Abstract
Single molecule fluorescence resonance energy transfer (smFRET) methods were used to study the assembly pathway and DNA unwinding activity of the bacteriophage T4 helicase-primase (primosome) complex. The helicase substrates used were surface-immobilized model DNA replication forks ‘internally’ labeled in the duplex region with opposed donor/acceptor (iCy3/iCy5) chromophore pairs in the lagging and leading strands. The time-dependence of the smFRET signals was monitored during the unwinding process and helicase rates and processivities were measured as a function of GTP concentration. This smFRET approach was also used to investigate the subunit stoichiometry of the primosome and the assembly pathway required to form functional and fully active primosome-DNA complexes. We confirmed that gp41 helicase monomer subunits form stable hexameric helicases in the presence of GTP and that the resulting (gp41)6 complexes bind only weakly at DNA fork junctions. The addition of a single subunit of gp61 primase stabilized the resulting primosome complex at the fork and resulted in fully active and processive primosome helicases with gp41:gp61 subunit ratios of 6:1, while higher and lower subunit ratios substantially reduced the primosome unwinding activity. The use of alternative assembly pathways resulted in loss of helicase activity and formation of metastable DNA-protein aggregates, which were easily detected in our smFRET experiments as intense light-scattering foci. These single molecule experiments provide a detailed real time visualization of the assembly pathway and duplex DNA unwinding activity of the T4 primosome and are consistent with more indirect equilibrium and steady state results obtained in bulk solution studies.
Introduction
DNA replication helicases are ATP-dependent molecular motors that unwind double-stranded (ds) DNA at replication forks to expose the single-stranded (ss) templates for leading and lagging strand DNA synthesis, and as such are central components of the DNA replication systems of all organisms. The DNA replication complex coded by bacteriophage T4 contains eight different types of subunits, some present in multiple copies, and can be reconstituted in vitro from its components. The reconstituted system performs DNA synthesis at rates and with processivity and fidelity comparable those demonstrated by the in vivo complex.1, 2 The reconstituted T4 replication system serves as an excellent model system for the study of the replication mechanisms of higher organisms, because it is the simplest system to use both a sliding replication clamp-clamp loader sub-assembly to control the processivity of the replication DNA polymerases and a hexameric helicase-primase (primosome) sub-assembly to unwind the base pairs ahead of the polymerases at the replication fork in a processive fashion and to catalyze the synthesis of the RNA primers required for re-initiation of lagging strand synthesis at the 3′-ends of newly formed Okazaki fragments. In this study we use single molecule FRET methodology to investigate the assembly and unwinding reaction pathways of the T4 primosome helicase.
Materials and Methods
DNA substrates
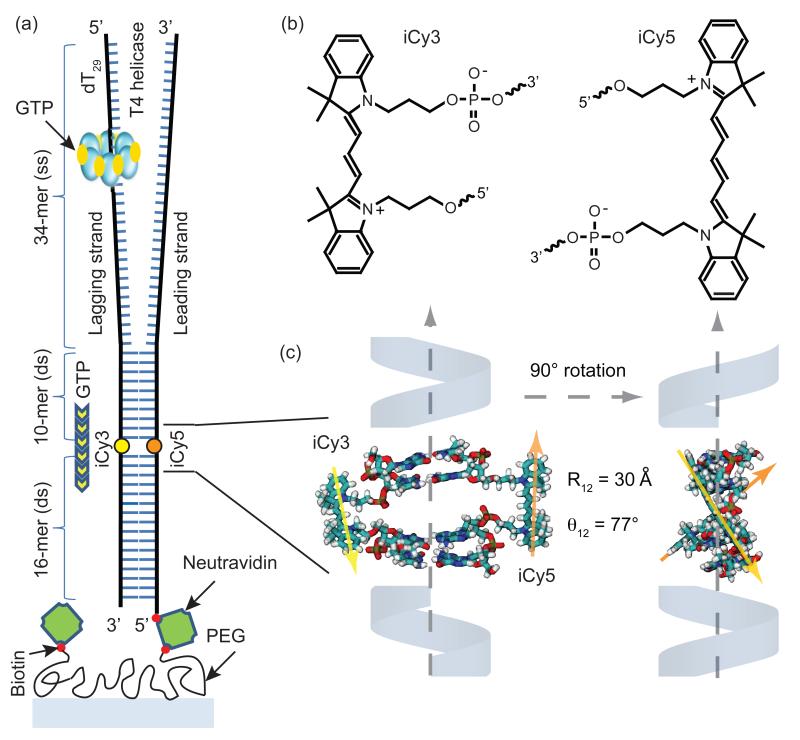
The bacteriophage T4 gp41 helicase unwinds oligomeric double-stranded (ds) DNA substrates in the 5′→3′ direction, with the reaction coupled to GTP hydrolysis.3,4 We designed a model DNA replication fork for single molecule (sm) helicase unwinding smFRET experiments that contains a donor-acceptor iCy3/iCy5 fluorophore pair ‘internally’ labeled on opposing strands within the duplex region, and a single-stranded (ss) 5′ dT29 ‘loading sequence’ upstream of the duplex region (see Fig. 1 and Table S1). As detailed below, bulk helicase unwinding assays were performed to show that the T4 helicase activity is not significantly inhibited by the incorporation of the internally-labeled Cy3/Cy5 FRET complex (see SI and Fig. S1). Functionalized DNA strands, which were assembled using phosphoramidite chemistry, were purchased from Integrated DNA Technologies (Coralville, IA) and are designated as ‘leading’ and ‘lagging’ strands in Fig. 1a (see SI for DNA sequences). These DNA fork substrates were prepared by mixing 100 nM concentrations of the biotinylated leading strand and the non-biotinylated lagging strand in a 1:1.5 ratio in standard imaging buffer (10 mM Tris at pH 8.0, 100 mM NaCl, and 6 mM MgCl2). The DNA construct was assembled and annealed by heating to 90°C for 3 to 4 min and then cooling slowly to room temperature (~22°C). The annealed construct was then diluted to a concentration of 50 to 100 pM, and a 50 JL aliquot was introduced into the sample chamber and incubated for 2 to 3 min. This protocol resulted in the formation of a surface-immobilized model replication fork construct with the FRET donor-acceptor iCy3/iCy5 pair placed within the sugar-phosphate backbone of the duplex region, as shown in Fig. 1a. Unbound DNA molecules were washed away using the standard buffer solution.
Figure 1. The DNA replication fork construct used in this work.
(a) The T4 helicase binds to the d(T)29 loading sequence on the lagging strand, and unwinds the double-stranded (ds) region in the presence of GTP. The strands within the dsDNA region are internally labeled with the FRET donor-acceptor chromophores iCy3 and iCy5, respectively. (b) The donor-acceptor iCy3/iCy5 chromophores are incorporated into the sugar-phosphate backbone using a phosphoramidite oligonucleotide synthesis procedure (see text). (c) Molecular model showing the three-dimensional structure of the iCy3/iCy5 labeled duplex region of the DNA constructs. The chromophores are rigidly positioned within the sugar-phosphate backbone with transition dipole center-to-center distance R12 ~ 30 Å and relative angle θ12 ~ 77°. When the primosome complex unwinds the duplex the distance between the donor and acceptor chromophores becomes greater than the Förster distance (~ 50 Å), disrupting the FRET efficiency and leading to a decrease in acceptor emission and a concomitant increase in donor emission.
Protein purification
The T4-coded DNA replication helicase (gp41) was cloned and overexpressed in E. coli OR1264/pDH518 cells5 and purified as previously described.6 The T4 primase (gp61) protein carries a his-tag and was prepared and purified, as reported previously.7 The concentrations of purified gp41 and gp61 were determined by UV absorbance at 280 nm, using molar (per subunit) extinction coefficients (εM, 280) of 7.6 × 104 M−1-cm−1 for gp41 and 6.9 × 104 M−1-cm−1 for gp61. These extinction coefficients were calculated from amino acid composition data as described elsewhere.8, 9
Slide cleaning and sample chamber preparation
We constructed microfluidic sample chambers for our SM-FRET experiments using the following multistep procedure.10-12 Holes were drilled into quartz slides (GM Associates, Inc. #7525-01, 76.2 mm × 25.4 mm × 1 mm) using 1.0 mm diameter diamond tipped bits (1-0501-100, Kingsley North Inc., Norway, MI). The slides were placed in a petri dish containing 10% (w/v) aqueous Alconox (detergent) solution and sonicated for 20 min, followed by 5 min of sonication in double-distilled water (ddH2O), 15 min in acetone and finally 20 min in 1 M KOH. Each slide was hand-scrubbed with ddH2O, passed through a propane torch flame to remove organic impurities and residual water and immersed in a basic ‘piranha’ solution (1:1:5 NH4OH : H2O2 : ddH2O) at 75 to 80°C for 20 to 30 min. The slides were then rinsed with ddH2O and passed again through a propane torch flame. To avoid possible nonspecific protein or DNA adsorption to the slide surface, the slides were then amine-functionalized with a solution of 1 mL of N-(2-aminoethyl)-3-aminopropyl-trimethoxysilane (97%) (A0700, United Chemical Technologies, Bristol, PA) mixed with an acidic methanol solution (5 mL of acetic acid, 100 mL of methanol), and treated for ~4 hrs with a lightly functionalized polyethylene glycol (PEG) solution consisting of a mixture of 8 mg methoxy-PEG-succinimidyl valerate (mPEG-SVA-5000, Laysan Bio, Huntsville, AL) and 0.1 to 0.2 mg biotin-PEG-SVA-5000 (Laysan Bio, Huntsville, AL) dissolved in 64 JL of 100 mM NaHCO3. Glass coverslips (48393-230, No. 1 ½, 24 mm × 40 mm, VWR) were sonicated in 1 M KOH for 20 min, rinsed with ddH2O, treated with amino-silane, and then immersed for ~4 hours in an mPEG solution not containing biotin. These PEG-coated slides and coverslips were thoroughly rinsed with ddH2O, blown dry with N2, and stored in vacuum-sealed bags at −20°C until used.10
Micro-fluidic sample chambers were constructed by pipetting a 1.5 JL drop of standard buffer solution onto a PEG-coated microscope slide. A cover slip (10 mm × 24 mm) was placed on top of the slide with the PEG-coated side facing inward and the surfaces were sealed using 5-min epoxy cement. Two polyethylene tubes (PE50, 0.965 mm O.D., Becton Dickinson, Sparks, MD) were connected to the pre-drilled holes in the slide and sealed with epoxy. After the sample chamber was dry, a 50 JL aliquot of 0.1-0.2 mg/ml neutravidin (Thermo Scientific, Rockford, IL) was injected into the inlet tube and incubated for 2 to 3 min before flushing with standard buffer. 5′ biotin-labeled DNA substrates were introduced into the sample chamber between the coverslip and the neutravidin-coated slide (see Fig. 1a and Fig. S2a). Using this procedure we were able to image hundreds (~200 to 650, see Fig. S3) of surface-immobilized fluorescent molecules within an imaging area of ~30 Jm × 60 Jm. We found that quartz slides prepared in this way could be recycled multiple times (≥10) by immersing the entire sample chamber in acetone for a few days to dissolve the epoxy and using a razor blade to remove residual debris. The standard cleaning procedure outlined above was then repeated, excluding the treatment with basic piranha solution.
