The transcriptional regulator cyAbrB2 in Synechocystis sp. PCC 6803 is indispensable for coordination of cellular metabolism upon the changes in trophic conditions.
Abstract
cyAbrB is a transcriptional regulator unique to and highly conserved among cyanobacterial species. A gene-disrupted mutant of cyabrB2 (sll0822) in Synechocystis sp. PCC 6803 exhibited severe growth inhibition and abnormal accumulation of glycogen granules within cells under photomixotrophic conditions. Within 6 h after the shift to photomixotrophic conditions, sodium bicarbonate-dependent oxygen evolution activity markedly declined in the ΔcyabrB2 mutant, but the decrease in methyl viologen-dependent electron transport activity was much smaller, indicating inhibition in carbon dioxide fixation. Decreases in the transcript levels of several genes related to sugar catabolism, carbon dioxide fixation, and nitrogen metabolism were also observed within 6 h. Metabolome analysis by capillary electrophoresis mass spectrometry revealed that several metabolites accumulated differently in the wild-type and mutant strains. For example, the amounts of pyruvate and 2-oxoglutarate (2OG) were significantly lower in the mutant than in the wild type, irrespective of trophic conditions. The growth rate of the ΔcyabrB2 mutant was restored to a level comparable to that under photoautotrophic conditions by addition of 2OG to the growth medium under photomixotrophic conditions. Activities of various metabolic processes, including carbon dioxide fixation, respiration, and nitrogen assimilation, seemed to be enhanced by 2OG addition. These observations suggest that cyAbrB2 is essential for the active transcription of genes related to carbon and nitrogen metabolism upon a shift to photomixotrophic conditions. Deletion of cyAbrB2 is likely to deregulate the partition of carbon between storage forms and soluble forms used for biosynthetic purposes. This disorder may cause inactivation of cellular metabolism, excess accumulation of reducing equivalents, and subsequent loss of viability under photomixotrophic conditions.
All known cyanobacterial genomes possess multiple copies of genes encoding cyAbrB transcriptional regulators. cyAbrBs are structurally different from other AbrB-type regulators in that they have a DNA-binding domain in the C-terminal region instead of the usual N-terminal region (Ishii and Hihara, 2008). Larsson et al. (2011) reported that cyAbrB is one of the two protein families unique to the cyanobacterial phylum among 404 protein families conserved in all sequenced cyanobacterial genomes. Notably, multiple copies of cyAbrB genes exist in every cyanobacterial genome. Multiple copies in a single organism tend to belong to clades A and B or marine clades A and B (Ishii and Hihara, 2008). Here, we propose to call clade A genes cyabrB1 and clade B genes cyabrB2.
Synechocystis sp. PCC 6803 possesses two cyAbrB genes, cyabrB1 (sll0359) and cyabrB2 (sll0822), as well as several abrB genes that encode AbrB-type regulators with an N-terminal DNA-binding domain (ssl8028, ssl7048, and ssr7040; according to InterPro, http://www.ebi.ac.uk/interpro/). Although cyabrB1 is an essential gene in Synechocystis sp. PCC 6803, cyabrB2 can be deleted, and the gene-disrupted mutant has been characterized by several research groups. The following information has been obtained to date concerning the target genes and physiological roles of cyAbrB2 in Synechocystis sp. PCC 6803. (1) cyAbrB2 binds to its own promoter region to achieve negative autoregulation in both the Glc-tolerant strain (Ishii and Hihara, 2008; Yamauchi et al., 2011) and the Glc-sensitive original strain (Dutheil et al., 2012). (2) cyAbrB2 positively regulates nitrogen-regulated genes, such as urtA, amt1, glnB, sigE, and the nrt operon, under ambient CO2 conditions. Binding of cyAbrB2 to the upstream regions of the nrt operon and amt1 has been demonstrated (Ishii and Hihara, 2008). (3) cyAbrB2 functions as a repressor of inorganic carbon (Ci)-uptake-related genes under 5% CO2. The ΔcyabrB2 mutant exhibits the elevated apparent photosynthetic affinity for Ci that is typical in wild-type cells, but only under low CO2 conditions. Binding of cyAbrB2 to the upstream region of sbtA, which encodes a Na+/HCO3– symporter, has been demonstrated (Lieman-Hurwitz et al., 2009). (5) The expression levels of genes encoding cell division-related proteins such as FtsZ and FtsW are lower in the ΔcyabrB2 mutant. The ΔcyabrB2 cellular volume is 5 times that of the wild type, and the proportion of dividing cells is notably higher in the mutant culture, indicating a cell division defect (Yamauchi et al., 2011). (5) cyAbrB2 represses expression of the hox operon, which encodes the bidirectional nickel-iron hydrogenase. Binding of cyAbrB2 to the upstream region of the hox operon has been demonstrated in the Glc-sensitive strain (Dutheil et al., 2012). (6) Decrease in the transcript level of an antisense RNA to the flv4 gene, which encodes the flavodiiron protein, was observed in the ΔcyabrB2 mutant (Eisenhut et al., 2012).
As listed above, cyAbrB2 is involved in the regulation of various cellular processes, such as uptake of nitrogen and carbon, cell division, hydrogen metabolism, and expression of antisense RNAs, although it is not clear if cyAbrB2 itself directly regulates these processes or not. Nevertheless, cyAbrB2 is dispensable under photoautotrophic conditions, and its physiological significance remains elusive. Here, we report that growth of the ΔcyabrB2 mutant of the Glc-tolerant wild-type strain was severely inhibited under photomixotrophic conditions with Glc. We investigated the phenotypic changes in the ΔcyabrB2 mutant upon a shift from photoautotrophic to photomixotrophic conditions to clarify the impact of the lack of cyAbrB2 on cellular metabolism and the cause of the growth inhibition. Our data suggest that cyAbrB2 is essential for the maintenance of active cellular metabolism upon a change in trophic conditions. Deletion of cyAbrB2 is likely to deregulate the partition of carbon between storage forms and soluble forms used for biosynthetic purposes. Complementation of Glc-sensitive phenotype of the cyAbrB2 mutant by addition of 2-oxoglutarate (2OG) suggests that active consumption of the reducing power provided by photosynthetic electron transport seemed particularly important for survival under photomixotrophic conditions.
RESULTS
Growth Inhibition of the ΔcyabrB2 Mutant under Photomixotrophic Conditions
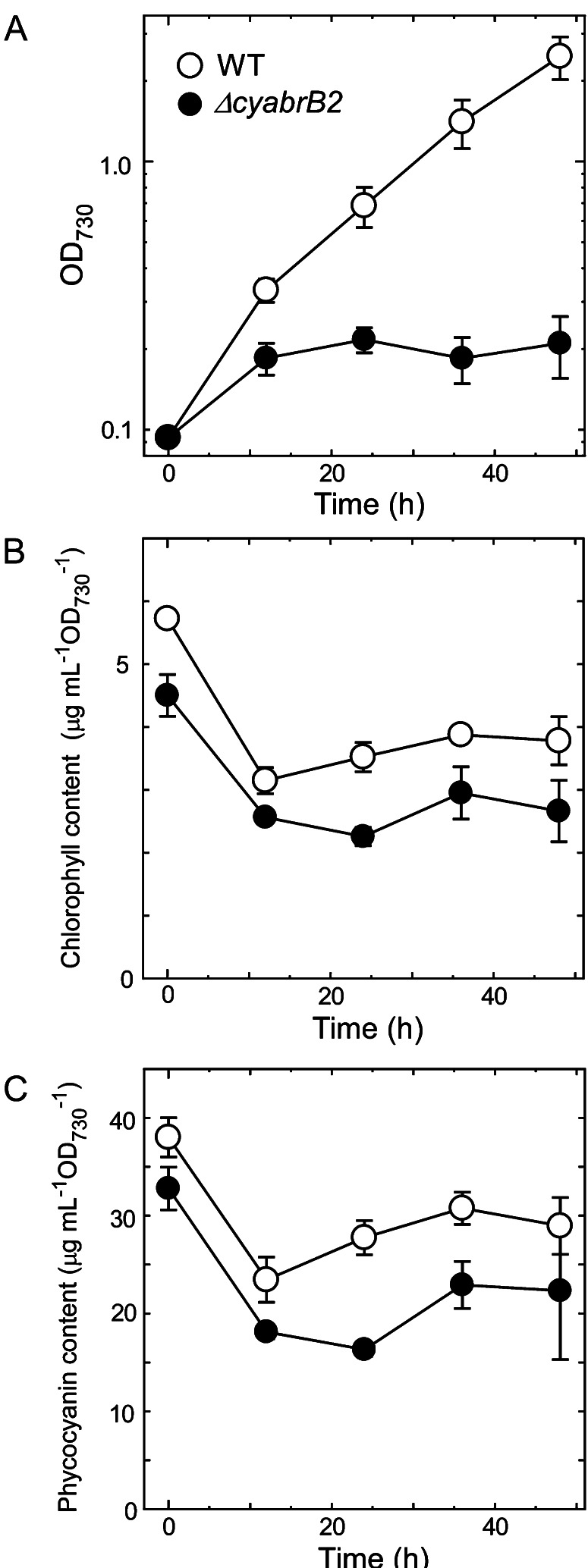
Addition of 5 mm Glc to the ΔcyabrB2 mutant under normal growth conditions (20 μmol photons m–2 s–1, ambient CO2) caused severe growth inhibition (Fig. 1A). The doubling time of wild-type cells was decreased following the addition of Glc, from 20.8 h under photoautotrophic conditions to 6.7 h under photomixotrophic conditions. The doubling time of the ΔcyabrB2 mutant was 34 h under photoautotrophic conditions. Upon Glc addition, optical density at 730 nm (OD730) of mutant cells doubled during the first 24 h, but subsequently, its growth almost completely stopped. The amounts of chlorophyll and phycocyanin were lower in the mutant than in the wild type under photoautotrophic conditions (Fig. 1, B and C). Upon a shift to photomixotrophic conditions, the pigment contents of both strains decreased to about 60% of the initial level within 12 h. Although growth of the mutant stopped after 24 h, no further decline in pigment content was observed. Changing the nitrogen source from nitrate to ammonium did not mitigate the inhibited growth under photomixotrophic conditions (Supplemental Fig. S1). Interestingly, growth inhibition was observed not only in the mutant but also in wild-type cultures upon simultaneous addition of ammonium and Glc.
