Abstract
Variation in voluntary exercise behavior is an important determinant of long-term human health. Increased physical activity is used as a preventative measure or therapeutic intervention for disease, and a sedentary lifestyle has generally been viewed as unhealthy. Predisposition to engage in voluntary activity is heritable and induces protective metabolic changes, but its complex genetic/genomic architecture has only recently begun to emerge. We first present a brief historical perspective and summary of the known benefits of voluntary exercise. Second, we describe human and mouse model studies using genomic and transcriptomic approaches to reveal the genetic architecture of exercise. Third, we discuss the merging of genomic information and physiological observations, revealing systems and networks that lead to a more complete mechanistic understanding of how exercise protects against disease pathogenesis. Finally, we explore potential regulation of physical activity through epigenetic mechanisms, including those that persist across multiple generations.
Keywords: wheel running, physical activity, quantitative trait loci (QTL), expression quantitative trait loci (eQTL), genomics, collaborative cross
Brief history of exercise in human society
Movement or locomotion is perhaps the most meaningful behavior animals engage in. Humans are no exception, and approximately 2 million years ago evolved an ability to run sustained long distances [1,2]. Independent of the necessity to procure food or find reproductive partners, it has been well documented that humans and mammals frequently engage in play behavior [3], or specifically within human populations - leisure time activity or “sport.” The need for or social implications of play behavior are beyond the scope of this review [but see 4]. A more relevant aspect is the importance of play behavior in facilitating exercise training, which may allow individuals to acclimatize to physical activity [5]. Regardless of the specific benefits of physical activity or exercise, organized sport has perhaps always had a place in human society [6].
Although the relative importance of sport or leisure physical activity and its perceived impact on the body has fluctuated over time, exercise, and specifically jogging, became a cultural phenomenon in the late 1960s and 1970s [7]. This sensation was initiated and perpetuated by promotion of the health benefits of physical activity, programs accessible to all ages, and marketing and profits related to promotion of sport [e.g., 8,9]. In addition to running, over the past four decades enormous volumes of heath-centered programs (both psychological and physiological) based on a variety of activities have seen gains in popularity. Remarkably, as insights into the benefits of regular physical activity have increased dramatically since inception of the jogging revolution, total physical activity has been on a global downward trend since 1965 [10,11]. Regardless of this global trend, many people remain largely inactive, some are only irregularly active, whereas others do engage in enough activity to realize the health benefits [12,13]. Here we review current literature aimed at dissecting genetic mechanisms underlying variation in the predisposition to voluntary exercise behavior, as depicted in Figure 1.
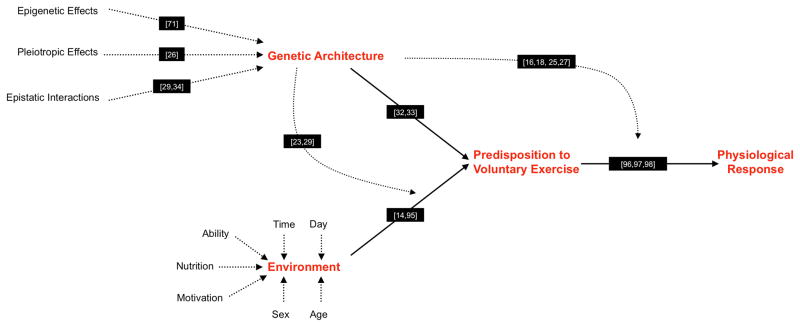
Figure 1.
The predisposition to engage in voluntary exercise is complex and simultaneously effected by genetic architecture, the environment, and gene-environment and gene-gene interactions. Both genetic architecture and the environment are comprised of multiple components with the relative influence of each varying. Here, we have attempted to depict only a fraction of the environmental components, with the primary focus intended to be the genetic regulation of physical activity as described in the text. We have provided example references (black boxes) of each of the effects shown.
We do not attempt to cover all “types” of physical activity, but emphasize that there is a continuum between obligatory and voluntary exercise. The parsing of physical activity, based on necessity and behavior type, is reviewed at length elsewhere [14], and some operational definitions and accompanying references are provided in Table 1. Below, we reference studies examining a broad range of human physical activities, but with regard to mouse models we focus almost exclusively on studies utilizing voluntary wheel running as primary means to uncover the genetic determinants of voluntary exercise. Reviewing the use of a running wheel to measure activity in rodents, others have hypothesized “that wheel-running behavior is not solely reflective of the tendency to be physically active, but is a complex and dynamic behavior that interacts with genetics and the environment” and contend that “wheel-running behavior represents factors in addition to rodents’ tendency to be physically active, engaging additional neural and physiological mechanisms which can then independently alter energy balance and behavior” [15]. For these reasons we believe voluntary wheel running to appropriately model voluntary exercise in human populations, a complex behavior simultaneously affected by central and peripheral mechanisms that interact to regulate duration, intensity, and adherence (e.g., [16]).
Table 1.
Defining voluntary exercise and its components
| Traita | Operational Definitionb | Subject | Reference |
|---|---|---|---|
| Home Cage Activity | “The activity of mice in their home cage …such as horizontal locomotion, climbing the cage lid, hanging on the lid, and jumping…” | Rodent | [77], Pp. 477, 478 |
| Voluntary Wheel Running | “…mice were exposed to running wheels (model 80850, Lafayette Instruments, Lafayette, IN; circumference = 1.1 m) for 6 days. Voluntary wheel running was recorded electronically in 1-min intervals for 23–24 h of each of the 6 days of wheel access.” | Rodent | [26], p. 113 |
| Open Field Activity | “We used total distance traveled during 5 min in a darkened and sound-attenuated chamber as our measure of activity.” | Rodent | [78], p. 112 |
| Treadmill Running | “…a 10 m/min starting speed that increased 1 m/min every 2 min at a constant 15° grade. Rats were removed from the treadmill at the point of exhaustion, operationally defined as the third time a rat would sustain 3 s on a shock grid (1.2 mAmp at 3 Hz)…” | Rodent | [79], p. 92 |
| Spontaneous Physical Activity (SPA) | “…activity that is non-volitional, or subconscious, such as fidgeting and shifting in one’s seat, and time spent moving (standing and ambulating).” | Human or Rodent | [80], p. 294 |
| Voluntary Exercise | “…locomotor activity that is not directly required for survival or homeostasis and not directly motivated by any external factor.” | Human or Rodent | [14], p. 207 |
| Nonexercise Activity Thermogenesis (NEAT) | “…EE (energy expenditure) of performing all of our daily tasks such as walking, talking, yard work, and fidgeting.” | Human | [81], p. 169 |
| “For a laboratory rat, NEAT principally includes grooming, walking around the cage, rearing, the locomotion of feeding, and other less de- finable movements such as head shaking.” | Rodent | [81], p. 169 |
Activity traits are not necessarily mutually exclusive. And, in the case of NEAT, traits are defined by the result of the activity as opposed to the activity itself.