Imaging buffer
All our single-molecule experiments were conducted in standard imaging buffer (10 mM Tris at pH 8.0, 100 mM NaCl, 6 mM MgCl2), and stock solutions of gp41 and gp61 proteins were made up in this buffer as well. In addition, these solutions contained the oxygen-scavenging and triplet-quenching system [165 U/ml glucose oxidase (Sigma), 0.8% (w/v) D-glucose (Sigma), 2170 U/ml Catalase (Sigma) in saturated (≥2 mM) Trolox (Sigma)], which has been shown to work well with the Cy3/Cy5 smFRET chromophore system used in our experiments.10, 13
Total internal reflection fluorescence (TIRF) microscopy and single molecule (sm) FRET
For our smFRET experiments we employed the TIRF illumination geometry in which an evanescent field is used to excite a very thin region of the sample located 100 to 200 nm from the surface to which the fluorescently-labeled substrates had been bound.14-16 This technique greatly enhances signal contrast by reducing background noise due to scattered excitation light and fluorescence from unbound chromophores. We performed smFRET measurements using a prism-type TIRF microscope, which we constructed using an inverted microscope stage (TE2000-U, Nikon, Tokyo, Japan) (see Fig. S2a). A fused silica prism (325-3206, Pellin Broca Prism, Eksma, Lithuania) was mounted onto a XYZ-micro-translation stage (PT3, Thorlabs, Newton, NJ, 1 inch travel), which was used to position the prism on the top surface of the microscope slide as shown in Fig. S2a. A thin layer of immersion oil (Cargille, Cedar Grove, NJ) was used to index-match the interface between the prism and the quartz slide surfaces, and also the glass coverslip surfaces and the objective lens. A diode-pumped Nd:YAG laser (Coherent Compass™ 215M-50, wavelength 532 nm) served as the excitation source for the FRET iCy3/iCy5 chromophore pair. The incident laser intensity was adjusted using a half-wave plate (λ/2 450 – 800 nm, AHWP05M-630, Thorlabs, Newton, NJ) and a polarizing beam-splitting cube (10FC16PB.3, Newport). The laser was focused into the prism at the TIRF critical angle using a 100-mm focal length lens (LA1207, Thorlabs, Newton, NJ). Fluorescence was imaged using a 100× NA = 1.4 oil-immersion objective (Plan Apo, Nikon), and filtered using a long-pass filter (BLP01-532R-25, Semrock, Rochester, NY).
Fluorescence from the iCy3 (donor) and iCy5 (acceptor) chromophores was spatially separated and simultaneously imaged onto a split screen digital CCD camera using short beam paths and a minimal number of transmissive optical components. This was accomplished by directing the fluorescence image onto a dichroic mirror (FF650-Di01-25×36, Semrock, Rochester, NY), which had been placed at the input of the beam-separation optics arranged as a Sagnac interferometer. The dichroic mirror was placed at the beam-splitter position and produced two counter-propagating light paths with two broadband dielectric mirrors (BB1-E02, Thorlabs, Newton, NJ), as shown in Fig. S2b. The dichroic mirror reflected the shorter wavelength emission from the iCy3 donor chromophores, while transmitting the longer wavelength emission from the iCy5 chromophores. The separation between the iCy3 and iCy5 beam paths was adjusted by fine-tuning the angles of the dichroic beam-splitter and the broadband mirrors so that each image was projected onto one-half of the active area of an electron-multiplied charge-coupled-device camera (EM-CCD, iXon DV897-BB, 512 × 512 pixels, Andor Technology, Belfast), as shown in Figs. S2 and S3.
For a typical data-acquisition run, sequential images were recorded at 100-ms intervals for a total duration of 2 min. Individual image frames contained, on average, ~400 iCy3 (donor)/iCy5 (acceptor) single molecule features, which were analyzed using software written in IDL programming language. These data were used to construct single-molecule time-dependent trajectories for the donor-acceptor chromophore intensities ID and IA, permitting calculation of the FRET efficiency, defined as IA/(ID + IA). We have designated the occurrence of a precipitous drop in the value of the FRET efficiency, corresponding to a simultaneous decrease in IA and increase in ID, as a ‘FRET-conversion event’, which we have found to be diagnostic of a helicase-driven DNA fork construct unwinding event (see Results).
ATPase assays monitored by inorganic phosphate release
ATPase assays (Fig. S1) were performed in a reaction buffer containing 33 mM Tris-OAc (pH 7.8), 125 mM KOAc, and 6 mM Mg(OAc)2 in addition to 3.5 mM ATP and 50 nM [γ-32P] rATP. Unless otherwise indicated, the reaction mixtures contained 300 nM gp41 and 50 nM gp61 (concentrations of protein subunits) and 50 nM of ssDNA constructs in a 20-μL reaction volume. The reaction mixtures were pre-incubated at 37°C for 2 min. Reactions were started by the addition of the protein components and quenched by spotting an aliquot of reaction mixture onto a PEI-F cellulose TLC plate at various times. The PEI cellulose plates were then developed in 35 mM potassium phosphate buffer (pH 7.0). The dried plates were exposed on a PhosphorImage plate, and radioactive ADP and ATP were quantified using an Image Quantum method.
Molecular modeling calculations
We performed an equilibrium geometry optimization calculation on a fragment of the iCy3/iCy5 dsDNA construct used in our single molecule experiments based on the molecular mechanics force field (MMFFaq). The MMFFaq model is parameterized to predict geometries and conformations of organic molecules and biological polymers, and it includes an aqueous solvent energy correction in determining stability.17 For our calculations, we used the same local sequence in the region containing the chromophore residues as for our experiments, with lagging strand sequence 3′-CT-iCy3-CG-5′ and leading strand sequence 5′-GA-iCy5-GC-3′. The flanking bases about the iCy3 and iCy5 residues were constrained to adopt their B-form duplex structure, and the conformation of the probe residues were allowed to relax to their most stable conformation (see Fig. 1c). All calculations were performed using the Spartan ‘10 suite of programs (Wavefunction Inc., Irvine, CA).
Results
The assembly pathway of the T4 primosome complex
Assembly of functional T4 helicase hexamers depends on the concentrations of gp41 subunits and the availability of NTP ligands. At low concentrations the gp41 helicase protein exists in aqueous solution primarily as monomer subunits that can be driven to assemble into ring-shaped hexameric helicases by the addition of ATP or GTP.4, 6, 18 In previous bulk solution studies,18 we used sedimentation velocity methods to show that solutions containing 3 JM gp41 require ~60 JM GTPγS (guanosine-5′-O-gamma-thiotriphosphate) to drive gp41 assembly fully to the hexameric state, apparently via the sequential formation of dimers, tetramers and then hexamers, with GTPγS ligands bound at the interfaces between each pair of gp41 subunits and thus serving to stabilize the smaller intermediates and the resulting final hexagonal (gp41)6 structure. Lower concentrations of either gp41 subunits or GTPγS resulted in incomplete hexamer formation and significant concentrations of dimer and tetramer gp41 intermediate structures.
We note that GTPγS is a useful ligand in this context because it stabilizes the hexameric form of the T4 replication helicase and permits it to bind to a model DNA replication fork, and to subsequently form a complex with a single T4 primase (gp61) subunit as a stable initial primosome-DNA fork complex (such as that shown in Fig. 1a). Further DNA unwinding is inhibited due to the absence of an NTP substrate carrying a hydrolyzable β–γ phosphate-phosphate bond.18, 19 In the smFRET activity studies described here, we used lower concentrations of gp41 and hydrolyzable GTP (0.3 JM gp41 and 6 JM GTP) in order to maintain the above concentration ratio of gp41 to nucleotide triphosphate while avoiding the occurrence of metastable protein-protein and protein-DNA aggregates in our single molecule sample cells, which did occur (see below) at higher gp41 concentrations. In addition, the dilution factor was necessary to maintain the concentration of trolox (≥ 2mM) and oxygen scavenger enzymes (described in Materials and Methods) high enough for stable single-molecule detection. However, based on our bulk solution results, we would expect that gp41 oligomerization should not go to completion at the lower component concentrations used in these single molecule studies. Rather, we might expect to see a mixture of GTP-stabilized gp41 dimers and tetramers in labile equilibrium with fully formed and active gp41 hexamer helicase molecules.
Our recent bulk solution studies, using GTPγS to avoid GTP hydrolysis, have also shown that a ‘correct’ T4 primosome assembly pathway involving both gp41 and gp61 primase can be defined that leads to the formation of stable and monodisperse equilibrium complexes comprising single DNA fork constructs bound to single primosome complexes.18 In those studies we also demonstrated that the addition of components in incorrect (‘off-pathway’) orders resulted in formation of metastable protein-DNA aggregates. We note that those experiments were all conducted with ‘GTPγS-locked’ primosome complexes, which permitted stable ‘initiation’ binding of the primosome helicase to DNA fork constructs, but no further helicase-catalyzed unwinding of the replication fork until the non-hydrolyzable GTPγS ligands had been exchanged with GTP. In the following section we determine the effects of order of addition changes on the assembly of a fully functional primosome helicase.
