Abstract
TpbA is a periplasmic dual specificity phosphatase (DUSP) that controls biofilm formation in the pathogenic bacterium, Pseudomonas aeruginosa. While DUSPs are known to regulate important cellular functions in both prokaryotes and eukaryotes, very few structures of bacterial DUSPs are available. Here, we present the solution structure of TpbA in the ligand-free open conformation, along with an analysis of the structural and dynamic changes that accompany ligand/phosphate binding. While TpbA adopts a typical DUSP fold, it also possesses distinct structural features that distinguish it from eukaryotic DUSPs. These include additional secondary structural elements, β0 and α6, and unique conformations of the variable insert, the α4-α5 loop and helix α5 that impart TpbA with a flat active site surface. In the absence of ligand, the protein tyrosine phosphatase (PTP) loop is disordered and the general acid loop adopts an open conformation, placing the catalytic aspartate, Asp105, more than 11 Å away from the active site. Furthermore, the loops surrounding the active site experience motions on multiple timescales, consistent with a combination of conformational heterogeneity and fast (ps-ns) timescale dynamics, which are significantly reduced upon ligand binding. Taken together, these data structurally distinguish TpbA and possibly other bacterial DUSPs from eukaryotic DUSPs, and provide a rich picture of active site dynamics in the ligand-free state that are lost upon ligand binding.
Keywords: Dual specificity phosphatase, NMR spectroscopy, open/closed state active site dynamics, bacterial signaling, Pseudomonas aeruginosa
Introduction
The dual specificity phosphatases (DUSPs) are cysteine-based phosphatases that catalyze the dephosphorylation of substrates on phosphorylated tyrosine, serine and threonine residues. As a subset of the protein tyrosine phosphatase (PTP) superfamily, DUSPs contain the highly conserved signature motif HCXXXXXR and a conserved aspartic acid residue that is important for catalysis1. Eukaryotic DUSPs play critical regulatory roles in numerous cellular processes including proliferation, apoptosis, differentiation and stress response2; 3; 4. In bacteria, DUSPs are intimately involved in mechanisms controlling pathogenicity, including manipulation of host cell proteins 5 and polysaccharide production, which is crucial for biofilm growth6. Importantly, outside of the conserved signature motif, there is low sequence homology between bacterial and eukaryotic DUSPs. Thus, despite the wealth of information available for eukaryotic DUSPs, little is known about the structural features specific to bacterial DUSPs.
Tyrosine phosphatase related to biofilm formation A (TpbA) is a DUSP from the pathogenic bacterium, Pseudomonas aeruginosa. Infection by P. aeruginosa is one of the most common and life-threatening diseases faced by patients suffering from cystic fibrosis (CF)7. The persistence of P. aeruginosa infections is linked to its ability to form biofilms, organized communities of microorganisms encased in a polymeric matrix8. Critically, TpbA reduces biofilm formation in response to acyl homoserine lactone, a quorum sensing molecule6. TpbA is targeted to the periplasm and is a negative regulator of TpbB, a diguanylate cyclase (GGDEF). TpbB catalyzes the synthesis of 3,5-cyclic diguanylic acid (c-di-GMP), an important second messenger that increases transcription across the pel operon, which, in turn, leads to the formation of pellicles, one of the major biofilms in P. aeruginosa. By attenuating the activity of TpbB through dephosphorylation of both tyrosine and serine/threonine residues, TpbA reduces the intracellular concentration of c-di-GMP, which leads to a reduction in biofilm formation9. The essential role of TpbA in regulating P. aeruginosa biofilm formation makes it a potential drug target for P. aeruginosa infections.
PTPs, LMW-PTPs and DUSPs, like TpbA, share a common catalytic mechanism10. However, apart from the loops that define the active site, they have very low sequence homology. The active site is defined by the P-loop, which includes the conserved cysteine that functions as a catalytic nucleophile, and the general acid loop, which contains the catalytic aspartic acid that functions as an acid/base during the dephosphorylation reaction. In PTPs, substrate binding is accompanied by rotation of the general acid loop, resulting in a movement of the catalytic acid/base by up to 10 Å11. This converts the PTP active site from an open catalytically inactive to a ‘closed’ catalytically active state. In contrast, far less is known about the changes that occur in DUSPs during the catalytic cycle, especially DUSPs from bacteria. This is because only a handful of DUSPs have been studied in the open conformation and even less using nuclear magnetic resonance (NMR) spectroscopy. Thus, very little is known about the dynamics of the loops that define the active site between the ligand-free and ligand-bound states and the role of loop dynamics in ligand binding and catalysis.
Here, we report the solution NMR structure of TpbA, the first structure of a bacterial periplasmic DUSP. We show that TpbA adopts a canonical DUSP fold, similar to eukaryotic DUSPs. However, TpbA also has a number of structural features which distinguish it from its eukaryotic counterparts, including additional secondary structural elements and distinct loop conformations. In addition, because the structure of TpbA was determined in the ligand-free state, it is in an open conformation, with an open general acid loop and a disordered PTP loop. Most importantly, we performed ligand titrations using inorganic phosphate to identify all residues of TpbA that respond to ligand binding, which include the PTP loop, the general acid loop and the α4-α5 loop. Finally, we provide the first detailed description of changes in the motions of these functionally important loops in both the absence and presence of ligand, revealing that ligand binding “locks” out conformational dynamics that occur on multiple timescales in loops surrounding the active site.
Results
The first structure of a bacterial periplasmic DUSP, TpbA
TpbA (residues 29-218, 21 kDa) showed high levels of soluble overexpression in E. coli and behaves as a monomer in solution as verified by size-exclusion chromatography. It can be concentrated to 1 mM without precipitation or signs of aggregation and yields a high quality 2D [1H,15N] HSQC spectrum. Out of 183 expected non-proline amide backbone cross peaks, 164 could be assigned with high confidence12. To overcome the lack of NOE distance constraints in areas with unassigned amide backbone NH pairs, all non-exchangeable side chain hydrogen atoms were assigned using different HCCH-based and a 3D 13C-resolved [1H,1H] NOESY experiments. A total of 2504 NOE-based distance restraints and 270 dihedral angle restraints were used for the 3-dimensional structure calculation of TpbA29-218. The 20 conformers from the final CYANA cycle with the lowest residual CYANA target function values were energy-minimized in a water shell (13; 14; Table 1). The core of TpbA, which is composed of residues 40-194, is well-defined and adopts a compact fold. It consists of a central 6-stranded β-sheet with a folding topology +1, +1, +2x, +1x, −2x, flanked by 5 α-helices on one side, and 2 α-helices on the other (Fig. 1A). Residues 29-39 and 195-218 are flexible and unstructured in solution based on chemical shift index (CSI) calculations derived from Cα and Cβ chemical shifts and 15N[1H]-NOE analysis (12; Supp. Fig. 5C). As a result, these regions lack NOE-based distance restraints (Supp. Fig. 1A) and are poorly defined in the final structural bundle of TpbA (Supp. Fig. 1B).
