ASF1A and ASF1B genes are regulated by cell cycle progression and are involved in DNA repair after UV-B irradiation.
Abstract
ANTI-SILENCING FUNCTION1 (ASF1) is a key histone H3/H4 chaperone that participates in a variety of DNA- and chromatin-related processes, including DNA repair, where chromatin assembly and disassembly are of primary relevance. Information concerning the role of ASF1 proteins in the post-ultraviolet (UV) response in higher plants is currently limited. In Arabidopsis (Arabidopsis thaliana), an initial analysis of in vivo localization of ASF1A and ASF1B indicates that both proteins are mainly expressed in proliferative tissues. In silico promoter analysis identified ASF1A and ASF1B as potential targets of Elongation Factor2 (E2F) transcription factors. These observations were experimentally validated, both in vitro, by electrophoretic mobility shift assays, and in vivo, by chromatin immunoprecipitation assays and expression analysis using transgenic plants with altered levels of different E2F transcription factors. These data suggest that ASF1A and ASF1B are regulated during cell cycle progression through E2F transcription factors. In addition, we found that ASF1A and ASF1B are associated with the UV-B-induced DNA damage response in Arabidopsis. Transcript levels of ASF1A and ASF1B were increased following UV-B treatment. Consistent with a potential role in UV-B response, RNA interference-silenced plants of both genes showed increased sensitivity to UV-B compared with wild-type plants. Finally, by coimmunoprecipitation analysis, we found that ASF1 physically interacts with amino-terminal acetylated histones H3 and H4 and with acetyltransferases of the Histone Acetyl Transferase subfamily, which are known to be involved in cell cycle control and DNA repair, among other functions. Together, we provide evidence that ASF1A and ASF1B are regulated by cell cycle progression and are involved in DNA repair after UV-B irradiation.
Plants, because of their sessile condition and their requirement of sunlight for photosynthesis, are inevitably exposed to UV-B radiation (290–315 nm), which causes direct damage to DNA, proteins, lipids, and RNA (Britt, 1996; Jansen et al., 1998; Gerhard et al., 1999; Casati and Walbot, 2004a). Thus, plants have not only developed mechanisms that filter or absorb UV-B to protect them against DNA damage (Mazza et al., 2000; Bieza and Lois, 2001) but also have different DNA repair systems to remove or tolerate DNA lesions (Hays, 2002; Bray and West, 2005; Kimura and Sakaguchi, 2006).
Absorption of UV-B by DNA induces the formation of cyclobutane pyrimidine dimers (CPDs) and, to a lesser extent, pyrimidine (6-4) pyrimidone photoproducts (Friedberg et al., 1995). These lesions disrupt base pairing and block DNA replication and transcription if photoproducts persist, or they result in mutations if photoproducts are bypassed by error-prone DNA polymerases (Britt, 1996). Therefore, accumulation of such lesions must be prevented to maintain genome integrity, plant growth, and seed viability. At the genome level, the accessibility of DNA sequences is determined by the structure of chromatin, which is subjected to epigenetic regulation. The structure of the chromatin can be remodeled in several ways, including nucleosome assembly/disassembly: replacement of canonical histones with histone variants, covalent modifications of histones, such as phosphorylation, acetylation, methylation, ubiquitylation, sumoylation; ATP-dependent reorganization and positioning of DNA histones; and DNA methylation (Verbsky and Richards, 2001; Eberharter and Becker, 2002; Pfluger and Wagner, 2007; Vaillant and Paszkowski, 2007).
The efficient spontaneous assembly of nucleosomes is precluded by the strong electrostatic interactions between DNA and histones. Consequently, proteins known as histone chaperones facilitate the assembly and disassembly of nucleosomes by interacting with the corresponding histones (Park and Luger, 2008; Avvakumov et al., 2011). Histone chaperones are conserved in eukaryotes and are classified as either H3-H4 or H2A-H2B type, according to their activity. In Arabidopsis (Arabidopsis thaliana), the best studied are the H3-H4 chaperones CHROMATIN ASSEMBLY FACTOR1 (CAF1), HISTONE REGULATORY HOMOLOG A (HIRA), and ANTI-SILENCING FUNCTION1 (ASF1) and the H2A-H2B chaperones NUCLEOSOME ASSEMBLY PROTEIN1 (NAP1), NAP1-RELATED PROTEINS, and FACILITATES CHROMATIN TRANSCRIPTION (Zhu et al., 2012).
ASF1 was originally identified by its ability to derepress transcriptional silencing when overexpressed in yeast (Saccharomyces cerevisiae; Le et al., 1997; Singer et al., 1998). In yeast and animals, ASF1 proteins play important roles in chromatin-related processes, such as transcription and DNA replication and repair. They participate both in the replication-dependent and the replication-independent chromatin assembly pathways, as ASF1 copurifies with the replication-specific histone H3.1 and with the transcription-specific HISTONE H3.3 and HIRA, respectively (Tyler et al., 1999; Myung et al., 2003; Adkins et al., 2004; Prado et al., 2004; Ramey et al., 2004; Tagami et al., 2004; Franco et al., 2005; Zhang et al., 2005; Tang et al., 2006). In Arabidopsis, there are two genes encoding ASF1 homologs, AtASF1A and AtASF1B (At1g66740 and At5g38110, respectively; Zhu et al., 2011). Both proteins bind histone H3 and are localized in the cytoplasm and the nucleus. Mutants in either AtASF1A or AtASF1B show no obvious defects, while the double mutant shows inhibition of plant growth and abnormal vegetative and reproductive organ development. In addition, asf1a/asf1b plants exhibit cell number reduction, S-phase delay, and reduced endopolyploidy levels (Zhu et al., 2011). Double mutants also show selective increased expression of CYCB1;1 (a gene involved in the G2/M transition) and genes required for S-phase checkpoint and for DNA damage checkpoint and repair, suggesting that these histone chaperones are involved in cell cycle regulation. However, reports on Arabidopsis ASF1 are still limited. Even more, there is no information on the role of ASF1 in the post-UV response in higher plants.
In this work, we have addressed the cell cycle regulation of ASF1 expression and its potential role in the post-UV-B response in relation to its known function as a histone chaperone. First, we analyzed the in vivo localization of ASF1A and ASF1B, showing that both proteins are mainly expressed in proliferative tissues. We then analyzed their regulation by Elongation Factor2 (E2F) transcription factors and experimentally validated ASF1A and ASF1B as targets of these transcription factors, which have a pivotal role in controlling cell cycle progression. In addition, using transgenic plants with decreased transcript levels of both ASF1A and ASF1B, we demonstrate that ASF1A and ASF1B contribute to the UV-B-induced DNA damage response in Arabidopsis. In fact, ASF1A and ASF1B transcripts increased following a UV-B treatment, and asf1a/asf1b RNA interference (RNAi) transgenic seedlings accumulated more DNA damage after UV-B exposure compared with wild-type plants. Finally, by coimmunoprecipitation analysis, we found that ASF1 interacts with N-terminal acetylated H3 and HAM1/HAM2 histone acetyltransferases. HAM1/HAM2 are related to human Tat-Interacting Protein 60 kD (TIP60), which is involved in cell cycle control, regulation of apoptosis, and DNA repair as well as acting as a coactivator for a wide range of transcription factors (Sapountzi et al., 2006). Together, our data provide evidence that both ASF1A and ASF1B are regulated during cell cycle progression and participate in UV-B-induced DNA damage repair.
RESULTS
ASF1A and ASF1B Are Expressed in Actively Dividing Cells
It was previously demonstrated, by reverse transcription-PCR analysis, that both AtASF1A and AtASF1B genes were ubiquitously expressed in most of the Arabidopsis plant tissues analyzed (Zhu et al., 2011). To study the spatiotemporal expression of ASF1A and ASF1B in more detail, we constructed transgenic plants expressing the GUS gene under the control of the ASF1A (ASF1A-GUS) and ASF1B (ASF1B-GUS) promoters, as described in “Materials and Methods.” At least four independent transgenic lines with comparable GUS activity levels were analyzed.