Single molecule helicase unwinding experiments as a function of primosome assembly pathway
We used two distinct order-of-addition protocols to test whether varying the primosome-DNA assembly pathway and the helicase loading procedure also affects DNA fork unwinding activity at the component input concentrations we used in these single molecule experiments. In the first protocol we used a sequential two-step process modeled after the procedure we had found to be optimal for stable primosome complex assembly and binding to DNA constructs in bulk solution with nonhydrolyzable GTPγS ligands.18 A solution containing gp41 subunits and GTP was injected into the sample chamber, followed by the injection of gp61 primase after a short incubation period. This sequential procedure allowed the hexameric helicase to assemble initially in a labile (component-concentration-dependent) fashion and establish a dynamic (weak binding) equilibrium with the gp41 hexamer at the DNA fork junction. This process was then followed by the addition of primase, stabilizing the resultant gp41-gp61 complex due to binding of the gp61 primase and formation of a stable primosome complex with the DNA fork construct.
In the second protocol the gp41 helicase and gp61 primase subunits were mixed in the presence of GTP prior to injecting the mixed solution into the sample chamber. In bulk solution ultracentrifugation studies with GTPγS we had previously demonstrated that such direct mixing protocols result in the formation of protein-DNA aggregates.18 As we show below, the use of this direct-mixing procedure in smFRET studies also resulted in reduced duplex DNA unwinding activity and the formation of metastable aggregates, as manifested by the appearance of light-scattering foci in the smFRET images (see below). We note that the observation of such light-scattering foci in single molecule experiments may represent a much easier way to detect the formation of metastable aggregates (and thus to test for inappropriate assembly pathways) than do the conventional bulk solution approaches we have used before. The single molecule experiments presented below provide a real-time visualization of T4 primosome assembly and helicase function. Moreover, our results establish definitively, and at the single molecule functional level, that the component stoichiometry of the T4 primosome complex that was previously shown by bulk solution equilibrium methods to be (gp41)6-gp61-DNA in ‘GTPγS-locked’ primosome helicase initiation complexes also applies in actual unwinding experiments.
Unwinding assays with ‘internally’ and ‘externally’ labeled DNA fork constructs
As described in the Methods section, we used DNA replication fork substrates that had been internally labeled with donor-acceptor iCy3/iCy5 chromophore pairs (Fig. 1a) for our single molecule helicase experiments. The chromophores were incorporated into the sugar-phosphate backbones of the oligonucleotide strands as an intermediate reaction step in the sequential-step phosphoramidite synthesis of the DNA strands (Fig. 1b). Upon assembly of the DNA replication fork substrate, our geometry optimization calculations suggest that the iCy3/iCy5 chromophores are likely positioned within the duplex structure of the dsDNA region and held there with fixed donor-acceptor separation and orientation [R12 ≈ 30 Å, θ12 ≈ 77°, see Fig. 1c]. If indeed the iCy3/iCy5 chromophores are buried in this way, as our calculations suggest, this would also be consistent with our observation that these internal iCy3 and iCy5 chromophores are much less subject to dynamic collisional quenching than are their externally labeled counterparts (see below). Further evidence for a buried chromophore probe conformation in the duplex region is provided by our CD measurements of the ssDNA components and the fully annealed dsDNA replication fork construct. Fig. S4 shows that the CD spectra of the separate ssDNA strands of the fork construct display no measurable CD signal in the vicinity of the probe absorption maximum, while the CD spectrum of the chromophore complex of the annealed duplex over this wavelength range is significant. This rigid conformation serves to minimize the effects of thermal fluctuations on the Förster coupling strength, which depends sensitively on the values of R12 and θ12. Thus our observations of changes in the FRET signal using the iCy3/iCy5 chromophores can be attributed solely to the helicase activity of the primosome complex.
We also tested DNA unwinding substrates that had been externally labeled; i.e., fork constructs that carried eCy3/eCy5 chromophores attached by flexible amine-functionalized linkers at the C6 positions of specific thymine bases. These externally labeled unwinding substrates have often been used in smFRET-detected helicase assays because the chromophores in these positions should not perturb the sequence of the DNA fork construct to the same extent as insertion of iCy3/iCy5 chromophores directly into the DNA backbones, and thus might be expected to interfere less with helicase function.20, 21 However, our control measurements indicate that DNA substrates using externally labeled chromophores exhibit a significant background of false positive FRET conversion events, even in the absence of helicase proteins. This residual FRET activity may reflect increased collisional quenching events or photochemical damage due to the increased solvent exposure of these external fluorophores. In either case this increase in false positive FRET conversion events with the externally labeled DNA fork constructs significantly decreased the signal-to-noise ratios of our helicase unwinding experiments. To avoid these problems we used the internally labeled DNA fork constructs for all of our measurements, because with these fluorophores very little residual FRET conversion background could be detected and unwinding proceeded very efficiently. Control bulk solution unwinding assays (SI text and Fig. S1) showed that the presence of the iCy3/iCy5 labels within the fork construct helicase substrate reduced the rate of the T4 primosome helicase unwinding activity by less than 50%, and had little if any effect on the extent of the unwinding process. We further note that our single molecule measurements did not exhibit the quasi-periodic FRET conversion patterns that have been reported in previous single molecule helicase-mediated dsDNA unwinding experiments by others.20-22 Thus, the use of internally labeled DNA fork constructs permitted us to eliminate a significant source of background noise in our helicase unwinding experiments.
Helicase-dependent ‘FRET conversion events’
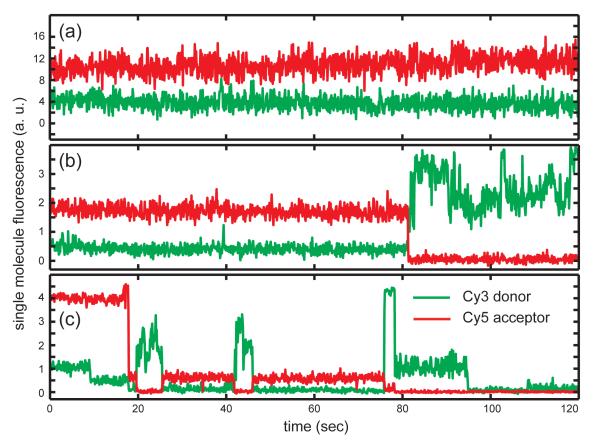
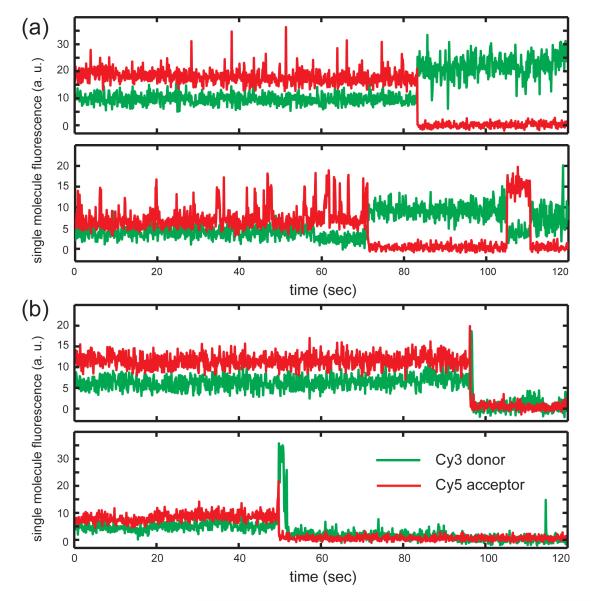
We monitored single molecule dsDNA unwinding activity by injecting solutions containing helicase proteins into the sample chamber and detecting the ensuing variation of the iCy3/iCy5 fluorescence. In Fig. 2, we show examples of three time-dependent single molecule trajectories, each recorded over a data-acquisition period of two minutes. At the beginning of a typical trajectory the acceptor fluorescence was observed at high levels, while the donor fluorescence was low. In many instances these relative intensities were maintained at constant levels throughout the data run (see Fig 2a). A ‘FRET conversion event’ was associated with an abrupt drop in the acceptor chromophore emission and a simultaneous rise in the donor emission. Often just one FRET conversion event was observed per DNA construct (Fig. 2b), while in other instances multiple FRET conversion events were observed for a single construct (Fig. 2c). Soon after a FRET conversion event we often observed the donor emission fluorescence drop irreversibly to zero, which we took to indicate completion of the dsDNA unwinding process accompanied by the disassociation and diffusion out of Förster energy transfer range of the iCy3-containing lagging strand (see Fig. 1a). In order to monitor the helicase-catalyzed DNA unwinding activity quantitatively, we determined the percentage of single molecules observed within each illuminated area that experienced one or more FRET conversion events.
Figure 2. Representative single molecule donor-acceptor FRET trajectories.
(a) The DNA replication fork construct in the absence of gp41 or gp61 protein. Under these conditions, ~99% of the molecules observed (excluding the FRET pairs showing donor-only signals) exhibited constant FRET signals during the 120-second data acquisition period. (b) A representative single molecule trajectory exhibiting a single FRET conversion event. (c) A representative single molecule trajectory exhibiting multiple FRET conversion events. For the experiments shown in both panels (b) and (c) the functional (gp41)6-gp61-DNA primosome complex was assembled using a sequential two-step injection protocol in the presence of 6 JM GTP (see text).