Table 1.
NMR refinement statistics for ligand-free TpbA
| Protein | |
|---|---|
| NMR distance and dihedral constraints | |
| Distance constraints | |
| Total NOE | 2504 |
| Intra-residue | 581 |
| Inter-residue | |
| Sequential (∣i – j∣ = 1) | 701 |
| Medium-range (∣i – j∣ < 4) | 515 |
| Long-range (∣i – j∣ > 5) | 707 |
| Intermolecular | 0 |
| Hydrogen bonds | 0 |
| Total dihedral angle restraints | 270 |
| ϕ | 135 |
| ψ | 135 |
| Structure statistics | |
| Violations (mean and s.d.) | |
| Distance constraints (Å) | 0.0175 ± 0.0010 |
| Dihedral angle constraints (°) | 0.481 ± 0.044 |
| Max. dihedral angle violation (°) | 3.658 ± 0.709 |
| Max. distance constraint violation (Å) | 0.226 ± 0.041 |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.0106 ± 0.0002 |
| Bond angles (°) | 1.26 ± 0.03 |
| Impropers (°) | 1.46 ± 0.06 |
| Average pairwise r.m.s. deviation* (Å) | |
| Heavy | 7.85 ± 2.86 |
| Backbone | 7.56 ± 2.85 |
| Secondary Structure Heavy** | 1.02 ± 0.09 |
| Secondary Structure Backbone** | 0.56 ± 0.08 |
Pairwise r.m.s. deviation was calculated among 20 refined structures using MolMol.
Pairwise r.m.s. deviation was calculated among secondary structure residues only, excluding helix α6 (residues 40-44, 49-51, 56-59, 64-73, 77-79, 87-90, 96-98, 109-122, 128-131, 136-149, 155-164, and 175-182).
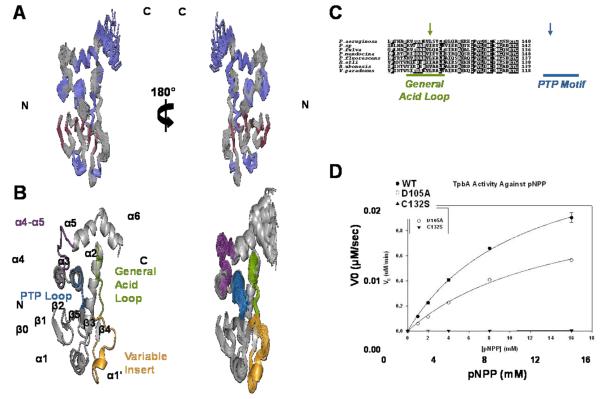
Figure 1. TpbA adopts a canonical DUSP fold.
(A) Bundle of the 20 lowest energy structures of TpbA. The superposition is the best fit of the structured regions, excluding helix α6, which are colored in blue for α-helices and red for β-strands. (B) Left, The lowest energy structure of TpbA is shown as a cartoon, with functionally important regions highlighted, including the PTP loop (blue), the general acid loop (green), the variable insert (yellow) and the α4-α5 loop (purple). Right, Bundle in A shown as a cartoon using the same color scheme as in B. (C) Sequence conservation of the general acid loop and PTP motif across bacterial species. Identical residues are highlighted black, while similar residues are highlighted gray. The catalytic residues, C132 and D105, are marked with blue and green arrows, respectively. (D) Dephosphorylation of pNPP by WT (•), D105A (○) and C132S (▼) TpbA mutants.
TpbA adopts a eukaryotic-like DUSP fold, confirming bioinformatics predictions and previous experiments showing phosphatase activity against phosphotyrosine (pTyr), phosphoserine (pSer) and phosphothreonine (pThr)6; 9. The active site architecture of the DUSP catalytic domain is defined by four loops (Fig. 1B): (1) the PTP loop, which contains the conserved cysteine that functions as the catalytic nucleophile, (2) the general acid loop, which contains a conserved aspartic acid residue that functions as a catalytic acid/base (3), the variable insert, which varies in sequence and length among DUSPs and may be involved in substrate recognition, and (4) loop α4-α5, which is in the position of the Q-loop in PTPs. Also, DUSPs have a characteristically shallow active site (~6 Å) that allows access of both pTyr and pSer/pThr to the active site cysteine, whereas the deeper active site in specific PTPs (~9 Å) is selective for pTyr. As expected for a DUSP, TpbA has a shallow active site, consistent with its ability to dephosphorylate pTyr, pSer and pThr residues. While the structures of numerous eukaryotic DUSPs are available, TpbA is only the fourth DUSP from bacteria whose structure has been determined15; 16; 17. Most importantly, this is the first structure of a bacterial DUSP solved in a ligand-free-state, which adopts an open general acid and disordered PTP loop, giving valuable insights into the active site architecture in the absence of ligand as well as the molecular basis for substrate binding.
Cys132 and Asp105 mediate catalysis in TpbA
The consensus catalytic sequence that defines Type-1 cysteine-based PTPs, which includes DUSPs, is HCXXXXXR1. In TpbA, this sequence is 131HCKHGNNR138, with Cys132 predicted to function as the catalytic nucleophile during catalysis (Fig. 1C). Although three Asp residues are present in the general acid loop, only Asp105 and Asp108 are conserved among multiple Pseudomonas species. Furthermore, our structure suggests that only Asp105 is capable of accessing the active site without requiring large, global structural rearrangements.
To test the role of Asp105 and Cys132 in catalysis, we measured the catalytic activity of WT TpbA, a D105A and a C132S mutant against the general PTP and DUSP model substrate, p-nitrophenyl phosphate (pNPP; Fig. 1D). The Cys to Ser mutation is a well-characterized mutation that typically results in a complete loss of catalytic activity in PTPs and DUSPs18; 19; 20; 21. Mutation of the conserved Asp also leads to a loss of catalytic activity, although a subset of DUSPs/PTPs are only mildly affected17; 22; 23; 24. We found that while WT TpbA effectively dephosphorylated pNPP, the C132S mutant had no detectable activity, and the D105A mutant resulted in a 53% decrease in catalytic efficiency (Table 2). These data, in combination with the structure, support the role of Asp105 as the general acid/base and Cys132 as the catalytic cysteine in the TpbA-catalyzed dephosphorylation reaction.
Table 2.
