM-ALK protein contains 12 tyrosine residues (www.phosphosite.org), and some of these residues are used for associations and interactions with other proteins. The tyrosine residues that associate NPM-ALK with IRS-1, Vav3, pp60c-src, and Shc have been identified as Tyr156, Tyr343, Tyr418, and Tyr567, respectively [11,49,63]. In addition, the Grb2 protein was shown to associate with several amino acid residues of NPM-ALK including Tyr156 and Tyr567 [50]. Of important note is that the sites of the association between NPM-ALK and some of its most important downstream targets such as PI3K/AKT and STAT3 are not known. In relevance to our findings related to Tyr664, it was previously reported that PLC-γ contributes to the mitogenicity of NPM-ALK through its association with Tyr664 [39]. The conclusion that PLC-γ contributes to the mitogenicity of NPM-ALK was largely based on two indirect observations: 1) overexpression of PLC-γ partially rescued Ba/F3 cells transfected with NPM-ALKY664F from interleukin-3 (IL-3) dependence, and 2) unlike NPM-ALKY664F, the expression of NPM-ALK enhanced the production of inositol phosphates in Ba/F3 cells [39]. In our study, specific down-regulation of PLC-γ by siRNA neither decreased the proliferation of NPM-ALK+ T cell lymphoma cells nor decreased the expression levels of pSTAT3Y705, pAKTS473, and pERKT202/Y204. Although a role of PLC-γ in NPM-ALK+ T cell lymphoma cannot be ruled out, the exact mechanisms of this role remain to be delineated.
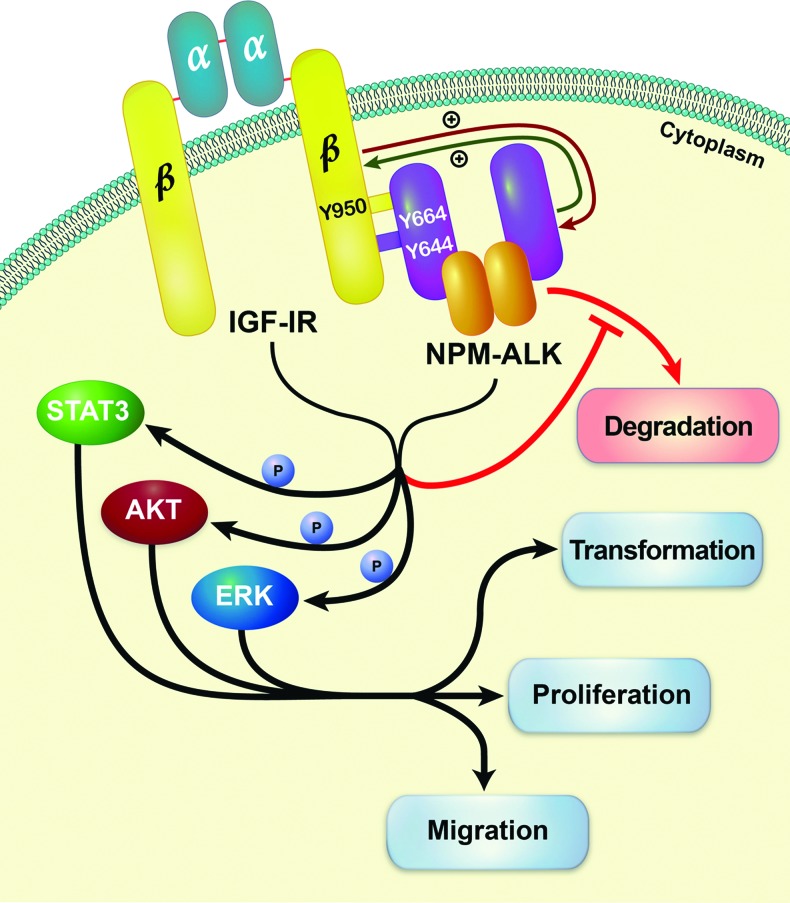
In conclusion, in this work we present novel data showing that the oncogenic role of NPM-ALK is facilitated through its physical association and interactions with another oncogenic tyrosine kinase, namely, IGF-IR. The association with IGF-IR supports high levels of kinase activity and tyrosine phosphorylation levels of NPM-ALK, which appears to enhance the phosphorylation of common oncogenic downstream targets. Our results suggest that the functional interplay between NPM-ALK and IGF-IR stimulates cellular transformation, proliferation, and migration; induced by NPM-ALK, as well as hinders the degradation of NPM-ALK protein (proposed model is depicted in Figure 7). These findings expand current knowledge of the biology of NPM-ALK and may lead to improved therapeutic approaches to the treatment of clinically aggressive NPM-ALK+ T cell lymphoma.
