MICAL-L1 and the BAR-domain protein syndapin2 bind to phosphatidic acid (PA), a novel lipid component of recycling endosomes (REs). Interactions between these proteins stabilize their association with membranes and allow nucleation of tubules by syndapin2. A new role is highlighted for PA in recycling, suggesting a mechanism for tubular RE formation.
Abstract
Endocytic transport necessitates the generation of membrane tubules and their subsequent fission to transport vesicles for sorting of cargo molecules. The endocytic recycling compartment, an array of tubular and vesicular membranes decorated by the Eps15 homology domain protein, EHD1, is responsible for receptor and lipid recycling to the plasma membrane. It has been proposed that EHD dimers bind and bend membranes, thus generating recycling endosome (RE) tubules. However, recent studies show that molecules interacting with CasL-Like1 (MICAL-L1), a second, recently identified RE tubule marker, recruits EHD1 to preexisting tubules. The mechanisms and events supporting the generation of tubular recycling endosomes were unclear. Here, we propose a mechanism for the biogenesis of RE tubules. We demonstrate that MICAL-L1 and the BAR-domain protein syndapin2 bind to phosphatidic acid, which we identify as a novel lipid component of RE. Our studies demonstrate that direct interactions between these two proteins stabilize their association with membranes, allowing for nucleation of tubules by syndapin2. Indeed, the presence of phosphatidic acid in liposomes enhances the ability of syndapin2 to tubulate membranes in vitro. Overall our results highlight a new role for phosphatidic acid in endocytic recycling and provide new insights into the mechanisms by which tubular REs are generated.
INTRODUCTION
Mammalian cells constantly sample the extracellular milieu and internalize receptors, ligands, nutrients, and plasma membrane constituents, which include proteins and lipids. Accordingly, a high level of endocytic regulation is required to maintain the fine balance between internalized molecules and those returned to the plasma membrane through the process of recycling. In addition to maintaining cell shape by the return of lipids and receptors to the plasma membrane, endocytic recycling has specific effects on a variety of cellular processes. For example, studies show that vesicles involved in furrow cleavage and cytokinesis originate in part from recycling endosomes (Montagnac et al., 2008), as well as from the process of abscission, in which the cytokinetic bridge is severed (Skop et al., 2001; Fielding et al., 2005). Recycling also plays a crucial role in cell adhesion, spreading, and migration (Caswell and Norman, 2008) and has specialized roles in maintaining epithelial polarization (Wang et al., 2000), regulating cell fusion in myoblasts (Doherty et al., 2008), and controlling α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptors in neurons (Park et al., 2004).
A wide array of proteins regulate endocytic recycling; notably, the Rab-family proteins Rab11 and Rab4, as well as Rab21, Rab22, Rab8, Rab15, and Rab35 (Grant and Donaldson, 2009; Hsu and Prekeris, 2010). Various Rab effectors also control recycling (Grosshans et al., 2006), some of which interact with the C-terminal Eps15 homology domain protein 1 (EHD1), another known regulator of endocytic recycling (Naslavsky and Caplan, 2011). Studies demonstrated that EHD1 depletion affects the recycling of multiple receptors that are internalized both through clathrin-coated pits and independent of clathrin, including transferrin receptor (Lin et al., 2001), major histocompatibility complex class I proteins (Caplan et al., 2002), GLUT4 transporters, and other channels (Guilherme et al., 2004). β1 integrins are also subject to EHD1 regulation (Jovic et al., 2007), suggesting a rationale for the significance of recycling in cell migration.
Recent studies provided molecular and atomic mechanisms explaining the function of EHD1. The nuclear magnetic resonance solution structure of the EHD1 EH domain (Kieken et al., 2007) led to the identification of novel interaction partners containing asparagine–proline–phenylalanine (NPF) motifs followed by acidic residues that selectively interact with the positively charged EH domain electrostatic surface area (Henry et al., 2010; Kieken et al., 2010). Perhaps most striking are recent studies providing evidence that EHD proteins serve as dynamin-like ATPases (Lee et al., 2005; Daumke et al., 2007; Jakobsson et al., 2011; Cai et al., 2012), suggesting an important role for the protein in the scission of vesicles destined to be recycled back to the plasma membrane from the endocytic recycling compartment (ERC).
EHD1 and its Caenorhabditis elegans orthologue, Rme1, control the morphology of the ERC (Grant et al., 2001), which is a complex network of tubular and vesicular membranes, concentrated in some cells to the juxtanuclear region (Maxfield and McGraw, 2004). EHD1 localizes to and is the best-known marker of large recycling tubules, whose diameter can reach up to 200 nm (Caplan et al., 2002). Tubular intermediates play important roles in sorting and cargo selection (reviewed in Maxfield and McGraw, 2004). It was suggested that due to the high surface-to-volume ratio of tubular endosomes, membranes and recycling receptors can be preferentially segregated from soluble cargo in early/sorting endosomes (Maxfield and McGraw, 2004). Similar mechanisms are predicted for exit of receptors from the ERC to the plasma membrane.
Although evidence supports the notion that EHD1 localizes to tubular recycling endosomes and functions as a “pinchase” in membrane constriction and scission, the mode of biogenesis of these unique tubular membranes has remained an enigma. Although in vitro experiments show that EHD proteins are capable of inducing lipid tubulation (Daumke et al., 2007; Pant et al., 2009), studies in cells are not entirely consistent with this notion. For example, the recent identification of Molecules Interacting with CAsL-Like1 (MICAL-L1; see Figure 1A) as a novel tubular recycling endosome (RE) marker, EHD1 interaction partner (Sharma et al., 2009), and membrane hub (Rahajeng et al., 2012) has challenged this view. MICAL-L1, originally identified as an NPF-containing protein that binds to the EHD1 EH domain through its acidic-flanked NPF motif (Kieken et al., 2010), recruits both EHD1 and Rab8 to tubular REs (Sharma et al., 2009). However, depletion of EHD1 has no effect on the association of MICAL-L1 with tubular REs, demonstrating that they are generated and maintained even in the absence of EHD1 (Sharma et al., 2009).
FIGURE 1:
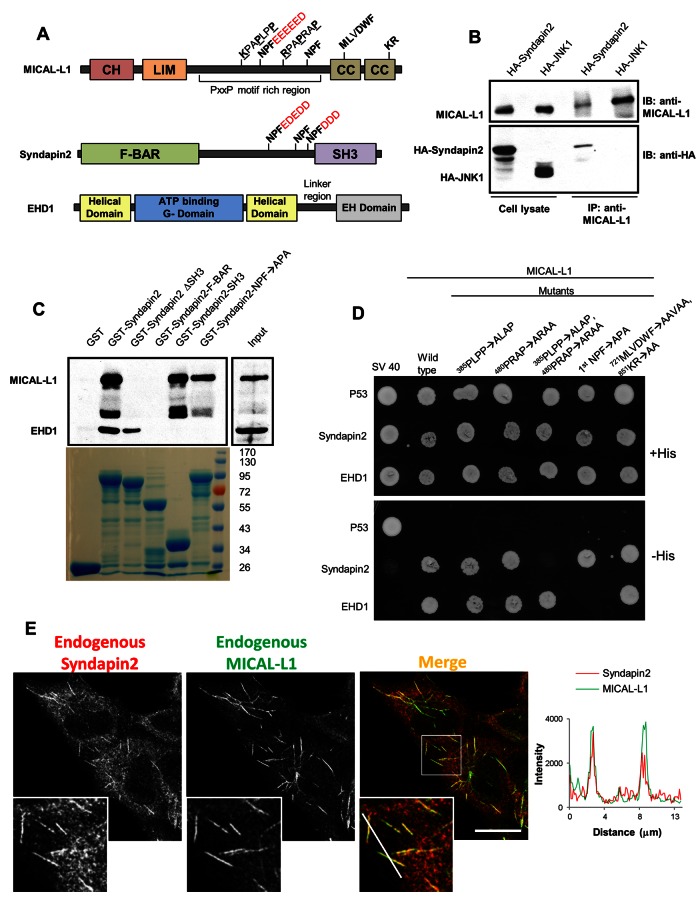
MICAL-L1 and Synd2 are direct binding partners that localize to tubular endosomes. (A) Domain architecture of MICAL-L1, Synd2, and EHD1. CH, calponin homology domain; LIM, Lin11, Isl-1, Mec-3 domain; CC, coiled coil domain; F-BAR, FCH-Bin–amphiphysin–Rvs domain; SH3: Src homology 3 domain; EH, Eps15 homology domain. (B) Endogenous MICAL-L1 was immunoprecipitated from HeLa cells transfected either with HA-tagged Synd2 or JNK1. Coimmunoprecipitation of HA-tagged proteins was examined by immunoblotting with anti-HA antibodies. (C) GST pull-down experiments were performed with GST, GST-fusion Synd2, or its truncation/mutants (as indicated) using bovine brain cytosol. Precipitates were immunoblotted with antibodies for MICAL-L1 and EHD1. Coomassie blue staining was used to visualize GST and GST-fusion proteins (bottom). (D) Selective yeast two-hybrid assays were performed with MICAL-L1 or mutants (as indicated) cotransformed with either Synd2 or EHD1. p53 and SV40 serve as controls. (E) HeLa cells were immunostained for endogenous Synd2 and MICAL-L1. A profile scan for the region transected by the white line in the merged inset is shown on the right. Bar, 10 μm.
For this study, we proposed that the recruitment of protein(s) containing membrane-sensing and curvature-inducing BAR domains may result in RE tubulation. Such BAR domain–containing proteins induce membrane bending and curvature by mechanisms of hydrophobic insertion and/or scaffolding mechanisms (McMahon and Gallop, 2005; Zimmerberg and Kozlov, 2006). Two candidate BAR domain–containing proteins that interact with EHD1 are the N-BAR protein amphiphysin/Bin1 (Pant et al., 2009) and the F-BAR protein syndapin2 (Synd2, also known as PACSIN2; see Figure 1A; Braun et al., 2005).
To gain insight into the mechanism of RE tubule biogenesis, we identified Synd2 as a third marker protein of RE tubules (colocalizing with EHD1 and MICAL-L1). We discovered that the Synd2 SH3 domain interacts directly with MICAL-L1 through two of the latter's 14 proline-rich domains (PRDs) and that this interaction is needed for the stable association of proteins on membranes and the nucleation of membrane tubules. We also identified MICAL-L1 and Synd2 as two of only a handful of known phosphatidic acid (PA)–binding proteins. Our study indicates that PA is an essential lipid component of RE tubules and that the presence of PA enhances Synd2 F-BAR tubulation activity. In addition, impairment of PA synthesis with inhibitors significantly delays endocytic recycling. These results promote a new model suggesting that 1) local lipid concentration of PA in membranes facilitates the independent binding of MICAL-L1 and Synd2 to these membranes, 2) association of MICAL-L1 and Synd2 with membranes is stabilized upon their interaction, and 3) this interaction on membranes leads to tubule biogenesis by the Synd2 F-BAR domain. We propose that EHD1 is recruited to tubular RE by MICAL-L1 and/or Synd2, where it performs scission to facilitate vesicle transport and recycling to the plasma membrane.