In some cases text was added (in italics) for clarification and is not attributable to the authors cited.
Genetics of voluntary exercise
It has been well established that genetics plays an important role in regulation of voluntary activity levels and ability to respond to exercise training [17,18]. Empirical support is rooted in the demonstration of heritability in humans, mice, and rats utilizing several different approaches. A study of 1022 pairs of twins estimated broad-sense heritability of total physical activity to be near 50%, and heritabilities of individual components of total activity (occupational, leisure time excluding sport, sport during leisure time) ranged from 40% to 65% [19]. Additional human studies have estimated the heritability of physical activity to be anywhere between 18% and 92%, with variable environmental contributions [20,21]. Perhaps the most convincing test of whether or not a trait, such as voluntary exercise behavior, exhibits significant narrow sense heritability is its response to direct selection. An ongoing artificial selection experiment for voluntary wheel running behavior in 4 replicate lines of mice resulted in a dramatic increases in activity [22]. By generation 16, and continuing through generation 50 and beyond, the high-running (HR) lines had diverged significantly from the control lines with an approximate 3-fold increase in running on days 5 and 6 of a 6 day test [22]. Narrow-sense heritability, representing the additive genetic component of phenotypic variation, was calculated to be 0.28 [22].
Given that exercise propensity has a heritable component and is an important component of positive health-related outcomes, investigations into the location or identity of specific genetic markers linked to exercise are becoming increasingly common. Below, we discuss several approaches utilizing human and mouse populations leading to the identification of chromosomal regions and individual genes underlying variation in exercise propensity.
Linkage and GWAS studies
Quantitative trait loci (QTL), chromosomal regions harboring genetic variation contributing to phenotypic variation, have been mapped for a variety of exercise phenotypes (e.g., distance, speed, and duration as well as home-cage activity) in a number of different mouse [23] and human [24] populations. Moreover, a subset of these studies has identified additional QTL responsible for body composition changes as a result of exercise that did not co-localize with QTL for running traits [16,25–27].
Compared to the traditional F2 or backcross mapping populations, an advanced intercross line (AIL) of mice (G4), generated from a line selectively bred for high voluntary wheel running and the widely used inbred line C57BL6/J, yielded an approximate 3-fold genetic map expansion and provided increased mapping resolution for exercise QTL [23]. Utilizing this AIL, 32 significant and 13 suggestive QTL were identified underlying running distance, duration, average speed, and maximum speed [23]; however, the largest QTL effect accounted for only 6.6% of the total phenotypic variation, indicating the presence of a complex genetic architecture for voluntary activity likely involving many genes with relatively small effects. Further illustrating this complexity was the identification of QTL underlying the slope (i.e., trajectory of running on a temporal scale) and intercept (i.e., starting point of temporal trajectory) of a linear regression across all 6 days of running. A portion of these loci represented a combination of daily exercise traits, but often they did not colocalize with regions controlling the behavior on individual days (Figure 2). These results indicated that the underlying genetic architecture of running on any single day was quite different than that underlying running patterns on a larger temporal scale. A possible explanation may be the interplay between two components that almost certainly comprise voluntary exercise behavior in mice and humans – ability and motivation. That is, ability may influence motivation (or “liking”) and if the former changes with training, so may the latter. This phenomenon has previously been termed self-induced adaptive plasticity [28]. A group [29] utilizing an F2 population (C57L/J X C3H/HeJ) also reported temporally fluctuating QTL. These examples may be useful genetic targets when attempting to understand the variation underlying willingness to initiate, adhere to, and complete an exercise regime among human populations.
Figure 2.
Data from [23] depicting QTL underlying the temporal pattern of voluntary activity as well as the trajectory of activity across the entire 6 day wheel access period. Suggestive (P ≤ 0.1) or significant (P ≤ 0.05) peaks imply unique genomic regions are least partially responsible for the initiation, continuation, and temporal pattern of exercise. For example, regions on chromosome 1 have been shown to harbor genes involved in anxiety-like behavior in rodents. Therefore, fear, or general anxiety may contribute to wheel running during initial exposure to wheels (days 1–2), with the daily routine of exercise governed by distinct loci (days 4–6). And, finally, the exercise trajectory over time (slope across days 1–6) is regulated by still another unique genomic region. Pictorially, the background photographs represent each of these phenomena. Photographs provided by Jason Smith, University of North Carolina – Chapel Hill.
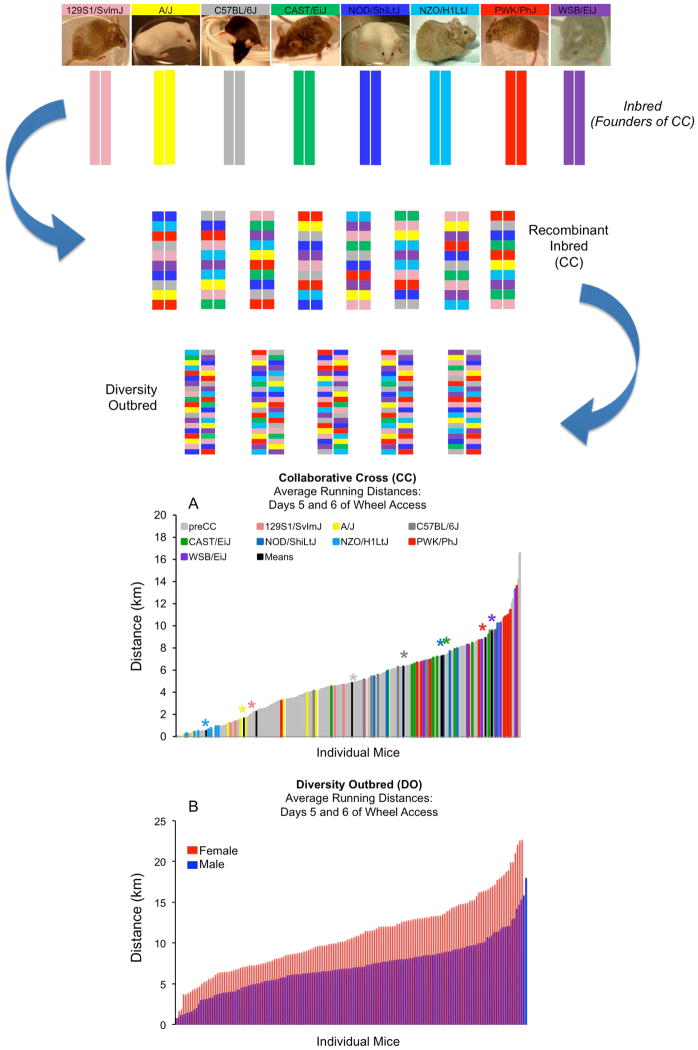
Using mice from emerging lines of the Collaborative Cross (CC) (Figure 3), a large panel of recombinant inbred mouse lines derived from a genetically diverse set of eight founder strains, it was observed that wheel running was highly variable with individuals running, on average, from just a few km per night up to nearly 20 km daily [16]. Similar findings were observed in the Mouse Diversity (DO) outbred population [30] (Figure 3). Using the CC, researchers identified QTL for wheel-running distance on days 5 and 6 and days 11 and 12 (of a 12-day test), with the latter accounting for 17% of the total phenotypic variation [16]. Separately, haplotype association mapping across 38 inbred strains identified a total of 12 significant QTL for wheel-running traits (distance, duration, and speed) on Chromosomes 5, 6, 8, 11, 12, 13, and 18 [31]. Of these 12, only 3 were common to both males and females, and despite the presence of six of the eight founder CC strains in this study, there was no overlap with the other findings [16]. Additional studies utilizing F2 and backcross populations have revealed multiple additional QTL, with little colocalization across populations [29,32,33]. Although there is little apparent overlap in QTL across mouse studies, direct comparisons are difficult to make due to methodological differences (running wheel utilized, running traits examined, location of study, etc.) and the apparently significant role of gene-gene (epistatic) interactions in the regulation of physical activity ([34] and Figure 4). The evidence presented above re-emphasizes the infinitesimal model (control by a very large number of loci each having a relatively small effect) [35], with many epistatic interactions, as the likely genetic architecture for exercise in mice.
