Abstract
Human embryonic stem cells and induced pluripotent stem cells proliferate rapidly and divide symmetrically producing equivalent progeny cells. In contrast, lineage committed cells acquire an extended symmetrical cell cycle. Self-renewal of tissue-specific stem cells is sustained by asymmetric cell division where one progeny cell remains a progenitor while the partner progeny cell exits the cell cycle and differentiates. There are three principal contexts for considering the operation and regulation of the pluripotent cell cycle: temporal, regulatory andstructural. The primary temporal context that the pluripotent self-renewal cell cycle of human embryonic stem cells (hESCs) is a short G1 period without reducing periods of time allocated to S phase, G2, and mitosis. The rules that govern proliferation in hESCs remain to be comprehensively established. However, several lines of evidence suggest a key role for the naïve transcriptome of hESCs, which is competent to stringently regulate the ESC cell cycle. This supports the requirements of pluripotent cells to self propagate while suppressing expression of genes that confer lineage commitment and/or tissue specificity. However, for the first time, we consider unique dimensions to the architectural organization and assembly of regulatory machinery for gene expression in nuclear microenviornments that define parameters of pluripotency. From both fundamental biological and clinical perspectives, understanding control of the abbreviated embryonic stem cell cycle can provide options to coordinate control of proliferation versus differentiation. Wound healing, tissue engineering, and cell-based therapy to mitigate developmental aberrations illustrate applications that benefit from knowledge of the biology of the pluripotent cell cycle.
Keywords: Embryonic stem cells, Pluripotency, Cell Cycle
Human embryonic stem cells (hESCs) are uniquely dedicated to rapid, unrestricted proliferation (Figure 1)(Thomson et al., 1998;Amit et al., 2000;Zwaka and Thomson, 2005). Within the blastocyst of the embryo as well as in culture, hESCs repeatedly traverse the cell cycle and undergo successive symmetrical cell divisions to provide structurally and functionally equivalent progeny cells to retain pluripotency and refrain from gene expression and epigenetic control associated with lineage commitment (Savatier et al., 1994;White et al., 2005;Galderisi et al., 2006;Singh and Dalton, 2009). It is particularly striking that hESCs utilize oncogenes which include retinoblastoma (Rb) and Myc to traverse the cell cycle, yet do not undergo oncogenic transformation. There are unique architectural features to the organization and assembly of genetic and epigenetic regulatory machinery in the nuclei of pluripotent cells that, for the first time are presented in relation to the biology of stem cell proliferation and differentiation. A relevant observation is that the cells divide symmetrically, with equivalent distribution of the genome and structural and regulatory machinery.
Figure 1. Differences between pluripotent and lineage committed cell cycle length.
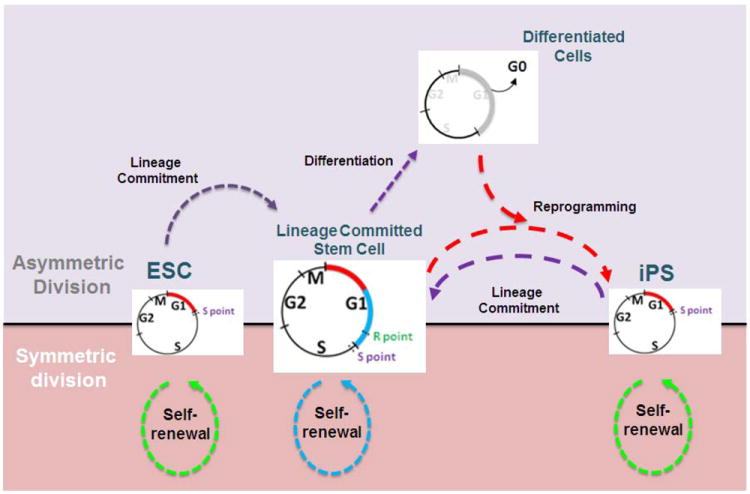
The four stages of the somatic cell cycle (G1: gap 1, S: synthesis of DNA, G2: gap 2, and M: mitosis) support duplication of the genome and subsequent segregation of a diploid set of chromosomes into two progeny cells. Post-fertilization, during early development, embryonic stem cells (ESC), derived from the inner cell mass (ICM) of blastocysts, have an abbreviated cell cycle, as a consequence of a very short G1 phase (red continuous segment; 2-3 h). During lineage commitment, ESCs undergo asymmetric division to produce more stem cells and new precursor cells (lineage committed stem cells and lineage specific progenitors) that will support growth, differentiation, and organogenesis. These precursor cells continue dividing with a normal cell cycle, but extended G1 phase (red/blue continuous segments; 8-12 h). The reprogramming of pre-committed somatic cells to a pluripotent state results in the reacquisition of a shortened cell cycle with a short G1 phase (red continuous segment; 2-3 h) and constant S, G2 and M phases. Upon differentiation, the cell exits the cell cycle after mitosis and goes to G0 (gray continuous segment). All pluripotent cells have the potential to undergo symmetric or asymmetric division. This equilibrated state (central black line) allows the cells to be responsive to signals for self-renewal or lineage commitment. The green and blue dotted lines depict cell divisions during self-renewal. The purple dotted lines represent the intermediate steps necessary for lineage commitment. The red dotted lines symbolize the chain of events necessary to reprogram cells (iPS cells). For simplicity, the intermediary states between lineage-committed stem cells and differentiated cells are not shown. R and S points represent the restriction points controlling the G1/S transition. Note, that the R point is only present in committed cells. Both restriction points are more extensively described in Figure 5.
Two compelling questions are evident: What mechanism(s) allow hESCs to divide and execute required functions without transforming? What is the significance of structural and regulatory machinery retention during cell division? These are questions that go beyond the characterization of regulatory molecules, pathways and networks. With a short G1 period in hESCs the possibilities include accelerated progression of regulatory cascades, elimination of components to regulatory networks that are operative during G1 and constituitive expression of cell cycle regulatory factors that are confined to G1 following lineage commitment.
Although there are exceptions, in general, hESCs divide rapidly with generation times of 8-16 hours. 65% of the time, pluripotent cells reside in S phase and 15% of the time in G1 (Becker et al., 2006). Somatic cells that are reprogrammed to pluripotency adhere to these trends, suggesting that rapid cell division and shortening of the cell cycle are requirements for pluripotency (Ghule et al., 2011;Koledova et al., 2010;Ruiz et al., 2011). In support of this concept, committed cells have generation times greater than 16 hours and 40% of the cell cycle is allocated to G1. Overexpression and knockdown of cell cycle regulatory factors induces cell proliferation and increases efficiency of reprogramming. In contrast, cell cycle arrest inhibits reprogramming. Consistent with these findings, cell cycle arrest is sufficient to promote differentiation of hESCs (Neganova and Lako, 2008;Wang and Blelloch, 2009).
This review addresses these ideas with the realization that important mechanisms that regulate the pluripotent cell cycle remain to be established.
A. Accommodating Cell Cycle Regulatory Cascades
Complex and inter-dependent signaling networks that modulate genetic and epigenetic regulation are operative throughout the cell cycle in both hESCs and somatic cells to promote proliferation, control movement through successive stages of the cell cycle, and provide surveillance mechanisms that monitor genomic integrity, chromatin packaging, and mitotic division (Becker et al., 2007; Stein and Pardee, 2004; Pardee and Stein, 2009) The biochemical and molecular parameters of control for the pluripotent and lineage committed cell cycles are illustrated and described in Figures 1 and 2, in Table 1, and are detailed in Pardee et al. (Stein and Pardee 2004; Pardee and Stein 2009) Effectiveness of these processes are essential for immediate detection of errors in genome replication or control at checkpoints (Figure 2). During the G1 period of the cell cycle, a series of regulatory events are orchestrated that support proliferation (Neganova and Lako, 2008). These include cell cycle progression and initiation of DNA replication. The activities of cyclins, cyclin-dependent kinases (CDKs), cyclin-dependent kinase regulators, and signaling factors that monitor and control the regulatory cascades that are operative during G1 are summarized in Table 1.
Figure 2. Multiple checkpoints control cell cycle progression.
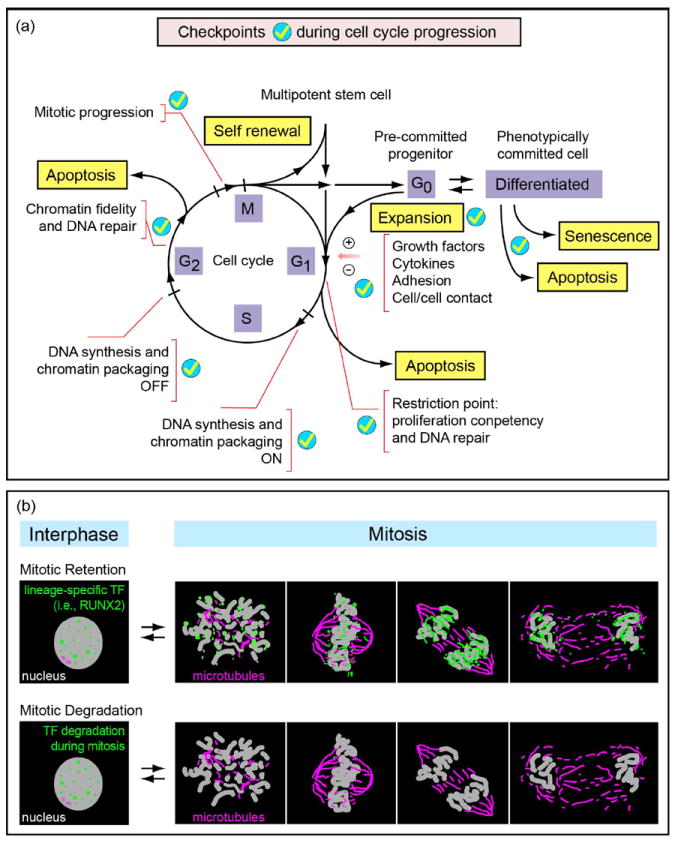
(a) The cell cycle is regulated by several critical cell cycle checkpoints (ticks) at which competency for cell cycle progression is monitored. Entry into and exit from the cell cycle (black lines and lettering) is controlled by growth regulatory factors (e.g. cytokines, growth factors, cell adhesion and/or cell–cell contact), which determine self-renewal of stem cells and expansion of pre-committed progenitor cells. The biochemical parameters associated with each cell cycle checkpoint are indicated by red lettering. Options for defaulting to apoptosis (yellow lettering) during G1 and G2 are evaluated by surveillance mechanisms that assess fidelity of structural and regulatory parameters of cell cycle control. (b) Transcription factors (green) are organized in distinct foci in the interphasic nuclei. Although some lineage-specific transcription factors (i.e. RUNX2) are retained on target gene promoters in chromosomes at all stages of mitosis, others do not associate with chromosomes and are degraded. This retention of transcription factors in addition to the occurrence of certain histone modifications indicate that certain genes are bookmarked for expression after mitosis.
Table 1.
