Abstract
While metalloprotein design has now yielded a number of successful metal-bound and even catalytically active constructs, the question of where to put a metal site along a linear, repetitive sequence has not been thoroughly addressed. Often several possibilities in a given sequence may exist that would appear equivalent, but may in fact differ for metal affinity, substrate access or protein dynamics. We present a systematic variation of active site location for a hydrolytically active ZnHis3O site contained within a de novo designed three-stranded coiled coil. We find that the maximal rate, substrate access, and metal-binding affinity is dependent on the selected position, while catalytic efficiency for p-nitrophenyl acetate hydrolysis can be retained regardless of the location of the active site. This achievement demonstrates how efficient tailor-made enzymes which control rate, pKa, substrate and solvent access (and selectivity), and metal-binding affinity may be realized. These findings may be applied to the more advanced de novo design of constructs containing secondary interactions, such as hydrogen bonding channels. We are now confident that changes to location for accommodating such channels can be achieved without location-dependent loss of catalytic efficiency. These findings bring us closer to our ultimate goal of incorporating the secondary interactions we believe will be necessary in order to improve both active site properties and the catalytic efficiency to be competitive with the native enzyme, carbonic anhydrase.
INTRODUCTION
Metal ions are an essential component of more than one-third of characterized proteins, with varied functions from hydrolytic to redox chemistry. Protein design is a powerful strategy for understanding how metal ions are incorporated into and function within these proteins.1–3 De novo or “from scratch” protein design is one approach, involving both incorporating a new metal-binding site and applying first principles of protein structure to preparing a well-folded protein scaffold with a unique primary amino acid sequence. Although there are many examples of de novo designed metal sites4–8 (some catalyti-cally active), no study thoroughly examines protein matrix effects on the primary metal site. The question of whether the specific location of a metal site along the primary sequence matters, while important, has not been addressed. Insights into how the placement of an active site in differing environments around a protein affects active site properties (rate, substrate access, binding affinity) should greatly assist design efforts. Understanding these effects is important for deciding whether the location of a metal site matters for the properties one is interested in optimizing in a given system and, if so, where to engineer the desired activity within a protein.
Studies in our research group focus on de novo designed metallopeptides that aggregate to form α-helical three-stranded coiled coils (3SCC’s) above pH 5.5.9 Previously, we reported the structure and hydrolytic activity of [Hg(II)]S[Zn(II)(H2O/OH−)]N(TRIL9CL23H)3n+ towards p-nitrophenyl acetate (pNPA, 23.3 M−1 s−1 at pH 9.5) and CO2 (1.8 × 105 M−1 s−1 at pH 9.5).10 Our goal was to prepare a hydrophobic protein microenvironment containing a metal catalyst in order to carry out reactions in water, which are not limited by unwanted dimerization or product inhibition. Other de novo designed Zn(II)His3 proteins have been reported4,10–15, but few with structures and catalytic activities.10,11 There are also examples of Zn(II)His3 sites (analogous to the active site of the zinc metalloenzyme carbonic anhydrase (CA)) resulting from the redesign of natural scaffolds, but no activity has been reported.16,17 There is one related redesigned protein with a Zn(II)His3Asp center that supports organophosphate hydrolysis.18 Another designed metallohydrolase consists of a de novo designed Zn(II)His3 site at the interface of a dimer protein (MID1-Zn), which catalyzes pNPA hydrolysis (630 M−1 s−1).11 No other designed proteins have reported CO2 hydration activity. Our model is the fastest de novo designed metalloenzyme for a physiological reaction relative to a native enzyme (within a factor of 500 of the fastest CA19). The efficiency of our model is even more competitive with mutant CA’s, where important secondary interactions such as hydrogen bonding to the coordinated solvent molecule of the active site Zn(II) have been removed (this T199A mutant suffers from a loss of ~100-fold catalytic efficiency for both pNPA hydrolysis and CO2 hydration20,21). Given that these interactions are not currently included in our well-defined minimal model, the most important next step is to begin incorporating hydrogen bonding networks with the goal of improving reactivity. However, the design of hydrogen bonding channels could be strongly dependent on location of the metal site in the 3SCC and factors such as solvent and substrate access and metal-binding affinities. The effects of changing the location to accommodate such a channel in an α-helical system on the catalytic efficiency are not known. The aim of this study is to address how catalytic efficiency may be affected when moving the active site to different locations to control rate, substrate access, and binding affinity, depending on the desired secondary interactions.
The general sequences for the peptides used in the present work (Table 1) retains the Hg(II)-bound tris-thiolate site and the Zn(II)(His)3(Solvent) site but alters the structures in a systematic way. We have previously reported that there are differences in selectivity for heavy metal binding to thiols in a versus d sites in each heptad.22–24 We have also demonstrated how the position of a metal site along the sequence of a peptide that folds into a 3SCC can define the physical properties of Cd(II) binding.25 One might imagine that factors such as catalyst proximity to the frayed ends of the helix, orientation of the active site with respect to the helical dipole or the location near the center versus the end of the coiled coil might influence catalytic efficiency, rate, substrate access, and/or binding affinity in these systems. In addition, the presence of a second, non-reactive metal site (i.e., Hg(II)), could influence the kinetic parameters either directly through an electrostatic influence or by impacting the breathing motions/fraying of the coiled coil. A major benefit of our architecture is that it is very straightforward to move the metal site along the primary sequence in order to interrogate the chemistry in alternative positions.
Table 1.
Peptide Sequences Used in This Study
| Peptidea | Sequence |
|---|---|
| TRIL2W | Ac-G WKALEEK LKALEEK LKALEEK LKALEEK G-NH2 |
| TRIL2WL23H | Ac-G WKALEEK LKALEEK LKALEEK HKALEEK G- NH2 |
| TRIL9CL23H | Ac-G LKALEEK CKALEEK LKALEEK HKALEEK G- NH2 |
| TRIL9HL23C | Ac-G LKALEEK HKALEEK LKALEEK CKALEEK G- NH2 |
| TRIL9CL19H | Ac-G LKALEEK CKALEEK LKAHEEK LKALEEK G- NH2 |
C- and N-termini are capped by acetyl (Ac) and NH2 groups, respectively.
There is one example of a redesigned system in which a mononuclear non-heme iron site was incorporated into six different positions of thioredoxin (within three different types of surface locations: groove, shallow pocket, and deep pocket) for superoxide dismutase function.26 This study demonstrated that one could see significant functional differences in the metal center without “prior adaptations” for the protein structure. Since systematic variation of a catalytic center has never been examined within a helical (or any de novo designed) structure this allows us, for the first time, to be able to address how orientation and distance relative to the helical dipole and frayed termini matter, specifically with respect to metal-binding affinities, catalytic activity, and kinetic pKa’s. The results will have significant implications for the selection of active site location in future de novo design studies, particularly since to date the majority of systems incorporate metal sites into α-helical scaffolds.
METHODS
Peptide Synthesis and Purification
Peptides were synthesized on an Applied Biosystems 433A peptide synthesizer using standard protocols27 and purified and characterized as described previously.28 The concentrations for peptides containing Cys sites were determined as previously reported28, and those of TRIL2WL23H solutions were based on the tryptophan absorbance at 280 nm using ε = 5500 cm−1 M−1.
Ultraviolet-Visible (UV-Vis) and Circular Dichroism (CD) Spectroscopy
CD and UV-Vis spectra were recorded in quartz cuvettes at 25°C on an Aviv 62DS spectrometer and Cary 100 Bio UV-Vis spectrometer, respectively. Guanidine hydrochloride CD titrations were performed at pH 8.5 as described previously.9 The UV-Vis spectra of Hg(II) binding to the Cys sites in each peptide were obtained as previously described at pH 8.5.10,29,30 All solutions were purged with argon prior to use in order to minimize oxidation of peptides and formation of disulfide bonds.