Figure 3.
Schematic illustration of creation of the Collaborative Cross (CC) and Diversity Outbred (DO) mouse populations, two next-generation mouse models developed through balanced crossing of 8 progenitor inbred strains, including several of wild origin. Whereas CC lines represent new recombinant inbred strains, the DO [39] is derived from progenitor lines of the Collaborative Cross [93] and is maintained by a randomized outbreeding strategy. Thus the DO and CC populations capture the same set of natural allelic variants derived from a common set of eight founder strains, but the DO is maintained as an outbred population (with heterozygosity) while the CC is maintained as an inbred population. Although the DO is an ideal resource for high-resolution genetic mapping, the CC can provide predictive validation of mapping results obtained with the DO, as well as a source of reproducible genotypes for mechanistic studies [30]. Both of these populations exhibit dramatic phenotypic variability for voluntary exercise with extensive transgressive variation. Average wheel-running distances for A) male pre-CC and founder line males on days 5 and 6. Group means for each founder line are indicated in black with appropriate color-coded asterisk above to indicate strain being depicted. B) Average wheel-running distances for DO male and female mice on days 5 and 6. The original figure showing the color coded mosaic structure of the DO and CC is from Gary Churchill and Karen Svenson (The Jackson Laboratory). The original figure (A) is from [16]. Data used to generate the figure (B) is unpublished (provided by Daniel Pomp).
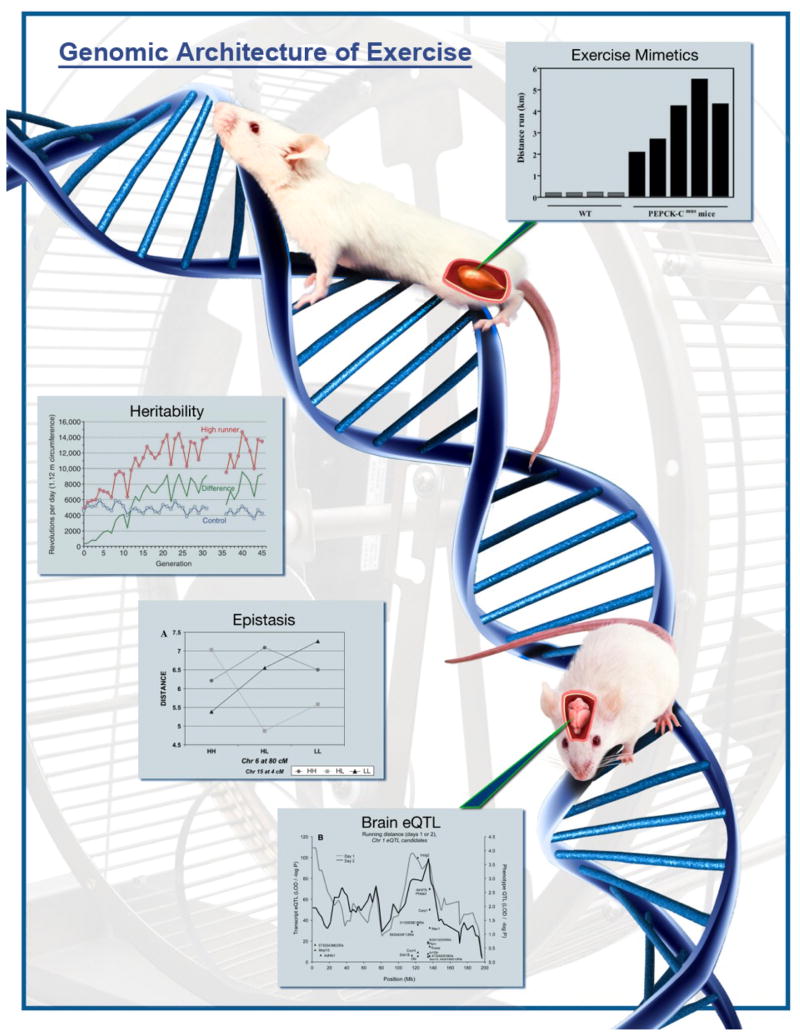
Figure 4.
Murine models have been utilized to show heritability of the predisposition to engage in voluntary exercise (Fig. 1 in [94]; see also [22]) with epistatic interactions accounting for a considerable amount of genetic variation within a population (Fig. 1A in [34]). Furthermore, it has been demonstrated that the biological basis of voluntary exercise is composed of both ability, as illustrated through exercise mimetics (Fig. 3 in [75]), and motivation, supported via the identification of brain expression quantitative trait loci, or brain eQTL) (Fig. 3B in [45]). The original individual figures are from [34,45,75,94]. Composite image was designed and created by Mark A. Schmitter, Ohio Wesleyan University.
In human populations, only four linkage studies have been performed to date (see Table 3 in [36]). Suggestive or significant loci were identified for the following phenotypic measures: total physical activity, moderate-to-vigorous physical activity, time spent in physical activity, low physical activity, moderate physical activity, vigorous physical activity, exercise participation, and athlete status. In all of the studies (with the exception of [24]) activity parameters were measured using self-assessment or recall surveys (often the only feasible method), an approach where considerable measurement error may be introduced especially for moderate to low levels of activity [37]. Regardless of the lack of phenotypic measurement precision, these four studies collectively mapped regions on chromosomes 3, 4, 7, 9, 11, 13, 18, and 19 as harboring QTL for physical activity in humans. Interestingly, one group [24] mapped a locus on chromosome 18 associated with variation in physical activity, a region previously shown to harbor the melanocortin 4 receptor gene (MC4R), a gene associated with obesity. Furthermore, another study [38] revealed suggestive evidence of sex-specific loci underlying exercise participation. Overall, no loci in the four studies colocalized with one another. This lack of colocalization across studies is similar to what has collectively been revealed using mouse models, and there also appears to be little colocalization between mouse and human studies. However, interspecies comparisons are difficult for reasons already mentioned: methodological differences (running wheel utilized, running traits examined, location of study, etc.) and epistatic interactions, among many others. Comparisons will likely become more feasible once specific candidate genes begin to emerge in both human and mouse populations.
