Abstract
Phosphorylation offers a dynamic way to regulate protein activity, subcellular localization, and stability. The majority of signaling pathways involve an extensive set of protein-protein interactions, and phosphorylation is widely used to regulate protein-protein binding by affecting the stability, kinetics and specificity of interactions. Previously it was found that phosphorylation sites tend to be located on protein-protein binding interfaces and may orthosterically modulate the strength of interactions. Here we studied the effect of phosphorylation on protein binding in relation to intrinsic disorder for different types of human protein complexes with known structure of binding interface. Our results suggest that the processes of phosphorylation, binding and disorder-order transitions are coupled to each other, with about one quarter of all disordered interface Ser/Thr/Tyr sites being phosphorylated. Namely, residue site disorder and interfacial states significantly affect the phosphorylation of serine and to a lesser extent of threonine. Tyrosine phosphorylation might not be directly associated with binding through disorder, and is often observed in ordered interface regions which are not predicted to be disordered in the unbound state. We analyze possible mechanisms of how phosphorylation might regulate protein-protein binding via intrinsic disorder, and specifically focus on how phosphorylation could prevent disorder-order transitions upon binding.
Introduction
Intrinsically disordered proteins (IDPs) play many functions in a cell 1. They lack a single well-defined structure and are characterized by specific amino acid composition, a propensity for post-translational modifications, and the ability to bind to many different partners. The abundance of disordered proteins inside the cell is tightly controlled at the synthesis and degradation levels 2. Many studies have suggested that intrinsically disordered regions may undergo disorder-order transitions, i.e. folding upon binding 1, 3. At the same time, disorder may play an important functional role in protein complexes 4, 5, especially in homooligomers 6 without evident disorder-order transition. Nevertheless the relationship between disorder and binding is not fully understood.
The importance of disorder in protein–protein interactions is apparent from analysis of protein-protein interaction networks. Several studies have shown that hub proteins in interaction networks have more disordered residues than non-hubs 7–9 and that there may be a weak correlation between the disorder of a protein and the number of its partners 8, 10, 11. By investigating disorder directly at binding interfaces, other studies showed that hubs binding multiple partners using the same interface region might have a higher fraction of disordered residues than non-hub proteins 12. However, binding interfaces were found to be less disordered than non-interface positions within both full-chain proteins 13 and domain regions 11.
The functional diversity of disordered proteins, their multi-binding properties, and their propensity for posttranslational modifications allow them to play a unique role in signaling networks 9, 14. Indeed, signaling proteins were previously found to have significantly greater disorder than proteins with other functions 15, 16. One way to regulate protein activity, subcellular localization, and stability is through protein covalent modifications, and phosphorylation is one of the most abundant classes of protein covalent modifications 17. Indeed, recent phosphoproteomic analyses have revealed that the majority of proteins in a mammalian cell are phosphorylated 18so regulatory mechanisms involving phosphorylation are very widespread. The dynamic regulation of cellular processes can be achieved through reversibility and the fast kinetics of phosphorylation. Adding or removing a dianionic phosphate group on a protein changes its physico-chemical properties and can affect the stability, kinetics, and dynamics of the protein 19.
Recently, we studied the coupling between phosphorylation and protein-protein binding by examining binding interfaces and locations of phosphosites in protein structural complexes 20. We found an association between phosphorylation sites and binding interfaces. Since phosphorylation has been previously linked to intrinsic disorder, one possible mechanism of coupling between phosphorylation and protein binding might involve disorder-order or order-disorder transitions. Indeed several examples of disorder-order as well as order-disorder transitions upon phosphorylation were reported previously 2, 21, 22. Here we study the effect of phosphorylation on protein binding in relation to intrinsic disorder for all available protein structural complexes from the human proteome with known phosphorylation sites. Our results suggest that the processes of phosphorylation and binding via disordered regions might be coupled to each other, with one quarter of all disordered interface sites being phosphorylated. Residue disorder, interfacial states and the type of protein complex (homo or heterooligomer) are significantly correlated with the phosphorylation of threonine and, to an even greater extent, of serine. We suggest that serine is especially important for disorder-order or order-disorder transitions upon binding. However, tyrosine phosphorylation does not appear to be directly associated with binding through disorder-to-order transitions based on our dataset. We discuss possible functional roles of phosphorylation in regulating the coupling between disorder and binding.