Figure 1.
Growth properties and pigment contents of the wild type and the ΔcyabrB2 mutant under photomixotrophic conditions. At time 0, the wild type and the ΔcyabrB2 mutant grown under photoautotrophic conditions (20 μmol photons m–2 s–1, ambient CO2) were transferred to photomixotrophic conditions with 5 mm Glc, and changes at OD730 (A), chlorophyll content (B), and phycocyanin content (C) were monitored. Data and error bars were calculated from the results of three independent experiments. WT, Wild type.
When His-cyAbrB2 was introduced into the ΔcyabrB2 mutant, growth and viability under photomixotrophic conditions were significantly improved (Supplemental Fig. S2, A and B). Full recovery to wild-type level was not attained probably due to the low expression level of His-cyAbrB2 in the complementation strain (60% of wild-type level; Yamauchi et al., 2011). Recovery of transcript levels in the ΔcyabrB2 mutant was also observed by introduction of His-cyAbrB2 (Supplemental Fig. S2C). These results confirm that observed sensitivity to photomixotrophic conditions in the ΔcyabrB2 mutant is caused by the lack of the cyAbrB2 protein.
We evaluated changes in the cell morphology of the wild type and the ΔcyabrB2 mutant after the shift to photomixotrophic conditions using differential interference contrast microscopy (Supplemental Fig. S3). As reported previously (Yamauchi et al., 2011), the diameter of the ΔcyabrB2 cells was 1.7 times that of wild-type cells under photoautotrophic conditions. After 12 h of incubation under photomixotrophic conditions, slight increases in cell size and intracellular accumulation of particles were observed in both the wild-type and mutant strains. After 48 h, mutant cells were morphologically heterogeneous, and some were breached and swollen, whereas wild-type cells did not exhibit further morphological changes.
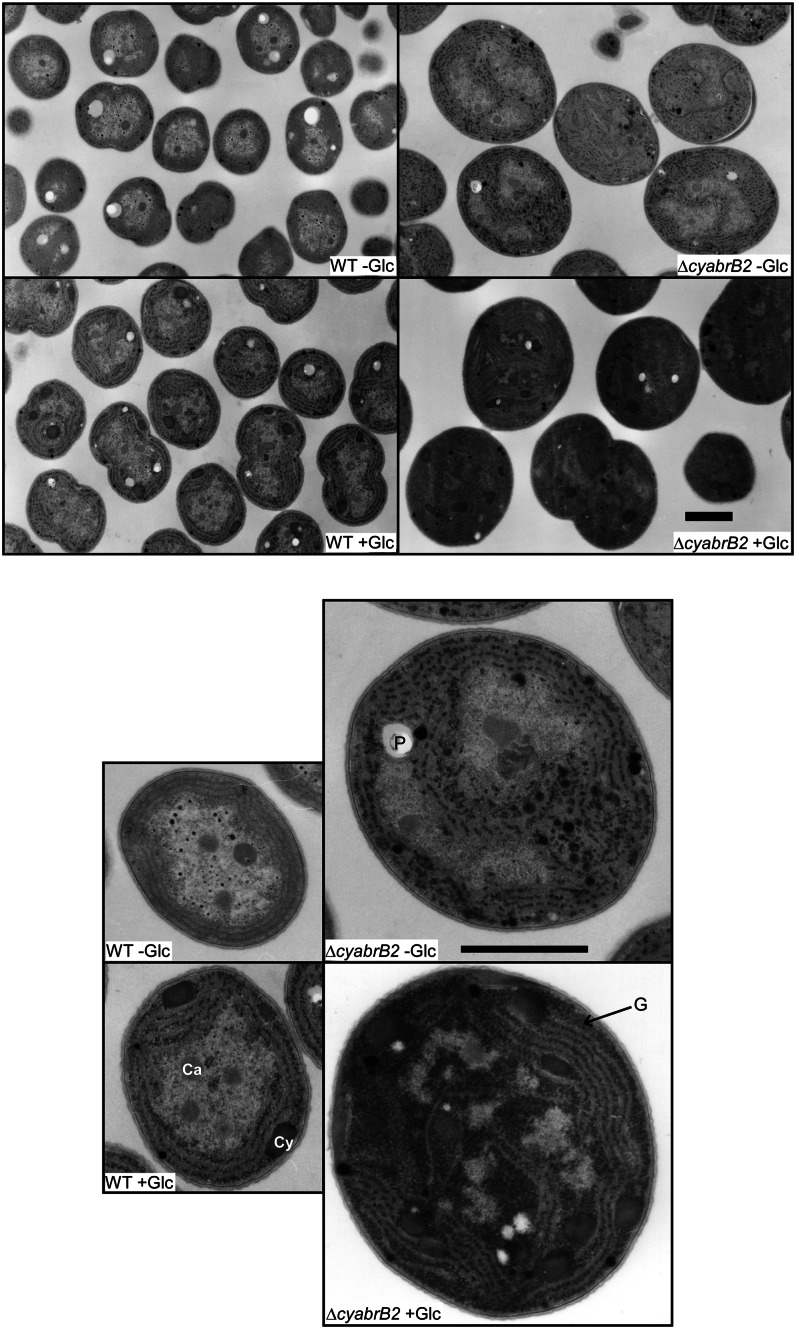
Figure 2 shows electron micrographs of ultrathin sections of wild-type and ΔcyabrB2 cells cultured under the different trophic conditions. We previously reported that glycogen granules accumulate in the spaces between the thylakoid membranes in the mutant but not in the wild type under photoautotrophic conditions (Yamauchi et al., 2011). When incubated under photomixotrophic conditions for 12 h, accumulation of glycogen granules in the interthylakoid spaces was observed also in wild-type cells. It is notable that the ΔcyabrB mutant appeared blackish due to the glycogen granules that had accumulated throughout the cell. Accumulation of cyanophycin granules, a nonprotein nitrogen storage polymer composed of Asp and Arg, was observed in both strains under photomixotrophic, but not photoautotrophic, conditions. Accumulation of cyanophycin under photomixotrophic conditions, where addition of Glc decreases the nitrogen/carbon ratio, was quite unexpected, because cyanophycin has been thought to accumulate under high nitrogen/carbon conditions. Similarly, decrease in 2OG level upon the shift to photomixotrophic conditions mentioned later was unexpected, because 2OG has been considered as a signaling molecule of low nitrogen/carbon ratio. Considering that accumulation of cyanophycin and decrease in 2OG level can be observed also in photomixotrophically grown wild-type cells, they cannot be a consequence of failure in metabolic regulation.
Figure 2.
Electron micrographs of ultrathin sections of wild-type and ΔcyabrB2 mutant cells before and after incubation under photomixotrophic conditions for 12 h. Magnified images of one cell are shown in the lower panels. WT, Wild type; Ca, carboxysomes; G, glycogen granules; Cy, cyanophycins; P, residual holes of polyphosphate bodies. Bars = 1 μm.