Molecular differences in cell cycle control among ESCs, lineage committed cells, and iPSCs.
| ESCs | Lineage Committed Cells | iPS Cells | References | |
|---|---|---|---|---|
| Cell Cycle Length | Mice: 10 h | 24-32 h | 16-18 h | 1, 2, 7, 13, 15 |
| Humans: 15-16 h | ||||
|
| ||||
| G1 Length | Mice: 1 – 2 h | 8-12 h | 2-3 h | 1, 2, 7, 10 |
| Human: 2.5 -3 h | ||||
|
| ||||
| CDK Activity | Most CDKs are active throughout the cell cycle, as a consequence of the relatively stable levels of cyclins, and the absence (or very low expression) of CDKIs. | Periodic activation of CDKs, during the cell cycle. This is regulated by expression of CDKs and by cyclins and CDKIs. | Cell cycle-dependent expression of cyclin E and cyclin B1. | 1, 2, 3, 5, 6, 7, 9, 10, 11, 12, 14 |
| Very high expression of CDK2. | CDK activity is counteracted by CDKIs, p15, p16, p18, p19, p21, p27 and p57. | Constitutively high levels of CDK2. | ||
| In mice, cyclins D1, D3, E and A2 are expressed at comparable levels throughout the cell cycle. | Cyclin D1 levels are higher than in ESCs. Down-regulation of cyclin E1/D2 decreases the efficiency of reprogramming. | |||
| The CDK4/cyclin D complex is not present in mouse, | Down-regulation of CDK2, CDK4, or cyclin D1 does not affect reprogramming efficiency. | |||
| CDKIs, p16, p21, p27, and p57 are silenced or expressed at very low levels. | Overexpression of p15, p16, or p21 blocks reprogramming. | |||
| Some genes show cell cycle dependent expression: CDC25a, cyclin E, cyclin D2, CDK4, CDK6, cyclin A, c-Myc, CDK1, and cyclin B1. | Overexpression of cyclin D1, D2, or E2, increases reprogramming efficiency. | |||
|
| ||||
| Check Points | Lack normal somatic cell cycle checkpoint controls at the G1/S transition. | Strong cell cycle checkpoints in G1 and G2. | Lack normal somatic cell cycle checkpoint controls in the G1/S transition. | 5,10,14, 16, 18 |
| After exposure to radiation, cells arrest in G2 Following activation of the ATM-dependent checkpoint signaling cascade, double stranded breaks are repaired. | After exposure radiation, cells arrest in G2. Following activation of the ATM-dependent checkpoint signaling cascade, double stranded breaks are repaired. | |||
|
| ||||
| G1/S Transition | Exogenous growth factor independent. | Exogenous growth factor dependent. | Exogenous growth factor independent. | 1, 4, 5, 6, 7, 8, 10, 14, 15, 17, 19 |
| Independent of the pRB/E2F switch at R point. | Dependent on the pRb/E2F switch and c-Myc at R-point | Down-regulation of pRb increases the efficiency of reprogramming. | ||
| Hinf-P/NPAT dependent (S-point). | Hinf-P/NPAT dependent (S-point). | Hinf-P/NPAT dependent (S-point). | ||
| Insensitive to cyclin D/CDK regulation and to the CDK inhibitor, p16Ink4a. | Sensitive to cyclin D/CDK regulation and to the CDK inhibitor, p16Ink4a. | ND: Insensitive to CDK regulation. | ||
| p53 does not induce G1 arrest in response to DNA damage. | p53-dependent cell cycle arrest in response to DNA damage. | p53-dependent cell cycle arrest in G1/S, mediated by p21, leads to senescence, which inhibits reprogramming. | ||
|
| ||||
| Nuclear Structure | NAPT foci double prior to the onset of S phase. | NAPT foci double only upon entry into S phase. | NAPT foci double prior to the onset of S phase. | 2, 19 |
DSB: Double strand breaks; ND: Not directly experimentally validated. However, evidence suggests this is likely the case. iPS cells: inducible pluripotent stem cells
References
Becker et al. J. Cell. Phys. Vol. 209:883–893, 2006.
Ghule et al. J. Cell. Phys. Vol.226: 1149–1156, 2011.
Zhang et al., J. Cell Biol. Vol. 184(1): 67–82, 2009.
Ghule et al. J. Cell. Phys. Vol.213: 9–17, 2007.
He et al. Annu. Rev. Cell Dev. Biol. Vol. 25:377–406, 2009.
Becker et al. J. Cell. Physiol. 222: 456–464, 2010.
Stein GS et al. Control of the Human Pluripotent Cell Cycle. In, “Stem Cells: From Bench to Bedside” (2nd edition). Bongso A and Lee EH, eds. World Scientific Press, 2010.
Zhao and Xu. Trends in Cell Biology, Vol. 20: 170-175, 2010.
Edel et al. Genes Dev. Vol. 24: 561-573, 2010.
Boheler, J. Cell. Phys. Vol. 221: 10–17, 2009.
Ruiz et al. Current Biology. Vol. 21: 45–52, 2011.
Lange and Calegari. Cell Cycle, Vol. 9:1893-1900, 2010.
Liu et al. J. Cell. Phys. Vol. 211: 279–286, 2007.
Neganova and Lako. J. Anat. Vol. 213: 30-44, 2008
Orford and Scodel. Nature Reviews Genetic. Vol. 9:115, 2009.
Momcilovic et al. PloS one, Vol. 5 (10): e13410, 2010.
Solozobova, V. World Journal of Biological Chemistry, Vol. 2 (9): 202, 2011.
Filion et al. Journal of cellular physiology, Vol. 220 (3): 586-92, 2009.
Ghule et al. PNAS, Vol. 105 (44): 16964-9, 2008.
The initial component of G1 is dedicated to establishing engagement in the proliferative process. Responsiveness to a broad spectrum of physiological cues as well as to intrinsic and extrinsic stress occur early during G1. Late in G1, the restriction point marks acquisition of growth factor independence for cell cycle progression (Pardee, 1974;Pardee, 1989). At this time, genes, including thymidine kinase, thymidylate synthetase, and dihydrofolatereductase, that encode the enzymology for deoxynucleotide metabolism are upregulated. Subsequently, at the G1/S phase transition (the S point), the initiation of DNA replication is accompanied by and functionally coupled with the activation of histone gene expression. The relationship between histone gene expression and DNA replication ensures the availability of histone proteins to package newly replicated DNA as chromatin. The magnitude of this requirement is strikingly illustrated by biosynthesis of ~2.3 meters of DNA during the 8-9 hour S phase. The G1/S phase transition is equally relevant to the mechanisms that monitor fidelity of DNA replication and support compensatory processes, such as editing, if necessary. This transition is associated with a series of surveillance checkpoints that, if required, delay the onset of proliferation, invoke DNA repair and/or default to apoptosis (Figure 3) (Becker et al., 2007).
Figure 3. Surveillance and editing mechanisms mediating checkpoint control.
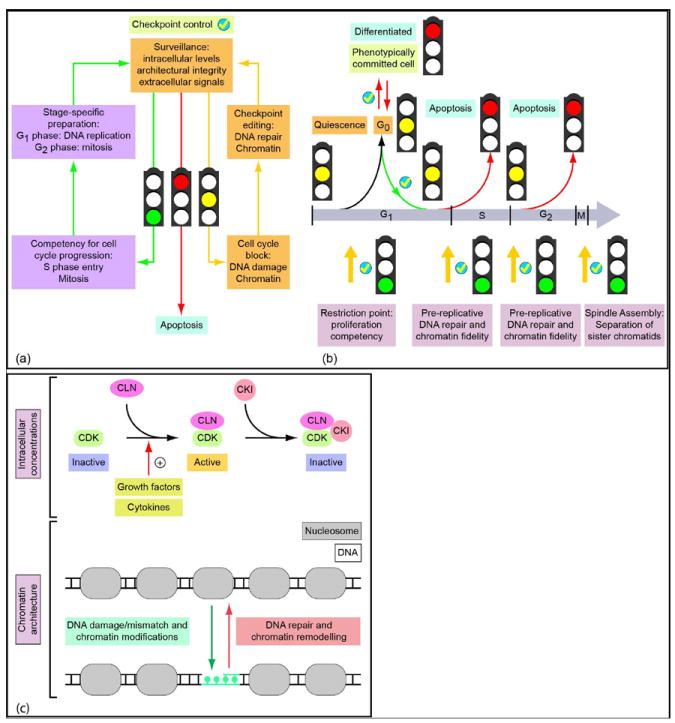
(a) Surveillance mechanisms monitor multiple biochemical and architectural parameters that control cell cycle progression. These parameters include the intracellular levels of regulatory proteins, structural and informational integrity of the genome, as well as extracellular signals governing cell cycle progression. The integration of this regulatory input can result in (i) competency for cell cycle progression (green traffic light and arrows), (ii) cell cycle inhibition and activation of editing mechanisms (yellow traffic light and arrows) or (iii) the active and regulated destruction of the cell in response to apoptotic signals (red traffic light and red arrow). (b) Traverse of the cell cycle is regulated by a series of checkpoints at strategic positions within the cell cycle. Several major checkpoints (yellow arrows with ticks and light purple lettering) only allow a cell to commit to a subsequent cell cycle stage upon satisfying essential biochemical and architectural criteria governing competency for cell cycle progression (green traffic lights). For example, at the ‘restriction point’ surveillance mechanisms (yellow traffic lights) integrate cell growth stimulatory and inhibitory signals, including growth factors, cell adhesion and nutrient status (light purple lettering). Checkpoints in G1 and G2 are necessary to ensure the integrity of the genome and, if necessary, activate chromatin editing mechanisms (light purple lettering). The spindle assembly checkpoint ensures equal chromosome segregation. (c) Checkpoint control mechanisms monitor intracellular levels of cell cycle regulatory factors, as well as parameters of chromatin architecture. For example, the activation of cyclin-dependent kinases reflects the sensing of intracellular concentrations of the cognate cyclins. CDK activation is attenuated by CDK inhibitor proteins (CKIs) which inactivate CDK/cyclin complexes. Competency for cell cycle progression requires that cyclin levels reach a threshold (e.g. by exceeding the levels of available CKIs, or phosphorylation events altering the affinities of cyclins and CKIs for CDKs). As a consequence, activated CDK/cyclin complexes phosphorylate transcription factors that regulate expression of cell cycle stage-specific genes. Furthermore, key checkpoints in G1 and G2 monitor chromatin integrity and perform essential editing functions. DNA damage activates DNA-repair mechanisms that fix informational errors in the genome and restore nucleosomal organization by chromatin remodeling.
The 2.5 – 3 hour abbreviated G1 phase in hESCs, compared to the 8-12 hour G1 period in human somatic cells, appears to require the activity of the full complement of cell cycle regulatory factors (Figure 1). However, the signaling cascades that control critical steps during G1 that culminate in genome replication are confined to a brief period. Mapping the temporal sequence of events that occurs during G1 in hESCs is imperative. Results to date establish that regulatory activity, which is restricted to the interval between completion of mitosis and the restriction point, may be modified. In contrast, the regulatory events that support competency for genome replication, histone gene expression, and DNA synthesis appear unaltered but occur rapidly following exit from mitosis. Perhaps the most compelling evidence for a functional relationship between the abbreviated cell cycle and pluripotency is transition from an abbreviated to an extended G1 period with the initiation of hESC differentiation and reversion to an abbreviated cell cycle in human induced pluripotent stem cells (hIPSCs) (Becker et al., 2006;Becker et al., 2007;Ghule et al., 2007;Ghule et al., 2008;Ghule et al., 2009;Xie et al., 2009;Becker et al., 2010).
B. Molecular Components of the Pluripotent Cell Cycle
1. Proliferation-differentiation relationships
Because of the distinct architecturally organized nuclear signature of the pluripotent cell cycle, it can be informative to consider which molecular parameters of control support the relationship between competency for pluripotency and unrestricted proliferation (summarized in Table 1). For example, it is well established that the Myc, Nanog, Sox2, and Oct4 transcription factors promote pluripotency by transcriptional regulation of cell cycle genes and miRNAs that promote pluripotency (Lee et al., 2010;Neganova and Lako, 2008;Wang and Blelloch, 2009). In contrast to pluripotency transcription factors, lineage commitment factors control cell cycle progression in adult stem cells. This component of control is demonstrated by the contribution of Sox17 to the maintenance of human fetal and neonatal stem cells. Runx1 contributes to the G1/S transition in adult stem cells (He et al., 2009;Friedman, 2009). Transcription factors assume independent architectural influences on regulation of transcription. It will be instructive to determine if the functions of these and other transcription factors are linked with established requirements for nuclear architecture during the cell cycle.