Material and Methods
Identification of phosphorylation sites on homo- and heterooligomers
We compiled a data set consisting of all human protein complexes of known three-dimensional structure from the Protein Data Bank (PDB) as described previously 20. We then removed redundant proteins using BLAST23, using a p-value threshold of 10e-07. A complex was considered homooligomeric if pairwise sequence identity was higher than 90% for all pairs of chains in the complex , otherwise it was defined as heterooligomeric. Phosphorylation sites were derived from PhosphoSitePlus 24, Phospho.ELM 25, and PHOSIDA 26 databases. Then, phosphorylation sites identified by high-throughput methods were additionally verified by the GPS 2.1 program 27. All phosphorylation sites were mapped onto protein structures in the PDB using alignments calculated by the MUSCLE program 28. The oligomeric states and binding interfaces were defined by PISA 29, the solvent accessible surface area (ASA) of each residue was provided by PISA, and converted into relative ASA based on accessible surface area on Gly-X-Gly tripeptides. As a result we obtained 1983 phosphorylation sites on 382 homooligomeric and 551 heterooligomeric human complexes, only two of which were kinase-substrate complexes. Some of the complexes contained largely disordered proteins with a relatively short structured region bound to a structured partner. A larger group consisted of cases where both partners had well-defined structures. The majority of protein complexes in our study (and in PDB in general) did not have the actual phosphate group in the crystallized structure.
Prediction of intrinsically disordered regions
Disordered regions were predicted using the Disopred 30 and PONDR-FIT 31 programs. We defined disordered sites as those predicted as disordered by both methods. We sped up Disopred calculations by generating sequence profiles using PSI-BLAST against the Uniref50 database (instead of the default non-redundant database), otherwise retained the default parameters. PONDR-FIT predictions were obtained by submitting sequences to the webserver (http://www.disprot.org/pondr-fit.php). These predictors are expected to be complementary as they measure somewhat different properties of a protein. Disopred is a support vector machine method trained on sequence profiles of disordered and ordered regions in crystallized structures. PONDR-FIT is a meta-predictor that combines and improves on the results of PONDR-VLXT, PONDR-VSL2, PONDR-VL3, older meta-predictors 32; FoldIndex (uses pairwise inter-residue energy) 33; IUPred (uses hydrophobicity and net charge properties) 34; and TopIDP (uses amino acid propensities for disorders) 35. PONDER-FIT and DISOPRED identified 474 and 435 phosphorylation sites as disordered respectively. Overall, 248 out of 1,983 phosphorylation sites and 996 out of 31,071 Ser, Thr, and Tyr non-phosphorylation sites were identified on disordered regions by both methods.
Statistical tests, log-linear analysis
We performed log-linear analysis which examines the distribution of frequencies in the contingency tables involving several variables and tries to explain the variation observed between the cells of the table as a function of (in)dependence between variables 36. For example, consider three variables X, Y and Z which might take values between (1,…,I), (1,…,J), and (1,…,K), respectively. The log-linear model would involve all possible one-way, two way and three-way associations between the variables. Most of our variables are binary and we represent the two categories as values 0 or 1. Then the logarithm of expected frequency for cell ijk can be written as:
Where µ is the mean of the logarithms of the expected frequencies, λ are parameters characterizing the effect of categories of different variables. If parameter corresponding to interaction XY is equal to zero, then no interaction between variables is observed. The goal is to construct a model such that the cell frequencies in a contingency table are accounted for by the minimum number of terms. Each model is evaluated based on its ability to reproduce the observed cell counts in the table using maximum likelihood. The null hypothesis is that the model generates predicted data that is not significantly different from the observed table. Therefore, good models have p-values larger than 0.05.