Figure 7.
Our data propose a novel model in which NPM-ALK and IGF-IR are physically associated through Tyr644 and Tyr664 of NPM-ALK and Tyr950 of IGF-IR. Through this association, the phosphorylation and tyrosine kinase activity of the two oncogenic proteins are maintained at high levels, and this leads to the up-regulation of the phosphorylation of common downstream targets including STAT3, AKT, and ERK. Such effects enhance the transformation, proliferation, and migration of NPM-ALK+ T cell lymphoma cells. In addition, the physical association between NPM-ALK and IGF-IR, in the presence of Hsp90 (not shown), appears to sustain the stability of NPM-ALK protein.
Supplementary Material
Acknowledgments
The authors thank Dawn Chalaire for outstanding assistance with the preparation of this paper.
Abbreviations
- ALK
anaplastic lymphoma kinase
- CHX
cycloheximide
- FBS
fetal bovine serum
- GST
glutathione S-transferase
- IGF-IR
type I insulin-like growth factor receptor
- IP
immunoprecipitation
- NPM
nucleophosmin
- PDGFR
platelet-derived growth factor receptor
- PLA
proximity ligation assay
- PLC-γ
phospholipase C-γ
- WB
Western blot
- WT
wild type
Footnotes
The study is supported by grant R01 CA151533 from the National Cancer Institute and by a Bridge Funding grant from MD Anderson Cancer Center to H.M.A. The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. The authors declare no competing financial interests.
This article refers to supplementary materials, which are designated by Figures W1 to W6 and are available online at www.neoplasia.com.
References
- 1.Morris SW, Naeve C, Mathew P, James PL, Kirstein MN, Cui X, Witte DP. ALK, the chromosome 2 gene locus altered by the t(2;5) in non-Hodgkin's lymphoma, encodes a novel neural receptor tyrosine kinase that is highly related to leukocyte tyrosine kinase (LTK) Oncogene. 1997;14:2175–2188. doi: 10.1038/sj.onc.1201062. [DOI] [PubMed] [Google Scholar]
- 2.Bowden ET, Stoica GE, Wellstein A. Anti-apoptotic signaling of pleiotrophin through its receptor, anaplastic lymphoma kinase. J Biol Chem. 2002;227:35862–35868. doi: 10.1074/jbc.M203963200. [DOI] [PubMed] [Google Scholar]
- 3.Stoica GE, Kuo A, Powers C, Bowden ET, Sale EB, Riegel AT, Wellstein A. Midkine binds to anaplastic lymphoma kinase (ALK) and acts as a growth factor for different cell types. J Biol Chem. 2002;277:35990–35998. doi: 10.1074/jbc.M205749200. [DOI] [PubMed] [Google Scholar]
- 4.Lorente M, Torres S, Salazar M, Carracedo A, Hernandez-Tiedra S, Rodriguez-Fornes F, Garcia-Taboada E, Melendez B, Mollejo M, Campos-Martin Y, et al. Stimulation of the midkine/ALK axis renders glioma cells resistant to cannabinoid antitumoral action. Cell Death Differ. 2011;18:959–973. doi: 10.1038/cdd.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iwahara M, Fujimoto J, Wen D, Cupples R, Bucay N, Arakwa T, Mori S, Ratzkin B, Yamamoto T. Molecular characterization of ALK, a receptor tyrosine kinase expressed specifically in the nervous system. Oncogene. 1997;14:439–449. doi: 10.1038/sj.onc.1200849. [DOI] [PubMed] [Google Scholar]
- 6.Amin HM, Lai R. Pathobiology of ALK+ anaplastic large-cell lymphoma. Blood. 2007;110:2259–2267. doi: 10.1182/blood-2007-04-060715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiarle R, Voena C, Ambrogio C, Piva R, Inghirami G. The anaplastic lymphoma kinase in the pathogenesis of cancer. Nat Rev Cancer. 2008;8:11–23. doi: 10.1038/nrc2291. [DOI] [PubMed] [Google Scholar]