The expression of ASF1A is high in cotyledons and in the shoot apical region (Fig. 1A). ASF1A is also expressed in the roots of 10-d-old plants (Fig. 1B). In 10-d-old seedlings, ASF1A is restricted to the shoot apical meristem, roots, and proliferating leaves (Fig. 1, C and D), while in mature leaves, ASF1A expression is restricted to the hydatodes at the leaf margin (Fig. 1E). In flowers, ASF1A expression is mainly detected in developing anthers and pistils (Fig. 1G). Siliques also show GUS activity, mainly in the dehiscence region (Fig. 1F). The reporter activity of ASF1B-GUS is similar but weaker than that of ASF1A-GUS (Fig. 1, H–Q). Taken together, our results indicate that both ASF1A and ASF1B are expressed in highly dividing tissues, and their expression seems to be redundant, at least in unstressed conditions, consistent with the phenotype of single asf1 mutants (Zhu et al., 2011).
Figure 1.
Histochemical localization of GUS activity in transgenic plants carrying the ASF1A (A–G) and ASF1B (H–Q) promoters. A, Three-day-old whole seedlings. H, Six-day-old whole seedlings. B and K, Roots from 10-d-old seedlings. C and I, Ten-day-old wild-type whole seedlings. J, Magnification of I. D, Proliferating leaves. E and L, Mature leaves. F and P, Siliques. Q, Magnification of P. G, M, and O, Flowers at different stages of development. N, Magnification of M. [See online article for color version of this figure.]
ASF1A and ASF1B Are Regulated by E2F Transcription Factors
The E2F transcription factors are key components of the cyclin/retinoblastoma/E2F pathway that control cell cycle transitions in multicellular organisms (Gutierrez et al., 2002). In humans, ASF1B is regulated by E2F transcription factors during cell cycle progression (Hayashi et al., 2007). Moreover, in plants, Vandepoele et al. (2005) were able to identify that ASF1A and ASF1B are among the 181 putative E2Fa-DPa target genes. To validate these observations, we investigated the regulation of ASF1A and ASF1B genes by the E2F family.
Arabidopsis contains six functional E2F genes that, according to their structural and functional characteristics, can be divided into two different groups. The first group includes AtE2Fa (At2g36010), AtE2Fb (At5g22220), and AtE2Fc (At1g47870), which possess a conserved DNA-binding site, a heterodimerization domain, and a transactivation domain embracing a retinoblastoma-related (RBR) binding site. These E2F factors associate with a DP protein (AtDPa or AtDPb) to form a functional heterodimer that can specifically recognize E2F cis-elements, transactivate E2F-responsive reporter genes, and be negatively regulated by RBR protein (Mariconti et al., 2002; Shen, 2002). The second group includes AtE2Fd/DEL2 (At5g14960), AtE2Fe/DEL1 (At3g48160), and AtE2Ff/DEL3 (At3g01330), which contain two conserved DNA-binding domains and lack a dimerization domain (Mariconti et al., 2002; Shen, 2002).
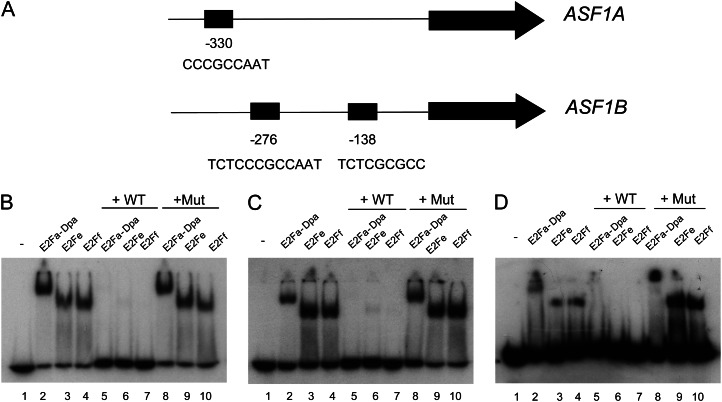
We first searched for putative E2F binding sites 1 kb upstream of the start codon of ASF1A and ASF1B genes (Fig. 2A). Our analysis revealed the presence of one consensus E2F binding site for the ASF1A gene, TAACCGCCC (at −330 bp from the putative ATG), in reverse orientation, and two consensus E2F binding sites for the ASF1B gene, TCTCCCGCCAAT (at −276 bp), containing a double palindromic sequence, and TCTCGCGCC (at −138 bp). These data are consistent with previous reports that identified ASF1A and ASF1B as putative targets of E2Fa-DPa (Vandepoele et al., 2005; Naouar et al., 2009).
Figure 2.
E2F in vitro binding to ASF1A and ASF1B proximal promoters. A, Schemes of the ASF1A and ASF1B proximal promoters showing the positions and sequences of E2F-binding sites. B to D, EMSA analysis of protein-DNA complexes performed with recombinant E2Fa/DPa, E2Fe, and E2Ff proteins and a fragment of the promoter containing the E2F sites as a probe. B, Site at −330 bp from the ATG translation start codon of the ASF1A promoter. C, Site at −138 bp from the ATG translation start codon of the ASF1B promoter. D, Site at −276 bp from the ATG translation start codon of the ASF1B promoter. Lane 1, free probe; lanes 2 to 4, binding of E2Fa/DPa, E2Fe, and E2Ff, respectively, to the promoter; lanes 5 to 7, protein-DNA complexes were competed out with a 100-fold molar excess of unlabeled oligonucleotide containing a consensus E2F site (wild type [WT]); lanes 8 to 10, specific protein-DNA binding was challenged with a 100-fold molar excess of a mutated oligonucleotide (Mut).
To experimentally validate whether the bioinformatically identified E2F sites in the ASF1A and ASF1B promoters mediate E2F binding, we carried out electrophoretic mobility shift assay (EMSA) using labeled double-stranded oligonucleotides corresponding to each of the E2F sites found in the ASF1A (Fig. 2B; Supplemental Table S1) and ASF1B (Fig. 2, C and D; Supplemental Table S1) promoters. Recombinant Arabidopsis E2Fa-DPa, E2Fe/DEL1, and E2Ff/DEL3 proteins bound to all these probes in a specific and E2F site-dependent manner (Fig. 2, B–D). Thus, addition of a competitor oligonucleotide containing a general E2F consensus site (TTTCGCGC; Fig. 2, B–D; Supplemental Table S1) eliminated the specific complex, but the yield of preformed complexes was unaffected when the reaction was performed with a mutated version of the consensus sequence (TTTCGATC; Fig. 2, B–D; Supplemental Table S1). Therefore, E2F members of the two groups are able to bind in vitro to each of the three sites analyzed, suggesting that these transcription factors could regulate the in vivo expression of both genes.
