Abstract
Mutations on the LMNA gene are responsible for an heterogeneous group of diseases. Overlapping syndromes related to LMNA gene alterations have been extensively reported. Study scope is to perform a systematic analysis of the overlapping syndromes so far described and to try to correlate the clinical features to the associated genetic alterations. We evaluated all the dominant overlapping syndromes reported by means of a PubMed search and by the analysis of the main databases containing the pathogenic LMNA gene variations and the associated diseases.
Metabolic alterations in association to skeletal and/or cardiac alterations proved to be the most frequent overlap syndrome. Overlapping syndromes are mostly associated to inframe mutations in exons 1, 2, 8 and 9. These data further improve the understanding of the pathogenesis of laminopathies.
Key words: Lamin A/C, laminopathies, LMNA overlapping syndromes
Introduction
The LMNA gene, placed on chromosome 1q21-22, spans 12 exons and codes via alternative splicing for the A type lamins (1). A type lamins, which belong to the type V intermediate filaments and include lamins A, C, (the major isoforms), C2 and A 10 (the minor isoforms) (2), are characterized by an N-terminal head domain, a central α-helical rod domain, and a COOH-terminal ''tail domain'' (3). The rod domain is constituted by 4 regions with a typical α-helical organization (1A, 1B, 2A, 2B), that are interconnected by 3 intervening regions with the role of linkers (L1, L12, and L2). The portion of A type lamins with an α-helical organization presents the repeated sequence a-b-c-d-e-f-g with a and d being predominantly apolar and e and g polar residues; the heptad repeat sequence facilitates the interaction between lamins monomers and the formation of dimers via non covalent interactions among apolar residues located on the rod domain of different lamins (4). A type lamins dimers are also predicted to interact in a "head to tail" fashion, via non covalent interactions between regions of lamins with a different charge (4); the regions of lamin molecules predicted to allow the head to tail interaction, include two positively charged segments (the first from 1 to 28 residue, the second from residue 386 to residue 402) and two or three negatively charged segments (essentially, the N terminal and C terminal parts of the ROD domain) (4). The LMNA gene exon 1 yields the head domain and the first tract of the rod domain; exons 2-6 encode for what remains of the rod domain; exons 7-9 code for the portion of COOH-tail domain shared by both A and C lamins, including the region of nuclear localization signal (NLS) and the portions of lamins binding directly to DNA; the exon 10 contains the splicing site alternatively activated/ silenced for the production of A and C lamins; also, exon 10 codes for the remaining portion of the COOH terminal head domain of lamins C whilst part of exon 10 and the whole exons 11 and 12 yield for the lamins A terminus portion (5).
These proteins take part in the constitution of the nuclear lamina, a complex network of proteins located underneath the inner nuclear membrane (1). Lamins interact with several partners including nuclear envelope constituents, nucleoplasmic actin, chromatin, DNA, regulators of genes expression and molecules implicated in signal transduction (6). Such a plethora of interactions explains why A type lamins play a central role in the physiologic processes of cell life, including formation and homeostasis of the nucleus (7), apoptosis (8), repair (9), replication and transcription of DNA (10), regulation of chromosomal positioning (10). They are also involved in other important processes including metabolic, biochemical and signal transduction pathways (11, 12). Mutations on the Lamin A/C gene cause several defined clinical conditions, commonly termed as laminopathies, consisting in a heterogeneous group of diseases which include: the autosomal dominant and recessive forms of Emery Dreifuss muscular dystrophy (EDMD2 and EDMD3); the limb girdle muscular dystrophy 1B (LGMD1B); the congenital muscular dystrophy-L (CMDL); the dilated cardiomyopathy with conduction defects (DCM1A); the heart hand syndrome of Slovenian type (HHS); a recessive form of sensory-motor peripheral neuropathy (CMT2B); the familial partial lipodystrophy of the Dunnigan type (FPLD2); the Hutchinson- Gilford progeria syndrome (HGPS); the atypical form of Werner syndrome (WS); the restrictive dermopathy (RD) and the mandibuloacral dysplasia (MADA) (13). Several clinical complex entities, obtained by the concomitant presence in the same subject of different diseases related to LMNA gene mutations, have also been reported (14-60). Diseases characterized by the compromise of skeletal muscles and/or the heart are associated to mutations spread throughout the gene (14), while diseases primarily affecting the peripheral nerves, the metabolism, the bones or causing alterations of the ageing mechanisms tend to be associated to particular mutations and to cluster to peculiar regions of the gene (62-65). A full correlation between genetic alterations and clinical manifestations has not been established; however, genetic studies demonstrated the presence of a non random association between clinical manifestations and Lamin A/C gene alterations (66), and the presence of a clustering among neuromuscular phenotypes (46); in particular, phenotypes characterized by skeletal and cardiac compromise tend to be associated to LMNA gene alterations placed upstream of the NLS, while clinical entities affecting the metabolism, the bones or causing premature ageing syndromes tend to be caused by alterations located downstream of the NLS (66). It has also been reported that frameshift and nonsense mutations are frequently associated to late onset cardiac and skeletal phenotypes; the possible pathogenic mechanism invoked is haploinsufficiency due to non-sense mediated mRNA decay or a rapid degradation of the aberrant transcript (46). On the other hand, early onset phenotypes affecting the skeletal muscles are mostly associated to alterations of the LMNA gene maintaining the reading frame; in this case, the pathogenic mechanism hypothesized is the poison peptide effect caused by the altered properties of mutated lamins (46). In the present paper, the authors showed the results of a meta-analysis study aimed at evaluating the pathogenic bases and the clinical manifestations of the overlapping syndromes related to Lamin A/C gene and identifying a possible relationship between the complex phenotypes producing the overlapping syndromes and the mutations of LMNA gene.