Changes in the Photosynthetic and Respiratory Activities of the Wild Type and the ΔcyabrB2 Mutant upon a Shift to Photomixotrophic Conditions
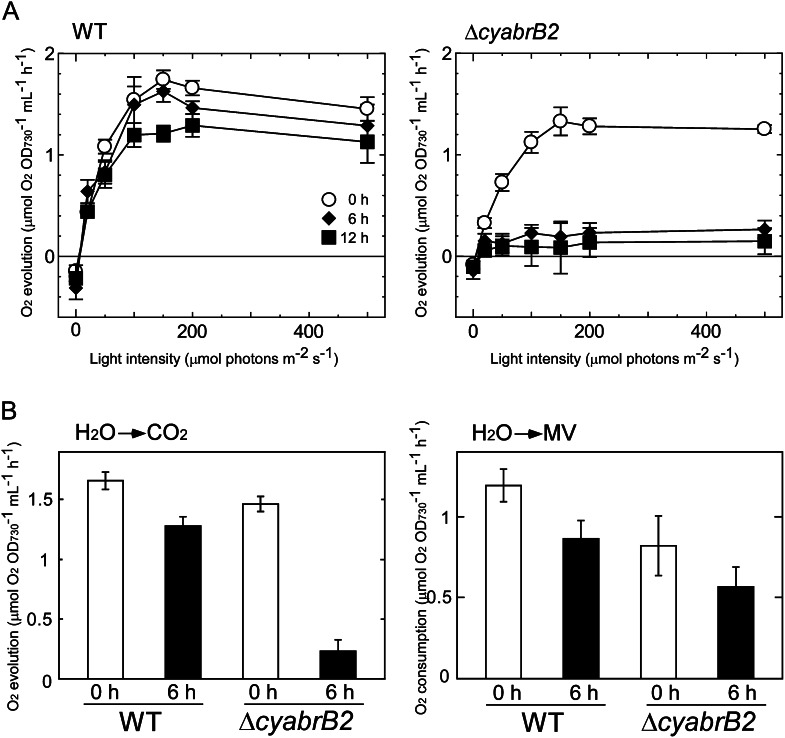
The photosynthetic and respiratory activities of the wild type and the ΔcyabrB2 mutant were examined after 0, 6, and 12 h of incubation under photomixotrophic conditions. Figure 3A shows the light saturation curves of oxygen evolution activity supported by 2 mm NaHCO3. In the wild type, the maximal photosynthetic activity decreased to 69% of the initial level after 12 h of incubation under photomixotrophic conditions. When measured under light-limiting conditions, the photosynthetic activity of the wild type was negligibly affected, even after 12 h, indicating that CO2 fixation activity, but not photosynthetic electron transport, is down-regulated under photomixotrophic conditions. In the ΔcyabrB2 mutant, photosynthetic activities measured under saturating and growth light conditions (20 μmol photons m–2 s–1) markedly declined to 14% and 46% of the initial levels, respectively, within 6 h of incubation under photomixotrophic conditions. Respiratory activity, measured as the rate of oxygen consumption in the dark with 5 mm Glc, was similar in the wild type and the mutant, and was not so much affected by the change in trophic conditions (0 h: wild type, 0.11 ± 0.02 and ΔcyabrB2, 0.11 ± 0.01; 6 h: wild type, 0.12 ± 0.04 and ΔcyabrB2, 0.08 ± 0.01; and 12 h: wild type, 0.11 ± 0.03 and ΔcyabrB2, 0.08 ± 0.02 μmol oxygen consumed OD730–1 mL–1 h–1). This suggests that the decline in oxygen evolution activity of the mutant under photomixotrophic conditions was not due to enhanced respiratory activity.
Figure 3.
Changes in photosynthetic activities of wild-type and ΔcyabrB2 mutant cells upon a shift to photomixotrophic conditions. A, The light saturation curves of photosynthesis in wild-type and ΔcyabrB2 mutant cells incubated under photomixotrophic conditions for 0, 6, or 12 h. Rates of CO2-dependent O2 evolution on a per OD730 basis were measured as a function of actinic light intensity. Data and error bars were calculated from the results of at least three independent experiments. B, Comparison of CO2-dependent photosynthetic activity and methyl viologen-dependent whole-chain electron transport activity before and after incubation under photomixotrophic conditions for 6 h. Measurements were performed under saturated light conditions (200 μmol photons m–2 s–1). Methyl viologen-dependent electron transport activity was measured as O2 consumption rate on a per OD730 basis. Data and error bars were calculated from the results of at least three independent experiments. WT, Wild type; MV, methyl viologen.
When whole-chain photosynthetic electron transport activity was measured using methyl viologen as an electron acceptor, the decrease in activity of the mutant upon the addition of Glc was not obvious compared with the decrease in NaHCO3-dependent oxygen evolution activity (Fig. 3B). This observation supports the hypothesis that not photosynthetic electron transport but CO2 fixation is down-regulated in the mutant under photomixotrophic conditions. The concentration of NaHCO3 added to the oxygen electrode chamber was increased to 10 mm to determine whether low Ci-uptake efficiency was the cause of the low CO2 fixation activity in the mutant. This 5-fold increase in NaHCO3 concentration did not enhance the photosynthetic activity of the mutant (data not shown), indicating that Ci uptake was not the limiting factor.
Changes in Expression Levels of Genes Related to Carbon and Nitrogen Metabolism upon a Shift to Photomixotrophic Conditions
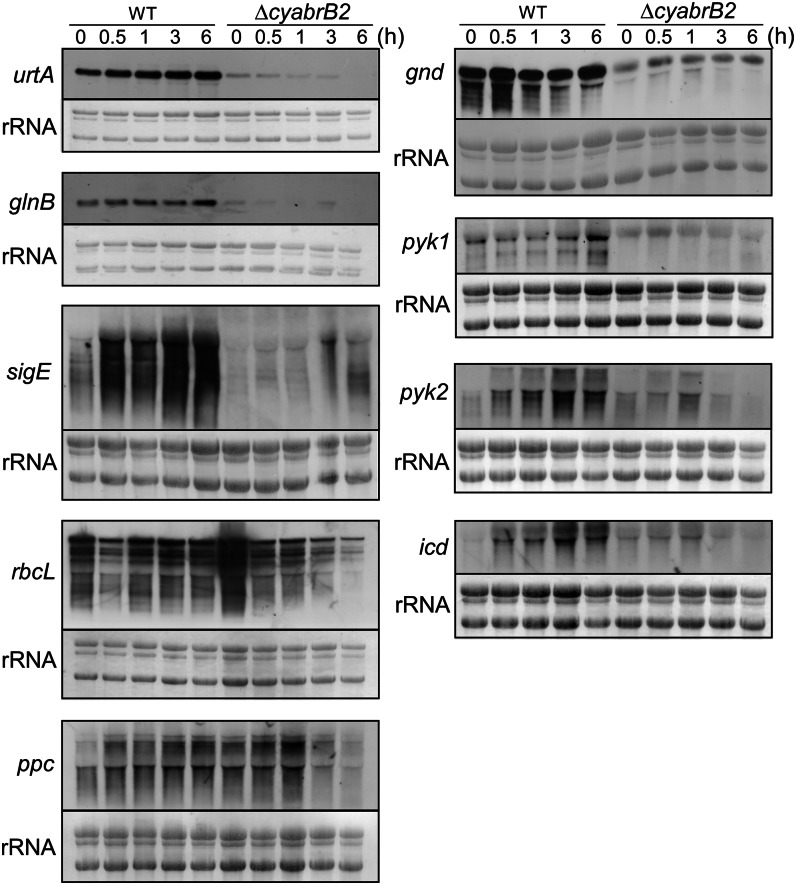
Changes in transcript levels were compared between the wild type and the ΔcyabrB2 mutant upon a shift to photomixotrophic conditions (Fig. 4). Expression levels of nitrogen-regulated genes, such as urtA, which encodes a subunit of the urea transporter glnB, which encodes the nitrogen regulatory protein PII, and sigE, which encodes a group 2 σ factor, were markedly lower in the mutant than in the wild type under photoautotrophic conditions, as reported previously (Ishii and Hihara, 2008). Upon a shift to photomixotrophic conditions, gradual accumulation of these transcripts was observed in the wild type, whereas they remained at low level in the mutant.
Figure 4.
Changes in transcript levels of the wild type and the ΔcyabrB2 mutant upon the shift to photomixotrophic conditions as determined by RNA gel-blot analysis. The amount of total RNA loaded per lane was as follows: 2 μg for rbcL, urtA, and glnB, 4 μg for gnd, and 6 μg for sigE, ppc, pyk1, pyk2, and icd. Total RNA was stained with methylene blue to compare within each blot. WT, Wild type.