Competitive Zn(II)-Binding Titrations
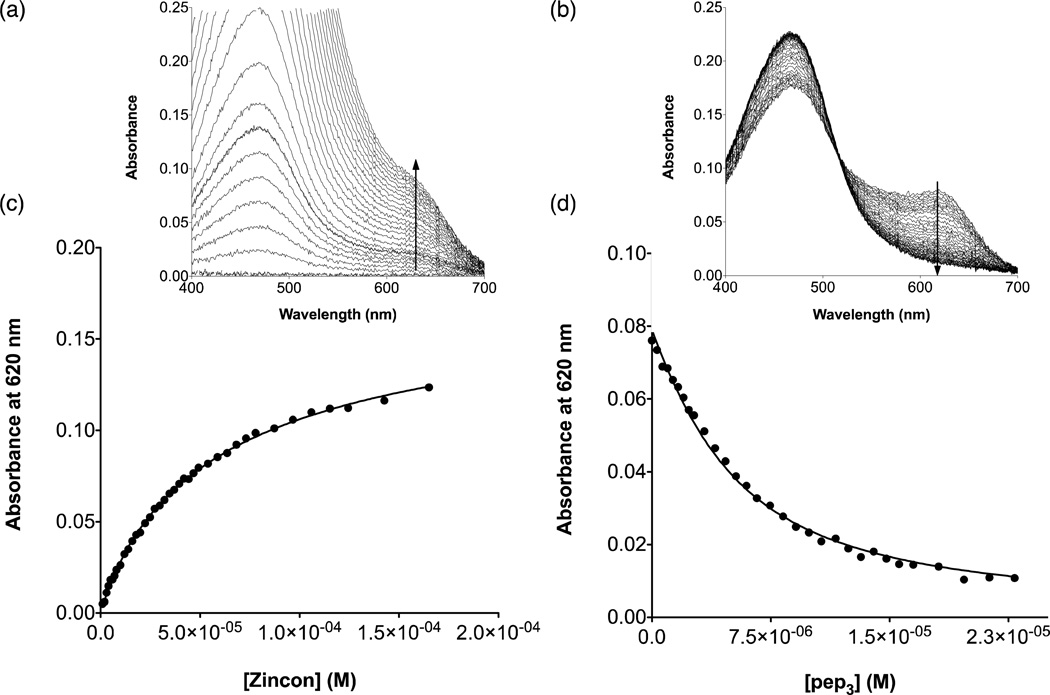
The apparent binding constants were determined by competition assay with the colorimetric Zn(II) chelator, Zincon (2-carboxy-2’-hydroxy-5’-(sulfoformazyl)benzene).31,32 Zn(II) forms a 1:1 complex with Zincon (Zi) with a distinct absorption band at 620 nm (ε ~16000 cm−1 M−1) at pH 7.5 and apparent dissociation constant (Kd,app) 2.80 × 10−6 M (consistent with the range of reported values).31,33–37 These parameters were evaluated for each pH and each buffer used in our experiments (pH 7.5 in 50 mM HEPES, 0.1 M Na2SO4 and pH 9.0 in 50 mM CHES, 0.1 M Na2SO4) by titrating Zn(II) into a solution of 10 µM Zi. For pH 9.0, ε620 ~27500 cm−1 M−1 and Kd,app = 5 × 10−8 M (a 5 cm pathlength cell was used for the calibration titration at this pH). For experiments involving peptide, an excess of Zi over ZnSO4 (10 µM Zi and 5 µM Zn(II) to ensure most Zn(II) is bound to ligand) was used as the starting point of the titration. Analogously, Zn(II) was bound to an excess of peptide (20 µM pep3 and 10 µM Zn(II)) for the reverse titration (in the case for [Hg(II)]S(TRIL9HL23C)3−, 5 µM Zi/2.5 µM Zn(II)and 30 µM pep3/15 µM Zn(II)were used for the forward and reverse titrations, respectively, because this peptide had a weaker Kd,app compared to the others). Both approaches should give similar apparent binding constants for Znpep3, indicating equilibrium had been reached.
The binding equilibrium for this competition experiment can be expressed by eq 1.
| (1) |
The apparent dissociation constant of Znpep3 can be calculated using eq 2.
| (2) |
For the forward titration, where a solution of Zi is titrated into Znpep3, the absorption band at 620 nm is due to the ZnZi complex, and increases as Zi is added to Znpep3, reflecting transfer of Zn(II) from pep3 to Zi and therefore yielding [ZnZi] and [Znpep3] for eq 2. [Znpep3] can be defined as the total amount of Zn(II) present ([Zn]T) minus the Zn(II)-bound fraction ([ZnZi]), under the conditions of this experiment. The amount of unbound peptide can then be defined as the total peptide minus the Zn(II)-bound fraction, [pep3] = [pep3]T – [Znpep3] = [pep3]T – ([Zn]T – [ZnZi]). The amount of free Zi is then related to the total amount of Zi minus the Zn(II)-bound fraction, [Zi] = [Zi]T - [ZnZi]. Substituting all of the above into eq 2 yields a quadratic expression, eq 3, which can be solved for [ZnZi]. Since this quantity is directly related to the absorbance at 620 nm by the Beer-Lambert law, the real solution to eq 3 can be inserted into eq 4 to give an equation which can be fitted in Prism 5 (GraphPad Software) to yield the dissociation constant for Znpep3.
| (3) |
| (4) |
An analogous approach was used for the reverse titration, in which pep3 is added to ZnZi. Representative titrations and fitted data are shown in Figure 2 and S4–6.
Figure 2.
Competitive Zincon binding titrations at pH 7.5 for [Hg(II)]S(TRIL9CL23H)3− in the forward (Zn(II)pep3 + Zincon) and reverse (Zn(II)Zi + pep3) directions. a) Representative UV-Vis spectra for the titration in the forward direction and b) in the reverse direction. c) Plot of absorbance at 620 nm as a function of increasing [Zincon] for the forward titration and d) as a function of increasing [pep3] for the reverse titration.
Esterase Activity Assays
The esterase activities of Zn(II)-bound peptides were determined spectrophotometrically with p-nitrophenyl acetate (pNPA, 200–1400 µM) substrate at 25°C. Measurements were made at 348 nm (ε = 5000 cm−1 M−1).38 The procedure is similar to that which was previously described10, with some modifications. The substrate solution was prepared by quickly diluting a 0.1 M pNPA acetone solution into doubly-distilled water (ddH2O) to a concentration of 3 mM. The procedure for measuring esterase activity is as follows: in a 1 mm pathlength quartz cuvette, buffer (50 mM, HEPES if pH 7.5 or 8, CHES for pH 8.5–9.65), ddH2O, and metal-peptide solution were mixed. pNPA was added, mixed, and the absorbance increase recorded every 25 seconds for 7–20 minutes. Metal-peptide solutions contained either excess peptide or excess ZnSO4 in order to ensure all Zn(II) was bound to the peptide. Controls then contained either apo-peptide or ZnSO4, respectively, and their initial rates were subtracted from those of Zn(II)-bound peptide samples. The controls containing ZnSO4 do not exhibit any activity over blank controls, but there is some activity from the free peptide, due to unbound His residues.10 Initial rates determined from linear fits of the first 2–10% of the reaction were plotted as a function of pNPA concentration and fitted to the Michaelis-Menten equation in Prism 5 (GraphPad Software). The concentration of enzyme is 10 or 20 µM and is accounted for in all reported values.