RESULTS
Synd2/MICAL-L1/EHD1 exists as a complex decorating tubular recycling endosomes
Initially, we evaluated the ability of EHD1 to localize to tubular REs in the absence of both Bin1 and Synd2 (with MICAL-L1 as a positive control). Although EHD1 remained closely associated with tubular REs in the absence of Bin1, depletion of either Synd2 or MICAL-L1 induced the loss of EHD1 localization to tubules (Supplemental Figure S1, A and B). Of the three syndapins, only Synd2 is ubiquitously expressed and was therefore chosen to assess whether it interacts with MICAL-L1 and forms a complex. In support of this idea, a recent large-scale proteomic study found a potential interaction between the SH3 domain of Synd2 and MICAL-L1 (Linkermann et al., 2009). To see whether Synd2 and MICAL-L1 interact in vivo, we performed coimmunoprecipitation experiments. HeLa cells were transfected either with hemagglutinin (HA)-Synd2 or HA-JNK1 (control) and pulled down with antibodies to endogenous MICAL-L1. We found that Synd2, but not JNK1, coimmunoprecipitated with MICAL-L1, indicating that MICAL-L1 and Synd2 can interact in vivo (Figure 1B). We next assessed whether the MICAL-L1-Synd2 interaction occurs via EHD1. Because MICAL-L1 and Synd2 both have NPF motifs that interact with EHD1 (Braun et al., 2005; Sharma et al., 2009), glutathione S-transferase (GST) pull-down and yeast two-hybrid experiments were done to determine whether EHD1 mediates this interaction. GST pull-down experiments with GST alone (control), GST fused to full-length Synd2, or GST fused to a series of Synd2 truncation/deletions mutants was done using bovine brain cytosol. Whereas GST-Synd2 precipitated both MICAL-L1 and EHD1 (Figure 1C), GST alone did not pull down either protein. Removal of the Synd2 SH3 domain (Synd2 ΔSH3) but not its NPF motifs led to a loss of interaction with MICAL-L1 but not EHD1, indicating that Synd2 interacts with MICAL-L1 through its SH3 domain. Furthermore, when pull-downs were done with only the Synd2 SH3 domain, MICAL-L1 but not EHD1 was precipitated. Mutation of all Synd2 NPF motifs to APA (GST-Synd2-NPF → APA) or a Synd2 truncation containing only its F-BAR domain (GST-Synd2 F-BAR, which lacks NPF motifs) prevented its interaction with EHD1, as expected. Our studies lead us to conclude that the interactions between Synd2, MICAL-L1, and EHD1 are mutually exclusive and direct.
We next determined which of the 14 PRDs contained by MICAL-L1 are required to interact with the Synd2 SH3 domain. As demonstrated, Synd2 associates with both MICAL-L1 and the four EHD proteins by selective yeast two-hybrid (Y2H) assays (Supplemental Figure S1C). Y2H experiments were then performed with serial truncation mutants of MICAL-L1 to narrow down the PXXP motifs used by MICAL-L1 for binding to Synd2 (Supplemental Figure S1, D–F). We identified two MICAL-L1 class I PXXP motifs that are preceded by basic residues (380KKKPAPLPP and 472KTKKRPAPRAP) required for interaction with Synd2 (denoted in Figure 1D as 385PLPP-ALAP and 480PRAP-ARAA). These PXXP motifs displayed similarity with syndapin-binding PXXP motifs in dynamin1 (Supplemental Figure S1H). On loss of either PXXP motif in the context of full-length MICAL-L1, an interaction with Synd2 was observed; however, loss of both PXXP motifs abrogated MICAL-L1–Synd2 binding (Figure 1D). Consistent with our pull-down studies, the MICAL-L1 double PXXP mutant nonetheless bound to EHD1, whereas a MICAL-L1 NPF mutant (which does not bind EHD1) was still capable of binding to Synd2. Mutations in the MICAL-L1 CC domain (721MLVDWF and 851KR) that affect its ability to bind to membranes (Sharma et al., 2009) had no effect on MICAL-L1 binding to Synd2 and EHD1, indicating that membrane association is not a prerequisite for MICAL-L1 to interact with either EHD1 or Synd2. Because MICAL-L1 is capable of homo-oligomerization (Supplemental Figure S1G), as are Synd2 (Qualmann et al., 2011) and EHD1 (Lee et al., 2005), this further supports the possibility of tripartite MICAL-L1–Synd2–EHD1 complexes by three-way direct binding. Moreover, despite the transient nature of MICAL-L1 interactions with other proteins, we did observe small bands of Synd2 and EHD1 that coprecipitated with MICAL-L1 (Supplemental Figure S1I), whereas the highly abundant actin did not coprecipitate. In addition, EHD3, which displays 86.5% identity with EHD1, was precipitated by GST-Synd2 (Supplemental Figure S1J) and bound to Synd2 in yeast two-hybrid experiments (Supplemental Figure S1C), in agreement with the studies of Braun et al. (2005).
We next investigated whether Synd2 localizes to tubular REs. Using MICAL-L1 as a known marker for these structures, we stained HeLa cells for both endogenous Synd2 and MICAL-L1. In addition to its localization to the cytoplasm and vesicular membranes, Synd2 also localized to tubular structures that highly colocalized with MICAL-L1 (Figure 1E). The degree of colocalization between the endogenous proteins on tubular REs is highlighted with a profile scan. When Tomato-Synd2 was exogenously expressed with green fluorescent protein (GFP)–MICAL-L1, a high level of colocalization was observed on tubular membranes (Supplemental Figure S2B, bottom). Even expression of Tomato-Synd2 alone led to its localization to tubular membranes, although this distribution was partially masked by its cytoplasmic pool (Supplemental Figure S2B, top left).
We previously showed that the BAR domain–containing protein Bin1, also known as amphiphysin1, regulates tubular localization of the worm EHD1 orthologue, RME1 in C. elegans (Pant et al., 2009). Although Bin1 localizes to recycling endosomes, its loss in mammalian cells did not affect the tubular localization of MICAL-L1, Synd2, or EHD1 (Supplemental Figures S1, A and B, and S2A). Thus endogenous Synd2 and MICAL-L1 continue to display colocalization to RE tubules, even in the absence of Bin1 (Supplemental Figure S2A), further supporting the notion that Bin1 is not required for RE tubule biogenesis. These data lead us to suggest that in mammalian cells, the localization of the BAR-domain protein Synd2 to tubular RE and its function in a complex with MICAL-L1 and EHD1 are crucial for RE tubule biogenesis.
In previous studies, we determined that both EHD1 and MICAL-L1 are dynamically recruited onto tubular membranes from a cytoplasmic pool (Sharma et al., 2009). To assess whether Synd2 also alternates between tubular membranes and the cytoplasm, we performed fluorescence recovery after photobleaching (FRAP) experiments on live cells expressing Tomato-Synd2 and GFP-MICAL-L1 (Supplemental Figure S3). A region of interest (ROI; boxed region) containing a tubular membrane decorated by both Tomato-Synd2 and GFP–MICAL-L1 was photobleached using high laser intensity, and images were obtained every 30 s by dual-channel time-lapse confocal microscopy to monitor recovery of fluorescence signal in the ROI. Simultaneous recovery of both MICAL-L1 and Synd2 was seen on the tubular membrane within 2 min of photobleaching, and >50% of recovery was seen within 4 min. These data provide a strong indication that Synd2 and MICAL-L1 display similar dynamics of association with tubular membranes.
The interaction between MICAL-L1 and Synd2 is required for their localization to tubular recycling endosomes
We previously showed that MICAL-L1 is required for the recruitment and/or stabilization of EHD1 on tubular recycling endosomes (Supplemental Figure S1A; Sharma et al., 2009). The identification of Synd2 as a third “marker” of EHD1-MICAL-L1–containing recycling tubules and an interaction partner of both MICAL-L1 and EHD1 raised new questions regarding the recruitment of Synd2 to these structures. For example, how does Synd2 localize to tubular endosomes, and what is the role of each of these three proteins in regulating the localization of the other two partners to the tubular membranes? To answer these questions, we initially used a depletion approach. As shown by immunoblotting, individual small interfering RNA (siRNA) knockdown of MICAL-L1, Synd2, and EHD1 caused a depletion of >90% of each protein (with actin as a loading control; Figure 2A). Although depletion of either EHD1 or MICAL-L1 did not affect the expression levels of the remaining two binding partners, loss of Synd2 significantly reduced cellular MICAL-L1 levels (Figure 2A, top, right lane).
FIGURE 2:
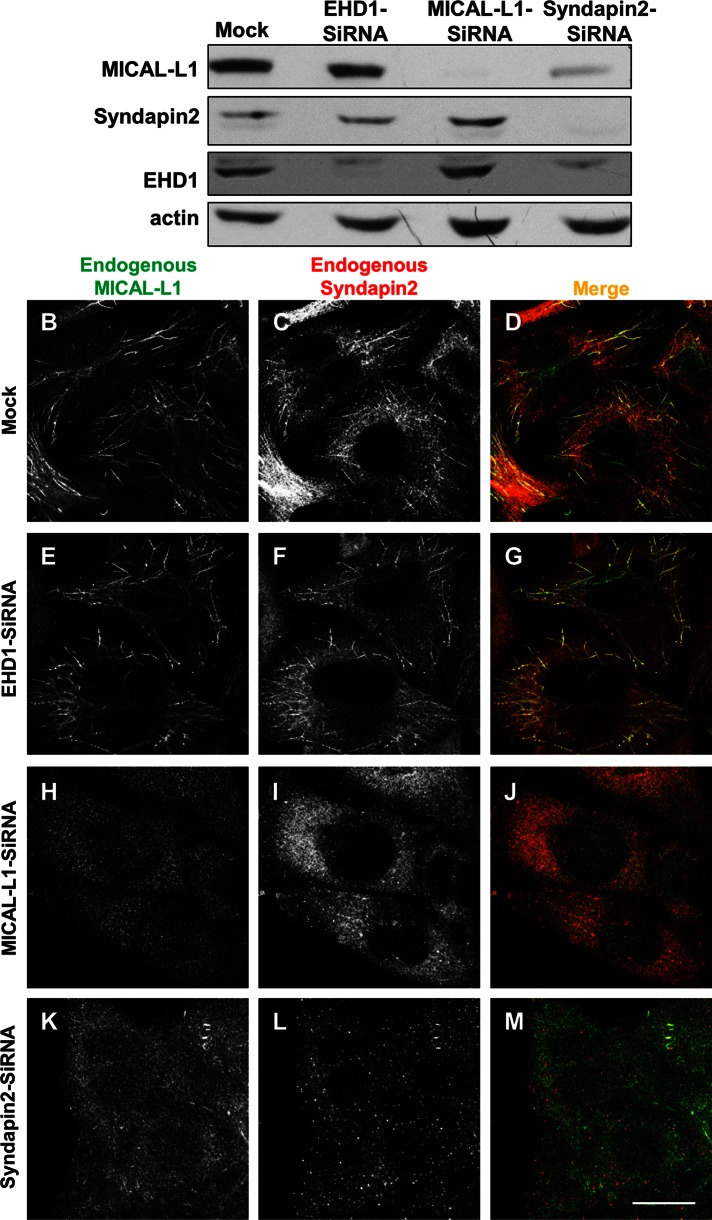
Loss of either MICAL-L1 or Synd2 causes the absence of all known tubular endosomal markers from these structures. Mock-treated or siRNA-treated cells were examined 72 h after treatment. (A) siRNA- and mock-treated cells were immunoblotted for endogenous MICAL-L1, Synd2, EHD1, and actin (loading control). (B–M) siRNA- and mock-treated cells were immunostained for endogenous MICAL-L1 and Synd2. Bar, 10 μm.