Materials and methods
We searched, by indicating in PubMed as keywords LMNA and Lamin A/C, for all papers reporting the overlapping syndromes related to LMNA gene mutations. We also looked at the UMD-LMNA mutations databases (14) [http://www.umd.be/LMNA/ (Universal Mutation Database The UMD-LMNA mutations database)] and Leiden muscular Dystrophy database (15) [http://www.dmd.nl/ (Leiden Muscular Dystrophy pages©)] in order to identify all the dominant LMNA gene mutations associated to overlapping syndromes and the papers cited in the references. We prepared a database containing the mutations identified and the complex phenotypes associated to the mutations, specifying the tissues and organs compromised; we also indicated any alterations of metabolisms or signs of premature ageing. Then, we considered the type of mutation, its position on the gene and on the protein, the effect on the aminoacidic sequence and the possible pathogenic role (haploinsufficiency, poison peptide effect) exerted by the mutations. We also calculated the frequency of the mutations per exon, associated to the overlapping syndromes. Finally, COILS software was applied to predict the coiled-coil forming and the heptad position for each aminoacidic substitution evaluated. Coils software gives a score from 0 to 1 (0: no possibility of coiled coil; 1: highest probability of coiled coil), according to the probability for the aminoacid to belong to the coiled-coil region (67).
Results
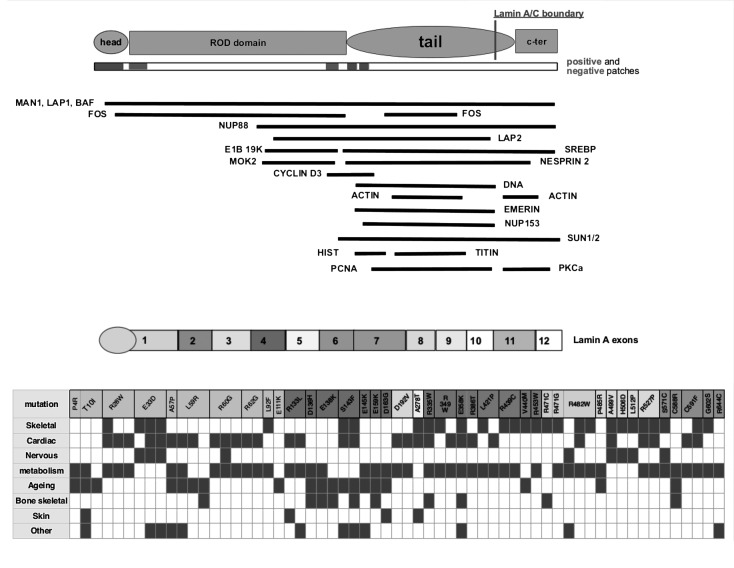
Table 1 shows the complex phenotypes related to dominant LMNA gene mutations and the characteristics of the genetic alterations. Of the identified syndromes, 69 cases are associated to 46 dominant mutations, 41 of them proved to be unique missense mutations located in 41 different positions; 31 of the 41 missense mutations involve a polar aminoacid residue, which is mutated in an apolar aminoacid in about 50% of cases; the remaining 10 missense mutations involve an apolar residue and determine in half of the cases a substitution with an aminoacid with the same polarity. Among the missense mutations, we decided to include c. 1698+13 C > T, p. Arg566 +5Cys observed in exon10; we considered the mutation position as a terminal part of the gene region coding for C lamin. A higher frequency of mutations causing overlapping syndromes per exon was observed in exons 1-2, 8 and 9 (Table 2). About half of the missense mutations are located in coiled coils regions (predicted by COILS with a probability higher than 0.5), involving in about 20% of cases the positions a and d of the heptad repeat. Six missense mutations are predicted to occur within the head-to- tail interaction region as defined by Strelkov (P4R, T101, R28W, E33D, E358K, R386T). Figure 1 also summarizes the clinical phenotypes of the overlapping syndromes associated to the reported LMNA A/C gene missense mutations, related to lamin structure and its main partners.
Table 1.
Characteristics of complex phenotypes caused by dominant LMNA gene mutations and of the related genetic alterations.
| Overlapping syndrome | LMNA Exon | Gene mutation | Protein mutation | Mutation type | Position in protein | Aminoacid substitution | COILS probability | Heptad position | Skeletal muscle phenotype | Cardiac phenotype | Nervous system (peripheral or central) | Metabolism | Ageing mechanisms | Bone/skeletal | Skin | Other | Mutation position |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 1 | c. 3-12 del | Deletion | 2 | 0 | Yes | Neuropathy | - | Head | ||||||||
| 1 | 1 | c.11C>G | P4R | Missense | 4 | P=apolare R=polare |
0 | Metabolic disturbances | Progerioid features | Bones abnormalities | Yes | Head | |||||
| 2 | 1 | c.29C>T | T10I | Missense | 10 | T=polare I=apolare |
0 | - | High triglycerides + glycemia, lipoatrophy | Progerioid features | Thinned skin | Short stature | Head | ||||
| 2 | 1 | c.29C>T | T10I | Missense | 10 | T=polare I=apolare |
0 | High triglycerides + glycemia, lipoatrophy | Progerioid features | Thinned skin | Short stature | Head | |||||
| 3 | 1 | c.82C>T | R28W | Missense | 28 | R=polare W=apolare |
0,997 | a | Yes | Yes | FLPD2 | Head | |||||
| 3 | 1 | c.82C>T | R28W | Missense | 28 | R=polare W=apolare |
0,997 | a | Yes | FLPD2 | - | Head | |||||
| 3 | 1 | c.82C>T | R28W | Missense | 28 | R=polare W=apolare |
