Abstract
Several organisms have retained methyltransferase 2 (Dnmt2) as their only candidate DNA methyltransferase gene. However, information about Dnmt2-dependent methylation patterns has been limited to a few isolated loci and the results have been discussed controversially. In addition, recent studies have shown that Dnmt2 functions as a tRNA methyltransferase, which raised the possibility that Dnmt2-only genomes might be unmethylated. We have now used whole-genome bisulfite sequencing to analyze the methylomes of Dnmt2-only organisms at single-base resolution. Our results show that the genomes of Schistosoma mansoni and Drosophila melanogaster lack detectable DNA methylation patterns. Residual unconverted cytosine residues shared many attributes with bisulfite deamination artifacts and were observed at comparable levels in Dnmt2-deficient flies. Furthermore, genetically modified Dnmt2-only mouse embryonic stem cells lost the DNA methylation patterns found in wild-type cells. Our results thus uncover fundamental differences among animal methylomes and suggest that DNA methylation is dispensable for a considerable number of eukaryotic organisms.
Keywords: RNA methylation, epigenetics
DNA methylation is an epigenetic modification with important functions in cellular differentiation, organismal development, and human disease (1, 2). DNA methylation is widely conserved in the animal and plant kingdoms and established and maintained by a family of enzymes termed DNA methyltransferases (3, 4). In animals, DNA methyltransferase 3 (Dnmt3) enzymes act as de novo methyltransferases that establish DNA methylation patterns, most prominently during early stages of development. Established methylation patterns are then maintained by DNA methyltransferase 1 (Dnmt1) enzymes, which copy methylation marks from methylated CpG dinucleotides on the parental DNA strand to complementary CpG dinucleotides on the daughter strand.
DNA methyltransferase 2 (Dnmt2), which is also known as tRNA aspartic acid methyltransferase 1 (Trdmt1), is the second member of the DNA methyltransferase family and shows strong sequence conservation to the catalytic motifs of established DNA methyltransferases (5–7). However, the actual DNA methyltransferase activity of Dnmt2 was found to be substantially weaker than for other DNA methyltransferases (8, 9). It was later shown that Dnmt2 has a robust methyltransferase activity toward cytosine 38 in the anticodon loop of tRNAAsp and other tRNAs (9, 10) and that the enzyme uses the conserved DNA methyltransferase mechanism to methylate tRNA (11). In agreement with this notion, several independent phylogenetic analyses have suggested that Dnmt2 is an ancient DNA methyltransferase that has switched its substrate specificity from DNA to tRNA (12–14).
The number of Dnmt genes can vary greatly between genomes, and various organisms have been shown to encode different sets of Dnmt enzymes (15). Mammalian genomes encode one Dnmt1 gene and three paralogs of Dnmt3. This contrasts, for example, with the genome of the parasitic wasp Nasonia vitripennis, which encodes three paralogs of Dnmt1 and a single Dnmt3 homolog (16). These variations have been interpreted to reflect multiple versions of a tool kit for phenotypic adaptation (17). During evolution, parts of this tool kit could have been contracted or expanded to facilitate specific requirements for genome regulation (17). In this context, Dnmt2 has been shown to provide a link between environmental cues, such as stress and nutrition signals, to tRNA stability and the regulation of protein synthesis (10, 18, 19).
Interestingly, there is a diverse group of animal species that has retained Dnmt2 as their only candidate DNA methyltransferase. This group includes several highly relevant model organisms, such as Schizosaccharomyces pombe, Dictyostelium discoideum, Entamoeba histolytica, Schistosoma mansoni, and Drosophila melanogaster. Global DNA methylation levels in these Dnmt2-only systems have been found to be very low and have often been discussed controversially (20–22). More recently, however, two prominent studies have provided support for a biologically important function of Dnmt2-dependent DNA methylation. For example, it has been suggested that Dnmt2-dependent DNA methylation regulates oviposition in S. mansoni (23), the causative agent of bilharziosis. In addition, Dnmt2-dependent methylation of transposons has been linked to genome stability in D. melanogaster (24). These results have necessitated a more detailed analysis of genome methylation patterns in Schistosoma and Drosophila.