- 8.Morris SW, Kirstein MN, Valentine MB, Dittmer KG, Shapiro DN, Saltman DL, Look AT. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science. 1994;263:1281–1284. doi: 10.1126/science.8122112. [DOI] [PubMed] [Google Scholar]
- 9.Drexler HG, Gignac SM, von Wasielewski R, Werner M, Dirks WG. Pathobiology of NPM-ALK and variant fusion genes in anaplastic large cell lymphoma and other lymphomas. Leukemia. 2000;14:1533–1559. doi: 10.1038/sj.leu.2401878. [DOI] [PubMed] [Google Scholar]
- 10.Bischof D, Pulford K, Mason DY, Morris SW. Role of the nucleophosmin (NPM) portion of the non-Hodgkin's lymphoma-associated NPM-anaplastic lymphoma kinase fusion protein in oncogenesis. Mol Cell Biol. 1997;17:2312–2325. doi: 10.1128/mcb.17.4.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujimoto J, Shiota M, Iwahara T, Seki N, Satoh H, Mori S, Yamamoto T. Characterization of the transforming activity of p80, a hyperphosphorylated protein in a Ki-1 lymphoma cell line with chromosomal translocation t(2;5) Proc Natl Acad Sci USA. 1996;93:4181–4186. doi: 10.1073/pnas.93.9.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiarle R, Gong JZ, Guasparri I, Pesci A, Cai J, Liu J, Simmons WJ, Dhall G, Howes J, Piva R, et al. NPM-ALK transgenic mice spontaneously develop T-cell lymphomas and plasma cell tumors. Blood. 2003;101:1919–1927. doi: 10.1182/blood-2002-05-1343. [DOI] [PubMed] [Google Scholar]
- 13.Giuriato S, Foisseau M, Dejean E, Felsher DW, Al Saati T, Demur C, Ragab A, Kruczynski A, Schiff C, Delsol G, et al. Conditional TPM3-ALK and NPM-ALK transgenic mice develop reversible ALK-positive early B-cell lymphoma/leukemia. Blood. 2010;115:4061–4070. doi: 10.1182/blood-2008-06-163386. [DOI] [PubMed] [Google Scholar]
- 14.Bai RY, Ouyang T, Miething C, Morris SW, Peschel C, Duyster J. Nucleophosmin-anaplastic lymphoma kinase associated with anaplastic large-cell lymphoma activates the phosphatidylinositol 3-kinase/Akt antiapoptotic signaling pathway. Blood. 2000;96:4319–4327. [PubMed] [Google Scholar]
- 15.Zamo A, Chiarle R, Piva R, Howes J, Fan Y, Chilosi M, Levy DE, Inghirami G. Anaplastic lymphoma kinase (ALK) activates Stat3 and protects hematopoietic cells from cell death. Oncogene. 2002;21:1038–1047. doi: 10.1038/sj.onc.1205152. [DOI] [PubMed] [Google Scholar]
- 16.Amin HM, McDonnell TJ, Ma Y, Lin Q, Fujio Y, Kunisada K, Leventaki V, Das P, Rassidakis GZ, Cutler C, et al. Selective inhibition of STAT3 induces apoptosis and G1 cell cycle arrest in ALK-positive anaplastic large cell lymphoma. Oncogene. 2003;23:5426–5434. doi: 10.1038/sj.onc.1207703. [DOI] [PubMed] [Google Scholar]
- 17.Rassidakis GZ, Feretzaki M, Atwell C, Grammatikakis I, Lin Q, Lai R, Claret FX, Medeiros LJ, Amin HM. Inhibition of Akt increases p27Kip1 levels and induces cell cycle arrest in anaplastic large cell lymphoma. Blood. 2005;105:827–829. doi: 10.1182/blood-2004-06-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Staber PB, Vesely P, Haq N, Ott RG, Funato K, Bambach I, Fuchs C, Schauer S, Linkesch W, Hrzenjak A, et al. The oncoprotein NPM-ALK of anaplastic large-cell lymphoma induces JUNB transcription via ERK1/2 and JunB translation via mTOR signaling. Blood. 2007;110:3374–3383. doi: 10.1182/blood-2007-02-071258. [DOI] [PubMed] [Google Scholar]