In comparison with the volume of human and rodent linkage studies, there is only one published genome-wide association study examining the genetic architecture of voluntary activity. This study [39] found SNPs significantly associated with exercise participation in six candidate genes, highlighting two genes (SH3-domain GRB2-like (endophilin) interacting protein 1, SGIP1; leptin receptor, LEPR) whose effects were independent of individuals’ BMI. SGIP1 and LEPR, which are expressed in the hypothalamus, have previously been reported to be involved in the regulation of energy homeostasis [41,41], leading the authors [39] to hypothesize that there is a “direct role of hypothalamic factors in the drive to exercise.”
Expression approaches
Linkage studies have been useful in mapping chromosomal regions containing QTL, but identification of the underlying individual genes and specific responsible genetic variation contributing to predisposition for voluntary exercise has been elusive. Although there are some limitations [42], combining QTL mapping with large-scale gene expression analysis [43], or expression quantitative trait loci (eQTL) mapping, is a strategy used to dissect complex traits and aid in the selection and prioritization of candidate genes underlying predisposition loci [44].
The same G4 population as described above was used to examine the transcriptional landscape relevant to neurobiological aspects of voluntary exercise by means of global mRNA expression profiles from brain tissue [45]. Additionally, subsets of cis-acting eQTL that mapped to previously identified loci for exercise traits were used to produce a list of potential candidate genes. It should be noted, however, that the number of cis-acting eQTL colocalizing with previously identified QTL was still quite high. For example, 30 positional candidate genes colocalized with a single QTL (Chr. 7) for running distance. Although filtering the list of 30 candidate genes in this example may be feasible, narrowing the list colocalizing with all 32 significant and 13 suggestive previously identified exercise-related QTL [23] will be much more daunting. Aside from individual candidate genes, a potential master regulatory region (i.e., an eQTL trans-regulating hundreds of transcripts) was observed on the distal end of Chr. 1 (~170–180 Mb), a region that has been previously implicated in anxiety-like behavior in rodents [46], and the corresponding region in humans has been associated with panic disorder [47]. Previously, exercise-related QTL mapped on Chr. 1 were only implicated during the initial exposure to running wheels [23]. These QTL and the potential master regulatory eQTL trans-band region on Chr. 1 may underlie behaviors related to fear, or lack thereof, of the running wheel, general anxiety resulting from solitary housing, or the “learning” process involved with wheel running.
In comparison with eQTL investigations, using global transcriptome profiling to examine differential gene expression in animal models of exercise has been more commonplace. This approach has proven especially powerful in the context of selective breeding experiments. For example, significant differential gene expression was observed that was associated with changes in the dopamine pathway (D1 and D2 receptors), G-proteins, and adenylate cyclase in mice selectively bred for either high-running or obesity relative to a non-selected outbred strain of mice [58]. Rats selectively bred for high- and low-aerobic capacity (treadmill endurance capacity), were used to [49] generate a transcriptional map of the impact of endurance exercise training on skeletal muscle. Similar approaches have been used in human cohorts (e.g., studies reviewed in [50,51]). Additionally, combining results from an F2 intercross population and interval-specific congenic strains, as well as brain expression profiles, identified Tcfap2a as a regulator of physical activity via the dopaminergic pathway [52]. Gene expression approaches to study exercise have been successful and are increasingly becoming commonplace, however the ability to simultaneously examine the transcriptional landscape across large groups of genes and connect their integration to higher-level mechanistic function is critical.
Genetic networks – implications for human health
Recent investigations have highlighted the complexity of the molecular effects of exercise. For example, it has been demonstrated that muscle tissue has the potential to act as an endocrine organ, communicating with other tissues via myokines [53–55]. Specifically, a myokine called irisin has been shown to be stimulated during exercise and initiates the transformation of white adipose cells into cells that more closely resemble brown adipose cells (“brite” cells) [56]. Following injections of either peroxisome proliferator-activated receptor δ agonist GW501516 or AICAR (an AMPK agonist), mouse spatial memory was shown to be significantly improved in a Morris water maze [57]. Additionally, injections resulted in elevated dentate gyrus neurogenesis.
Given the complexity of the underlying genomic architecture of voluntary exercise, we believe systems approaches (e.g., see Figure 4 in [58]) are crucial to the continued progression of understanding the biological basis of physical activity. Because exercise has such broad physiological implications, these systems approaches would be most beneficial when simultaneously coupled with investigations into physiological systems that also directly underlie disease risk. For instance, examination of the linkage between aerobic exercise capacity and the risk for metabolic syndrome utilizing rats selectively bred for high or low treadmill endurance revealed that the two lines of rats also differed in their risk for metabolic syndrome as indicated by levels of blood glucose, insulin, triglycerides, free fatty acids, and visceral adiposity [59, 60]. Similar correlated responses to selection for high voluntary wheel running in mice have also been observed [61,62]. By examining skeletal muscle gene expression in the same rats, it was observed that unregulated genes in the high endurance lines were also associated with oxidative energy metabolism and correlated significantly with exercise and disease risk phenotypes [59, 60].
Finding and subsequently untangling networks of genes that affect not only exercise propensity and capacity but also disease risk phenotypes will ultimately provide a more precise target for exercise prescriptions among human populations [63]. For example, among twin pairs that differed in leisure time physical activity for 30 years, muscle and adipose gene expression revealed that energy metabolism pathways (e.g., oxidative phosphorylation and lipid metabolism) were chronically up regulated [64]. Moreover, up regulation of these particular pathways was associated with aerobic fitness and HDL cholesterol levels. These are but a few examples of the synergistic power of approaches that investigate not only the genetic architecture of physical activity (directly or indirectly) but concurrently examine the underlying physiology of the disease states that it affects.
Epigenetic regulation of physical activity
The idea that an individual’s exercise habits may be regulated, at least partially, by early life environmental exposures resulting in epigenetic modifications, and that these modifications can be inherited, is tantalizing. This is especially true given the implications of lifestyle modification on the prevention of chronic disease [65,66]. Consequently, the epigenetic regulation of behavioral and physiological traits has recently become of increasing interest, especially with regard to obesity [67 and references therein]. The idea that an individual’s nutrition and the resulting physiological modifications not only affect their own phenotype but may also impact that of their offspring, has added complexity to the search for the underlying genetic architecture of complex traits. Furthermore, it has been demonstrated that maternal nutritional choices may also yield transgenerational effects [68].