Enzymatic activity of WT and TpbA variants
| Vmax (uM/min) | kcat (s−1) | Km (mM) | kcat/Km (M−1s−1) | Fold difference in kcat/Km |
|
|---|---|---|---|---|---|
| WT | 1.55 ± 0.05 | 1.62×10−3 | 11.21 ± 0.72 | 0.14 | 1.00 |
| D105A | 1.12 ± 0.11 | 1.17 ×10−3 | 15.22 ± 2.62 | 0.08 | 0.53 |
| C132S | ND | ND | ND | NA | NA |
TpbA is Unique among DUSPs
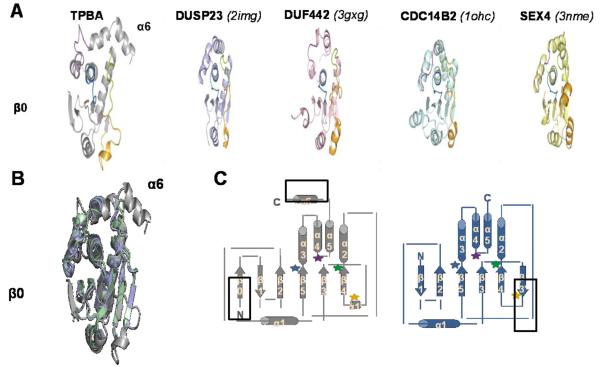
A search for homologous structures using the DALI server identified DUSP2325, a eukaryotic atypical DUSP, as the structure most similar to TpbA, with a DALI Z-score of 10.1. The next most similar structures are DUF442, a bacterial protein predicted to be a DUSP from Shewanella putrefaciens (Z-score = 9.5), CDC14B226, a eukaryotic DUSP in the PTEN family (Z-score = 9.5) and SEX427, a DUSP from Arabidopsis thaliana (Z-score = 9.4).
A comparison of TpbA with these and other DUSP structures reveals that, while TpbA adopts a canonical DUSP architecture, it is also unique (Fig. 2A). First, compared with other DUSPs, TpbA has an additional helix, helix α6. This helix does not make any stable contacts with the rest of the phosphatase domain and no NOEs are observed between it and the remainder of the protein. However, helix α6 is required for TpbA solubility, as deletion of this helix results in insoluble expression of TpbA in E. coli. Second, most DUSPs have 5 β-strands that form the central β-sheet. In contrast, the central β-sheet of TpbA is composed of 6 β-strands (Fig. 2B). The only other DUSPs that have a 6-stranded central β-sheet are DUSP6 (PYST/MKP3) and DUSP9 (MKP4). However, in these DUSPs, the additional strand is located in the variable insert (Fig. 2C). In contrast, the additional β-strand in TpbA is located at the N-terminus of the protein, capping the β-sheet, a feature that, to our knowledge, is unique to TpbA.
Figure 2. TpbA is unique among DUSPs.
(A) TpbA and its four closest structural homologs, DUSP23 (PDB ID 2IMG; Z-score 10.1, light purple), DUF442 (PDB ID 3GXG, Z-score 9.5, pink), CDC14B2 (PDB ID 1OHC, Z-score 9.5, light cyan) and SEX4 (3NME, Z-score 9.4, yellow); coloring scheme of loops same as in Fig 1B. (B) Overlay of TpbA with DUSP6 (blue, PYST/MKP3) and DUSP9 (green, MKP4). Structural features unique to TpbA, strand β0 and helix α6, are labeled. (C) Cartoon illustrating the secondary structure topology of TpbA (grey) and DUSP6/DUSP9 (blue). The additional secondary structural elements in these DUSPs compared to canonical DUSPs are boxed. Colored stars indicate the locations of the loops colored in A.
Not only does TpbA possess additional secondary structural features, but other conserved regions of the protein adopt conformations distinct from other DUSPs. First, in some DUSPs, a residue in the variable insert makes polar contacts with the conserved Arg in the PTP motif, positioning the Arg sidechain in the active site. In TpbA, the orientation of the variable insert is disengaged from the active site, leaving the active site face of TpbA flat and open. This is significant because the variable insert contributes to the depth of the active site27. Second, the conformation of the α4-α5 loop and helix α5 is unique to TpbA. In most DUSPs, this loop defines the upper boundary of the PTP pocket (see for example DUSP23, CDC14B2 and SEX4 in Fig. 2A), and in human DUSPs this loop is also conserved in sequence (known as the R-motif28). In contrast, in TpbA, this loop folds away from the PTP loop, adopting a conformation more similar to that of another bacterial DUSP, DUF442. Finally, the AYLM motif, which forms part of the extended signature sequence of the DUSP family28, is also not conserved in TpbA. This motif has been replaced by the sequence MYRIV in helix α3, resulting in an alternate orientation of helix α5 that allows it to bridge helix α2 and helix α3. This in turn enables helix α6 to point away from the catalytic domain, a conformation unique to TpbA.
Stabilization of the TpbA active site upon ligand binding
In the ligand-free form of TpbA, the majority of the unassigned amide backbone NH pairs (Ser80, Phe81, Ile82, Lys83, Ala104, Glu118, Lys133, His134, Gly135, Asn136, Thr139, Gly140, Phe142, Ala158, Asp172, Met173, Arg174, Leu204 and His206) belong to the variable insert, the PTP loop and the α4-α5 loop. Due to the lack of NOE distance constraints, these loops do not converge as well as the rest of the core phosphatase domain. In TpbA there are, on average, 35 NOE-derived distance restraints per residue for all well-defined regions of the structure, whereas there are only 17 restraints per residue in the loop regions mentioned above (Supp. Fig. 1A). This lack of convergence in loop conformations has also been observed for other phosphatases, including the DUSP PRL-329, a bacterial acylphosphatase30 and DUSP PAC-131.
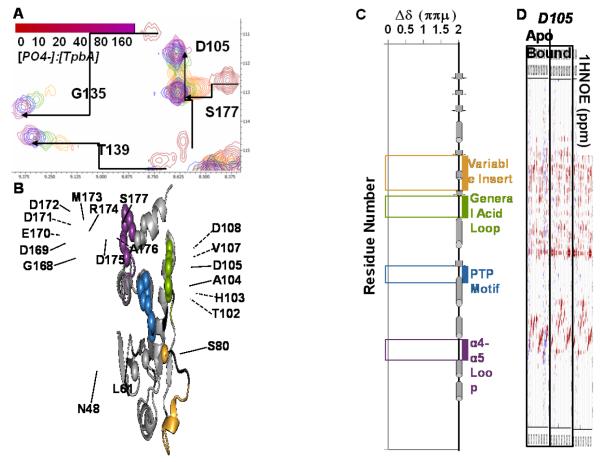
To determine whether the conformation of the active site is stabilized upon ligand binding, we performed titration experiments of TpbA with sodium phosphate using protein:phosphate ratios ranging from 1:10 to 1:320, the latter of which resulted in full saturation (Fig. 3; Supp. Fig. 2). The number of NH cross peaks in a 2D [1H,15N] HSQC spectrum of TpbA increases from 164 in the ligand-free-state to 172 in the phosphate-bound state. The increased number of NH cross peaks indicate that these residues undergo a significant change in their backbone dynamics from the intermediate timescale to the fast timescale exchange regime. Indeed, inspection of the 2D [1H,15N] HSQC spectra reveals increases in peak intensities for residues in the general acid loop, the PTP motif, helix α3, and the α4-α5 loop in phosphate-bound versus ligand-free TpbA, consistent with a shift from the intermediate to the fast exchange regime (Fig. 3A; Supp. Fig. 2). Asp105 experiences the greatest relative change in intensity, as it is 6.7 times more intense in the phosphate-bound TpbA 2D [1H,15N] HSQC spectrum than in the ligand-free TpbA spectrum. Additionally, Gly135 and Thr139 in the PTP motif, and Asp171 and Ala176 in the α4-α5 loop all have 3 to 3.5 times greater intensity in the phosphate-bound 2D [1H,15N] HSQC spectrum. Other residues in these regions (His103, Asn136, Phe142 and Asp172) are broadened beyond detection in the 2D [1H,15N] HSQC spectrum of ligand-free TpbA, but show average cross peaks intensities in the phosphate-bound 2D [1H,15N] HSQC TpbA spectrum. Interestingly, this behavior is distinct from that observed for the low molecular tyrosine phosphatase MptpA, where many more NH cross peaks become broadened in the phosphate-bound form in the P-loop and those amino acids flanking the D-and W-loops32.