The CO2 fixation activity of the mutant decreased drastically upon a shift to photomixotrophic conditions (Fig. 3). Thus, we investigated transcript levels of rbcL and ppc, which encode the CO2 fixation enzymes Rubisco and phosphoenolpyruvate (PEP) carboxylase, respectively. In the wild type, rbcL transcript levels transiently decreased, while those of ppc increased upon a shift to photomixotrophic conditions. Notably, the expression levels of both genes were higher in the mutant than in the wild type under photoautotrophic conditions. However, their expression in the mutant decreased gradually during incubation under photomixotrophic conditions.
The expression levels of genes involved in sugar catabolism were also examined. The gnd gene, which encodes 6-phosphogluconate dehydrogenase of the oxidative pentose phosphate pathway (OPP), was highly expressed in the wild type, but its expression level was considerably lower in the mutant irrespective of trophic conditions. The pyk1 (sll0587) and pyk2 (sll1275) genes, which encode the glycolytic enzyme pyruvate kinase, were up-regulated in the wild type upon addition of Glc. The pyk1 transcript level in the mutant was constitutively low, and that of pyk2 was slightly up-regulated after 1 h but decreased below the detection limit after 6 h. The icd gene, which encodes the tricarboxylic acid (TCA) cycle enzyme isocitrate dehydrogenase, was up-regulated in the wild type upon a shift to photomixotrophic conditions. The icd expression level in the mutant was higher than that in the wild type under photoautotrophic conditions but decreased to near the detection limit after 6 h of incubation under photomixotrophic conditions.
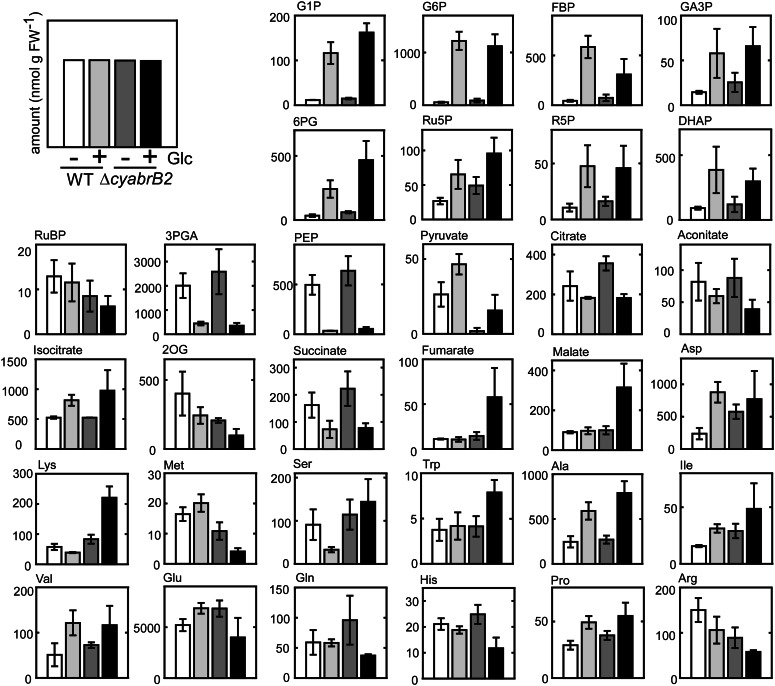
Metabolome Analysis of the Wild-Type and ΔcyabrB2 Mutant Cells under Different Trophic Conditions
We performed metabolome analysis using capillary electrophoresis mass spectrometry (CE/MS) to investigate the effect of a lack of cyAbrB2 on central carbon metabolism (Fig. 5; Supplemental Table S1; Supplemental Figs. S4 and S5). Metabolite levels were compared before and after 12-h incubation under photomixotrophic conditions. A marked increase in the metabolite levels in glycolysis and the OPP together with a decrease in the amount of 3-phosphoglycerate, PEP, 2OG, and succinate were observed in both wild-type and mutant cells. These changes in metabolite levels upon a shift to photomixotrophic conditions are consistent with our previous observations (Takahashi et al., 2008).
Figure 5.
Metabolome analysis of the wild type and ΔcyabrB2 mutant under different trophic conditions. The wild type and ΔcyabrB2 mutant were grown photoautotrophically (Glc–) or photomixotrophically (Glc+) for 12 h, and then metabolites were extracted and analyzed by CE/MS. Data and error bars were calculated from the results of four independent experiments (nmol g–1 fresh weight). WT, Wild type.
Notably, there were several differences in metabolite levels between the wild type and mutant. When grown under photoautotrophic conditions, the amounts of sugar phosphates were 1.5 to 2 times higher in the mutant than in the wild type. By contrast, the amounts of pyruvate and 2OG were significantly lower in the mutant than in the wild type, irrespective of trophic condition. Upon a shift to photomixotrophic conditions, increases in Ser, Trp, Lys, malate, and fumarate and decreases in Met, Gln, and Glu levels were detected specifically in the mutant. Although a sharp decline in CO2 fixation activity upon a shift to photomixotrophic conditions was suggested in the mutant (Fig. 3), it was not reflected in the amounts of sugar phosphates. It is likely that an increase in carbon flux through glycolysis and the OPP upon addition of Glc masked the changes in metabolite levels caused by the decrease in the Calvin cycle activity.
Effects of Addition of Metabolites on the ΔcyabrB2 Mutant under Photomixotrophic Conditions
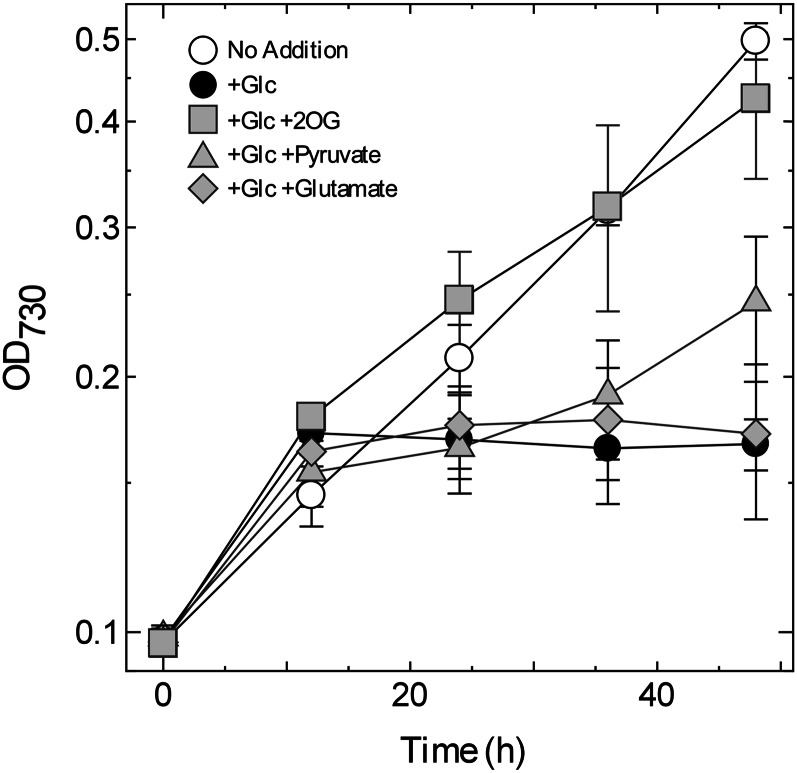
The metabolome analysis revealed that the amounts of pyruvate and 2OG were significantly lower in the ΔcyabrB2 mutant than in the wild type, irrespective of trophic conditions (Fig. 5). To assess the effect of a shortage of these metabolites on the viability of the ΔcyabrB2 mutant under photomixotrophic conditions, the growth medium of the mutant was supplemented with pyruvate, 2OG, or Glu to a final concentration of 40 mm (Fig. 6). Addition of Glu did not compensate for the growth defect in the mutant, despite the fact that this cyanobacterium possesses a Glu transporter (Quintero et al., 2001). Pyruvate mitigated the inhibition of growth to some extent. Surprisingly, when 2OG was added, the growth capacity of the mutant was restored to the level observed under photoautotrophic conditions. Microscopic observations showed that the morphology of the ΔcyabrB2 mutant was not recovered by addition of 2OG (Supplemental Fig. S6), suggesting no correlation between viability and cell morphology.
Figure 6.
Effects of addition of metabolites to the culture medium on growth of the ΔcyabrB2 mutant under photomixotrophic conditions. At time 0, the ΔcyabrB2 mutant grown under photoautotrophic conditions (20 μmol photons m–2 s–1, ambient CO2) was supplemented with 5 mm Glc and 40 mm metabolites. Growth profiles of the mutant under photoautotrophic conditions (no addition), under photomixotrophic conditions without addition of any metabolites (+Glc), under photomixotrophic conditions with 2OG (+Glc, +2OG), with pyruvate (+Glc, +pyruvate), and with Glu (+Glc, +Glu) were determined. Data and error bars were calculated from the results of three independent experiments.