We used several other tests to study the (in) dependence of different factors and their effect on the fraction of phosphorylation sites. To analyze the dependence of phosphorylation on other factors we used ANOVA (“Analysis of Variance”). We supported our conclusions using Kruskal–Wallis analysis of variance which is a non-parametric method for testing whether samples originate from the same distribution and if they are independent. Unlike ANOVA, Kruskal–Wallis analysis does not assume the normal distribution of each feature. Finally we applied the Duncan's multiple range test to compare multiple means and determine which mean values differ from each other. To perform statistical tests we used Splus, R and Statistica packages37.
Results
Correlation and cluster analyses
We represent each variable in a binary form and assign zero or one if a given feature is present or absent at a given site (phosphorylated/non-phosphorylated, disordered/ordered, interface/non-interface). We use five different categorical variables which might take value “1” if a feature is present in a given site and “0” otherwise: phosphorylation (P), disorder (D), interface (I), type of residue (T), and homo/hetero oligomerization state (H). In addition, all sites are separated into solvent accessible and buried sites. Regions that have coordinates in structures and are not predicted to be disordered are referred to as “ordered.”
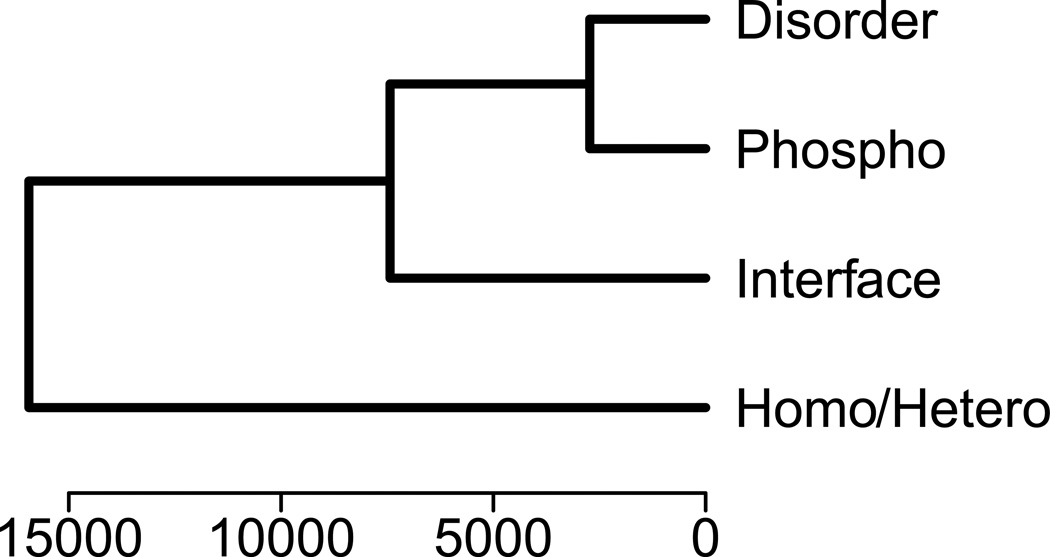
Here we are trying to understand which of these features contribute significantly to the observed distribution of our data, and whether this is a result of interactions between features or the result of their independent effects. First we calculated Spearman rank correlation coefficients between our different variables and found statistically significant positive correlations for all three types of residues (Ser, Thr, and Tyr) between phosphorylation and disorder (the strongest correlation); between phosphorylation and interface; and between disorder and interface (Table S1). We also performed cluster analysis (Figure 1) using Manhattan distance as a distance measure and a weighted pair-group clustering algorithm (where the choice of distance metric or clustering algorithm does not affect the shape of the cluster tree). As one can see from the cluster tree, disorder and phosphorylation are clustered together at the first step, then interface, and homo/heterooligomer features merged at the successive stages.
Figure 1. Cluster tree showing the association between phosphorylation, disorder, interface and homo/ heterooligomeric states.
The clustering was done with the Manhattan distance used as a distance measure and weighted pair-group clustering algorithm.
Examining concurrent relationships between features by log –linear analysis
Frequency distributions of different variables were compared by categories using Fisher’s exact test (Table S2). Since a significant fraction of phosphorylation sites (30%) are directly located on binding interfaces and (de)phosphorylation on binding interfaces might be associated with the intrinsic disorder through the disorder-order or order-disorder transitions, we performed the multivariate analyses outlined below.