Figure 3. Phosphate-binding involves the general acid loop, the PTP and the α4-α5 loops.
(A) Portion of the 2D [1H,15N] HSQC spectrum of 15N-TpbA from increasing titrations of sodium phosphate. Chemical shift perturbations (CSPs, indicated by arrows) and intensity changes are observed for residues surrounding the active site, including G135 and T139 in the PTP loop and D105 in the general acid loop. (B) Residues with CSP ≥ 0.2 ppm are mapped onto the structure of TpbA and shown as spheres using the same color scheme as in Fig 1B. (C) Histogram showing the CSPs between ligand-free and phosphate-bound TpbA. 15NH resonances that are only assigned in either ligand-free or phosphate-bound TpbA are set to an arbitrary value of 2 ppm. Functionally important loops are boxed, labeled and colored as in Fig 1B. (D) The 1H NOESY pattern of D105 changes upon phosphate binding. Strips corresponding to the D105 backbone HN spin system of ligand-free (left) and phosphate-bound TpbA (right). In ligand-free TpbA, only intra-residue NOEs are observed. Upon phosphate binding, 11 new inter-residue NOEs appear, consistent with D105 closing over the TpbA active site upon ligand binding.
To better understand these changes, we performed the sequence-specific backbone assignment of TpbA in the phosphate-bound state. CSI values show that the secondary structure elements are identical in ligand-free and phosphate-bound TpbA (Supp. Fig. 3). Chemical shift perturbation (CSP) mapping of ligand-free and phosphate-bound TpbA shows that the PTP, the general acid and the α4-α5 loops are most affected by phosphate binding (Fig. 3B; Fig. 3C). Interestingly, the variable insert is not affected, except for a single residue S80. This differs from the human DUSP PRL-3 where all variable insert residues have CSPs upon phosphate binding29.
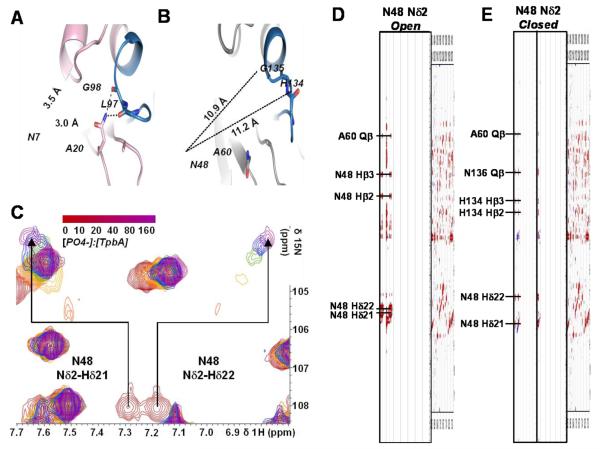
We also measured a 3D 15N-resolved [1H,1H] NOESY spectrum of phosphate-bound TpbA. Changes in NOEs of loop residues, particularly D105, are consistent with a conformational change in which the general acid loop is closed over the TpbA active site (Fig. 3D; Supp. Fig. 4). Additionally, the side chain NH2 group of N48 in the β1-β2 loop experiences large CSPs and dramatic change of NOEs upon phosphate binding (Fig. 4). In the crystal structure of DUSP23, which has a closed general acid loop, residue N7 in the β1-β2 loop makes polar contacts with the backbone carbonyls of L97 and G98 in the PTP loop (Fig. 4A). If the PTP loop of TpbA adopted a conformation similar to DUSP23, NOEs between N48 Hδ21/Hδ22 and the PTP loop should be observed. However, the structure of open conformation TpbA shows that N48 is 11 Å away from H134 and G135 in the PTP loop (Fig. 4B). Yet, N48 is clearly affected by ligand binding as the NH2 side chain protons experience large CSPs in response to phosphate binding (Fig. 4C). Thus, while no NOEs are detected in the ligand-free open conformation (Fig. 4D), in the phosphate-bound state, N48 Hδ21/Hδ22 protons have many medium-range NOEs with H134 Hβ2/Hβ3 and N136 Qβ, consistent with the conformation of the PTP motif observed in the closed structure of DUSP23 (Fig. 4E). Taken together, these data show that phosphate-bound TpbA adopts the expected active site architecture observed in numerous structures of DUSPs in the closed conformation25; 33; 34; 35.
Figure 4. Ligand binding causes structural changes in the PTP loop that involve N48.
(A) The crystal structure of DUSP23 (PDB ID 2IMG) bound to malate (omitted for clarity). N7 makes polar contacts with the backbone carbonyls of PTP loop residues G98 and L97. Polar interactions are indicated by dashed lines. (B) The lowest energy structure of TpbA with residues homologous to those in A (N48, G135 and H134, respectively) shown as sticks. The distances between N48 and the backbone carbonyls of G135 and H134 are shown as dashed lines. (C) CSPs of the N48 side chain protons, Hδ21 and Hδ22, in increasing concentrations of phosphate. (D) In the absence of substrate (Open) intra-residue NOEs are identified for N48 Hδ21 and Hδ22 side chain protons. (E) Upon phosphate binding (Closed) NOEs to H134 and N136 are identified for N48 Hδ21 and Hδ22.