0,997 | a | Yes | FPLD | - | Head | |||||
| 4 | 1 | c.99G>T | E33D | Missense | 33 | E=polare D=polare |
1 | f | Yes | Yes | Neuropathy | - | Head | ||||
| 4 | 1 | c.99G>T | E33D | Missense | 33 | E=polare D=polare |
1 | f | Yes | Yes | Axonal neuropathy |
Leukonichia | Head | ||||
| 4 | 1 | c.99G>T | E33D | Missense | 33 | E=polare D=polare |
1 | f | Yes | Yes | Neuropathy | - | Leukonichia | Head | |||
| 5 | 1 | c.169G>C | A57P | Missense | 57 | A=polare P=apolare |
1 | b | Yes | Partial lipodystrophy | Atypical WS | Slopping shoulders hypogonadism (ovarian failure) | c-Fos binding domain 1 | ||||
| 5 | 1 | c.176T>G | L59R | Missense | 59 | L=apolare R=polare |
1 | d | Yes | Partial lipodystrophy | Atypical WS | Slopping shoulders hypogonadism (ovarian failure) | c-Fos binding domain 1 |
||||
| 6 | 1 | c.176T>G, de novo | L59R | Missense | 59 | L=apolare R=polare |
1 | d | Yes | - | Werner S | - | c-Fos binding domain 1 |
||||
| 7 | 1 | c.176T>G | L59R | Missense | 59 | L=apolare R=polare |
1 | d | Progerioid features | MADA | c-Fos binding domain 1 |
||||||
| 3 | 1 | c.178C>G | R60G | Missense | 60 | R=polare G=apolare |
1 | e | Yes | Fat accumulation on face and neck and lipoatrophy on limbs | - | c-Fos binding domain 1 |
|||||
| 13 | 1 | c.178C>G | R60G | Missense | 60 | R=polare G=apolare |
1 | e | Yes | Axonal peripheral neuropathy | Fat accumulation on face and neck and lipoatrophy on limbs | - | c-Fos binding domain 1 |
||||
| 3 | 1 | c.178C>G | R60G | Missense | 60 | R=polare G=apolare |
1 | e | Yes | FPLD2 | - | c-Fos binding domain 1 |
|||||
| 3 | 1 | c.184C>G | R62G | Missense | 62 | R=polare G=apolare |
0,998 | g | Yes | FPLD2 | - | c-Fos binding domain 1 |
|||||
| 3 | 1 | c.184C>G | R62G | Missense | 62 | R=polare G=apolare |
0,998 | g | Yes | FPLD2 | c-Fos binding domain 1 |
||||||
| 3 | 1 | c.274C>T | L92F | Missense | 92 | L=apolare F=apolare |
1 | a | Yes | - | FPLD | - | coil 1b | ||||
| 1 | 1 | c.331G>A | E111K | Missense | 111 | E=polare K=polare |
1 | f | Metabolic disturbances | Progerioid features | Bones abnormalities | Yes | coil 1b | ||||
| 3 | 2 | c. 398 G>T | R133L | Missense | 133 | R=polare L=apolare |
1 | g | Yes | Lipodystrophy+ hepatic steatosis+ high triglycerides | - | Skin changes | coil 1b | ||||
| 3 | 2 | c.398G>T | R133L | Missense | 133 | R=polare L=apolare |
1 | g | Yes | Lipoatrophy, diabetes, liver steatosis | - | Leukomelanodermic papules | coil 1b | ||||
| 1 | 2 | c.406G>C | D136H | Missense | 136 | D=polare K=polare |
1 | c | Metabolic disturbances | Progerioid features | Bones abnormalities | coil 1b | |||||
| 1 | 2 | c.412G>A | E138K | Missense | 138 | E=polare K=polare |
1 | e | Metabolic disturbances | Progerioid features | Bones abnormalities | coil 1b | |||||
| 7 | 2 | c. 412 G>A | E138K | Missense | 138 | E=polare K=polare |
1 | e | Progeria syndrome | MADA | coil 1b | ||||||
| 6 | 2 | 428 C>T de novo | S143F | Missense | 143 | S=polare F=apolare |
0,999 | c | Yes | Yes | - | Progeria | - | Contractures | coil 1b | ||
| 8 | 2 | 428 C>T de novo | S143F | Missense | 143 | S=polare F=apolare |
0,999 | c | Yes | Yes | - | Progeria | Ospteolysis, ospeopenia, midface hypoplasia | Leukomelanodermic papules | coil 1b | ||
| 2 | 2 | c.433 G>A | E145K | Missense | 145 | E=polare K=polare |
0,999 | e | Alterations of subcutaneous fat distribution | Atypical HGPS | - | Persisting coarse hair | coil 1b | ||||
| 4 | 2 | c. 471 G>A | T157T | Synonymous | 157 | 1 | c | Yes | Neuropathy | - | coil 1b | ||||||
| 1 | 2 | c.475G>A | E159K | Missense | 159 | E=polare K=polare |
1 | e | Metabolic disturbances | Progerioid features | Bones abnormalities | coil 1b | |||||
| 2 | 2 | c. 407A>G | D163G | Missense | 163 | D=polare G=apolare |
1 | b | Lipodystrophy, insulin resistence | Progeroid facies | Achantosis nigricans | coil 1b | |||||
| 4 | 5 | c.864-867del; fs*190 | H289fsX | Frameshift | 190 | H=polare A=apolare |
1 | e | Yes | Yes | Myopathic and neurogenic features, at muscle biopsy | - | E 1B 19K | ||||
| 3 | 3 | c. 575 A>T | D192V | Missense | 192 | D=polare V=apolare |
1 | c | Yes | FPLD2 | - | coil 1b | |||||
| 3 | 3 | c. 575 A>T | D192V | Missense | 192 | D=polare V=apolare |
1 | c | Yes | FPLD | - | coil 1b | |||||
| 9 | 5 | c.832 G>A | A278T | Missense | 278 | A=apolare T=polare |
0,63 | a | Yes | Yes | Achantosis nigricans | E 1B 19K | |||||