Acetate Inhibition Assays
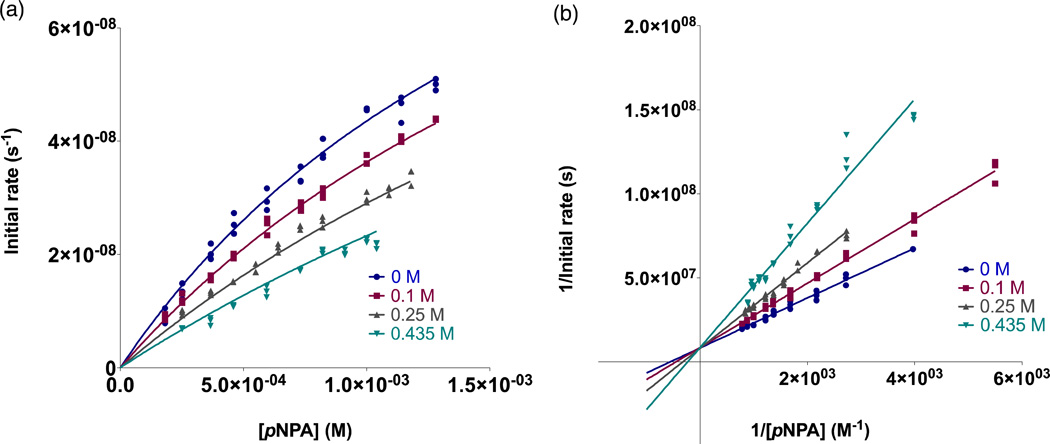
Inhibition experiments were performed under the same buffer conditions and with the same general procedure as above. Potassium acetate (KOAc) stock solutions (~7–10 M) were prepared by dissolving KOAc in a small amount of water, adjusting pH to 8.5 with glacial acetic acid (and vigorous stirring), followed by dilution to the final volume with ddH2O. The acetate concentration is determined by combining the concentration from the amount of solid and from the acetic acid added. For running the assays, buffer, ddH2O, KOAc, and enzyme were mixed together, then pNPA added and the absorbance monitored as above. Metal-peptide solutions were either 20 or 50 µM Znpep3 with 4× excess ZnSO4 present. Controls contained all components except peptide and the difference between initial rates was taken. Initial rates were plotted and fitted as described above. For each peptide, at least three different concentrations of KOAc were tested, and plots of initial rates vs. [pNPA] were prepared and fitted to competitive, non-competitive, and mixed inhibition equations in Prism 5 (GraphPad Software). The individual kinetic parameters for each concentration of acetate were examined in order to estimate the applicable inhibition model (all peptides had increasing KM and decreasing kcat values as a function of increasing substrate concentration supporting a competitive inhibition model).39 Lineweaver-Burke plots, in addition to examination of the α value from a mixed model fit for each complex, were also used to conclude that the inhibition observed was competitive for all peptide complexes containing a ZnHis3 site (Lineweaver-Burke plots intersect on the y-axis and α > 1). Figures 4 and S8–10 show the non-linear competitive fits and Lineweaver-Burke analysis for each complex.
Figure 4.
Inhibition of 20 µM [Zn(II)(H2O/OH)]N(TRIL2WL23H)3n+-catalyzed pNPA hydrolysis by acetate. a) Initial rates as a function of substrate concentration in the presence of 0 M (•), 0.1 M (■), 0.25 M (▲), and 0.435 M (▼) potassium acetate fitted to a competitive inhibition model in Prism 5 (GraphPad Software). The global data yields the reported KI and corresponding error. Data shown consists of each individual measured initial rate and does not represent averages. Fitting the same data to a mixed inhibition model yielded α ≈ 6 × 1019 supporting the assignment of a competitive inhibition model39 b) Lineweaver-Burke (double-reciprocal) plots corresponding to the data in a). Visual inspection of the intersection of linear fits to each dataset (at the y-axis) also supports a competitive inhibition model.
The apo-peptides can also catalyze pNPA hydrolysis due to the presence of free histidine residues.10 A control experiment in which we attempted to inhibit this activity using 0.5 M acetate showed some inhibition (it is possible that acetate may block pNPA from entering the 3SCC to some extent), but it was less than half of that observed in the presence of Zn(II) and did not strictly fit any inhibition model. Therefore, we conclude that the observed competitive inhibition is due to acetate competing for the ZnN3O site.
RESULTS AND DISCUSSION
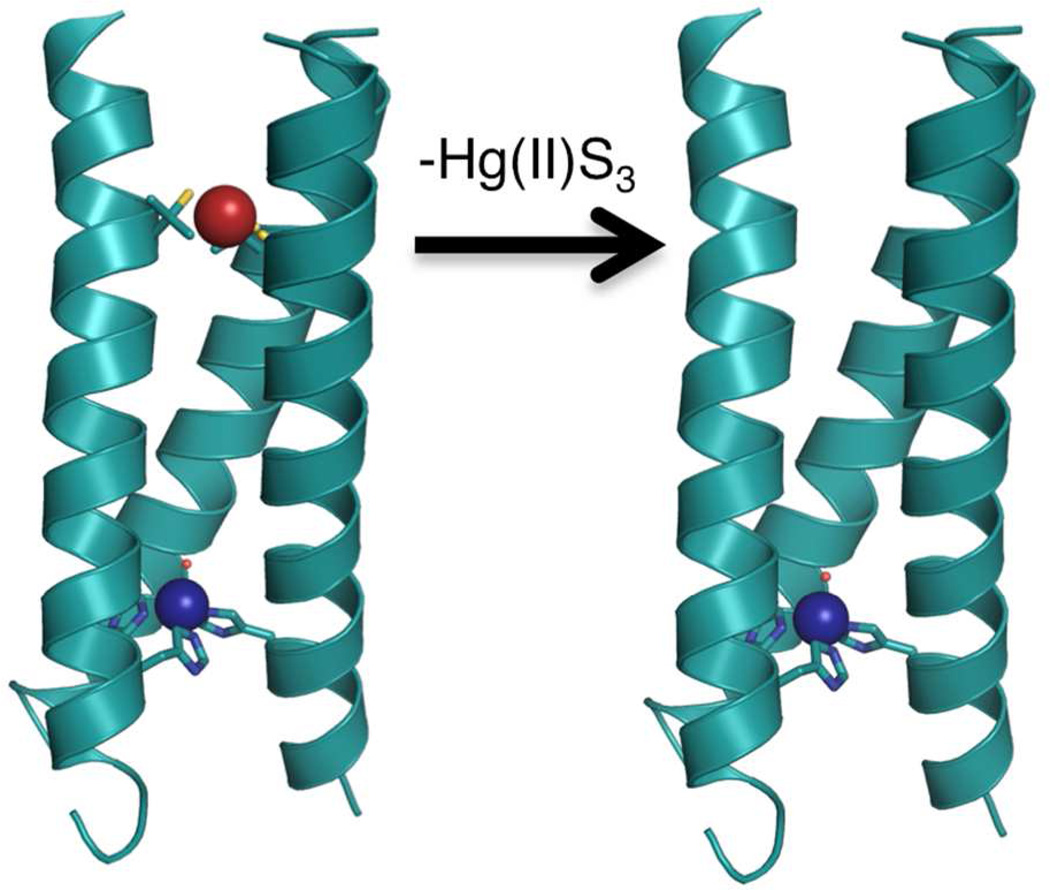
Our first example of catalyst modification was to remove the heavy metal-binding site at the 9 position of the peptide (Figure 1). The site had been developed in order to stabilize the peptide, especially at high pH, since the addition of three histidines in the same layer of the 3SCC potentially could lead to increased fraying or even dissociation of the third α-helix. However, by incorporating an Hg(II)(SR)3− center into the peptide, we have introduced both an additional divalent cation into the construct and the presence of a negatively charged center in relatively close proximity to the catalytic site. By removing the cysteines in TRIL2WL23H, we may assess whether this mercury-based stabilizing site influences Zn(II) binding affinity or the kinetic parameters for the pNPA hydrolysis reaction. We previously assessed the stabilities of (TRIL23H)3 and (TRIL9CL23H)33− through their guanidine hydrochloride-induced unfolding titrations.10 We concluded that the Hg(II) binding had the desired stabilization effect, which was particularly important at higher pH values (>9).
Figure 1.
Comparison of the X-ray crystal structure of [Hg(II)]S[Zn(II)(H2O/OH−)]N(CSL9PenL23H)3n+ (3PBJ)10on left with a Pymol model of [Zn(II)(H2O/OH−)]N(TRIL2WL23H)3n+ lacking the Hg(II)S3 structural site (right, based on 3PBJ).