2. The CDK Family: Major Regulators of Cell Cycle Progression
The primary function of cyclin-dependent kinases (CDKs) is promotion of cell cycle entry. Equally important, CDKs support cell cycle progression which includes transition from G1 to S phase, DNA replication, chromosome separation, and cell division. CDK activation is accomplished by binding of cyclins that vary in expression levels periodically during the cell cycle. In addition, CDK activity is modulated by CDK kinases, that include the CDK activating complex, CAK (a complex of CDK7, cyclin H), and Wee/Myt1, by CDK phosphatases (CDC25 phosphatases), and by CDK inhibitors (CDKIs), including the Ink4 (p15Ink4b, p16Ink4a, p18Ink4c, and p19Ink4d) and Cip/Kip (p21Cip1, p27Kip1, and p57Kip2) families (Koledova et al., 2010). Although these molecular processes are highly regulated in somatic cells (at restriction points), ESCs have been shown to be refractory to these canonical control mechanisms. There is an explanation for the apparent restriction point-independent behavior of ESCs. They possess constitutively hyperphosphorylated pRB, and almost all of the CDKs, with the exception of CDK1-cyclin B which becomes selectively activated before mitosis, are active throughout the cell cycle (He et al., 2009). The high levels of CDK activity in ESCs are a consequence of the absence or very weak expression of CDKIs, which are associated with the relatively high and constant levels of the cyclins throughout the cell cycle (Stein et al., 2010;Koledova et al., 2010;He et al., 2009).
CDK2 is considered the principle CDK controlling the G1-S transition. The inhibition of CDK2 activity delays the G1-S transition, and knockdown of CDK2 leads to G1 arrest in ESCs. Interestingly, these events are sufficient to induce differentiation of ESCs, which reveal the existence of regulatory connections between cell-cycle and self-renewal for pluripotency (Neganova et al., 2009) (Neganova et al., 2011). This linked relationship is further supported by the observation that core pluripotency factors OCT4, SOX2, and NANOG regulate the expression of key cell-cycle regulatory proteins such as CDK1, cyclin D1, CDK6, CDC25A, and CDC7 (Neganova and Lako, 2008;Koledova et al., 2010;Zhang et al., 2009).
It appears to be significant that the induction of cell proliferation increases reprogramming efficiency, whereas cell cycle arrest inhibits successful reprogramming (Ruiz et al., 2011). Consistently, the reprogrammed cells possess high CDK2 levels and have a short cell cycle length, reinforcing the idea that rapid division and shortening of the cell cycle is required for pluripotency.
3. Myc: A persuasive influence on proliferation
Myc plays a pivotal role in regulating the pluripotent cell cycle. Although Myc contributes to control of proliferation, it remains unclear how Myc specifically maintains the pluripotent, self-renewing state of stem cells. Some studies are beginning to provide clues to the mechanism of Myc-mediated pluripotency. Upon Myc inactivation, there is a decreased rate of cell division and cell cycle remodeling (Smith et al., 2010). Several genome-wide chromatin immunoprecipitation studies have identified direct Myc targets. Many of these genes participate in cell cycle and metabolic control, including cyclins and cyclin dependent kinases (CDKs) (Chen et al., 2008;Kidder et al., 2008;Kim et al., 2008;Sridharan et al., 2009).
Myc is mechanistically versatile because it can both activate and repress gene expression. This dual role is attributed to the ability of Myc to control histone H3 and H4 modifications across the genome. Activation of Myc target genes occurs in response to histone acetylation, while repression occurs when histones are deacetylated (McMahon et al., 2000;Vervoorts et al., 2003;Kleine-Kohlbrecher et al., 2006)((Iraci et al., 2011;Kurland and Tansey, 2008;Frank et al., 2003;Knoepfler et al., 2006). Myc binds and recruits LSD1, a flavin dependent demethylase, and favors local and transient H3K4me3 demethylation at Myc genomic targets. This mechanism may facilitate unwinding chromatin to initiate Myc-mediated transcription (Amente et al., 2010b;Amente et al., 2010a). Recently, LSD1 has been found to regulate the balance between self-renewal and differentiation of hESCs by modulating the physiological balance between H3K4me2/me3 and H3K27me3 at target developmental genes that are poised for expression in hESCs(Adamo et al., 2011). These data are consistent with a contribution by Myc to LSD1-mediated gene expression in hESCs.
Myc is a dominant determinant for reprogramming lineage-committed cells to a pluripotent-like phenotype. Although it is possible to generate iPSCs without ectopic Myc (Nakagawa et al., 2008;Yu et al., 2007), there is a plethora of reports that Myc increases competency for stem cell reprogramming and maintenance of pluripotency (Takahashi and Yamanaka, 2006;Okita et al., 2007;Sridharan et al., 2009;Cartwright et al., 2005;Varlakhanova et al., 2010). During reprogramming, dramatic changes in the cell cycle are observed and may be due to regulation of gene expression by Myc (Singh and Dalton, 2009).
In addition to promoting pluripotency directly, Myc may concomittantly repress differentiation. A characteristic of pluripotency is an open chromatin conformation. A surprising observation from these studies is that in reprogrammed cells, the chromatin state resembles that of embryonic stem cells (Park et al., 2008;Takahashi and Yamanaka, 2006;Kim et al., 2010;Chen et al., 2008). Myc may contribute to sustaining the euchromatic chromatin organization by globally regulating histone acetylation and methylation (Lin et al., 2009;Knoepfler, 2008). Further, Myc upregulates cell cycle genes and represses fibroblast-specific gene expression (Sridharan et al., 2009). Myc can impact self-renewal by influencing the cell cycle regulatory network and simultaneously maintain pluripotency by repression of GATA6, a master regulator of primitive endoderm differentiation (Smith et al., 2010). In mESCs, the GATA6 locus is inactive, but poised for activation; it is marked bivalently with H3K4me3 and H3K27me3 (Bernstein et al., 2006;Mikkelsen et al., 2007). Nanog, another direct repressor of GATA6 (Hough et al., 2006;Singh et al., 2007), may cooperate with Myc to repress expression and thereby contribute to promoting and/or sustaining the pluripotent state. Such mechanisms may be a crucial switch that determines whether a cell will perpetuate pluripotency or differentiate.
4. microRNAs: Fine-tuning gene expression to promote pluripotency
In addition to transcription factors, specific microRNAs (miRNAs) regulate pluripotency and the expression of cell cycle genes. miRNAs are short, (~21 nucleotide) non-coding RNAs that directly, negatively regulate gene expression by binding to sequences in the target mRNA, usually in the 3’ untranslated region (UTR) (Kapinas and Delany, 2011; Lian et al., 2012; Hassan et al., 2011). Several lines of evidence implicate microRNAs in establishing and/or perpetuating pluripotency in hESCs during development as well as supporting restoration of pluripotency by reprogramming lineage committed cells. The miRNA family studied most extensively within the context of pluripotency is the miR-302 cluster on human chromosome 4 (Barroso-del et al., 2009;Stadler et al., 2010). Overexpression of this cluster, particularly miR-367, efficiently reprograms cells to a pluripotent iPSC state (Anokye-Danso et al., 2011;Card et al., 2008;Lin et al., 2008;Lin et al., 2011). In addition, miR-302 expression decreases with differentiation (Card et al., 2008;Stadler et al., 2010), suggesting this cluster promotes pluripotency. miR-367 activates Oct4 gene expression and suppress HDAC2 for generation of iPSCs (Anokye-Danso et al., 2011). Oct4, Sox2, and Nanog bind the promoter region of the miR-302 cluster in hESCs, but only Oct4 and Sox2 are required for expression of the miR-302 cluster (Card et al., 2008;Liu et al., 2011). An indication of a relationship between miR-302 and cell cycle control is provided by evidence that expression of the miR-302 cluster is associated with a short G1 phase, and longer S and G2/M phases, in HeLa cells (Card et al., 2008). This may be attributable to miR-302 targeting cyclin D1 and/or CDK2. As differentiation progresses, cyclin D1 and CDK2 protein levels increase (Card et al., 2008). Additional support for regulation of the cell cycle (Chen et al., 2008) are findings that overexpression attenuates the normal cell cycle rate without causing apoptosis. The antiproliferative effect has been attributed to suppression of the CDK2 and CDK4/6 cell cycle pathways (Lin et al., 2010).
Another family of miRNAs that has a prominent role in ESCs is the miR-17-92 family. This cluster is regulated by Myc. In stem cells, the cluster is marked by H3K4me3 and H3K36me3 activation modifications (Smith 2011). Although the miR-17-92 family has been studied extensively in numerous cancers, its role in ESCs is not as clearly defined. However, this family may target cell cycle regulators that include the E2F transcription factors, cyclins, and the retinoblastoma (Rb) family of tumor suppressors (Mendell, 2008;Wang et al., 2008).
C. Architectural Organization of Cell Cycle Regulatory Machinery in a Minimally Organized Pluripotent Cell Nucleus
1. The Pluripotent Nuclear Landscape
a. Hierarchical organization of nuclear regulatory machinery
Multiple levels of nuclear organization contribute to genetic and epigenetic control of transcription in pluripotent as well as lineage committed cells (Figure 4). From a hierarchical perspective, there is linear organization of genes with regulatory elements strategically residing upstream, downstream and within mRNA coding sequences. Chromatin structure, nucleosome organization and higher order conformations include long range protein-DNA, protein-protein interactions, nuclear matrix association, cross talk between gene loci residing in independent chromosomes and organization of regulatory machinery in punctate nuclear microenvironments.
Figure 4. (A) Multiple Levels of Nuclear Organization.
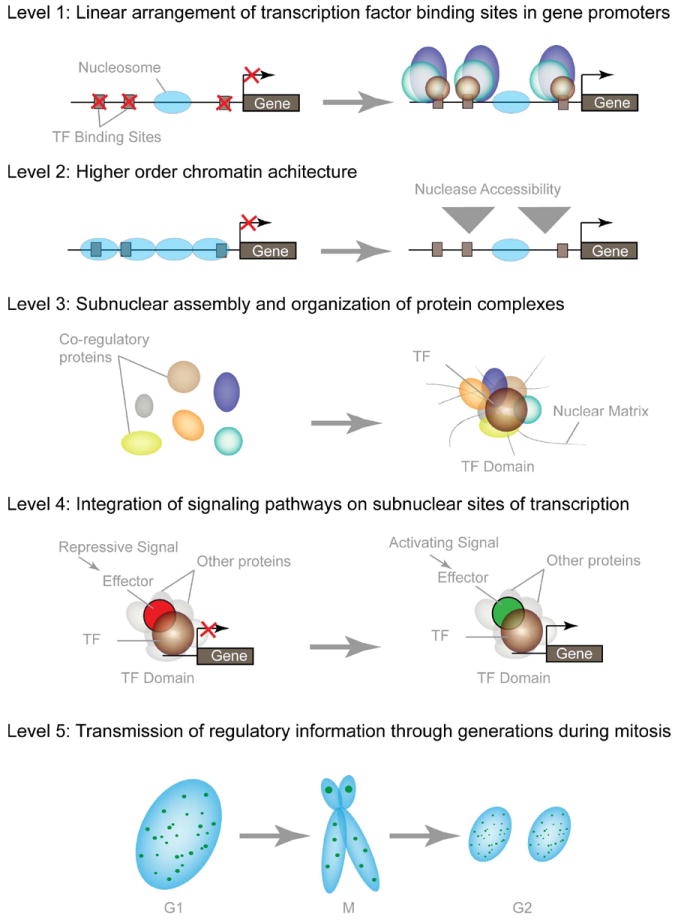
The organization of cognate DNA-regulatory elements in a linear fashion within gene promoters comprises the primary level of nuclear organization. The distance between these regulatory sites is additionally regulated by the folding of DNA into nucleosomes and higher order chromatin structures. Scaffolding nuclear proteins, usually transcription factors (TF), assemble multiprotein complexes to facilitate the combinatorial control of gene expression within the context of nuclear structure, thus forming dynamic microenvironments within the nucleus. Typical nuclear microenvironments contain various co-regulatory proteins that are involved in combinatorial control of gene activation, as well as repression, chromatin remodeling and cellular signaling. In addition, many nuclear microenvironments are equally partitioned during mitosis to epigenetically regulate cell growth and phenotypic properties.
