Abstract
Among 594 Streptococcus pneumoniae serotype 19A invasive pneumococcal disease (IPD) isolates collected from 1993 to 2011, we identified 85 sequence types by multilocus sequence typing. CC320 was associated with multidrug resistance and reduced susceptibility to penicillin and ceftriaxone and still predominated among declining serotype 19A IPD isolates following PCV13 introduction.
TEXT
The epidemiology of Streptococcus pneumoniae is constantly changing because of normal fluctuations and the selective pressure caused by antibiotic and vaccine use. Serotype 19A, which is not included in the seven-valent pneumococcal conjugate vaccine (PCV7), emerged as the most prevalent serotype in several studies from the United States and was associated with multidrug resistance (1–4). Pediatric invasive pneumococcal disease (IPD) caused by serotype 19A has been reported in European and Asian countries (5–7). Serotype 19A has a propensity to cause mastoiditis (2), and mastoiditis caused by serotype 19A infections was more likely to require intraoperative mastoidectomy than mastoiditis caused by non-19A infections (8). Serotype 19A was also common among nasopharyngeal and noninvasive disease isolates in some studies (9, 10).
The United States Pediatric Multicenter Pneumococcal Surveillance Group has continuously identified patients and collected isolates from all cases of IPD at eight children's hospitals since 1993. The present study investigated all of the serotype 19A IPD isolates obtained from 1993 to 2011, with a focus on disease presentation, genotypes as determined by multilocus sequence typing (MLST), and antimicrobial susceptibility patterns. This study was approved by the institutional review boards of all of the participating institutions. Patients with serotype 19A IPD were identified from a surveillance database. DNA was isolated from the corresponding isolates and MLST performed as described previously (11). Sequence types (STs) were compared by using eBURST V3 (www.mlst.net). Susceptibilities to the antimicrobials penicillin and ceftriaxone were determined by broth microdilution assay and susceptibilities to clindamycin, erythromycin, and trimethoprim-sulfamethoxazole (TMP-SMX) were determined by the Kirby-Bauer test (12). Isolates were divided into four groups: CC199, CC320, CC172, and other STs. Each ST group was compared to all of the other ST groups by using STATA11 (StataCorp LP, College Station, TX). Fisher's exact test was employed for dichotomous variables, and the Kolmogorov-Smirnov test was used to analyze continuous variables. Median penicillin and ceftriaxone MICs were compared between groups by using a K sample equality-of-medians test.