Dynamics of ligand-free and phosphate-bound TpbA
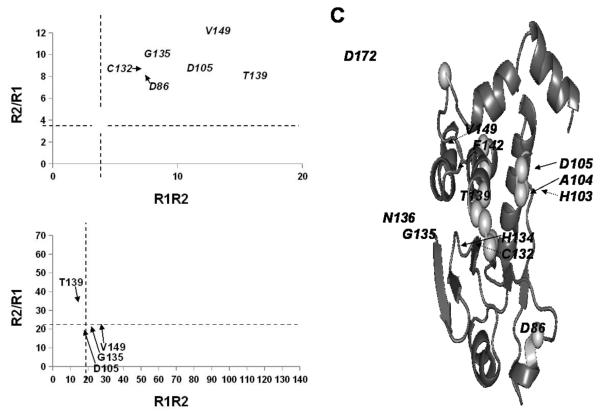
To better understand the dynamics changes that occur upon ligand binding in TpbA, we performed 15N-based auto-correlated relaxation analysis of the open and closed states of TpbA (Supp. Fig. 5 and 6). The average R1 relaxation rate of the core phosphatase domain (residues 40-194, including the PTP and general acid loops) changes between ligand-free open TpbA (1.15 ± 0.08 s−1) and phosphate-bound closed TpbA (1.00 ± 0.09 s−1), indicating a loss of ns relaxation in closed TpbA. All of the core residues, for which the difference in R1 between apo- and phosphate-bound states exceeds twice the propagated error in the measurements, have decreases in R1, suggesting that this effect, though modest, is significant. Furthermore, the transverse relaxation rates for backbone 15NH cross peaks in the PTP motif and the general acid loop differ in the open and closed states. A plot of R2/R1 versus R1R2 highlights these differences (36; Fig. 5). Residues in the PTP and general acid loops, namely Asp105, Cys132, Gly135 and Thr139, are clear outliers in ligand-free TpbA, which is primarily attributed to elevated R2 rates (Fig. 5A). The average R2 value for these residues is 56.6 ± 7.2 s−1, which is nearly twice that of the core phosphatase domain (29.6 ± 9.6 s−1). This difference disappears in the phosphate-bound state (Fig. 5B) where the distribution of the 15N relaxation states is tightly clustered in the R2/R1 versus R1R2 plot, with an average R2 of 24.8 ± 4.0 s−1. Again, while the R2 averages show only a modest effect, in direct comparisons, 78% of core residues with significant differences in relaxation rates between apo- and bound states experienced a decline in R2.
Figure 5. The TpbA PTP and general acid loops experience a loss of μs/ms motions upon ligand binding.
R2/R1 is plotted as a function of the product of R1R2 for (A) 1 mM 15N-TpbA in the open conformation, and (B) 0.97 mM 15N-TpbA bound to phosphate (1:320 protein:ligand molar ratio) in the closed conformation. Error bars represent a propagation of errors from the relaxation measurements. Dashed lines show the averages of R2/R1 and R1R2. (C) TpbA residues that experience a change in dynamics between the ligand-free and the phosphate-bound state are plotted as spheres on the lowest energy structure of TpbA and labeled.
The reduced dynamic range for both R2/R1 and R1R2 indicates a loss of slower μs-ms motions associated with Rex. This is consistent with the observation that exchange broadening impeded assignment of the backbone 15NH pairs for many of these residues in the open conformation. For the same reason, quantitative relaxation measurements for His103 and Ala104 in the general acid loop, His134 and Asn136 in the PTP loop, Phe142 in helix α3, and Asp172 in the α4-α5 loop could not be obtained for ligand-free TpbA, but were readily measured in phosphate-bound TpbA (Supp. Fig. 7). Upon titration of phosphate, these resonances sharpen significantly, indicating that they undergo a change in their dynamics from the intermediate exchange to the fast exchange regime. This is consistent with the overall decrease in R1 for the PTP and general acid loops, supporting a loss of ns motions in addition to the loss of μs-ms motions in these regions. These data, together with the sharply increased number of NOEs in phosphate-bound TpbA for many residues in the PTP, the general acid and other active site loops (Supp. Fig. 4), strongly suggest that phosphate and/or ligand binding leads to a narrower range of conformational possibilities of the PTP and general acid loops in phosphate-bound TpbA (Fig. 5C).
Discussion
Our structure of TpbA is the first of a bacterial periplasmic DUSP and one of the few ligand-free open DUSPs. Equally important, our studies also provide the first detailed description of functionally important loop dynamics in a DUSP in the absence of ligand and show how they change in response to ligand binding. Taken together, this work significantly extends the small but growing body of evidence suggesting cysteine-based phosphatases experience conformational flexibility in the open ligand-free state29; 30; 31; 37; 38. Furthermore, this is the first time that such an observation has been reported for a bacterial DUSP, showing that bacterial DUSPs may share not only structural but also dynamic similarities with their eukaryotic counterparts.
The catalytic activity and substrate specificity differs between DUSPs, depending on the enzyme itself and the substrate. For example, although the catalytic activity of the human DUSP VHZ against the nonspecific substrate pNPP is low (kcat = 0.0094 s−1)39, it increases ~50-fold when the substrate is a phosphorylated peptide40. Similar to VHZ, we show that TpbA has a low catalytic activity against pNPP (kcat = 0.00162 s−1) and has likely a higher activity against a specific substrate. Presently, TpbB is the only known substrate of TpbA, and the primary amino acid sequence that is recognized by TpbA is currently unknown. Thus, identification of the TpbA-binding sequence in TpbB will be necessary to test TpbA catalytic activity against a specific substrate.
To date, only a small number of cysteine-based phosphatases have been characterized in the open state. This is because these proteins generally do not crystalize in the absence of ligand, and, because of their size, are also more difficult targets for structure determination using NMR spectroscopy. Moreover, even when open LMW-PTPs and DUSPs have been studied using NMR spectroscopy, NH cross-peaks for functionally relevant residues in the active loops are often missing. These observations suggest that these proteins are dynamic in the ligand-free open state, with the active site loops likely adopting multiple conformations29; 30; 31; 37; 38. Here, we show that for TpbA, addition of the simplest ligand, inorganic phosphate, is sufficient to induce a significant change in both the structure and dynamics of its active site loops, as evidenced by an increase of the number of NH crosspeaks in the ligand-bound 2D [1H,15N] HSQC spectrum of TpbA. The phenomenon whereby ligand binding causes PTP loop resonances to appear in the NMR spectra has been observed in some phosphatases outside the DUSP family, such as LWM-PTPs30; 37; 38. However, the degree to which these active site loops experience dynamics, and, furthermore, on which timescales, have not yet been reported. This is in part because these important regions are generally not detectable in the absence of ligand. Importantly, while many NH crosspeaks are weak in ligand-free TpbA, they can be detected. Thus, TpbA provides an unprecedented opportunity to further probe the nature of the dynamics that define the DUSP active site, and, critically, to characterize how they change in response to ligand binding. Here, we report both fast (ns) and slow (μs-ms) timescale dynamics for TpbA, the first time these motions have been examined in a DUSP. We discovered that ligand binding significantly changes the active site loops dynamics. Our analysis shows that both fast and intermediate time scale dynamics change upon ligand binding, resulting in a narrower range of conformational possibilities for the PTP and general acid loops. That is, ligand-binding reduces the conformational dynamics that occur on multiple timescales in the loops at the active site. Further investigation into the relationship between these dynamic motions in both phosphatase activity and substrate selectivity will facilitate the discovery of targeted inhibitors and activators, as well as further our understanding of the DUSP family.