We then assessed the effect of siRNA knockdown on the localization of the proteins. Similar to mock treatment (Figure 2, B–D), EHD1-siRNA treatment did not alter Synd2 or MICAL-L1 localization to tubular membranes (Figure 2, E–G). We previously showed that MICAL-L1 depletion causes the absence of EHD1 from membrane tubules. To determine the mutual relationship between MICAL-L1 and Synd2, we examined Synd2 localization upon MICAL-L1 depletion. As expected, on MICAL-L1 siRNA treatment, levels of the endogenous protein were undetectable by immunofluorescence (Figure 2H). Under these conditions, Synd2 no longer decorated tubular membranes but instead appeared dispersed in the cytoplasm (Figure 2I). On the other hand, Synd2 depletion (which led to nearly undetectable Synd2 levels by immunofluorescence) likely led to MICAL-L1 instability and degradation, as observed by both immunoblot and immunostaining (Figure 2, A and K–M). The data support the idea that Synd2 and MICAL-L1 are required for each other's stable localization to tubular recycling endosomes and raise the possibility that Synd2 recruitment to membranes is necessary for the generation of these structures.
Although we hypothesized that Synd2 regulates receptor recycling in concert with MICAL-L1 and EHD1, it was not possible to test this directly, as we found that Synd2 depletion impaired receptor internalization (data not shown). This was not surprising, given the previously described role for Synd2 in controlling microfilament and microtubule networks (reviewed in Kessels and Qualmann, 2004) and considering the range of effects we observed upon Synd2 depletion on early endosomes, Golgi, and lysosomes (Supplemental Figure S4A).
Consistent with the functions described for EHD1 and EHD3 in regulation of retromer-based retrograde traffic (Gokool et al., 2007; Zhang et al., 2012a, b), endogenous Synd2 displayed a partial but clear colocalization to sorting nexin-1 (SNX1)–containing vesicles (Supplemental Figure S4B; mock, see inset). However, Synd2 depletion did not have a major effect on SNX1 localization and did not appear to diminish the small, SNX1-decorated tubules observed in both mock and Synd2-depleted cells (Supplemental Figure S4B; see insets). Because EHD1 also interacts with the macropinocytosis regulator Rabankyrin-5 (Zhang et al., 2012b), we tested the effect of Synd2 depletion on Rabankyrin-5. In mock-treated cells, a degree of colocalization was noted between Synd2 and Rabankyrin-5 on vesicular structures (Supplemental Figure S4B; mock, see inset). On Synd2 depletion, we observed a modest change in Rabankyrin-5 distribution to a more peripheral pattern (Supplemental Figure S4B; Synd2-siRNA), suggesting a potential relationship between these two proteins, possibly through EHD1.
EHD1 stabilizes MICAL-L1/Synd2 interaction on recycling tubules
Because depletion of MICAL-L1 causes a failure of Synd2 to localize to tubular REs, we predicted that generation of a MICAL-L1 protein that lacks the PRD motifs necessary for its interaction with Synd2 would lead to the absence of both proteins from recycling tubules. To test this idea, we used siRNA to deplete cells of MICAL-L1 and then reintroduced siRNA-resistant versions of MICAL-L1 as we did previously (Jovic et al., 2009; Sharma et al., 2009). As a control we first demonstrated that in ∼90% of MICAL-L1–depleted cells, Synd2 did not localize to tubular membranes; however, >85% of cells with siRNA-resistant wild-type MICAL-L1 reintroduced exhibited tubules decorated with both exogenous MICAL-L1 and endogenous Synd2, confirming that MICAL-L1 is required for Synd2 localization to the tubular membranes (Figure 3A; quantified in Figure 3E).
FIGURE 3:
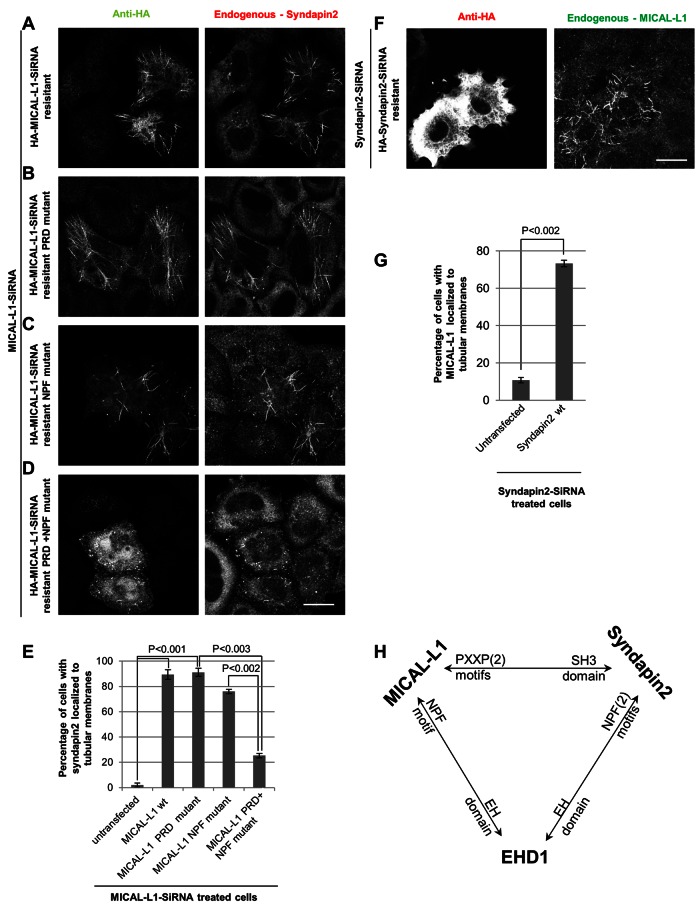
Complex interactions between MICAL-L1, Synd2, and EHD1 in tubular localization and biogenesis. (A–D) Cells treated for 72 h with MICAL-L1 siRNA were transfected for the last 48 h with either siRNA-resistant, wild-type, HA-tagged MICAL-L1 or mutant constructs. The cells were then immunostained with anti-HA and anti-Synd2 antibodies. Bar, 10 μm. (E) HeLa cells treated as in A were quantified to assess the percentage of cells with Synd2 localized to tubular membranes. Error bars, SE. (F) Cells treated for 72 h with Synd2 siRNA were transfected for the last 48 h with siRNA-resistant, wild-type, HA-tagged Synd2. Cells were then immunostained with anti-HA and anti–MICAL-L1 antibodies. Bar, 10 μm. (G) HeLa cells treated as in F were quantified to assess the percentage of cells with MICAL-L1 localized to tubular membranes. Error bars, SE. (H) Model illustrating the interaction profile between MICAL-L1, Synd2, and EHD1.
We then reintroduced an siRNA-resistant MICAL-L1 PRD mutant incapable of direct interaction with Synd2. Surprisingly, ∼90% of cells expressing the MICAL-L1 mutant did display both the mutant MICAL-L1 protein and endogenous Synd2 on tubular endosomes (Figure 3B; quantified in Figure 3E). We then hypothesized that endogenous EHD1 might link the mutant MICAL-L1 and endogenous Synd2 on the tubular membranes through its homo-oligomerization and association with both proteins by EH-domain/NPF motif interactions (illustrated in Figure 3H). To examine this idea, we first made a MICAL-L1 siRNA–resistant mutant with a mutation in its first NPF motif that is no longer able to bind EHD1 (Sharma et al., 2009). As depicted (Figure 3C; quantified in Figure 3E), this mutation still enabled Synd2 to localize to tubular endosomes (in ∼80% of the transfected cells).
We then took MICAL-L1–depleted cells and transfected them with a MICAL-L1 siRNA–resistant double mutant (PRD plus NPF) incapable of interacting with either Synd2 or EHD1. As demonstrated, in ∼80% of transfected cells the double mutant MICAL-L1 neither localized to tubular endosomes nor recruited Synd2 to these structures (Figure 3D; quantified in Figure 3E), suggesting that EHD1 can link MICAL-L1 and Synd2 on these membranes via its EH domain. Similar knock-in studies were performed on Synd2 siRNA–treated cells. Whereas <15% of Synd2-depleted cells exhibited MICAL-L1 localized to tubular membranes, reintroducing siRNA-resistant Synd2 significantly increased MICAL-L1 associated with tubular membranes in >70% of transfected cells (Figure 3F; quantified in Figure 3G). On the basis of these data, we suggest that a tripartite interaction between MICAL-L1, Synd2, and EHD1 (Figure 3H) controls association with recycling tubules and their biogenesis.
MICAL-L1 and Synd2 are phosphatidic acid–binding proteins
Little is known about the lipid selectivity that regulates MICAL-L1 and Synd2 binding to lipid bilayers. Whereas Synd2 binds to lipids through its BAR domain (reviewed in Qualmann et al., 2011), MICAL-L1 requires its C-terminal coiled-coil (CC) region to localize to tubular REs (Figure 4A; Sharma et al., 2009). To determine the preference of lipid binding, we purified the Synd2 F-BAR domain and the MICAL-L1 CC domain. Because the MICAL-L1 CC domain was highly insoluble, denaturation and renaturation steps were required (Supplemental Figure S5, A–C), and the renatured histidine (His)-tagged CC domain was analyzed by circular dichroism to ensure proper folding of the amphipathic α-helical structure (Supplemental Figure S5D).