(These results were presented in part at the Infectious Diseases Society of America Annual Meeting, Philadelphia, PA, October 2010, and at ID Week 2012, San Diego, CA.)
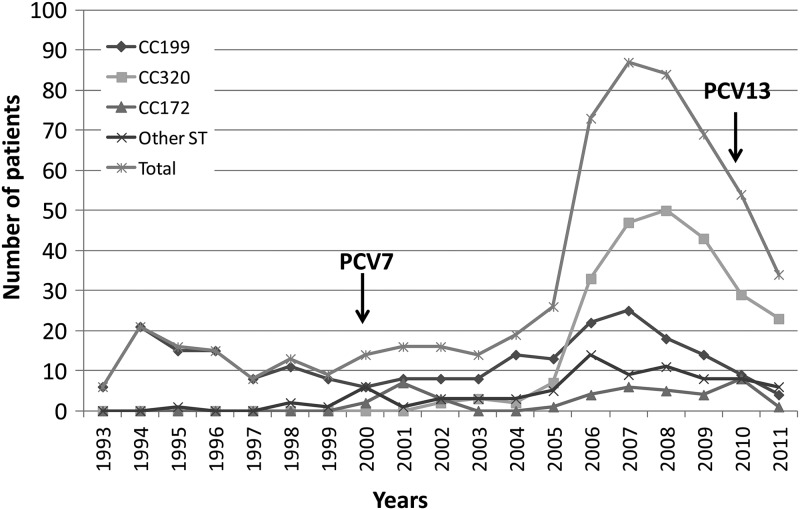
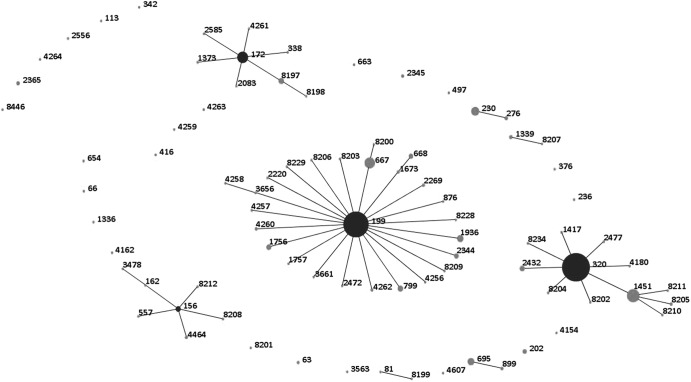
A total of 594 invasive serotype 19A isolates from patients with completed case report forms from 1993 to 2011 were analyzed. Patient demographics and the strain collection per study site are presented in Table 1. From 1993 to 2005, fewer than 30 serotype 19A isolates were obtained per year, ranging from 6 to 26 isolates annually (Fig. 1). A peak was reached in 2007, after which the annual number of invasive serotype 19A isolates declined. A total of 85 STs were identified, 55 of which were previously known. Clonal clusters included CC156 (n = 13), CC172 (n = 41), CC199 (n = 233), CC230 (n = 19), CC320 (n = 239), and CC695 (n = 13) (Fig. 2). For subsequent comparisons, isolates were divided into four major ST groups: CC199 (n = 233, 39.2%), CC320 (n = 239, 40.2%), CC172 (n = 41, 6.9%), and other STs (n = 81, 13.6%). CC199 isolates were observed throughout the study period and predominated among serotype 19A isolates through 2005, similar to previous reports (4). The genotypic variation increased over time, and in accordance with other studies, the increase in serotype 19A was associated with the introduction of multidrug-resistant ST320 and related isolates (1, 4, 13, 14). The first isolates of CC320 were observed in 2002, and by 2006, 45% of the serotype 19A isolates belonged to this cluster. The reason for the introduction and expansion of this multidrug resistance genotype is unclear; it is likely that the introduction of PCV7 in combination with antibiotic pressure contributed to the opportunity for ST320 to emerge in the United States. In South Korea, multiresistant serotype 19AST320 emerged prior to the introduction of PCV7, and antimicrobial drug use was thought to be the possible cause of the increase (7). The potential genetic causes of the selective advantage of serotype 19AST320 was investigated by Pillai et al., who compared the whole genome sequence of a multidrug resistant serotype 19AST320 isolate to that of a multidrug-resistant serotype 19FST320 isolate and found unique single-nucleotide polymorphisms related to serotype 19AST320; 61 of these were found within antimicrobial resistance genes (14). Contrary to the experience of the United States and several other countries, Norway, a country with a prevailing strict antibiotic prescription practice, experienced a postvaccination increase of serotype 19A driven by the expansion of CC199 isolates (15). Thus, it appears that antibiotic pressure may be the most important, but not the only, factor in the successful establishment of serotype 19AST320 in many parts of the world.
Table 1.