Methods and Materials
Protein Expression and Purification
TpbA29-218 was expressed in E. coli and purified as previously described 12. TpbA29-184, which lacks helix α6, was insolubly expressed in E. coli. Briefly, TpbA29-218 was purified by Ni2+-affinity chromatography and size exclusion chromatography (SEC, Superdex 75 26/60) using 10 mM Tris-HCl pH 7.8, 100 mM NaCl and 0.5 mM TCEP as the final buffer. Partially 2H,15N,13C-labeled TpbA29-218 was expressed in E. coli cultures grown in M9 minimal media containing 4 g/L 13C-D-glucose, 1 g/L 15N-NH4Cl and 25% H2O/75% D2O. Cultures were grown at 37°C and 250 rpm to a final OD600 of 0.9. Protein expression was induced with the addition of 1 mM IPTG and cultures were incubated for ~20 hours (18°C, 250 rpm). The protein yield was ~30 mg protein/ L cell culture. Mass spectrometry (MALDI-TOF and ESI) was used to determine the overall percentage of 2H incorporation into the protein, which was ~45%.
NMR Spectroscopy
NMR data were collected on Bruker Avance 500 and 800 MHz spectrometers equipped with TCI HCN Z-gradient cryoprobes at 298 K. NMR measurements of TpbA29-218 were recorded using either 15N- or 15N,13C-labeled protein at a final concentration of 1 mM in 10 mM Tris pH 7.8, 100 mM NaCl, 0.5 mM TCEP and 90% H2O/10% D2O. Sequence-specific backbone and side chain assignments were obtained as previously described12. Briefly, side chain assignments for all aliphatic residues were obtained from a 3D HC(C)H-TOCSY spectrum (Tm = 12 ms, 800 MHz 1H Larmor frequency), a 3D (H)CCH-TOCSY spectrum (Tm = 12 ms, 500 MHz 1H Larmor frequency) and a 3D HC(C)H-COSY spectrum (500 MHz 1H Larmor frequency). NOE-based distance restraints were assigned from a 3D 15N-resolved [1H,1H] NOESY (Tm = 65 ms, recorded at 800 MHz 1H Larmor frequency), a 3D 13C-resolved [1H,1H] NOESY (Tm = 65 ms, recorded at 800 MHz 1H Larmor frequency), a 3D 13C-resolved [1H,1H] NOESY (Tm = 65 ms, recorded at 800 MHz 1H Larmor frequency; 45% 2H/55% 1H,15N,13C-TpbA) and a 2D [1H,1H] NOESY (Tm = 65 ms, recorded at 800 MHz 1H Larmor frequency; 100% D2O solution). The ATNOS/CANDID software package41; 42 was used for automated NOESY peak picking and NOE assignment. Spectra were processed with Topspin 2.1/3.0/3.1 (Bruker, Billerica, MA) and data were evaluated using CARA (http://cara.nmr.ch).
Structure Calculation and Refinement
2054 unambiguous NOESY-derived distance restraints along with 270 total dihedral angle restraints derived from 13C-chemical shifts using TALOS+43 were used in the initial structure calculations using CYANA 41. Final energy-minimization and structure refinement was performed in explicit solvent using CNS 1.313 along with the RECOORD script package14. 200 structures were generated for each cycle and the 20 conformers with the lowest restraint violation energies were selected as the final representative model. The quality of the final ensemble of lowest-energy structures was assessed by the programs WHATCHECK44, AQUA45 and NMR-PROCHECK45, which are part of the iCing suite (http://nmr.cmbi.ru.nl/cing/iCing.html), and MOLMOL46. Ramachandran analysis showed that the TpbA structure has excellent stereochemistry with 98.4% of residues in the most favored and allowed region, 0.8% in the generously allowed region and 0.8% in the disallowed region of the Ramachandran plot.
Phosphatase Activity Assay
Activity of TpbA against the general PTP/DUSP substrate, p-nitrophenyl phosphate (pNPP), was tested in 50 mM sodium acetate pH 5.5, 100 mM NaCl and 5 mM DTT. 40 μl of enzyme at a 16 μM concentration was added to 60 μl of pNPP substrate resuspended in the reaction buffer. The final concentrations of pNPP used in this experiment were 0, 1, 2, 4, 8 and 16 mM. The enzyme was incubated with the substrate at 37°C for 1 hour, after which 100 μl of 1 M NaOH was added to a final concentration of 0.5 M NaOH to quench the reaction. Absorbance was measured at 405 nm and the final concentration of p-nitrophenol (pNP) was calculated using the molar extinction coefficient of 18000 M−1cm−1. A control reaction in which substrate was incubated in the absence of enzyme was subtracted from all other experimental values. All experiments were performed in triplicate, and plotted as the average of the three experiments (error bars are the standard deviation). Km and kcat values were obtained by data fitting to the Michaelis-Menten model in Sigma Plot (Systat Software).
NMR Analysis of Phosphate Binding
Sodium phosphate, pH 7.8 was titrated into 500 μM 15N-TpbA at final concentrations of 5, 10, 20, 40, 80 and 160 mM, and 2D [1H,15N] HSQC spectra were recorded for each titration point. Chemical shift differences (Δδ) between ligand-free TpbA (0 mM phosphate) and phosphate-bound TpbA (160 mM phosphate) spectra were calculated using:
The sequence-specific backbone assignment of phosphate-bound TpbA (1 mM 15N,13C-TpbA, 320 mM sodium phosphate, 10 mM Tris pH 7.8, 100 mM NaCl, 0.5 mM TCEP) was achieved using the following experiments at 500 MHz 1H Larmor frequency: 2D [1H,15N] HSQC, 3D HNCA, 3D HNCACB, 3D CBCA(CO)NH and a 3D HBHA(CO)NH. A 3D 15N-resolved [1H,1H]-NOESY spectrum (Tm = 65 ms) was also recorded for phosphate-bound TpbA (1 mM 15N-TpbA, 320 mM sodium phosphate, 10 mM Tris pH 7.8, 100 mM NaCl, 0.5 mM TCEP) at 800 MHz 1H Larmor frequency.
Relaxation Measurements and Analysis
The same experiments were used to measure fast-timescale backbone dynamics for ligand-free TpbA (1 mM 15N-TpbA, 10 mM Tris pH 7.8, 100 mM NaCl, 0.5 mM TCEP) and phosphate-bound TpbA (0.97 mM 15N-TpbA, 310 mM sodium phosphate, 10 mM Tris pH 7.8, 100 mM NaCl, 0.5 mM TCEP; 1:320 molar ratio) at 500 MHz 1H Larmor frequency.15N longitudinal (R1) and transverse (R2) relaxation rates and 15N[1H]-NOE (hetNOE) measurements were acquired using sensitivity-enhanced experiments. T1 and T2 experiments were acquired with a recycle delay of 3 seconds between experiments, and the following relaxation delays for T1: 5, 100, 400, 600, 800, 1000, 1200, and 1500 ms; and T2: 17.36, 34.72, 69.44, 104.16, 121.52, 156.24, 173.6, 190.96, 208.32, and 243.04 ms. The total length of the T2 relaxation delay is determined by the length of one Carr-Purcell-Meiboom-Gill (CPMG) cycle multiplied by the number of cycles (1-14 cycles). Systematic error in both T1 and T2 experiments was estimated from variance averaging of repetition experiments, which were acquired with delays of 100 and 800 ms for T1, and 69.44 and 173.6 ms for T2. The hetNOE measurements were determined from a pair of interleaved spectra acquired with or without presaturation, and a recycle delay of 5 seconds. R1 and R2 relaxation rates and the hetNOE were analyzed using Bruker Dynamics Center 2.0 (Bruker, Billerica, MA).