Compartmentalization of the components for transcriptional control is dynamic and supports integration of regulatory signals. A synergistic relationship between nuclear organization and epigenetic control is reflected by localization of factors that support DNA methylation and histone modifications at promoter regulatory domains. Here, conformational modifications support and influence transcriptional activation and/or suppression by mitotic retention of regulatory complexes at target gene loci during mitosis (e.g. bookmarking) to sustain competency for gene expression in progeny cells (Zaidi et al., 2010; Pockwinse et al., 2011).
b. A dominant “open” genomic organization
There are unique features to the nuclear organization of regulatory machinery in hESCs that are distinct from the nuclear regulatory landscape of lineage committed cells (Bakshi et al., 2010; Young et al., 2007a;Stein et al., 1989;Young et al., 2007b;Kouzarides, 2007;Li et al., 2007;Rao et al., 2004;Bhattacharya et al., 2004;Wei et al., 2005;Boyer et al., 2005). The hESC genome is primarily packaged as euchromatin with minimal presence of heterochromatin. Although the functional significance remains to be mechanistically established, considerations include: 1) a genomic organization that is poised to support initial lineage-specific gene expression and tissue-specific gene expression, 2) a genomic organization that is essential for pluripotency, but is compatible with a limited extent of architectural remodeling that can be accommodated during an abbreviated cell cycle, and 3) limited compartmentalization of regulatory machinery in nuclear microenvironments that characterize differentiated cells.
The genomic organization in pluripotent cells is a component of transcriptional control. There are low levels of transcripts from phenotypic genes, but an absence of the encoded proteins. Some parameters of subnuclear organization (e.g., X chromosome inactivation (Hall and Lawrence, 2010;Hall et al., 2009;Clemson et al., 2006;Hall and Lawrence, 2003;Chow et al., 2003;Chow et al., 2002;Hall et al., 2002)) are established during the initial stages of development, while other parameters are in place and functionally operative during the earliest stages of development in hESCs. This includes nucleoli and histone locus body (HLB) organization. The significance of the relationships between functionally-organized, temporally staged and architecturally-associated regulatory complexes with biological control is evident, as described in the following sections.
c. Expeditious genomic reorganization in the pluripotent cell cycle
There are fundamental architectural modifications in genome configurations during the abbreviated cell cycle of hESCs that establish competency for DNA replication. Chromosome decondensation as cells exit mitosis and immediate assembly of chromatin-related nuclear microenvironments essential for gene expression (HLBs) are expedited during hESC self-renewal (Ghule et al., 2008). The mechanisms that render genes required for S phase selectively and rapidly accessible to regulatory factors during the abbreviated G1 phase of hESCs must be understood.
Maintenance of an open chromatin structure is essential for the pluripotent state. For example, depletion of the chromatin remodeling factor Chd1 in mouse ESCs results in accumulation of heterochromatin and loss of pluripotency (Gaspar-Maia et al., 2009). The transcription factors Oct4, Sox2, Nanog, and Myc constitute the core regulatory circuitry of ESCs and sustain pluripotency by activating cohorts of genes (Cole and Young, 2008;Marson et al., 2008;Silva and Smith, 2008;Chen et al., 2008). These pluripotency factors also repress cell-lineage-specific regulators to maintain the undifferentiated state (Bilodeau et al., 2009;Pasini et al., 2008;Lee et al., 2006;Loh et al., 2006) (Boyer et al., 2005;Boyer et al., 2006b;Boyer et al., 2006a).
To retain options for differentiation into all cell types, the chromatin of undifferentiated ESCs is transcriptionally permissive, with pronounced sensitivity to nucleases, limited heterochromatinization, as well as highly dynamic binding of structural proteins (e.g., histones H2A and H2B, HP1), general transcription factors (e.g., GTF2a1, GTF2b) and chromatin remodeling factors (e.g., Smarca4, Chd1) (Efroni et al., 2008;Mattout and Meshorer, 2010). Upon differentiation of ES cells, chromatin structure becomes more compact and repressive (Ahmed et al., 2010;Efroni et al., 2008;Schaniel et al., 2009). Thus, in contrast to the gene-selective chromatin remodeling that occurs during the cell cycle on a ‘mixed background’ of euchromatin and heterochromatin in committed cells, S phase related changes in chromatin architecture in ESCs must be achieved on a predominantly euchromatin background.
This open chromatin structure of proliferating pluripotent cells must be rapidly re-established following mitosis by global decondensation while selectively generating accessibility to cell cycle stage specific genes. The very short G1 phase of ESCs temporally compresses chromatin remodeling of genes that are transcriptionally induced at the G1/S-phase transition. Therefore, it is important to establish how chromatin structure is locally re-organized to support gene activation of a limited number of genomic loci that are essential for S phase progression, while cells decondense chromatin following mitotic division in anticipation of accelerated S-phase entry. It is necessary to define molecular mechanisms that control the selective activation of the genes most highly expressed at the G1/S-phase transition of pluripotent stem cells. Modulation of histone gene expression as the most prominent gene regulatory program at S-phase entry is mediated by active remodeling of chromatin, dynamic recruitment of gene regulatory factors and epigenetic marking of nucleosomes that are distinctly organized. It is critical to establish mechanisms that support gene-selective reconfiguration of chromatin architecture that proceeds in parallel with decondensation of chromatin as cells exit from mitosis through an abbreviated G1 phase in preparation for S-phase in pluripotent hESCs.
2. Nuclear Compartmentalization of Histone Gene Regulatory Machinery for the Pluripotent Cell Cycle
The architectural events necessary for initiation of histone gene expression contributes to control of the G1/S phase transition in pluripotent cells (Figure 5). Histone gene expression in hESCs is coupled to architectural localization of regulatory machinery in nuclear microenvironments (Figure 6). Further, the S-phase specific expression of histone genes is temporally and functionally coupled with DNA replication. This was the initial example of cell cycle dependent gene regulation and provides a paradigm for understanding regulatory mechanisms at the G1/S transition (Stein et al., 1975; Stein et al., 1996;Stein et al., 1975;Osley, 1991;Stein et al., 1984;Marzluff, 1992;Prescott, 1966). The induction of histone mRNA and protein synthesis at the initiation of S phase supports packaging newly replicated DNA into chromatin (Stein et al., 1975; Chrysogelos et al., 1985; Chrysogelos et al., 1989; Osley, 1991;Marashi et al., 1982;Morris et al., 1986;Plumb et al., 1983;Baumbach et al., 1987). Histone H4 gene transcription is activated at the G1/S phase transition and down-regulated during quiescence or differentiation (Shakoori et al., 1995;van Wijnen et al., 1997;van Wijnen et al., 1991;van den Ent et al., 1993;Ramsey-Ewing et al., 1995;Stein et al., 1989;Hovhannisyan et al., 2003;Cho et al., 2002;Wright et al., 1992;Pauli et al., 1987;Pauli et al., 1989;Pauli et al., 1988;Kroeger et al., 1995). These transcriptional modulations occur concomitant with dynamic modifications in chromatin structure and in vivo occupancy of histone gene promoters in somatic cells (Hovhannisyan et al., 2003;Pauli et al., 1988;Chrysogelos et al., 1989;Moreno et al., 1986;Chrysogelos et al., 1985). The increase in histone gene transcription early in S phase is mediated by transcription factors that have been identified in somatic cells (Mitra et al., 2003;Miele et al., 2005;Holmes et al., 2005;van Wijnen et al., 1992;Vaughan et al., 1995;van Wijnen et al., 1994;van den Ent et al., 1994;van Wijnen et al., 1996;Guo et al., 1995;Guo et al., 1997;Birnbaum et al., 1995;Mitra et al., 2001;van Wijnen et al., 1989;Ramsey-Ewing et al., 1994).
Figure 5. Transcriptional control at the G1/S phase transition.
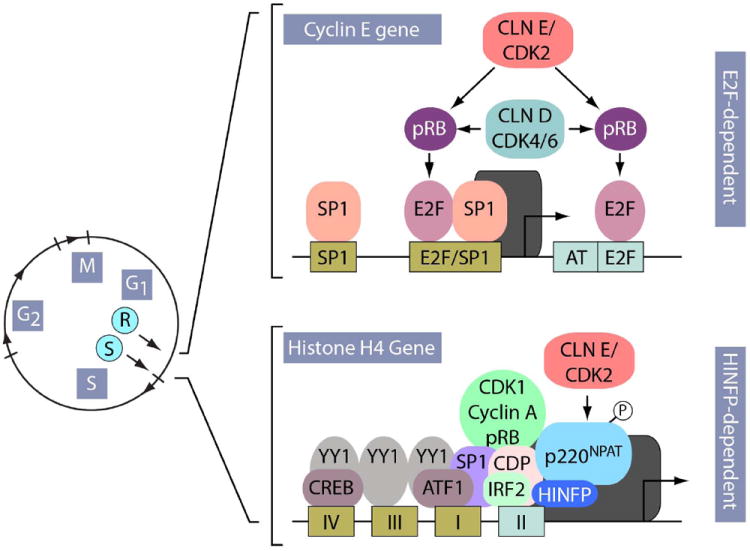
The genes encoding cell cycle regulatory subunits (e.g. cyclin E) and histone biosynthesis (e.g. H4) are each controlled by intricate arrays of promoter regulatory elements that influence transcriptional initiation by RNA polymerase II. E2F elements in the promoter of the cyclin E gene interact with E2F factors that associate with CDKs, cyclins and pRB-related proteins (R-point). In contrast, histone genes are controlled by the site II cell cycle regulatory element, which interacts with CDP-cut and IRF2 proteins, and the HINFP/p220NPAT complex. Analogous to E2Fdependent mechanisms, CDP-cut interacts with CDK1, cyclin A and pRB, whereas IRF2 performs an activating function similar to ‘free’ E2F. HINFP binds to this cell cycle regulatory element (site II) and recruits p220NPAT, thus integrating signals from the cyclin E/CDK2 kinase pathway (S-point). The presence of SP1 in the promoters of G1/S phase-related genes provides a shared mechanism for further enhancement of transcription at the onset of S phase.
Figure 6. The cell cycle control of histone gene expression is required to support the abbreviated cell cycle in pluripotent stem cells.
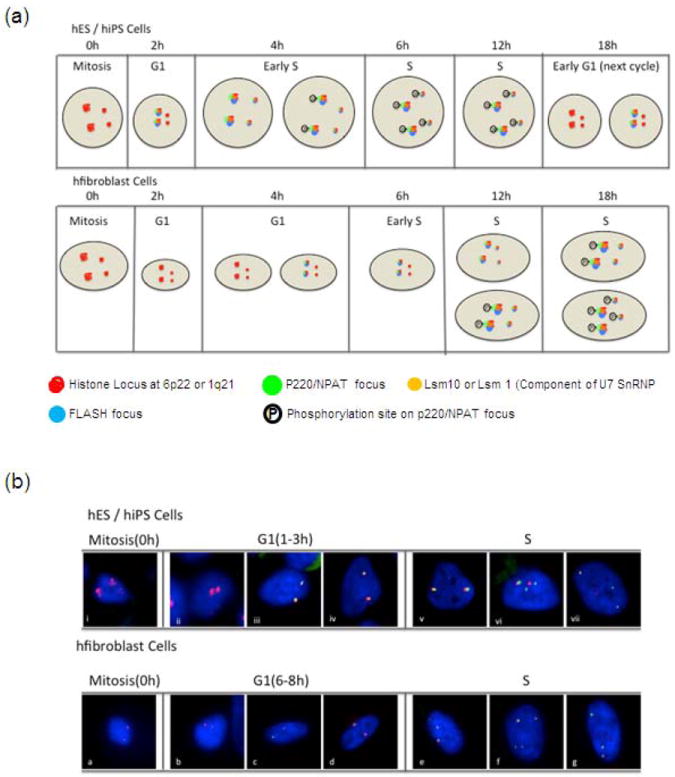
In pluripotent stem cells, as in lineage committed cells, histone genes are not regulated by an E2F/RB switch but by a HiNF-P/p220NPAT co-activation complex. However, the cell cycle dependent organization of p220NPAT foci is different between somatic and pluripotent stem cells. For example, in pluripotent stem cells, the number of p220NPAT foci increases in G1 prior to the CDK dependent phosphorylation of p220NPAT in the S phase (a, b; top panel). In contrast, the number of p220NPAT foci double from two to four upon entry into S phase in lineage committed cells (a, b; bottom panel). The increase in the kinetic of HLBs formation in pluripotent stem cells may render the p220NPAT/HiNF-P/histone gene regulatory complex poised for rapid activation by cyclin/CDK complexes to induce histone gene expression at the onset of DNA synthesis. (a) Diagrammatic representation of temporally different assembly of Histone Locus Bodies (HLBs) during G1/S transition in pluripotent vs lineage committed cells. (b) Immunofluorescence microscopy demonstrating temporal differences in the assembly of HLBs during G1/S transition in pluripotent vs lineage committed cells. Mitotically synchronized hES cells at various cell cycle stages were monitored by immunofluorescence (IF) microscopies for association of NPAT (green), showing spatial and temporal linkage of the histone gene cluster at 6p22 (red). DAPI staining (blue) was used to visualize the nucleus.