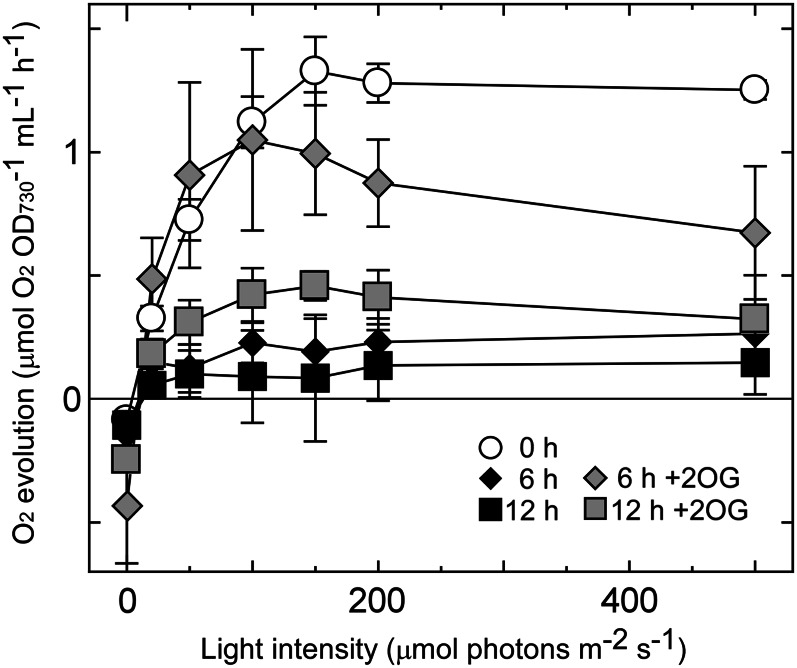
Measurement of NaHCO3-dependent oxygen evolution activity revealed that 2OG prevented the decline in photosynthetic activity of the mutant after 6 h of incubation under photomixotrophic conditions (Fig. 7). Photosynthetic activity decreased after 12 h incubation with 2OG, but remained higher than that without 2OG. Respiratory activity was also enhanced by addition of 2OG and was higher at 12 h than at 6 h (6 h: –2OG, 0.08 ± 0.01 and +2OG, 0.15 ± 0.01 and 12 h: –2OG, 0.08 ± 0.02 and +2OG, 0.20 ± 0.01 μmol oxygen consumed OD730–1 mL–1 h–1).
Figure 7.
Effects of addition of 2OG to the culture medium on the photosynthetic activity of the ΔcyabrB2 mutant under photomixotrophic conditions. The light saturation curve of photosynthesis in ΔcyabrB2 mutant cells incubated under photomixotrophic conditions with or without 2OG for 0, 6, or 12 h is presented. Rates of CO2-dependent O2 evolution on a per OD730 basis were measured as a function of actinic light intensity. Data and error bars were calculated from the results of at least three independent experiments.
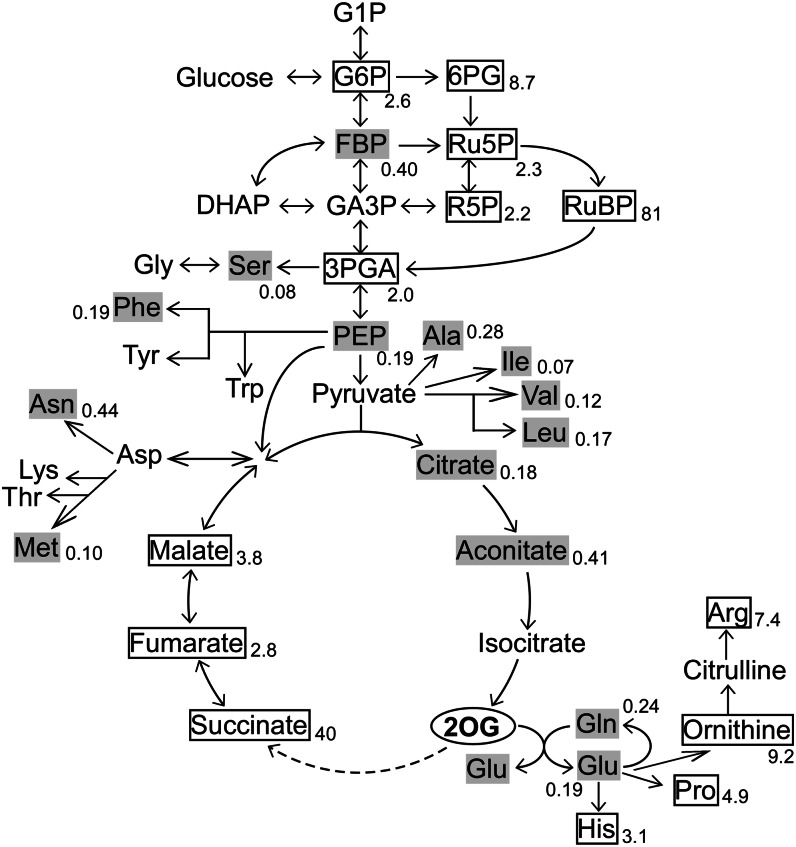
Next, we investigated the effect of 2OG addition on cellular metabolism (Fig. 8; Supplemental Table S1; Supplemental Figs. S7 and S8). The metabolite levels of the mutant with or without 2OG were compared at 24 h, because the advantageous effect of 2OG addition on growth became obvious at about 24 h incubation under photomixotrophic conditions. The CE/MS analysis revealed that 2OG addition had profound effects on the metabolism of Synechocystis. The levels of nitrogenous compounds such as His, Pro, Orn, and Arg increased, whereas those of Gln and Glu decreased. Citrate and aconitate decreased, whereas metabolites located downstream of 2OG, particularly succinate, showed marked increases. Surprisingly, the amount of ribulose 1,5-bisphosphate increased by more than 80-fold. Other notable changes were increases in the amounts of sugar phosphates, such as Glc-6-P, 6-phosphogluconate, ribulose-5-P, and ribose-5-P, and decreases in the levels of amino acids.
Figure 8.
Effects of addition of 2OG to the culture medium on cellular metabolism of the ΔcyabrB2 mutant under photomixotrophic conditions. The ΔcyabrB2 mutant was incubated under photomixotrophic conditions with or without 40 mm 2OG for 24 h, and then metabolites were extracted and analyzed by CE/MS. Extent of induction or repression upon addition of 2OG is shown as a fold-change value. Values are means ± sd of three independent and duplicate experiments (n = 6). Increased (more than 2-fold, P < 0.05) and decreased (less than one-half, P < 0.05) metabolite levels are indicated by boxes and shading, respectively.
To assess whether accumulation level of glycogen is affected by the addition of 2OG, we quantified glycogen content of both strains. Under photoautotrophic conditions, glycogen content of ΔcyabrB2 cells was 3.4 times that of wild-type cells (wild type, 4.3 ± 1.6 and ΔcyabrB2, 14.6 ± 3.9 μg mL–1 OD730–1). More than 6 times increase in glycogen content was observed in both strains after 24 h of incubation under photomixotrophic conditions (wild type, 26.9 ± 5.5 and ΔcyabrB2, 88.4 ± 7.4 μg mL–1 OD730–1). When 2OG was added upon the shift to photomixotrophic conditions, glycogen content of the mutant after 24 h was 94.3 ± 1.1 μg mL–1 OD730–1, indicating that sugar anabolism was not promoted by the addition of 2OG.
DISCUSSION
The Physiological Significance of cyAbrB2 in the Regulation of Cellular Metabolism under Different Trophic Conditions
In this study, we observed that a lack of cyAbrB2 had profound effects on transcript levels and cellular metabolism upon a shift from photoautotrophic to photomixotrophic conditions. The RNA gel-blot analysis (Fig. 4) revealed that the expression level of gnd, a gene involved in the OPP, was markedly lower in the ΔcyabrB2 mutant than in the wild type, irrespective of trophic conditions. The pyk genes, which are involved in the glycolytic pathway, and the icd gene, which is involved in the TCA cycle, were not up-regulated in the mutant upon a shift to photomixotrophic conditions. Pyruvate kinase, which is encoded by the pyk gene, catalyzes the conversion of PEP to pyruvate and is one of the key regulatory enzymes in the glycolytic pathway (Knowles et al., 2001). Isocitrate dehydrogenase, encoded by the icd gene, plays an important role in controlling nitrogen and carbon metabolism, because 2OG produced in the isocitrate dehydrogenase reaction is used as a carbon skeleton for nitrogen assimilation in cyanobacteria (Muro-Pastor et al., 1996). Thus, failure to activate these genes has a serious impact on cellular metabolism. In fact, the metabolome analysis revealed that the amounts of pyruvate and 2OG were significantly lower in the mutant than in the wild type (Fig. 5). By contrast, amounts of sugar phosphates were 1.5 to 2 times higher in the mutant than in the wild type under photoautotrophic conditions, suggesting that the reduced activity of the downstream pathways causes accumulation of sugars within mutant cells. After addition of Glc, the difference in the amounts of sugar phosphates between strains became negligible, probably due to the large increase in sugar influx. Based on these transcriptomic and metabolomic data, it can be assumed that sugar catabolic activity is lower in the ΔcyabrB2 mutant than in the wild type, which causes abnormal accumulation of glycogen under both photoautotrophic and photomixotrophic conditions (Fig. 2).