Over the past few years, whole-genome bisulfite sequencing has been established as a method to characterize genome-wide DNA methylation patterns at single-base resolution (25). This method has been successfully used to characterize the methylomes of various animal organisms that are known to establish and maintain their methylation patterns by Dnmt1 and/or Dnmt3 enzymes. The results revealed a certain degree of diversity among animal methylomes, but also identified a number of conserved features, which include the specificity for CpG dinucleotides and an enrichment of methylation in defined genetic elements (26, 27). We have now used whole-genome bisulfite sequencing for an unbiased characterization of the Schistosoma and Drosophila methylomes. Our results fail to reveal any evidence for defined DNA methylation patterns in these organisms and thus uncover fundamental differences between Dnmt1/3-dependent and Dnmt2-dependent methylomes.
Results
Whole-Genome Bisulfite Sequencing of Three Independent Dnmt2-Only Models.
We investigated Dnmt2-dependent DNA methylation in three independent models: (i) S. mansoni, (ii) D. melanogaster, and (iii) a triple knockout (TKO) mouse embryonic stem cell line, which is deficient for Dnmt1, Dnmt3a, and Dnmt3b but has retained an intact Dnmt2 gene (28). All samples were obtained from the same strains and developmental stages that were used for previous studies (Table 1). Furthermore, Dnmt2 activity was confirmed by methylation analysis of tRNAAsp, which showed high levels of methylation for the established C38 target site in all three samples (Fig. S1).
Table 1.
Sequencing data
| Sample | Comment | No. of reads (pairs) | Mapping efficiency, % | Coverage | Conversion, % |
| Schistosoma | Mixed adult Puerto Rican | 254,347,320 | 73 | 13.4× | 99.02 |
| Drosophila | w1118 0–2 h embryos | 174,821,591 | 57 | 32.0× | 99.75 |
| TKO | Mouse TKO ES cells | 43,583,131 | 74 | 0.8× | 98.22 |
| PCR fragment | Spiked into TKO | 1,593 | ND | ND | 98.38 |
| Human sperm DNA | Spiked into Drosophila | 1,637,070 | ND | ND | 99.71 |
| Drosophila Dnmt2−/− | Dnmt2149 0- to 2-h embryos | 129,094,743 | 52 | 23.6× | 99.42 |
Sequencing coverages are indicated per strand. Conversion rates were calculated as the average conversion ratio of all cytosine residues that were covered by the dataset. For human sperm DNA, the conversion rate was calculated as the average conversion ratio of all non-CpG dinucleotides that were covered by the dataset. ND, not determined.
We then used whole-genome bisulfite sequencing to comprehensively analyze genomic DNA methylation patterns in all three models. After bisulfite deamination, DNA libraries were prepared and subjected to paired-end Illumina sequencing. Read pairs were subsequently mapped to the corresponding reference genomes using BSMAP 2.0 (29). This resulted in average strand-specific genome coverages of 13× for Schistosoma, 32× for Drosophila, and 1× for TKO cells (Table 1). The conversion rate of mapped cytosine residues was >98.0% in all cases (Table 1), which suggested that bisulfite deamination had been efficient and that the sequence information could be used for methylation analysis.
Methylome of S. mansoni.
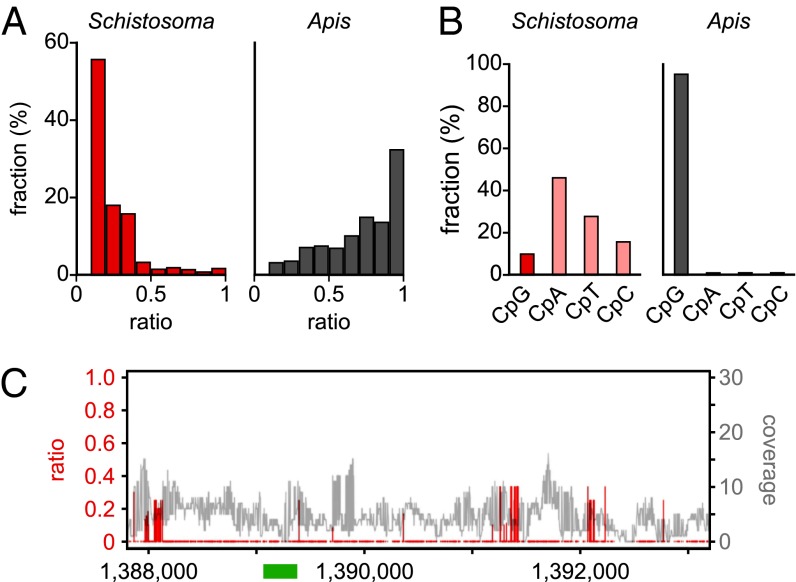
As an initial step toward a comprehensive analysis of the Schistosoma methylome, nonconversion ratios were determined for all cytosine residues with a sequence coverage greater than three. This revealed that 97% of the cytosines were completely converted (ratio <0.1). When the remaining cytosines were distributed into bins with increasing nonconversion ratios, the majority (93%) showed ratios <0.5, whereas only a minor fraction (7%) had ratios >0.5 (Fig. 1A). This distribution strongly contrasts the results from honey bees, which have an established Dnmt1/3-dependent methylome with very low levels of DNA methylation (30). Here, only 21% of all not completely converted cytosines showed a nonconversion (methylation) ratio <0.5, whereas 79% had a ratio of >0.5 (Fig. 1A). These results suggest that the Schistosoma genome contains either extremely low levels or no DNA methylation at all.