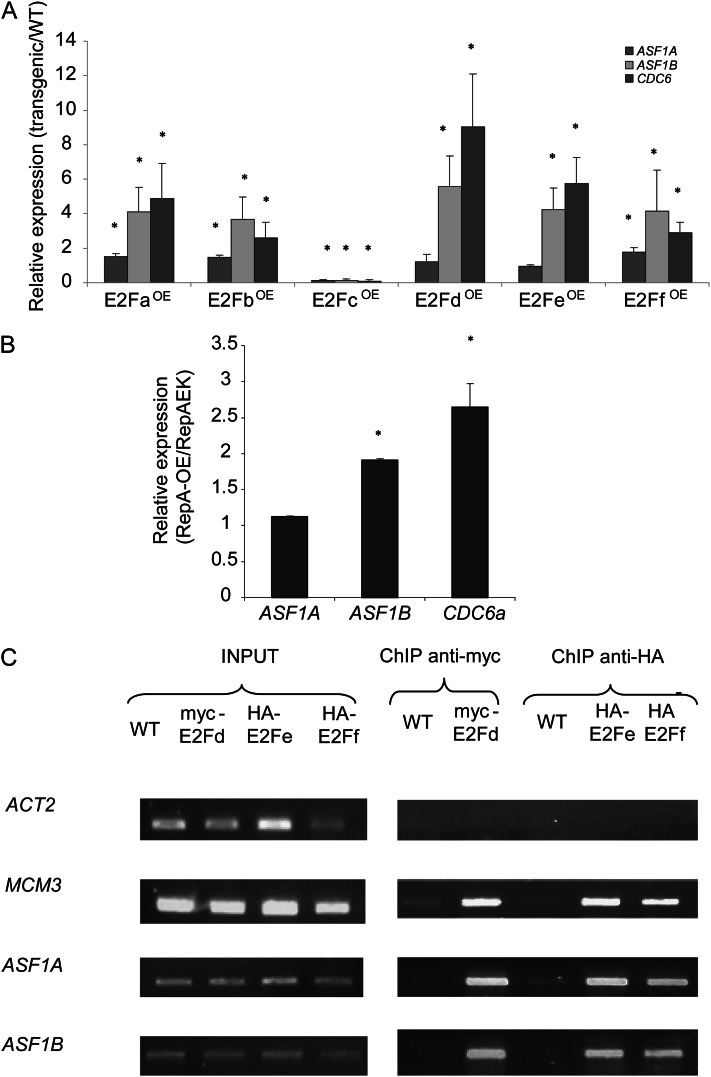
To test this hypothesis, we used a set of transgenic Arabidopsis seedlings with altered E2F expression (Ramirez-Parra and Gutierrez, 2007) and analyzed ASF1A and ASF1B mRNA levels in these plants by quantitative reverse transcription-PCR (qRT-PCR; Supplemental Table S2). CELL DIVISION CYCLE 6a (CDC6a; At2g29680), a well-characterized E2F target gene (Castellano et al., 2001), was used as a positive control. Figure 3A shows increases in both ASF1A and ASF1B transcript levels in plants that overexpress E2Fa, E2Fb, and E2Ff/DEL3. Overexpression of the transcriptional repressor E2Fc contributed to repress the expression of both ASF1A and ASF1B genes. Moreover, mRNA levels for ASF1B are also increased in plants that overexpress the atypical E2Fs: E2Fd/DEL2 and E2Fe/DEL1 (Fig. 3A). It is interesting that the ASF1B levels were always higher than those of ASF1A in all the transgenic plants analyzed (Fig. 3A).
Figure 3.
E2F-regulated expression of ASF1A and ASF1B genes. A, Expression levels of ASF1A, ASF1B, and CDC6 genes determined by qRT-PCR analysis in transgenic plants overexpressing each of the six Arabidopsis E2F transcription factors (E2FOE). Measurements are relative to the amount in wild-type (WT) plants. Asterisks indicate statistical differences applying Student’s t test (P < 0.05). B, Relative expression levels of ASF1A, ASF1B, and CDC6 genes determined by qRT-PCR analysis in transgenic plants overexpressing RepA protein compared with plants overexpressing RepA-E198K protein. C, ChIP assays using anti-myc or anti-HA antibodies with nuclei prepared from wild-type or transgenic plants expressing myc-E2Fd, HA-E2Fe, or HA-E2Ff. The immunoprecipitates and input DNA before immunoprecipitation were analyzed by PCR for the presence of promoter sequences of ASF1A and ASF1B; ACT2, a control gene that is not regulated by E2F factors; and MCM3, a control gene that is regulated by E2F factors. Three PCR experiments were done with each sample.
Finally, ASF1A and ASF1B transcript levels were analyzed in plants overexpressing RepA, a viral protein that increases endogenous E2F activity by inactivating the RBR protein through physical interaction, and in plants overexpressing a mutated version of the RepA protein (RepA E198K), in which RBR interaction is abolished (Desvoyes et al., 2006). Figure 3B shows that ASF1B mRNA levels are higher in the RepA-expressing plants relative to the RepA mutant plants, while ASF1A levels are similar in both plants.
In addition, chromatin immunoprecipitation (ChIP) experiments were performed using commercial antibodies that recognize hemagglutinin (HA) or anti-myc antibodies with transgenic Arabidopsis plants that overexpress HA-E2Fe (E2FeOE), HA-E2Ff (E2FfOE; Ramirez-Parra et al., 2004), or myc-E2Fd (E2FdOE; Ramirez-Parra and Gutierrez, 2007) fusion proteins. We chose to use these plants because (1) they show high induction (Fig. 3A), (2) these E2Fs do not need a DP, and (3) they do not interact with RBR, which may simplify the interpretation of data (Mariconti et al., 2002; Lammens et al., 2008). Arabidopsis wild-type plants were used as a negative control. Genomic immunoprecipitated DNA was screened by PCR for the presence of promoter regions of ASF1A and ASF1B. Primers for the promoter region of ACTIN2 (ACT2; At3g18780), a non-E2F-regulated gene, were used as a negative control, while primers designed to amplify a promoter region of the MCM3 gene (At5g46280), which is regulated by E2F factors (Stevens et al., 2002), were used as a positive control (Supplemental Table S3). After 34 PCR cycles, total input DNA from sonicated nuclei generated positive amplification signals with all the primers used (Fig. 3C). Likewise, promoter fragments of ASF1A and ASF1B and the positive control MCM3 that contains E2F-binding sites were significantly amplified from the anti-HA immunoprecipitates of E2FfOE and E2FeOE plant extracts and from the anti-myc immunoprecipitates of E2FdOE extract (Fig. 3C). However, no or very low amplification was obtained when primers specific for the promoter of ACT2 were used (Fig. 3C). Taken together, these results demonstrate that E2F family members can bind to both ASF1A and ASF1B promoters in vitro and in vivo and regulate the expression of both genes.
To further validate the cell cycle regulation of ASF1A and ASF1B, their expression was investigated in microarray data generated using the Genevestigator tool package. Data were obtained from experiments done using Arabidopsis cultured cells synchronized by Suc starvation for 24 h (Hennig et al., 2003). Samples were collected at the indicated times after release from the Suc block (Supplemental Fig. S1). ASF1A and, in particular, ASF1B transcript levels were compared at each time, together with MCM3 and CYCB1.4 (At2g26760), as markers for up-regulated genes at the G1/S and G2/M transitions, respectively, and with ACT2, which remains constant during the cell cycle. These results demonstrate that both ASF1A and ASF1B are regulated during the cell cycle and show higher expression during S phase (Supplemental Fig. S1). These results are consistent with these genes being targets of E2F transcription factors.
Transgenic Plants with Decreased ASF1A and ASF1B mRNA Levels Show Decreased Rosette Area Compared with Wild-Type Plants
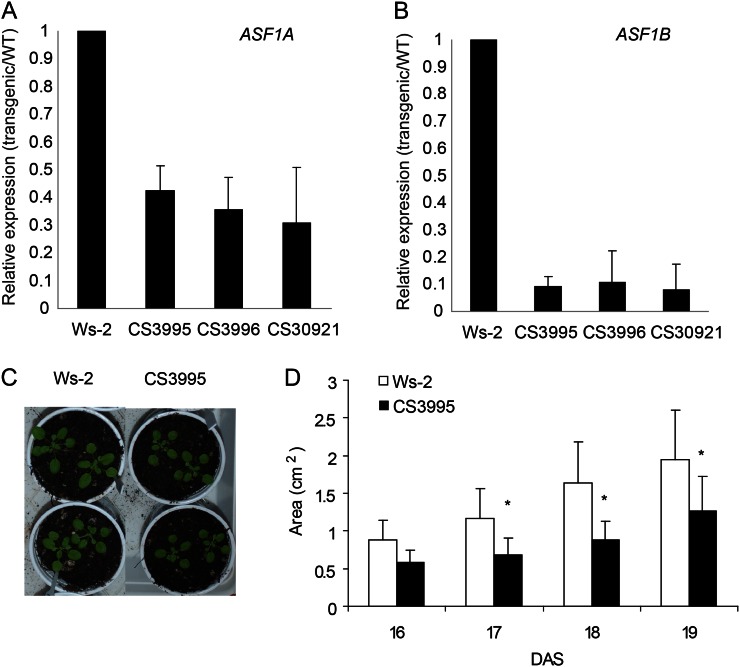
To gain insight into AtASF1A and AtASF1B function, seeds of RNAi plants with decreased levels of both ASF1A and ASF1B (lines CS3995, CS3996, and CS30921) were obtained from the Plant Chromatin Consortium (http://www.chromdb.org). By qRT-PCR, we found that the expression of ASF1A is decreased about 3-fold in three independent transgenic lines (Fig. 4A), while ASF1B is decreased about 10-fold (Fig. 4B). It is interesting that the RNAi construction was made to target ASF1B; however, as the sequences of both genes are highly similar (Zhu et al., 2011), the RNAi construct is able to silence both genes, at least partially. Figure 4, C and D, shows that RNAi transgenic plants show decreased rosette area compared with wild-type plants, indicating probable defects in cell proliferation or cell expansion. Interestingly, Zhu et al. (2011) previously reported that loss of function of either AtASF1A or AtASF1B did not show obvious defects, but the average surface area of leaf blade of the double mutant asf1a/asf1b was about 60% of that of the wild type because of a reduction in the numbers of palisade and pavement cells, S-phase delay/arrest, and reduced polyploidy levels, suggesting that ASF1 proteins are involved in the control of cell cycle progress.