| 3 | 6 | c. 1001-1003 del GCC p.Ser334- Ser334 del | S334del | Deletion | 334 | 1 | d | Yes | FPLD | - | Local interaction site | ||||||
| 10 | 6 | c. 1003 C>T | R335W | Missense | 335 | R=polare W=apolare |
1 | e | Yes | Yes | High triglycerides | Acro-osteolysis | Local interaction site | ||||
| 3 | 6 | c.1045 C>T | R349W | Missense | 349 | R=polare W=apolare |
1 | e | Yes | Lipodystrophy | - | Local interaction site | |||||
| 3 | 6 | c. 1045 C>T | R349W | Missense | 349 | R=polare W=apolare |
1 | e | Yes | FPLD | - | Local interaction site | |||||
| 10 | 6 | c. 1072 G>A | E358K | Missense | 358 | E=polare K=polare |
1 | c | Yes | Yes | FPLD2 like phenotype | Midfacial hypoplasia; short stature | Broad nasal bridge, limited eye closure, uterine fibroids; Respiratory failure | Local interaction site | |||
| 3 | 6 | c.1157G >C | R386T | Missense | 386 | R=polare T=polare |
0,638 | g | Yes | FPLD | - | Emerin binding domain | |||||
| 3 | 7 | c.1262 T>C | L421P | Missense | 421 | L=apolare P=apolare |
0 | Yes | IRS | - | NLS | ||||||
| 3 | 7 | c.1262 T>C | L421P | Missense | 421 | L=apolare P=apolare |
0 | Yes | Met syndrome | - | NLS | ||||||
| 3 | 7 | c.1315 C>T | R439C | Missense | 439 | R=polare C=polare |
0 | Yes | FPLD | - | PCNA interaction site | ||||||
| 3 | 7 | c.1315 C>T | R439C | Missense | 439 | R=polare C=polare |
0 | Yes | Met syndrome and fat distribution abnormalities | - | PCNA interaction site | ||||||
| 14 | 7 | c. 1318 G>A | V440M | Missense | 440 | V=apolare M=apolare |
0 | Yes | MADA | PCNA interaction site | |||||||
| 3 | 7 | c.1357 C>T | R453W | Missense | 453 | R=polare W=apolare |
0 | Yes | FPLD | - | c-Fos binding domain 2 | ||||||
| 14 | 8 | c. 1411 C>T | R471C | Missense | 471 | R=polare C=polare |
0 | Yes | MADA | Actin binding domain (1) | |||||||
| 3 | 8 | c.1411C>G | R471G | Missense | 471 | R=polare G=apolare |
0 | Yes | FPLD2 | - | Actin binding domain (1) | ||||||
| 11 | 8 | c.1444 C>T | R482W | Missense | 482 | R=polare W=apolare |
0 | - | Akinetohypertonic syndrome | FPLD2 | - | Multinodular goiter, primary hyperaldosteronism | Actin binding domain (1) | ||||
| 3 | 8 | c.1444 C>T | R482W | Missense | 482 | R=polare W=apolare |
0 | Yes | Lipodystrophy | - | Actin binding domain (1) | ||||||
| 3 | 8 | c.1444 C>T | R482W | Missense | 482 | R=polare W=apolare |
0 | Yes | FPLD | - | Actin binding domain (1) | ||||||
| 2 | 8 | c.1454C>G | P485R | Missense | 485 | P=apolare R=polare |
0 | FPLD | WS Actin binding domain | - | Actin binding domain (1) | ||||||
| 4 | 9 | c. 1496delC fsX49 | A499V | Missense | 499 | A=apolare V=apolare |
0 | Yes | Yes | Neuropathy | PKC Alpha Binding site | ||||||
| 3 | 9 | c. 1516 C>G | H506D | Missense | 506 | H=polare D=polare |
0 | Yes | - | FPLD | - | PKC Alpha Binding site | |||||
| 4 | 9 | c. 1535 T>C | L512P | Missense | 512 | L=apolare P=apolare |
0 | Yes | HNPP + Axonal Loss | PKC Alpha Binding site | |||||||
| 12 | 9 | c.1551G>A | Q517Q | Synonymous | 517 | 0 | Neuropathy | FPLD2 | PKC Alpha Binding site | ||||||||
| 12 | 9 | c. 1551G>A | Q517Q | Synonymous | 517 | 0 | Neuropathy | PLD | PKC Alpha Binding site | ||||||||
| 3 | 9 | c.1580G>C | R527P | Missense | 527 | R=polare P=apolare |
0 | Yes | Yes | FPLD2 | PKC Alpha Binding site | ||||||
| 3 | 9 | c.1580G>C | R527P | Missense | 527 | R=polare P=apolare |
0 | Yes | Yes | Lipoatrophy of trunk and proximal limbs | PKC Alpha Binding site | ||||||
| 12 | 10 | c. 1683 G>C | L561L | Synonymous | 561 | 0 | Neuropathy | FPLD2 | PKC Alpha Binding site | ||||||||
| 13 | 11 | c. 1711 A >T | S571C | Missense | 571 | S=polare C=apolare |
0 | Yes | Neuropathy | PLD | Lamin A tail | ||||||
| 1 | 11 | c.1762T>C | C588R | Missense | 588 | C=polare R=polare |
0 | Metabolic disturbances | Progerioid features | Bones abnormalities | Lamin A tail | ||||||
| 3 | 11 | c. 1772 G>T | C591F | Missense | 591 | C=polare F=apolare |
0 | Yes | FPLD2, liver steatosis | - | Lamin A tail | ||||||
| 3 | 11 | c.1772G>T | C591F | Missense | 591 | C=polare F=apolare |
0 | Yes | Yes | FPLD2 | Polymenorrhea | Lamin A tail | |||||
| 3 | 11 | c. 1804 G>A | G602S | Missense | 602 | G=apolare S=polare |
0 | Yes | IRS | Lamin A tail | |||||||
| 1 | 11 | c.1930C>T | p.R644C | Missense | 644 | R=polare C=polare |
0 | Metabolic disturbances | Progerioid features | Bones abnormalities | Lamin A tail |
Table 2.