- 19.Ullrich A, Gray A, Tam AW, Yang-Feng T, Tsubokawa M, Collins C, Henzel W, LeBon T, Kathuria S, Chen E, et al. Insulin-like growth factor I receptor primary structure: comparison with insulin receptor suggests structural determinants that define functional specificity. EMBO J. 1986;5:2503–2512. doi: 10.1002/j.1460-2075.1986.tb04528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaleko M, Butter WJ, Miller AD. Overexpression of the human insulinlike growth factor I receptor promotes ligand-dependent neoplastic transformation. Mol Cell Biol. 1990;10:464–473. doi: 10.1128/mcb.10.2.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pietrzkowski Z, Lammers R, Carpenter G, Soderquist AM, Limardo M, Phillips PD, Ullrich A, Baserga R. Constitutive expression of insulin-like growth factor 1 and insulin-like growth factor 1 receptor abrogates all requirements for exogenous growth factors. Cell Growth Differ. 1992;3:199–205. [PubMed] [Google Scholar]
- 22.Baserga R, Sell C, Porcu P, Rubini M. The role of the IGF-I receptor in the growth and transformation of mammalian cells. Cell Prolif. 1994;27:63–71. doi: 10.1111/j.1365-2184.1994.tb01406.x. [DOI] [PubMed] [Google Scholar]
- 23.Mitsiades CS, Mitsiades NS, McMullan CJ, Poulaki V, Shringarpure R, Akiyama M, Hideshima T, Chauhan D, Joseph M, Libermann TA, et al. Inhibition of the insulin-like growth factor receptor-1 tyrosine kinase activity as a therapeutic strategy for multiple myeloma, other hematologic malignancies, and solid tumors. Cancer Cell. 2004;5:221–230. doi: 10.1016/s1535-6108(04)00050-9. [DOI] [PubMed] [Google Scholar]
- 24.Samani AA, Yakar S, LeRoith D, Brodt P. The role of the IGF system in cancer growth and metastasis: overview and recent insights. Endocr Rev. 2007;28:20–47. doi: 10.1210/er.2006-0001. [DOI] [PubMed] [Google Scholar]
- 25.Shi P, Chandra J, Sun X, Gergely M, Cortes JE, Garcia-Manero G, Arlinghaus RB, Lai R, Amin HM. Inhibition of IGF-IR tyrosine kinase induces apoptosis and cell cycle arrest in imatinib-resistant chronic myeloid leukemia cells. J Cell Mol Med. 2010;14:1777–1792. doi: 10.1111/j.1582-4934.2009.00795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vishwamitra D, Shi P, Wilson D, Manshouri R, Vega F, Schlette EJ, Amin HM. Expression and effects of inhibition of type I insulin-like growth factor receptor tyrosine kinase in mantle cell lymphoma. Haematologica. 2011;96:871–880. doi: 10.3324/haematol.2010.031567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sell C, Rubini M, Rubin R, Liu JP, Efstratiadis A, Baserga R. Simian virus 40 large tumor antigen is unable to transform mouse embryonic fibroblasts lacking type 1 insulin-like growth factor receptor. Proc Natl Acad Sci USA. 1993;90:11217–11221. doi: 10.1073/pnas.90.23.11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sell C, Dumenil G, Deveaud C, Miura M, Coppola D, DeAngelis T, Rubin R, Estratiadis A, Baserga R. Effect of a null mutation of the insulin-like growth factor I receptor gene on growth and transformation of mouse embryo fibroblasts. Mol Cell Biol. 1994;14:3604–3612. doi: 10.1128/mcb.14.6.3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coppola D, Ferber A, Miura M, Sell C, D'Ambrosio C, Rubin R, Baserga R. A functional insulin-like growth factor I receptor is required for the mitogenic and transforming activities of the epidermal growth factor receptor. Mol Cell Biol. 1994;14:4588–4595. doi: 10.1128/mcb.14.7.4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeAngelis T, Ferber A, Baserga R. Insulin-like growth factor I receptor is required for the mitogenic and transforming activities of the platelet-derived growth factor receptor. J Cell Physiol. 1995;164:214–221. doi: 10.1002/jcp.1041640126. [DOI] [PubMed] [Google Scholar]