Patient and isolate details per clonal cluster group
| Parameter | All (n = 594) | CC199 (n = 233) | CC320 (n = 239) | CC172 (n = 41) | Other STs (n = 81) | P value |
|---|---|---|---|---|---|---|
| Median age in mo (range) | 15.0 (0–250.5)a | 15.5 (0–250.5) | 15.3 (0.2–186.7) | 8.4 (0.32–121.2) | 16.4 (0.23–218.5)a | <0.002d |
| Race | ||||||
| Black | 167 (28.4)h | 66 (28.8) | 47 (19.8) | 26 (63.4) | 28 (34.6) | |
| Caucasian | 259 (44.1) | 81 (35.4) | 141 (59.0) | 11 (26.8) | 26 (32.1) | <0.0001e |
| Hispanic | 96 (16.3) | 56 (24.5) | 26 (11.0) | 1 (2.4) | 13 (16.1) | |
| Other/unknown | 72 (12.1) | 30 (11.4) | 25 (9.7) | 3 (7.3) | 14 (17.3) | |
| No. (%) of males | 337 (57.0)c | 138 (59.2) | 123 (51.9)b | 29(72.5)a | 47/81 (58.0) | 0.07 |
| Diagnosis | ||||||
| Meningitis | 58 (9.8) | 25 (10.7) | 21 (8.8) | 3 (7.3) | 9 (11.1) | |
| Bacteremia | 198 (33.3) | 83 (35.6) | 58 (24.3) | 20 (48.8) | 37 (45.7) | |
| Pneumonia | 214 (36.0) | 86 (36.9) | 88 (36.8) | 14 (34.2) | 26 (32.1) | |
| Bone and joint infections | 24 (4.0) | 9 (3.9) | 10 (4.2) | 2 (4.9) | 3 (3.7) | |
| Mastoiditis | 74 (12.5) | 19 (8.2) | 50 (20.9) | 1 (2.4) | 4 (4.9) | <0.0001f |
| Cellulitis/abscess | 17 (2.9) | 7 (3.0) | 9 (3.8) | 1 (2.4) | 0 | |
| Other | 9 (1.5) | 4 (1.7) | 3 (1.3) | 0 | 2 (2.5) | |
| Center | ||||||
| Chicago | 38 (6.4) | 16/38 (42.1) | 19/38 (50.0) | 0 | 3/38 (7.9) | |
| Columbus | 98 (16.5) | 30/98 (30.6) | 54/98 (55.1) | 0 | 14/98 (14.3) | |
| Houston | 182 (30.6) | 83/182 (45.6) | 71/182 (39.0) | 3/182 (1.6) | 25/182 (13.7) | |
| Little Rock | 90 (15.2) | 20/90 (22.2) | 32/90 (35.6) | 32/90 (35.6) | 6/90 (6.7) | <0.0001g |
| Los Angeles | 22 (3.7) | 13/22(59.1) | 4/22 (18.2) | 1/22 (4.5) | 4/22 (1.8) | |
| Pittsburgh | 69 (11.6) | 19/69 (27.5) | 33/69 (47.8) | 4/69 (5.8) | 16/69 (23.2) | |
| San Diego | 54 (9.1) | 31/54 (57.4) | 12/54 (22.2) | 1/54 (1.9) | 10/54 (18.5) | |
| Winston-Salem | 41 (6.9) | 21/41 (51.2) | 14/41 (34.1) | 0 | 6/41 (14.6) |
One missing data point.
Two missing data points.
Three missing data points.
Patients with CC172 isolates (8.4 [range, 0.32 to 121.2] months) versus patients with other isolates (15.4 [range, 0 to 250.5] months) (Kolmogorov-Smirnov test).
Caucasian patients with CC320 isolates (141/239, 59.0%) versus other isolates (118/355, 33.2%) (Fisher's exact test).
Patients with mastoiditis caused by CC320 isolates (50/239, 20.9%) versus other isolates (24/355, 6.8%) (Fisher's exact test).
CC172 isolates obtained from patients from Arkansas Children's Hospital, Little Rock (32/90, 35.6%), versus other patients (9/504, 1.8%) (Fisher's exact test).
Unless noted otherwise, all values are number (percentages) of patients.
Fig 1.
Number of serotype 19A isolates per year for the period 1993 to 2011, in total and for the most common clonal clusters. The introductions of pneumococcal conjugate vaccines PCV7 and PCV13 are indicated.
Fig 2.
eBURST of 594 serotype 19A IPD isolates obtained from 1993 to 2011.