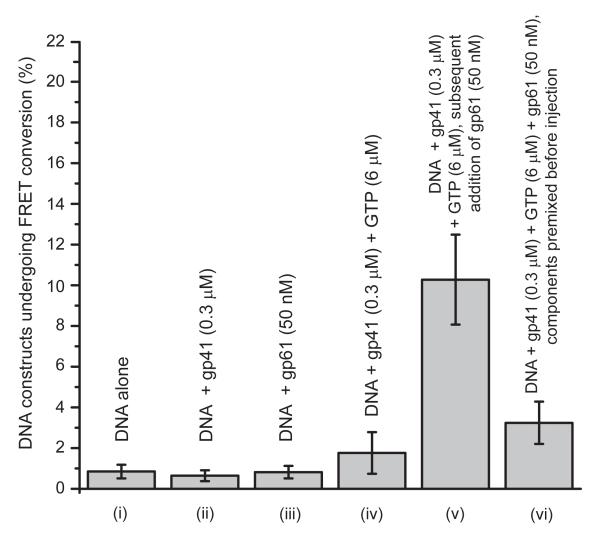
In Fig. 3, we summarize the results, obtained in standard imaging buffer, of our experiments on surface-immobilized DNA replication fork substrates. We compared the dsDNA unwinding activity under a variety of experimental protocols in which the different protein components were introduced separately and in combination, as well as in the presence and absence of GTP. Our control measurements with DNA substrates alone indicate the existence of a ~1% background level of residual FRET conversion events. We interpret this background as a reflection of the sum of fairly rare processes, such as a spontaneous change in conformation of the DNA backbone or a photochemical reaction that disrupts the stability or optical properties of the iCy5 acceptor chromophore. This ~1% background FRET conversion level was unchanged upon the injection into the sample chamber of either a 0.3 JM solution of gp41 helicase subunits or a 50 nM solution of gp61 primase subunits in the absence of GTP. However, we observed a marginal increase in the FRET conversion activity (~2% ±1%) when a 0.3 JM solution of gp41 helicase subunits containing 6 JM GTP was introduced into the sample chamber.
Figure 3. Percentage of DNA constructs that undergo one or more FRET conversion events per imaging area within 120 seconds as a function of protein components and GTP ligands present and their concentrations.
The bar heights represent the percentage of DNA constructs that have undergone one or more FRET conversion events per imaging area in 120 sec. The structure of the DNA construct is shown in Fig. 1a (and the DNA sequence in the SI text). n corresponds to the number of independent experiments of each type conducted, and N to the total detected number of FRET pairs in the multiple image areas analyzed (n). (i) surface immobilized DNA replication fork constructs (rfDNA) without helicase (n = 5, N = 2849); (ii) rfDNA + gp41 (0.3 JM) (n = 3, N = 3162); (iii) rfDNA + gp61 (50 nM) (n = 3, N = 1107); (iv) rfDNA + gp41 (0.3 JM) + GTP (6 JM) (n = 5, N = 3162); (v) rfDNA + gp41 (0.3 JM) + GTP (6 JM) + gp61 (50 nM) (n = 7, N = 4416); (vi) rfDNA + premixed gp41 (0.3 JM) + GTP (6 JM) + gp61 (50 nM) (n = 8, N = 4401). All measurements were performed in the presence of 6 μM GTP.
As described above, in the presence of GTP the gp41 helicase subunits assemble to form an equilibrium mixture of GTP-stabilized oligomers, including significant concentrations of the active (but weakly binding in isolation) (gp41)6 hexameric helicase form. The presence of these hexameric helicases induced only very weak dsDNA unwinding activity as observed by smFRET (Fig. 3, column iv). However, upon addition to this sample of a solution containing 50 nM gp61 primase in the presence of 6 JM GTP, we observed a significant increase in the dsDNA unwinding activity (~10% ±2.5%). These results show that: (i) hexameric T4 (gp41)6 helicase, which in the absence of gp61 binds only weakly to the DNA replication fork,18 also shows only minimal processive DNA fork unwinding activity; (ii) this duplex DNA unwinding activity is greatly enhanced in the presence of the fully assembled and primase-stabilized primosome helicase complex; and (iii) in both situations the observed levels of FRET conversion may be lower than expected because, at the protein and NTP component-concentration levels tested, the helicase hexamer, either alone or in the presence of primase, may not be fully assembled.
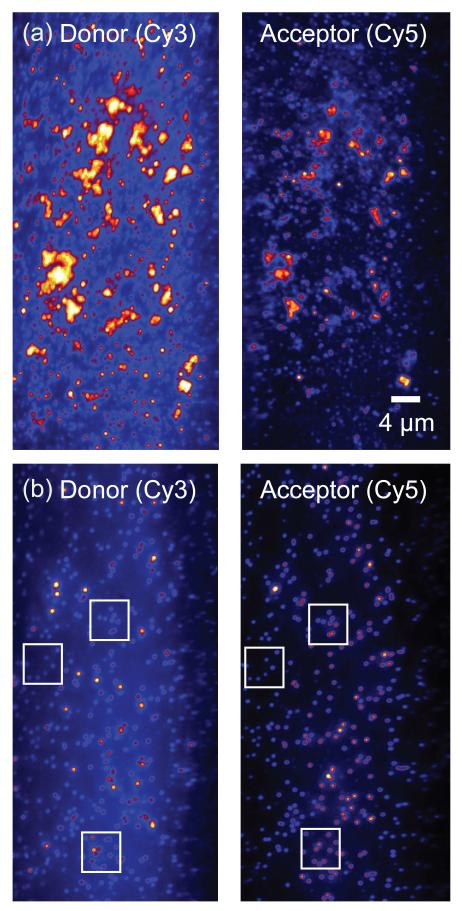
To investigate the effects of varying the primosome complex assembly pathway we performed measurements in which a ‘premixed’ solution containing 0.3 JM gp41, 6 JM GTP and 50 nM gp61 was injected into the sample chamber. In this situation the observed the DNA fork unwinding activity was ~3% (see Fig. 3, column vi). The diminished capacity of the ‘premixed’ primosome complex to carry out duplex DNA unwinding – in comparison to the results with the two-step assembly pathway described above – is consistent with the results of recent ensemble measurements in which the same premixing conditions resulted in the formation of nonfunctional metastable aggregates.18 Furthermore, we were able to observe what appeared to be aggregated structures within the imaging area of these samples, manifested by enhanced light scattering foci, as illustrated in Fig. 4a. Such light-scattering foci were not observed under the conditions of our two-step assembly pathway measurements (Fig. 4b). These results demonstrate, by direct single molecule visualization, that the order of addition of gp41, gp61, GTP, and DNA components is also critical to the correct assembly of stable and functional T4 primosome complexes in our smFRET unwinding assays, and also provide a convenient single-molecule assay of potentially general utility for detecting the occurrence of metastable aggregate formation when assembling macromolecular complexes.
Figure 4. ‘Off-pathway’ primosome assembly results in the formation of metastable aggregates.
(a) In this experiment gp41 (0.3 JM), GTP (6 JM) and gp61 (50 nM) were premixed and injected into the sample chamber. These mixing conditions resulted in scattered excitation light from aggregated structures in the illuminated sample area. (b) Samples prepared using the two-step assembly pathway showed no evidence of aggregation throughout all the measurements, indicating that the two-step assembly pathway (gp41+GTP and then gp61) avoids the formation of metastable aggregates. The image is split into donor (left) and acceptor (right) channels. A donor molecule and its paired acceptor molecule are separated by 255 pixels on the CCD camera (represented as three paired boxes in Fig. 4b).
The hexameric helicase alone unwinds DNA fork constructs very inefficiently at physiological GTP concentrations, while the full primosome complex shows significantly increased duplex DNA unwinding activity
We next examined the smFRET unwinding activities of the gp41 helicase and gp41-gp61 helicase-primase (primosome) complexes as a function of GTP concentration. Previous ensemble measurements had shown that the highest rates of unwinding with gp41 helicase alone were obtained at ATP or GTP concentrations approaching ~1 mM23 and that the addition of gp61 primase subunits increased the observed unwinding rate.3
We performed smFRET experiments with the same sequential two-step order-of-addition protocol used in the previous section to test the effect of changes in GTP concentration in single molecule unwinding assays driven both by the T4 helicase hexamer alone and the fully assembled helicase-primase (primosome) complex. The concentrations of gp41 (0.3 JM) and gp61 (50 nM) were held fixed, while the concentration of GTP was varied. When we injected 0.3 JM gp41 solution containing 3 mM GTP into the sample chamber we observed the dsDNA unwinding activity (as measured by FRET conversion events) to be ~5%, about twice the level observed when 0.3 JM gp41 and 6 JM GTP were used. We next introduced into the same sample a 50 nM gp61 primase solution in the presence of 3 mM GTP. Immediately after this injection, we observed that nearly all of the iCy3/iCy5 donor-acceptor signals within the illumination area rapidly disappeared, suggesting that primosome-catalyzed unwinding of the DNA fork constructs had gone to completion and also that the unwinding rate at these elevated GTP concentrations was too high to be measured quantitatively using this direct injection approach.
If the smFRET-detected fork unwinding activity of the gp41 helicase is indeed enhanced in the presence of the gp61 primase, the number of duplex DNA molecules detected after gp61 addition would be expected to decrease with time, because the complete unwinding of the DNA replication fork by the primosome will fully displace the lagging strand (which contains the donor fluorophore, iCy3) from the leading strand that contains the acceptor fluorophore (iCy5) and is attached to the slide surface (Fig. 1a), and as a consequence the fluorescence signal of the completely unwound fork construct illuminated at donor absorbance wavelengths will effectively disappear. Furthermore, if the reduction in the number of detectable molecules with time is indeed due to the enhanced (and processive) helicase activity of the fully assembled primosome complex, we would expect the remaining intact duplex DNA molecules to exhibit a time-dependent increase in the percentage of FRET conversion events. Thus observation of a correlation between the total number of detectable molecules and the percentage of molecules exhibiting FRET conversion events (see Fig. 5, below) supports the view that the fully assembled primosome complex is responsible for the dsDNA unwinding activity detected in our experiments.
