Abstract
In order to assess the frequency of clinically relevant linezolid-resistant staphylococcal isolates, and the role of linezolid in maintaining and coselecting multiple resistance mechanisms (cfr, 23S rRNA, L3/L4 mutations), a prospective Italian study was performed from 2010 to 2011 to confirm the diffusion of three major multidrug-resistant clones (ST2, ST5, ST23).
TEXT
Linezolid-resistant strains appeared soon after the approval of linezolid (1, 2) and were first associated with mutations in domain V of the 23S rRNA genes (G2576T); over time, a variety of mutations in this region have been identified (3), and, to date, they remain the most commonly reported class of mutations affecting linezolid. Recently, mutations in L3 and L4 ribosomal proteins have been described in linezolid-resistant staphylococci (3, 4). The only known transferable mechanism of linezolid resistance is conferred by the methyltransferase mechanism codified by the cfr gene, acquired horizontally from strains of veterinary origin (5). Linezolid resistance in staphylococci is still a rare epidemiological phenomenon. Various authors have documented the onset of this resistance in numerous strains of Staphylococcus epidermidis but less frequently in Staphylococcus aureus, often due to outbreaks and cross-transmission (6) or clonal expansion (7, 8).
We have already described the first appearance of linezolid resistance in coagulase-negative staphylococci (CoNS) in different clones in Italy from 2008 to 2009 (9). This new study increases our previous knowledge on the nature of linezolid resistance and emphasizes the leading role of linezolid in maintaining and, especially, coselecting multiple mechanisms of resistance in only two more years of clinical use.
In the period from January 2010 to December 2011, 50 clinically relevant linezolid-resistant staphylococci were collected from 7 clinical institutions in Italy. Forty-seven strains were from bloodstream infection (BSI), and 3 were from cerebrospinal fluid (A. Vena, M. Falcone, E. Comandini, M. Meledandri, A. Novelli, F. Campanile, S. Stefani, M. Venditti, submitted for publication). Forty-five strains were Staphylococcus epidermidis, 3 were Staphylococcus hominis, 1 was Staphylococcus capitis, and 1 was Staphylococcus aureus. All strains were tested for their susceptibility profiles, in accordance with Clinical and Laboratory Standards Institute 2012 guidelines (10) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines (11).
Molecular analysis, oligonucleotide sequences, and PCR conditions used were described elsewhere (9). Sequence alignments were performed by using BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) and UniProt programs (http://www.uniprot.org/blast/uniprot/) (UniProtKB/Swiss-Prot database). A reconstruction of the peptidyl transferase center (PTC) center and the L3 and L4 riboproteins was performed with the Swiss-Model ExPASy program (http://swissmodel.expasy.org/workspace).
The molecular typing was performed by pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST), as previously described (9), and the eBURST version 3 program (www.sepidermidis.mlst.net/eburst) was used to elaborate these data.
Our results pointed out that all strains were selected mainly under antibiotic pressure, including linezolid: in fact, 31 out of the 50 patients (62%) in which a linezolid-resistant strain was isolated had received this drug as previous treatment, together with other antimicrobial agents. One important point is that, due the coexistence of different genes and mechanisms, this resistance can be easily maintained by resistant strains even in the absence of specific pressure.
All linezolid-resistant isolates were methicillin resistant and resistant to erythromycin and lincomycin (89 and 100%, respectively); they showed a multidrug-resistant (MDR) profile, including resistance to levofloxacin (100%), gentamicin (75.5%), and cotrimoxazole (55.5%). With respect to glycopeptides, the MIC90 was 4 mg/liter for vancomycin and 16 mg/liter for teicoplanin; 57.7% of strains had MICs of >4 mg/liter and thus were resistant to teicoplanin. All strains were fully susceptible to daptomycin (MIC90 of 0.25 mg/liter) and quinupristin-dalfopristin (MIC90 of 2 mg/liter).
Table 1 shows the phenotypic and genotypic characterization of the sample in the study, subdivided by their mechanisms of linezolid resistance. In our Italian scenario, 9 presumptive mechanisms of resistance were found as a single mechanism or due to a variable combination; only 1 had no apparently specific mechanism of resistance.
Table 1.