Expression levels of rbcL, which encodes the large subunit of Rubisco, and ppc, which encodes PEP carboxylase, were higher in the mutant than in the wild type under photoautotrophic conditions. We have reported previously that the expression levels of Ci-uptake-related genes such as ndhF3 and sbtA (Lieman-Hurwitz et al., 2009; Yamauchi et al., 2011) and photosynthetic affinity to external Ci (Lieman-Hurwitz et al., 2009) are lower in the mutant under ambient CO2 conditions. Nevertheless, the photosynthetic activity of the photoautotrophically grown mutant was comparable to that of the wild type (Fig. 3). The low activity of Ci concentration mechanism may be compensated for by the high expression level of CO2 fixation enzymes. However, rbcL and ppc transcript levels decreased gradually in the mutant upon a shift to photomixotrophic conditions (Fig. 4). After 6 h, rbcL and ppc transcripts together with CO2 fixation activity (Fig. 3) significantly declined. The reaction catalyzed by PEP carboxylase contributes about 25% of the assimilated CO2 in photomixotrophically grown wild-type cells (Yang et al., 2002). In the case of the mutant, the activation of PEP carboxylase under photomixotrophic conditions is unlikely due to (1) the low transcript level, (2) low CO2 fixation activity, and (3) marked accumulation of malate and fumarate, which could inhibit PEP carboxylase activity (Owttrim and Colman, 1986). Metabolic flux analysis performed by Young et al. (2011) suggested that PEP carboxylase and malic enzyme cooperate in Synechocystis sp. PCC 6803 (i.e. carbon fixed via PEP carboxylase is released by malic enzyme). This cycle is supposed to replenish pyruvate, without any net effect on the carbon balance. The low amount of pyruvate in the ΔcyabrB2 mutant may be partly caused by the low activity of this cycle.
The expression levels of genes related to sugar catabolism, CO2 fixation, and nitrogen metabolism all declined in the mutant during 6-h incubation under photomixotrophic conditions. This was not the consequence of a loss of the viability, because the mutant continued to propagate at this time point (Fig. 1). It is likely that cyAbrB2 is essential for active transcription of genes related to carbon and nitrogen metabolism under photomixotrophic conditions.
Then, which gene product regulated by cyAbrB2 is primarily responsible for the pleiotrophic effects observed in morphology, metabolism, and photosynthesis? The most possible candidate could be SigE, a group 2 σ factor for RNA polymerase. Osanai et al. (2005) reported that SigE is involved in activation of genes related to sugar catabolism, including pyk1 and gnd. Furthermore, they reported a decrease in transcript levels of glycogen catabolism-related genes and aberrant accumulation of glycogen in the sigE mutant. We observed that the sigE expression level is significantly lower in the ΔcyabrB2 mutant irrespective of trophic conditions (Fig. 4). This may, in part, be responsible for the insufficient sugar catabolism in the mutant. On the other hand, the failure in activation of nitrogen-related genes and insufficient nitrogen metabolism cannot be caused by SigE. Although SigE was first reported as a nitrogen-regulated σ factor (Muro-Pastor et al., 2001), DNA microarray analysis revealed that transcript levels of most nitrogen-regulated genes were unaffected by the disruption of sigE (Osanai et al., 2005). It may be possible that the low expression level of glnB in the ΔcyabrB2 mutant (Fig. 4) causes the disorder in nitrogen metabolism. PII, the gene product of glnB, is a well-known regulatory protein working as both a sensor and a regulator of carbon and nitrogen balance (Forchhammer, 2004). Although the role of the PII protein under photomixotrophic conditions is unclear, the increase in phosphorylation level of PII by the addition of Glc has been reported (Kloft and Forchhammer, 2005). In addition to SigE and PII, the direct regulation of cellular metabolism by cyAbrB2 is also plausible. We observed the direct binding of cyAbrB2 to the promoter regions of the nrt operon and amt1 (Ishii and Hihara, 2008). Onizuka et al. (2002) reported binding of cyAbrB to the upstream region of the rbc operon in Synechococcus sp. PCC 7002. cyAbrB2 may directly bind to the promoter region of rbc operon also in Synechocystis sp. PCC 6803.
Cause of Growth Inhibition of the ΔcyabrB2 Mutant under Photomixotrophic Conditions
Photosynthetic electron transport activity in the ΔcyabrB2 mutant did not decrease markedly compared with that of CO2 fixation under photomixotrophic conditions (Fig. 3B). Moreover, the amount of photosynthetic pigments (i.e. antenna size) did not decline in the mutant even after cessation of growth (Fig. 1). These observations indicate that active production of reducing equivalents continues in photomixotrophically grown mutants. By contrast, the activities of the Calvin cycle, nitrogen assimilation, and other biosynthetic pathways must have decreased significantly, judging from the photosynthetic activity (Fig. 3A) and metabolite levels (Fig. 5). Because these pathways are major consumers of the reducing equivalents under photomixotrophic conditions (Yang et al., 2002), it is plausible that the supply of reducing equivalents greatly exceeded consumption in photomixotrophically grown mutant cells, which led to the loss of viability.
The fact that addition of 2OG but not Glu rescued the lethal ΔcyabrB2 mutant phenotype (Fig. 6) seems an important clue to understanding the nature of the growth inhibition in the mutant. Ammonium is incorporated into carbon skeletons of cyanobacteria mainly by the combined action of Gln synthetase and Glu synthase (GS-GOGAT cycle; Muro-Pastor et al., 2005). GS catalyzes the ATP-dependent amidation of Glu to yield Gln. Then, GOGAT catalyzes the transfer of the amide group from Gln to 2OG to yield two molecules of Glu using two electrons provided by NADH or ferredoxin. The successful rescue by 2OG, but not Glu, suggests that either 2OG itself or the GOGAT reaction, but not the resulting product, is critical for survival of the mutant under photomixotrophic conditions. 2OG is a well-known signaling molecule for a high carbon/nitrogen ratio, and its accumulation is thought to cause the activation of the nitrate assimilatory pathway in Synechocystis sp. PCC 6803 (Kobayashi et al., 2005; Ohashi et al., 2011). In our rescue experiment, addition of 2OG had a marked positive effect on the nitrogen assimilation activity of the ΔcyabrB2 mutant, in which the expression level of nitrogen uptake-related genes, such as nrt, amt, and urt, was very low (Ishii and Hihara, 2008). Levels of the nitrogenous compounds located downstream of the GS-GOGAT cycle increased significantly, whereas those of Gln, Glu, and metabolites located upstream of 2OG in the TCA cycle decreased after addition of 2OG (Fig. 8). We hypothesized that increased consumption of reducing equivalents by nitrogen assimilation was a major contributor to the rescue of the mutant.
Nitrogen assimilation is highly dependent on the reducing equivalents produced by photosynthetic electron transport. Nitrate taken up by Synechocystis sp. PCC 6803 cells is reduced to nitrite by nitrate reductase using two electrons and subsequently to ammonium by nitrite reductase using six electrons. In addition, incorporation of ammonium into the carbon skeleton requires two electrons at the GOGAT reaction step. Several observations demonstrate the physiological significance of nitrogen assimilation under photomixotrophic conditions. Terauchi et al. (1996) reported that a ferredoxin-dependent GOGAT mutant cannot grow under photomixotrophic conditions. The growth inhibition observed in both the wild type and the ΔcyabrB2 mutant in the presence of ammonium and Glc (Supplemental Fig. S1) may be due to blockage of nitrate assimilation by ammonium. Kloft and Forchhammer (2005) reported that nitrate uptake activity in the wild type was doubled by addition of Glc. Consumption of reducing equivalents by nitrogen assimilation seems important for maintenance of the cellular redox balance under photomixotrophic conditions.