In pluripotent stem cells, as in somatic cells, histone genes are not regulated by an E2F/RB switch but by a HiNF-P/p220NPAT co-activation complex (Stead et al., 2002). The number of p220NPAT foci increases in G1 prior to the CDK dependent phosphorylation of p220NPAT in S phase. This increase may render the p220NPAT/HiNF-P/histone gene regulatory complex poised for rapid activation by cyclin/CDK complexes to induce histone gene expression at the onset of DNA synthesis. The CLNE/CDK2/NPAP/HINFP pathway defines a novel cell cycle transition point, designated the ‘S-point’. S point-related cell cycle control mechanisms in the context of subnuclear organization can provide an understanding of the assembly of the histone gene expression machinery at dedicated subnuclear domains (p220NPAT foci or HLBs) in both naïve and pre-committed hESCs.
Spatial mechanisms for synthesis and processing of histone gene transcripts are different between hESCs (Becker et al., 2007) and lineage-committed somatic cells (Mitra et al., 2003;Miele et al., 2005; Zhao et al., 1998;Zhao et al., 2000;Ma et al., 2000;Ye et al., 2003;Wei et al., 2003). For example, the number of p220NPAT foci double from two to four upon entry into S phase in somatic cells (Frey and Matera, 1995;Shopland et al., 2001;Ma et al., 2000;Zhao et al., 2000;Miele et al., 2005). In contrast, in hESCs, p220NPAT forms two subnuclear foci in G1 that double to four foci prior to the onset of S phase (Ghule et al., 2007). The biological basis of how hESCs expedite G1 progression to accelerate the self-renewal cell cycle is of extreme importance for the development of potential therapeutic measures.
In another example, while Cajal bodies and p220NPAT subnuclear foci are relatively stable, they exhibit fluctuations in their resident components depending on the species, cell type, and/or cell cycle stage. The p220NPAT foci detected in G1 of hESCs do not colocalize with coilin. Although a subset of p220NPAT foci co-localizes with coilin as S phase progresses, a minor subset of foci contain only one of the proteins. Therefore, p220NPAT foci and Cajal bodies containing coilin may be related, but are distinct subnuclear entities, likely with a variety of functions. In somatic cells, p220NPAT and HiNF-P are associated with the two large human histone gene clusters on Chromosomes 1 and 6, as well as the unique U7 snRNP that cleaves the 3’ end of nascent histone gene transcripts to generate mature non-polyadenylated mRNAs. The prototypical Cajal body component coilin interacts with U7snRNP, thereby providing structural linkage between Cajal bodies and the histone pre-mRNA processing machinery (Bellini and Gall, 1998). A subset of p220NPAT foci may coincide with Cajal bodies. These bodies contain an integrated architectural complex of histone gene transcription factors, p220NPAT, histone gene clusters, and the U7 snRNP related 3’ end processing machinery. p220NPAT and FLASH may be necessary to maintain this structure during the cell cycle (Ye et al., 2003;Barcaroli et al., 2006).
Lastly, the restoration of an abbreviated G1 period in iPSCs, with organization of HLBs immediately following completion of mitosis, supports the functional relationship between the intranuclear localization of transcriptional regulatory machinery and cell cycle control that is retained in hESCs but modified to occur immediately in G1 for accommodation of the abbreviated G1 period. It remains unknown as to what extent the nuclear organization of regulatory machinery that characterizes hESCs is restored with reprogramming. However, it appears that critical components of cell cycle control exhibit conserved interrelationships between nuclear structure and function.
3. Biological Relevance for an Abbreviated G1 in the Pluripotent Cell Cycle
Cell growth is a principle metabolic process in the G1 period of the cell cycle. The abbreviated G1 period in pluripotent cells may be linked to the reduced size compared with specialized cells that have an extended G1 period. The physiological relevance of the abbreviated G1 period in pluripotent cells remains to be experimentally determined. However, division of pluripotent versus differentiated cells may involve different molecular mechanisms for cell cycle control that reflect the unique requirements of these “primitive” cells. For example, it is necessary to mechanistically accommodate the priority of an ESC to proliferate and maintain pluripotency, in contrast to the requirement of a differentiated cell to acquire and support a specialized role.
Cells preferentially initiate differentiation from G1. Therefore, a short G1 limits the time during which a cell can be influenced by and responsive to external differentiation cues. Inhibition of G1 progression compromises pluripotency. An extended S phase supports maintenance of euchromatin, that can be conducive to rapid regulation of gene transcription. During mitosis, the displacement of some but not all transcription factors from condensing chromatin may “reset” transcriptional programs for the next G1, which requires significant time to remodel. In the absence of mitogenic signals, cells exit cycle in G1 and become quiescent (G0), but in response to mitogenic signaling, can re-enter the cycle in response to mitogenic signaling (R restriction point). Subsequent to the R point, signaling is no longer required for cell cycle progression.
Organization of the regulatory machinery at punctate foci designated HLBs (Ghule et al. 2008) at the onset of DNA synthesis provides a benchmark for experimentally establishing functional linkage of nuclear microenvironments with control of the pluripotent cell cycle. The restoration of an abbreviated G1 period in iPSCs, with organization of histone locus bodies immediately following completion of mitosis, supports the functional relationship between the intranuclear localization of transcriptional regulatory machinery and cell cycle control that is retained in hESCs but modified to occur immediately in G1 for accommodation of the abbreviated G1 period. It remains unknown as to what extent the nuclear organization of regulatory machinery that characterizes hESCs is restored with reprogramming. However, it appears that critical components of cell cycle control exhibit conserved interrelationships between nuclear structure and function.
4. Regulatory Infrastructure in Pluripotent Cell Nucleus: Options and Obligations
The abbreviated cell cycle in hESCs provides a streamlined process for proliferation during initial stages of development where pluripotency is required. From a regulatory perspective, genes that mediate competency to proliferate and cell cycle progression, that are selectively expressed in a cell cycle dependent manner, appear to be constitutive. Superimposed, there is retention of mechanisms that control execution of M/G1, G1/S, S/G2 and G2/M transitions as well as fidelity of R-point and S-point regulation. Other components of cell cycle control that are specific for hESCs include preferential expression of regulatory factors.
These characteristics of cell cycle-regulated pluripotency raise a series of mechanistic questions. What is the extent that regulatory events which are confined to specific stages of the cell cycle become constitutive in pluripotent cells? How extensive is regulatory machinery for DNA replication organized and assembled immediately following mitotic division? Are unique cell cycle regulatory proteins involved at the transition from an abbreviated pluripotent cell cycle to an extended cell cycle in lineage committed cells? Can parameters of cell cycle and growth control be identified to account for properties of lineage committed stem cells which exhibit extended periods of quiescence and mitotic divisions that are generally asymmetric rather than symmetric? How does proliferative dormancy of cancer stem cells contribute to resistance to both radiation and chemotherapy? A provocative answer, with a positive outlook, to the latter question is reviewed in Clevers 2011.
Beyond the biochemical mechanisms that are hallmarks of pluripotent cells, the parameters of nuclear organization that distinguish these primitive cells from lineage committed cells may offer insight into nuclear structure-gene expression relationships that can facilitate exploitation of pluripotent cells for tissue engineering and provide therapeutic targets in cancer stem cells. The minimal organization of regulatory machinery in nuclei of pluripotent cells may identify components of nuclear architecture that are fundamental to governing proliferation and cellular function in a broader context.
Architectural organization and compartmentalization of cell cycle regulatory complexes can be instructive for discriminating between regulatory processes that are operative in biology and pathology. The assembly and activity of HLBs when S phase is initiated rapidly following completion of mitosis in hESCs points to cellular dependence on compartmentalization of cell cycle regulatory machinery. Assembly happens after an extended G1 period in lineage committed cells, and after completion of mitosis in reprogrammed pluripotent stem cells, which have an abbreviated G1 period,. Whether there are variations in the mechanisms for such architectural organization of genes and cognate factors that support cell cycle and growth control that are operative in pluripotent and lineage committed cells remains to be established. However, it is realistic to anticipate that further insight into linkage of nuclear structure and gene expression will be mechanistically informative. From both fundamental regulatory and clinical perspectives, further understanding of the pluripotent cell cycle is relevant to applications of stem cells for regenerative medicine and new dimensions to therapy where traditional drug discovery strategies have been minimally effective.
Acknowledgments
We thank Patricia Jamieson and Priscilla Vazquez for editorial assistance in the preparation of the manuscript. Studies described in this review were supported by grants from the National Institutes of Health (AR039588, AR48818, AG035886, CA82834, and CA139322).
Footnotes
The authors have no conflicts of interest.