FIGURE 4:
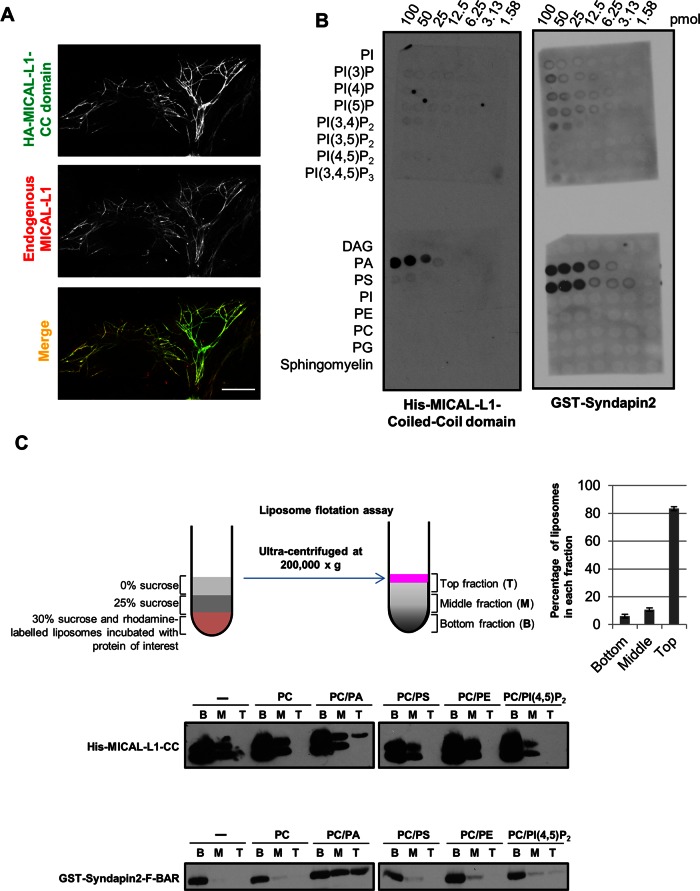
MICAL-L1 and Synd2 are phosphatidic acid–binding proteins. (A) HeLa cells transfected with HA–MICAL-L1 CC domain were immunostained for endogenous MICAL-L1 and HA tag. Bar 10 μm. (B) His–MICAL-L1 CC domain and GST-Synd2 were subjected to a lipid overlay assay and immunoblotted with antibodies for their respective tags. The concentration of the lipids is indicated on the top. (C) His–MICAL-L1 CC and GST-Synd2 F-BAR domains were subjected to liposome flotation assays (depicted schematically). Liposome fractions (bottom [B], middle [M], top [T]) were immunoblotted with antibodies for the respective protein tag. The distribution of liposomes in each fraction postcentrifugation from six experiments is displayed as a graph. Error bars, SE.
We then performed lipid overlay assays with His-tagged MICAL-L1 CC domain and the GST-Synd2 F-BAR domain, as well as with the full-length GST-Synd2 protein (Figure 4B and Supplemental Figure S6, A and B). Both proteins showed a degree of selectivity for PA under these conditions. Synd2 also bound to phosphatidylserine (PS; Figure 4B, and Supplemental Figure S6, A and B), but its isolated F-BAR domain displayed less selectivity to PS than to PA (Supplemental Figure S6A, middle and right).
To examine the PA binding of both proteins under more physiological conditions, we used a liposome flotation assay. In these experiments, the tagged proteins were incubated with rhodamine-labeled liposomes comprising varying lipids and subjected to a stepwise sucrose gradient and ultracentrifugation. A linear sucrose gradient was established, with >80% of the liposomes present in the top layer (Figure 4C, schematic diagram and graph). Under these conditions, we observed that both Synd2 F-BAR and the MICAL-L1 CC domain were present in the top fraction in PA-containing liposomes but not in liposomes lacking PA. This supports the notion that Synd2 and MICAL-L1 are PA-binding proteins.
Phosphatidic acid is a key component of tubular REs
On characterizing both Synd2 and MICAL-L1 as PA-binding proteins, we hypothesized that the presence of PA on recycling endosomes might enable the binding of these proteins to membranes. To test this in vivo, we transfected cells with the well-characterized PA probe Spo20p (residues 51–91) tagged to GFP (Zeniou-Meyer et al., 2007). This probe partially colocalized with MICAL-L1 on tubular REs, whereas a mutant incapable of PA binding (Spo20p-L67P-PABD) was mostly cytosolic and displayed no colocalization with MICAL-L1 (Supplemental Figure S6C). Overexpression of the MICAL-L1 CC domain, which retains its localization to tubular endosomes (Sharma et al., 2009) and is unable to bind to Synd2 or EHD1, did not compete with binding of either endogenous MICAL-L1 or endogenous Synd2 to tubular recycling endosomes (Supplemental Figure S6D). Similarly, it did not affect the internalization or recycling of transferrin and its receptor (Supplemental Figure S7). Overall these data lend support to the idea that phosphatidic acid is an in vivo component of tubular recycling endosomes.
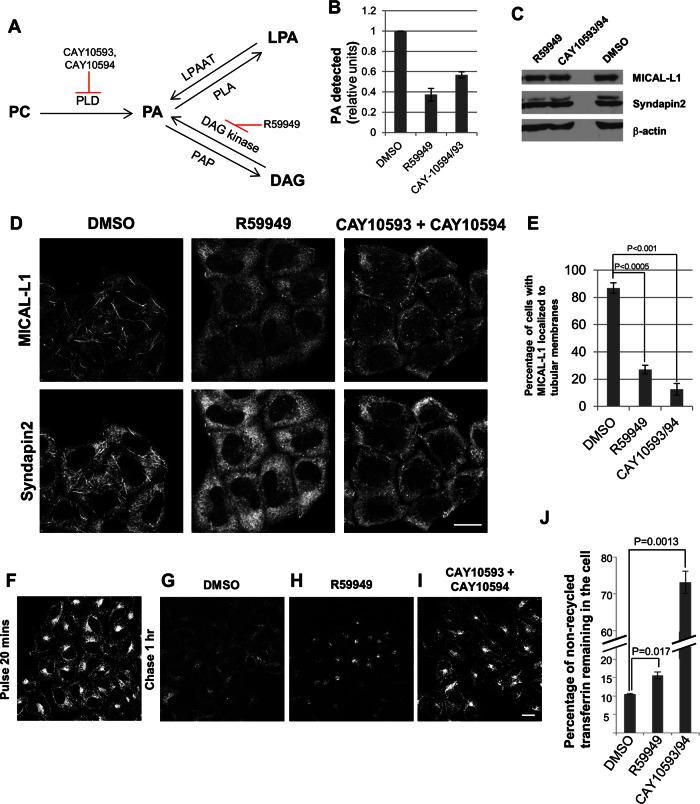
To determine whether recycling endosomes require PA for their tubular shape and function in recycling, we used several inhibitors expected to reduce cellular PA levels. There are three described pathways for PA production (Figure 5A): 1) conversion of phosphatidylcholine (PC) to PA by the lipase activity of phospholipase D (PLD), 2) conversion of diacylglycerol (DAG) to PA by the kinase activity of DAG kinase, and 3) conversion of lysophosphatidic acid (LPA) to PA by the acyltransferase activity of LPA acyltransferase. On the other hand, PA is catabolized to LPA and DAG by phospholipase A and PA phosphatase, respectively.
FIGURE 5:
Phosphatidic acid controls generation and trafficking through tubular recycling endosomes. (A) Metabolic pathways leading to phosphatidic acid synthesis. (B) PA levels were quantified enzymatically in HeLa cells treated with the indicated drugs for 1 h (compared with DMSO control treatment) from three independent experiments. Error bars, SE. (C) Cells treated as in B were immunoblotted for MICAL-L1, Synd2, and actin (loading control). (D) Cells treated as in B were immunostained for endogenous MICAL-L1 and Synd2. Bar, 10 μm. (E) Quantification of the percentage of cells from D with MICAL-L1 localized to tubular membranes from three independent experiments. Error bars, SE. (F–I) Cells incubated with Alexa Fluor 568–labeled transferrin for 20 min were either fixed (pulse) or fixed after 1 h treatment with the indicated drugs (chase). Bar, 10 μm. (J) HeLa cells incubated with Alexa Fluor 633–labeled transferrin for 30 min were fixed after 1 h treatment with DMSO (control) or the indicated drugs. The percentage of nonrecycled transferrin remaining in the cell was analyzed by flow cytometry from three independent experiments. Error bars, SE.
We used the drug R59949 and a combination of the inhibitors CAY10593 and CAY10594 (which block DAG kinase and PLD activity, respectively). We then measured intracellular PA as described (Morita et al., 2009) and found that in comparison to dimethyl sulfoxide (DMSO)–treated control cells, R59949 or CAY treatment reduced cellular PA levels to <40 and 60%, respectively (Figure 5B). However, the drugs did not alter either MICAL-L1 or Synd2 cellular expression (Figure 5C). On the other hand, PA reduction by the drugs dramatically altered the localization of both endogenous MICAL-L1 and Synd2 (Figure 5, D and E). Whereas >80% of DMSO-treated cells displayed MICAL-L1 localized to tubular endosomes, this was decreased to <30 and 20% of the R59949 and CAY drug-treated cells, respectively Figure 5E). These data led us to conclude that PA is required for the normal localization of both MICAL-L1 and Synd2 to recycling endosomes and likely a significant factor for RE tubule biogenesis.
Because both MICAL-L1 and Synd2 interact with EHD1 and are involved in membrane recycling, we next tested whether decreased levels of cellular PA affect receptor recycling. Cells were incubated with dye-labeled transferrin (Tf) for 20 min to allow internalization and accumulation in the ERC (Figure 5F). These cells were then mock treated with DMSO or either R59949 or the CAY drugs for 1 h in serum-containing media (chase) to assess the recycling of Tf. As expected, over this time the DMSO-treated cells exhibited active recycling and retained <11% of the internalized Tf within the cell (Figure 5, G and J). On the other hand, there was >50% increase in the nonrecycled Tf in R59949-treated cells, representing a statistically significant difference (Figure 5, H and J). More dramatically, we found that cells treated with the two CAY drugs retained >70% of their internalized Tf, indicating a severe delay in recycling (Figure 5, I and J). Although both sets of inhibitors induce relatively similar decreases in overall cellular PA levels (Figure 5B), differences in their effect on recycling might stem from the localized action of the enzymes (PLD and DAG kinase) or potentially be due to additional, as-yet-uncharacterized effects of the inhibitors.