Figure 4.
A and B, Relative transcript levels of Arabidopsis ASF1A (A) and ASF1B (B) measured by qRT-PCR in wild-type (WT) plants (Wassilewskija [Ws-2]) and in asf1a/asf1b RNAi transgenic plants (lines CS3995, CS3996, and CS30921). C and D, Comparison of the rosette area in wild-type and asf1a/asf1b RNAi transgenic plants from 16 to 19 DAS. Asterisks indicate the statistical differences applying Student’s t test (P < 0.05). [See online article for color version of this figure.]
ASF1 Interacts with Acetylated H3 and H4 Histones and with HAM1/HAM2 Histone Acetylase
Histone acetylation is mediated by histone acetyltransferases/deacetylases. In Arabidopsis, there are four different families of histone acetyltransferases and three families of histone deacetylases (Pandey et al., 2002). The histone acetyltransferase families (http://www.chromdb.org) include the p300/CREB-binding protein family (HAC; five genes), the GCN5-related N-terminal acetyltransferase superfamily (HAG; three genes), the TAFII250 family (for TATA-binding protein-associated factors; HAF; two genes), and the MYST (for MOZ, Ybf2/Sas3, Sas2, and TIP60) family (HAM; two genes). Each of these enzymes can acetylate different amino acid residues of the histones, giving them specificity. In humans, the best characterized member of the MYST family is TIP60. TIP60 has important roles during DNA repair, gene transactivation in response to DNA damage, and, more importantly, histone H4 acetylation when DNA is damaged (Squatrito et al., 2006). In addition, TIP60 not only acetylates histones but also nonhistone proteins, such as the ATM kinase, a key regulator of the DNA double-strand break (DSB) repair pathway and checkpoint activation (Sun et al., 2010).
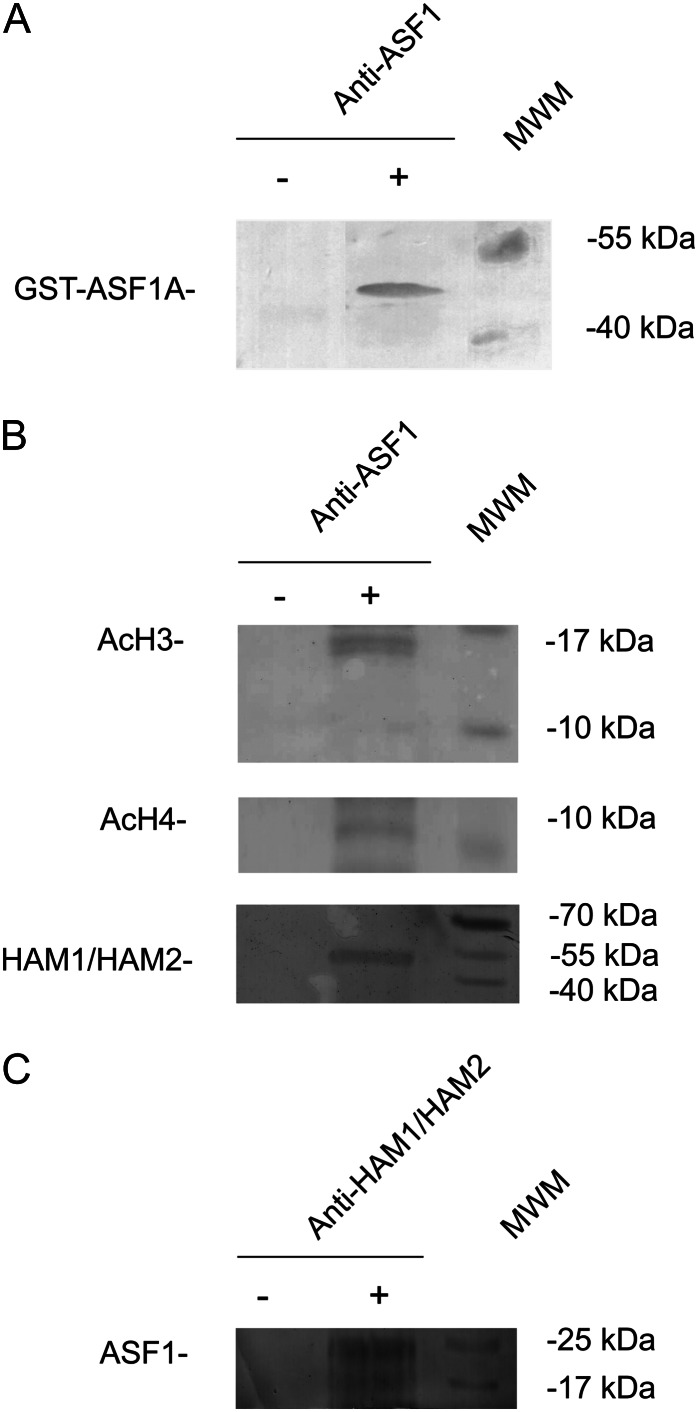
A physical interaction in yeast between Asf1 and SAS2, a member of the MYST subfamily of acetyltransferase (Osada et al., 2001), has been reported previously. To further investigate these observations in plants, we carried out coimmunoprecipitation experiments using commercial antibodies against human ASF1A + ASF1B proteins (ABCAM ab53608). These antibodies recognized a unique band when western blot analysis was performed with Escherichia coli crude protein extracts expressing the recombinant GST-ASF1A fusion protein (Fig. 5A). Figure 5B shows that ASF1 proteins coimmunoprecipitated with N-terminal acetylated forms of H3 and H4 histones. These results validate the proposed function of AtASF1 as a histone-binding protein.
Figure 5.
A, Western-blot analysis using anti-ASF1 antibodies and E. coli protein extracts expressing GST-ASF1A fusion protein (+). As a control, an E. coli protein extract before induction was used (−). B and C, Coimmunoprecipitation experiments using anti-ASF1 (B) or anti-HAM (C) antibodies. As a control, a nuclear protein extract not incubated with any antibody was used (−). Western blots were developed using anti-N-terminal acetylated H3 (AcH3) or H4 (AcH4) or anti-TIP60 (HAM1/HAM2) antibodies. Prestained molecular weight markers (MWM) and their corresponding molecular masses are included at the right side of each gel.
Next, we analyzed the interaction of ASF1 proteins with the Arabidopsis TIP60 homologs HAM1 and HAM2. Figure 5B shows that ASF1 binds in vivo to a protein with the predicted molecular mass of HAM1/HAM2. This interaction was also observed when coimmunoprecipitation was carried out using antibodies against human TIP60 protein (ABCAM ab23886), and immunoblotting was revealed using the anti-ASF1 antibodies (Fig. 5C). Thus, these results confirm the physical interaction between ASF1 and N-terminal acetylated histones H3 and H4 and HAM in plants.