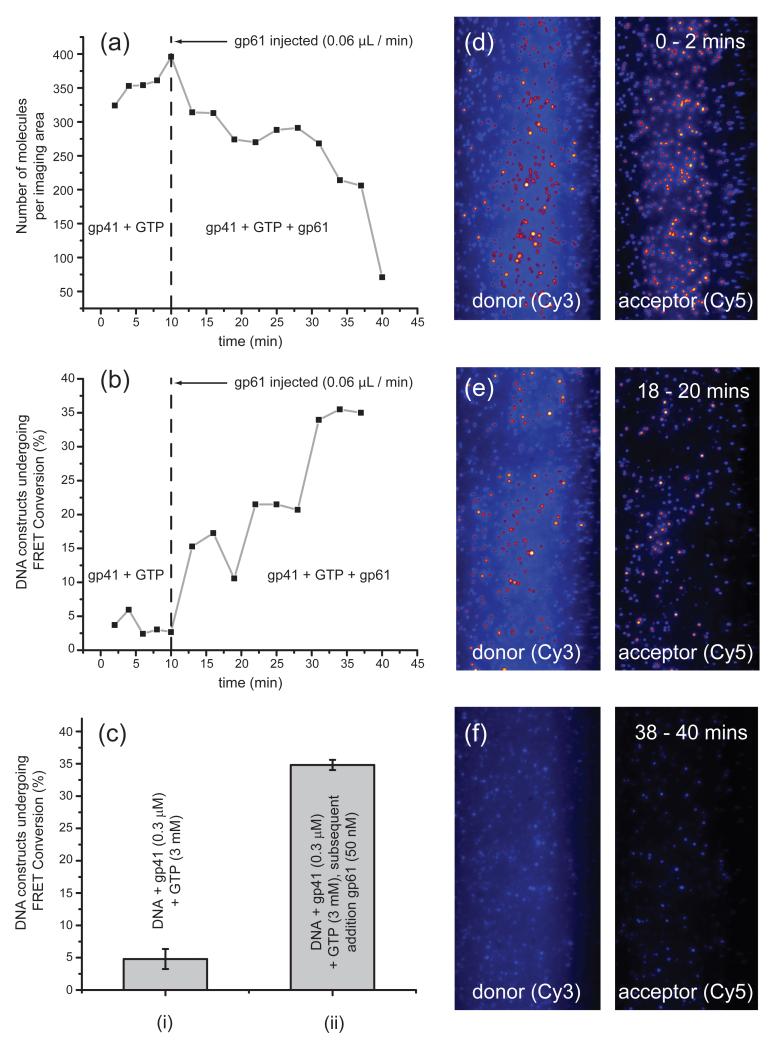
Figure 5. ‘Timed-release’ experiments.
A 50 nM solution of gp61 was injected into a sample chamber containing 0.3 JM gp41 and 3 mM GTP. (a) Plot of the number of DNA constructs detected within the imaging area as a function of time before and after the slow introduction of gp61 using a syringe pump (0.06 JL min−1); and (b) percentage of DNA constructs that undergo one or more FRET conversion event(s) per imaging area within 120 seconds as a function of time before and after the slow introduction of gp61. The vertical line at 10 minutes indicates the time at which sample injection was initiated. (c) GTP concentration-dependent unwinding activity of helicase and helicase-primase plotted as a percentage of DNA constructs that undergo one or more FRET conversion event(s) per imaging area within 120 seconds as a function of time before and after the slow introduction of gp61. The bar graph shows the percentage of observed FRET conversion events under different sample conditions using the surface immobilized DNA replication fork (rfDNA): (i) rfDNA + gp41 (0.3 JM) + GTP (3 mM) (n = 4; N = 1224); and (ii) rfDNA + gp41 (0.3 JM) + GTP (3 mM) + gp61 (50 nM) (n = 3; N = 979). (d) A representative single molecule image obtained during the 0 to 2 min data acquisition period in panels (a) and (b). (e) Same as (d), but during the 18 to 20 min interval. (f) Same as (d), but during the 38 to 40 min interval. The image scales are the same as those in Fig. 4.
‘Timed-release’ studies involving the slow addition of gp61 primase
To achieve a more precise level of control over the addition of the gp61 primase to the gp41 helicase-DNA system in the presence of 3 mM GTP, we modified our experimental set-up to permit the introduction of the 50 nM gp61 solution into the sample chamber slowly with the aid of a programmable syringe pump. For this purpose we employed a 100 cL glass syringe (1710RN; Hamilton, Reno, NA) fitted to a syringe pump (NE-501, New Era Pump Systems, Farmingdale, NY) that was operated at a flow rate of 0.06 JL/min. We carried out such ‘timed-release’ experiments over a total duration of 40 minutes. Successive data sets were acquired as described above, with a new imaging area selected every two minutes.
In Fig. 5, we present the results of such ‘timed-release’ smFRET unwinding experiments. As in our previous measurements, we first injected a solution containing 0.3 JM gp41 helicase and 3 mM of GTP into the sample chamber. The donor-acceptor iCy3/iCy5 signal was calibrated by recording successive 2 minute data sets over a 10 minute data collection period, after which we initiated the timed injection of the 50 nM gp61 solution. As shown in Fig. 5a, the detected number of DNA fork constructs per imaging area was relatively constant (~350) during the 10-minute period prior to primase addition, which likely reflects both the relatively low unwinding activity of the helicase hexamer in the absence of primase and also the reduced processivity (relative to primosome-induced unwinding) of the (gp41)6 helicase in the absence of primase. Such less processive unwinding may also favor the formation of ‘incompletely’ unwound DNA constructs (see below), which are then more likely to re-anneal to form native DNA constructs when the helicase has dissociated. After injection of the gp61 primase solution (vertical dashed line) the number of fork constructs per imaging area gradually decreased to zero over the 40-minute period, indicating that the duplex DNA constructs were being completely (and irreversibly, due to loss into solution of the donor-containing strand) unwound. Furthermore, we observed a concomitant increase in the number of FRET conversion events (Fig. 5b), which we interpreted as a direct measure of the primosome-catalyzed dsDNA unwinding activity.
We determined the limiting values of the dsDNA unwinding activity with 3 mM GTP for the gp41 helicase protein alone (~5%), and for the fully assembled primosome complex (~35%), from the asymptotic behavior of the time-dependent data (Fig. 5b). Fig. 5b shows that before adding gp61 the (gp41)6-driven unwinding process at 3 mM GTP resulted in about 5% FRET conversion events within each 120 sec time window, while after adding gp61 at the same GTP concentration the FRET conversion events per 120 sec window plateaued at ~35%. As indicated above, the gradual increase in the percentage of FRET conversion events seen in Fig. 5b likely reflects the multiple equilibria involved in the primosome assembly process as the concentration of gp61 increases during the slow injection process. Our results again indicate that the hexameric gp41 helicase binds only weakly to the DNA replication fork (and may also be only partially assembled), even at the elevated 3 mM GTP concentration level. However, as shown in bar (ii) of Fig. 5c, the efficiency of the dsDNA unwinding activity is greatly enhanced in the presence of the fully assembled primosome complex under these same conditions. Selected image frames taken over the course of the 40-minute data acquisition period are shown in Figs. 5d-f. We note that in the frame after 38-40 min of gp61 injection (Fig. 5f) little donor fluorescence (and thus also acceptor) fluorescence remains, indicating that virtually all the DNA fork constructs had been completely unwound by the primosome helicase and the donor strands lost.
The probability that a duplex DNA molecule will undergo ‘complete’ primosome-facilitated unwinding increases with GTP concentration
We next asked whether the concentration of GTP influences the assembly and the processivity, and hence the duplex DNA unwinding efficiency, of the fully assembled primosome complex. The pattern of single molecule FRET trajectories was sensitive to the total GTP concentration. At relatively low and intermediate GTP concentrations (≤ 1 mM) we observed a combination of two types of behaviors. In general, immediately after a FRET conversion event, the donor fluorescence intensity was high and the acceptor signal was low. In many instances the donor intensity retained its high level over the duration of the 2-minute trajectory, indicating that the lagging strand remained at least partially annealed to the leading strand during this period (Fig. 6a). We refer to these single molecule trajectories as representing ‘incomplete’ helicase-driven unwinding events. In the alternative situation we observed the donor fluorescence intensity decrease to zero soon after the FRET conversion event (Fig. 6b), suggesting that the lagging strand had fully disassociated from the construct. We refer to the latter as ‘complete’ helicase-driven unwinding events.
Figure 6. The smFRET pattern as a function of GTP concentration using a properly assembled (gp41)6-gp61-DNA primosome complex in a sequential two-step injection.
The experiments were performed using the same concentrations of gp41 (0.3 JM) and gp61 (50 nM), but varying GTP concentrations. Note that the donor fluorophore lies within the lagging (displaced) strand, and the acceptor within the leading strand. (a) Representative traces of a molecule exhibiting ‘incomplete’ unwinding FRET pattern. Following a high-to-low FRET conversion event, the donor signals for the lagging strand remained on the surface and also within the Förster distance of the acceptor, exhibiting a pattern reflecting repeated partial helicase unwinding events followed by rapid rewinding (reannealing) of the duplex portion of the fork construct. At a low concentration of GTP (6 JM) with a properly assembled (gp41)6-gp61-DNA primosome complex, ~83% of the fork constructs that we observed showed FRET conversion events with this incomplete unwinding pattern. (b) Representative traces of a fork construct exhibiting ‘complete’ unwinding FRET pattern, leading to a donor signal on the lagging strand that disappears abruptly right after the high-to-low FRET conversion event.