Fig. 1.
Characterization of the Schistosoma mansoni methylome. (A) Average methylation levels were determined for all cytosine residues with a methylation ratio >0.1 and then distributed into bins with increasing methylation ratios (red bars). For comparison, the corresponding data are also shown for honey bee worker brains (gray bars), an established Dnmt1/3-dependent methylome with a very low DNA methylation level (30). (B) Dinucleotide sequence contexts of unconverted cytosines in Schistosoma (red) and in honey bees (gray). (C) Position-specific nonconversion ratios (red) and coverages (gray) of the Schistosoma forkhead gene. The specific region previously reported to be methylated (23) is indicated as a green bar. Sequence position numbers refer to GenBank accession JF781495.
A conserved feature of all known animal methylomes is their high degree of CpG-specificity (26, 27), which is related to the stable transmission of symmetric CpG methylation marks by maintenance methylation mechanisms. Therefore, we also determined the dinucleotide sequence context of nonconverted cytosines in our Schistosoma dataset and could not detect any evidence for CpG specificity (Fig. 1B). In contrast, unconverted cytosines from the honey bee methylome dataset were strongly (>95%) enriched for CpG dinucleotides (Fig. 1B). Finally, we also used our data for a detailed analysis of the Schistosoma forkhead gene that had previously been reported to be methylated in a Dnmt2-dependent manner (23). Our results failed to reveal any evidence for DNA methylation at the previously analyzed region (Fig. 1C). We did, however, observe dense clusters of incompletely converted cytosines outside of this region (Fig. 1C), preferentially in sequences with low complexity. Similar patterns have not been described in any of the published methylomes, but correspond to known characteristics of bisulfite deamination artifacts (31). Taken together, these results strongly suggest that the Schistosoma genome is unmethylated.
Methylome of D. melanogaster.
To characterize the methylome of a second Dnmt2-only organism, we obtained genome-wide methylation profiles from D. melanogaster embryos. A previous phylogenetic analysis of DNA methylation patterns had already suggested that DNA from Drosophila embryos is unmethylated (27). However, the data were obtained from an unspecified strain and the sequencing coverage was comparably low. To further characterize the Drosophila methylome, we therefore obtained an independent methylation profile from 0- to 2-h-old wildtype (w1118) embryos. This is the same strain and developmental stage that was used for a previous analysis describing DNA methylation of Invader4 elements (24). As a reference, 1% of human sperm DNA was spiked into the Drosophila sample before bisulfite conversion. Human sperm DNA is known to be highly methylated (32) and the spiked-in DNA sample thus served as an important internal control.
A detailed analysis of the Drosophila data showed that the vast majority (99.7%) of cytosine residues appeared completely unmethylated (ratio <0.1), whereas only 0.003% showed a nonconversion ratio >0.5 (Fig. 2A). This distribution was substantially different for the spiked-in human sperm DNA, which showed complete methylation (ratio >0.9) for 4.3% of the cytosine residues that were analyzed (Fig. 2A). Pronounced differences between the Drosophila and the control sample were also detectable for the dinucleotide sequence context of nonconverted cytosine residues. In the Drosophila dataset, only 11% of the nonconverted cytosine residues were found in CpG dinucleotides (Fig. 2B). This distribution strongly contrasted the control sample, which showed a high degree (98%) of CpG specificity (Fig. 2B). Finally, we also used our data for a detailed analysis of Drosophila Invader4 elements, which have previously been reported to be methylated in a Dnmt2-dependent manner (24). Our results failed to reveal any evidence for DNA methylation at the previously analyzed region and at other Invader4 sequences (Fig. 2C). Together, these results strongly suggest that the Drosophila genome is unmethylated.