Reference List
- Adamo A, Sese B, Boue S, Castano J, Paramonov I, Barrero MJ, Izpisua Belmonte JC. LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat Cell Biol. 2011;13:652–659. doi: 10.1038/ncb2246. [DOI] [PubMed] [Google Scholar]
- Ahmed K, Dehghani H, Rugg-Gunn P, Fussner E, Rossant J, Bazett-Jones DP. Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PLoS ONE. 2010;5:e10531. doi: 10.1371/journal.pone.0010531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amente S, Bertoni A, Morano A, Lania L, Avvedimento EV, Majello B. LSD1-mediated demethylation of histone H3 lysine 4 triggers Myc-induced transcription. Oncogene. 2010a;29:3691–3702. doi: 10.1038/onc.2010.120. [DOI] [PubMed] [Google Scholar]
- Amente S, Lania L, Avvedimento EV, Majello B. DNA oxidation drives Myc mediated transcription. Cell Cycle. 2010b;9:3002–3004. doi: 10.4161/cc.9.15.12499. [DOI] [PubMed] [Google Scholar]
- Amit M, Carpenter MK, Inokuma MS, Chiu CP, Harris CP, Waknitz MA, Itskovitz-Eldor J, Thomson JA. Clonally derived human embryonic stem cell lines maintain pluripotency and proliferative potential for prolonged periods of culture. Dev Biol. 2000;227:271–278. doi: 10.1006/dbio.2000.9912. [DOI] [PubMed] [Google Scholar]
- Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, Morrisey EE. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcaroli D, Bongiorno-Borbone L, Terrinoni A, Hofmann TG, Rossi M, Knight RA, Bakshi, et al. 2010 doi: 10.1073/pnas.0604227103. [DOI] [PMC free article] [PubMed] [Google Scholar]; Matera AG, Melino G, De LV. FLASH is required for histone transcription and S-phase progression. Proc Natl Acad Sci U S A. 2006;103:14808–14812. doi: 10.1073/pnas.0604227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso-del JA, Lucena-Aguilar G, Menendez P. The miR-302-367 cluster as a potential stemness regulator in ESCs. Cell Cycle. 2009;8:394–398. doi: 10.4161/cc.8.3.7554. [DOI] [PubMed] [Google Scholar]
- Baumbach LL, Stein GS, Stein JL. Regulation of human histone gene expression: transcriptional and posttranscriptional control in the coupling of histone messenger RNA stability with DNA replication. Biochemistry. 1987;26:6178–6187. doi: 10.1021/bi00393a034. [DOI] [PubMed] [Google Scholar]
- Becker KA, Ghule PN, Therrien JA, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J Cell Physiol. 2006;209:883–893. doi: 10.1002/jcp.20776. [DOI] [PubMed] [Google Scholar]
- Becker KA, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Establishment of histone gene regulation and cell cycle checkpoint control in human embryonic stem cells. J Cell Physiol. 2007;210:517–526. doi: 10.1002/jcp.20903. [DOI] [PubMed] [Google Scholar]
- Becker KA, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Human embryonic stem cells are pre-mitotically committed to self-renewal and acquire a lengthened G1 phase upon lineage programming. J Cell Physiol. 2010;222:103–110. doi: 10.1002/jcp.21925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellini M, Gall JG. Coilin can form a complex with the U7 small nuclear ribonucleoprotein. Mol Biol Cell. 1998;9:2987–3001. doi: 10.1091/mbc.9.10.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bhattacharya B, Miura T, Brandenberger R, Mejido J, Luo Y, Yang AX, Joshi BH, Ginis I, Thies RS, Amit M, Lyons I, Condie BG, Itskovitz-Eldor J, Rao MS, Puri RK. Gene expression in human embryonic stem cell lines: unique molecular signature. Blood. 2004;103:2956–2964. doi: 10.1182/blood-2003-09-3314. [DOI] [PubMed] [Google Scholar]
- Bilodeau S, Kagey MH, Frampton GM, Rahl PB, Young RA. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev. 2009;23:2484–2489. doi: 10.1101/gad.1837309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum MJ, Wright KL, van Wijnen AJ, Ramsey-Ewing AL, Bourke MT, Last TJ, Aziz F, Frenkel B, Rao BR, Aronin N, Stein GS, Stein JL. Functional role for Sp1 in the transcriptional amplification of a cell cycle regulated histone H4 gene. Biochemistry. 1995;34:7648–7658. doi: 10.1021/bi00023a011. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Mathur D, Jaenisch R. Molecular control of pluripotency. Curr Opin Genet Dev. 2006a;16:455–462. doi: 10.1016/j.gde.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006b;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Card DA, Hebbar PB, Li L, Trotter KW, Komatsu Y, Mishina Y, Archer TK. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol. 2008;28:6426–6438. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright P, McLean C, Sheppard A, Rivett D, Jones K, Dalton S. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development. 2005;132:885–896. doi: 10.1242/dev.01670. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung WK, Clarke ND, Wei CL, Ng HH. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Cho B, Hovhannisyan H, Montecino M, Stein JL, van Wijnen AJ, Stein GS. Cell cycle stage-specific chromatin modifications of the histone H4 gene locus 2002 [Google Scholar]
- Chow JC, Hall LL, Clemson CM, Lawrence JB, Brown CJ. Characterization of expression at the human XIST locus in somatic, embryonal carcinoma, and transgenic cell lines. Genomics. 2003;82:309–322. doi: 10.1016/s0888-7543(03)00170-8. [DOI] [PubMed] [Google Scholar]
- Chow JC, Hall LL, Lawrence JB, Brown CJ. Ectopic XIST transcripts in human somatic cells show variable expression and localization. Cytogenet Genome Res. 2002;99:92–98. doi: 10.1159/000071579. [DOI] [PubMed] [Google Scholar]
- Chrysogelos S, Pauli U, Stein G, Stein J. Fine mapping of the chromatin structure of a cell cycle- regulated human H4 histone gene. J Biol Chem. 1989;264:1232–1237. [PubMed] [Google Scholar]
- Chrysogelos S, Riley DE, Stein G, Stein J. A human histone H4 gene exhibits cell cycle-dependent changes in chromatin structure that correlate with its expression. Proc Natl Acad Sci U S A. 1985;82:7535–7539. doi: 10.1073/pnas.82.22.7535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hall LL, Byron M, McNeil J, Lawrence JB. The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences. Proc Natl Acad Sci U S A. 2006;103:7688–7693. doi: 10.1073/pnas.0601069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. The cancer stem cell: premises, promises and challenges. Nature Medicine. 2011;17:313. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- Cole MF, Young RA. Mapping key features of transcriptional regulatory circuitry in embryonic stem cells. Cold Spring Harb Symp Quant Biol. 2008;73:183–193. doi: 10.1101/sqb.2008.73.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, Bazett-Jones DP, Le GS, McKay RD, Buetow KH, Gingeras TR, Misteli T, Meshorer E. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:437–447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SR, Parisi T, Taubert S, Fernandez P, Fuchs M, Chan HM, Livingston DM, Amati B. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 2003;4:575–580. doi: 10.1038/sj.embor.embor861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey MR, Matera G. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc Natl Acad Sci USA. 1995;92:5915–5919. doi: 10.1073/pnas.92.13.5915. erratum in: Proc Natl Acad Sci U S A 1995 Aug 29;92(18):8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman AD. Cell cycle and developmental control of hematopoiesis by Runx1. J Cell Physiol. 2009;219:520–524. doi: 10.1002/jcp.21738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galderisi U, Cipollaro M, Giordano A. The retinoblastoma gene is involved in multiple aspects of stem cell biology. Oncogene. 2006;25:5250–5256. doi: 10.1038/sj.onc.1209736. [DOI] [PubMed] [Google Scholar]
- Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A, Ramalho-Santos J, McManus MT, Plath K, Meshorer E, Ramalho-Santos M. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460:863–868. doi: 10.1038/nature08212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghule PN, Becker KA, Harper JW, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Cell cycle dependent phosphorylation and subnuclear organization of the histone gene regulator p220NPAT in human embryonic stem cells. J Cell Physiol. 2007;213:9–17. doi: 10.1002/jcp.21119. [DOI] [PubMed] [Google Scholar]
- Ghule PN, Dominski Z, Lian JB, Stein JL, van Wijnen AJ, Stein GS. The subnuclear organization of histone gene regulatory proteins and 3’ end processing factors of normal somatic and embryonic stem cells is compromised in selected human cancer cell types. J Cell Physiol. 2009;220:129–135. doi: 10.1002/jcp.21740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghule PN, Dominski Z, Yang XC, Marzluff WF, Becker KA, Harper JW, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Staged assembly of histone gene expression machinery at subnuclear foci in the abbreviated cell cycle of human embryonic stem cells. Proc Natl Acad Sci U S A. 2008;105:16964–16969. doi: 10.1073/pnas.0809273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghule PN, Medina R, Lengner CJ, Mandeville M, Qiao M, Dominski Z, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Reprogramming the pluripotent cell cycle: Restoration of an abbreviated G1 phase in human induced pluripotent stem (iPS) cells. J Cell Physiol. 2011;226:1149–1156. doi: 10.1002/jcp.22440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Odgren PR, van Wijnen AJ, Last TJ, Nickerson J, Penman S, Lian JB, Stein JL, Stein GS. The nuclear matrix protein NMP-1 is the transcription factor YY1. Proc Natl Acad Sci USA. 1995;92:10526–10530. doi: 10.1073/pnas.92.23.10526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Stein JL, van Wijnen AJ, Stein GS. ATF1 and CREB trans-activate a cell cycle regulated histone H4 gene at a distal nuclear matrix associated promoter element. Biochemistry. 1997;36:14447–14455. doi: 10.1021/bi971781s. [DOI] [PubMed] [Google Scholar]
- Hall LL, Byron M, Pageau G, Lawrence JB. AURKB-mediated effects on chromatin regulate binding versus release of XIST RNA to the inactive chromosome. J Cell Biol. 2009;186:491–507. doi: 10.1083/jcb.200811143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Byron M, Sakai K, Carrel L, Willard HF, Lawrence JB. An ectopic human XIST gene can induce chromosome inactivation in postdifferentiation human HT-1080 cells. Proc Natl Acad Sci U S A. 2002;99:8677–8682. doi: 10.1073/pnas.132468999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Lawrence JB. The cell biology of a novel chromosomal RNA: chromosome painting by XIST/Xist RNA initiates a remodeling cascade. Semin Cell Dev Biol. 2003;14:369–378. doi: 10.1016/j.semcdb.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Hall LL, Lawrence JB. XIST RNA and Architecture of the Inactive X Chromosome: Implications for the Repeat Genome. Cold Spring Harb Symp Quant Biol. 2010;75:345–356. doi: 10.1101/sqb.2010.75.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Nakada D, Morrison SJ. 2009 [Google Scholar]; Hassan, et al. Mechanisms of stem cell self-renewal. Annu Rev Cell Dev Biol. 2010;25:377–406. doi: 10.1146/annurev.cellbio.042308.113248. [DOI] [PubMed] [Google Scholar]
- Holmes WF, Braastad CD, Mitra P, Hampe C, Doenecke D, Albig W, Stein JL, van Wijnen AJ, Stein GS. Coordinate control and selective expression of the full complement of replication-dependent histone H4 genes in normal and cancer cells. J Biol Chem. 2005;280:37400–37407. doi: 10.1074/jbc.M506995200. [DOI] [PubMed] [Google Scholar]
- Hough SR, Clements I, Welch PJ, Wiederholt KA. Differentiation of mouse embryonic stem cells after RNA interference-mediated silencing of OCT4 and Nanog. Stem Cells. 2006;24:1467–1475. doi: 10.1634/stemcells.2005-0475. [DOI] [PubMed] [Google Scholar]