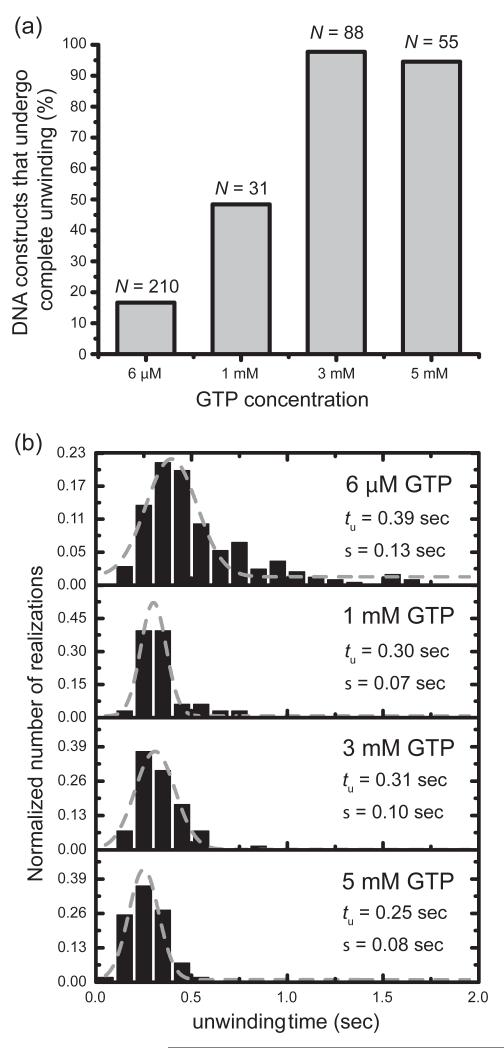
We used the same two-step assembly protocol described above to prepare the fully assembled primosome complex, first introducing a 0.3 μM gp41 solution into the sample chamber in the presence of GTP, and then adding a 50 nM solution of gp61. We compared the populations of complete and incomplete duplex unwinding events (as defined above) as a function of GTP concentration, and our results are summarized in Fig. 7a. In the lowest GTP concentration that we used (6 JM), the majority of DNA constructs (~83%) experienced incomplete duplex unwinding. For samples using 1 mM GTP, the relative proportion of complete and incomplete unwinding events was equal. At the highest GTP concentrations examined (3 mM and 5 mM), nearly all of the constructs (>97%) experienced complete unwinding events.
Figure 7. At elevated GTP concentration (3 mM), 97% of the fork constructs observed exhibited a ‘complete’ unwinding pattern.
(a) Percentage of DNA constructs showing a ‘complete’ unwinding pattern, taken from the total number (N) of FRET-active single molecule trajectories, increased at high GTP concentration. (b) Histogram of unwinding times (number of trajectories N ranged from 33 to 210). Grey curves are Gaussians fits described by exp[−(t − tu)2/2σ2]. The best-fit values of the mean unwinding time tu and standard deviation σ are given in the panels. All histograms are normalized to the number of traces N.
Using only the single molecule trajectories that we identified as complete unwinding events, we constructed histograms of the ‘unwinding time.’ We defined the unwinding time as the period between the FRET conversion event and the time that the donor fluorescence intensity dropped to zero, indicating full disassociation of the lagging strand. We summarize these results in Fig. 7b. At low GTP concentration (6 JM) the unwinding times were broadly distributed, with a mean value ~390 msec. The apparent unwinding time distribution narrowed as the GTP concentration was increased, with the mean unwinding time decreasing to ~250 msec in 5 mM GTP. Based on these numbers we estimate the range of processive unwinding rates catalyzed by the primosome helicase to be ~40-60 bps sec−1, corresponding to primosome translocation rates along the duplex DNA construct of ~15-20 nm sec−1.
Functional and fully assembled gp41-gp61 primosome complexes display a helicase (gp41) to primase (gp61) subunit stoichiometry of 6:1
Previous bulk solution investigations have demonstrated that duplex DNA unwinding activity of the T4 gp41 hexameric helicase is significantly enhanced by the addition of gp61 primase subunits. The results of our smFRET experiments described in the preceding section also show that the addition of primase increases the RNA-primer-synthesizing activity of the resulting complexes. In order to achieve maximal processivity and activity, a functional helicase must bind as a fully formed stoichiometric complex to the DNA fork construct that serves as the unwinding substrate. Although previous bulk solution studies had concluded that the subunit stoichiometry of the fully assembled primosome complex is ~6:1,3, 4, 18 other reports had suggested that the ratio might be as high as 6:6.24-26 For a complete discussion of the relevant literature see ref.18
We have used our SM-FRET approach to make a direct functional measurement of the dependence of the observed primosome-driven helicase unwinding rate on the subunit stoichiometry of the T4 gp41-gp61 primosome complex. To this end we performed a series of experiments with our DNA unwinding substrate construct in which we systematically varied the relative concentrations of the protein components. We assembled the primosome complex onto DNA replication fork substrates using a protocol designed to achieve a proper ‘on-pathway’ loading sequence of the gp41 and gp61 components while maintaining constant subunit ratios. For this purpose we prepared the DNA fork constructs at 100 pM strand concentrations from 100 nM concentrations of annealed constructs in 10 mM Tris (pH 8.0), 100 mM NaCl and 6 mM MgCl2, and then added 0.3 JM gp41 and 6 JM non-hydrolyzable GTPγS to this solution. This procedure forms weakly associating DNA-(gp41-GTPγS)6 complexes in the sample tube. Then, after 2 min of incubation, various amounts of gp61 were introduced to achieve gp41:gp61 subunit ratios in the solution ranging from 6:0.1 to 6:6. After two minutes of pre-incubation, the resulting DNA-primosome initiation complexes were immobilized onto the surface of the sample chamber using biotin-neutravidin linkages as described in Materials and Methods.
The above procedure resulted in the generation of ‘arrested’ and stable helicase initiation complexes bound at the DNA fork junction in an ‘activated state’ due to the use of non-hydrolyzable GTPγS in the assembly of the hexameric gp41 helicase.19 Upon subsequent injection of ~200 JL of standard imaging buffer we were able to confirm the stable presence of the DNA substrates at the sample surface by monitoring the iCy3/iCy5 fluorescence.
We note that very little FRET conversion activity was observed from samples prepared in this way. However, these ‘dormant’ pre-bound primosome complexes could be readily converted into their active duplex DNA unwinding forms by injecting 1 mM GTP into the sample chamber. After a short lag-time, presumably reflecting the relatively slow rate of exchange with GTP of the non-hydrolyzable GTPγS ligands with which the bound complex had been constructed,18 we observed the initiation of FRET conversion events associated with duplex DNA unwinding activity. In addition to initiating the unwinding process, the introduction of the 1 mM GTP solution also served to displace unbound proteins (e.g., weakly bound helicase, etc.) from the sample chamber so that only the ‘GTPγS-locked’ primosome complexes that were initially bound to the DNA constructs as stable activated initiation complexes could contribute to the single molecule unwinding trajectories (FRET conversion events) observed following GTP solution addition.
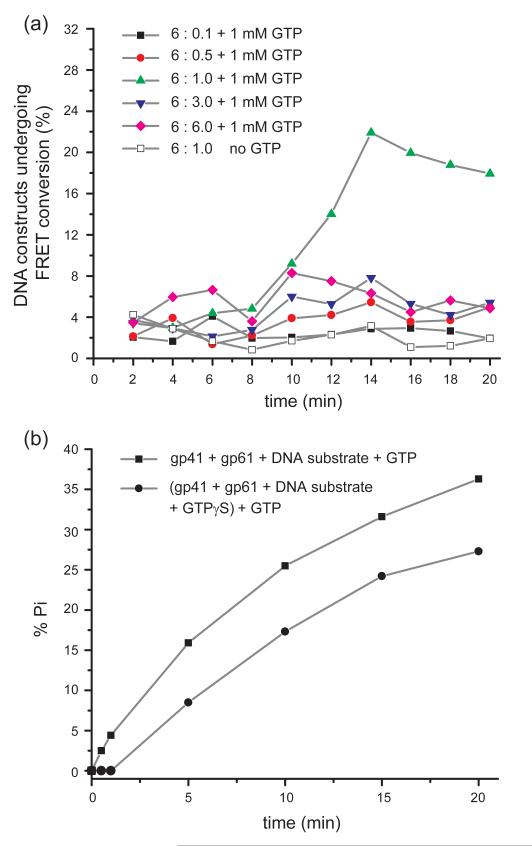
We prepared a number of samples following the above procedure, with various gp41-gp61 subunit ratios. A solution of 1 mM GTP was injected into the sample chamber at time zero, and sequential images were recorded for 2-minute intervals over a 20-minute period. The results of these experiments are summarized in Fig. 8a. As a control measurement, the DNA-primosome complex with a gp41:gp61 subunit input ratio of 6:1 was monitored without introducing GTP (open white squares). During the 20-minute data acquisition period, the FRET conversion activity for this sample fluctuated between 1 and 4%. When a 1 mM GTP solution was injected into the sample chamber, we observed, after an initial lag, that the duplex DNA unwinding activity reached a maximum value of ~22% for primosome complexes prepared with a gp41-gp61 subunit ratio 6:1. No other subunit ratios approached this level of efficiency. The next highest performers, with subunit ratios 6:6 and 6:3, only reached maximal unwinding activities of ~8%, while the unwinding rates observed in experiments with sub-optimal gp41:gp61 subunit ratios (6:0.5 and 6:0.1) were lower yet.
Figure 8. Comparison of smFRET and GTPase unwinding assays at various gp41:gp61 ratios as a function of time.
(a) Percentage of DNA constructs that undergo one or more FRET conversion events per imaging area within 120 seconds as a function of time after 1 mM GTP addition. FRET conversion events were initiated with pre-formed ‘GTPγS-locked’ helicase initiation complexes [(gp41)6-gp61-DNA]. GTPγS-locked helicase initiation complexes were prepared with helicase:primase subunit input ratios ranging from 6:0.1 to 6:6 and then the unwinding of the DNA fork constructs was monitored by injecting 1 mM GTP into the sample chamber at zero-time. (b) Ensemble measurements of dsDNA unwinding by the primosome complex were obtained by monitoring inorganic phosphate (Pi) release resulting from GTP hydrolysis during DNA fork unwinding. Two types of experiments were performed. In the first, the helicase hexamers were pre-formed with non-hydrolyzable GTPγS (60 JM) and then a 700-fold excess of GTP was added (filled circles). In the second, the solution did not contain any GTPγS, and GTP (3.5 mM) was directly added to the complex, so that GTP hydrolysis could begin immediately after the gp41 helicase hexamer and gp61 primase monomer had bound to the DNA fork construct (filled squares). The time lag observed in the single-molecule experiments is consistent with the time lag measured in the ensemble studies, and presumably corresponds to the slow initial exchange of GTPγS by GTP. Both the single-molecule and the ensemble experiments used the same DNA unwinding constructs and were conducted under identical experimental conditions.