Supplementary Material
The NMR structure of TpbA is the first structure of a bacterial periplasmic DUSP.
pbA has unique structural features that distinguish it from eukaryotic DUSPs.
Ligand-free TpbA shows active site motions at multiple timescales.
Ligand binding significantly reduces active site dynamics.
TpbA structure and dynamics provide new insights into enzyme regulation of DUSPs.
Figure 6.
Acknowledgments
The authors thank Drs. David A. Critton and Thusitha B. Jayasundera for help at early stages of the project, Dr. Joshua Wand (U. Penn) for helpful discussions, Dr. Torsten Herrmann (CNRS Lyon, France) for help with the structure calculation and Dr. Tun-Li Shen (Brown University, Chemistry) for recording MS data. This work was supported by NSF CAREER (MCB0952550) to R.P. and NIH (GM089999) to T. K. W. 800 MHz NMR data were recorded at Brandeis University (NIH S10-RR017269).
Footnotes
Accession Numbers All chemical shifts for ligand-free and phosphate-bound TpbA were deposited in the BioMagResBank (http://www.bmrb.wisc.edu) under accession numbers 18228 and 18977. Atomic coordinates for the ligand-free TpbA structure have been deposited in the Protein Data Bank under the PDB code 2M3V.
Supplementary Data Supplementary data associated with this article can be found, in the online version, at doi.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Alonso A, Sasin J, Bottini N, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 2.Diamond RH, Cressman DE, Laz TM, Abrams CS, Taub R. PRL-1, a unique nuclear protein tyrosine phosphatase, affects cell growth. Mol Cell Biol. 1994;14:3752–62. doi: 10.1128/mcb.14.6.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu JJ, Bennett AM. Essential role for mitogen-activated protein (MAP) kinase phosphatase-1 in stress-responsive MAP kinase and cell survival signaling. J Biol Chem. 2005;280:16461–6. doi: 10.1074/jbc.M501762200. [DOI] [PubMed] [Google Scholar]
- 4.Dickinson RJ, Keyse SM. Diverse physiological functions for dual-specificity MAP kinase phosphatases. J Cell Sci. 2006;119:4607–15. doi: 10.1242/jcs.03266. [DOI] [PubMed] [Google Scholar]
- 5.Reiterer V, Grossniklaus L, Tschon T, Kasper CA, Sorg I, Arrieumerlou C. Shigella flexneri type III secreted effector OspF reveals new crosstalks of proinflammatory signaling pathways during bacterial infection. Cell Signal. 2011;23:1188–96. doi: 10.1016/j.cellsig.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Ueda A, Wood TK. Connecting quorum sensing, c-di-GMP, pel polysaccharide, and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885) PLoS Pathog. 2009;5:e1000483. doi: 10.1371/journal.ppat.1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hassett DJ, Sutton MD, Schurr MJ, Herr AB, Caldwell CC, Matu JO. Pseudomonas aeruginosa hypoxic or anaerobic biofilm infections within cystic fibrosis airways. Trends Microbiol. 2009;17:130–8. doi: 10.1016/j.tim.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 8.Costerton W, Veeh R, Shirtliff M, Pasmore M, Post C, Ehrlich G. The application of biofilm science to the study and control of chronic bacterial infections. J Clin Invest. 2003;112:1466–77. doi: 10.1172/JCI20365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pu M, Wood TK. Tyrosine phosphatase TpbA controls rugose colony formation in Pseudomonas aeruginosa by dephosphorylating diguanylate cyclase TpbB. Biochem Biophys Res Commun. 2010;402:351–5. doi: 10.1016/j.bbrc.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denu JM, Dixon JE. A catalytic mechanism for the dual-specific phosphatases. Proc Natl Acad Sci U S A. 1995;92:5910–4. doi: 10.1073/pnas.92.13.5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barford D, Das AK, Egloff MP. The structure and mechanism of protein phosphatases: insights into catalysis and regulation. Annu Rev Biophys Biomol Struct. 1998;27:133–64. doi: 10.1146/annurev.biophys.27.1.133. [DOI] [PubMed] [Google Scholar]
- 12.Koveal D, Jayasundera TB, Wood TK, Peti W, Page R. Backbone and sidechain (1)H, (15)N and (13)C assignments of Tyrosine Phosphatase related to Biofilm formation A (TpbA) of Pseudomonas aeruginosa. Biomol NMR Assign. 2012 doi: 10.1007/s12104-012-9376-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 14.Nederveen AJ, Doreleijers JF, Vranken W, Miller Z, Spronk CA, Nabuurs SB, Guntert P, Livny M, Markley JL, Nilges M, Ulrich EL, Kaptein R, Bonvin AM. RECOORD: a recalculated coordinate database of 500+ proteins from the PDB using restraints from the BioMagResBank. Proteins. 2005;59:662–72. doi: 10.1002/prot.20408. [DOI] [PubMed] [Google Scholar]
- 15.Gruninger RJ, Selinger LB, Mosimann SC. Structural analysis of a multifunctional, tandemly repeated inositol polyphosphatase. J Mol Biol. 2009;392:75–86. doi: 10.1016/j.jmb.2009.05.079. [DOI] [PubMed] [Google Scholar]
- 16.Chu HM, Guo RT, Lin TW, Chou CC, Shr HL, Lai HL, Tang TY, Cheng KJ, Selinger BL, Wang AH. Structures of Selenomonas ruminantium phytase in complex with persulfated phytate: DSP phytase fold and mechanism for sequential substrate hydrolysis. Structure. 2004;12:2015–24. doi: 10.1016/j.str.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 17.Puhl AA, Gruninger RJ, Greiner R, Janzen TW, Mosimann SC, Selinger LB. Kinetic and structural analysis of a bacterial protein tyrosine phosphatase-like myo-inositol polyphosphatase. Protein Sci. 2007;16:1368–78. doi: 10.1110/ps.062738307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Myers MP, Pass I, Batty IH, Van der Kaay J, Stolarov JP, Hemmings BA, Wigler MH, Downes CP, Tonks NK. The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proc Natl Acad Sci U S A. 1998;95:13513–8. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slack DN, Seternes OM, Gabrielsen M, Keyse SM. Distinct binding determinants for ERK2/p38alpha and JNK map kinases mediate catalytic activation and substrate selectivity of map kinase phosphatase-1. J Biol Chem. 2001;276:16491–500. doi: 10.1074/jbc.M010966200. [DOI] [PubMed] [Google Scholar]