First, we performed log-linear analysis, which examines the distribution of frequencies in the contingency tables and tests the conditional relationships between two or more discrete, categorical variables with no distinction between independent and dependent variables. We tested several models corresponding to different sets of variables, and for each set of variables the best model was reported. The interactions between phosphorylation and disorder variables (denoted as “PD”), between phosphorylation and interface (“PI”), disorder and interface (“DI”) and oligomerization state and interface (“IH”) were examined. As can be seen from Table 1, some of the best models included only pairwise interaction terms between variables. The null hypothesis is that the model generates predicted data that is not significantly different from the observed table, therefore good models have higher p-values and require p-values larger than 0.05. The interactions between phosphorylation and disorder variables (PD), and interactions PI, DI and IH were observed for all three residues types (Ser/Thr/Tyr), while interactions PH and DH were found only for two out of three residues. The model for Tyr was of lower quality compared to Ser and Thr (Table 1), and overall the models did not change their form if only solvent-accessible residues or all residues (accessible and buried) were considered. In fact 87% of phosphorylation sites in our dataset were either solvent accessible or interfacial.
Table 1. The results of log-linear analysis.
The following notations are used: PD – interaction between phosphorylation and disorder features; PI – phosphorylation and interface; DI – disorder and interface; PH – phosphorylation and homooligomer; IH – interface and homooligomer; DH – disorder and homooligomer. “T” is a residue type. Good models have p-values larger than 0.05. Here we used all residues but the models did not change their form if only solvent-accessible residues were considered. The exception is the model for all three residues (last row of the table) which, in case of solvent accessible residues, included only three interaction terms (PIH, DI, PD).
| Model | Residue type |
Pearson chi- square |
Degrees of freedom |
p-value |
|---|---|---|---|---|
| PD+PI+DI+PH+IH | Ser | 3.83 | 6 | 0.70 |
| PD+PI+DI+IH+DH+PH | Thr | 3.43 | 5 | 0.64 |
| PD + PI+ DI+ IH + DH | Tyr | 7.16 | 6 | 0.31 |
| DI+PDT+DTH+ITH+PIH | Ser+Thr+Tyr | 9.98 | 20 | 0.97 |
Dependence of phosphorylation on other variables
Next we chose phosphorylation as a dependent variable to assess its dependence on several other variables. We introduce here a new categorical joint variable with four different levels depending on the disorder and interface state of a given site. First level of joint variable corresponds to the cases of interface sites which are at the same time predicted intrinsically disordered (D=1, I=1), these regions might undergo disorder-to-order transition while binding to other proteins. Similarly other levels correspond to disordered non-interface (D=1, I=0), ordered interface (D=0, I=1) and ordered non-interface (D=0, I=0) regions. Non-interface sites are defined as all residues in a given category excluding interface residues. To analyze the dependence of phosphorylation on other variables we used ANOVA, and non-parametrical methods (Kruskal-Wallis test). Figure 2 shows the influence of different levels on residue phosphorylation for those residues which are solvent accessible in the unbound state; the overall results did not change if we analyzed all protein residues instead. The results obtained by the non-parametric Kruskal-Wallis test were consistent with the results obtained by the ANOVA. We found that, overall, disorder and interface significantly influenced phosphorylation of serine and threonine (p-value << 0.01) and to a lesser extent of tyrosine (p-value = 0.02). The largest fraction of phosphorylated residues on disordered binding regions was obtained for Ser (28% of all serines on disordered interfaces were phosphorylated), followed by Tyr (27%) and Thr (20%).
Figure 2. Dependence of fraction of phosphorylation sites on different factors.
Fraction of phosphorylated sites on solvent accessible regions is calculated for each level of joint variable: D=1, I=1: Disordered Interface; D=1, I=0: Disordered nonInterface; D=0, I=1: Ordered Interface and D=0, I=0: Ordered nonInterface. Error bars indicate 95% confidential interval. Homo and heterooligomers are shown in red and blue, respectively.