- Hovhannisyan H, Cho B, Mitra P, Montecino M, Stein GS, van Wijnen AJ, Stein JL. Maintenance of open chromatin and selective genomic occupancy at the cell-cycle-regulated histone H4 promoter during differentiation of HL-60 promyelocytic leukemia cells. Mol Cell Biol. 2003;23:1460–1469. doi: 10.1128/MCB.23.4.1460-1469.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iraci N, Diolaiti D, Papa A, Porro A, Valli E, Gherardi S, Herold S, Eilers M, Bernardoni R, Della VG, Perini G. A SP1/MIZ1/MYCN repression complex recruits HDAC1 at the TRKA and p75NTR promoters and affects neuroblastoma malignancy by inhibiting the cell response to NGF. Cancer Res. 2011;71:404–412. doi: 10.1158/0008-5472.CAN-10-2627. [DOI] [PubMed] [Google Scholar]
- Kapinas K, Delany AM. MicroRNA biogenesis and regulation of bone remodeling. Arthritis Res Ther. 2011;13:220. doi: 10.1186/ar3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidder BL, Yang J, Palmer S. Stat3 and c-Myc genome-wide promoter occupancy in embryonic stem cells. PLoS ONE. 2008;3:e3932. doi: 10.1371/journal.pone.0003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Kohlbrecher D, Adhikary S, Eilers M. Mechanisms of transcriptional repression by Myc. Curr Top Microbiol Immunol. 2006;302:51–62. doi: 10.1007/3-540-32952-8_3. [DOI] [PubMed] [Google Scholar]
- Knoepfler PS. Why myc? An unexpected ingredient in the stem cell cocktail. Cell Stem Cell. 2008;2:18–21. doi: 10.1016/j.stem.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. EMBO J. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koledova Z, Kramer A, Kafkova LR, Divoky V. Cell-cycle regulation in embryonic stem cells: centrosomal decisions on self-renewal. Stem Cells Dev. 2010;19:1663–1678. doi: 10.1089/scd.2010.0136. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kroeger PE, van Wijnen AJ, Pauli U, Wright KL, Stein GS, Stein JL. In vivo occupancy of histone gene proximal promoter elements reflects gene copy number-dependent titratable transactivation factors and cross-species compatibility of regulatory sequences. J Cell Biochem. 1995;57:191–207. doi: 10.1002/jcb.240570204. [DOI] [PubMed] [Google Scholar]
- Kurland JF, Tansey WP. Myc-mediated transcriptional repression by recruitment of histone deacetylase. Cancer Res. 2008;68:3624–3629. doi: 10.1158/0008-5472.CAN-07-6552. [DOI] [PubMed] [Google Scholar]
- Lee J, Go Y, Kang I, Han YM, Kim J. Oct-4 controls cell-cycle progression of embryonic stem cells. Biochem J. 2010;426:171–181. doi: 10.1042/BJ20091439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. 2007 [Google Scholar]; Lian, et al. The role of chromatin during transcription. Cell. 2012;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Lin CH, Lin C, Tanaka H, Fero ML, Eisenman RN. Gene regulation and epigenetic remodeling in murine embryonic stem cells by c-Myc. PLoS ONE. 2009;4:e7839. doi: 10.1371/journal.pone.0007839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SL, Chang DC, Chang-Lin S, Lin CH, Wu DT, Chen DT, Ying SY. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA. 2008;14:2115–2124. doi: 10.1261/rna.1162708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SL, Chang DC, Lin CH, Ying SY, Leu D, Wu DT. Regulation of somatic cell reprogramming through inducible mir-302 expression. Nucleic Acids Res. 2011;39:1054–1065. doi: 10.1093/nar/gkq850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SL, Chang DC, Ying SY, Leu D, Wu DT. MicroRNA miR-302 inhibits the tumorigenecity of human pluripotent stem cells by coordinate suppression of the CDK2 and CDK4/6 cell cycle pathways. Cancer Res. 2010;70:9473–9482. doi: 10.1158/0008-5472.CAN-10-2746. [DOI] [PubMed] [Google Scholar]
- Liu H, Deng S, Zhao Z, Zhang H, Xiao J, Song W, Gao F, Guan Y. Oct4 regulates the miR-302 cluster in P19 mouse embryonic carcinoma cells. Mol Biol Rep. 2011;38:2155–2160. doi: 10.1007/s11033-010-0343-4. [DOI] [PubMed] [Google Scholar]
- Liu JL, Murphy C, Buszczak M, Clatterbuck S, Goodman R, Gall JG. The Drosophila melanogaster Cajal body. J Cell Biol. 2006;172:875–884. doi: 10.1083/jcb.200511038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CW, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei CL, Ruan Y, Lim B, Ng HH. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Ma T, Van Tine BA, Wei Y, Garrett MD, Nelson D, Adams PD, Wang J, Qin J, Chow LT, Harper JW. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 2000;14:2298–2313. doi: 10.1101/gad.829500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marashi F, Baumbach L, Rickles R, Sierra F, Stein JL, Stein GS. Histone proteins in HeLa S3 cells are synthesized in a cell cycle stage specific manner. Science. 1982;215:683–685. doi: 10.1126/science.7058333. [DOI] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, Calabrese JM, Dennis LM, Volkert TL, Gupta S, Love J, Hannett N, Sharp PA, Bartel DP, Jaenisch R, Young RA. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF. Histone 3’ ends: essential and regulatory functions. Gene Expr. 1992;2:93–97. [PMC free article] [PubMed] [Google Scholar]
- Mattout A, Meshorer E. Chromatin plasticity and genome organization in pluripotent embryonic stem cells. Curr Opin Cell Biol. 2010;22:334–341. doi: 10.1016/j.ceb.2010.02.001. [DOI] [PubMed] [Google Scholar]
- McMahon SB, Wood MA, Cole MD. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol. 2000;20:556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele A, Braastad CD, Holmes WF, Mitra P, Medina R, Xie R, Zaidi SK, Ye X, Wei Y, Harper JW, van Wijnen AJ, Stein JL, Stein GS. HiNF-P directly links the cyclin E/CDK1/p220NPAT pathway to histone H4 gene regulation at the G1/S phase cell cycle transition. Mol Cell Biol. 2005;25:6140–6153. doi: 10.1128/MCB.25.14.6140-6153.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra P, Vaughan PS, Stein JL, Stein GS, van Wijnen AJ. Purification and functional analysis of a novel leucine-zipper/nucleotide-fold protein, BZAP45, stimulating cell cycle regulated histone H4 gene transcription. Biochemistry. 2001;40:10693–10699. doi: 10.1021/bi010529o. [DOI] [PubMed] [Google Scholar]
- Mitra P, Xie RL, Medina R, Hovhannisyan H, Zaidi SK, Wei Y, Harper JW, Stein JL, van Wijnen AJ, Stein GS. Identification of HiNF-P, a key activator of cell cycle controlled histone H4 genes at the onset of S phase. Mol Cell Biol. 2003;23:8110–8123. doi: 10.1128/MCB.23.22.8110-8123.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno ML, Chrysogelos SA, Stein GS, Stein JL. Reversible changes in the nucleosomal organization of a human H4 histone gene during the cell cycle. Biochemistry. 1986;25:5364–5370. doi: 10.1021/bi00367a003. [DOI] [PubMed] [Google Scholar]
- Morris T, Marashi F, Weber L, Hickey E, Greenspan D, Bonner J, Stein J, Stein G. Involvement of the 5’-leader sequence in coupling the stability of a human H3 histone mRNA with DNA replication. Proc Natl Acad Sci USA. 1986;83:981–985. doi: 10.1073/pnas.83.4.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Neganova I, Lako M. G1 to S phase cell cycle transition in somatic and embryonic stem cells. J Anat. 2008;213:30–44. doi: 10.1111/j.1469-7580.2008.00931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neganova I, Vilella F, Atkinson SP, Lloret M, Passos JF, von ZT, O’Connor JE, Burks D, Jones R, Armstrong L, Lako M. An important role for CDK2 in G1 to S checkpoint activation and DNA damage response in human embryonic stem cells. Stem Cells. 2011;29:651–659. doi: 10.1002/stem.620. [DOI] [PubMed] [Google Scholar]
- Neganova I, Zhang X, Atkinson S, Lako M. Expression and functional analysis of G1 to S regulatory components reveals an important role for CDK2 in cell cycle regulation in human embryonic stem cells. Oncogene. 2009;28:20–30. doi: 10.1038/onc.2008.358. [DOI] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Osley MA. The regulation of histone synthesis in the cell cycle. Annu Rev Biochem. 1991;60:827–861. doi: 10.1146/annurev.bi.60.070191.004143. [DOI] [PubMed] [Google Scholar]
- Pardee AB. A restriction point for control of normal animal cell proliferation. Proc Natl Acad Sci U S A. 1974;71:1286–1290. doi: 10.1073/pnas.71.4.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee AB. G1 events and regulation of cell proliferation. Science. 1989;246:603–608. doi: 10.1126/science.2683075. [DOI] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Pardee AB, Stein GS. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Agger K, Christensen J, Hansen K, Cloos PA, Helin K. Regulation of stem cell differentiation by histone methyltransferases and demethylases. Cold Spring Harb Symp Quant Biol. 2008;73:253–263. doi: 10.1101/sqb.2008.73.009. [DOI] [PubMed] [Google Scholar]
- Pauli U, Chrysogelos S, Nick H, Stein G, Stein J. In vivo protein binding sites and nuclease hypersensitivity in the promoter region of a cell cycle regulated human H3 histone gene. Nucl Acids Res. 1989;17:2333–2350. doi: 10.1093/nar/17.6.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli U, Chrysogelos S, Stein G, Stein J, Nick H. Protein-DNA interactions in vivo upstream of a cell cycle- regulated human H4 histone gene. Science. 1987;236:1308–1311. doi: 10.1126/science.3035717. [DOI] [PubMed] [Google Scholar]
- Pauli U, Chrysogelos S, Stein J, Stein G. Native genomic blotting: high-resolution mapping of DNase I- hypersensitive sites and protein-DNA interactions. Proc Natl Acad Sci USA. 1988;85:16–20. doi: 10.1073/pnas.85.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumb M, Stein J, Stein G. 1983 [Google Scholar]; Pockwinse, et al. Coordinate regulation of multiple histone mRNAs during the cell cycle in HeLa cells. Nucl Acids Res. 2011;11:2391–2410. doi: 10.1093/nar/11.8.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott DM. The syntheses of total macronuclear protein, histone, and DNA during the cell cycle in Euplotes eurystomus. J Cell Biol. 1966;31:1–9. doi: 10.1083/jcb.31.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey-Ewing A, van Wijnen AJ, Stein GS, Stein JL. Delineation of a human histone H4 cell cycle element in vivo: the master switch for H4 gene transcription. Proc Natl Acad Sci USA. 1994;91:4475–4479. doi: 10.1073/pnas.91.10.4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey-Ewing AL, Bortell R, Stein GS, Stein JL. Histone H4 proximal promoter mediates a complex transcriptional response during differentiation of 3T3L1 adipocytes. J Cell Physiol. 1995;163:312–320. doi: 10.1002/jcp.1041630212. [DOI] [PubMed] [Google Scholar]
- Rao RR, Calhoun JD, Qin X, Rekaya R, Clark JK, Stice SL. Comparative transcriptional profiling of two human embryonic stem cell lines. Biotechnol Bioeng. 2004;88:273–286. doi: 10.1002/bit.20245. [DOI] [PubMed] [Google Scholar]
- Ruiz S, Panopoulos AD, Herrerias A, Bissig KD, Lutz M, Berggren WT, Verma IM, Izpisua Belmonte JC. A high proliferation rate is required for cell reprogramming and maintenance of human embryonic stem cell identity. Curr Biol. 2011;21:45–52. doi: 10.1016/j.cub.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savatier P, Huang S, Szekely L, Wiman KG, Samarut J. Contrasting patterns of retinoblastoma protein expression in mouse embryonic stem cells and embryonic fibroblasts. Oncogene. 1994;9:809–818. [PubMed] [Google Scholar]
- Schaniel C, Ang YS, Ratnakumar K, Cormier C, James T, Bernstein E, Lemischka IR, Paddison PJ. Smarcc1/Baf155 couples self-renewal gene repression with changes in chromatin structure in mouse embryonic stem cells. Stem Cells. 2009;27:2979–2991. doi: 10.1002/stem.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schul W, van Der Kraan I, Matera AG, van Driel R, de Jong L. Nuclear domains enriched in RNA 3’-processing factors associate with coiled bodies and histone genes in a cell cycle-dependent manner. Mol Biol Cell. 1999;10:3815–3824. doi: 10.1091/mbc.10.11.3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakoori AR, van Wijnen AJ, Cooper C, Aziz F, Birnbaum M, Reddy GP, Grana X, De Luca A, Giordano A, Lian JB, Stein JL, Quesenberry P, Stein GS. Cytokine induction of proliferation and expression of CDC2 and cyclin A in FDC-P1 myeloid hematopoietic progenitor cells: regulation of ubiquitous and cell cycle-dependent histone gene transcription factors. J Cell Biochem. 1995;59:291–302. doi: 10.1002/jcb.240590302. [DOI] [PubMed] [Google Scholar]