- 31.Toretsky JA, Kalebic T, Blakesley V, LeRoith D, Helman LJ. The insulin-like growth factor-I receptor is required for EWS/FLI-1 transformation of fibroblasts. J Biol Chem. 1997;272:30822–30827. doi: 10.1074/jbc.272.49.30822. [DOI] [PubMed] [Google Scholar]
- 32.Shi P, Lai R, Lin Q, Iqbal AS, Young LC, Kwak LW, Ford RJ, Amin HM. IGF-IR tyrosine kinase interacts with NPM-ALK oncogene to induce survival of T-cell ALK+ anaplastic large-cell lymphoma cells. Blood. 2009;114:360–370. doi: 10.1182/blood-2007-11-125658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drexler HG. Guide to Leukemia-Lymphoma Cell Lines. Braunschweig, Germany: 2010. [Google Scholar]
- 34.Liu J-P, Baker J, Perkins AS, Robertson EJ, Efstratiadis A. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r) Cell. 1993;75:59–72. [PubMed] [Google Scholar]
- 35.Söderberg O, Gullberg M, Jarvius M, Ridderstråle K, Leuchowius K-J, Wester K, Hydbring P, Bahram F, Larsson LG, Landegren U. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 36.Bonvini P, Dalla Rosa H, Vignes N, Rosolen A. Ubiquitination and proteasomal degradation of nucleophosmin-anaplastic lymphoma kinase induced by 17-allylamino-demethoxygeldanamycin: role of the cochaperone carboxyl heat shock protein 70-interacting protein. Cancer Res. 2004;64:3256–3264. doi: 10.1158/0008-5472.can-03-3531. [DOI] [PubMed] [Google Scholar]
- 37.Han Y, Amin HM, Franko B, Frantz C, Shi X, Lai R. Loss of SHP1 enhances JAK3/STAT3 signaling and decreases proteosome degradation of JAK3 and NPM-ALK in ALK+ anaplastic large-cell lymphoma. Blood. 2006;108:2796–2803. doi: 10.1182/blood-2006-04-017434. [DOI] [PubMed] [Google Scholar]
- 38.Bonvini P, Gastaldi R, Falini B, Rosolen A. Nucleophosmin-anaplastic lymphoma kinase (NPM-ALK), a novel Hsp90-client tyrosine kinase: down-regulation of NPM-ALK expression and tyrosine phosphorylation in ALK+ CD30+ lymphoma cells by the Hsp90 antagonist 17-allylamino,17-demethoxygeldanamycin. Cancer Res. 2002;62:1559–1566. [PubMed] [Google Scholar]
- 39.Bai R-Y, Dieter P, Peschel C, Morris SW, Duyster J. Nucleophosmin-anaplastic lymphoma kinase of large-cell anaplastic lymphoma is a constitutively active tyrosine kinase that utilizes phospholipase C-γ to mediate its mitogenicity. Mol Cell Biol. 1998;18:6951–6961. doi: 10.1128/mcb.18.12.6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laimer D, Dolzing H, Kollmann K, Vesely PW, Schlederer M, Merkel O, Schiefer A-I, Hassler MR, Heider S, Amenitsch L, et al. PDGFR blockade is a rational and effective therapy for NPM-ALK-driven lymphomas. Nat Med. 2012;18:1699–1704. doi: 10.1038/nm.2966. [DOI] [PubMed] [Google Scholar]
- 41.Miura M, Li S, Baserga R. Effect of a mutation at tyrosine 950 of the insulin-like growth factor I receptor on the growth and transformation of cells. Cancer Res. 1995;55:663–667. [PubMed] [Google Scholar]