Then we used multiple comparison methods (Dunkan and Kruskal tests) and found that phosphorylation on disordered interface exceeded the phosphorylation on disordered non-interface, ordered interface and ordered non-interface regions (p-value=0.005, Figure 2A). However, if Ser, Thr and Tyr are examined separately, the null hypothesis that phosphorylation on disordered interface exceeds phosphorylation on disordered non-interface regions (first and second levels on Figure 2) holds true only for serine (p-value=0.009) but not for threonine (p-value = 0.08) or tyrosine (p-value = 0.14). For all three residues, phosphorylation on ordered interface regions was higher than on ordered non-interface region (p-values = 0.0001, third and fourth levels on Figure 2).
Heterooligomers showed higher fraction of phosphorylation sites compared to homooligomers for disordered interfaces (D=1, I=1) and for ordered interfaces (O=1, I=1) for all three residues taken together (p-value=0.04 and p-value << 0.01 respectively, Figure 2A). Similar trends were observed if solvent-accessible residues separately or consider all residues (Figure S2). In summary, while only a small fraction of binding interfaces in structured complexes contains predicted disordered regions, and most phosphorylation sites are located on ordered interface regions (Figure S1), there is a strong association between phosphorylation and disordered interface: while 25% of disordered interface Ser/Thr/Tyr sites are phosphorylated, only 8% of ordered interface Ser/Thr/Tyr sites are phosphorylated.
Functional importance of phosphorylation on disordered interfaces
As we mentioned previously there are examples where phosphorylation might happen on ordered flexible or inflexible regions 20. Here we focus instead on functional mechanisms of regulation of disorder-order or order-disorder transitions upon binding. We provide examples that show how phosphorylation might prevent binding and possible disorder-order transition and, in some cases, facilitate binding to another partner, which competes with the first one. For all discussed complexes, binding interfaces are predicted to be disordered and phosphorylated, though ordered and unphosphorylated in the bound state.
p21 is an important protein in cell cycle arrest in response to DNA damage. It blocks the progress of cell-cycle by binding to cyclin-dependent kinases (CDKs). It can also inhibit DNA replication directly by binding to proliferating cell nuclear antigen (PCNA) which is an accessory protein of DNA polymerases delta and epsilon. The full length p21 protein does not have any stable structure in solution but can form an ordered stable conformation upon binding to the target proteins, such as CDKs 38 or PCNA 39. Residue Thr145 of p21 is located in the PCNA binding region (region from 144 to 151 residues, Figure 3A) and its phosphorylation may inhibit the association with PCNA, which allow PCNA bind to other components of the polymerase 40.
Figure 3. Examples of phosphorylation sites on disordered interface regions.
Phosphorylation site are shown in red stick models. A: p21/PCNA complex (PDBID: 1AXC, chain A, B). p21 (colored in yellow) forms an ordered conformation upon binding to PCNA (green). B: RhoGDI-Rac1 complex (PDBID: 1HH4, chain B, E). RhoGDI has a C-terminal domain (shown in cyan) and a flexible N-terminal region (yellow) which become ordered upon binding to Rac1 (pink).
Another example is the Rho GDP-dissociation inhibitor 1 (RhoGDI1) which regulates Rho family GTPase in several ways (Figure 3B). RhoGDIs can inhibit Rho proteins by preventing the release of GDP and the loading of GTP and can transfer inactive Rho proteins from cell membrane to prevent their degradation and inappropriate activation. Phosphorylation of RhoGDI Ser, Thr and Tyr residues is very important for its function, and phosphorylation of multiple sites can promote the release of multiple Rho proteins simultaneously 41. The key functional region is its N-terminal domain which is disordered. However, it can form two helices and bind to switch I and switch II regions of GTPase to restrain the conformational changes required for exchange of GDP and GTP 42. Tyr27 on the disordered N-terminal domain of RhoGDI1 can be phosphorylated and is located on the binding interface. It has been shown that its phosphorylation promotes dissociation of RhoA, Rac1, and cdc42 from RhoGDI1 and make GTPases available for activation43.