We note (Fig. 8a) that the maximal duplex DNA unwinding activity in these single molecule experiments was attained only after a significant time lag, presumably reflecting the time required for the exchange of the pre-bound non-hydrolysable GTPγS with hydrolysable GTP. Similar results were also obtained with ensemble measurements in which we measured construct unwinding indirectly by monitoring the GTPase activity of the primosome (Fig. 8b). In these experiments we monitored the release of the inorganic phosphate (Pi) resulting from the hydrolysis of GTP to GDP. The maximal duplex DNA unwinding activity that we determined with our single molecule measurements was ~22%, which is comparable to the maximal GTP hydrolysis rate of ~25% observed in the corresponding ensemble measurements. We further note that the maximum activity for the initially GTPγS-locked DNA replication fork substrates (~22%) was always lower than the unwinding rate obtained when GTPγS was not initially contained in the reaction (~35%, see Fig. 5c). This reduced activity in the presence of GTPγS, as well as the initial lag in the ATPase rate, is in agreement with the results of ensemble measurements in which 3.5 mM GTP was introduced to initiate unwinding of ‘GTPγS-locked’ DNA replication fork substrates. Our results suggest that the initial GTPγS-GTP exchange process at the GTP binding sites of the hexameric gp41 helicase is very slow under these conditions. Possible molecular mechanisms that might account for these effects are considered in the Discussion.
IV. Discussion
The single molecule experiments presented in this work represent a detailed examination of the functional duplex DNA unwinding activity and assembly pathway of the bacteriophage T4 helicase-primase (primosome) complex. We note that we used ‘internally labeled’ Cy3/Cy5 constructs (see Fig. 1) throughout, and that our results show that the T4 primosome helicase is apparently able to ‘unwind past’ these internal labels without significant difficulty (Fig. S1, SI text and Table S1). Our smFRET results show that dsDNA unwinding activity depends sensitively on the assembly pathway, the gp41 helicase, gp61 primase and GTP concentrations and the gp41:gp61 subunit ratio. The following results, which are relevant to the assembly pathway of the T4 primosome complex, have been established or confirmed by these experiments.
Duplex DNA unwinding activity is significantly enhanced in the presence of the gp61 primase
Ensemble studies of the T4 helicase have shown that dsDNA unwinding activity with 5′-to-3′ polarity is significantly stimulated by binding of the gp61 primase to the gp41 hexamer.5, 7, 23, 27 Liu et al.27 suggested that the T4 helicase simultaneously interacts with the gp61 primase to make the penta-ribonucleotide primers that initiate Okazaki fragments at specific primer initiation sites. Venkatesan et al.23 showed that the helicase activity of gene 41 protein alone is low, but is increased 6 to 18-fold by the addition of gp61, as measured by RNA primer synthesis. Jing et al.7 suggested that gp61 interacts in binary complexes with both ssDNA and the gp41 hexamer and that these interactions in both binary complexes involve both termini of the gp61 protein. Furthermore, the interaction affinities between the components are significantly increased within the ternary (primosome) complex. Hinton et al.5 estimated that 62% of the DNA hybrid was unwound with gp41 in the presence of gp61, while 2.3% was unwound by gp41 alone at 500 JM ATP. In addition Lionnet et al.28 performed single molecule experiments involving magnetic tweezers to vary the force destabilizing the DNA substrate and also proposed that the T4 helicase hexamer alone cannot efficiently unwind the T4 genome.
Recently, Jose et al.18 showed that hexamers of gp41 assembled with GTPγS in the absence of gp61 do not form stable complexes with ssDNA in bulk solution, as measured by sedimentation velocity ultracentrifugation and fluorescence anisotropy experiments. They proposed a specific binding role for the gp61 primase subunit to both gp41 and to the DNA fork construct in the context of a rotating mechanism for the translocation of the hexameric helicase into the DNA fork, coupled with the concommitant movement of a single ‘mobile’ primase subunit from one gp41 subunit to the next.
Our single molecule observations in the present work show also that, in the absence of the gp61 primase, the gp41 hexameric helicase binds only weakly to the DNA replication fork. However, in the presence of the gp61 primase, a stable primosome complex is formed at the fork junction that is also a much more active helicase than is (gp41)6 alone. The duplex DNA unwinding activity of the hexameric gp41 protein in the presence of gp61 and 6 JM GTP was ~10% (in terms of FRET conversion events per observation period), compared to the ~2% level seen in the absence of the gp61 protein (see Fig. 3). At higher GTP concentration (3 mM), the duplex DNA unwinding activity was ~35% in the presence of the gp61 protein (Fig. 5c). These observations confirm that the T4 gp41 protein alone cannot bring about processive and efficient unwinding of the replication fork.
GTP activates the duplex DNA unwinding activity of the gp41 helicase and the gp41-gp61 helicase-primase complex
It has previously been shown that the highest rates of unwinding of the bacteriophage T4 gene 41 helicase, as measured in ensemble experiments, were obtained at ATP or GTP concentrations greater than 1 mM.3, 23 Richardson et al.3 also reported that when the GTP concentration was increased to 1 mM (with 0.5 mM ATP and 0.2 mM CTP and UTP), the rate of DNA unwinding by gp41 protein alone, or by gp41 and gp61 proteins together, were each increased about a factor of 2, as measured in ensemble experiments. Venkatesan et al.23 also found that the highest rates of DNA unwinding by the gp41 protein alone was achieved at 5 mM ATP. Similarly, single molecule unwinding studies of the T4 gp41 helicase using magnetic tweezers showed that the DNA unwinding rates were significantly influenced by ATP concentration when a destabilizing force (F = 3 – 12 pN) was applied to the duplex DNA.28
Our single molecule FRET studies have shown that the dsDNA unwinding activity of the gp41 helicase hexamer alone is very low, with only ~2% of the fork constructs demonstrating FRET conversion events in the presence of 6 JM GTP (Fig. 3), and only ~5% in the presence of 3 mM GTP (Fig. 5c). However, upon addition of the gp61 primase the dsDNA unwinding activity of the helicase-primase complex increased to ~35% in the presence of 3 mM GTP (Fig. 5c). This activity level of the primosome complex should be compared to the ~10% level observed in our single molecule studies with 6 JM GTP (Fig. 3). Our observations indicate that the hexameric gp41 helicase binds and unwinds DNA fork substrates more effectively at higher GTP concentrations, in accord with our previous demonstration using bulk solution experiments that the extent of gp41 helicase hexamer formation is a function of both total GTP (or GTPγS) and of gp41 and gp61 subunit concentrations.
The functional dsDNA unwinding activity of the fully assembled gp41-gp61 primosome complex is strongly dependent on GTP concentration
FRET-based single molecule DNA unwinding studies of non-ring-shaped helicases, such as the tetrameric HCV NS320 and E. coli Rep22 exhibited a quasi-periodic unwinding pattern when the helicases encountered the biotin-streptavidin linkages that immobilize the DNA substrates to the sample surface – a so-called ‘biotin-streptavidin-blockade.’ Myong et al.20 reported that when the dsDNA substrate was tethered to the sample surface via a biotin-streptavidin linkage, duplex DNA unwinding by the NS3 helicase could not be completed due to steric hindrance by the biotin-streptavidin complex. In their experiments, the majority of molecules (75%) exhibited a quasi-periodic dsDNA unwinding pattern, in which the NS3 helicase appeared to snap back to the primer-template junction upon encountering a biotin-streptavidin blockade.
Although our DNA fork constructs were tethered to the sample surface via a chemically identical biotin-streptavidin linkage (see Fig. 1a), we did not observe -- under any of the conditions we investigated -- a quasi-periodic helicase unwinding pattern. However, at low and intermediate GTP concentrations (6 μM and 1 mM), we observed that a substantial fraction of molecules underwent ‘incomplete dsDNA unwinding’, in which the lagging strand remained at least partially bound to the tethered leading strand after the primosome complex had made its first pass through the duplex region (Figs. 7a and b). At higher GTP concentrations (3 mM and 5 mM), almost all of the observed FRET conversion events corresponded to ‘complete’ DNA construct unwinding.
The incomplete dsDNA unwinding events that we observed at low GTP concentrations are likely due to a decreased helicase processivity rate or a decreased level of complete primosome assembly, leading to an increased likelihood of the occurrence of ‘paused’ states during unwinding of the DNA-primosome complex. During such paused states a variety of processes that compete with translocation may occur, including disassociation of the primosome and re-annealing of the exposed DNA template strands. Because our control experiments revealed only a baseline activity of FRET conversion events (~1%, Fig. 3), we associate our observations of incomplete fork construct unwinding with the dynamic properties of the primosome complex. Conversely we associate our observations of ‘complete’ duplex DNA unwinding at elevated GTP concentrations with increased primosome, stability coupled with an increased rate and processivity of primosome translocation (Figs. 7a and b).
The functional and fully assembled gp41-gp61 primosome complex has a subunit stoichiometry of 6:1
In previous ensemble experiments, it was demonstrated that the gp61 primase activity of gp41-gp61 primosome complexes prepared in vitro reached a maximum value at a gp41:gp61 number ratio somewhat in excess of 6:1 – i.e., approximately one gp61 primase subunit per six gp41 helicase subunits. It was found that the addition of more gp61 subunits resulted in a progressive decrease in the rate of primase activity, which showed that primase concentrations beyond the level required to reach 6:1 stoichiometry resulted in the inhibition of RNA primer synthesis.18
Our current single molecule studies are also consistent with the a 6:1 helicase:primase subunit stoichiometry for the formation of stable and functional gp41-gp61 primosome complexes. This is evident from our comparison of the duplex DNA unwinding activities of a series of samples in which the gp41:gp61 number ratio was systematically varied (Fig. 8a). The maximum dsDNA unwinding activity of ~25% FRET conversion was reached at a stoichiometric subunit ratio of 6:1, while the subunit ratios 6:6 and 6:3 subunit ratios resulted in much lower dsDNA unwinding activities (~8%). This reduced functional helicase activity at non-stoichiometric protein subunit concentrations is likely a consequence of the formation of metastable aggregates of gp61 and DNA, as also observed in sedimentation velocity ultracentrifugation and RNA primer synthesis studies.18 For helicase:primase subunit ratios below 6:1 (6:0.1 and 6:0.5), the observed dsDNA fork unwinding activities were even lower than those observed for the 6:3 and 6:6 ratios. This may reflect a combination of the weak binding affinity of the gp41 hexameric helicase lacking a gp61 primase subunit with the likely tendency of the labile functioning primosome helicase complexes containing a mobile primase subunit to dissociate into smaller (and inactive) gp41 oligomers between reaction cycles as the limiting gp61 subunits may dissociate and move from one gp41 hexamer to another during function.