ASF1A and ASF1B Expression Is Induced by UV-B Radiation through E2F Transcription Factors, and Both Proteins Participate in UV-B-Induced DNA Damage Repair
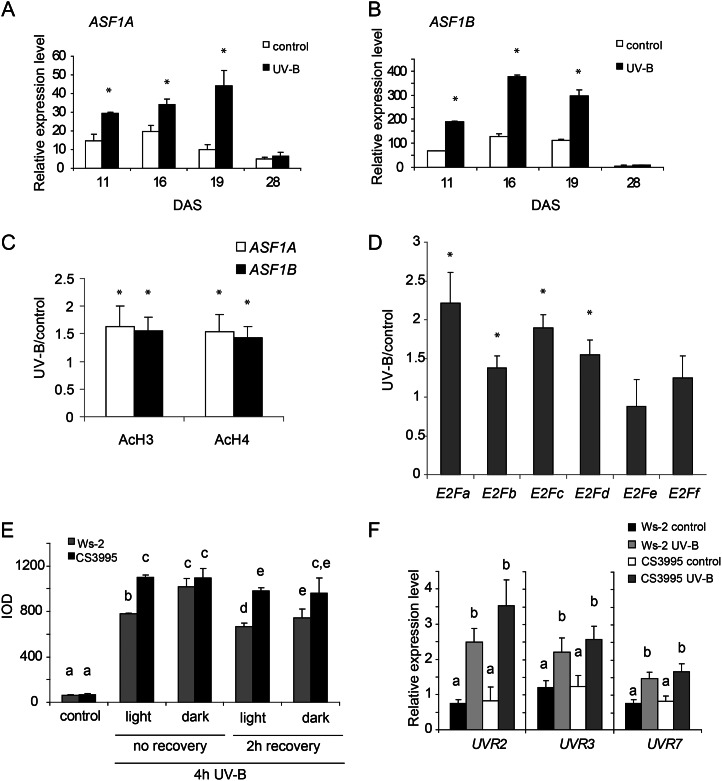
In plants, E2F factors contribute to the transcriptional induction of genes upon DNA damage (Mariconti et al., 2002; Shen, 2002; Ramirez-Parra and Gutierrez, 2007; Lincker et al., 2008). In addition, chromatin remodeling-deficient plants show increased DNA damage by UV-B (Campi et al., 2012). Thus, it is possible that AtASF1 proteins may have a role during DNA damage and repair. To investigate if this is the case, we focused on the potential role of AtASF1 proteins in the UV-B-induced DNA damage response pathway in plants. We first analyzed their expression after UV-B by qRT-PCR analysis in plants exposed under UV-B lamps for 4 h (2 W m−2) in growth chamber conditions. After the treatment, rosettes from plants at 11, 16, 19, and 28 d after sowing (DAS) were collected for RNA extraction. Figure 6, A and B, shows that ASF1A and ASF1B transcript levels significantly increased after UV-B irradiation. Moreover, transgenic plants expressing GUS under the control of the ASF1A and ASF1B promoters also show differences in the intensity of GUS histochemical staining after UV-B treatment (Supplemental Fig. S2).
Figure 6.
UV-B-regulated expression of ASF1A and ASF1B genes. A and B, Relative expression levels of ASF1A (A) and ASF1B (B) were determined by qRT-PCR analysis. Arabidopsis seedlings at 11, 16, 19, and 28 DAS were irradiated with a 4-h UV-B treatment or kept under control conditions without UV-B. Each reaction was normalized using the cycle threshold values corresponding to the CPK3 transcript, which is not regulated by UV-B. Asterisks indicate the statistical differences applying Student’s t test (P < 0.05). C, ChIP assays performed using anti-N-terminal acetylated H3 (AcH3) or H4 (AcH4) antibodies and nuclei prepared from wild-type seedlings after a 4-h UV-B treatment or kept under control conditions without UV-B. Immunoprecipitated and input DNA before immunoprecipitation were analyzed for the presence of ASF1A and ASF1B promoter sequences. Three PCR experiments were carried out with each sample. Asterisks indicate statistical differences applying Student’s t test (P < 0.05). D, UV-B-regulated expression of the E2F genes. Wild-type plants (Wassilewskija [Ws-2]) were irradiated with UV-B light for 4 h or kept under control conditions as indicated in “Materials and Methods.” Expression levels of UV-B-irradiated versus control samples were determined by qRT-PCR analysis. Data show mean values ± sd of at least three independent experiments. Asterisks indicate statistical differences applying Student’s t test (P < 0.05). E, CPD levels in the DNA of wild-type and asf1a/asf1b RNAi transgenic seedlings (CS3995 line) under control conditions without UV-B immediately after or 2 h after a 4-h UV-B treatment. Experiments were done under conditions that allowed photorepair in the light or under dark conditions. CPD levels are indicated as integrated optical density (IOD) values. Results represent averages ± se of three independent biological replicates. F, Relative expression of UVR2, UVR3, and UVR7 transcripts by qRT-PCR. Wild-type and asf1a/asf1b RNAi transgenic (CS3995 line) seedlings were irradiated with UV-B for 4 h or kept under control conditions without UV-B. Data show mean values ± sd of at least three independent experiments. For each transcript analyzed, different letters indicate significant statistical differences (P < 0.05).
To further analyze the presence of acetylated histones H3 and H4 in ASF1A and ASF1B promoters after irradiation with UV-B, we carried out ChIP assays. Previously, we demonstrated that acetylated histones, typical marks of transcriptionally active chromatin, contribute to the transcriptional response to UV-B (Casati et al., 2008, Qüesta et al., 2010). In maize (Zea mays), UV-B-tolerant lines exhibit greater acetylation on N-terminal tails of histones H3 and H4 after irradiation, and these acetylated histones are enriched in the promoter and transcribed regions of UV-B-up-regulated genes. Thus, ChIP analysis was performed using commercially available antibodies specific for acetylated Lys residues in the N-terminal tails of histones H3 and H4. DNA recovered after immunoprecipitation was screened via quantitative PCR (qPCR) for the presence of promoter regions of ASF1A and ASF1B (−533 to +71 and −532 to +80 from the ATG translation start sites, respectively). qRT-PCR was also performed with samples incubated in the absence of antibody to evaluate nonspecific binding. All ChIP samples were normalized to total input DNA from sonicated nuclei for determining the selective recovery of gene segments. Figure 6C shows that promoter regions of both ASF1A and ASF1B were enriched significantly in the fractions immunoprecipitated with anti-acetylated H3 and anti-acetylated H4 from UV-B-irradiated samples Therefore, the increase in H3 and H4 acetylation at the promoter region correlates with the increase in ASF1A and ASF1B transcript abundance.
In parallel, the regulation of different E2F factors by UV-B radiation was analyzed to determine if UV-B induction of ASF1A and ASF1B could be mediated, at least in part, by one or more of these transcription factors. For these experiments, 12-DAS wild-type Arabidopsis seedlings were irradiated with UV-B during 4 h, and mRNA levels of E2F factors were assayed under control conditions and after the different UV-B treatments. Figure 6D shows that E2Fa, E2Fb, E2Fc, and E2Fd are induced after 4 h of UV-B. However, E2Fe and E2Ff are not regulated by this radiation. Therefore, UV-B regulation of ASF1A and ASF1B may be mediated by some or all of the UV-B-regulated E2F transcription factors.
To investigate the hypothesis that ASF1 proteins participate in DNA damage repair by UV-B radiation, we grew Arabidopsis wild-type and RNAi transgenic plants in the growth chamber in the absence of UV-B for 12 DAS. Plants were then exposed for 4 h to UV-B radiation (2 W m−2), both under light conditions that allow photoreactivation (light) or in the absence of white light to analyze dark repair (dark). As a control, different plants were irradiated with the same lamps covered with a polyester plastic that absorbs UV-B (see “Materials and Methods”). Leaf samples from control and treated plants maintained under light and dark conditions were collected immediately or 2 h after the end of the UV-B treatment, DNA was extracted, and the CPD accumulation was compared in the RNAi plants relative to that in wild-type plants using monoclonal antibodies specifically raised against them. A comparison of the CPD accumulation in samples from wild-type and RNAi plants after the different UV-B treatments and in control conditions in the absence of UV-B, as described in “Materials and Methods,” is shown in Figure 6E. In the absence of UV-B, the steady-state levels of CPDs in wild-type and RNAi plants were similar. However, after 4 h of exposure to UV-B radiation, more unrepaired lesions accumulated in the RNAi plants than in wild-type plants when plants were irradiated with UV-B in the presence of white light (Fig. 6E). This difference was not observed when plants were kept under dark conditions. The same results were obtained after 2 h of recovery in the absence of UV-B. Although photoreactivation is evident in both wild-type and RNAi plants after 2 h of recovery in the light, the RNAi plants still show more CPDs than the wild-type plants (Fig. 6E). On the other hand, CPD accumulation under dark conditions was similar in both plants. Therefore, this result demonstrates that the photorepair of UV-B-induced DNA lesions is less efficient in plants that are deficient in the expression of both ASF1A and ASF1B, while dark repair is not affected in the RNAi plants.