The third example is Na(+)/H(+) exchange regulatory cofactor NHERF1, an adaptor protein which connects plasma membrane proteins to other membrane-cytoskeleton proteins, and hence regulates cell shape and migration. NHERF1 consists of three regions: ordered PDZ domains (PDZ1 and PDZ2) interacting with Cystic fibrosis transmembrane conductance regulator (CFTR) and disordered C-terminal ERM binding region which associates with ERM domains in membrane-cytoskeleton proteins. Additionally this disordered C-terminal region is used for auto-inhibition mechanism of NHERF1; the region interacts with the PDZ2 domain and prevents PDZ2 from binding to CFTR. In both cases, the C-terminal region becomes ordered upon binding to other proteins 44. An earlier experimental study revealed that phosphorylation on Ser339 and Ser340 in the C-terminal region disrupts the binding between the C-terminal region and PDZ2 domain, hence increasing the binding affinity of PDZ2 to CFTR 45.
Discussion
Our previous analysis of the intrinsic disorder of human proteins participating in certain relationships in biochemical pathways showed that gene expression, phosphorylation and protein–protein binding/association relations are consistently enriched in disorder 16. In our later study we analyzed regulatory roles of phosphorylation in protein-protein association or dissociation by focusing on the structural complexes with known binding interfaces 20. Here, we analyze different factors that might contribute to the regulation of protein binding by phosphorylation and intrinsic disorder.
First, we found a significant association between phosphorylation, disorder and interface states for residue sites in human protein complexes. Disordered interface and homo/heteroligomer states significantly correlate with the phosphorylation of all three residues (Ser, Thr and Tyr). This signal is most pronounced for serine and diminishing for threonine and tyrosine in this order. We also found that for serine the phosphorylation on disordered interfaces exceeded the phosphorylation on disordered non-interface regions.
Short structured protein regions within longer disordered sequences that enhance molecular recognition and binding to larger proteins (MoRFs) were found to contain 45%, 36%, and 19% of pSer, pThr, and pTyr respectively46. Here we analyzed possible disorder-order transitions in structural complexes, not completely disordered proteins and found similar but not identical fractions of pSer, pThr and pTyr on disordered binding interfaces: 59%, 26% and 15% respectively. These fractions were quite different for ordered structured interfaces, where pTyr occupied half of all phosphorylated residues followed by pSer (28%) and pThr (22%).
Indeed, serine is commonly found in disordered regions of proteins, especially in disordered protein hubs in protein-protein interaction networks 8. Here we suggest that this might be driven by the exceptional functional role of serine for disorder-order transition type binding. While Ser and Thr are frequently found in disordered and flexible regions, Tyr is more likely to be found in structured regions. Unlike serine and threonine, phosphorylation of nearly half of pTyr is associated with protein structural domains 47. In contrast to serine and threonine, tyrosine does not have a tendency to be in disordered compared to ordered regions 22, is characteristic for thermostable complexes48 and is depleted among MoRFs 46.
Binding mediated by disorder has certain advantages. It allows a protein structure to adapt to multiple interaction interfaces and may provide large interface regions exposed by disordered regions. Disorder may also play a key role in molecular adaptation to different environmental constraints: highly unstructured, rapidly evolving viral proteins on the one hand and highly structured proteins from thermophilic organisms on the other hand 49, 50. Since disordered proteins and disordered binding regions lack structures, the relationship between disorder and binding is very difficult to decipher.
There is a limited though growing number of examples of experimentally verified disorder-order and order-disorder transitions, and several attempts have been made to understand the underlying principles of molecular recognition by disordered regions. For example, physicochemical features of interfaces formed by disordered proteins were found to be quite different from those formed by structured proteins: disordered protein interfaces had a large number of contacts per residue, exhibited prominent preference for hydrophobic residues and were localized linearly on the primary sequence 4, 51. Moreover, the high number of inter-chain compared to intra-chain contacts was shown to be a signature of disordered binding segments 52. In addition, disorder-order transition might allow uncoupling of binding affinity from specificity and provide kinetic advantages through fly-casting mechanisms 53.