Fig. 2.
Characterization of the Drosophila melanogaster methylome. (A) Average methylation levels were determined for all cytosine residues and then distributed into bins with increasing methylation ratios (blue bars). For comparison, the corresponding data are also shown for human sperm DNA that was spiked into the Drosophila sample before bisulfite conversion (black bars). The actual numerical values of the first bins are 99.7% (Drosophila) and 92.9% (human sperm). (B) Dinucleotide sequence context of unconverted cytosines in Drosophila (blue) and in human sperm (black). (C) Position-specific nonconversion ratios (red) and coverage (gray) of the Drosophila Invader4 element. Results are shown for the sequence with the lowest conversion rate among genomic Invader4 elements. The specific region previously reported to be methylated (24) is indicated as a green bar. Sequence position numbers refer to GenBank accession AE014135.3.
Methylome of Mouse TKO ES Cells.
To further confirm the lack of DNA methylation in Dnmt2-only systems we also used bisulfite sequencing to analyze DNA methylation patterns in the mouse TKO ES cell line (Fig. 3A). A detailed analysis of the sequencing data showed that the vast majority (96.5%) of the 75,778,741 genomic cytosine residues that were covered by at least three reads appeared completely unmethylated (ratio <0.1), whereas only 0.03% showed a nonconversion ratio >0.5 (Fig. 3B). A substantially different distribution was observed in a published dataset from wild-type mouse ES cells (33), which showed complete methylation (ratio >0.9) for 2.5% of the cytosine residues (Fig. 3B). A more detailed analysis showed a high proportion of CpG methylation and significant levels of non-CpG methylation in wild-type ES cells (Fig. 3C), consistent with earlier observations (34, 35). Even with a low-stringency cutoff (ratio >0.1), the TKO cells showed substantially reduced signals (Fig. 3C), which suggests that the TKO genome is unmethylated.
Fig. 3.
Characterization of DNA methylation in the TKO mouse ES cell model. (A) Schematic illustration of Dnmt genotypes in wild-type and TKO mouse ES cells. (B) Average methylation levels were determined for all cytosine residues and then distributed into bins with increasing methylation ratios (orange bars). For comparison, the corresponding data are also shown for wild-type mouse ES cells (gray bars). The actual numerical values of the first bins are 86.9% (wild type) and 96.5% (TKO). (C) Fractions of nonconverted (ratio >0.1) CpN dinucleotides in wild-type cells (gray bars) and TKO cells (orange bars).
Analysis of Control Datasets.
To further determine the significance of the observed nonconverted cytosine residues, several controls were used (Table 1). (i) An unmethylated PCR fragment that had been spiked into the TKO sample before bisulfite conversion, (ii) human sperm DNA that was spiked into the Drosophila sample before bisulfite conversion, and (iii) a Dnmt2-deficient Drosophila strain. Further data analysis showed that the difference in the conversion efficiencies between the TKO dataset and the spiked-in control was small (0.12%, Table 1), but statistically significant (P = 0.012, Fisher’s exact test). This could be related to an intrinsically higher conversion efficiency of the PCR fragment, relative to genomic DNA. We therefore also analyzed the difference between the Drosophila sample and the spiked-in human sperm control. Indeed, the difference in the deamination rates was very small (0.04%, Table 1) and not statistically significant (P = 1.0, Fisher’s exact test). These results provide important confirmation for the absence of defined DNA methylation patterns in Dnmt2-only organisms.