Synd2 preferentially tubulates liposomes containing PA
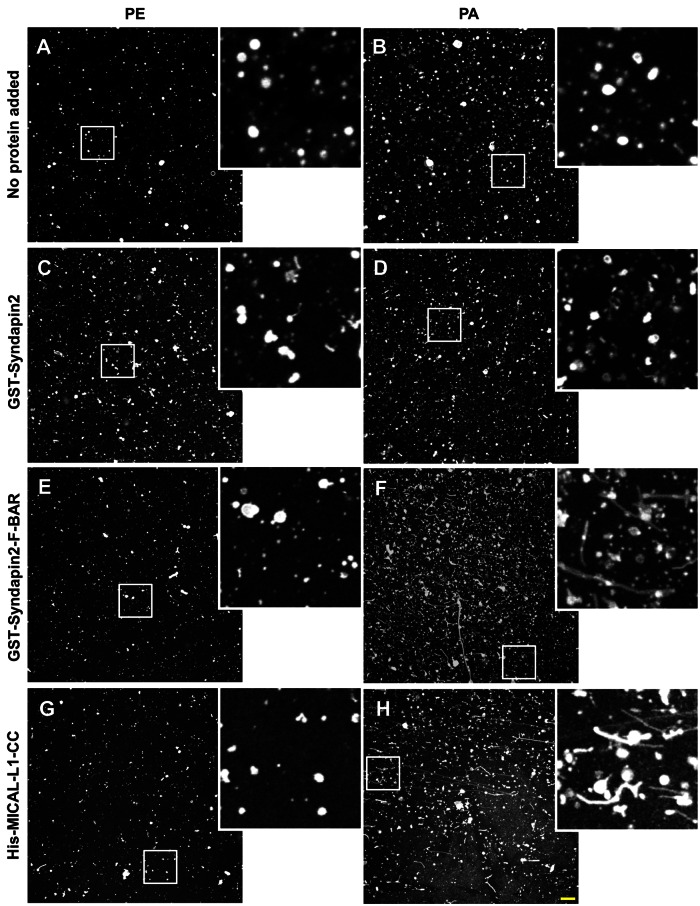
Given the role of PA in recruitment of MICAL-L1 and Synd2 to recycling endosomes and the ability of BAR domain–containing proteins such as Synd2 to tubulate membranes, we next calibrated an in vitro assay to test whether Synd2 preferably tubulates large multilamellar vesicles (LMVs) that are enriched in PA. Without addition of any protein to the liposomes, no tubulation was observed with liposomes containing PC and PE or PC and PA (Figure 6, A and B). As depicted, and consistent with previous studies (Wang et al., 2009), the full-length Synd2 displayed little or no tubulation of LMVs composed of PC/PE (Figure 6C). Indeed, changing the LMV composition from PC/PE to PC/PA did not enhance tubulation by full-length Synd2 (Figure 6D). However, the isolated F-BAR domain of Synd2 (known to be active in membrane tubulation; Wang et al., 2009) induced extensive tubulation of LMVs containing PC/PA as compared with PC/PE (Figure 6, compare F and E). Although the MICAL-L1 CC domain does not display homology to BAR domains, we nevertheless tested its ability to tubulate LMVs. As shown, no tubulation was observed in PC/PE LMVs (Figure 6G); however, we did detect the generation of tubular structures when the MICAL-L1 CC was incubated with LMVs containing PC/PA (Figure 6H). Both phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) and PS were also tested in PC LMVs (Supplemental Figure S8). PS-containing LMVs displayed some spontaneous tubulation, which was enhanced by the Synd2 F-BAR domain (Supplemental Figure S8, A, C, E, and G). However, in our hands PI(4,5)P2-containing LMVs displayed no tubulation under any conditions (Supplemental Figure S8, B, D, F, and H). Taken together, these data show that the PA-binding proteins, MICAL-L1, and Synd2 are capable of generating tubules from PA-containing membranes.
FIGURE 6:
Phosphatidic acid–rich LMVs undergo tubulation in the presence of F-BAR domain of Synd2 and CC domain of MICAL-L1. In vitro tubulation assays were performed with (A) PE-containing liposomes only, (B) PA-containing liposomes only, (C) PE-containing liposomes and GST-Synd2, (D) PA-containing liposomes and GST-syndapin, (E) PE-containing liposomes and GST-Synd2-F-BAR, (F) PA-containing liposomes and GST-Synd2-F-BAR, (G) PE-containing liposomes and His-MICAL-L1-CC, or (H) PA-containing liposomes and His-MICAL-L1-CC. LMVs comprised a mass ratio of 70% PC, 10% NBD-PE, and 20% PE or PA. Insets depict the region in the white box. Bar, 10 μm.
DISCUSSION
RE tubules are important for the efficient sorting and recycling of internalized cargo and lipids back to the plasma membrane (Jovic et al., 2009). Despite the existence of this unique network of intertwined tubules and vesicles that comprise the ERC (Maxfield and McGraw, 2004), little is known about the biogenesis of this complex organelle or even about the biogenesis of individual tubular REs. Since identifying MICAL-L1 as an EHD1 interaction partner and RE tubule marker, our previous studies have demonstrated that EHD1 is not required for the biogenesis of these tubules in vivo (Sharma et al., 2009), suggesting the involvement of other proteins in this process.
The BAR domain–containing protein Bin1 interacts with both the C. elegans Rme1/EHD1 orthologue and the human EHD1 protein and in vitro is capable of inducing tubulation of lipid monolayers (Pant et al., 2009). It is also necessary for Rme1 tubulation in worms. However, our experiments suggest that in human cells Bin1 is not essential for the biogenesis of RE tubules decorated by EHD1 and MICAL-L1 (Supplemental Figures S1A and S2A). On the other hand, our discovery that the EHD1 interaction partner and F-BAR–containing protein Synd2 directly interacts with MICAL-L1 has led to the notion that Synd2 may be essential for RE tubule biogenesis. It is noteworthy that mammalian Bin1 lacks the EH domain–binding NPF motif that is found in the worm orthologue, whereas the worm syndapin lacks the NPF motif present in mammalian orthologues. Thus it is possible that different BAR domain proteins play variable roles in the process of RE tubule biogenesis in different species.
Syndapin proteins, including the ubiquitously expressed Synd2 (also known as PACSIN2), interact with EHD1 (Xu et al., 2004; Braun et al., 2005) and are implicated in the regulation of endocytic events (Modregger et al., 2000; Qualmann and Kelly, 2000). More recently, Synd2 was identified as an important protein in the generation of caveolar membranes (Hansen et al., 2011; Senju et al., 2011) and the regulation of Rac1-mediated cell spreading and migration (de Kreuk et al., 2011). However, the potential role of Synd2 in RE tubule biogenesis has not been examined.
Given that all three proteins, EHD1, MICAL-L1, and Synd2, interact with one another and decorate tubular RE, a key issue is how they are recruited to REs. Given that MICAL-L1 recruits EHD1 to these membranes (Sharma et al., 2009), our goal was to determine whether MICAL-L1 or Synd2 is first recruited to REs or they simultaneously associate with the membranes. Indeed, depletion of either protein caused the removal of its partner from these structures, and FRAP experiments display nearly identical dynamics of recruitment to tubules for each protein. Because the recovery of these proteins occurs along the entire length of the tubule, this suggests that there is stable MICAL-L1–Synd2 binding even after tubule generation, hinting that the complex may play an additional function in these membrane structures, such as the recruitment of EHD1.
Synd2 depletion results in decreased MICAL-L1 expression, suggesting that the latter is destabilized when not associated with Synd2 on membranes. As we anticipated, the reintroduction of wild-type MICAL-L1 into depleted cells results in the recruitment and stabilization of Synd2 on RE tubules with MICAL-L1. We were initially surprised that a MICAL-L1 PRD mutant (incapable of interacting directly with Synd2) nonetheless localized to tubular membranes and induced the recruitment of Synd2 to these structures. However, we reasoned that this may be due to an indirect interaction between MICAL-L1 and Synd2 through EHD1; indeed, a double MICAL-L1 mutant incapable of interacting with either Synd2 or EHD1 no longer localized to RE tubules, nor did it recruit Synd2 to these structures.
Given that both MICAL-L1 and Synd2 appear to be recruited to RE membranes with similar kinetics, we determined the lipid binding of each protein. Under the conditions we used, our data led to the notion that both proteins bind PA. Because there are only a handful of known PA-binding proteins (reviewed in Stace and Ktistakis, 2006; Kassas et al., 2012), the identification of a PA-binding consensus is problematic. However, the well-characterized yeast protein Spo20p PA-binding domain clearly localized to MICAL-L1–containing tubular RE, indicating that these tubules indeed contain PA in vivo. Moreover, in vitro tubulation assays indicate that the Synd2 F-BAR domain (and the MICAL-L1 CC) displays preference for tubulating liposomes containing PA. Although Synd1 and Synd2 can bind liposomes comprising PC/PS (Dharmalingam et al., 2009), in contrast to our study, in these experiments the conditions of Bigay et al. (2005) were modified so that low-salt conditions were used. We propose that due to the symmetrical nature of PA and its tendency to eliminate spontaneous curvature (as opposed to lysophospholipids; reviewed in (Graham and Kozlov, 2010), the local concentration of PA in membranes may further facilitate Synd2 F-BAR domain–induced tubulation.
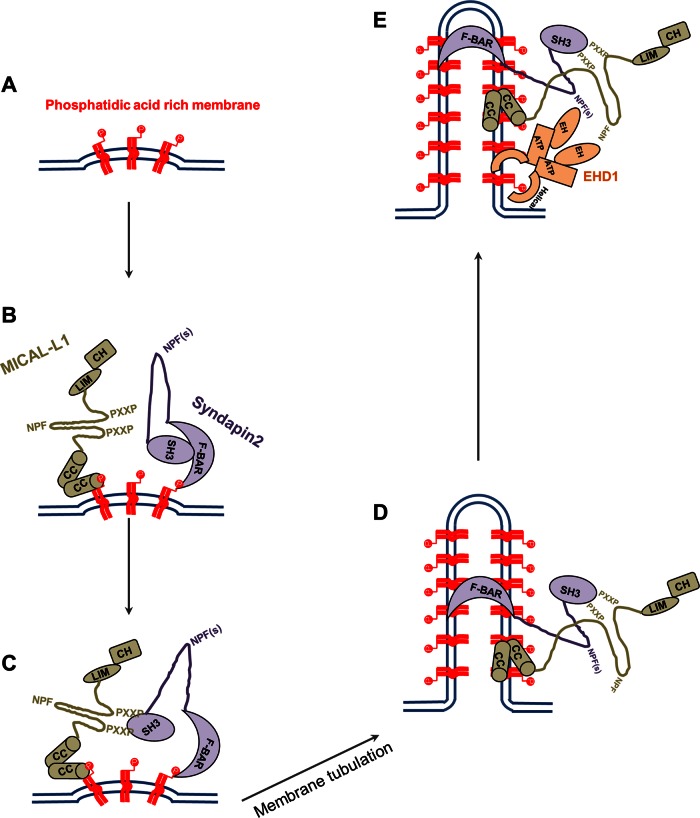
Our data support a novel model for the biogenesis of RE tubules (Figure 7). Through the generation and/or concentration of PA in membranes (Figure 7A), MICAL-L1 and Synd2 are recruited via their coiled-coil and BAR domains, respectively (Figure 7B). The direct interaction between MICAL-L1 and Synd2 (via 2 of the former's 14 PRDs and the latter's SH3 domain) stabilizes their localization on membranes (Figure 7C). This in turn would allow the amphipathic BAR domain to insert within the membrane and induce curvature that drives the tubulation (Farsad and De Camilli, 2003; Drin and Antonny, 2010; Rao et al., 2010) that is typical for RE membranes (Figure 7D). We expect that the recruitment of EHD1 and other factors later drive the fission of vesicles from these tubular REs (Figure 7E), generating recycling carriers of receptors and membranes bound for the plasma membrane (Daumke et al., 2007; Jakobsson et al., 2011; Cai et al., 2012).
FIGURE 7:
Model for biogenesis of tubular recycling endosomes. (A) Phosphatidic acid is generated or enriched on membranes. (B) MICAL-L1 (via its CC domain) and Synd2 (via its F-BAR domain) are recruited to PA-enriched membranes. (C) The MICAL-L1 PXXP motifs interact with the SH3 domain of Synd2 to stabilize both proteins on the membranes and (D) facilitate the generation of tubular endosomes by Synd2. (E) Synd2 and MICAL-L1 bind to the EH domain of EHD1 via their NPF motifs and recruit EHD1 to these tubular membranes, potentially facilitating vesiculation.