- 42.Dey BR, Frick K, Lopaczynski W, Nissley SP, Furlanetto RW. Evidence for the direct interaction of the insulin-like growth factor I receptor with IRS-1, Shc, and Grb10. Mol Endocrinol. 1996;10:631–641. doi: 10.1210/mend.10.6.8776723. [DOI] [PubMed] [Google Scholar]
- 43.Jiang Y, Chan JL-K, Zong CS, Wang L-H. Effect of tyrosine mutations on the kinase activity and transforming potential of an oncogenic human insulin-like growth factor I receptor. J Biol Chem. 1996;271:160–167. doi: 10.1074/jbc.271.1.160. [DOI] [PubMed] [Google Scholar]
- 44.Sasaki N, Rees-Jones RW, Zick Y, Nissley SP, Rechler MM. Characterization of insulin-like growth factor I-stimulated tyrosine kinase activity associated with the β-subunit of type I insulin-like growth factor receptors of rat liver cells. J Biol Chem. 1985;260:9793–9804. [PubMed] [Google Scholar]
- 45.Yu K-T, Peters MA, Czech MP. Similar control mechanisms regulate the insulin and type I insulin-like growth factor receptor kinases. J Biol Chem. 1986;261:11341–11349. [PubMed] [Google Scholar]
- 46.Kalebic T, Blakesley V, Slade C, Plasschaert S, LeRoith D, Helman LJ. Expression of a kinase-deficient IGF-I-R suppresses tumorigenicity of rhabdomyosarcoma cells constitutively expressing a wild type IGF-I-R. Int J Cancer. 1998;76:223–227. doi: 10.1002/(sici)1097-0215(19980413)76:2<223::aid-ijc9>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 47.Tartari CJ, Gunby RH, Coluccia AML, Sottocornola R, Cimbro B, Scapozza L, Donella-Deana A, Pinna LA, Gambacorti-Passerini C. Characterization of some molecular mechanisms governing autoactivation of the catalytic domain of the anaplastic lymphoma kinase. J Biol Chem. 2008;283:3743–3750. doi: 10.1074/jbc.M706067200. [DOI] [PubMed] [Google Scholar]
- 48.Tartare-Deckert S, Sawka-Verhelle D, Murdaca J, Van Obberghen E. Evidence for a differential interaction of SHC and the insulin receptor substrate-1 (IRS-1) with the insulin-like growth factor-I (IGF-I) receptor in the yeast two-hybrid system. J Biol Chem. 1995;270:23456–23460. doi: 10.1074/jbc.270.40.23456. [DOI] [PubMed] [Google Scholar]
- 49.Cussac D, Greenland C, Roche S, Bai R-Y, Duyster J, Morris SW, Delsol G, Allouche M, Payrastre B. Nucleophosmin-anaplastic lymphoma kinase of anaplastic large-cell lymphoma recruits, activates, and uses pp60c-src to mediate its mitogenicity. Blood. 2004;103:1464–1471. doi: 10.1182/blood-2003-04-1038. [DOI] [PubMed] [Google Scholar]
- 50.Riera L, Lasorsa E, Ambrogio C, Surrenti N, Voena C, Chiarle R. Involvement of Grb2 adaptor protein in nucleophosmin-anaplastic lymphoma kinase (NPM-ALK)-mediated signaling and anaplastic large cell lymphoma growth. J Biol Chem. 2010;285:26441–26450. doi: 10.1074/jbc.M110.116327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Borer RA, Lehner CF, Eppenberger HM, Nigg EA. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 1989;56:379–390. doi: 10.1016/0092-8674(89)90241-9. [DOI] [PubMed] [Google Scholar]
- 52.Simonitsch I, Polgar D, Hajek M, Duchek P, Skrzypek B, Fassl S, Lamprecht A, Schmidt G, Krupitza G, Cerni C. The cytoplasmic truncated receptor tyrosine kinase ALK homodimer immortalizes and cooperates with ras in cellular transformation. FASEB J. 2001;15:1416–1418. doi: 10.1096/fj.00-0678fje. Published online April 6, 2001. [DOI] [PubMed] [Google Scholar]