Distribution and frequency of the mutations causing the complex phenotypes distributed per exon.
| Exon | Unique mutations | % unique mutations | Total mutations | % total mutations | Protein exon length | Total frequency normalized by exon length |
|---|---|---|---|---|---|---|
| 1 | 11 | 23.91 | 21 | 30.43 | 119 | 25.58 |
| 2 | 8 | 17.39 | 11 | 15.94 | 52 | 30.66 |
| 3 | 1 | 2.17 | 2 | 2.90 | 42 | 6.90 |
| 4 | 0 | 0 | 0 | 0 | 57 | 0 |
| 5 | 2 | 4.35 | 2 | 2.90 | 42 | 6.90 |
| 6 | 4 | 8.70 | 5 | 7.25 | 73 | 9.93 |
| 7 | 4 | 8.70 | 6 | 8.70 | 73 | 11.91 |
| 8 | 4 | 8.70 | 7 | 10.14 | 36 | 28.18 |
| 9 | 5 | 10.87 | 7 | 10.14 | 40 | 25.36 |
| 10 | 2 | 4.35 | 2 | 2.90 | 30 | 9.66 |
| 11 | 5 | 10.87 | 6 | 8.70 | 80 | 10.87 |
| 12 | 0 | 0 | 0 | 0 | 9 | 0 |
| TOT | 46 | 69 |
Figure 1.
Causative missense mutations in the context of the lamin A/C protein organization and related overlapping syndromes.
Discussion
We report a meta-analysis describing the clinical features of all overlapping syndromes related to dominant LMNA gene mutations so far published and the possible relationship with the underlying genetic alterations. We identified at least 14 different overlapping syndromes due to dominant mutations on the Lamin A/C gene. As shown in tables 1 and 2, LMNA gene mutations may be associated to complex phenotypes obtained by the variable association of different phenotypes including metabolism disturbances, premature ageing syndromes, dermatologic changes, skeletal and cardiac compromise, nervous system alterations. The most frequent overlapping syndrome linked to LMNA gene alterations is the association between metabolic alterations and skeletal and/or cardiac involvement caused by inframe mutations spread throughout the gene. It is likely that the pathogenic mechanism underlying this condition is the poison peptide effect: as a matter of fact, all the mutations so far identified alter the biochemical properties of A type lamins, either perturbing their stability or modifying the possible interactions with the numerous binding partners (54).
The overlapping syndrome characterized by the association of skeletal and/or cardiac compromise with neuropathy and inconstant dermatologic abnormalities are caused by mutations spread throughout the gene; a possible pathogenic effect should be either a dominant negative or even a haploinsufficiency secondary to the production of un unstable mRNA or of a mutated protein, lacking the typical structure of intermediate filaments. For the third and fourth group of complex phenotypes, obtained by the variable association among muscle and/or heart disease, peripheral neuropathy, metabolism disturbances and concomitant presence of lipodystrophy, the few reports so far published do not consent any final correlation. However, the presence of either missense or silent mutations suggest that a dominant negative effect may play a major role in the pathogenesis of these two entities. For overlapping syndromes with variable association of MADA/bones alterations, metabolism abnormalities and premature ageing syndromes and other clinical entities such as dermatologic abnormalities, skeletal and/or cardiac diseases, the paucity of reports again do not consent any correlation with the mutation's position. Furthermore any direct correlation between clinical manifestations and LMNA gene mutations is hampered by the pleiotropic effect possibly exerted by Lamin A/C gene mutations (17-18, 36, 39, 53, 55, 69-70).
However, we can speculate that overlapping syndromes are mostly associated to inframe mutations able to alter the stability of A type lamins and the interactions with the numerous partners (54), causing a perturbation of the physiologic processes regulated by lamins on the different tissues. These data contribute to further improve the understanding of the pathogenic mechanisms of laminopathies.
References
- 1.Bonne G. Karpati G. Structural and Molecular Basis of Skeletal Muscle Diseases, chapter 3. Basel, Switzerland: ISN Neuropath Press; 2002. Disease associated with myonuclear abnormalities: defects of nuclear membrane related proteins (emerin, lamins a/c) pp. 48–56. [Google Scholar]
- 2.Dittmer T, Misteli T. The lamin protein family. Genome Biology. 2011;2011:222–222. doi: 10.1186/gb-2011-12-5-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strelkov SV, Herrmann H, Aebi U. Molecular architecture of intermediate filaments. Bio Essays. 2003;25:243–251. doi: 10.1002/bies.10246. [DOI] [PubMed] [Google Scholar]
- 4.Strelkov SV, Schumacher J, Burkhard P, et al. Crystal structure of the human lamin a coil 2B dimer: implications for the head-to-tail association of nuclear lamins. J Mol Biol. 2004;343:1067–1080. doi: 10.1016/j.jmb.2004.08.093. [DOI] [PubMed] [Google Scholar]
- 5.Brown CA, Lanning RW, McKinney KQ, et al. Novel and recurrent mutations in lamin A/C in patients with Emery–Dreyfuss muscular dystrophy. Am J Med Genet. 2001;102:359–367. doi: 10.1002/ajmg.1463. [DOI] [PubMed] [Google Scholar]