To discard the possibility that decreased expression of ASF1A and ASF1B genes may be affecting the expression of DNA repair enzymes of other repair systems, UVR2 (encoding a CPD photolyase; At1g12370), UVR3 (encoding a 6-4 photoproduct photolyase; At3g15620), and UVR7 (encoding ERCC1, a DNA excision repair protein of the nucleotide excision repair system; At3g05210) transcript levels were analyzed by qRT-PCR in wild-type and RNAi plants. Similar levels of both transcripts were measured in wild-type and transgenic plants, both under control conditions and after the 4-h UV-B treatment (Fig. 6F). These results indicate that major CPD removal mechanisms are unaffected in mutant plants. Collectively, these results suggest that ASF1 activities participate in CPD photorepair in Arabidopsis, because in the RNAi plants, chromatin is more accessible to damage accumulation, and not because ASF1 proteins regulate the expression of repair enzymes.
DISCUSSION
The deposition of histones H3/H4 onto DNA to give the tetrasome, and the removal of H3/H4 from DNA, are the first and the last steps in nucleosome assembly and disassembly, respectively. ASF1 has been shown to be an H3/H4 chaperone that functions in both of these processes in yeast and other eukaryotes (Yuan and Zhu, 2012). In yeast, it was demonstrated that Asf1 shields H3/H4 from unfavorable DNA interactions and aids the formation of favorable histone-DNA interactions through the formation of disomes (Donham et al., 2011). In addition, yeast cells lacking Asf1 display an increased frequency of genome instability and spontaneous genome rearrangement (Myung et al., 2003; Prado et al., 2004; Ramey et al., 2004). ASF1 is also required to efficiently complete DNA replication in the presence of DNA-damaging agents or compromised replication machinery (Franco et al., 2005). In Drosophila melanogaster, ASF1 was shown to assemble chromatin onto newly replicated DNA in vitro in synergy with CAF1 (Tyler et al., 1999) and to colocalize with active replication forks (Schulz and Tyler, 2006). Even though information on Arabidopsis ASF1 is limited, the results obtained so far suggest that this histone chaperone is involved in cell cycle regulation. In this species, loss of function of the two Arabidopsis ASF1 genes, AtASF1A and AtASF1B, caused S-phase delay/arrest and an increased level of DNA damage (Zhu et al., 2011). Even more, it was demonstrated that ASF1B is a target of TOUSLED cell cycle-related kinase (Ehsan et al., 2004). In this work, we provide new evidence that the expression of ASF1 proteins is coordinated with cell cycle progression and participates in UV-induced DNA damage repair in Arabidopsis plants. Here, we first constructed transgenic plants expressing the GUS reporter gene directed by ASF1A and ASF1B basal promoters. The results show that both ASF1A and ASF1B proteins are mainly localized in proliferative tissues (Fig. 1) and that their expression is redundant, thus suggesting that both proteins have a role during cell proliferation.
We then investigated the regulation of ASF1 genes by E2F transcription factors, which are key components of the RBR/E2F pathway that controls cell cycle transitions in multicellular organisms, including plants (Gutierrez et al., 2002). Our experiments show that ASF1A and ASF1B are targets of these transcription factors, both in vitro and in vivo. First, we demonstrated by both EMSA and ChIP analysis that E2F factors bind to ASF1A and ASF1B promoters; second, transgenic plants that overexpress different E2F factors show altered levels of both ASF1A and ASF1B transcripts. Both ASF1A and ASF1B have been previously identified as putative targets of the E2F transcription factors (Vandepoele et al., 2005). The ASF1A promoter has one E2F-binding sequence in its basal promoter, while ASF1B has two E2F-binding sites (Fig. 2A). All three of these sites were shown to be bound in vitro by E2Fa-DPa, E2Fe/DEL1, and E2Ff/DEL3 (Fig. 2, B and C). On the contrary, both ASF1A and ASF1B show decreased expression in plants overexpressing E2Fc, similar to the E2F-regulated gene CDC6a (Fig. 3A). Arabidopsis E2Fa transcripts peak shortly before the S phase, while E2Fb mRNAs are higher at the G1/S transition. Both E2Fc and E2Fd increase during the progression into S phase and show maximum expression after the passage into G2, while E2Fe and E2Ff are expressed at the G1/S and S/G2 transitions (Mariconti et al., 2002). Therefore, our results suggest that E2F transcription factors regulate the expression of ASF1A and ASF1B genes through cell cycle progression.
We have confirmed that ASF1 interacts with histone H3, as reported previously (Fig. 4; Zhu et al., 2011). In addition, we here demonstrate that ASF1 proteins bind to the acetylated form of this histone and also with the N-terminal acetylated histone H4. In yeast, it was reported that Asf1 is required for the acetylation of Lys-9 and Lys-56 (K56), and newly synthesized H3 K56 modification is predicted to contribute to chromatin assembly (Masumoto et al., 2005; Xu et al., 2005). In addition, Adkins et al. (2007) previously proposed that Asf1 presents the newly synthesized H3 K56 for acetylation by the histone acetyltransferase Gcn5 prior to chromatin assembly.
On the other hand, in this species, Asf1 functions with a Clr6 histone deacetylase complex to silence heterochromatic repeats by promoting histone deacetylation (Yamane et al., 2011). Therefore, yeast Asf1 has a role both in histone acetylation and deacetylation. Thus, it is possible that this role may also be conserved in plants. We also found that ASF1 binds to Arabidopsis HAM1/HAM2 acetyltransferases in vivo (Fig. 5). In yeast, it was also demonstrated that Asf1 interacts with a SAS complex, which is a member of the MYST acetyltransferase family (Osada et al., 2001). In this species, the SAS complex promotes silencing at telomeres, providing evidence for an important role of the acetyltransferase activity of the SAS complex in silencing. Even more, yeast asf1 mutants show silencing defects similar to mutants in the SAS complex (Osada et al., 2001). Thus, Asf1-dependent chromatin assembly may mediate the silencing role of the SAS complex. On the other hand, TIP60 interacts in various eukaryotes with multiple protein partners besides histones and promotes their acetylation. In this way, TIP60 is a highly connecting protein, controlling the acetylation of a wide range of cellular proteins that are required to maintain cell viability (for review, see Sun et al., 2010). One substrate of TIP60 is the ATM protein kinase, which is a key regulator of the DSB repair pathway. Following DSB production, inactive ATM-Tip60 complex is recruited to the DSB by the mre11-rad50-nbs1 complex. Tip60 chromodomain then interacts with histone H3 trimethylated on Lys-9, activating Tip60 acetyltransferase activity and stimulating the subsequent acetylation and activation of ATM kinase activity. Active ATM kinase phosphorylates proteins involved in both checkpoint activation and DNA repair. Thus, chromatin structure regulates DNA damage signaling and histone modifications coordinate DNA repair (Sun et al., 2010). In accordance with these data, plant ham1 and ham2 mutants show increased DNA damage after UV-B irradiation (Campi et al., 2012). Therefore, the interaction between ASF1 and HAM proteins may be crucial to regulate cell cycle progression after a genotoxic stress.