The colorimetric chelator Zincon was used in competitive binding titrations to determine the binding affinity of Zn(II) to the His3 sites. The reaction of (TRIL2WL23H)3 with Zn(II) to form [Zn(II)(H2O/OH−)]N(TRIL2WL23H)3n+ yielded dissociation constants of 0.6 ± 0.1 µM and 0.24 ± 0.02 µM at pH 7.5 and 9.0, respectively (Table 2, Figure 2) and are consistent with designed Zn(II) binding sites with three protein ligands.12,13,15–17,40–42 This compares with the corresponding Kd = 0.8 ± 0.1 µM and 0.22 ± 0.06 µM at pH 7.5 and 9.0, respectively, for Zn(II) binding to [Hg(II)]S(TRIL9CL23H)3−. Clearly, there are no significant differences between these values, indicating that while the overall peptide association is stabilized by Hg(II), the affinity of the protein for Zn(II) is unaffected. This observation does not follow the previous report that the binding of Cd(II) to trigonal thiolate sites in this class of designed proteins exhibited a linear free energy correlation between peptide self-association and Cd(II) dissociation constants.43
Table 2.
Apparent Dissociation Constants for Zn(II) Binding to His3 Sites in the TRI Peptides
| Peptide complex |
Kd,app at pH 7.5 [µM]a |
Kd,app at Ph 9.0 [µM]b |
|---|---|---|
| [ZnII(H2O/OH−)]N(TRIL2WL23H)3n+ | 0.6 ± 0.1 | 0.24 ± 0.02 |
| [HgII]S[ZnII(H2O/OH−)]N(TRIL9CL23H)3n+ | 0.8 ± 0.1 | 0.22 ± 0.06 |
| [ZnII(H2O/OH−)]N[HgII]S(TRIL9HL23C)3n+ | ~8 | 0.8 ± 0.3 |
| [HgII]S[ZnII(H2O/OH−)]N(TRIL9CL19H)3n+ | 3.7 ± 1.3 | 0.4 ± 0.2 |
pH 7.5 measured in 50 mM HEPES buffer.
pH 9.0 measured in 50 mM CHES buffer.
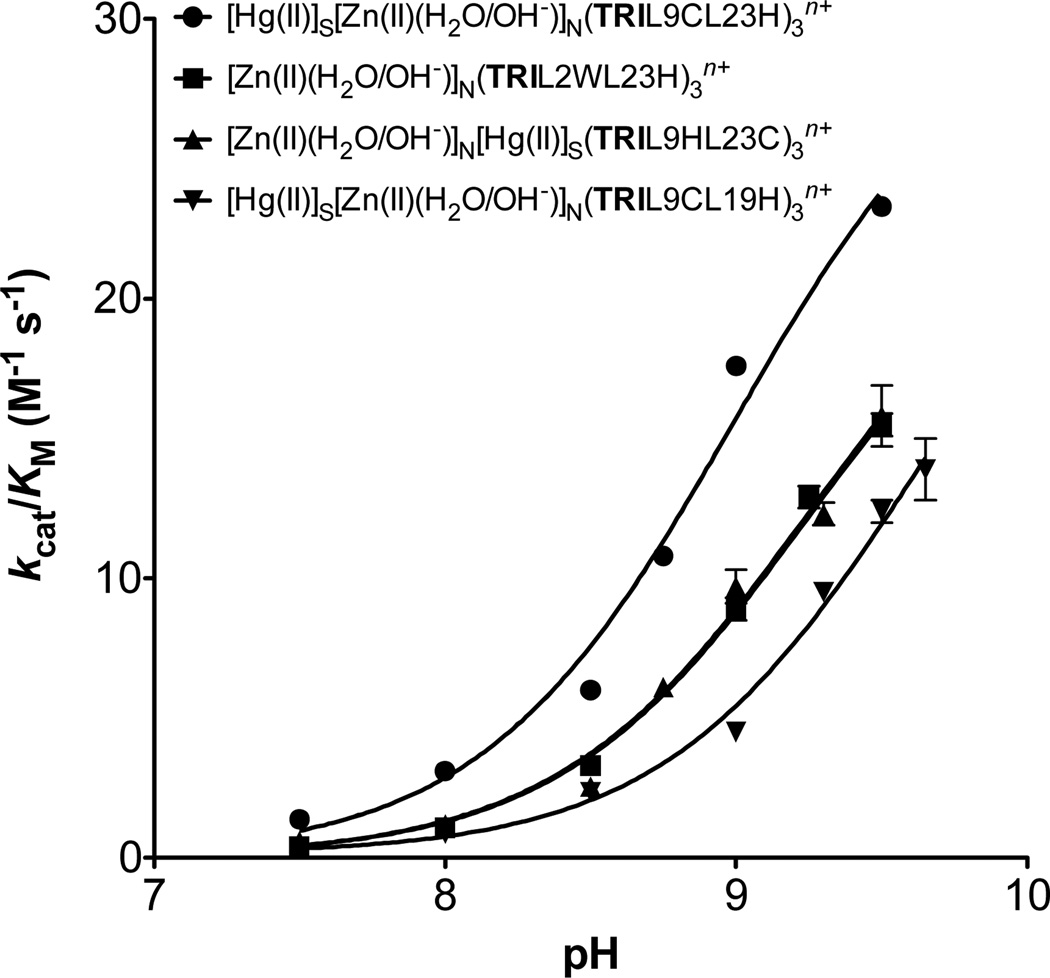
We next compared the hydrolytic activity of [Zn(II)(H2O/OH−)]N(TRIL2WL23H)3n+ towards pNPA to that of [Hg(II)]S([Zn(II)(H2O/OH−)]N(TRIL9CL23H)3n+. The data provided in Table 3 demonstrate that with respect to kcat, Km and kcat/Km there are no significant differences between the two constructs, except possibly at pH 9.5 where [Zn(II)(H2O/OH−)]N(TRIL2WL23H)3n+ has a slightly decreased catalytic efficiency which may be attributed to the decreased stability of this construct in basic media. Moreover, to analyze the kinetics of these complexes, plots of catalytic efficiencies vs. pH have been prepared and fitted to eq 5 in Prism 5 (GraphPad Software) to determine a kinetic pKa (Table 4, Figure 3).
| (5) |
Table 3.
pH-Dependent Kinetic Parameters for pNPA hydrolysis by Zn(II)-Bound TRI Peptidesa
| Peptide complexb | pHc |
kcat/KM [M−1 s−1] |
kcat [s−1] | KM [mM] |
|---|---|---|---|---|
| [Zn(II)(H2O/OH−)]N | 7.5 | 0.41 ± 0.03 | 0.0011 ± 0.0002 | 2.7 ± 0.8 |
| (TRIL2WL23H)3n+ | 8.0 | 1.07 ± 0.06 | 0.0029 ± 0.0005 | 2.7 ± 0.6 |
| 8.5 | 3.3 ± 0.2 | 0.0060 ± 0.0007 | 1.8 ± 0.3 | |
| 9.0 | 8.9 ± 0.4 | 0.016 ± 0.001 | 1.8 ± 0.2 | |
| 9.25 | 12.9 ± 0.4 | 0.022 ± 0.001 | 1.7 ± 0.2 | |
| 9.5 | 15.5 ± 0.4 | 0.033 ± 0.002 | 2.1 ± 0.2 | |
| [Hg(II)]S | 7.5 | 1.38 ± 0.04 | 0.0022 ± 0.0005 | 1.6 ± 0.4 |
| [Zn(II)(H2O/OH−)]N | 8.0 | 3.1 ± 0.1 | 0.0054 ± 0.0015 | 1.7 ± 0.5 |
| (TRIL9CL23H)3n+(d) | 8.5 | 6.0 ± 0.1 | 0.012 ± 0.004 | 1.9 ± 0.6 |
| 8.75 | 10.8 ± 0.3 | 0.021 ± 0.010 | 2.0 ± 0.9 | |
| 9.0 | 17.6 ± 0.3 | 0.038 ± 0.010 | 2.1 ± 0.6 | |
| 9.5 | 23.3 ± 0.3 | 0.040 ± 0.012 | 1.7 ± 0.5 | |
| [Zn(II)(H2O/OH−)]N | 7.5 | 0.6 ± 0.1 | 0.0005 ± 0.0001 | 0.9 ± 0.4 |
| [Hg(II)]S | 8.0 | 1.2 ± 0.2 | 0.0009 ± 0.0002 | 0.7 ± 0.3 |
| (TRIL9HL23C)3n+ | 8.5 | 1.9 ± 0.2 | 0.0027 ± 0.0007 | 1.4 ± 0.5 |
| 8.75 | 6.1 ± 0.2 | 0.0062 ± 0.0003 | 1.0 ± 0.1 | |
| 9.0 | 9.7 ± 0.6 | 0.011 ± 0.001 | 1.1 ± 0.1 | |
| 9.3 | 12.3 ± 0.4 | 0.016 ± 0.001 | 1.3 ± 0.1 | |
| 9.5 | 15.8 ± 1.1 | 0.020 ± 0.002 | 1.2 ± 0.2 | |
| [Hg(II)]S | 7.5 | 0.33 ± 0.01 | 0.0014 ± 0.0002 | 4.2 ± 0.9 |
| [Zn(II)(H2O/OH−)]N | 8.0 | 0.86 ± 0.02 | 0.0034 ± 0.0004 | 3.9 ± 0.5 |
| (TRIL9CL19H)3n+ | 8.5 | 2.35 ± 0.08 | 0.0062 ± 0.0007 | 2.5 ± 0.4 |
| 9.0 | 4.5 ± 0.2 | 0.010 ± 0.001 | 2.3 ± 0.4 | |
| 9.3 | 9.5 ± 0.3 | 0.022 ± 0.002 | 2.4 ± 0.3 | |
| 9.5 | 12.4 ± 0.4 | 0.035 ± 0.004 | 2.8 ± 0.4 | |
| 9.65 | 13.9 ± 1.1 | 0.04 ± 0.01 | 2.9 ± 1.0 |
Error bars result from fitting all individual initial rates measured (three per concentration of substrate, without averaging) to the Michaelis Menten equation in Prism 5 (GraphPad Software).