Few investigations have attempted to understand epigenetic regulation of predisposition to engage in voluntary exercise. One group [69] measured global DNA methylation in peripheral blood in response to different levels of physical activity in 45–75 year old individuals. They did not find a significant effect of physical activity on methylation but did discover weak evidence of an elevated risk of global hypothmethylation associated with low levels of physical activity [69]. Another group [70] found dose-dependent expression and promoter hypomethylation of peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α), pyruvate dehydrogenase kinase, isoenzyme 4 (PDK4), and peroxisome proliferator-activated receptor δ (PPAR-δ) in human skeletal muscle in response to acute exercise. This finding that acute exercise leads to transient changes in DNA methylation in adult skeletal muscle was interesting not only because it potentially links exercise to epigenetic modifications, but also because of the gene-specificity identified in such exercise-induced hypomethylation. This suggests that methylation may serve as a selective mechanism to orchestrate the activation of specific genes in response to exercise [70].
A study in mice using chromosome substitution strains derived from lines displaying high (C57BL/6J) and low (A/J) movement distances in an open-field activity identified a QTL on chromosome 1 underlying horizontal distance that was dependent on grandparent genetic origin [71]. Additionally, using the G4 described above, parent-of-origin effects (in some cases interacting with sex) were observed in the regulation of voluntary activity levels and body composition traits that had persisted across several generations [72]. This study also discussed a variety of phenomena potentially facilitating these findings including genetic (i.e., X-linked or mtDNA variations), epigenetic (i.e., genomic imprinting), and environmental (i.e., in utero environment or maternal care) [72].
Exercise mimetics
Knockout, knockdown, and overexpression studies have also been successfully used to investigate the genetics of voluntary exercise in rodent models. A particularly revealing series of studies [73] implicated the NHLH2 (nescient helix loop helix 2) gene in the regulation of physical activity and adult-onset obesity in rodents. Furthermore, the human homologue has been implicated in the transcriptional control of MC4R (melanocortin receptor 4), a gene associated with a lower exercise level in humans [24,74]. Another group [75] overexpressed the cytosolic form of phosphoenolpyruvate carboxykinase (GTP) (PEPCK-C) in skeletal muscle in mice. This overexpression led to increased home-cage activity, improved endurance capacity, and extended life span relative to control mice. Additionally, the PEPCK-Cmus exhibited decreased body mass relative to control mice despite consuming 60% more food.
Although understanding the genetic architecture underlying voluntary exercise is critical to gaining a comprehensive view of these complex behaviors, the location of QTL and the knowledge of candidate genes are not necessary to artificially simulate exercise. Consequently, pathway-specific drugs are being increasingly employed in the search for exercise mimetics that can replicate some fraction of the beneficial effects of physical activity. For example, agonists for both PPARdelta and AMPK have previously been shown to increase mouse treadmill running endurance synergistically with and in the absence of exercise training, respectively [76]. These experiments demonstrated that exercise-induced muscle activation may underlie positive neurobiological effects of physical activity. It should be noted that while the examples described above present tantalizing therapeutic targets, no reported QTL to date have harbored these genes. This disparity only further highlights the complexity of the connection between the underlying genetic architecture and the manifestation of exercise phenotypes.
Concluding remarks
Clearly, the predisposition to engage in exercise is a highly complex trait. Although heritability has been well established, QTLs identified, and candidate genes proposed, individual effects of QTLs and genes are generally modest, and epistasis appears to be important. Furthermore, the genetic architecture underlying voluntary exercise behavior is often sex-, population-, and age-dependent, shifts or manifests itself differently over the course of an exercise regime, and is influenced by diet and body composition prior to the initiation of exercise. This overall complexity may in part explain why, despite its overwhelming documented benefits, many people do not engage in this beneficial behavior for extended periods of time. This may be because of a low propensity to engage in exercise, and/or to sustain an exercise program, and/or to obtain benefits in terms of weight loss, all of which could change depending on environmental context. Further gains in our understanding of the genetic underpinnings of exercise and its downstream molecular networks may potentially clarify the importance of these components, and how they interact with each other, in regulating the overall predisposition for voluntary exercise.
Such an understanding will require a better catalog of the relevant genes. Although many genomic regions have been implicated in regulating exercise (especially in rodent models), narrowing these regions to plausible candidate genes remains a major challenge. Furthermore, connecting genetic variants to specific physiological functions that regulate exercise behavior will be a major advancement. Box 2 discusses additional outstanding questions. In light of the recent surge in interest in the health ramifications of physical inactivity, we feel the most important future direction will be to determine if the underlying genetic architecture of physical activity is the same as the underlying genetic architecture of physical inactivity.
Box 2. Outstanding questions.
| What genetic factors contribute to the temporal stability of voluntary exercise levels? |
| Is there an imbalance in the number of genes that induce elevated vs. suppressed exercise levels? |
| How much variation in voluntary activity can be accounted for by epigenetic regulation? |
| What environmental conditions result in these epigenetic modifications (e.g., in utero exposure to exercise, exercise exposure early in development, reinforced exercise regimes throughout life, etc.)? Are these epigenetic mechanisms heritable/transgenerational? |
| What are the genes (and specific genetic variants) underlying QTL for exercise, and how do they function physiologically? |
| Do genes that control variation in voluntary exercise also directly impact disease through pleiotropy? |
Box 1. Health benefits of exercise.
The benefits of regular exercise include positive health outcomes associated with prevention of cardiovascular disease [82], stroke [83], cancer [84], diabetes [85], obesity [86], cognitive decline [87], and learning deficits [88]. Although this list is not exhaustive, prevention and treatment of these disease states in particular would help to alleviate a substantial healthcare and economic burden. Importantly, although disease prevention is consistent, changes resulting from exercise remain considerably variable within populations of both humans and rodents, especially with regard to weight and adiposity [16, 89]. The variability in response to exercise makes dosage recommendations [90] difficult to prescribe at the level of the individual and highlights the complexity of the mechanistic basis of exercise effects on physiological and psychological health. Given a recent example of the power of integrative personal “omics” profiling [91], we view the genomic nature of the predisposition to voluntarily exercise as a critical component in designing personalized activity based therapies for health, and in particular for diseases that exercise is known to effect. We want to emphasize that the health related effects of exercise can often be variable depending on sex, age, and other contextual variables, and in some cases exercise may even be detrimental (e.g., [92]). This is especially relevant in human cohorts where results are often correlative and depend on self-reporting, and thus much yet remains to be understood.
Acknowledgments
We thank Tim Lightfoot for useful comments on a previous version of this manuscript. We are appreciative of Ted Garland Jr., Larry Leamy, Tim Lightfoot, Derrick Nehrenberg and Kunjie Hua for their collaborative efforts on research described in this paper. Some research described in the present review was supported by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant DK076050. Some phenotypes were collected using the Animal Metabolism Phenotyping core facility within the University of North Carolina’s Clinical Nutrition Research Center (funded by NIDDK grant DK056350).