The possibility that activation of metabolic pathway(s) other than nitrogen assimilation also contributes to rescue of the mutant cannot be excluded. A marked enhancement of photosynthetic and respiratory activities was identified in the ΔcyabrB2 mutant supplemented with 2OG (Fig. 7). Moreover, the CE/MS analysis revealed many unexpected effects on cellular metabolism by addition of 2OG (Fig. 8). The increase in sugar phosphates may reflect the up-regulation of the OPP and/or the Calvin cycle. Two explanations seem possible for the increase in malate, fumarate, and succinate: (1) PEP carboxylase activity was enhanced by addition of 2OG, or (2) the 2OG was converted to succinate. Although the TCA cycle has been thought to be incomplete in cyanobacteria due to a lack of the 2OG dehydrogenase complex (Smith et al., 1967), existence of a pathway to convert 2OG to succinate in vivo was suggested in Synechocystis sp. PCC 6803 (Cooley et al., 2000). Two novel enzymes involved in this process were recently identified in Synechococcus sp. PCC 7002 (Zhang and Bryant, 2011). Notably, levels of most amino acids, except those located downstream of the GS-GOGAT cycle, decreased upon 2OG addition. Our observation raises the possibility that 2OG is involved in a much wider range of metabolic regulation than expected.
Taken together, the rescue experiment with 2OG suggests that activation of metabolic processes and the resulting mitigation of the over-reduced state are essential for the survival of the mutant under photomixotrophic conditions. Laurent et al. (2008) reported that the inhibited growth of the SpkD Ser/Thr kinase mutant under low-Ci conditions or in the presence of ammonium can be rescued by addition of 2OG. Also in this case, consumption of reducing equivalents may be enhanced by 2OG, leading to alleviation of the growth inhibition in the over-reduced state. Adjustment of metabolic activities to maintain the cellular redox balance is important for survival under different environmental conditions, and cyAbrB2 must be a key factor in such regulation.
MATERIALS AND METHODS
Strains and Culture Conditions
A Glc-tolerant strain of Synechocystis sp. PCC 6803 was grown at 32°C in BG-11 medium containing 20 mm HEPES-NaOH, pH 7.0, under continuous illumination at 20 μmol photons m–2 s–1 with bubbling of air. The ΔcyabrB2-disrupted mutant (Ishii and Hihara, 2008) was grown under the same conditions, except that 20 μg mL–1 kanamycin was added to the medium. Glc was added at a final concentration of 5 mm for the photomixotrophic conditions. Metabolites dissolved in BG-11 medium were buffered at pH 7.0 for the complementation analysis and added to the culture at a final concentration of 40 mm. Cell density was estimated by measuring OD730 using a spectrophotometer (model UV-160A, Shimadzu). As cells of the wild type and the ΔcyabrB2 mutant strains differ in size, equal OD730 does not mean equal cell number. For example, 1 mL of wild-type culture at OD730 = 1.0 contains 1.3 × 108 cells, whereas the mutant culture of the same turbidity contains only 3.8 × 107 cells under photoautotrophic conditions (Yamauchi et al., 2011).
Determination of Pigment Contents
In vivo absorption spectra of whole cells suspended in BG-11 medium were measured at room temperature using a spectrophotometer (model 557, Hitachi) with an end-on photomultiplier. Chlorophyll and phycocyanin contents were calculated from the peak heights of absorption spectra using the equations of Arnon et al. (1974).
Optical and Electron Microscopy
Optical and electron microscopy were performed as described previously (Yamauchi et al., 2011).
RNA Gel-Blot Analysis
Isolation of total RNA by the hot phenol method and RNA gel-blot analyses, using a DIG RNA Labeling and Detection Kit (Roche), were performed as described previously (Muramatsu and Hihara, 2003). Template DNA fragments were prepared by PCR using the following primers to generate RNA probes by in vitro transcription:
urtA-F (5′-ATGACTAACCCTTTTGGA-3′) and SP6-urtA-R (5′-ATTTAGGTGACACTATAGAATACAGCAACCAATCCACCGCC-3′), PglnB-F (5′-AGAGGAAAAGTTTTTCGA-3′) and T7-glnB-R (5′-TAATACGACTCACTATAGGGCGATTAAATAGCTTCGGTATC-3′), sigE-F (5′-ATGAGCGATATGTCTTCC-3′) and T7-sigE-R (5′-TAATACGACTCACTATAGGGCGAATCGCCCCTTCTTGGATC-3′), gnd-F (5′-GTCCACAACGGCATTGAG-3′) and SP6-gnd-R (5′-ATTTAGGTGACACTATAGAATACTTTGCTTGAATTC-3′), pyk1-F (5′-ATTGGCGATCGCCATCCT-3′) and SP6-pyk1-R (5′-ATTTAGGTGACACTATAGAATACAGCGGTGATGACCGGAAT-3′), pyk2-F (5′-GACGAAGTACCTTCTGGG-3′) and SP6-pyk2-R (5′-ATTTAGGTGACACTATAGAATACAGGAATGCCCAACCGGTT-3′), icd-F (5′-TATCAGTATTTACCGGAA-3′) and SP6-icd-R (5′-ATTTAGGTGACACTATAGAATACAATGTTGCCCTTATGGAC-3′), rbcL-F (5′-TGGACCACCGTTTGGACT-3′) and T7-rbcL-R (5′-TAATACGACTCACTATAGGGCGAGATCGCCTCTTGAACGAA-3′), and ppc-F (5′-GAACGGGACCGCATTCAA-3′) and SP6-ppc-R (5′-ATTTAGGTGACACTATAGAATACACTGGCATCGTTGGTCAT-3′).
The underlining indicates the T7 polymerase recognition sequence (TAATACGACTCACTATAGGGCGA) or the SP6 polymerase recognition sequence (ATTTAGGTGACACTATAGAATAC) added to the reverse primers to use the PCR products directly as templates for in vitro transcription reactions.
Measurement of Photosynthetic and Respiratory Activities
A 1-mL aliquot of cultures grown photoautotrophically or photomixotrophically to an OD730 of 0.5 was placed in a Clark-type oxygen electrode chamber and stirred gently at 30°C. Whole-cell photosynthetic activity was measured as oxygen evolution supported by 2 mm NaHCO3. Whole-chain photosynthetic electron transport activity was measured as oxygen consumption by adding 1 mm methyl viologen and 1 mm KCN. For measurement of respiratory activity, cultures were concentrated about 5-fold by centrifugation, and oxygen consumption was measured in the presence of 5 mm Glc in the dark. Oxygen evolution and consumption rates were calculated in terms of μmol of oxygen evolved or consumed OD730–1 mL–1 h–1.
Metabolome Analysis
The metabolome analysis was performed using a CE/MS system (Agilent Technologies) with the method described by Takahashi et al. (2008) and Miyagi et al. (2010) with minor modification. Briefly, 50 mL of culture at an OD730 of 0.5 was harvested, and the metabolites were extracted with ice-cold 50% (v/v) methanol containing internal standards (50 μm PIPES and 50 μm MES). After filtration through a 3-kD-cutoff membrane (Amicon Ultra-0.5 3K, Millipore), anionic metabolites (sugar phosphates and organic acids) were separated on a polyethylene glycol-coated capillary (100 cm × 50 µm i.d., DB-WAX, Agilent Technologies) using 20 mm ammonium acetate, pH 8.5, as a running buffer. For cationic metabolites (amino acids), uncoated fused silica capillary (100 cm × 50 µm i.d., silica capillary tube, GL Sciences) was used with 1 m formic acid, pH 1.9, as a running buffer.
Determination of Glycogen Content
Cultures grown photoautotrophically or photomixotrophically to an OD730 of 0.2 to 0.5 were harvested. The glycogen content of each sample was determined by the method described in Suzuki et al. (2010). In short, cell pellet was resuspended in absolute methanol and kept at –20°C overnight. After centrifugation, the dried pellet was resuspended and incubated at 100°C for 40 min and then at 40°C for 1 h with glucoamylase. Glycogen content was determined enzymatically by addition of hexokinase and Glc-6-P dehydrogenase. The amount of Glc moieties derived from glycogen was determined as the increase of optical density at 340 nm.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Growth properties of the wild type and the ΔcyabrB2 mutant under photoautotrophic and photomixotrophic conditions in the presence of ammonium.
Supplemental Figure S2. Complementation experiments by expression of His-cyAbrB2 protein in the ΔcyabrB2 mutant.
Supplemental Figure S3. Changes in cell morphology of the wild type and the ΔcyabrB2 mutant upon a shift to photomixotrophic conditions observed by differential interference contrast microscopy.
Supplemental Figure S4. Principal component analysis of metabolites expressed as nmol g–1 fresh weight in the wild type and the ΔcyabrB2 mutant under photoautotrophic and photomixotrophic conditions.
Supplemental Figure S5. Hierarchical classification of metabolite profiles in the wild type and the ΔcyabrB2 mutant under photoautotrophic and photomixotrophic conditions.
Supplemental Figure S6. Cell morphology of the ΔcyabrB2 mutant after 48 h of incubation with various metabolites under photomixotrophic conditions as observed by differential interference contrast microscopy.
Supplemental Figure S7. Principal component analysis of metabolites expressed as nmol g–1 fresh weight in the ΔcyabrB2 mutant under photomixotrophic conditions with or without 2OG.