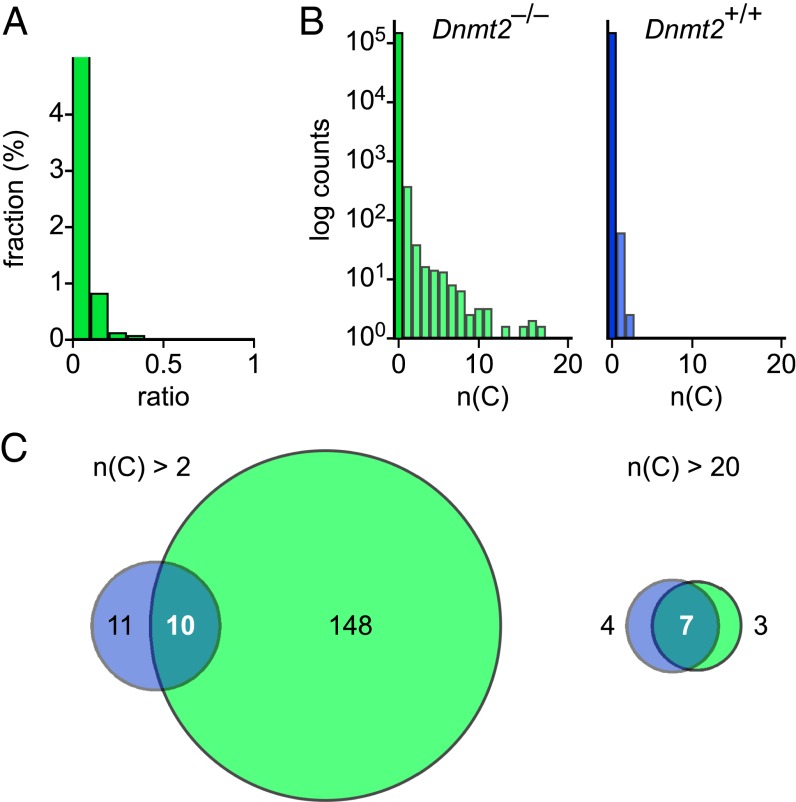
Finally, to directly investigate the significance of Dnmt2 for the remaining nonconverted cytosine residues in Drosophila, we generated genome-wide methylation profiles from 0- to 2-h-old homozygous Dnmt2 mutant embryos. We obtained an average strand-specific genome coverage of 24× and a conversion rate of 99.4% (Table 1). Further data analysis showed that the vast majority (99.0%) of cytosine residues appeared completely unmethylated (Fig. 4A). Notably, nonconverted cytosine residues appeared to be slightly more frequent than in the wild-type Drosophila sample (Fig. 4A), which further suggests that the remaining cytosine residues represent bisulfite deamination artifacts, rather than Dnmt2-mediated DNA methylation. We also used a sliding window approach to identify sequences with an increased density of nonconverted cytosines. The results showed that the vast majority (>99.9%) of 1-kb windows completely lacked nonconverted cytosines (Fig. 4B). Among the remaining windows, many contained a single nonconverted cytosine residue (Fig. 4B). Notably, a substantial fraction of windows with more than two nonconverted cytosine residues in wild-type embryos also showed inefficient conversion in Dnmt2 mutant embryos (Fig. 4C). These regions were often characterized by a high CG content and a low base complexity, which would render them relatively resistant to denaturation during the bisulfite conversion step. These results further argue against genuine DNA methylation in Drosophila embryos and provide additional support for the notion that Dnmt2-only organisms lack DNA methylation.
Fig. 4.
Characterization of the Dnmt2 mutant Drosophila methylome. (A) Average methylation levels were determined for all cytosine residues that were covered by more than three sequence reads and then distributed into bins with increasing methylation ratios (green bars). The actual numerical value of the first bin is 99.0%. (B) Histograms showing the number of nonconverted (ratio >0.5) cytosine residues in 1-kb windows. For comparison, the corresponding data are also shown for the wild-type Drosophila methylome (blue). (C) Venn diagram showing overlapping windows with >2 (Left) or >20 (Right) nonconverted cytosine residues in wild-type (blue) and Dnmt2 mutant (green) embryos.
Discussion
The DNA methylation status of Dnmt2-only organisms has been a controversial topic for a long time. This may be related to the fact that the reported methylation levels were often close to the detection limits of the various methods that were used for DNA methylation analysis (22). The results from chromatographic analyses (23, 36, 37) may also have been affected by contamination with methylated DNA from other organisms, including bacteria. Similarly, immunological detection approaches of 5-methylcytosine in Drosophila embryos (38) could have been affected by low antibody specificity. Also, many previous bisulfite sequencing analyses were limited to isolated genomic loci (23, 24), which made them more susceptible to false positive results. Finally, it is also possible that the conserved catalytic mechanism of Dnmt2 (11) permits a limited “star activity,” i.e. a low enzymatic activity with relaxed substrate specificity, on DNA substrates. This star activity could be responsible for residual amounts of genuine DNA methylation and might become increased under certain experimental conditions (8, 38). However, because we could not detect any relevant DNA methylation patterns in our analyses, we would interpret these spurious methylation marks as biological artifacts.