- Shopland LS, Byron M, Stein JL, Lian JB, Stein GS, Lawrence JB. Replication-dependent histone gene expression is related to Cajal body (CB) association but does not require sustained CB contact. Mol Biol Cell. 2001;12:565–576. doi: 10.1091/mbc.12.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Smith A. Capturing pluripotency. Cell. 2008;132:532–536. doi: 10.1016/j.cell.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AM, Dalton S. The cell cycle and Myc intersect with mechanisms that regulate pluripotency and reprogramming. Cell Stem Cell. 2009;5:141–149. doi: 10.1016/j.stem.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AM, Hamazaki T, Hankowski KE, Terada N. A heterogeneous expression pattern for Nanog in embryonic stem cells. Stem Cells. 2007;25:2534–2542. doi: 10.1634/stemcells.2007-0126. [DOI] [PubMed] [Google Scholar]
- Smith KN, Lim JM, Wells L, Dalton S. Myc orchestrates a regulatory network required for the establishment and maintenance of pluripotency. Cell Cycle. 2011;10:592–597. doi: 10.4161/cc.10.4.14792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KN, Singh AM, Dalton S. Myc represses primitive endoderm differentiation in pluripotent stem cells. Cell Stem Cell. 2010;7:343–354. doi: 10.1016/j.stem.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler B, Ivanovska I, Mehta K, Song S, Nelson A, Tan Y, Mathieu J, Darby C, Blau CA, Ware C, Peters G, Miller DG, Shen L, Cleary MA, Ruohola-Baker H. Characterization of microRNAs involved in embryonic stem cell states. Stem Cells Dev. 2010;19:935–950. doi: 10.1089/scd.2009.0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanek D, Rader SD, Klingauf M, Neugebauer KM. Targeting of U4/U6 small nuclear RNP assembly factor SART3/p110 to Cajal bodies. J Cell Biol. 2003;160:505–516. doi: 10.1083/jcb.200210087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead E, White J, Faast R, Conn S, Goldstone S, Rathjen J, Dhingra U, Rathjen P, Walker D, Dalton S. Pluripotent cell division cycles are driven by ectopic Cdk2, cyclin A/E and E2F activities. Oncogene. 2002;21:8320–8333. doi: 10.1038/sj.onc.1206015. [DOI] [PubMed] [Google Scholar]
- Stein G, Lian J, Stein J, Briggs R, Shalhoub V, Wright K, Pauli U, van Wijnen A. Altered binding of human histone gene transcription factors during the shutdown of proliferation and onset of differentiation in HL-60 cells. Proc Natl Acad Sci USA. 1989;86:1865–1869. doi: 10.1073/pnas.86.6.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein G, Park W, Thrall C, Mans R, Stein J. 1975 doi: 10.1038/257764a0. [DOI] [PubMed] [Google Scholar]; Stein GS, Pardee AB. Regulation of cell cycle stage-specific transcription of histone genes from chromatin by non-histone chromosomal proteins. Nature. 2004;257:764–767. doi: 10.1038/257764a0. [DOI] [PubMed] [Google Scholar]
- Stein GS, Stein JL, Marzluff WF. Histone Genes. John Wiley & Sons, Inc.; New York: 1984. [Google Scholar]
- Stein GS, Stein JL, van Wijnen AJ, Lian JB. Transcriptional control of cell cycle progression: the histone gene is a paradigm for the G1/S phase and proliferation/differentiation transitions. Cell Biol Int. 1996;20:41–49. doi: 10.1006/cbir.1996.0007. [DOI] [PubMed] [Google Scholar]
- Stein GS, van Wijnen AJ, Stein JL, Lian JB, Ghule P, Medina R, Becker KA, Montecino M, Zaidi SK. Control of the Human Pluripotent Cell Cycle. In: Bongso A, Lee EH, editors. Stem Cells: From Bench to Bedside. World Scientific Press; 2010. [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- van den Ent FM, van Wijnen AJ, Last TJ, Bortell R, Stein JL, Lian JB, Stein GS. Concerted control of multiple histone promoter factors during cell density inhibition of proliferation in osteosarcoma cells: reciprocal regulation of cell cycle-controlled and bone-related genes. Cancer Res. 1993;53:2399–2409. [PubMed] [Google Scholar]
- van den Ent FM, van Wijnen AJ, Lian JB, Stein JL, Stein GS. Cell cycle controlled histone H1, H3, and H4 genes share unusual arrangements of recognition motifs for HiNF-D supporting a coordinate promoter binding mechanism. J Cell Physiol. 1994;159:515–530. doi: 10.1002/jcp.1041590316. [DOI] [PubMed] [Google Scholar]
- van Wijnen AJ, Aziz F, Grana X, De Luca A, Desai RK, Jaarsveld K, Last TJ, Soprano K, Giordano A, Lian JB, et al. Transcription of histone H4, H3, and H1 cell cycle genes: promoter factor HiNF-D contains CDC2, cyclin A, and an RB-related protein. Proc Natl Acad Sci USA. 1994;91:12882–12886. doi: 10.1073/pnas.91.26.12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijnen AJ, Cooper C, Odgren P, Aziz F, De Luca A, Shakoori RA, Giordano A, Quesenberry PJ, Lian JB, Stein GS, Stein JL. Cell cycle-dependent modifications in activities of pRb-related tumor suppressors and proliferation-specific CDP/cut homeodomain factors in murine hematopoietic progenitor cells. J Cell Biochem. 1997;66:512–523. doi: 10.1002/(sici)1097-4644(19970915)66:4<512::aid-jcb10>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- van Wijnen AJ, Owen TA, Holthuis J, Lian JB, Stein JL, Stein GS. Coordination of protein-DNA interactions in the promoters of human H4, H3, and H1 histone genes during the cell cycle, tumorigenesis, and development. J Cell Physiol. 1991;148:174–189. doi: 10.1002/jcp.1041480120. [DOI] [PubMed] [Google Scholar]
- van Wijnen AJ, van den Ent FM, Lian JB, Stein JL, Stein GS. Overlapping and CpG methylation-sensitive protein-DNA interactions at the histone H4 transcriptional cell cycle domain: distinctions between two human H4 gene promoters. Mol Cell Biol. 1992;12:3273–3287. doi: 10.1128/mcb.12.7.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijnen AJ, van Gurp MF, de Ridder MC, Tufarelli C, Last TJ, Birnbaum M, Vaughan PS, Giordano A, Krek W, Neufeld EJ, Stein JL, Stein GS. CDP/cut is the DNA-binding subunit of histone gene transcription factor HiNF-D: a mechanism for gene regulation at the G1/S phase cell cycle transition point independent of transcription factor E2F. Proc Natl Acad Sci USA. 1996;93:11516–11521. doi: 10.1073/pnas.93.21.11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijnen AJ, Wright KL, Lian JB, Stein JL, Stein GS. Human H4 histone gene transcription requires the proliferation- specific nuclear factor HiNF-D. Auxiliary roles for HiNF-C (Sp1- like) and HiNF-A (high mobility group-like) J Biol Chem. 1989;264:15034–15042. [PubMed] [Google Scholar]
- Varlakhanova NV, Cotterman RF, deVries WN, Morgan J, Donahue LR, Murray S, Knowles BB, Knoepfler PS. myc maintains embryonic stem cell pluripotency and self-renewal. Differentiation. 2010;80:9–19. doi: 10.1016/j.diff.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan PS, Aziz F, van Wijnen AJ, Wu S, Harada H, Taniguchi T, Soprano KJ, Stein JL, Stein GS. Activation of a cell-cycle-regulated histone gene by the oncogenic transcription factor IRF-2. Nature. 1995;377:362–365. doi: 10.1038/377362a0. [DOI] [PubMed] [Google Scholar]
- Vervoorts J, Luscher-Firzlaff JM, Rottmann S, Lilischkis R, Walsemann G, Dohmann K, Austen M, Luscher B. Stimulation of c-MYC transcriptional activity and acetylation by recruitment of the cofactor CBP. EMBO Rep. 2003;4:484–490. doi: 10.1038/sj.embor.embor821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Li YC, Wang J, Kong J, Qi Y, Quigg RJ, Li X. miR-17-92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proc Natl Acad Sci U S A. 2008;105:2889–2894. doi: 10.1073/pnas.0800178105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Blelloch R. Cell cycle regulation by MicroRNAs in embryonic stem cells. Cancer Res. 2009;69:4093–4096. doi: 10.1158/0008-5472.CAN-09-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CL, Miura T, Robson P, Lim SK, Xu XQ, Lee MY, Gupta S, Stanton L, Luo Y, Schmitt J, Thies S, Wang W, Khrebtukova I, Zhou D, Liu ET, Ruan YJ, Rao M, Lim B. Transcriptome profiling of human and murine ESCs identifies divergent paths required to maintain the stem cell state. Stem Cells. 2005;23:166–185. doi: 10.1634/stemcells.2004-0162. [DOI] [PubMed] [Google Scholar]
- Wei Y, Jin J, Harper JW. The cyclin E/Cdk2 substrate and Cajal body component p220(NPAT) activates histone transcription through a novel LisH-like domain. Mol Cell Biol. 2003;23:3669–3680. doi: 10.1128/MCB.23.10.3669-3680.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Stead E, Faast R, Conn S, Cartwright P, Dalton S. Developmental activation of the Rb-E2F pathway and establishment of cell cycle-regulated cyclin-dependent kinase activity during embryonic stem cell differentiation. Mol Biol Cell. 2005;16:2018–2027. doi: 10.1091/mbc.E04-12-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright KL, Dell’Orco RT, van Wijnen AJ, Stein JL, Stein GS. Multiple mechanisms regulate the proliferation-specific histone gene transcription factor HiNF-D in normal human diploid fibroblasts. Biochemistry. 1992;31:2812–2818. doi: 10.1021/bi00125a023. [DOI] [PubMed] [Google Scholar]
- Xie R, Medina R, Zhang Y, Hussain S, Colby J, Ghule P, Sundararajan S, Keeler M, Liu L-J, van der Deen M, Mitra P, Lian JB, Rivera-Perez JA, Jones SN, Stein JL, van Wijnen AJ, Stein GS. The histone gene activator HINFP is a non-redundant cyclin E/CDK2 effector during early embryonic cell cycles. Proc Natl Acad Sci, USA. 2009;106:12359–12364. doi: 10.1073/pnas.0905651106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Wei Y, Nalepa G, Harper JW. The cyclin E/Cdk2 substrate p220(NPAT) is required for S-phase entry, histone gene expression, and Cajal body maintenance in human somatic cells. Mol Cell Biol. 2003;23:8586–8600. doi: 10.1128/MCB.23.23.8586-8600.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DW, Hassan MQ, Pratap J, Galindo M, Zaidi SK, Lee SH, Yang X, Xie R, Javed A, Underwood JM, Furcinitti P, Imbalzano AN, Penman S, Nickerson JA, Montecino MA, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature. 2007a;445:442–446. doi: 10.1038/nature05473. [DOI] [PubMed] [Google Scholar]
- Young DW, Hassan MQ, Yang X-Q, Galindo M, Javed A, Zaidi SK, Furcinitti P, Lapointe D, Montecino M, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic retention of gene expression patterns by the cell fate determining transcription factor Runx2. Proc Natl Acad Sci USA. 2007b;104:3189–3194. doi: 10.1073/pnas.0611419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Montecino M, Lian JB, van Wijnen AJ, Stein JL, Stein GS. Mitotic bookmarking of genes: a novel dimension to epigenetic control. Nat Rev Genet. 2010;11:583–589. doi: 10.1038/nrg2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Neganova I, Przyborski S, Yang C, Cooke M, Atkinson SP, Anyfantis G, Fenyk S, Keith WN, Hoare SF, Hughes O, Strachan T, Stojkovic M, Hinds PW, Armstrong L, Lako M. A role for NANOG in G1 to S transition in human embryonic stem cells through direct binding of CDK6 and CDC25A. J Cell Biol. 2009;184:67–82. doi: 10.1083/jcb.200801009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Dynlacht B, Imai T, Hori T, Harlow E. Expression of NPAT, a novel substrate of cyclin E-CDK2, promotes S- phase entry. Genes Dev. 1998;12:456–461. doi: 10.1101/gad.12.4.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Kennedy BK, Lawrence BD, Barbie DA, Matera AG, Fletcher JA, Harlow E. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 2000;14:2283–2297. [PMC free article] [PubMed] [Google Scholar]
- Zwaka TP, Thomson JA. Differentiation of human embryonic stem cells occurs through symmetric cell division. Stem Cells. 2005;23:146–149. doi: 10.1634/stemcells.2004-0248. [DOI] [PubMed] [Google Scholar]
- Stein GS, Pardee AB, editors. Cell Cycle and Growth Control: Biomolecular Regulation and Cancer. John Wiley & Sons, Inc.; Hoboken NJ: 2004. [Google Scholar]
- Pardee AB, Stein GS, editors. The Biology and Treatment of Cancer: Understanding Cancer. John Wiley & Sons, Inc.; Hoboken NJ: 2009. [Google Scholar]