The role of PA in RE tubule biogenesis is ambiguous. Several studies showed that the lysophosphatidic acid acyl transferase inhibitor (see Figure 5A) CI-976 induces tubulation of Golgi, COP1 vesicles, and even recycling endosomes (Chambers et al., 2005; Yang et al., 2011). In HeLa cells, we measured only a ∼27% decrease in PA levels upon CI-976 treatment (compared with 43 and 62% reduction for CAY and R59949 inhibitors, respectively); however, this does lead to decreased MICAL-L1–containing recycling tubules (unpublished observations). Moreover, the studies by Chambers et al. (2005) address the generation of much shorter, thinner, and labile tubules visualized with internalized transferrin as a marker. It is possible that PA supports the generation of long, wide, and stable MICAL-L1–containing tubules, whereas LPA (increased upon CI-976 treatment) promotes the generation of shorter, thinner tubules to which transferrin localizes.
A recent study determined that reduction of PA levels in cells leads to decreased clathrin-coated pit formation, slower budding from the plasma membrane, and reduced rate of epidermal growth factor receptor internalization (Antonescu et al., 2010). Although it was reported that phospholipase D2 (PLD2) affects transferrin receptor recycling (Padron et al., 2006), and PLD2 indirectly controls cellular PA levels, little is known about the role of PA in controlling endocytic recycling. Given that PA also stimulates PIP5 kinase, which in turn generates PI(4,5)P2 (Moritz et al., 1992; Jenkins et al., 1994), it is possible that PLD inhibition might affect recycling by decreasing PI(4,5)P2 levels, as well as PA levels (Padron et al., 2006). Indeed, PI(4,5)P2 is also an essential phospholipid for RE tubules (Jovic et al., 2009). In fact, although our enzymatic PA measurement showed similar levels of PA decrease with either PLD or diacylglycerol kinase inhibitors, the more robust inhibition of endocytic recycling that we observed with the PLD inhibitors might be due to indirect effects on PI(4,5)P2, phosphatidylinositol 4-phosphate, and other phosphoinositides. Moreover, given that Arf6 activation stimulates PA generation (Brown et al., 1993; Jovanovic et al., 2006), it is noteworthy that Arf6 depletion causes the removal of MICAL-L1 from tubular RE (Rahajeng et al., 2012), leading to the notion that this occurs indirectly as a result of PA inhibition.
In summary, we provide evidence promoting a new model in which MICAL-L1 and Synd2 are recruited to RE by direct interactions with PA, which we now demonstrate is an important lipid for RE tubule biogenesis and efficient endocytic recycling. EHD1 is likely recruited onto preexisting tubules, where it acts as part of the fission machinery to allow vesicular transport onto the plasma membrane.
MATERIALS AND METHODS
Antibodies and reagents
The following primary antibodies were used: mouse polyclonal anti–MICAL-L1 (Novus Biologicals, Littleton, CO); rabbit polyclonal anti-Synd2 purchased from Abgent (San Diego, CA; for immunofluorescence) and Proteintech (Chicago, IL; for Western blot); rabbit polyclonal anti-Bin1 (Proteintech); mouse anti-HA epitope (Covance, Princeton, NJ); mouse anti-EEA1 (BD Biosciences, San Jose, CA); mouse anti-hexahistidine tag, mouse anti-actin, and rabbit anti-giantin (Abcam, Cambridge, MA); goat anti-GST conjugated to horseradish peroxidase (HRP; GE Life Sciences, Piscataway, NJ); mouse anti-lamp1 (Developmental Studies Hybridoma Bank, Iowa City, IA); mouse anti-SNX1 (BD Biosciences); mouse anti–Rabankyrin-5 (Abnova, Walnut, CA); rabbit anti-HA (Signalway Antibody, Pearland, TX); and polyclonal rabbit anti-EHD1 (Naslavsky et al., 2004). Secondary antibodies Alexa Fluor 568–conjugated goat anti-mouse, Alexa Fluor 488–conjugated goat anti-mouse, and Alexa Fluor 568–conjugated goat anti-rabbit antibodies were purchased from Invitrogen. HRP conjugated to goat anti-mouse and donkey anti-rabbit were obtained from Jackson ImmunoResearch Laboratories (West Grove, PA) and GE Life Sciences, respectively. Tf-568 and Tf-633 was purchased from Invitrogen. Brij 98 and R59949 were obtained from Sigma-Aldrich (St. Louis, MO). CAY10593 and CAY10594 were purchased from Cayman Chemical Company (Ann Arbor, MI).
Constructs
Cloning of EHD1, EHD2, EHD3, EHD4, and MICAL-L1 into yeast two-hybrid vectors pGBKT7 and pGADT7, as well as cloning of HA–MICAL-L1, HA–MICAL-L1 resistant to siRNA treatment, and GFP-Myc–EHD1, was as described (Caplan et al., 2002; Sharma et al., 2009). Two-hybrid control vectors (Gal4ad-SV40 large T-antigen and Gal4bd-p53) were purchased from Clontech (Palo Alto, CA). Human Synd2 (longer isoform) was PCR amplified from HeLa cDNA and cloned into pGBKT-7 and the hemagglutinin fusion mammalian expression vector pHA-CMV (Clontech), using standard procedures. Synd2 was subcloned into the GST fusion bacterial expression vector pGEX-6P2 (GE Life Sciences), which was later truncated at residues 280 (F-BAR) and 430 (ΔSH3) or mutated at all of its NPF motifs to APA (NPF → APA) using QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). siRNA-resistant Synd2 was created similarly by generating CDNA mutation at the oligonucleotide-binding region. The SH3 domain of Synd2 (residues 426–486) was cloned into pGEX-6p2. MICAL-L1 residues 700–863 constituting the CC domain was cloned into the His-tag fusion bacterial expression vector pET-14b (Novagen, Madison, WI). MICAL-L1 residues 368–468 and 468–863 were cloned into pGADT7. Truncation and mutations in MICAL-L1 full length, MICAL-L1 resistant to siRNA treatment, and residues 368–468 and 468–863 (present in mammalian and yeast-two hybrid vectors) were generated using QuikChange Site-Directed Mutagenesis Kit. HA-JNK1 was a kind gift from Sylvio Gutkind (National Institutes of Health, Bethesda, MD).
Cell culture, transfections, and siRNA treatment
HeLa cells were grown in DMEM containing with 10% fetal bovine serum (Sigma-Aldrich). HeLa cells were transfected with either FuGENE 6 or FuGENE HD (Roche Diagnostics, Indianapolis, IN). On-Target SMART Pool siRNA from Dharmacon (Lafayette, CO) for MICAL-L1, Synd2, and Bin1 and siRNA duplexes for EHD1 (base pairs gaaagagatgcccaatgtc, synthesized by Dharmacon) were transfected using Dharmafect (Dharmacon) to perform knockdown studies as previously described (Naslavsky et al., 2006).
Immunofluorescence
Cells were fixed with 4% paraformaldehyde and immunostained as previously described (Naslavsky et al., 2006). Images were acquired using a Zeiss LSM 5 Pascal confocal microscope (Carl Zeiss, Jena, Germany) by using a 63×/1.4 numerical aperture objective with appropriate filters.
Transferrin uptake assays
To study the role of PA in endocytic trafficking (Figure 5, F–J), HeLa cells were starved in DMEM media containing 0.5% bovine serum albumin (BSA) for 1 h and pulsed with 1 μg/ml of either Alexa Fluor 568–labeled transferrin (Tf-568) for 20 min for microscopy studies or Alexa Fluor 633–labeled transferrin (Tf-633) for 30 min for quantification by flow cytometry. Cells were either fixed immediately or fixed after incubation with DMEM containing 10% serum and the indicated drug for 1 h. At least 10,000 cells were analyzed for internal Tf-633 by flow cytometry analysis. To study the effect of exogenous expression of the MICAL-L1-CC domain on endocytic recycling (Supplemental Figure S7), HeLa cells were transiently transfected with HA–MICAL-L1 CC domain for 16 h and incubated with Tf-568 for 5 min. Cells were fixed either immediately or after incubation with DMEM containing 10% serum for the indicated time period.
Phosphatidic acid measurement
Cellular PA levels were measured using an enzymatic assay described by Morita et al. (2009). Briefly, cells were treated with drugs (as indicated) for 1 h, and lipids were extracted from cells using the Bligh–Dyer lipid extraction method (Bligh and Dyer, 1959). Lipids were then dried under flow of nitrogen and suspended in 2% Triton X-100. Lipids were then incubated with lipoprotein lipase generated from Burkholderia sp. (Sigma-Aldrich), followed by incubation with glycerol-3-phosphate oxidase (from Aerococcus viridans; Sigma-Aldrich), horseradish peroxidase (Sigma-Aldrich), and Amplex red (Invitrogen). Fluorescent resorufin, the end product of this enzymatic assay, was analyzed for its fluorescence with excitation at 544 nm and emission at 590 nm. A standard curve was generated for each experiment using egg-PA (Avanti Polar Lipids, Alabaster, AL) to check whether PA measurements made were within the linear range. Protein content measured by Bradford assay was used for normalization. Finally, PA values in treated cells were normalized to that of DMSO-treated cells for each experiment.
Immunoprecipitation and GST pull-down
HeLa cells transiently transfected with either HA-Synd2 or HA-JNK1 (Figure 1B) or coexpressed with both HA-Synd2 and GFP-Myc-EHD1 (Supplemental Figure S1I) were lysed in buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% Brij 98, 10 mM iodoacetamide, 250 μM 4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride, 10 μM leupeptin, and 10 μM aprotonin. Cell lysates were incubated with MICAL-L1 antibodies overnight, followed by incubation with protein L agarose beads (Fisher Thermo Scientific, Waltham, MA) for 2 h at 4°C. The beads were washed three times in wash buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1% Brij 98, 1.8 mg/ml iodoacetamide), eluted with 4× sample buffer, and subjected to immunoblotting.
To address the binding of Synd2 to MICAL-L1 and EHD1 (Figure 1C), GST-fusion proteins of Synd2 (50 μg each) immobilized onto GST beads were incubated with bovine brain cytosol (BBC; prepared in 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], 100 mM NaCl, 1 mM EDTA, pH 7.4, 0.5% Triton X-100, and protease inhibitors) overnight and with two additional rounds of BBC incubation, each for 2 h at 4°C. Beads were then washed 3× in phosphate-buffered saline containing 0.1% TX-100, eluted with 4× sample buffer, and analyzed by immunoblotting.
To address the binding of Synd2 to EHD3 (Supplemental Figure S1J), HeLa cells were transfected with either GFP or GFP-EHD3 for 48 h, lysed in 1% Brij lysis buffer, and incubated with 50 μg of Synd2 immobilized to GST beads overnight. The beads were then washed 3× with 0.1% Brij wash buffer, eluted with 4× sample buffer, and subjected to immunoblotting.