Footnotes
S.A Kelly and D. Pomp both contributed to the writing of the present review.
There are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bramble DM, Lieberman DE. Endurance running and the evolution of Homo. Nature. 2004;432:345–352. doi: 10.1038/nature03052. [DOI] [PubMed] [Google Scholar]
- 2.Lieberman DE, Bramble DM. The evolution of marathon running: capabilities in humans. Sports Med. 2007;37:288–290. doi: 10.2165/00007256-200737040-00004. [DOI] [PubMed] [Google Scholar]
- 3.Iwaniuk AN, et al. Do big-brained animals play more? Comparative analyses of play and relative brain size in mammals. J Comp Psychol. 2001;115:29–41. doi: 10.1037/0735-7036.115.1.29. [DOI] [PubMed] [Google Scholar]
- 4.Fox J. The ball. Harper Collins Publishers; 2012. [Google Scholar]
- 5.Kelly SA, Garland T., Jr Phenotypic plasticity and experimental evolution. J Exp Biol. 2006;209:2344–2361. doi: 10.1242/jeb.02244. [DOI] [PubMed] [Google Scholar]
- 6.Mechikoff R. A history and philosophy of sport and physical education: from ancient civilizations to the modern world. New York, New York: McGraw Hill; 2010. [Google Scholar]
- 7.Gotaas T. Running: A global history. Reaktion Books, Ltd; 2009. The jogging revolution; pp. 240–261. [Google Scholar]
- 8.Bowerman WJ. Jogging: a physical program for all ages. Ace Books; 1967. [Google Scholar]
- 9.Cooper KH. Aerobics. Bantam Books; 1977. [Google Scholar]
- 10.Ng SW, Popkin BM. Time use and physical activity: a shift away from movement across the globe. Obes Rev. 2012 Jun 14; doi: 10.1111/j.1467-789X.2011.00982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Westerterp KR, Speakman JR. Physical activity energy expenditure has not decline since the 1980s and matches energy expenditures wild mammals. Int J Obes. 2008;32:1256–1263. doi: 10.1038/ijo.2008.74. [DOI] [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention CDC. Physical activity levels among children aged 9–13 years – United States, 2002. MMWR Morb Mortal Wkly Rep. 2003;52:785–788. [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention CDC. Physical activity levels of high school students – United States, 2010. MMWR Morb Mortal Wkly Rep. 2011;60:773–777. [PubMed] [Google Scholar]
- 14.Garland T, Jr, et al. The biological control of voluntary exercise, spontaneous physical activity and daily energy expenditure in relation to obesity: human and rodent perspectives. J Exp Biol. 2011;214:206–229. doi: 10.1242/jeb.048397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Novak CM, et al. The use of a running wheel to measure activity in rodents: relationship to energy balance, general activity, and reward. Neurosci Biobehav Rev. 2012;36:1001–1014. doi: 10.1016/j.neubiorev.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathes WF, et al. Architecture of energy balance traits in emerging lines of the Collaborative Cross. Am J Physiol Endocrinol Metab. 2011;300:E1124–1134. doi: 10.1152/ajpendo.00707.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Moor MH, et al. Genome-wide association study of exercise behavior in Dutch and American adults. Med Sci Sports Exerc. 2009;41:1887–1895. doi: 10.1249/MSS.0b013e3181a2f646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bouchard C. Genomic predictors of trainability. Exp Physiol. 2012;97:347–352. doi: 10.1113/expphysiol.2011.058735. [DOI] [PubMed] [Google Scholar]
- 19.Eriksson M, et al. Genetic factors in physical activity and the equal environment assumption – the Swedish young male twins study. Behav Genet. 2006;36:238–247. doi: 10.1007/s10519-005-9018-7. [DOI] [PubMed] [Google Scholar]
- 20.Joosen AM, et al. Genetic analysis of physical activity in twins. Am J Clin Nutr. 2005;82:1253–1259. doi: 10.1093/ajcn/82.6.1253. [DOI] [PubMed] [Google Scholar]
- 21.Stubbe JH, de Geus EJC. Genetics of exercise behavior. In: Kim Y-K, editor. Handbook of Behavior Genetics. New York: Springer Science and Business Media; 2009. pp. 343–358. [Google Scholar]
- 22.Swallow JG, et al. Artificial selection for increased wheel-running behavior in house mice. Behav Genet. 1998;28:227–237. doi: 10.1023/a:1021479331779. [DOI] [PubMed] [Google Scholar]
- 23.Kelly SA, et al. Genetic architecture of voluntary exercise in an advanced intercross line of mice. Physiol Genomics. 2010;42:190–200. doi: 10.1152/physiolgenomics.00028.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai G, et al. A quantitative trait locus on chromosome 18q for physical activity and dietary intake in Hispanic children. Obesity. 2006;14:1596–1604. doi: 10.1038/oby.2006.184. [DOI] [PubMed] [Google Scholar]
- 25.Leamy LJ, et al. Genetic variation for body weight change in mice in response to physical exercise. BMC Genet. 2009;10:58. doi: 10.1186/1471-2156-10-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leamy LJ, et al. Genetic variation in the pleiotropic association between physical activity and body weight in mice. Genet Sel Evol. 2009;41:41. doi: 10.1186/1297-9686-41-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelly SA, et al. Exercise, weight loss, and changes in body composition in mice: phenotypic relationships and genetic architecture. Physiol Genomics. 2011;43:199–212. doi: 10.1152/physiolgenomics.00217.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swallow JG, et al. Phenotypic and evolutionary plasticity of organ masses in response to voluntary exercise in house mice. Integr Comp Biol. 2005;45:426–437. doi: 10.1093/icb/45.3.426. [DOI] [PubMed] [Google Scholar]
- 29.Leamy LJ, et al. A search for quantitative trait loci controlling within-individual variation of physical activity traits in mice. BMC Genet. 2010;11:83. doi: 10.1186/1471-2156-11-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Svenson KL. High-resolution genetic mapping using the Mouse Diversity outbred population. Genetics. 2012;190:437–447. doi: 10.1534/genetics.111.132597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lightfoot JT, et al. Strain screen and haplotype association mapping of wheel running in inbred mouse strains. J Appl Physiol. 2010;109:623–634. doi: 10.1152/japplphysiol.00525.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lightfoot JT, et al. Quantitative trait loci for physical activity traits in mice. Physiol Genomics. 2008;32:401–408. doi: 10.1152/physiolgenomics.00241.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nehrenberg DL, et al. QTL underlying voluntary exercise in mice: interactions with the ‘mini muscle’ locus and sex. J Hered. 2010;101:42–53. doi: 10.1093/jhered/esp066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leamy LJ, et al. An epistatic genetic basis for physical activity traits in mice. J Hered. 2008;99:639–646. doi: 10.1093/jhered/esn045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fisher RA. The Genetical Theory of Natural Selection. Oxford University Press; Oxford: 1930. [Google Scholar]