Differences in the distribution of STs among the eight children's hospitals in this study were observed. In particular, the majority of the CC172 isolates, including a new single locus variant (SLV) of 172 (ST8197), was obtained from the Arkansas Children's Hospital (P < 0.0001). CC199 and CC320 isolates were obtained from all of the centers and represented 22 to 59% and 18 to 55%, respectively, of the isolates obtained from the individual centers (Table 1). Compared to all of the other isolates, CC320 was associated with the Caucasian race (59.0 versus 33.2%, P < 0.0001). Patients with CC172 tended to be younger than patients with other isolates, with a median age of 8.4 months (range, 0.32 to 121.2 months) compared to 15.4 months (range, 0 to 250.5 months) for all of the other isolates (Kolmogorov-Smirnov test, P = 0.002).
Disease presentations included pneumonia (n = 214), bacteremia (n = 198), mastoiditis (n = 74), meningitis (n = 58), bone and joint infections (n = 24), and cellulitis/abscess (n = 17) (Table 1). Nine other infections included peritonitis (n = 7), ventriculitis (n = 1), and necrotizing fasciitis (n = 1) (Table 1). Disease presentations were evenly distributed among the clonal clusters, with the exception of mastoiditis, which was more commonly associated with CC320 than with other STs (50/239 [20.9%] versus 24/355 [6.8%]; P < 0.0001). In fact, two-thirds of the mastoiditis cases were caused by CC320. Why ST320 has a propensity to cause mastoiditis remains unknown.
In 2011, a total of 34 serotype 19A isolates were identified that belonged to clonal clusters CC199 (2 ST199 and 2 ST667) and CC320 (17 ST320, 4 ST1451, 1 ST2432 and 1 8205) and 6 other groups (1 ST156, 1 CC172, 2 ST230, 1 ST276, 1 ST695, and 1 ST8446). Disease presentations included pneumonia (n = 15), bacteremia (n = 7), meningitis (n = 6), peritonitis (n = 3), bone and joint infections (n = 2), and mastoiditis (n = 1).
Antimicrobial susceptibility patterns differed between clonal clusters (Table 2; see Fig. S1 and S2 in the supplemental material). Thirty (51.7%) of 58 meningitis isolates had penicillin MICs of >0.06 μg/ml, and 15 (25.9%) had ceftriaxone MICs of >0.5 μg/ml, meeting the current CLSI criteria for antibiotic nonsusceptibility. Among the serotype 19A isolates from 2011, the median MIC of penicillin (range) was 1 μg/ml (0.008 to 2 μg/ml) and that of ceftriaxone was 0.5 μg/ml (0.008 to 2 μg/ml).
Table 2.
Antimicrobial susceptibility per clonal cluster group
| Antibiotic susceptibility | All (n = 594) | CC199 (n = 233) | CC320 (n = 239) | CC172 (n = 41) | Other ST (n = 81) |
|---|---|---|---|---|---|
| Median MIC (range) of: | |||||
| Penicillin | 0.25 (0.008–8) | 0.03 (0.008–4) | 2 (0.008–8) | 0.25 (0.008–0.5) | 0.125 (0.008–4) |
| Ceftriaxone | 0.125 (0.008–8)a | 0.015 (0.008–1)a | 1 (0.008–8) | 0.06 (0.008–1) | 0.06 (0.008–4) |
| No. (%) of isolates nonsusceptible to: | |||||
| Erythromycin | 333 (56.1)a | 64 (27.6)a | 234 (96.7) | 4 (9.8) | 31 (38.3) |
| Clindamycin | 242 (40.1)a | 3 (1.3)a | 225 (94.1) | 1 (2.4) | 13 (16.0) |
| TMP/SMX | 440 (74.7)a | 121 (52.2)a | 235 (98.3) | 29 (70.7) | 55 (67.9) |
One missing data point.