Yeast two-hybrid study
Yeast two-hybrid study was performed as previously described (Naslavsky et al., 2006). The Saccharomyces cerevisiae strain AH109 was cotransformed with the mentioned constructs using lithium acetate procedure and allowed to grow at 30°C after streaking on plates lacking leucine and tryptophan. Once the colonies grew large enough, an average of three to four colonies were chosen, suspended in water, equilibrated to the same optical density at 600 nm, and replated on plates lacking leucine and tryptophan (+His), as well as plates also lacking histidine (–His).
Lipid overlay assay
Lipid overlay assay was as previously described (Naslavsky et al., 2007). Briefly, phosphatidylinositol (PIP) strips, PIP arrays, SphingoStrips, and membrane arrays (Echelon Biosciences, Salt Lake City, UT) were blocked in TBST-BSA (10 mM Tris, pH 8.0, 150 mM NaCl, 0.1% Tween-20 [vol/vol]) supplemented with 3% BSA for 1 h at room temperature and then incubated with 1 μg/ml GST or His-tag fusion protein diluted in TBST-BSA overnight at 4°C. After washes with TBST-BSA, the strips were immunoblotted for their respective tags.
Liposome flotation assay
Liposome preparation was performed as described by Pant et al. (2009). Lipids solubilized in chloroform were purchased from Avanti Polar Lipids. Liposomes were prepared in mass ratio composition of 85% PC, 5% rhodamine-labeled phosphatidyl ethanolamine, and 10% PC, PE, PA, PS, or PI(4,5)P2. Lipids were mixed in glassware that was washed in chloroform and dried under nitrogen gas. Lipids were dried in a stream of nitrogen gas and incubated in vacuum overnight. Dried lipids were then resuspended to a concentration of 1.65 mg/ml in liposome-binding buffer (LBB; 20 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM MgCl2) and were extruded on Mini-Extruder (Avanti Polar Lipids) through 200-nm polycarbonate track-etched membrane filters to produce liposomes of 200-nm diameter.
Liposome flotation assay was adapted from protocol described by Bigay et al. (2005). Briefly, 5 μg of protein was incubated with 150 μl of rhodamine-labeled liposomes (at a 1 mg/ml concentration) or LBB buffer at room temperature for 15 min. This suspension was mixed with 100 μl of LBB containing ice-cold 75% sucrose, resulting in a 30% sucrose solution, which was overlaid with 200 μl of 25% sucrose and 150 μl of 0% sucrose solution prepared in ice-cold LBB. This sample was centrifuged at 200,000 × g for 1 h at 4°C in a Beckman swinging rotor (SW 60 Ti). The top 150 μl (T), middle 200 μl (M), and bottom 250 μl (B) were manually collected from the top and analyzed for fluorescence at excitation and emission of 544 and 590 nm, respectively, to quantitate the average percentage of liposome present in each fraction. Equal volumes of fractions were run on SDS–PAGE and immunoblotted for their respective tags.
Lipid tubulation assay
The lipid tubulation assay was adapted from Quan et al. (2012). Liposomes were prepared as described in the mass ratio of 70% PC, 10% NBD-PE (Invitrogen), and 20% PE, PA, PS, or PIP2. Dried lipids were hydrated in LBB for 1 h at 37°C and then vortexed and pipeted to form MVLs. Liposomes of 1 mg/ml concentration (in a volume of 100 μl) were incubated with 5 μg (Synd2 or Synd2–F-BAR) or 2 μg (MICAL-L1 CC) of the protein of interest at room temperature for 15 min. Later this liposome–protein mixture was diluted in LBB to a liposome concentration of 0.33 mg/ml, and 5 μl of this mixture was spotted onto a coverglass and hydrated at room temperature for 15 min. Liposomes on coverglasses were mounted and analyzed.
Statistical analysis
Data sets were analyzed by Student's one-tailed paired t test analysis, and data were considered significantly different for p < 0.05. The p values are shown for each experiment.
Supplementary Material
Acknowledgments
We thank Laura Allen, Fabien Kieken, and Juliati Rahajeng for their assistance. This work is supported by Nebraska Center for Cell Signaling Grant 5P20GM103489-10 from the National Institute of General Medical Sciences, Grant RO1GM087455 from the National Institute of General Medical Sciences, and the Nebraska Department of Health.
Abbreviations used:
- EHD1
C-terminal Eps15 homology domain protein 1
- LMV
large multilamellar vesicle
- MICAL-L1
molecules interacting with CasL-Like1
- PA
phosphatidic acid
- RE
recycling endosome
- Synd2
syndapin2
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-01-0026) on April 17, 2013.
REFERENCES
- Antonescu CN, Danuser G, Schmid SL. Phosphatidic acid plays a regulatory role in clathrin-mediated endocytosis. Mol Biol Cell. 2010;21:2944–2952. doi: 10.1091/mbc.E10-05-0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigay J, Casella JF, Drin G, Mesmin B, Antonny B. ArfGAP1 responds to membrane curvature through the folding of a lipid packing sensor motif. EMBO J. 2005;24:2244–2253. doi: 10.1038/sj.emboj.7600714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Braun A, Pinyol R, Dahlhaus R, Koch D, Fonarev P, Grant BD, Kessels MM, Qualmann B. EHD proteins associate with syndapin I and II and such interactions play a crucial role in endosomal recycling. Mol Biol Cell. 2005;16:3642–3658. doi: 10.1091/mbc.E05-01-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown HA, Gutowski S, Moomaw CR, Slaughter C, Sternweis PC. ADP-ribosylation factor, a small GTP-dependent regulatory protein, stimulates phospholipase D activity. Cell. 1993;75:1137–1144. doi: 10.1016/0092-8674(93)90323-i. [DOI] [PubMed] [Google Scholar]
- Cai B, Caplan S, Naslavsky N. cPLA2alpha and EHD1 interact and regulate the vesiculation of cholesterol-rich GPI-anchored protein-containing endosomes. Mol Biol Cell. 2012;23:1874–1888. doi: 10.1091/mbc.E11-10-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan S, Naslavsky N, Hartnell LM, Lodge R, Polishchuk RS, Donaldson JG, Bonifacino JS. A tubular EHD1-containing compartment involved in the recycling of major histocompatibility complex class I molecules to the plasma membrane. EMBO J. 2002;21:2557–2567. doi: 10.1093/emboj/21.11.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell P, Norman J. Endocytic transport of integrins during cell migration and invasion. Trends Cell Biol. 2008;18:257–263. doi: 10.1016/j.tcb.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Chambers K, Judson B, Brown WJ. A unique lysophospholipid acyltransferase (LPAT) antagonist, CI-976, affects secretory and endocytic membrane trafficking pathways. J Cell Sci. 2005;118:3061–3071. doi: 10.1242/jcs.02435. [DOI] [PubMed] [Google Scholar]
- Daumke O, Lundmark R, Vallis Y, Martens S, Butler PJ, McMahon HT. Architectural and mechanistic insights into an EHD ATPase involved in membrane remodelling. Nature. 2007;449:923–927. doi: 10.1038/nature06173. [DOI] [PubMed] [Google Scholar]
- de Kreuk BJ, Nethe M, Fernandez-Borja M, Anthony EC, Hensbergen PJ, Deelder AM, Plomann M, Hordijk PL. The F-BAR domain protein PACSIN2 associates with Rac1 and regulates cell spreading and migration. J Cell Sci. 2011;124:2375–2388. doi: 10.1242/jcs.080630. [DOI] [PubMed] [Google Scholar]
- Dharmalingam E, Haeckel A, Pinyol R, Schwintzer L, Koch D, Kessels MM, Qualmann B. F-BAR proteins of the syndapin family shape the plasma membrane and are crucial for neuromorphogenesis. J Neurosci. 2009;29:13315–13327. doi: 10.1523/JNEUROSCI.3973-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty KR, Demonbreun A, Wallace GQ, Cave A, Posey AD, Heretis K, Pytel P, McNally EM. The endocytic recycling protein EHD2 interacts with myoferlin to regulate myoblast fusion. J Biol Chem. 2008;283:20252–20260. doi: 10.1074/jbc.M802306200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drin G, Antonny B. Amphipathic helices and membrane curvature. FEBS Lett. 2010;584:1840–1847. doi: 10.1016/j.febslet.2009.10.022. [DOI] [PubMed] [Google Scholar]
- Farsad K, De Camilli P. Mechanisms of membrane deformation. Curr Opin Cell Biol. 2003;15:372–381. doi: 10.1016/s0955-0674(03)00073-5. [DOI] [PubMed] [Google Scholar]
- Fielding AB, Schonteich E, Matheson J, Wilson G, Yu X, Hickson GR, Srivastava S, Baldwin SA, Prekeris R, Gould GW. Rab11-FIP3 and FIP4 interact with Arf6 and the exocyst to control membrane traffic in cytokinesis. EMBO J. 2005;24:3389–3399. doi: 10.1038/sj.emboj.7600803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokool S, Tattersall D, Seaman MN. EHD1 interacts with retromer to stabilize SNX1 tubules and facilitate endosome-to-Golgi retrieval. Traffic. 2007;8:1873–1886. doi: 10.1111/j.1600-0854.2007.00652.x. [DOI] [PubMed] [Google Scholar]
- Graham TR, Kozlov MM. Interplay of proteins and lipids in generating membrane curvature. Curr Opin Cell Biol. 2010;22:430–436. doi: 10.1016/j.ceb.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B, Zhang Y, Paupard MC, Lin SX, Hall DH, Hirsh D. Evidence that RME-1, a conserved C. elegans EH-domain protein, functions in endocytic recycling. Nat Cell Biol. 2001;3:573–579. doi: 10.1038/35078549. [DOI] [PubMed] [Google Scholar]
- Grant BD, Donaldson JG. Pathways and mechanisms of endocytic recycling. Nat Rev. Mol Cell Biol. 2009;10:597–608. doi: 10.1038/nrm2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: achieving specificity in membrane traffic. Proc Natl Acad Sci USA. 2006;103:11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilherme A, Soriano NA, Furcinitti PS, Czech MP. Role of EHD1 and EHBP1 in perinuclear sorting and insulin-regulated GLUT4 recycling in 3T3-L1 adipocytes. J Biol Chem. 2004;279:40062–40075. doi: 10.1074/jbc.M401918200. [DOI] [PubMed] [Google Scholar]
- Hansen CG, Howard G, Nichols BJ. Pacsin 2 is recruited to caveolae and functions in caveolar biogenesis. J Cell Sci. 2011;124:2777–2785. doi: 10.1242/jcs.084319. [DOI] [PubMed] [Google Scholar]