We emphasize that the protocol we developed to examine the protein subunit stoichiometry of the primosome-DNA complex could be applied to many other protein-nucleic acid systems. Fully assembled (gp41-GTPγS)6-gp61-DNA complexes were prepared in solution, and then chemically bound to the sample surface. Subsequent injection of GTP, followed by the slow chemical exchange between the non-hydrolyzable GTPγS and hydrolysable GTP at the gp41 binding sites, served to initiate dsDNA unwinding by the primosome complex. This initializing injection of GTP solution also served to displace and flush out weakly bound species, perhaps including excess DNA-primosome complexes, and weakly bound gp41 helicases. We further note that a similar procedure could be applied to study the kinetics of the exchange of GTP with non-hydrolyzable GTPγS at the single molecule level, and thus obtain further insight into the role of GTP in stabilizing the hexameric helicase and of GTP hydrolysis in driving primase release and perhaps gp41 hexamer rotation during the helicase-driven dsDNA unwinding process.
Finally, given the paradigmic nature of the bacteriophage T4-coded DNA replication complex for the replication complexes of higher organisms (bacteria, yeast, humans), it is relevant to ask whether observation of the full unwinding activities of the respective replication helicases of these higher organisms in isolation might also require the presence of at least one bound primase subunit per helicase hexamer. We note that most published in vitro experiments on the unwinding rates and mechanisms catalyzed by the isolated helicases of these higher organisms have been performed in the absence of added primase subunits, and it might be worth considering whether assessing the fully processive and biologically relevant unwinding activities of the helicases of these organisms might also require the use of primosome helicase complexes rather than just hexameric helicases alone.
Supplementary Material
Acknowledgements
The authors thank Tongfei Bu and Jordan R. Sensibaugh for assistance in the preparation of the samples and sample cells used in our single molecule FRET experiments. We also thank Dr. Steve Weitzel for providing the T4 gp41 and gp61 preparations used in these studies. We are also grateful to Dr. Taekjip Ha, who introduced Dr. Wonbae Lee to biological single-molecule studies in the excellent NSF-sponsored workshop that he directs at the U. of Illinois Center for the Physics of Living Cells.
Funding: This work was supported by grants from the National Institutes of Health NIGMS-NIH (GM-15792 – to P.H.v.H.), from the Office of Naval Research (N00014-11-0193 – to AHM) and from the National Science Foundation, Chemistry of Life Processes Program (CHE-1105272 – to AHM). P.H.v.H. is an American Cancer Society Research Professor of Chemistry.
Footnotes
The authors declare no competing financial interest.
Associated Content.
Additional Supporting Information (SI) showing the sequences of our DNA unwinding assay constructs (Table S1), as well as bulk helicase unwinding assays using either unlabeled or iCy3/iCy5-labeled DNA fork constructs (Figure S1), further details of our experimental set-up and representative single molecule images obtained with our instrument (Figure S2 and S3, respectively), and control circular dichroism measurements on the DNA replication fork constructs containing the ‘internal’ Cy3 and Cy5 chromophores (Figure S4) is available. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Nossal N. Molecular Biology of Bacteriophage T4. Am. Soc. Microbiol; Washington, DC: 1994. pp. 43–53. [Google Scholar]
- (2).Alberts B. Prokaryotic DNA replication mechanisms. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 1987;317:395–420. doi: 10.1098/rstb.1987.0068. [DOI] [PubMed] [Google Scholar]
- (3).Richardson RW, Nossal N. Characterization of the bacteriophage T4 gene 41 DNA helicase. J. Biol. Chem. 1989;264:4725–4731. [PubMed] [Google Scholar]
- (4).Dong F, von Hippel PH. The ATP-activated hexameric helicase of bacteriophage T4 (gp41) forms a stable primosome with a single subunit of T4-coded primase (gp61) J. Biol. Chem. 1996;271:19625–19631. doi: 10.1074/jbc.271.32.19625. [DOI] [PubMed] [Google Scholar]
- (5).Hinton D, Silver L, Nossal N. Bacteriophage T4 DNA replication protein 41. Cloning of the gene and purification of the expressed protein. J. Biol. Chem. 1985;260:12851–12857. [PubMed] [Google Scholar]
- (6).Dong F, Gogol EP, von Hippel PH. The phage T4-coded DNA replication helicase (gp41) forms a hexamer upon activation by nucleoside triphosphate. J. Biol. Chem. 1995;270:7462–7473. doi: 10.1074/jbc.270.13.7462. [DOI] [PubMed] [Google Scholar]
- (7).Jing DH, Dong F, Latham GJ, von Hippel PH. Interactions of bacteriophage T4-coded primase (gp61) with the T4 replication helicase (gp41) and DNA in primosome formation. J. Biol. Chem. 1999;274:27287–27298. doi: 10.1074/jbc.274.38.27287. [DOI] [PubMed] [Google Scholar]
- (8).Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- (9).Young MC, Kuhl SB, von Hippel PH. Kinetic theory of ATP4 driven translocases on one-dimensional polymer lattices. J. Mol. Biol. 1994;235:1436–1446. doi: 10.1006/jmbi.1994.1099. [DOI] [PubMed] [Google Scholar]
- (10).Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat. Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Jain A, Liu R, Xiang YK, Ha T. Single-molecule pull-down for studying protein interactions. Nat. Protoc. 2012;7:445–452. doi: 10.1038/nprot.2011.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Ha T, Rasnik I, Cheng W, Babcock HP, Gauss GH, Lohman TM, Chu S. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 2002;419:638–641. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- (13).Rasnik I, McKinney SA, Ha T. Nonblinking and long-lasting single-molecule fluorescence imaging. Nat. Methods. 2006;3:891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- (14).Wazawa T, Ueda M. Total internal reflection fluorescence microscopy in single molecule nanobioscience. Adv. Biochem. Eng./Biotechnol. 2005;95:77–106. doi: 10.1007/b102211. [DOI] [PubMed] [Google Scholar]
- (15).Axelrod D, Burghardt TP, Thompson NL. Total internal reflection fluorescence. Annu. Rev. Biophys. Bio. 1984;13:247–268. doi: 10.1146/annurev.bb.13.060184.001335. [DOI] [PubMed] [Google Scholar]
- (16).Axelrod D. Total internal reflection fluorescence microscopy in cell biology. Traffic. 2001;2:764–774. doi: 10.1034/j.1600-0854.2001.21104.x. [DOI] [PubMed] [Google Scholar]
- (17).Halgren TA. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J. Comput. Chem. 1996;17:490–519. [Google Scholar]
- (18).Jose D, Weitzel SE, Jing D, von Hippel PH. Assembly and subunit stoichiometry of the functional helicase-primase (primosome) complex of bacteriophage T4. Proc. Natl. Acad. Sci. USA. 2012;109:13596–13601. doi: 10.1073/pnas.1210040109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Jose D, Weitzel SE, von Hippel PH. Breathing fluctuations in position-specific DNA base pairs are involved in regulating helicase movement into the replication fork. Proc. Natl. Acad. Sci. USA. 2012;109:14428–14433. doi: 10.1073/pnas.1212929109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Myong S, Bruno MM, Pyle AM, Ha T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science. 2007;317:513–516. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Yodh JG, Stevens BC, Kanagaraj R, Janscak P, Ha T. BLM helicase measures DNA unwound before switching strands and hRPA promotes unwinding reinitiation. The EMBO journal. 2009;28:405–416. doi: 10.1038/emboj.2008.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Repetitive shuttling of a motor protein on DNA. Nature. 2005;437:1321–1325. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- (23).Venkatesan M, Silver L, Nossal N. Bacteriophage T4 gene 41 protein, required for the synthesis of RNA primers, is also a DNA helicase. J. Biol. Chem. 1982;257:12426–12434. [PubMed] [Google Scholar]
- (24).Yang J, Xi J, Zhuang Z, Benkovic SJ. The oligomeric T4 primase is the functional form during replication. J. Biol. Chem. 2005;280:25416–25423. doi: 10.1074/jbc.M501847200. [DOI] [PubMed] [Google Scholar]
- (25).Valentine AM, Faoud T, Shier VK, Benkovic SJ. A zinc ribbon protein in DNA replication: primer synthesis and macromolecular interactions by the bacteriophage T4 primase. Biochem. 2001;40:15074–15085. doi: 10.1021/bi0108554. [DOI] [PubMed] [Google Scholar]
- (26).Norcum MT, Warrington JA, Spiering MM, Ishmael FT, Trakselis MA, Benkovic SJ. Architecture of the bacteriophage T4 primosome: electron microscopy studies of helicase (gp41) and primase (gp61) Proc. Natl. Acad. Sci. USA. 2005;102:3623–3626. doi: 10.1073/pnas.0500713102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Liu C, Alberts B. Characterization of the DNA-dependent GTPase activity of T4 gene 41 protein, an essential component of the T4 bacteriophage DNA replication apparatus. J. Biol. Chem. 1981;256:2813–2820. [PubMed] [Google Scholar]
- (28).Lionnet T, Spiering MM, Benkovic SJ, Bensimon D, Croquette V. Real-time observation of bacteriophage T4 gp41 helicase reveals an unwinding mechanism. Proc. Natl. Acad. Sci. USA. 2007;104:19790–19795. doi: 10.1073/pnas.0709793104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.