- 36.de Vilhena E, Santos DM, et al. Genetics of physical activity and physical inactivity in humans. Behav Genet. 2012 Mar 17; doi: 10.1007/s10519-012-9534-1. [DOI] [PubMed] [Google Scholar]
- 37.Shephard RJ, Vuillemin A. Limits to the measurement of habitual physical activity by questionnaires. Br J Sports Med. 2003;37:197–206. doi: 10.1136/bjsm.37.3.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Moor MH, et al. Genome-wide linkage scan for exercise participation in Dutch sibling pairs. Eur J Hum Genet. 2007;15:1252–1259. doi: 10.1038/sj.ejhg.5201907. [DOI] [PubMed] [Google Scholar]
- 39.De Moor MH, et al. Genome-wide association study of exercise behavior in Dutch and American adults. Med Sci Sports Exerc. 2009;41:1887–1895. doi: 10.1249/MSS.0b013e3181a2f646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trevaskis J, et al. Src homology 3-domain growth factor receptor-bound 2-like (endophilin) interacting protein 1, a novel neuronal protein that regulates energy balance. Endocrinology. 2005;146:3757–3764. doi: 10.1210/en.2005-0282. [DOI] [PubMed] [Google Scholar]
- 41.Park KS, et al. Polymorphisms in the leptin receptor (LEPR) - putative association with obesity and T2DM. J Hum Genet. 2006;51:85–91. doi: 10.1007/s10038-005-0327-8. [DOI] [PubMed] [Google Scholar]
- 42.Verdugo RA, et al. Serious limitations of the QTL/microarray approach for QTL gene discovery. BMC Biol. 2010;8:96. doi: 10.1186/1741-7007-8-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jansen RC, Nap JP. Genetical genomics: the added value from segregation. Trends Genet. 2001;17:388–391. doi: 10.1016/s0168-9525(01)02310-1. [DOI] [PubMed] [Google Scholar]
- 44.Davis RC, et al. Systems genetics of susceptibility to obesity-induced diabetes in mce. Physiol Genomics. 2012;44:1–13. doi: 10.1152/physiolgenomics.00003.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kelly SA, et al. Functional genomic architecture of predisposition to voluntary exercise in mice: expression QTL in the brain. Genetics. 2012;191:643–654. doi: 10.1534/genetics.112.140509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hitzemann R, et al. A strategy for the integration of QTL, gene expression, and sequence analysis. Mamm Genome. 2003;14:733–747. doi: 10.1007/s00335-003-2277-9. [DOI] [PubMed] [Google Scholar]
- 47.Leygraf A, et al. Rgs 2 gene polymorphisms as modulators of anxiety in humans? J Neural Transm. 2006;113:1921–1925. doi: 10.1007/s00702-006-0484-8. [DOI] [PubMed] [Google Scholar]
- 48.Mathes WF, et al. Dopaminergic dysregulation in mice selectively bred for excessive exercise or obesity. Behav Brain Res. 2010;210:155–163. doi: 10.1016/j.bbr.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keller P, et al. A transcriptional map of the impact of endurance exercise training on skeletal muscle phenotype. J Appl Physiol. 2011;110:46–59. doi: 10.1152/japplphysiol.00634.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bray MS, et al. The human gene map for performance and health related fitness phenotypes: the 2006–2007 update. Med Sci Sports Exerc. 2009;41:35–73. doi: 10.1249/mss.0b013e3181844179. [DOI] [PubMed] [Google Scholar]
- 51.Roth SM, et al. Advances in exercise, fitness, and performance genomics in 2011. Med Sci Sports Exerc. 2012;44:809–817. doi: 10.1249/MSS.0b013e31824f28b6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang HS, et al. High-resolution mapping of a novel genetic locus regulating voluntary physical activity in mice. Genes Brain Behav. 2012;11:113–124. doi: 10.1111/j.1601-183X.2011.00737.x. [DOI] [PubMed] [Google Scholar]
- 53.Pedersen BK. Muscles and their myokines. J Exp Biol. 2011;214(Pt 2):337–346. doi: 10.1242/jeb.048074. [DOI] [PubMed] [Google Scholar]
- 54.Pedersen BK, Febbraio MA. Muscles, exercise and obesity: skeletal muscle as a secretory organ. Nat Rev Endocrinol. 2012 Apr 3; doi: 10.1038/nrendo.2012.49.. [DOI] [PubMed] [Google Scholar]
- 55.Pedersen BK. A muscular twist on the fate of fat. N Engl J Med. 2012;366:1544–1545. doi: 10.1056/NEJMcibr1201024. [DOI] [PubMed] [Google Scholar]
- 56.Bostrom P, et al. A PGC1-α-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature. 2012;481:463–468. doi: 10.1038/nature10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kobilo T, et al. Endurance factors improve hippocampal neurogenesis and spatial memory in mice. Learn Mem. 2012;18:103–107. doi: 10.1101/lm.2001611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pomp D, et al. Complex genetics of obesity in mouse models. Ann Rev Nutr. 2008;28:331–45. doi: 10.1146/annurev.nutr.27.061406.093552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kivela R, et al. Gene expression centroids that link with low intrinsic aerobic exercise capacity and complex disease risk. FASEB J. 2010;24:4565–4574. doi: 10.1096/fj.10-157313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wisloff U, et al. Cardiovascular risk factors emerge after artificial selection for low aerobic capacity. Science. 2005;307:418–420. doi: 10.1126/science.1108177. [DOI] [PubMed] [Google Scholar]
- 61.Girard I, et al. Leptin levels and body composition of mice selectively bred for high voluntary activity. Physiol Biochem Zoo. 2007;80:568–579. doi: 10.1086/521086. [DOI] [PubMed] [Google Scholar]
- 62.Vaanholt LM, et al. Plasma adiponectin is increased in mice selectively bred for high wheel-running activity, but not by wheel running per sé. Horm Metab Res. 2007;39:377–383. doi: 10.1055/s-2007-976542. [DOI] [PubMed] [Google Scholar]
- 63.Pedersen BK, Saltin B. Evidence for prescribing exercise as therpy in chronic disease. Scand J Med Sports. 2006;16(Suppl 1):3–63. doi: 10.1111/j.1600-0838.2006.00520.x. [DOI] [PubMed] [Google Scholar]
- 64.Leskinen T, et al. Differences in muscle and adipose tissue gene expression and cardio-metabolic risk factors in the members of physical activity discordant twin pairs. PLoS One. 2010;5:e12609. doi: 10.1371/journal.pone.0012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alegria-Torres JA, et al. Epigenetics and lifestyle. Epigenomics. 2011;3:267–277. doi: 10.2217/epi.11.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Adamo KB, et al. Can we modify the intrauterine environment to halt the intergenerational cycle of obesity? Int J Environ Res Public Health. 2012;9:1263–1307. doi: 10.3390/ijerph9041263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Slomko H, et al. Minireview: Epigenetics of obesity and diabetes in humans. Endocrinology. 2012;153:1025–1030. doi: 10.1210/en.2011-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chong S, et al. Heritable germline epimutation is not the same as transgenerational epigenetic inheritance. Nat Genet. 2007;39:574–575. doi: 10.1038/ng0507-574. [DOI] [PubMed] [Google Scholar]