10–20 µM active Zn(II)-bound peptide complex.
pH 7.5–8.0 measured in 50 mM HEPES, pH 8.5–9.65 measured in 50 mM CHES.
Previously reported data included for comparison.10
Table 4.
Kinetic p Ka, Maximal Efficiency, and Maximal Rate Values forpNPA Hydrolysis by Zn(II)-Bound TRI Peptides
| Peptide complex | pKaa |
kcat/KM(max) [M−1S−1]b |
kcat(max) [s−1]c |
|---|---|---|---|
| [ZnII(H2O/OH−)]N(TRIL2WL23H)3n+ | 9.2 ± 0.1 | 25 ± 2 | ~0.055 |
| [HgII]S[ZnII(H2O/OH−)]N(TRIL9CL23H)3n+ | 9.0 ± 0.1 | 31 ± 4 | ~0.053 |
| [ZnII(H2O/OH−)]N[HgII]S(TRIL9HL23C)3n+ | 9.2 ± 0.1 | 24 ± 3 | ~0.030 |
| [HgII]S[ZnII(H2O/OH−)]N(TRIL9CL19H)3n+ | 9.6 ± 0.1 | 27 ± 5 | ~0.076 |
Determined by fitting individual kcat/KM values versus pH.
Maximal catalytic efficiency from the fitting of kcat/KM values versus pH (assuming that 100% active enzyme complex is present).
Estimated maximal rate determined as described in the text.
Figure 3.
pH-dependency of the catalytic efficiency for pNPA hydrolysis by Zn(II)-bound TRI peptides: [Hg(II)]S[Zn(II)(H2O/OH−)]N(TRIL9CL23H)3n+ (•), [Zn(II)(H2O/OH−)]N(TRIL2WL23H)3n+ (■), [Zn(II)(H2O/OH− )]N[Hg(II)]S(TRIL9HL23C)3n+ (▲), [Hg(II)]S[Zn(II)(H2O/OH−)]N(TRIL9CL19H)3n+ (▼).
The pH profiles for both peptides are essentially identical.
In addition to direct hydrolysis measurements, we also investigated product inhibition using acetate at pH 8.5. Both peptides exhibited competitive inhibition profiles with KI values of 0.34 ± 0.01 M and 0.32 ± 0.01 M for [Zn(II)(H2O/OH−)]N(TRIL2WL23H)3n+ and [Hg(II)]S([Zn(H2O/OH−)]N(TRIL9CL23H)3 n+, respectively (Table 5, Figure 4). These data taken together indicate that the presence of the Hg(II) has minimal impact on Zn(II)-binding affinity, or any of the kinetic parameters for substrate hydrolysis or product inhibition. Thus, we can conclude that while the Hg(II) ion serves to stabilize the system, it does not appear to diminish the kinetic parameters or metal-binding affinities. Because Hg(II) stabilizes the peptide at higher pH where activity is greatest and since Hg(II) allows us to use UV-Vis spectroscopy to monitor whether the peptide remains associated as a 3SCC,30 we subsequently carried out the remainder of the studies with Hg(II) present.
Table 5.
Kinetic Parameters for the Inhibition of pNPA Hydrolysis by Zn(II)-bound TRI Peptides at pH 8.5a
| Peptide complexb& KI (M) |
[OAc−] [M] |
kcat/KM [M−1 s−1] |
kcat [s−1] | KM [mM] |
|---|---|---|---|---|
| [Zn(II)(H2O/OH−)]N | 0 | 3.2 ± 0.1 | 0.0065 ± 0.0006 | 2.0 ± 0.3 |
| (TRIL2WL23H)3n+ | 0.1 | 2.51 ± 0.05 | 0.0068 ± 0.0004 | 2.7 ± 0.2 |
| KI (M) = 0.34 ± | ||||
| 0.01 | 0.25 | 2.01 ± 0.07 | 0.0058 ± 0.0006 | 2.9 ± 0.4 |
| 0.435 | 1.32 ± 0.07 | 0.0067 ± 0.0021 | 5.1 ± 1.9 | |
| [Hg(II)]S | 0 | 5.4 ± 0.1 | 0.011 ± 0.001 | 2.0 ± 0.2 |
| [Zn(II)(H2O/OH−)]N | 0.1 | 4.5 ± 0.2 | 0.012 ± 0.001 | 2.6 ± 0.4 |
| (TRIL9CL23H)3n+ | ||||
| KI (M) = 0.32 ± | 0.2 | 3.6 ± 0.2 | 0.010 ± 0.002 | 2.6 ± 0.6 |
| 0.01 | 0.4 | 2.5 ± 0.1 | 0.012 ± 0.003 | 5.0 ± 1.2 |
| 0.6 | 1.8 ± 0.1 | 0.010 ± 0.003 | 5.7 ± 1.8 | |
| [Zn(II)(H2O/OH−)]N | 0 | 2.54 ± 0.07 | 0.0039 ± 0.0002 | 1.5 ± 0.1 |
| [Hg(II)]S | 0.1 | 1.65 ± 0.02 | 0.0047 ± 0.0002 | 2.9 ± 0.2 |
| (TRIL9HL23C)3n+ | 0.25 | 1.09 ± 0.06 | 0.0036 ± 0.0008 | 3.3 ± 0.9 |
| KI (M) = 0.20 ± | ||||
| 0.01 | 0.5 | 0.72 ± 0.03 | 0.0042 ± 0.0012 | 5.8 ± 1.9 |
| [Hg(II)]S | 0 | 2.06 ± 0.06 | 0.0058 ± 0.0006 | 2.8 ± 0.4 |
| [Zn(II)(H2O/OH−)]N | 0.2 | 1.42 ± 0.04 | 0.0078 ± 0.0014 | 5.5 ± 1.1 |
| (TRIL9CL19H)3n+ | 0.4 | 0.94 ± 0.06 | 0.006 ± 0.003 | 6.5 ± 3.3 |
| KI (M) = 0.36 ± | ||||
| 0.01 | 0.6 | 0.73 ± 0.03 | 0.007 ± 0.003 | 9.1 ± 47 |
Error bars result from fitting all individual initial rates measured (three per concentration of substrate, without averaging) to the Michaelis Menten equation in Prism 5 (GraphPad Software).
20–50 µM active Zn(II)-bound peptide complex.