Subdivision of the 50 linezolid-resistant CoNS strains with respect to the mechanism of linezolid resistance, MICs of related drugs, and resistant gene contenta
| No. of strains: | ST/PFGE type | mec/ccr type | MIC range (mg/liter) |
Gene content (no. of strains with gene/total no.) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| LNZ | E | L | C | ermA | ermC | msrA | cat | |||
| With cfr alone | ||||||||||
| 13 | 23/A1-A2 | B/A2B2 | 8–≥256 | ≥256 | 32–≥256 | 64–256 | 12/13 | 1/13 | 6/13 | 13/13 |
| 2 | 2/C1 | A/A4B4 | 64–≥256 | 1–16 | ≥256 | 64 | 0/2 | 0/2 | 1/2 | 1/2 |
| 1 (Sh) | α | A/A2B2 | 8 | 64 | ≥256 | 64 | 0/1 | 1/1 | 0/1 | 1/1 |
| With cfr and L3/L4 (F147L and L94V in L3; G71D and N158S in L4) | ||||||||||
| 4 | 23/A2 | B/A2B2 | 64 | 128–≥256 | ≥256 | 64 | 4/4 | 0/4 | 4/4 | 4/4 |
| With cfr, 23S rRNA mutations, and L3 (G2447T in 23S; L94V in L3) | ||||||||||
| 2 | 2/C3 | A/A4B4 | ≥256 | 8–32 | ≥256 | 64 | 0/2 | 0/2 | 1/2 | 2/2 |
| With 23S rRNA mutations ± L3 (G2576T in 23S; L94V in L3) | ||||||||||
| 10 | 2/C1-C2- | A/A4B4 | 32–128 | 0.5–256 | 8–32 | 32–128 | 0/10 | 1/10 | 6/10 | 8/10 |
| 2 (Sh) | β | A/NT | 64 | 4 | 32 | 128 | 0/2 | 0/2 | 0/2 | 2/2 |
| 1 (Sa) | 5/USA100 | A/A2B2 | 32 | ≥256 | 2 | 16 | 1/1 | 0/1 | 0/1 | 0/1 |
| 1 (Sc) | NA | A/NT | 32 | 128 | 32 | 32 | 1/1 | 1/1 | 0/1 | 0/1 |
| With 23S rRNA mutations and L3 (G2576T in 23S; G137A and L94V in L3) | ||||||||||
| 2 | 2/C2 | A/A4B4+C | 32 | 2 | 32–≥256 | 64 | 0/2 | 0/2 | 0/2 | 2/2 |
| With 23S rRNA mutations and L3 (G2447T in 23S; L94V in L3) | ||||||||||
| 6 | 2/C3 | A/A4B4 | 64 | 8–32 | 4–16 | 4–8 | 0/6 | 0/6 | 6/6 | 0/6 |
| With L3/L4 (H146Q and L94V in L3; 71GGR72 and N158S in L4) | ||||||||||
| 3 | 5/F | B/A2B2 | 64 | 4 | 1–2 | 16–64 | 0/3 | 0/3 | 0/3 | 2/3 |
| With L3/L4 (F147L and L94V in L3; N158S in L4) | ||||||||||
| 2 | 23/A2 | B/A2B2 | 8–16 | 256 | ≥256 | 64 | 2/2 | 0/2 | 2/2 | 2/2 |
| Without specific linezolid-resistant mechanisms | ||||||||||
| 1 | 23/A1 | B/A2B2 | 32 | ≥256 | 16 | 64 | 1/1 | 0/1 | 0/1 | 1/1 |
LNZ, linezolid; E, erythromycin; L, lincomycin; C, cloramphenicol; Sh, S. hominis; Sc, S. capitis; Sa, S. aureus; NT, not typeable; NA, not applicable.