- 6.Goldman RD, Gruenbaum Y, Shumaker DK, et al. Nuclear lamins: building blocks of nuclear architecture. Genes Dev. 2002;1:533–547. doi: 10.1101/gad.960502. 16. [DOI] [PubMed] [Google Scholar]
- 7.Burke B, Stewart C. Life at the edge: the nuclear envelope and human disease. Nat Rev Mol Cell Biol. 2002;3:575–585. doi: 10.1038/nrm879. [DOI] [PubMed] [Google Scholar]
- 8.Cohen M, Lee K, Wilson K, et al. Transcriptional repression, apoptosis, human disease and the functional evolution of the nuclear lamina. Trends Biochem Sci. 2001;26:41–47. doi: 10.1016/s0968-0004(00)01727-8. [DOI] [PubMed] [Google Scholar]
- 9.Redwood AB, Perkins SM, Vanderwaal RP, et al. A dual role for A-type lamins in DNA double-strand break repair. Cell Cycle. 2011;10:2549–2560. doi: 10.4161/cc.10.15.16531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stierlé V, Couprie J, Ostlund C, et al. The carboxyl-terminal region common to lamins A and C contains a DNA binding domain. Biochemistry. 2003;42:4819–4828. doi: 10.1021/bi020704g. [DOI] [PubMed] [Google Scholar]
- 11.Cohen M, Lee K, Wilson K, et al. Transcriptional repression, apoptosis, human disease and the functional evolution of the nuclear lamina. Trends Biochem Sci. 2001;26:41–47. doi: 10.1016/s0968-0004(00)01727-8. [DOI] [PubMed] [Google Scholar]
- 12.Martelli AM, Bortul R, Tabellini G, et al. Molecular characterization of protein kinase C-alpha binding to lamin A. J Cell Biochem. 2002;86:320–330. doi: 10.1002/jcb.10227. [DOI] [PubMed] [Google Scholar]
- 13.Worman HJ. Nuclear lamins and laminopathies. J Pathol. 2012;226:316–325. doi: 10.1002/path.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. The UMD-LMNA mutations database Home. [Google Scholar]
- 15. Lamin A/C (LMNA) sequence variations. Leiden Muscular Dystrophy pages. [Google Scholar]
- 16.Kooi AJ, Bonne G, Duboc D, et al. Lamin A/C mutations with lipodystrophy, cardiac abnormalities, and muscular dystrophy. Neurology. 2002;59:620–623. doi: 10.1212/wnl.59.4.620. [DOI] [PubMed] [Google Scholar]
- 17.Carboni N, Porcu M, Mura M, et al. Evolution of the phenotype in a family with an LMNA gene mutation presenting with isolated cardiac involvement. Muscle Nerve. 2010;41:85–91. doi: 10.1002/mus.21443. [DOI] [PubMed] [Google Scholar]
- 18.Subramanyam L, Simha V, Garg A. Overlapping syndrome with familial partial lipodystrophy, Dunnigan variety and cardiomyopathy due to amino-terminal heterozygous missense lamin A/C mutations. Clin Genet. 2010;78:66–73. doi: 10.1111/j.1399-0004.2009.01350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duparc A, Cintas P, Somody E, et al. A cardioneurological form of laminopathy: dilated cardiomyopathy with permanent partial atrial standstill and axonal neuropathy. Pacing Clin Electrophysiol. 2009;32:410–415. doi: 10.1111/j.1540-8159.2008.02254.x. [DOI] [PubMed] [Google Scholar]
- 20.Goizet C, Yaou RB, Demay L, et al. A new mutation of the lamin A/C gene leading to autosomal dominant axonal neuropathy, muscular dystrophy, cardiac disease, and leuconychia. J Med Genet. 2004;41:e29–e29. doi: 10.1136/jmg.2003.013383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benedetti S, Bertini E, Iannaccone S, et al. Dominant LMNA mutations can cause combined muscular dystrophy and peripheral neuropathy. J Neurol Neurosurg Psychiatry. 2005;76:1019–1021. doi: 10.1136/jnnp.2004.046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agarwal AK, Kazachkova I, Ten S, et al. Severe mandibuloacral dysplasia associated to lipodystrophy and progeria in a young girl with a novel homozygous Arg527Cys LMNA mutation. J Clin Endocrinol Metab. 2008;93:4617–4623. doi: 10.1210/jc.2008-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Araujo-Vilar D, Lado-Abeal J, Palos-Paz F, et al. A novel phenotypic expression associated with a new mutation in LMNA gene, characterized by partial lipodystrophy, insulin resistance, aortic stenosis and hypertrophic cardiomyopathy. Clin Endocrinol (Oxf) 2008;69:61–68. doi: 10.1111/j.1365-2265.2007.03146.x. [DOI] [PubMed] [Google Scholar]
- 24.Vantyghem MC, Faivre-Defrance F, Marcelli-Tourvieille S, et al. Familial partial lipodystrophy due to the LMNA R482W mutation with multinodular goitre, extrapyramidal syndrome and primary hyperaldosteronism. Clin Endocrinol (Oxf) 2007;67:247–249. doi: 10.1111/j.1365-2265.2007.02870.x. [DOI] [PubMed] [Google Scholar]
- 25.Madej-Pilarczyk A, Kmieć T, Fidziańska A, et al. Progeria caused by a rare LMNA mutation S143F associated with mild myopathy and atrial fibrillation. Eur J Paediatr Neurol. 2008;12:427–430. doi: 10.1016/j.ejpn.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 26.Madej-Pilarczyk A, Rosińska-Borkowska D, Rekawek J, et al. Progeroid syndrome with scleroderma-like skin changes associated with homozygous R435C LMNA mutation. Am J Med Genet A. 2009;149A:2387–2392. doi: 10.1002/ajmg.a.33018. [DOI] [PubMed] [Google Scholar]
- 27.Kirschner J, Brune T, Wehnert M, et al. s143F mutation in lamin A/C gene leading to autosomal dominant axonal neuropathy, muscular dystrophy, cardiac disease and leuconychia. Ann Neurol. 2005;57:148–151. doi: 10.1002/ana.20359. [DOI] [PubMed] [Google Scholar]