Both ASF1A and ASF1B transcripts are increased following UV-B treatment, and this UV-B regulation may be, at least in part, mediated by some E2F factors, as E2Fa, E2Fb, E2Fc, and E2Fd mRNAs are rapidly increased by our UV-B treatment (Fig. 6D). In this respect, transgenic plants with decreased transcript levels of both ASF1A and ASF1B accumulate more DNA damage after UV-B exposure compared with wild-type plants at the seedling stage (Fig. 6D). Our results also demonstrate that the photorepair of UV-B-induced DNA lesions is less efficient in plants that are deficient in the expression of both ASF1A and ASF1B, while dark repair is not affected in the RNAi plants. Moreover, we here demonstrate that major CPD removal mechanisms are unaffected in mutant plants, suggesting that ASF1 activities participate in CPD photorepair in Arabidopsis, because in the RNAi plants, chromatin is more accessible to damage accumulation, and not because ASF1 proteins regulate the expression of repair enzymes. In yeast, Asf1 and the checkpoint kinase Rad53 are found in a complex in budding yeast cells in the absence of genotoxic stress (Jiao et al., 2012). Upon replication stress caused by hydroxyurea, the Asf1-Rad53 complex dissociates, suggesting that the complex is regulated by genotoxic stress conditions (Jiao et al., 2012). In addition, Asf1 is important for the transcriptional derepression of two DNA damage response genes during the S phase in response to hydroxyurea (Minard et al., 2011). The contribution of Asf1 to DNA damage response gene derepression depends on its ability to stimulate H3 K56 acetylation by the Lys acetyltransferase Rtt109 (Minard et al., 2011). In addition, as mentioned earlier, in Arabidopsis, ASF1B is phosphorylated by the TOUSLED protein kinase (Ehsan et al., 2004). Arabidopsis tls mutants are highly sensitive to genotoxic stress, including UV-B radiation (Wang et al., 2007). Even more, in humans, Sen and De Benedetti (2006) provided evidence that TLK1B protects against UV radiation via Asf1-mediated chromatin assembly at the sites of UV damage. Therefore, the participation of ASF1 proteins in DNA damage responses seems to be a mechanism to prevent cell cycle progression when damaged DNA accumulates, and this participation is probably regulated by the action of UV-B-regulated E2F transcription factors. In this way, organisms may adapt to environmentally harsh conditions by cell cycle reprogramming to ensure optimal growth.
In conclusion, in this work, we provide new evidence of the regulation of AtASF1A and AtASF1B by E2F factors and DNA damage. Our analysis shows that ASF1A and ASF1B are targets of the E2F transcription factors. In addition, we demonstrate that both ASF1A and ASF1B transcripts are present in proliferative tissues and are increased by UV-B irradiation in this species. We also observed that UV-B induced the accumulation of CPDs in asf1a/asf1b RNAi transgenic plants relative to wild-type plants. Finally, we found physical interaction between ASF1, N-terminal acetylated histones H3 and H4, and HAM acetyltransferases, proteins known to be involved in cell cycle control and DNA repair, among other functions. Together, our data provide evidence that both ASF1A and ASF1B proteins are regulated during cell cycle progression and participate in UV-induced DNA damage response.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana Columbia ecotype) seeds were sown on soil and placed at 4°C in the dark. After 3 d, pots were transferred to a greenhouse and plants were grown at 22°C under a 16-h/8-h light/dark regime. For in vitro growth of plants, seeds were sterilized and incubated at 4°C for 72 h before plating on Murashige and Skoog salt medium supplemented with 1.5% (w/v) Suc and 0.8% (w/v) agar.
RNAi lines (CS3995, CS3996, and CS30921) were obtained from the Arabidopsis Biological Resource Center. These lines confer silencing to the ASF1B target gene. Specific reduction of target mRNA was measured by qRT-PCR. Arabidopsis ecotype Wassilewskija was the background used for assays using RNAi lines. We also used plants expressing the geminivirus RepA, either the wild type or the E198K point mutant that abolishes interaction with RBR (Desvoyes et al., 2006), and plants with altered levels of different E2Fs (Ramirez-Parra and Gutierrez, 2007).
For expression analysis, a 780-bp region containing the AtASF1B promoter or a 655-bp region containing the AtASF1A promoter was amplified by PCR (using primers shown in Supplemental Table S4) and fused to the GUS coding sequence in pBI101.1 vectors (Jefferson et al., 1987). These fusion constructs (pAT-ASF1A:GUS and pAT-ASF1B:GUS) were transformed in Arabidopsis (Columbia ecotype) plants using Agrobacterium tumefaciens C58CRifR by the floral dip method (Clough and Bent, 1998). Transformed seedlings (T0 generation) were selected on Murashige and Skoog agar plates containing 50 mg mL−1 kanamycin and transferred to soil. T2 homozygous plants were selected for further analysis. Histochemical detection of GUS activity was carried out using 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (Jefferson et al., 1987).
EMSAs
DNA-binding assays were performed as described previously (Ramirez-Parra and Gutierrez, 2000). Briefly, reactions (20 µL) contained 20 mm HEPES, pH 7.9, 12% (v/v) glycerol, 50 mm KCl, 1 mm dithiothreitol, 1 mm EDTA, 1 mm MgCl2, double-stranded oligonucleotides containing E2F sites as probes (Supplemental Table S1), 1 μg of salmon sperm DNA as nonspecific competitor, and 200 ng of E2F proteins, as indicated. Proteins were obtained as described previously (Ramirez-Parra and Gutierrez, 2000). For competition experiments, 100-fold annealed unlabeled probe or unlabeled mutated probe (Supplemental Table S1) was included in the reactions. After incubation at 4°C for 20 min, DNA-protein complexes were loaded onto 4% native polyacrylamide gels and electrophoresed in 0.5× Tris-borate/EDTA buffer. Labeled DNA-protein complexes were visualized by autoradiography.
qRT-PCR
Total RNA was isolated from about 100 mg of tissue using the TRIzol reagent (Invitrogen) as described by the manufacturer’s protocol. The RNA was incubated with RNase-free DNase I (1 unit mL−1) following the protocol provided by the manufacturer to remove possible genomic DNA. Then, RNA was reverse transcribed into first-strand complementary DNA using SuperScript II reverse transcriptase (Invitrogen) and oligo(dT) as a primer. The resultant complementary DNA was used as a template for qPCR amplification in a MiniOPTICON2 apparatus (Bio-Rad), using the intercalation dye SYBR Green I (Invitrogen) as a fluorescent reporter and Platinum Taq DNA Polymerase (Invitrogen). Primers for each of the genes under study were designed using the PRIMER3 software (Rozen and Skaletsky, 2000) in order to amplify unique 150- to 250-bp products (Supplemental Table S2). Amplification conditions were carried out under the following conditions: 2 min of denaturation at 94°C; 40 to 45 cycles at 94°C for 15 s, 57°C for 20 s, and 72°C for 20 s; followed by a 10-min extension at 72°C. Three replicates were performed for each sample. Melting curves for each PCR were determined by measuring the decrease of fluorescence with increasing temperature (from 65°C to 98°C). PCR products were run on a 2% (w/v) agarose gel to confirm the size of the amplification products and to verify the presence of a unique PCR product. Gene expression was normalized to Arabidopsis ACT8 (At1g49240) or Calcium-Dependent Protein Kinase3 (CPK3; At4g23650). The expression of CPK3 has been reported previously to remain unchanged by UV-B (Casati and Walbot, 2004b; Ulm et al., 2004).
UV-B Irradiation Treatments
For analysis of gene expression, DNA damage, and ChIP experiments, Arabidopsis plants were exposed for 4 h to UV-B radiation from UV-B bulbs (Bio-Rad) in a growth chamber. UV-B lamps were covered with cellulose acetate filters (100-µm extra clear cellulose acetate plastic; Tap Plastics) and placed 30 cm above the plants in order to only exclude UV-C but not remove UV-B and UV-A radiation from the spectrum. The UV intensities measured with a UV-B/UV-A radiometer (UV203 AB radiometer; Macam Photometrics) were 2 and 0.65 W m−2 for UV-B and UV-A, respectively. Control plants (without supplemental UV-B radiation) were exposed for the same period of time to light sources described above but covered with polyester filters (100-µm clear polyester plastic; Tap Plastics). This polyester filter absorbs both UV-B (0.04 W m−2) and wavelengths less than 280 nm (UV-A radiation intensity was 0.4 W m−2). Immediately after irradiation, samples from at least three independent biological replicates were collected, frozen in liquid nitrogen, and stored at –70°C.