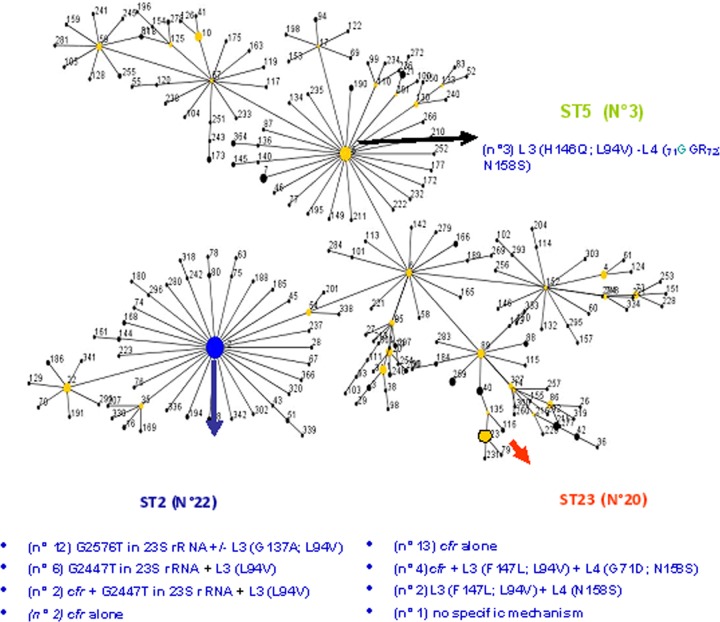
Among the 45 linezolid-resistant S. epidermidis strains, 3 different sequence types (STs) were found, namely, 23 (n = 20 strains), 2 (n = 22 strains), and 5 (n = 3 strains), related to 3 different PFGE types and subtypes, prevalently associated to the same ST. The genetic fingerprinting of some of these strains is closely related to those already characterized in our country (9).
The main mechanism of resistance was associated with the presence of the cfr gene, alone or combined with other mutational mechanisms, in 22 out of the 50 staphylococcal strains (44%) (21 S. epidermidis strains and 1 S. hominis strain), in which it confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilin, and streptogramin A (PhLOPSA); this mechanism was generally associated with higher linezolid MIC values. These results highlight the ability of the plasmid-mediated cfr gene to spread among diverse staphylococcal species and S. epidermidis clones, i.e., ST23/PFGEA1-A2 (17 strains) and ST2/PFGEC1-C3 (4 strains).
The spread of cfr, found mainly in ST23, suggests a role of the genetic background in the acceptance and mobilization of this mechanism, as already found in Italy (9), even if the recent acquisition of the cfr gene in 4 strains belonging to ST2 demonstrates the ability of this clone to acquire antibiotic resistance genes, making these strains more adapted to the hospital settings.
Mutations in domain V of 23S rRNA were similarly distributed among S. epidermidis strains, related to ST2 (n = 18), but also among S. hominis (n = 2), S. aureus (n = 1), and S. capitis (n = 1) strains. In S. epidermidis, PFGE typing was more sensitive than MLST to discriminate among strains carrying G2576T (n = 12 strains, PFGE C1-C2) or G2447T (n = 6 strains, PFGE C3) mutations, confirming the hypothesis that each molecular mechanism is always associated with the same clone. Moreover, this mechanism involving 23S rRNA not only showed high efficiency with regard to linezolid resistance but also caused moderate changes in the mechanism of action of lincosamides, suggesting an association between the two classes, also for this mechanism of resistance. This behavior was observed in various S. epidermidis strains without any other alternative mechanism linked to the presence of erm genes and in 1 S. hominis strain but not in S. aureus, probably due to the partial mutation of rrn operons. The latter strain was a multiple isolate, belonging to the ST5/USA100/staphylococcal cassette chromosome mec element (SCCmec) II clone, which also developed nonsusceptibility to daptomycin and the hVISA phenotype under clinical therapy.
In our study, 3 strains belonging to ST5/PFGE F were never associated to 23S rRNA mutations nor to the presence of the cfr gene but only to the H146Q substitution in L3 and, above all, the 71GGR72 insertion in L4; these results were also reported by Mendes et al. (8), hypothesizing that these features can compensate the emergence of other mechanisms of resistance, thus conferring a new fitness to this lineage. Moreover, it represents the only mechanism of resistance specifically directed toward oxazolidinones, as shown by linezolid MIC values of 64 mg/liter and full susceptibility to macrolides and low-level resistance to lincosamides.
In this study, several mutations in L3 and L4 riboproteins have been identified: some changes may have spontaneously appeared with no significant effects, while others are probably related to linezolid resistance: in fact, several of these L3 and L4 mutations have been identified interacting in the proximity of the PTC (12).