- 28.Vital A, Ferrer X, Goizet C, et al. Peripheral nerve lesions associated with a dominant missense mutation, E33D, of the lamin A/C gene. Neuromuscul Disord. 2005;15:618–621. doi: 10.1016/j.nmd.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 29.Garg A, Speckman RA, Bowcock AM. Multisystem dystrophy syndrome due to novel missense mutations in the amino-terminal head and alpha-helical rod domains of the Lamin A/C gene. Am J Med. 2002;112:549–555. doi: 10.1016/s0002-9343(02)01070-7. [DOI] [PubMed] [Google Scholar]
- 30.Kosho T, Takahashi J, Momose T, et al. Mandibuloacral dysplasia and a novel LMNA mutation in a woman with severe progressive skeletal changes. Am J Med Genet Part A. 2007;143A:2598–2603. doi: 10.1002/ajmg.a.31983. [DOI] [PubMed] [Google Scholar]
- 31.Caux F, Dubosclard E, Lascols O, et al. A new clinical condition linked to a novel mutation in lamins A and C with generalized lipoatrophy, insulin-resistant diabetes, disseminated leukomelanodermic papules, liver steat osis, and cardiomyopathy. J Clin Endocrinol Metab. 2003;88:1006–1013. doi: 10.1210/jc.2002-021506. [DOI] [PubMed] [Google Scholar]
- 32.Lattanzi G, Benedetti S, Bertini E, et al. Laminopathies: many diseases, one gene. Report of the first Italian Meeting Course on Laminopathies. Acta Myol. 2011;30:138–43. [PMC free article] [PubMed] [Google Scholar]
- 33.Araujo-Vilar D, Lado-Abeal J, Palos-Paz F, et al. A novel phenotypic expression associated with a new mutation in LMNA gene, characterized by partial lipodystrophy, insulin resistance, aortic stenosis and hypertrophic cardiomyopathy. Clin Endocrinol (Oxf) 2008;69:61–68. doi: 10.1111/j.1365-2265.2007.03146.x. [DOI] [PubMed] [Google Scholar]
- 34.Kirschner J, Brune T, Wehnert M, et al. p.S143F Mutation in Lamin A/C: A New Phenotype Combining Myopathy and Progeria. Ann Neurol. 2005;57:148–151. doi: 10.1002/ana.20359. [DOI] [PubMed] [Google Scholar]
- 35.Csoka AB, Cao H, Sammak PJ, et al. Novel lamin A/C gene (LMNA) mutations in atypical progeroid syndromes. J Med Genet. 2004;41:304–308. doi: 10.1136/jmg.2003.015651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rankin J, Auer-Grumbach M, Bagg W, et al. Extreme phenotypic diversity and non-penetrance in families with the LMNA gene mutation R644C. Am J Med Gene Part A. 2008;146A:1530–1542. doi: 10.1002/ajmg.a.32331. [DOI] [PubMed] [Google Scholar]
- 37.Genschel J, Schmidt H J. Mutations in the LMNA Gene Encoding Lamin A/C. Hum Mutat. 2000;16:451–459. doi: 10.1002/1098-1004(200012)16:6<451::AID-HUMU1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 38.Mercuri E, Brown SC, Nihoyannopoulos P, et al. Extreme Variability of Skeletal and cardiac muscle involvement in patients with mutations in exon 11 of the lamin A/C gene. Muscle Nerve. 2005;31:602–609. doi: 10.1002/mus.20293. [DOI] [PubMed] [Google Scholar]
- 39.Benedetti S, Bernasconi P, Bertini E, et al. The empowerment of translational research: lessons from laminopathies. Orphanet J Rare Dis. 2012;7:37–39. doi: 10.1186/1750-1172-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mercuri E, Poppe M, Quinlivan R, et al. Extreme Variability of Phenotype in Patients with an Identical Missense Mutation in the Lamin A/C Gene. Arch Neurol. 2004;61:690–694. doi: 10.1001/archneur.61.5.690. [DOI] [PubMed] [Google Scholar]
- 41.Brown CA, Lanning RW, McKinney KQ, et al. Novel and recurrent mutations in lamin A/C in patients with Emery-Dreifuss muscular dystrophy. Am J med Genet. 2001;102:359–367. doi: 10.1002/ajmg.1463. [DOI] [PubMed] [Google Scholar]
- 42.Politano L, Carboni N, Madej-Pilarczy A, et al. Advances in basic and clinical research in laminopathies. Acta Myol. 2013;32:18–22. [PMC free article] [PubMed] [Google Scholar]
- 43.Chen L, Lee L, Kudlow BA, et al. LMNA mutations in atypical Werner's syndrome. Lancet. 2003;362:440–445. doi: 10.1016/S0140-6736(03)14069-X. [DOI] [PubMed] [Google Scholar]
- 44.Carboni N, Floris M, Mateddu A, et al. Aberrant splicing in the LMNA gene caused by a novel mutation on the polypyrimidine tract of intron 5. Muscle Nerve. 2011;43:688–693. doi: 10.1002/mus.21937. [DOI] [PubMed] [Google Scholar]
- 45.Tintelen JP. High yield of LMNA mutations in patients with dilated cardiomyopathy and/or conduction disease referred to cardiogenetics outpatient clinics. Am Heart J. 2007;154:1130–1139. doi: 10.1016/j.ahj.2007.07.038. [DOI] [PubMed] [Google Scholar]
- 46.Benedetti S, Menditto I, Degano M, et al. Phenotypic clustering of lamin A/C mutations in neuromuscular patients. Neurology. 2007;69:1285–1292. doi: 10.1212/01.wnl.0000261254.87181.80. [DOI] [PubMed] [Google Scholar]
- 47.Nguyen D, Leistritz DF, Turner L, et al. Collagen expression in fibroblasts with a novel LMNA mutation. Biochem Biophys Res Commun. 2007;352:603–608. doi: 10.1016/j.bbrc.2006.11.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Decaudain A, Vantyghem MC, Guerci B, et al. New metabolic phenotypes in laminopathies: LMNA mutations in patients with severe metabolic syndrome. J Clin Endocrinol Metab. 2007;92:4835–4844. doi: 10.1210/jc.2007-0654. [DOI] [PubMed] [Google Scholar]