Here we attempted to decipher the principles of recognition through coupling of phosphorylation, binding and disorder. We showed that one of the reasons of association of phosphorylation sites with disordered regions could be their direct involvement in binding processes, and quantified this association for human protein complexes. Finally we suggested functional mechanisms for how phosphorylation can control disorder-order and order-disorder transitions and regulate protein-protein binding.
Supplementary Material
Acknowledgements
This work was supported by the Intramural Research Program of the National Library of Medicine at the U.S. National Institutes of Health. This work was partially supported by the Medical Research Council, UK, (MRC file reference number U105161047).
References
- 1.Wright PE, Dyson HJ. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 2.Gsponer J, Futschik ME, Teichmann SA, Babu MM. Science. 2008;322:1365–1368. doi: 10.1126/science.1163581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fink AL. Curr Opin Struct Biol. 2005;15:35–41. doi: 10.1016/j.sbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Meszaros B, Tompa P, Simon I, Dosztanyi Z. J Mol Biol. 2007;372:549–561. doi: 10.1016/j.jmb.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Lobanov MY, Shoemaker BA, Garbuzynskiy SO, Fong JH, Panchenko AR, Galzitskaya OV. Nucleic Acids Res. 2010;38:D283–D287. doi: 10.1093/nar/gkp963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fong JH, Shoemaker BA, Garbuzynskiy SO, Lobanov MY, Galzitskaya OV, Panchenko AR. PLoS Comput Biol. 2009;5:e1000316. doi: 10.1371/journal.pcbi.1000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J, Tan H, Rost B. J Mol Biol. 2002;322:53–64. doi: 10.1016/s0022-2836(02)00736-2. [DOI] [PubMed] [Google Scholar]
- 8.Patil A, Nakamura H. FEBS Lett. 2006;580:2041–2045. doi: 10.1016/j.febslet.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 9.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. FEBS J. 2005;272:5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 10.Schnell S, Fortunato S, Roy S. Proteomics. 2007;7:961–964. doi: 10.1002/pmic.200600455. [DOI] [PubMed] [Google Scholar]
- 11.Fong JH, Panchenko AR. Mol Biosyst. 2010;6:1821–1828. doi: 10.1039/c005144f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higurashi M, Ishida T, Kinoshita K. Protein Sci. 2008;17:72–78. doi: 10.1110/ps.073196308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim PM, Sboner A, Xia Y, Gerstein M. Mol Syst Biol. 2008;4:179. doi: 10.1038/msb.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turoverov KK, Kuznetsova IM, Uversky VN. Prog Biophys Mol Biol. 2010;102:73–84. doi: 10.1016/j.pbiomolbio.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. J Mol Biol. 2002;323:573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 16.Fong JH, Shoemaker BA, Panchenko AR. Mol Biosyst. 2012;8:320–326. doi: 10.1039/c1mb05274h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schlessinger J. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 18.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 19.Johnson LN. Biochem Soc Trans. 2009;37:627–641. doi: 10.1042/BST0370627. [DOI] [PubMed] [Google Scholar]
- 20.Nishi H, Hashimoto K, Panchenko AR. Structure. 2011;19:1807–1815. doi: 10.1016/j.str.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson LN, Lewis RJ. Chem Rev. 2001;101:2209–2242. doi: 10.1021/cr000225s. [DOI] [PubMed] [Google Scholar]
- 22.Iakoucheva LM, Radivojac P, Brown CJ, O'Connor TR, Sikes JG, Obradovic Z, Dunker AK. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B. Proteomics. 2004;4:1551–1561. doi: 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]