As concerns the L3 sequences (rplC) in 29 S. epidermidis strains, we found a silent mutation (L94V or L101V when aligned, respectively, with S. epidermidis ATCC 35984/RP62A or S. epidermidis ATCC 12228), which was not expected to influence linezolid resistance.
The F147L mutation associated with the cfr gene (n = 4 strains) or with other L3/L4 mutations (n = 2 strains) already found in the same species (13) and the H146Q mutation (n = 3 strains), always associated to the 71GGR72 insertion in L4 and other L3/L4 mutations, were of particular significance, given the high number of interactions that these portions of both proteins have with 23S rRNA.
As concerns the G71D and N158S mutations in the L4 protein (rplD), the former had already been found in Clostridium perfringens (14) and the latter in a strain of S. epidermidis (3).
In 2 S. epidermidis strains, we identified an additional mutation, G137A in the L3 ribosomal protein. A reconstruction performed with the Swiss-Model ExPASy program demonstrated that it resides close to the PTC, interfering with the linezolid binding site.
In addition, all strains were ermB, vga, vgb, vat, and vatB negative and did not possess any mutation in the L22 ribosomal protein (rplV).
We also analyzed the genetic relationships among STs 23, 2, and 5, detecting their correspondence to three different subclusters of CC2, as previously stated by Miragaia et al. (15) and revaluated by eBurst (Fig. 1). The spread of ST23, ST2, and ST5—clinically relevant MDR S. epidermidis clones—is important, as they are isolated worldwide but with geographical differences. ST2 has already been found in Europe (16), in Australia (17), in Brazil (18), in China (19), in Finland (20), and in the United States (7, 8); ST5 and ST23 have been described in the United States, in Brazil, and in Italy (7–9, 18).
Fig 1.
Snapshot of our isolates using eBurst version 3.
In conclusion, the use of linezolid together with many other antimicrobials as therapy preceding the staphylococcal recovery demonstrates the role of antibiotic pressure in maintaining resistant clones in the hospital environment. Paramount importance has to be given to infection control measures and continued surveillance, in order to preserve the clinical usefulness of linezolid and of the oxazolidinone class in development.
ACKNOWLEDGMENTS
This study was partially funded by the Italian Minister of Research MIUR protocol no. 20087SM5HM by S.S.
The authors wish to thank Antony Bridgewood for the language revision.
Stefania Stefani is on the board of speakers of Pfizer, Novartis Pharma, Astra Zeneca, DMG Italy, and she received research grants from the same company. Carlo Tascini and Francesco Menichetti, in the past two years, have been paid for talks on behalf of Pfizer, Merck Sharp and Dhome, Astellas, and Novartis Pharma. Antonella Repetto is on the board of speakers of Pfizer, Novartis, Merck Sharp and Dhome, and Angelini, receiving a money bonus for her work.
Footnotes
Published ahead of print 23 January 2013
REFERENCES
- 1. Gonzales RD, Schreckenberger PC, Graham MB, Kelkar S, DenBesten K, Quinn JP. 2001. Infections due to vancomycin-resistant Enterococcus faecium resistant to linezolid. Lancet 357:1179. [DOI] [PubMed] [Google Scholar]
- 2. Tsiodras S, Gold HS, Sakoulas G, Eliopoulos GM, Wennersten C, Venkataraman L, Moellering RC, Ferraro MJ. 2001. Linezolid resistance in clinical isolate of Staphylococcus aureus. Lancet 38:207–208 [DOI] [PubMed] [Google Scholar]
- 3. Stefani S, Bongiorno D, Mongelli G, Campanile F. 2010. Linezolid resistance in staphylococci. Pharmaceuticals 3:1988–2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Long KS, Vester B. 2012. Resistance to linezolid caused by modifications at its binding site on the ribosome. Antimicrob. Agents Chemother. 56:603–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schwarz S, Werckenthin C, Kehrenberg C. 2000. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob. Agents Chemother. 44:2530–2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Seral C, Sáenz Y, Algarate S, Duran E, Luque P, Torres C, Castillo FJ. 2011. Nosocomial outbreak of methicillin- and linezolid-resistant Staphylococcus epidermidis associated with catheter-related infections in intensive care unit patients. Int. J. Med. Microbiol. 301:354–358 [DOI] [PubMed] [Google Scholar]