- 20.Xie L, Zhang YL, Zhang ZY. Design and characterization of an improved protein tyrosine phosphatase substrate-trapping mutant. Biochemistry. 2002;41:4032–9. doi: 10.1021/bi015904r. [DOI] [PubMed] [Google Scholar]
- 21.Flint AJ, Tiganis T, Barford D, Tonks NK. Development of “substrate-trapping” mutants to identify physiological substrates of protein tyrosine phosphatases. Proc Natl Acad Sci U S A. 1997;94:1680–5. doi: 10.1073/pnas.94.5.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stewart AE, Dowd S, Keyse SM, McDonald NQ. Crystal structure of the MAPK phosphatase Pyst1 catalytic domain and implications for regulated activation. Nat Struct Biol. 1999;6:174–81. doi: 10.1038/5861. [DOI] [PubMed] [Google Scholar]
- 23.Kozlov G, Cheng J, Ziomek E, Banville D, Gehring K, Ekiel I. Structural insights into molecular function of the metastasis-associated phosphatase PRL-3. J Biol Chem. 2004;279:11882–9. doi: 10.1074/jbc.M312905200. [DOI] [PubMed] [Google Scholar]
- 24.Roma-Mateo C, Rios P, Tabernero L, Attwood TK, Pulido R. A novel phosphatase family, structurally related to dual-specificity phosphatases, that displays unique amino acid sequence and substrate specificity. J Mol Biol. 2007;374:899–909. doi: 10.1016/j.jmb.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 25.Agarwal R, Burley SK, Swaminathan S. Structure of human dual specificity protein phosphatase 23, VHZ, enzyme-substrate/product complex. J Biol Chem. 2008;283:8946–53. doi: 10.1074/jbc.M708945200. [DOI] [PubMed] [Google Scholar]
- 26.Gray CH, Good VM, Tonks NK, Barford D. The structure of the cell cycle protein Cdc14 reveals a proline-directed protein phosphatase. EMBO J. 2003;22:3524–35. doi: 10.1093/emboj/cdg348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vander Kooi CW, Taylor AO, Pace RM, Meekins DA, Guo HF, Kim Y, Gentry MS. Structural basis for the glucan phosphatase activity of Starch Excess4. Proc Natl Acad Sci U S A. 2010;107:15379–84. doi: 10.1073/pnas.1009386107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alonso A, Rojas A, Godzik A, Mustelin T. Topics in Current Genetics. Springer; Berlin: 2003. The dual-specific protein tyrosine phosphatase family; p. 5. [Google Scholar]
- 29.Kim KA, Song JS, Jee J, Sheen MR, Lee C, Lee TG, Ro S, Cho JM, Lee W, Yamazaki T, Jeon YH, Cheong C. Structure of human PRL-3, the phosphatase associated with cancer metastasis. FEBS Lett. 2004;565:181–7. doi: 10.1016/j.febslet.2004.03.062. [DOI] [PubMed] [Google Scholar]
- 30.Hu J, Li D, Su XD, Jin C, Xia B. Solution structure and conformational heterogeneity of acylphosphatase from Bacillus subtilis. FEBS Lett. 2010;584:2852–6. doi: 10.1016/j.febslet.2010.04.069. [DOI] [PubMed] [Google Scholar]
- 31.Farooq A, Plotnikova O, Chaturvedi G, Yan S, Zeng L, Zhang Q, Zhou MM. Solution structure of the MAPK phosphatase PAC-1 catalytic domain. Insights into substrate-induced enzymatic activation of MKP. Structure. 2003;11:155–64. doi: 10.1016/s0969-2126(02)00943-7. [DOI] [PubMed] [Google Scholar]
- 32.Stehle T, Sreeramulu S, Lohr F, Richter C, Saxena K, Jonker HR, Schwalbe H. The apo-structure of the low molecular weight protein-tyrosine phosphatase A (MptpA) from Mycobacterium tuberculosis allows for better target-specific drug development. J Biol Chem. 2012;287:34569–82. doi: 10.1074/jbc.M112.399261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim SJ, Jeong DG, Yoon TS, Son JH, Cho SK, Ryu SE, Kim JH. Crystal structure of human TMDP, a testis-specific dual specificity protein phosphatase: implications for substrate specificity. Proteins. 2007;66:239–45. doi: 10.1002/prot.21197. [DOI] [PubMed] [Google Scholar]
- 34.Jeong DG, Yoon TS, Kim JH, Shim MY, Jung SK, Son JH, Ryu SE, Kim SJ. Crystal structure of the catalytic domain of human MAP kinase phosphatase 5: structural insight into constitutively active phosphatase. J Mol Biol. 2006;360:946–55. doi: 10.1016/j.jmb.2006.05.059. [DOI] [PubMed] [Google Scholar]
- 35.Yuvaniyama J, Denu JM, Dixon JE, Saper MA. Crystal structure of the dual specificity protein phosphatase VHR. Science. 1996;272:1328–31. doi: 10.1126/science.272.5266.1328. [DOI] [PubMed] [Google Scholar]
- 36.Kneller JM, Lu M, Bracken C. An effective method for the discrimination of motional anisotropy and chemical exchange. J Am Chem Soc. 2002;124:1852–3. doi: 10.1021/ja017461k. [DOI] [PubMed] [Google Scholar]
- 37.Gustafson CL, Stauffacher CV, Hallenga K, Van Etten RL. Solution structure of the low-molecular-weight protein tyrosine phosphatase from Tritrichomonas foetus reveals a flexible phosphate binding loop. Protein Sci. 2005;14:2515–25. doi: 10.1110/ps.051618805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Logan TM, Zhou MM, Nettesheim DG, Meadows RP, Van Etten RL, Fesik SW. Solution structure of a low molecular weight protein tyrosine phosphatase. Biochemistry. 1994;33:11087–96. doi: 10.1021/bi00203a005. [DOI] [PubMed] [Google Scholar]
- 39.Alonso A, Burkhalter S, Sasin J, Tautz L, Bogetz J, Huynh H, Bremer MC, Holsinger LJ, Godzik A, Mustelin T. The minimal essential core of a cysteine-based protein-tyrosine phosphatase revealed by a novel 16-kDa VH1-like phosphatase, VHZ. J Biol Chem. 2004;279:35768–74. doi: 10.1074/jbc.M403412200. [DOI] [PubMed] [Google Scholar]
- 40.Kuznetsov VI, Hengge AC, Johnson SJ. New Aspects of the Phosphatase VHZ Revealed by a High-Resolution Structure with Vanadate and Substrate Screening. Biochemistry. 2012;51:9869–79. doi: 10.1021/bi300908y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–78. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 42.Herrmann T, Guntert P, Wuthrich K. Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J Biomol NMR. 2002;24:171–89. doi: 10.1023/a:1021614115432. [DOI] [PubMed] [Google Scholar]
- 43.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–23. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hooft RW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 45.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–86. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 46.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–5. 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.