The comprehensive nature of whole-genome bisulfite sequencing datasets allows additional quality control steps during data analysis and permits the identification of false positives with higher sensitivity (25). Also, whole-genome bisulfite sequencing protocols use substantially fewer PCR amplification cycles than locus-specific bisulfite sequencing protocols. This reduces the impact of PCR bias, another prominent source for false positive results in bisulfite sequencing (39). Our whole-genome bisulfite sequencing approach therefore allowed an unbiased characterization of the Schistosoma and Drosophila methylomes and revealed that both organisms lack defined DNA methylation patterns. Our results thus directly contradict previous reports (23, 24) and establish fundamental differences between Dnmt2-dependent and Dnmt1/3-dependent methylomes. We cannot presently exclude the possibility that Dnmt2 expression and/or Dnmt2-mediated DNA methylation vary according to unknown environmental cues (22). However, our analyses used the same strains and culture conditions as previously reported (23, 24). Furthermore, our results were derived from common laboratory strains of S. mansoni and D. melanogaster and can therefore be considered as a reference for future studies.
Schistosoma and Drosophila are important representatives from a phylogenetically diverse group of organisms that do not encode a canonical DNA methyltransferase enzyme (Dnmt1 or Dnmt3), but have retained a Dnmt2 gene. Our results suggest that the genomes of these Dnmt2-only organisms lack DNA methylation, which would significantly expand the number of eukaryotes with unmethylated genomes. A complete absence of DNA methylation was previously assumed for only a small group of organisms, including Saccharomyces cerevisiae and Caenorhabditis elegans, as these models apparently have lost all Dnmt genes and no experimental evidence for genomic DNA methylation has been obtained.
Importantly, the lack of DNA methylation in a considerable number of eukaryotes also suggests that this modification is not essential for the development of these organisms. This is consistent with the notion that DNA methylation has conserved adaptive functions that might be particularly relevant for lineages that are subject to high adaptive pressure (2, 15, 17). When DNA methylation is lost during the evolutionary history of a genome, other epigenetic mechanisms can be used to ensure the plasticity of gene expression programs. Besides chromatin-based mechanisms and small noncoding RNAs, these might also include the Dnmt2-dependent modification of RNAs, which has been implied in a variety of adaptive responses (10, 19, 40, 41).
Materials and Methods
DNA Samples.
For Schistosoma genomic DNA, adult male worms from the S. mansoni Puerto Rican strain were homogenized and DNA was isolated using the Qiagen DNeasy Blood and Tissue kit, according to the manufacturer’s instructions. For Drosophila genomic DNA, 0- to 2-h embryos were collected from w1118 and Dnmt2149 (24) strains. To extract genomic DNA, embryos were homogenized in 2× proteinase K (PK) buffer [200 mM Tris⋅HCl, pH 7.5, 25 mM EDTA, pH 8, 300 mM NaCl, 2% (wt/vol) SDS], followed by protein digestion with proteinase K for 2 h at 65 °C. DNA was extracted with an equal volume of phenol-chloroform, followed by ethanol precipitation. Samples were then treated with RNase A for 15 min at 37 °C and genomic DNA was purified by phenol-chloroform extraction. For human sperm DNA, sperm cells were purified and then sonicated. Genomic DNA was purified by double phenol-chloroform extraction and ethanol precipitation. For mouse TKO ES DNA, cells derived from the Dnmt1−/−; Dnmt3a−/−; Dnmt3b−/− clone 19 (28), kindly provided by Masaki Okano (Center for Developmental Biology, RIKEN, Kobe, Japan), were grown under feeder-free conditions on gelatin in complete ES medium (42). Before harvesting, the cells were stained by immunofluorescence against 5-methylcytosine and Dnmt1 for potential parental cell line contaminations. Genomic DNA was prepared using the Qiagen DNeasy Blood and Tissue kit, according to the manufacturer’s instructions. Additional details are provided in SI Materials and Methods.
Accession Numbers.