We next turned our attention to TRIL9HL23C, which inverts the sequence positions of the structural and catalytic sites. Provided that Zn(II) and coordinated solvent are oriented similarly toward the N-terminus of the coiled coil, as found in the structure of the previously reported10 [Hg(II)]S[Zn(II)(H2O/OH−)]N(CSL9PenL23H)3n+, the bound water/hydroxide should now be more exposed to bulk solvent and the site may better accommodate the substrate (Figure 5). The trimer containing a Cys3 site was evaluated both in the presence and absence of one equivalent of HgCl2 per trimer. The peptide has CD spectra characteristic of well-folded α-helical coiled coils at pH 8.5 with molar ellipticity values [Θ] obtained at 222 nm from –24000° dmol−1 cm2 to –27000° dmol−1 cm2 (Figure S1 and as previously reported10). As reported for (TRIL9CL23H)3−, the addition of Hg(II) to the Cys3 site further stabilizes (TRIL9HL23C)33− (Figure S1, S2). Hg(II) binding was evaluated using UV-Vis spectroscopy and confirms 1:1 binding in a trigonal geometry to the Cys3 site in (TRIL9HL23C)33− 10,29,30 Competitive binding titrations with Zincon yielded apparent dissociation constants in the µM range indicating that Zn(II) remains bound well, although still weak compared to native Zn(II) enzymes (often Kd = ~nM-pM).19,44–46 These values (Table 2, Figure S4, S6) are an order of magnitude weaker than determined for [Hg(II)]S[Zn(II)(H2O/OH− )]N(TRIL9CL23H)3n+ and [Zn(II)(H2O/OH−)]N(TRIL2WL23H)3n+ at pH 7.5 and a factor of four weaker at pH 9.0. Thus, while removal of Hg(II) does not alter Zn(II) affinity, reorientation of the Zn(II) site between the 9 and 23 positions (two a sites) leads to a ten-fold weaker binding to [Hg(II)]S(TRIL9HL23C)3− (Kd = ~8 µM) demonstrating that sequence position is important for metal affinity.
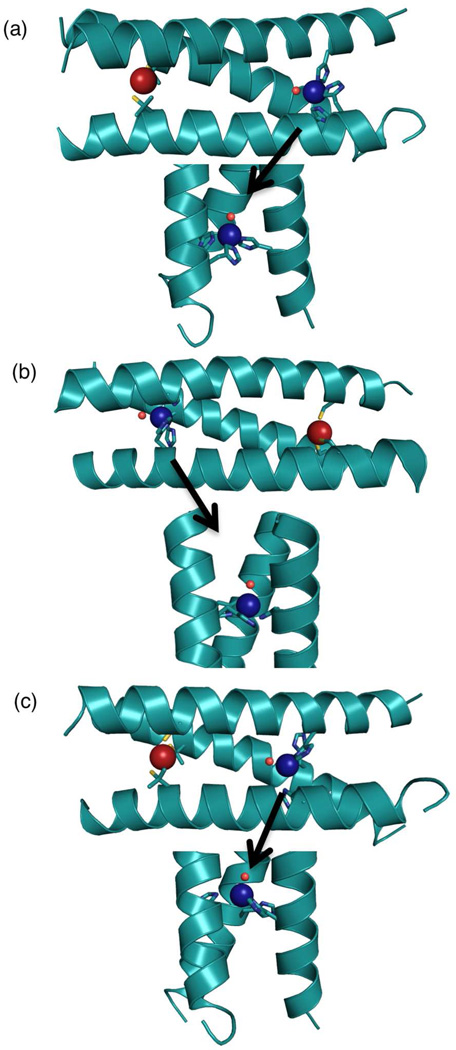
Figure 5.
Comparison of the X-ray crystal structure of a) [Hg(II)]S[Zn(II)(H2O/OH−)]N(CSL9PenL23H)3n+ (3PBJ)10with Pymol models of b) [Zn(II)(H2O/OH−)]N[Hg(II)]S(TRIL9HL23C)3n+ based on the coordinates of 2JGO48and c) [Hg(II)]S[Zn(II)(H2O/OH−)]N(TRIL9CL19H)3n+ based on the coordinates of 3PBJ. Models were prepared in Pymol using the mutagenesis option and Pymol’s rotamer library.49
We had previously shown that the pKa for thiolate binding to Cd(II) was critically dependent on whether cysteines were incorporated into a or d sites and subsequent X-ray crystallographic analysis indicated that the thiolate conformations were varied between the two types of positions.23,47 Reasoning that similar behavior might occur with Zn(II) imidazoles in a versus d sites along the sequence, we prepared TRIL9CL19H. Here, the structural site was held constant relative to the original model, but the catalytic site was moved one Leu layer closer to the N-terminus into a d site. Assessment of the stability of this complex in the presence and absence of Hg(II) revealed similar behavior to (TRIL9CL23H)33− and (TRIL9HL23C)33− (Figure S1, S2). Zn(II) binding to [Hg(II)]S(TRIL9CL19H)3− is about five-fold weaker (Kd = 3.7 ± 1.3 µM) than the L23H derivatives. This could be due to different orientation of the histidines in a vs. d sites or because the site is more constrained when located in the center of the coiled coil. All four peptides have stronger Kd’s (by three- to ten-fold) at pH 9.0 (Table 2), but follow the same trend with sequence position. All binding affinities are sufficiently strong to ensure Zn(II) is fully bound under the catalytic conditions described below. To our knowledge, this is the first systematic evaluation of changes in binding affinities of a catalytic site as it is varied along a helical structure.
We next evaluated the catalytic activities of these two designs. Hydrolysis of pNPA by [Zn(II)(H2O/OH− )]N[Hg(II)]S(TRIL9HL23C)3n+ and [Hg(II)]S[Zn(II)(H2O/OH− )]N(TRIL9CL19H)3n+ was monitored as a function of pH. Both of these peptide complexes show diminished catalytic efficiency (none more than two-fold) relative to our original model at pH 9.5 (Table 3).
The pKa’s of [Zn(II)(H2O/OH−)]N(TRIL2WL23H)3n+, [Hg(II)]S [Zn(II)(H2O/OH−)]N(TRIL9CL23H)3n+, and [Zn(II)(H2O/OH− )]N[Hg(II)]S(TRIL9HL23C)3n+ are similar (9.0–9.2 ± 0.1); however, that of [Hg(II)]S[Zn(II)(H2O/OH−)]N(TRIL9CL19H)3n+ increases to 9.6 ± 0.2 (Figure 3, Table 4). This increase might be due to the location of the metal site further within the core of the 3SCC relative to the other designs or simply because it is a d site and the His ligands may be oriented differently. Presumably, the pKa represents deprotonation of Zn(II)-bound water to form coordinated hydroxide, which is proposed to be the hydrolytically active species. It appears that the position of the catalytic site along the sequence of the peptide has a modest role in tuning this pKa, with the highest value for the least solvent accessible site. The pKa’s for the complexes remain ~2 units higher than that for CAII (6.8). This suggests that more drastic modification to the second sphere environment and beyond (probably through incorporation of hydrogen bonding) will be required to reduce the pKa to a more desirable value.
Regardless, since the pKa’s for these complexes are not identical, comparing their rates at pH 9.5 may not provide a complete picture. The nature of the designed peptide does not allow for the rates to be measured at pH’s much higher than 9.5 because salt bridge interactions (Glu-Lys) that stabilize the coiled coil will be interrupted. However, in addition to pKa’s, eq 5 also yields a value for the maximal catalytic efficiency (theoretical maximal efficiency if 100% of the Zn(II)-hydroxide complex were present). Although individual catalytic efficiencies are obviously different from one peptide to the next at pH 9.5, these extrapolated maximal values are not very different, within error (Table 4) suggesting that ester hydrolysis is independent of the Zn(II) position. However, significant differences are observed when the individual kcat and KM values are analyzed. For all peptides, the KM values are generally pH-independent (Figure S7), but differ relative to each other. The KM’s of [Hg(II)]S[Zn(II)(H2O/OH−)]N(TRIL9CL23H)3n+ and [Zn(II)(H2O/OH−)]N(TRIL2WL23H)3n+ are the same (~2 mM) but those of [Zn(II)(H2O/OH−)]N[Hg(II)]S(TRIL9HL23C)3n+ are ~1 mM. This result supports the notion that when the catalytic site is moved to the N-terminal end of the sequence, the substrate can more easily access the active site since it is expected to point towards the solvent. When the catalytic site is moved to the 19th position, there seems to be a modest increase in the KM (2.8 ± 0.4 mM, pH 9.5). As with catalytic efficiency, we can plot kcat vs. pH to fit for kinetic pKa’s and maximal kcat values, but given the larger errors in kcat due to poor solubility of the substrate, the extrapolated values obtained from these fits are not reliable (Figure S7). Therefore, we estimate maximal kcat by using maximal kcat/KM and the KM for that particular peptide. Based on analysis of these values (Table 4), it is apparent that the most solvent-exposed site in [Zn(II)(OH− )]N[Hg(II)]S(TRIL9HL23C)3 has the lowest rate, while that furthest in the interior of the 3SCC, [Hg(II)]S[Zn(II)(OH− )]N(TRIL9CL19H)3, has the highest. There is no significant difference in these values between [Hg(II)]S[Zn(II)(OH− )]N(TRIL9CL23H)3 and the peptide lacking the Hg(II)S3 site.