- 69.Zhang FF, et al. Physical activity and global genomic DNA methylation in a cancer free population. Epigenetics. 2011;6:293–299. doi: 10.4161/epi.6.3.14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barres R, et al. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metb. 2012;15:405–411. doi: 10.1016/j.cmet.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 71.de Mooji-van Malsen JG, et al. Evidence for epigenetic interactions for loci on mouse chromosome 1 regulating open field activity. Behav Genet. 2009;39:176–182. doi: 10.1007/s10519-008-9243-y. [DOI] [PubMed] [Google Scholar]
- 72.Kelly SA, et al. Parent-of-origin effects on voluntary exercise levels and body composition in mice. Physiol Genomics. 2010;40:111–120. doi: 10.1152/physiolgenomics.00139.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Good DJ, et al. Nhlh2: a basic helix-loop-helix transcription factor controlling physical activity. Exerc Sport Sci Rev. 2008;36:187–192. doi: 10.1097/JES.0b013e31818782dd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Loos RJ, et al. Melanocortin-4 receptor gene and physical activity in the Quebec Family Study. Int J Obes (Lond) 2005;29:420–428. doi: 10.1038/sj.ijo.0802869. [DOI] [PubMed] [Google Scholar]
- 75.Hakimi P, et al. Overexpression of the cytosolic form of phosphoenolpyruvate carboxykinase (GTP) in skeletal muscle repatterns energy metabolism in the mouse. J Biol Chem. 2007;282:32844–32855. doi: 10.1074/jbc.M706127200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Narkar VA, et al. AMPK and PPARdelta agonists are exercise mimetics. Cell. 2008;134:405–415. doi: 10.1016/j.cell.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nishi A, et al. QTL analysis of measures of mouse home-cage activity using B6/MSM consomic strains. Mamm Genome. 2010;21:477–485. doi: 10.1007/s00335-010-9289-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.de Ledesma AM, et al. Two new behavioral QTLs, Emo4 and Reb1, map to mouse Chromosome 1: Congenic strains and candidate gene identification studies. Mamm Genome. 2006;17:111–118. doi: 10.1007/s00335-005-0107-y. [DOI] [PubMed] [Google Scholar]
- 79.Sokoloff G, et al. Anxiety and fear in a cross of C57BL/6J and DBA/2J mice: mapping overlapping and independent QTL for related traits. Genes Brain Behav. 2011;10:604–614. doi: 10.1111/j.1601-183X.2011.00699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kotz CM. Integration of feeding and spontaneous physical activity: role for orexin. Physiol Behav. 2006;88:294–301. doi: 10.1016/j.physbeh.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 81.Levine JA, et al. Effect of hyperthyroidism on spontaneous physical activity and energy expenditure in rats. J Appl Physiol. 2003;94:165–170. doi: 10.1152/japplphysiol.00499.2002. [DOI] [PubMed] [Google Scholar]
- 82.Stensel D. Primary prevention of CVD: physical activity. Clin Evid (Online) 2009 pii, 0218. [PMC free article] [PubMed] [Google Scholar]
- 83.Gallanagh S, et al. Physical activity in the prevention and treatment of stroke. ISRN Neurol. 2011:953818. doi: 10.5402/2011/953818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Denlinger CS, Engstrom PF. Colorectal cancer survivorship: movement matters. Cancer Prev Res. 2011;4:502–511. doi: 10.1158/1940-6207.CAPR-11-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.The InterAct Consortium. Physical activity reduces the incidient type 2 diabetes in general and in abdominally lean and obese men and women: the EPIC-InterAct Study. Diabetologia. 2012;55:1944–1952. doi: 10.1007/s00125-012-2532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wadden TA, et al. Lifestyle modification for obesity: new developments in diet, physical activity, and behavior therapy. Circulation. 2012;125:1157–1170. doi: 10.1161/CIRCULATIONAHA.111.039453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Winchester J, et al. Walking stabilizes cognitive functioning in Alzheimer’s disease (AD) across one year. Arch Gerontol Geriatr. 2012 Sep 5; doi: 10.1016/j.archger.2012.06.016. http://dx.doi.org/10.1016/j.archger.2012.06.016. [DOI] [PMC free article] [PubMed]
- 88.Mustroph ML, et al. Aerobic exercise is the critical variable in an enriched environment that increases hippocampal neurogenesis and water maze learning in male C57BL/6J mice. Neuroscience. 2012;219:62–71. doi: 10.1016/j.neuroscience.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Church TS, et al. Changes in weight, waist circumference and compensatory responses with different doses of exercise among sedentary, overweight postmeno- pausal women. PLoS ONE. 2009;4:e4515. doi: 10.1371/journal.pone.0004515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tudor-Locke C, et al. How many steps/day are enough? For adults. Int J Behav Nutr Phys Act. 2011;8:79. [Google Scholar]
- 91.Chen R, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148:1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bouchard C, et al. Adverse metabolic response to regular exercise: is it a rare or common occurrence? PLoS One. 2012;7:e37887. doi: 10.1371/journal.pone.0037887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Collaborative Cross Consortium. The genome architecture of the Collaborative Cross mouse reference population. Genetics. 2012;190:389–401. doi: 10.1534/genetics.111.132639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kolb EK, et al. Erythropoietin elevates VO2,max but not voluntary wheel running in mice. J Exp Biol. 2010;213:510–519. doi: 10.1242/jeb.029074. [DOI] [PubMed] [Google Scholar]
- 95.Bowen RS, et al. Sex hormone effects on physical activity levels: why doesn’t Jane run as much as Dick? Sports Med. 2011;41:73–86. doi: 10.2165/11536860-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ricanati EHW, et al. Mitigating preventable chronic disease: progress report of the Cleveland Clinic’s Lifestyle 180 program. Nutr Metab (London) 2011;8:83. doi: 10.1186/1743-7075-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bravata DM, et al. Using pedometers to increase physical activity and improve health. JAMA. 2007;298:2296–2304. doi: 10.1001/jama.298.19.2296. [DOI] [PubMed] [Google Scholar]
- 98.Golbidi S, Laher I. Exercise and the cardiovascular system. Cardiol Res Pract. 2012:210852. doi: 10.1155/2012/210852. [DOI] [PMC free article] [PubMed] [Google Scholar]