Sequencing data have been deposited in the Gene Expression Omnibus database under accession nos. GSE39996 (D. melanogaster, together with human sperm spike-in), GSE39997 (S. mansoni), and GSE42170 (mouse TKO ES cell line, together with M13 PCR fragment spike-in).
Supplementary Material
Acknowledgments
We thank Stephan Wolf, Andre Leischwitz, and Kristina Tabbada for the Illumina sequencing services; Felix Krueger for bioinformatics support; and Tobias Reber for information technology support. This work was supported by grants from the Biotechnology and Biological Sciences Research Council (BBSRC), the Medical Research Council, and the Wellcome Trust (to W.R.); the National Institutes of Health (NIH) (5R01GM062534-12 to G.J.H.); and the Deutsche Forschungsgemeinschaft (FOR1082 to M.S. and F.L.). P.M.G. is an NIH trainee on Cold Spring Harbor Laboratory Watson School of Biological Sciences NIH Kirschstein–National Research Service Award predoctoral Award T32 GM065094, a William Randolph Hearst Scholar, and a Leslie Quick Junior Fellow. N.O. is a recipient of a BBSRC Collaborative Awards in Science and Engineering studentship and a Babraham Institute/Cambridge European Trust studentship.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE39996, GSE39997, and GSE42170).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1306723110/-/DCSupplemental.
References
- 1.Mohn F, Schübeler D. Genetics and epigenetics: Stability and plasticity during cellular differentiation. Trends Genet. 2009;25(3):129–136. doi: 10.1016/j.tig.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 2.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447(7143):433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 3.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 4.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11(3):204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoder JA, Bestor TH. A candidate mammalian DNA methyltransferase related to pmt1p of fission yeast. Hum Mol Genet. 1998;7(2):279–284. doi: 10.1093/hmg/7.2.279. [DOI] [PubMed] [Google Scholar]
- 6.Okano M, Xie S, Li E. Dnmt2 is not required for de novo and maintenance methylation of viral DNA in embryonic stem cells. Nucleic Acids Res. 1998;26(11):2536–2540. doi: 10.1093/nar/26.11.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong A, et al. Structure of human DNMT2, an enigmatic DNA methyltransferase homolog that displays denaturant-resistant binding to DNA. Nucleic Acids Res. 2001;29(2):439–448. doi: 10.1093/nar/29.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hermann A, Schmitt S, Jeltsch A. The human Dnmt2 has residual DNA-(cytosine-C5) methyltransferase activity. J Biol Chem. 2003;278(34):31717–31721. doi: 10.1074/jbc.M305448200. [DOI] [PubMed] [Google Scholar]
- 9.Goll MG, et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311(5759):395–398. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- 10.Schaefer M, et al. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24(15):1590–1595. doi: 10.1101/gad.586710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jurkowski TP, et al. Human DNMT2 methylates tRNA(Asp) molecules using a DNA methyltransferase-like catalytic mechanism. RNA. 2008;14(8):1663–1670. doi: 10.1261/rna.970408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sunita S, et al. Crystal structure of the Escherichia coli 23S rRNA:m5C methyltransferase RlmI (YccW) reveals evolutionary links between RNA modification enzymes. J Mol Biol. 2008;383(3):652–666. doi: 10.1016/j.jmb.2008.08.062. [DOI] [PubMed] [Google Scholar]
- 13.Iyer LM, Abhiman S, Aravind L. Natural history of eukaryotic DNA methylation systems. Prog Mol Biol Transl Sci. 2011;101:25–104. doi: 10.1016/B978-0-12-387685-0.00002-0. [DOI] [PubMed] [Google Scholar]
- 14.Jurkowski TP, Jeltsch A. On the evolutionary origin of eukaryotic DNA methyltransferases and Dnmt2. PLoS ONE. 2011;6(11):e28104. doi: 10.1371/journal.pone.0028104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zemach A, Zilberman D. Evolution of eukaryotic DNA methylation and the pursuit of safer sex. Curr Biol. 2010;20(17):R780–R785. doi: 10.1016/j.cub.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 16.Werren JH, et al. Nasonia Genome Working Group Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science. 2010;327(5963):343–348. doi: 10.1126/science.1178028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyko F, Maleszka R. Insects as innovative models for functional studies of DNA methylation. Trends Genet. 2011;27(4):127–131. doi: 10.1016/j.tig.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 18.Tuorto F, et al. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol. 2012;19(9):900–905. doi: 10.1038/nsmb.2357. [DOI] [PubMed] [Google Scholar]