Another method was needed to validate these conclusions on substrate binding access. Therefore, we evaluated product inhibition, which should follow a similar dependence to substrate binding. The products resulting from hydrolysis of pNPA are p-nitrophenol/p-nitrophenolate (pNP/pNP−) and acetate. No inhibition with either of these is observed under the conditions of the experiment (which only produces up to 1.4 mM product), demonstrating that product inhibition is not a concern during the course of our assays. This is an important benefit of our system since small molecule model complexes typically have extensive problems with-product inhibition. Further analysis was undertaken using potassium acetate as we reasoned that a smaller anion would have a better chance of binding in our complex (likely to Zn(II)) than pNP/pNP−. We found that higher concentrations of acetate (>0.1 M) can competitively inhibit pNPA hydrolysis by these complexes at pH 8.5 (Figure 4 and S8–10). No inhibition of the reaction was observed at pH 9.5, consistent with the trend for CA, where less inhibition by chloride is observed with increasing pH.50–52 This is presumably due to increasing competition with hydroxide for the Zn(II) center. Analogous to what was observed for the kinetic parameter KM, we found that the KI’s for all peptides were similar (~0.32–0.36 M, Table 5) except that for [Zn(II)(H2O/OH−)]N[Hg(II)]S(TRIL9HL23C)3n+ (0.20 M). This finding further supports the notion that moving the active site to the N-terminal end of the trimer leads to increased substrate and solvent access. This is an important finding for future design endeavors in which one may wish to control substrate access and selectivity or the level of product inhibition in a helical structure.
CONCLUSIONS
Herein we have presented the first detailed study of systematic movement of an active site along the sequence of an α-helical coiled coil in order to examine how changes in location of a metal site affect its properties. This work, combined with that for other designs12,13,15–17,40–42, suggests there may be a limit to the affinity that can be achieved solely with three protein ligands. Only a ~ten-fold variation in binding affinity is observed when the active site is moved along the 3SCC. Additionally, location of the active site at either end of a helical bundle does not appear to play a large role in tuning the Zn-OH2 pKa, thus allowing the desired metal site to be placed into the structure based on where it may be easiest to implement further secondary interactions to tune the pKa and catalytic activity, such as hydrogen bonding networks. However, because a change in pKa is observed upon going further into the interior of the coiled coil to a d site, one may exploit this to achieve effectively different catalytic efficiencies when designing models for specific pH conditions.
By moving the site from the C-terminus of the coiled coil to the N-terminus, solvent, substrate, and inhibitor accessibilities are significantly increased, whereas when the metal center is moved further into the core of the structure (and to a d site), access decreases modestly. As a consequence, the estimated maximal rate of the 19 site increases by a factor of 2.5 relative to the 9 site. Nevertheless, the fact that the catalytic efficiency (kcat/KM) is retained in all of these models implies that this minimal first-coordination-sphere only Zn(II) site is alone enough to confer significant hydrolytic activity. This finding means that, at least in these systems, the location of the minimal active site can be chosen to optimize substrate access and metal binding and for proximity to ideal positions for secondary interactions, all while retaining catalytic efficiency. The two complexes with Zn(II)His3 sites in the same sequence position (one with and one without the structural site) give similar results for almost all of the measured parameters. Thus, a separate stabilizing site is not detrimental to the properties of the catalytic site, demonstrating that future designs incorporating two separate metal centers with different or complementary functions can be realized.
The lack of any significant change in catalytic efficiency for the hydrolysis reactions in these coiled coils implies that future work will focus on refining the second coordination sphere environment around the metal center and defining solvent channels, but not be overly concerned about the effect of location on catalytic activity. This is contrary to the previous observation that location of an FeHis3 site in thioredoxin plays an important role in oxygen reactivity. Certainly, with coiled coils sequence position does not account for the orders-of-magnitude differences between designed metalloproteins and native enzymes. Overall, this work provides an excellent foundation for the engineering of a hydrolytically active minimal Zn(II) site into an α-helical coiled coil.
Supplementary Material
ACKNOWLEDGMENT
V.L.P. thanks the National Institutes of Health for support of this research (ES012236). M.L.Z. thanks the National Institutes of Health Chemistry-Biology Interface Training Program for support of this research.
Footnotes
ASSOCIATED CONTENT
Supporting Information.
Results from circular dichroism for folding of peptides and guanidine hydrochloride-induced unfolding; fitted data for competitive Zincon binding titrations at pH 9.0 for [Hg(II)]S[Zn(II)(OH−)]N(TRIL9CL23H)3 and at pH 7.5 and 9.0 for [Zn(II)(H2O/OH−)]N(TRIL2WL23H)3n+, [Zn(II)(H2O/OH−)]N[Hg(II)]S(TRIL9HL23C)3n+, and [Hg(II)]S[Zn(II)(OH−)]N(TRIL9CL19H)3; plots of pH-dependency of kcat and KM parameters for pNPA hydrolysis by Zn(II)-bound TRI peptides; fitted data for acetate inhibition of pNPA hydrolysis for [Hg(II)]S[Zn(II)(OH−)]N(TRIL9CL23H)3, [Zn(II)(H2O/OH−)]N[Hg(II)]S(TRIL9HL23C)3n+, and [Hg(II)]S[Zn(II)(OH−)]N(TRIL9CL19H)3. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The manuscript was written through contributions of all authors.
Notes
The authors declare no competing financial interest.