- 7. Wong A, Reddy SP, Smyth DS, Aguero-Rosenfeld ME, Sakoulas G, Robinson DA. 2010. Polyphyletic emergence of linezolid-resistant staphylococci in the United States. Antimicrob. Agents Chemother. 54:742–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mendes RE, Deshpande LM, Costello A, Farrell DJ. 2012. Molecular epidemiology of Staphylococcus epidermidis clinical isolates from U.S. hospitals. Antimicrob. Agents Chemother. 56:4656–4661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bongiorno D, Campanile F, Mongelli G, Baldi MT, Provenzani R, Reali S, Lo Russo C, Santagati M, Stefani S. 2010. DNA methylase modifications and other linezolid resistance mutations in coagulase-negative staphylococci in Italy. J. Antimicrob. Chemother. 65:2336–2340 [DOI] [PubMed] [Google Scholar]
- 10. Clinical and Laboratory Standard Institutes (CLSI) 2012. Performance standards for antimicrobial susceptibility testing; approved standard M100–S20. CLSI, Wayne, PA [Google Scholar]
- 11. European Committee on Antimicrobial Susceptibility Testing (EUCAST) 2012. Clinical breakpoints. http://www.eucast.org/clinical_breakpoints/
- 12. Locke JB, Hilgers M, Shaw KJ. 2009. Mutations in ribosomal protein L3 are associated with oxazolidinone resistance in staphylococci of clinical origin. Antimicrob. Agents Chemother. 53:5275–5278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pringle M, Poehlsgaard J, Vester B, Long KS. 2004. Mutations in ribosomal protein L3 and 23S ribosomal RNA at the peptidyl transferase centre are associated with reduced susceptibility to tiamulin in Brachyspira spp. isolates. Mol. Microbiol. 54:1295–1306 [DOI] [PubMed] [Google Scholar]
- 14. Holzel CS, Harms KS, Schwaiger K, Bauer J. 2010. Resistance to linezolid in a porcine Clostridium perfrigens strain carrying a mutation in the rplD gene encoding the ribosomal protein L4. Antimicrob. Agents Chemother. 54:1351–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miragaia M, Thomas JC, Couto I, Enright MC, de Lencastre H. 2007. Inferring a population structure for Staphylococcus epidermidis from multilocus sequence typing data. J. Bacteriol. 189:2540–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Widerstrom M, Monsen T, Karlsson C, Edebro H, Johansson A, Wistrom J. 2009. Clonality among multidrug-resistant hospital-associated Staphylococcus epidermidis in northern Europe. Scand. J. Infect. Dis. 41:642–649 [DOI] [PubMed] [Google Scholar]
- 17. Widerstrom M, McCullough CA, Coombs GW, Monsen T, Christiansen KJ. 2012. A multidrug-resistant Staphylococcus epidermidis clone (ST2) is an ongoing cause of hospital-acquired infection in a Western Australian hospital. J. Clin. Microbiol. 50:2147–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Iorio NL, Caboclo RF, Azevedo MB, Barcellos AG, Neves FP, Domingues RM, dos Santos KR. 2012. Characteristics related to antimicrobial resistance and biofilm formation of widespread methicillin-resistant Staphylococcus epidermidis ST2 and ST23 lineages in Rio de Janeiro hospitals, Brazil. Diagn. Microbiol. Infect. Dis. 72:32–40 [DOI] [PubMed] [Google Scholar]
- 19. Li M, Wang X, Gao Q, Lu Y. 2009. Molecular characterization of Staphylococcus epidermidis strains isolated from a teaching hospital in Shanghai, China. J. Med. Microbiol. 58(Pt 4):456–461 [DOI] [PubMed] [Google Scholar]
- 20. Ibrahem S, Salmenlinna S, Lyytikainen O, Vaara M, Vuopio-Varkila J. 2008. Molecular characterization of methicillin-resistant Staphylococcus epidermidis strains from bacteraemic patients. Clin. Microbiol. Infect. 14:1020–1027 [DOI] [PubMed] [Google Scholar]