- Henry GD, Corrigan DJ, Dineen JV, Baleja JD. Charge effects in the selection of NPF motifs by the EH domain of EHD1. Biochemistry. 2010;49:3381–3392. doi: 10.1021/bi100065r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu VW, Prekeris R. Transport at the recycling endosome. Curr Opin Cell Biol. 2010;22:528–534. doi: 10.1016/j.ceb.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson J, Ackermann F, Andersson F, Larhammar D, Low P, Brodin L. Regulation of synaptic vesicle budding and dynamin function by an EHD ATPase. J Neurosci. 2011;31:13972–13980. doi: 10.1523/JNEUROSCI.1289-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GH, Fisette PL, Anderson RA. Type I phosphatidylinositol 4-phosphate 5-kinase isoforms are specifically stimulated by phosphatidic acid. J Biol Chem. 1994;269:11547–11554. [PubMed] [Google Scholar]
- Jovanovic OA, Brown FD, Donaldson JG. An effector domain mutant of Arf6 implicates phospholipase D in endosomal membrane recycling. Mol Biol Cell. 2006;17:327–335. doi: 10.1091/mbc.E05-06-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovic M, Kieken F, Naslavsky N, Sorgen PL, Caplan S. Eps15 homology domain 1-associated tubules contain phosphatidylinositol-4-phosphate and phosphatidylinositol-(4,5)-bisphosphate and are required for efficient recycling. Mol Biol Cell. 2009;20:2731–2743. doi: 10.1091/mbc.E08-11-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovic M, Naslavsky N, Rapaport D, Horowitz M, Caplan S. EHD1 regulates beta1 integrin endosomal transport: effects on focal adhesions, cell spreading and migration. J Cell Sci. 2007;120:802–814. doi: 10.1242/jcs.03383. [DOI] [PubMed] [Google Scholar]
- Kassas N, Tryoen-Toth P, Corrotte M, Thahouly T, Bader MF, Grant NJ, Vitale N. Genetically encoded probes for phosphatidic acid. Methods Cell Biol. 2012;108:445–459. doi: 10.1016/B978-0-12-386487-1.00020-1. [DOI] [PubMed] [Google Scholar]
- Kessels MM, Qualmann B. The syndapin protein family: linking membrane trafficking with the cytoskeleton. J Cell Sci. 2004;117:3077–3086. doi: 10.1242/jcs.01290. [DOI] [PubMed] [Google Scholar]
- Kieken F, Jovic M, Naslavsky N, Caplan S, Sorgen PL. EH domain of EHD1. J Biomol NMR. 2007;39:323–329. doi: 10.1007/s10858-007-9196-0. [DOI] [PubMed] [Google Scholar]
- Kieken F, Sharma M, Jovic M, Giridharan SS, Naslavsky N, Caplan S, Sorgen PL. Mechanism for the selective interaction of C-terminal Eps15 homology domain proteins with specific Asn-Pro-Phe-containing partners. J Biol Chem. 2010;285:8687–8694. doi: 10.1074/jbc.M109.045666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DW, Zhao X, Scarselletta S, Schweinsberg PJ, Eisenberg E, Grant BD, Greene LE. ATP Binding regulates oligomerization and endosome association of RME-1 family proteins. J Biol Chem. 2005;280:280–290. doi: 10.1074/jbc.M412751200. [DOI] [PubMed] [Google Scholar]
- Lin SX, Grant B, Hirsh D, Maxfield FR. Rme-1 regulates the distribution and function of the endocytic recycling compartment in mammalian cells. Nat Cell Biol. 2001;3:567–572. doi: 10.1038/35078543. [DOI] [PubMed] [Google Scholar]
- Linkermann A, Gelhaus C, Lettau M, Qian J, Kabelitz D, Janssen O. Identification of interaction partners for individual SH3 domains of Fas ligand associated members of the PCH protein family in T lymphocytes. Biochim Biophys Acta. 2009;1794:168–176. doi: 10.1016/j.bbapap.2008.10.013. [DOI] [PubMed] [Google Scholar]
- Maxfield FR, McGraw TE. Endocytic recycling. Nat Rev Mol Cell Biol. 2004;5:121–132. doi: 10.1038/nrm1315. [DOI] [PubMed] [Google Scholar]
- McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- Modregger J, Ritter B, Witter B, Paulsson M, Plomann M. All three PACSIN isoforms bind to endocytic proteins and inhibit endocytosis. J Cell Sci. 2000;113:4511–4521. doi: 10.1242/jcs.113.24.4511. [DOI] [PubMed] [Google Scholar]
- Montagnac G, Echard A, Chavrier P. Endocytic traffic in animal cell cytokinesis. Curr Opin Cell Biol. 2008;20:454–461. doi: 10.1016/j.ceb.2008.03.011. [DOI] [PubMed] [Google Scholar]
- Morita SY, Ueda K, Kitagawa S. Enzymatic measurement of phosphatidic acid in cultured cells. J Lipid Res. 2009;50:1945–1952. doi: 10.1194/jlr.D900014-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz A, De Graan PN, Gispen WH, Wirtz KW. Phosphatidic acid is a specific activator of phosphatidylinositol-4-phosphate kinase. J Biol Chem. 1992;267:7207–7210. [PubMed] [Google Scholar]
- Naslavsky N, Boehm M, Backlund PS, Jr., Caplan S. Rabenosyn-5 and EHD1 interact and sequentially regulate protein recycling to the plasma membrane. Mol Biol Cell. 2004;15:2410–2422. doi: 10.1091/mbc.E03-10-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naslavsky N, Caplan S. EHD proteins: key conductors of endocytic transport. Trends Cell Biol. 2011;21:122–131. doi: 10.1016/j.tcb.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naslavsky N, Rahajeng J, Chenavas S, Sorgen PL, Caplan S. EHD1 and Eps15 interact with phosphatidylinositols via their Eps15 homology domains. J Biol Chem. 2007;282:16612–16622. doi: 10.1074/jbc.M609493200. [DOI] [PubMed] [Google Scholar]
- Naslavsky N, Rahajeng J, Sharma M, Jovic M, Caplan S. Interactions between EHD proteins and Rab11-FIP2: a role for EHD3 in early endosomal transport. Mol Biol Cell. 2006;17:163–177. doi: 10.1091/mbc.E05-05-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padron D, Tall RD, Roth MG. Phospholipase D2 is required for efficient endocytic recycling of transferrin receptors. Mol Biol Cell. 2006;17:598–606. doi: 10.1091/mbc.E05-05-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant S, Sharma M, Patel K, Caplan S, Carr CM, Grant BD. AMPH-1/amphiphysin/Bin1 functions with RME-1/Ehd1 in endocytic recycling. Nat Cell Biol. 2009;11:1399–1410. doi: 10.1038/ncb1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M, Penick EC, Edwards JG, Kauer JA, Ehlers MD. Recycling endosomes supply AMPA receptors for LTP. Science. 2004;305:1972–1975. doi: 10.1126/science.1102026. [DOI] [PubMed] [Google Scholar]
- Qualmann B, Kelly RB. Syndapin isoforms participate in receptor-mediated endocytosis and actin organization. J Cell Biol. 2000;148:1047–1062. doi: 10.1083/jcb.148.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualmann B, Koch D, Kessels MM. Let's go bananas: revisiting the endocytic BAR code. EMBO J. 2011;30:3501–3515. doi: 10.1038/emboj.2011.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan A, Xue J, Wielens J, Smillie KJ, Anggono V, Parker MW, Cousin MA, Graham ME, Robinson PJ. Phosphorylation of syndapin I F-BAR domain at two helix-capping motifs regulates membrane tubulation. Proc Natl Acad Sci USA. 2012;109:3760–3765. doi: 10.1073/pnas.1108294109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahajeng J, Panapakkam Giridharan SS, Cai B, Naslavsky N, Caplan S. MICAL-L1 is a tubular endosomal membrane hub that connects Rab35 and Arf6 with Rab8a. Traffic. 2012;13:82–93. doi: 10.1111/j.1600-0854.2011.01294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao Y, Ma Q, Vahedi-Faridi A, Sundborger A, Pechstein A, Puchkov D, Luo L, Shupliakov O, Saenger W, Haucke V. Molecular basis for SH3 domain regulation of F-BAR-mediated membrane deformation. Proc Natl Acad Sci USA. 2010;107:8213–8218. doi: 10.1073/pnas.1003478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senju Y, Itoh Y, Takano K, Hamada S, Suetsugu S. Essential role of PACSIN2/syndapin-II in caveolae membrane sculpting. J Cell Sci. 2011;124:2032–2040. doi: 10.1242/jcs.086264. [DOI] [PubMed] [Google Scholar]
- Sharma M, Giridharan SS, Rahajeng J, Naslavsky N, Caplan S. MICAL-L1 links EHD1 to tubular recycling endosomes and regulates receptor recycling. Mol Biol Cell. 2009;20:5181–5194. doi: 10.1091/mbc.E09-06-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skop AR, Bergmann D, Mohler WA, White JG. Completion of cytokinesis in C. elegans requires a brefeldin A-sensitive membrane accumulation at the cleavage furrow apex. Curr Biol. 2001;11:735–746. doi: 10.1016/s0960-9822(01)00231-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stace CL, Ktistakis NT. Phosphatidic acid- and phosphatidylserine-binding proteins. Biochim Biophys Acta. 2006;1761:913–926. doi: 10.1016/j.bbalip.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Wang E, Brown PS, Aroeti B, Chapin SJ, Mostov KE, Dunn KW. Apical and basolateral endocytic pathways of MDCK cells meet in acidic common endosomes distinct from a nearly-neutral apical recycling endosome. Traffic. 2000;1:480–493. doi: 10.1034/j.1600-0854.2000.010606.x. [DOI] [PubMed] [Google Scholar]
- Wang Q, Navarro MV, Peng G, Molinelli E, Goh SL, Judson BL, Rajashankar KR, Sondermann H. Molecular mechanism of membrane constriction and tubulation mediated by the F-BAR protein Pacsin/Syndapin. Proc Natl Acad Sci USA. 2009;106:12700–12705. doi: 10.1073/pnas.0902974106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Shi H, Wei S, Wong SH, Hong W. Mutually exclusive interactions of EHD1 with GS32 and Syndapin II. Mol Membrane Biol. 2004;21:269–277. doi: 10.1080/09687680410001716871. [DOI] [PubMed] [Google Scholar]
- Yang JS, et al. COPI acts in both vesicular and tubular transport. Nat Cell Biol. 2011;13:996–1003. doi: 10.1038/ncb2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeniou-Meyer M, et al. Phospholipase D1 production of phosphatidic acid at the plasma membrane promotes exocytosis of large dense-core granules at a late stage. J Biol Chem. 2007;282:21746–21757. doi: 10.1074/jbc.M702968200. [DOI] [PubMed] [Google Scholar]
- Zhang J, Naslavsky N, Caplan S. EHDs meet the retromer: complex regulation of retrograde transport. Cell Logist. 2012a;2:161–165. doi: 10.4161/cl.20582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Reiling C, Reinecke JB, Prislan I, Marky LA, Sorgen PL, Naslavsky N, Caplan S. Rabankyrin-5 interacts with EHD1 and Vps26 to regulate endocytic trafficking and retromer function. Traffic. 2012b;13:745–757. doi: 10.1111/j.1600-0854.2012.01334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.