REFERENCES
- 1.Lu Y, Yeung N, Sieracki N, Marshall NM. Nature. 2009;460:855–862. doi: 10.1038/nature08304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nanda V, Koder RL. Nat. Chem. 2010;2:15–24. doi: 10.1038/nchem.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zastrow ML, Pecoraro VL. Coordination Chemistry Reviews. 2013 doi: 10.1016/j.ccr.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Handel T, DeGrado WF. J. Am. Chem. Soc. 1990;112:6710–6711. [Google Scholar]
- 5.Klemba M, Regan L. Biochemistry. 1995;34:10094–10100. doi: 10.1021/bi00031a034. [DOI] [PubMed] [Google Scholar]
- 6.Lombardi A, Summa CM, Geremia S, Randaccio L, Pavone V, DeGrado WF. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6298–6305. doi: 10.1073/pnas.97.12.6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faiella M, Andreozzi C, De Rosales RTM, Pavone V, Maglio O, Nastri F, DeGrado WF, Lombardi A. Nat. Chem. Biol. 2009;5:882–884. doi: 10.1038/nchembio.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reig AJ, Pires MM, Snyder RA, Wu Y, Jo H, Kulp DW, Butch SE, Calhoun JR, Szyperski T, Szyperski TG, Solomon EI, DeGrado WF. Nat. Chem. 2012;4:900–906. doi: 10.1038/nchem.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dieckmann GR, McRorie DK, Lear JD, Sharp KA, DeGrado WF, Pecoraro VL. J. Mol. Biol. 1998;280:897–912. doi: 10.1006/jmbi.1998.1891. [DOI] [PubMed] [Google Scholar]
- 10.Zastrow ML, Peacock AFA, Stuckey JA, Pecoraro VL. Nat. Chem. 2012;4:118–123. doi: 10.1038/nchem.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Der BS, Edwards DR, Kuhlman B. Biochemistry. 2012;51:3933–3940. doi: 10.1021/bi201881p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiyokawa T, Kanaori K, Tajima K, Koike M, Mizuno T, Oku J-I, Tanaka T. J. Pept. Res. 2004;63:347–353. doi: 10.1111/j.1399-3011.2004.00109.x. [DOI] [PubMed] [Google Scholar]
- 13.Pessi A, Bianchi E, Crameri A, Venturini S, Tramontano A, Sollazzo M. Nature. 1993;362:367–369. doi: 10.1038/362367a0. [DOI] [PubMed] [Google Scholar]
- 14.Zhu C, Zhang C, Liang H, Lai L. Protein Cell. 2011;2:1006–1013. doi: 10.1007/s13238-011-1121-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Regan L. Trends Biochem. Sci. 1995;20:280–285. doi: 10.1016/s0968-0004(00)89044-1. [DOI] [PubMed] [Google Scholar]
- 16.Müller HN, Skerra A. Biochemistry. 1994;33:14126–14135. doi: 10.1021/bi00251a023. [DOI] [PubMed] [Google Scholar]
- 17.Vita C, Roumestand C, Toma F, Ménez A. Proc. Natl. Acad. Sci. U.S.A. 1995;92:6404–6408. doi: 10.1073/pnas.92.14.6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khare SD, Kipnis Y, Greisen PJ, Takeuchi R, Ashani Y, Goldsmith M, Song Y, Gallaher JL, Silman I, Leader H, Sussman JL, Stoddard BL, Tawfik DS, Baker D. Nat. Chem. Biol. 2012;8:294–300. doi: 10.1038/nchembio.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christianson DW, Fierke CA. Acc. Chem. Res. 1996;29:331–339. [Google Scholar]
- 20.Liang Z, Xue Y, Behravan G, Jonsson BH, Lindskog S. Eur. J. Biochem. 1993;211:821–827. doi: 10.1111/j.1432-1033.1993.tb17614.x. [DOI] [PubMed] [Google Scholar]
- 21.Krebs JF, Ippolito JA, Christianson DW, Fierke CA. J. Biol. Chem. 1993;268:27458–27466. [PubMed] [Google Scholar]
- 22.Peacock AFA, Iranzo O, Pecoraro VL. Dalton Trans. 2009;9226:2271–2280. doi: 10.1039/b818306f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matzapetakis M, Farrer BT, Weng T-C, Hemmingsen L, Penner-Hahn JE, Pecoraro VL. J. Am. Chem. Soc. 2002;124:8042–8054. doi: 10.1021/ja017520u. [DOI] [PubMed] [Google Scholar]
- 24.Matzapetakis M, Pecoraro VL. J. Am. Chem. Soc. 2005;127:18229–18233. doi: 10.1021/ja055433m. [DOI] [PubMed] [Google Scholar]
- 25.Iranzo O, Chakraborty S, Hemmingsen L, Pecoraro VL. J. Am. Chem. Soc. 2011;133:239–251. doi: 10.1021/ja104433n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benson DE, Wisz MS, Hellinga HW. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6292–6297. doi: 10.1073/pnas.97.12.6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan WC, White PD. In: Fmoc Solid Phase Peptide Synthesis: A Practical Approach. Chan WC, White PD, editors. Vol. 222. New York: Oxford University Press; 2000. [Google Scholar]
- 28.Farrer BT, Harris NP, Balchus KE, Pecoraro VL. Biochemistry. 2001;40:14696–14705. doi: 10.1021/bi015649a. [DOI] [PubMed] [Google Scholar]
- 29.Pecoraro VL, Peacock AFA, Iranzo O, Luczkowski M. ACS Symp. Ser. 2009;1012:183–197. [Google Scholar]
- 30.Iranzo O, Thulstrup PW, Ryu S-B, Hemmingsen L, Pecoraro VL. Chem.--Eur. J. 2007;13:9178–9190. doi: 10.1002/chem.200701208. [DOI] [PubMed] [Google Scholar]
- 31.Rush RM, Yoe JH. Analytical Chemistry. 1954;26:1345–1347. [Google Scholar]
- 32.Sadek FS, Schmid RW, Reilley CN. Talanta. 1959;2:38–51. [Google Scholar]
- 33.Talmard C, Bouzan A, Faller P. Biochemistry. 2007;46:13658–13666. doi: 10.1021/bi701355j. [DOI] [PubMed] [Google Scholar]
- 34.Shaw CF, Laib JE, Savas MM, Petering DH. Inorganic Chemistry. 1990;29:403–408. [Google Scholar]
- 35.Mekmouche Y, Coppel Y, Hochgräfe K, Guilloreau L, Talmard C, Mazarguil H, Faller P. ChemBioChem. 2005;6:1663–1671. doi: 10.1002/cbic.200500057. [DOI] [PubMed] [Google Scholar]
- 36.Laib J, Shaw CF, Petering DH, Eidsness MK, Elder RC, Garvey JS. Biochemistry. 1985;24:1977–1986. doi: 10.1021/bi00329a027. [DOI] [PubMed] [Google Scholar]
- 37.Säbel CE, Neureuther JM, Siemann S. Anal. Biochem. 2010;397:218–226. doi: 10.1016/j.ab.2009.10.037. [DOI] [PubMed] [Google Scholar]
- 38.Verpoorte JA, Mehta S, Edsall JT. J. Biol. Chem. 1967;242:4221–4229. [PubMed] [Google Scholar]
- 39.Marangoni AG. Enzyme Kinetics; reversible enzyme inhibition. Hoboken, NJ, USA: John Wiley & Sons, Inc; 2002. pp. 61–69. [Google Scholar]
- 40.Stewart JD, Roberts VA, Crowder MW, Getzoff ED, Benkovic SJ. J. Am. Chem. Soc. 1994;116:415–416. [Google Scholar]
- 41.Wade WS, Koh JS, Han N, Hoekstra DM, Lerner Ra. J. Am. Chem. Soc. 1993;115:4449–4456. [Google Scholar]
- 42.Handel TT, Williams SA, DeGrado WF. Science. 1993;261:879–885. doi: 10.1126/science.8346440. [DOI] [PubMed] [Google Scholar]
- 43.Ghosh D, Lee K-H, Demeler B, Pecoraro VL. Biochemistry. 2005;44:10732–10740. doi: 10.1021/bi0506674. [DOI] [PubMed] [Google Scholar]
- 44.Hitomi Y, Outten CE, O’Halloran TV. J. Am. Chem. Soc. 2001;123:8614–8615. doi: 10.1021/ja016146v. [DOI] [PubMed] [Google Scholar]
- 45.Fierke Ca, Thompson RB. Biometals. 2001;14:205–222. doi: 10.1023/a:1012980628412. [DOI] [PubMed] [Google Scholar]
- 46.Song H, Wilson DL, Farquhar ER, Lewis Ea, Emerson JP. Inorg. Chem. 2012;51:11098–11105. doi: 10.1021/ic301645j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chakraborty S, Touw DS, Peacock AFA, Stuckey J, Pecoraro VL. J. Am. Chem. Soc. 2010;132:13240–13250. doi: 10.1021/ja101812c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Touw DS, Nordman CE, Stuckey JA, Pecoraro VL. Proc. Natl. Acad. Sci. U.S.A. 2007;104:11969–11974. doi: 10.1073/pnas.0701979104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.The PyMOL Molecular Graphics System. Version 1.5.0.4. Schrödinger, LLC; [Google Scholar]
- 50.Pocker Y, Stone JT. Biochemistry. 1968;7:2936–2945. doi: 10.1021/bi00848a034. [DOI] [PubMed] [Google Scholar]
- 51.Pocker Y, Deits TL. J. Am. Chem. Soc. 1982;104:2424–2434. [Google Scholar]
- 52.Pocker Y, Stone JT. Biochemistry. 1967;6:668–678. doi: 10.1021/bi00855a005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.