- 49.Lupsa BC, Sachdev V, Lungu AO, et al. Cardiomyopathy in congenital and acquired generalized lipodystrophy: a clinical assessment. Medicine (Baltimore) 2010;89:245–250. doi: 10.1097/MD.0b013e3181e9442f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dutour A, Roll P, Gaborit B, et al. High prevalence of laminopathies among patients with metabolic syndrome. Hum Mol Genet. 2011;20:3779–3786. doi: 10.1093/hmg/ddr294. [DOI] [PubMed] [Google Scholar]
- 51.Lombardi F, Gullotta F, Columbaro M, et al. Compound heterozygosity for mutations in LMNA in a patient with a myopathic and lipodystrophic mandibuloacral dysplasia type A phenotype. J Clin Endocrinol Metab. 2007;92:4467–4471. doi: 10.1210/jc.2007-0116. [DOI] [PubMed] [Google Scholar]
- 52.Zirn B, Kress W, Grimm T, et al. Association of homozygous LMNA mutation R471C with new phenotype: mandibuloacral dysplasia, progeria, and rigid spine muscular dystrophy. Am J Med Genet A. 2008;146A:1049–1054. doi: 10.1002/ajmg.a.32259. [DOI] [PubMed] [Google Scholar]
- 53.Lanktree M, Cao H, Rabkin SW, et al. Novel LMNA mutations seen in patients with familial partial lipodystrophy subtype 2 (FPLD2; MIM 151660) Clin Genet. 2007;71:183–186. doi: 10.1111/j.1399-0004.2007.00740.x. [DOI] [PubMed] [Google Scholar]
- 54.Young J, Morbois-Trabut L, Couzinet B, et al. Type A insulin resistance syndrome revealing a novel lamin A mutation. Diabetes. 2005;54:1873–1878. doi: 10.2337/diabetes.54.6.1873. [DOI] [PubMed] [Google Scholar]
- 55.Scharner J, Brown CA, Bower M, et al. Novel LMNA mutations in patients with Emery-Dreifuss muscular dystrophy and functional characterization of four LMNA mutations. Hum Mutat. 2011;32:152–167. doi: 10.1002/humu.21361. [DOI] [PubMed] [Google Scholar]
- 56.Walter MC, Witt TN, Weigel BS, et al. Deletion of the LMNA initiator codon leading to a neurogenic variant of autosomal dominant Emery-Dreifuss muscular dystrophy. Neuromuscul Disord. 2005;15:40–44. doi: 10.1016/j.nmd.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 57.Csoka AB, Cao H, Sammak PJ, et al. Novel lamin A/C gene (LMNA) mutations in atypical progeroid syndromes. J Med Genet. 2004;41:304–308. doi: 10.1136/jmg.2003.015651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muschke P, Kölsch U, Jakubiczka S, et al. The heterozygous LMNA mutation p.R471G causes a variable phenotype with features of two types of familial partial lipodystrophy. Am J Med Genet A. 2007;143A:2810–2814. doi: 10.1002/ajmg.a.32046. [DOI] [PubMed] [Google Scholar]
- 59.Deconinck N, Dion E, Ben Yaou R, et al. Differentiating Emery- Dreifuss muscular dystrophy and collagen VI-related myopathies using a specific CT scanner pattern. Neuromuscul Disord. 2010;20:517–523. doi: 10.1016/j.nmd.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 60.Vigouroux C, Magré J, Vantyghem MC, et al. Lamin A/C gene: sex-determined expression of mutations in Dunnigan-type familial partial lipodystrophy and absence of coding mutations in congenital and acquired generalized lipoatrophy. Diabetes. 2000;49:1958–1962. doi: 10.2337/diabetes.49.11.1958. [DOI] [PubMed] [Google Scholar]
- 61.Sandre-Giovannoli A, Chaouch M, Kozlov S, et al. Gene defects in LMNA cause AR-CMT2 in human and mouse. Am J Hum Genet. 2002;70:726–736. doi: 10.1086/339274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shackleton S, Lloyd DJ, Jackson SN, et al. LMNA, encoding lamin A/C, is mutated in partial lipodystrophy. Nat Genet. 2000;24:153–156. doi: 10.1038/72807. [DOI] [PubMed] [Google Scholar]
- 63.Speckman RA, Garg A, Du F, et al. Mutational and haplotype analyses of families with familial partial lipodystrophy (Dunnigan variety) reveal recurrent missense mutations in the globular C-terminal domain of lamin A/C. Am J Hum Genet. 2000;66:1192–1198. doi: 10.1086/302836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Novelli G, Muchir A, Sangiuolo F, et al. Mandibuloacral dysplasia is caused by a mutation in LMNA-encoding lamin A/C. Am J Hum Genet. 2002;71:426–431. doi: 10.1086/341908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eriksson M, Brown WT, Gordon LB, et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:298–301. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hegele R. LMNA mutation position predicts organ system involvement in laminopathies. Clin Genet. 2005;68:31–34. doi: 10.1111/j.1399-0004.2005.00447.x. [DOI] [PubMed] [Google Scholar]
- 67.Lupas A, Dyke A, Stock J, et al. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 68.Zastrow MS, Vlcek S, Wilson KL. Proteins that bind A-type lamins: integrating isolated clues. J Cell Sci. 2004;117:979–987. doi: 10.1242/jcs.01102. [DOI] [PubMed] [Google Scholar]
- 69.Taylor MR, Fain PR, Sinagra G, et al. Natural history of dilated cardiomyopathy due to lamin A/C gene mutations. J Am Coll Cardiol. 2003;41:771–780. doi: 10.1016/s0735-1097(02)02954-6. [DOI] [PubMed] [Google Scholar]
- 70.Parks SB, Kushner JD, Nauman D, et al. Lamin A/C mutation analysis in a cohort of 324 unrelated patients with idiopathic or familial dilated cardiomyopathy. Am Heart J. 2008;156:161–169. doi: 10.1016/j.ahj.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]