- 25.Dinkel H, Chica C, Via A, Gould CM, Jensen LJ, Gibson TJ, Diella F. Nucleic Acids Res. 2011;39:D261–D267. doi: 10.1093/nar/gkq1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gnad F, Gunawardena J, Mann M. Nucleic Acids Res. 2011;39:D253–D260. doi: 10.1093/nar/gkq1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xue Y, Liu Z, Cao J, Ma Q, Gao X, Wang Q, Jin C, Zhou Y, Wen L, Ren J. Protein Eng Des Sel. 2011;24:255–260. doi: 10.1093/protein/gzq094. [DOI] [PubMed] [Google Scholar]
- 28.Edgar RC. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krissinel E, Henrick K. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 30.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 31.Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN. Biochim Biophys Acta. 2010;1804:996–1010. doi: 10.1016/j.bbapap.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng K, Radivojac P, Vucetic S, Dunker AK, Obradovic Z. BMC Bioinformatics. 2006;7:208. doi: 10.1186/1471-2105-7-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- 34.Dosztanyi Z, Csizmok V, Tompa P, Simon I. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 35.Campen A, Williams RM, Brown CJ, Meng J, Uversky VN, Dunker AK. Protein Pept Lett. 2008;15:956–963. doi: 10.2174/092986608785849164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brackley ME, De Boer JG, Glickman BW. Mutat Res. 1999;425:55–69. doi: 10.1016/s0027-5107(98)00249-8. [DOI] [PubMed] [Google Scholar]
- 37.Sa J. Applied Statistics Using Spss, STATISTICA, Matlab and R. Berlin: Springer; 2007. [Google Scholar]
- 38.Kriwacki RW, Hengst L, Tennant L, Reed SI, Wright PE. Proc Natl Acad Sci U S A. 1996;93:11504–11509. doi: 10.1073/pnas.93.21.11504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gulbis JM, Kelman Z, Hurwitz J, O'Donnell M, Kuriyan J. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 40.Li Y, Dowbenko D, Lasky LA. J Biol Chem. 2002;277:11352–11361. doi: 10.1074/jbc.M109062200. [DOI] [PubMed] [Google Scholar]
- 41.Garcia-Mata R, Boulter E, Burridge K. Nat Rev Mol Cell Biol. 2011;12:493–504. doi: 10.1038/nrm3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grizot S, Faure J, Fieschi F, Vignais PV, Dagher MC, Pebay-Peyroula E. Biochemistry. 2001;40:10007–10013. doi: 10.1021/bi010288k. [DOI] [PubMed] [Google Scholar]
- 43.DerMardirossian C, Rocklin G, Seo JY, Bokoch GM. Mol Biol Cell. 2006;17:4760–4768. doi: 10.1091/mbc.E06-06-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terawaki S, Maesaki R, Hakoshima T. Structure. 2006;14:777–789. doi: 10.1016/j.str.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 45.Li J, Poulikakos PI, Dai Z, Testa JR, Callaway DJ, Bu Z. J Biol Chem. 2007;282:27086–27099. doi: 10.1074/jbc.M702019200. [DOI] [PubMed] [Google Scholar]
- 46.Mohan A, Oldfield CJ, Radivojac P, Vacic V, Cortese MS, Dunker AK, Uversky VN. J Mol Biol. 2006;362:1043–1059. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- 47.Sugiyama N, Nakagami H, Mochida K, Daudi A, Tomita M, Shirasu K, Ishihama Y. Mol Syst Biol. 2008;4:193. doi: 10.1038/msb.2008.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma BG, Goncearenco A, Berezovsky IN. Structure. 2010;18:819–828. doi: 10.1016/j.str.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 49.Berezovsky IN. Phys Biol. 2011;8:035002. doi: 10.1088/1478-3975/8/3/035002. [DOI] [PubMed] [Google Scholar]
- 50.Mamonova TB, Glyakina AV, Kurnikova MG, Galzitskaya OV. J Bioinform Comput Biol. 2010;8:377–394. doi: 10.1142/s0219720010004690. [DOI] [PubMed] [Google Scholar]
- 51.Vacic V, Oldfield CJ, Mohan A, Radivojac P, Cortese MS, Uversky VN, Dunker AK. J Proteome Res. 2007;6:2351–2366. doi: 10.1021/pr0701411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meszaros B, Simon I, Dosztanyi Z. PLoS Comput Biol. 2009;5:e1000376. doi: 10.1371/journal.pcbi.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shoemaker BA, Portman JJ, Wolynes PG. Proc Natl Acad Sci U S A. 2000;97:8868–8873. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.