- 19.Becker M, et al. Pmt1, a Dnmt2 homolog in Schizosaccharomyces pombe, mediates tRNA methylation in response to nutrient signaling. Nucleic Acids Res. 2012;40(22):11648–11658. doi: 10.1093/nar/gks956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeltsch A, Nellen W, Lyko F. Two substrates are better than one: Dual specificities for Dnmt2 methyltransferases. Trends Biochem Sci. 2006;31(6):306–308. doi: 10.1016/j.tibs.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 21.Schaefer M, Lyko F. Solving the Dnmt2 enigma. Chromosoma. 2010;119(1):35–40. doi: 10.1007/s00412-009-0240-6. [DOI] [PubMed] [Google Scholar]
- 22.Krauss V, Reuter G. DNA methylation in Drosophila—a critical evaluation. Prog Mol Biol Transl Sci. 2011;101:177–191. doi: 10.1016/B978-0-12-387685-0.00003-2. [DOI] [PubMed] [Google Scholar]
- 23.Geyer KK, et al. Cytosine methylation regulates oviposition in the pathogenic blood fluke Schistosoma mansoni. Nat Commun. 2011;2:424. doi: 10.1038/ncomms1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phalke S, et al. Retrotransposon silencing and telomere integrity in somatic cells of Drosophila depends on the cytosine-5 methyltransferase DNMT2. Nat Genet. 2009;41(6):696–702. doi: 10.1038/ng.360. [DOI] [PubMed] [Google Scholar]
- 25.Lister R, Ecker JR. Finding the fifth base: Genome-wide sequencing of cytosine methylation. Genome Res. 2009;19(6):959–966. doi: 10.1101/gr.083451.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng S, et al. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA. 2010;107(19):8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328(5980):916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 28.Tsumura A, et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells. 2006;11(7):805–814. doi: 10.1111/j.1365-2443.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- 29.Xi Y, Li W. BSMAP: Whole genome bisulfite sequence MAPping program. BMC Bioinformatics. 2009;10:232. doi: 10.1186/1471-2105-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lyko F, et al. The honey bee epigenomes: Differential methylation of brain DNA in queens and workers. PLoS Biol. 2010;8(11):e1000506. doi: 10.1371/journal.pbio.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warnecke PM, et al. Identification and resolution of artifacts in bisulfite sequencing. Methods. 2002;27(2):101–107. doi: 10.1016/s1046-2023(02)00060-9. [DOI] [PubMed] [Google Scholar]
- 32.Molaro A, et al. Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell. 2011;146(6):1029–1041. doi: 10.1016/j.cell.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stadler MB, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480(7378):490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 34.Ramsahoye BH, et al. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci USA. 2000;97(10):5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gowher H, Leismann O, Jeltsch A. DNA of Drosophila melanogaster contains 5-methylcytosine. EMBO J. 2000;19(24):6918–6923. doi: 10.1093/emboj/19.24.6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lyko F, Ramsahoye BH, Jaenisch R. DNA methylation in Drosophila melanogaster. Nature. 2000;408(6812):538–540. doi: 10.1038/35046205. [DOI] [PubMed] [Google Scholar]
- 38.Kunert N, Marhold J, Stanke J, Stach D, Lyko F. A Dnmt2-like protein mediates DNA methylation in Drosophila. Development. 2003;130(21):5083–5090. doi: 10.1242/dev.00716. [DOI] [PubMed] [Google Scholar]
- 39.Warnecke PM, et al. Detection and measurement of PCR bias in quantitative methylation analysis of bisulphite-treated DNA. Nucleic Acids Res. 1997;25(21):4422–4426. doi: 10.1093/nar/25.21.4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thiagarajan D, Dev RR, Khosla S. The DNA methyltranferase Dnmt2 participates in RNA processing during cellular stress. Epigenetics. 2011;6(1):103–113. doi: 10.4161/epi.6.1.13418. [DOI] [PubMed] [Google Scholar]
- 41.Durdevic Z, et al. Efficient RNA virus control in Drosophila requires the RNA methyltransferase Dnmt2. EMBO Rep. 2013;14(3):269–275. doi: 10.1038/embor.2013.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ficz G, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473(7347):398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.