When a K sample equality-of-medians test was used to compare the distribution of MICs for each group to the median MIC for all of the samples (1 μg/ml), significantly more isolates of CC320 had MICs greater than the median than those of the other clonal clusters (P < 0.0001). Furthermore, comparisons to all of the other isolates showed CC320 isolates to have greater nonsusceptibility to erythromycin (234/239 [97.9%] versus 99/354 [28.0%]; P < 0.0001), clindamycin (225/239 [94.1%] versus 17/354 [4.8%]; P < 0.0001), and TMP-SMX (235/239 [98.3%] versus 205/354 [57.9%]; P < 0.0001).
Among the smaller clonal clusters observed, CC230 (ST230 and ST276) comprised 19 isolates from five of the participating hospitals, and approximately half of these isolates were obtained since 2009 with 3 isolates from 2011. CC230 was not usually associated with the increased MICs observed for CC320; one CC230 meningitis isolate was nonsusceptible to all five of the antibiotics tested. ST230 has been associated with other serotypes (14, 24F, and 23F) in addition to serotype 19A, while all of the ST276 isolates in the MLST database uniformly have been of serotype 19A. A French study reported ST276 as the emerging serotype 19A ST since the introduction of PCV7, causing 70/95 invasive and noninvasive infections (16). CC81 and CC156 include two STs previously associated with multidrug resistance, ST81 (PMEN1) and ST156 (PMEN3), that have both been associated with several different serotypes in the past (17). We found isolates of CC81 and CC156 to have reduced susceptibility to antibiotics (Table 2).
The increasing number of worldwide reports containing MLST data provide insight into certain STs' success in survival and pathogenesis, regardless of capsule production. In our study, disease-causing serotype 19A isolates declined in number prior to the introduction of PCV13, and this decline has continued. CC320 remains the dominant ST among serotype 19A isolates recovered from children since the introduction of PCV13. Since it is too early to predict the future serotype replacements and potential vaccine escape serotypes after the introduction of PCV13, further surveillance is necessary to determine the effectiveness of this new vaccine and whether capsule switches will allow genetically fit STs to emerge as important causes of nonvaccine serotype IPD.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by a grant from Pfizer Inc. The company was not involved in the study design, execution, or analysis or in the composition of the manuscript.
We acknowledge the use of the pneumococcal MLST database, which is located at Imperial College London and is funded by the Wellcome Trust. We thank Cynthia Bishop at Imperial College for her assistance with uploading the isolates to the database and assigning new STs. We also thank J. Chase McNeil for technical assistance.
Footnotes
Published ahead of print 6 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00058-13.
REFERENCES
- 1. Moore M, Gertz RE, Woodbury RL, Barkocy-Gallagher GA, Schaffner W, Lexau C, Gershman K, Reingold A, Farley M, Harrison LH, Hadler JL, Bennett NM, Thomas AR, McGee L, Pilishvili T, Brueggemann AB, Whitney CG, Jorgensen JH, Beall B. 2008. Population snapshot of emergent Streptococcus pneumoniae serotype 19A in the United States, 2005. J. Infect. Dis. 197:1016–1027 [DOI] [PubMed] [Google Scholar]
- 2. Kaplan SL, Barson WJ, Lin PL, Stovall SH, Bradley JS, Tan TQ, Hoffman JA, Givner LB, Mason EO., Jr 2010. Serotype 19A is the most common serotype causing invasive pneumococcal infections in children. Pediatrics 125:429–436 [DOI] [PubMed] [Google Scholar]