- 53.Lopez-Lopez C, LeRoith D, Torres-Aleman I. Insulin-like growth factor I is required for vessel remodeling in the adult brain. Proc Natl Acad Sci USA. 2004;101:9833–9838. doi: 10.1073/pnas.0400337101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meyn MA, Wilson MB, Abdi FA, Fahey N, Schiavone AP, Wu J, Hochrein JM, Engen JR, Smithgall TE. Src family kinases phosphorylate the Bcr-Abl SH3-SH2 region and modulate Bcr-Abl transforming activity. J Biol Chem. 2006;281:30907–30916. doi: 10.1074/jbc.M605902200. [DOI] [PubMed] [Google Scholar]
- 55.Peterson JE, Kulik G, Jelinek T, Reuter CWM, Shannon JA, Weber MJ. Src phosphorylates the insulin-like growth factor type I receptor on the autophosphorylation sites. J Biol Chem. 1996;271:31562–31571. doi: 10.1074/jbc.271.49.31562. [DOI] [PubMed] [Google Scholar]
- 56.Li S, Ferber A, Miura M, Baserga R. Mitogenicity and transforming activity of the insulin-like growth factor-I receptor with mutations in the tyrosine kinase domain. J Biol Chem. 1994;269:32558–32564. [PubMed] [Google Scholar]
- 57.Kato H, Faria TN, Stannard B, Roberts CT, Jr, LeRoith D. Essential role of tyrosine residues 1131, 1135, and 1136 of the insulin-like growth factor-I (IGF-I) receptor in IGF-I action. Mol Endocrinol. 1994;8:40–50. doi: 10.1210/mend.8.1.7512194. [DOI] [PubMed] [Google Scholar]
- 58.Zong CS, Zeng L, Jiang Y, Sadowski HB, Wang L-H. Stat3 plays an important role in oncogenic Ros- and insulin-like growth factor I receptor-induced anchorage-independent growth. J Biol Chem. 1998;273:28065–28072. doi: 10.1074/jbc.273.43.28065. [DOI] [PubMed] [Google Scholar]
- 59.Peruzzi F, Prisco M, Dews M, Salomoni P, Grassilli E, Romano G, Calabretta B, Baserga R. Multiple signaling pathways of the insulin-like growth factor 1 receptor in protection from apoptosis. Mol Cell Biol. 1999;19:7203–7215. doi: 10.1128/mcb.19.10.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dews M, Prisco M, Peruzzi F, Romano G, Morrione A, Baserga R. Domains of the insulin-like growth factor I receptor required for the activation of extracellular signal-regulated kinases. Endocrinology. 2000;141:1289–1300. doi: 10.1210/endo.141.4.7414. [DOI] [PubMed] [Google Scholar]
- 61.Zong CS, Chan J, Levy DE, Horvath C, Sadowski HB, Wang L-H. Mechanism of STAT3 activation by insulin-like growth factor I receptor. J Biol Chem. 2000;275:15099–15105. doi: 10.1074/jbc.M000089200. [DOI] [PubMed] [Google Scholar]
- 62.Shelton JG, Steelman LS, White ER, McCubrey JA. Synergy between PI3K/Akt and Raf/MEK/ERK pathways in IGF-1R mediated cell cycle progression and prevention of apoptosis in hematopoietic cells. Cell Cycle. 2004;3:372–379. [PubMed] [Google Scholar]
- 63.Colomba A, Courilleau D, Ramel D, Billadeau DD, Espinos E, Delsol G, Payrastre B, Gaits-Iacovoni F. Activation of Rac1 and the exchange factor Vav3 are involved in NPM-ALK signaling in anaplastic large cell lymphomas. Oncogene. 2008;27:2728–2736. doi: 10.1038/sj.onc.1210921. [DOI] [PubMed] [Google Scholar]
- 64.Dong Y, Jia L, Wang X, Tan X, Xu J, Deng Z, Jiang T, Rainov NG, Li B, Ren H. Selective inhibition of PDGFR by imatinib elicits the sustained activation of ERK and downstream receptor signaling in malignant glioma cells. Int J Cancer. 2011;38:555–569. doi: 10.3892/ijo.2010.861. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.