Supplemental Figure S8. Hierarchical classification of metabolite profiles in the ΔcyabrB2 mutant under photomixotrophic conditions with or without 2OG.
Supplemental Table S1. Metabolite levels of the wild type and the ΔcyabrB2 mutant before and after 12 h of incubation under photomixotrophic conditions and metabolite levels of the ΔcyabrB2 mutant after 24 h of incubation under photomixotrophic conditions with or without 2OG.
Acknowledgments
We thank Kintake Sonoike for advice on photosynthetic measurements, Tatsuo Omata for helpful discussion, and Akihito Kawahara for critical reading of the manuscript.
Glossary
- 2OG
2-oxoglutarate
- Ci
inorganic carbon
- OD730
optical density at 730 nm
- PEP
phosphoenolpyruvate
- OPP
oxidative pentose phosphate pathway
- TCA
tricarboxylic acid
- CE/MS
capillary electrophoresis mass spectrometry
- GS
Gln synthetase
- GOGAT
Glu synthase
- O2
superoxide
References
- Arnon DI, McSwain BD, Tsujimoto HY, Wada K. (1974) Photochemical activity and components of membrane preparations from blue-green algae. I. Coexistence of two photosystems in relation to chlorophyll a and removal of phycocyanin. Biochim Biophys Acta 357: 231–245 [DOI] [PubMed] [Google Scholar]
- Cooley JW, Howitt CA, Vermaas WF. (2000) Succinate:quinol oxidoreductases in the cyanobacterium synechocystis sp. strain PCC 6803: presence and function in metabolism and electron transport. J Bacteriol 182: 714–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutheil J, Saenkham P, Sakr S, Leplat C, Ortega-Ramos M, Bottin H, Cournac L, Cassier-Chauvat C, Chauvat F. (2012) The AbrB2 autorepressor, expressed from an atypical promoter, represses the hydrogenase operon to regulate hydrogen production in Synechocystis strain PCC6803. J Bacteriol 194: 5423–5433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhut M, Georg J, Klähn S, Sakurai I, Mustila H, Zhang P, Hess WR, Aro EM. (2012) The antisense RNA As1_flv4 in the cyanobacterium Synechocystis sp. PCC 6803 prevents premature expression of the flv4-2 operon upon shift in inorganic carbon supply. J Biol Chem 287: 33153–33162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forchhammer K. (2004) Global carbon/nitrogen control by PII signal transduction in cyanobacteria: from signals to targets. FEMS Microbiol Rev 28: 319–333 [DOI] [PubMed] [Google Scholar]
- Ishii A, Hihara Y. (2008) An AbrB-like transcriptional regulator, Sll0822, is essential for the activation of nitrogen-regulated genes in Synechocystis sp. PCC 6803. Plant Physiol 148: 660–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloft N, Forchhammer K. (2005) Signal transduction protein PII phosphatase PphA is required for light-dependent control of nitrate utilization in Synechocystis sp. strain PCC 6803. J Bacteriol 187: 6683–6690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles VL, Smith CS, Smith CR, Plaxton WC. (2001) Structural and regulatory properties of pyruvate kinase from the cyanobacterium Synechococcus PCC 6301. J Biol Chem 276: 20966–20972 [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Takatani N, Tanigawa M, Omata T. (2005) Posttranslational regulation of nitrate assimilation in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol 187: 498–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson J, Nylander JA, Bergman B. (2011) Genome fluctuations in cyanobacteria reflect evolutionary, developmental and adaptive traits. BMC Evol Biol 11: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent S, Jang J, Janicki A, Zhang CC, Bédu S. (2008) Inactivation of spkD, encoding a Ser/Thr kinase, affects the pool of the TCA cycle metabolites in Synechocystis sp. strain PCC 6803. Microbiology 154: 2161–2167 [DOI] [PubMed] [Google Scholar]
- Lieman-Hurwitz J, Haimovich M, Shalev-Malul G, Ishii A, Hihara Y, Gaathon A, Lebendiker M, Kaplan A. (2009) A cyanobacterial AbrB-like protein affects the apparent photosynthetic affinity for CO2 by modulating low-CO2-induced gene expression. Environ Microbiol 11: 927–936 [DOI] [PubMed] [Google Scholar]
- Miyagi A, Takahashi H, Takahara K, Hirabayashi T, Nishimura Y, Tezuka T, Kawai-Yamada M, Uchimiya H. (2010) Principal component and hierarchical clustering analysis of metabolites in destructive weeds; polygonaceous plants. Metabolomics 6: 146–155 [Google Scholar]
- Muramatsu M, Hihara Y. (2003) Transcriptional regulation of genes encoding subunits of photosystem I during acclimation to high-light conditions in Synechocystis sp. PCC 6803. Planta 216: 446–453 [DOI] [PubMed] [Google Scholar]
- Muro-Pastor AM, Herrero A, Flores E. (2001) Nitrogen-regulated group 2 sigma factor from Synechocystis sp. strain PCC 6803 involved in survival under nitrogen stress. J Bacteriol 183: 1090–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro-Pastor MI, Reyes JC, Florencio FJ. (1996) The NADP+-isocitrate dehydrogenase gene (icd) is nitrogen regulated in cyanobacteria. J Bacteriol 178: 4070–4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro-Pastor MI, Reyes JC, Florencio FJ. (2005) Ammonium assimilation in cyanobacteria. Photosynth Res 83: 135–150 [DOI] [PubMed] [Google Scholar]
- Ohashi Y, Shi W, Takatani N, Aichi M, Maeda S, Watanabe S, Yoshikawa H, Omata T. (2011) Regulation of nitrate assimilation in cyanobacteria. J Exp Bot 62: 1411–1424 [DOI] [PubMed] [Google Scholar]
- Onizuka T, Akiyama H, Endo S, Kanai S, Hirano M, Tanaka S, Miyasaka H. (2002) CO(2) response element and corresponding trans-acting factor of the promoter for ribulose-1,5-bisphosphate carboxylase/oxygenase genes in Synechococcus sp. PCC7002 found by an improved electrophoretic mobility shift assay. Plant Cell Physiol 43: 660–667 [DOI] [PubMed] [Google Scholar]
- Osanai T, Kanesaki Y, Nakano T, Takahashi H, Asayama M, Shirai M, Kanehisa M, Suzuki I, Murata N, Tanaka K. (2005) Positive regulation of sugar catabolic pathways in the cyanobacterium Synechocystis sp. PCC 6803 by the group 2 sigma factor sigE. J Biol Chem 280: 30653–30659 [DOI] [PubMed] [Google Scholar]
- Owttrim GW, Colman B. (1986) Purification and characterization of phosphoenolpyruvate carboxylase from a cyanobacterium. J Bacteriol 168: 207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero MJ, Montesinos ML, Herrero A, Flores E. (2001) Identification of genes encoding amino acid permeases by inactivation of selected ORFs from the Synechocystis genomic sequence. Genome Res 11: 2034–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AJ, London J, Stanier RY. (1967) Biochemical basis of obligate autotrophy in blue-green algae and thiobacilli. J Bacteriol 94: 972–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki E, Ohkawa H, Moriya K, Matsubara T, Nagaike Y, Iwasaki I, Fujiwara S, Tsuzuki M, Nakamura Y. (2010) Carbohydrate metabolism in mutants of the cyanobacterium Synechococcus elongatus PCC 7942 defective in glycogen synthesis. Appl Environ Microbiol 76: 3153–3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Uchimiya H, Hihara Y. (2008) Difference in metabolite levels between photoautotrophic and photomixotrophic cultures of Synechocystis sp. PCC 6803 examined by capillary electrophoresis electrospray ionization mass spectrometry. J Exp Bot 59: 3009–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terauchi K, Ikeuchi M, Ohmori M. (1996) Ferredoxin-dependent glutamate synthase is essential for the photomixotrophic growth of cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol 37: s38 [Google Scholar]
- Yamauchi Y, Kaniya Y, Kaneko Y, Hihara Y. (2011) Physiological roles of the cyAbrB transcriptional regulator pair Sll0822 and Sll0359 in Synechocystis sp. strain PCC 6803. J Bacteriol 193: 3702–3709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Hua Q, Shimizu K. (2002) Metabolic flux analysis in Synechocystis using isotope distribution from 13C-labeled glucose. Metab Eng 4: 202–216 [DOI] [PubMed] [Google Scholar]
- Young JD, Shastri AA, Stephanopoulos G, Morgan JA. (2011) Mapping photoautotrophic metabolism with isotopically nonstationary 13C flux analysis. Metab Eng 13: 656–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Bryant DA. (2011) The tricarboxylic acid cycle in cyanobacteria. Science 334: 1551–1553 [DOI] [PubMed] [Google Scholar]