- 3. Pelton SI, Huot H, Finkelstein JA, Bishop CJ, Hsu KK, Kellenberg J, Huang SS, Goldstein R, Hanage WP. 2007. Emergence of 19A as virulent and multidrug resistant Pneumococcus in Massachusetts following universal immunization of infants with pneumococcal conjugate vaccine. Pediatr. Infect. Dis. J. 26:468–472 [DOI] [PubMed] [Google Scholar]
- 4. Pai R, Moore MR, Pilishvili T, Gertz RE, Whitney CG, Beall B. 2005. Postvaccine genetic structure of Streptococcus pneumoniae serotype 19A from children in the United States. J. Infect. Dis. 192:1988–1995 [DOI] [PubMed] [Google Scholar]
- 5. Riva E, Salvini F, Garlaschi ML, Radaelli G, Giovannini M. 2012. The status of invasive pneumococcal disease among children younger than 5 years of age in north-west Lombardy, Italy. BMC Infect. Dis. 12:106 doi:10.1186/1471-2334-12-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shin J, Baek JY, Kim SH, Song JH, Ko KS. 2011. Predominance of ST320 among Streptococcus pneumoniae serotype 19A isolates from 10 Asian countries. J. Antimicrob. Chemother. 66:1001–1004 [DOI] [PubMed] [Google Scholar]
- 7. Choi EH, Kim SH, Eun BW, Kim SJ, Kim NH, Lee J, Lee HJ. 2008. Streptococcus pneumoniae serotype 19A in children, South Korea. Emerg. Infect. Dis. 14:275–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ongkasuwan J, Valdez TA, Hulten KG, Mason EO, Jr, Kaplan SL. 2008. Pneumococcal mastoiditis in children and the emergence of multidrug-resistant serotype 19A isolates. Pediatrics 122:34–39 [DOI] [PubMed] [Google Scholar]
- 9. McNeil JC, Hulten KG, Mason EO, Jr, Kaplan SL. 2009. Serotype 19A is the most common Streptococcus pneumoniae isolate in children with chronic sinusitis. Pediatr. Infect. Dis. J. 28:766–768 [DOI] [PubMed] [Google Scholar]
- 10. Xu Q, Pichichero ME, Casey JR, Zeng M. 2009. Novel type of Streptococcus pneumoniae causing multidrug-resistant acute otitis media in children. Emerg. Infect. Dis. 15:547–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Enright MC, Spratt BG. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049–3060 [DOI] [PubMed] [Google Scholar]
- 12. Clinical and Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing; 21st informational supplement, CLSI document M100-S21. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 13. Beall BW, Gertz RE, Hulkower RL, Whitney CG, Moore MR, Brueggemann AB. 2011. Shifting genetic structure of invasive serotype 19A pneumococci in the United States. J. Infect. Dis. 203:1360–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pillai DR, Shahinas D, Buzina A, Pollock RA, Lau R, Khairnar K, Wong A, Farrell DJ, Green K, McGeer A, Low DE. 2009. Genome-wide dissection of globally emergent multi-drug resistant serotype 19A Streptococcus pneumoniae. BMC Genomics 10:642 doi:10.1186/1471-2164-10-642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vestrheim DF, Steinbakk M, Aaberge IS, Caugant DA. 2012. Postvaccination increase in serotype 19A pneumococcal disease in Norway is driven by expansion of penicillin-susceptible strains of the ST199 complex. Clin. Vaccine Immunol. 19:443–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mahjoub-Messai F, Doit C, Koeck JL, Billard T, Evrard B, Bidet P, Hubans C, Raymond J, Levy C, Cohen R, Bingen E. 2009. Population snapshot of Streptococcus pneumoniae serotype 19A isolates before and after introduction of seven-valent pneumococcal vaccination for French children. J. Clin. Microbiol. 47:837–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hiller NL, Eutsey RA, Powell E, Earl JP, Janto B, Martin DP, Dawid S, Ahmed A, Longwell MJ, Dahlgren ME, Ezzo S, Tettelin H, Daugherty SC, Mitchell TJ, Hillman TA, Buchinsky FJ, Tomasz A, de Lencastre H, Sa-Leao R, Post JC, Hu FC, Ehrlich GD. 2011. Differences in genotype and virulence among four multidrug-resistant Streptococcus pneumoniae isolates belonging to the PMEN1 clone. PLoS One 6:e28850 doi:10.1371/journal.pone.0028850 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.