DNA Damage Analysis
CPD accumulation was measured as described previously (Lario et al., 2011). For the assay, leaf samples of 12 DAS were used. Samples were collected from control and UV-B-treated plants. UV-B treatments were performed both under light and dark conditions; plants irradiated under dark conditions were allowed to recover for 2 h under light or dark conditions. After the different treatments, plant samples (0.1 g) were collected, immediately immersed in liquid nitrogen, and stored at −70°C. DNA was extracted by a modified cetyl-trimetyl-ammonium bromide method, denatured in 0.3 m NaOH for 10 min, and 6-fold dot blotted onto a nylon membrane (Perkin-Elmer Life Sciences). The membrane was incubated for 2 h at 80°C and then blocked in Tris-buffered saline (TBS; 20 mm Tris-HCl, pH 7.6, 137 mm NaCl) containing 5% (w/v) dried milk for 1 h at room temperature. The blot was then washed with TBS and incubated with TDM-2 (1:2,000 in TBS) overnight at 4°C with agitation. Monoclonal antibodies specific to CPDs (TDM-2) were from Cosmo BioCo. Unbound antibody was washed away, and secondary antibody (Bio-Rad) conjugated to alkaline phosphatase (1:3,000) in TBS was added. The blot was then washed several times and subsequently developed with 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium. Quantification was achieved by densitometry of the dot blot using ImageQuant software version 5.2. DNA concentration was fluorometrically determined using the Qubit double-stranded DNA assay kit (Invitrogen) and checked on a 1% (w/v) agarose gel stained with SYBR Safe after quantification.
ChIP Assays
ChIP assays and data analysis were carried out basically as described previously (Ramirez-Parra et al., 2004). Briefly, whole 12-DAS wild-type or E2Fd (E2FdOE), E2Fe (E2FeOE), and E2Ff (E2FfOE) overexpressing plants were treated with 1% (v/v) formaldehyde under vacuum. Cross-linking reaction was then stopped with 0.125 m Gly. Nuclei were extracted, lysed in SDS buffer (50 mm Tris-HCl, pH 8, 10 mm EDTA, and 1% [w/v] SDS), and sonicated to shear DNA to an average size of 700 to 1,500 bp. Crude chromatin lysates were precleared with protein G agarose beads (Santa Cruz Biotech), blocked with salmon sperm DNA, and then incubated overnight at 4°C with anti-HA (Roche) or anti-myc 9E0 (Santa Cruz Biotech) antibodies, as indicated. Immunocomplexes were recovered using protein G agarose, extensively washed, and eluted from beads. Cross links were reversed, and samples were treated with proteinase K. DNA was then extracted by the phenol/chloroform method, ethanol precipitated, and resuspended in 50 µL of water. Aliquots (1 µL) were used for PCR. Sequences of the primers used are shown in Supplemental Table S3. To measure the levels of acetylation of histones in the promoter regions of ASF1A and ASF1B upon UV-B treatment, the following antibodies were used: 4 μL of anti-N-terminal acetylated H4 or 4 μL of anti-N-terminal acetylated H3 (06-598 and 06-599, respectively; Upstate Biotechnology). The antibodies used were previously tested for cross reactivity against maize (Zea mays) proteins (Casati et al., 2008; Qüesta et al., 2010). Three biological replicates of ChIP assays were performed from each sample type, and three qPCR experiments were done with each sample.
lsolation of Nuclei and Coimmunoprecipitation
Nuclei were isolated from 12-DAS wild-type Arabidopsis seedlings essentially as described by Gallagher and Ellis (1982). Seedlings (6 g) were ground with 25 mL of buffer 1 (0.4 m Suc, 10 mm Tris-HCl, pH 8, 10 mm MgCl2, 5 mm 2-mercaptoethanol, and 0.1 mm phenylmethylsulfonyl fluoride [PMSF]) in liquid nitrogen. The mixture was filtered through four layers of Miracloth and centrifuged at 5,000g for 20 min at 4°C. The pellet was gently resuspended in 7 mL of buffer 2 (0.25 m Suc, 10 mm Tris-HCl, pH 8, 10 mm MgCl2, 5 mm 2-mercaptoethanol, 0.1 mm PMSF, and 0.15% [v/v] Triton X-100) and then centrifuged at 12,000g for 10 min at 4°C. The pellet was resuspended in 4 mL of buffer 3 (0.44 m Suc, 25 mm Tris-HCl, pH 7.6, 10 mm MgCl2, 10 mm 2-mercaptoethanol, 2.5% [w/v] Ficoll F400, 5% [w/v] dextran T40, and 0.5% [v/v] Triton X-100) and layered onto a Percoll step gradient consisting of 2 mL of 40%, 60%, and 80% (v/v) Percoll. Percoll buffer contained 0.44 m Suc, 25 mm Tris-HCl, pH 7, and 10 mm MgCl2 (buffer 4). After centrifugation at 2,500g for 30 min, nuclei banded at the surface of the Suc pad. Nuclei were then washed twice with buffer 3 and suspended in buffer 5 (0.44 m Suc, 50 mm Tris-HCl, pH 7.0, 5 mm MgCl2, 10 mm 2-mercaptoethanol, and 20% [v/v] glycerol). After centrifugation at 12,000g for 10 min, the pellet was resuspended in lysis buffer (10 mm Tris-HCl, pH 7.5, 1% [v/v] Triton X-100, 500 mm NaCl, 1 mm PMSF, and 10% [v/v] glycerol) and sonicated. Finally, after centrifugation at 22,000g for 15 min, the supernatant was frozen at −20°C. For immunoprecipitation experiments, 250 µL of nuclear extract was combined with 1 μL of anti-ASF1A + ASF1B (ABCAM ab53608) or 2 μL of anti-TIP60 (ABCAM ab23886) antibody and rotated end-over-end at 4°C for 3 h. Protein G-agarose beads (20 μL; Boehringer Mannheim) were added, and the incubation was continued for 1 h. Immunocomplexes were washed four times with 1 mL of ice-cold extraction buffer (100 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% [v/v] Triton X-100, and 1 mm EDTA), resuspended in 50 μL of SDS-PAGE sample buffer (50 mm Tris-HCl, pH 6.8, 2% [w/v] SDS, 10% [v/v] glycerol, 2 mm β-mercaptoethanol, 12.5 mm EDTA, and 0.02% [w/v] bromphenol blue) heated to 70°C for 5 min, and analyzed by SDS-PAGE followed by immunodetection according to Burnette (1981). Commercial IgG fractions were used for the detection of N-terminal acetylated H4, N-terminal acetylated H3 (06-598 and 06-599, respectively; Upstate Biotechnology), and HAM. Bound antibodies were visualized by goat anti-rabbit or anti-mouse IgG conjugated to alkaline phosphatase according to the manufacturer’s instructions (Bio-Rad). The molecular masses of the polypeptides were estimated from a plot of the log of the molecular masses of marker standards (Bio-Rad) versus migration distance.
Rosette Area Quantification
Approximately, 20 seeds per tray were sown, leaving enough space between them to avoid superposition during plant growth. A group of 14-DAS plants were subjected to 4 h of UV-B treatment (2 W m−2), and another group was kept as control plants; after the treatment, all the plants were kept in the growth chamber under a 16-h/8-h light/dark regime until the end of the experiment. Every 3 d, photographs were taken and the rosette area of each plant was measured.
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following loci: At1g66740 (ASF1A), At5g38110 (ASF1B), At2g36010 (E2Fa), At5g22220 (E2Fb), At1g47870 (E2Fc), At5g14960 (E2Fd/DEL2), At3g48160 (E2Fe/DEL1), At3g01330 (E2Ff/DEL3), At3g12280 (RBR), At2g29680 (CDC6a), At4g23650 (CPK3), At3g18780 (ACT2), At5g46280 (MCM3), and At1g49240 (ACT8).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Cell cycle-regulated expression of genes encoding ASF1A and ASF1B.
Supplemental Figure S2. Histochemical localization of GUS activity in transgenic plants carrying the ASF1A and ASF1B promoters in the absence and presence of UV-B.
Supplemental Table S1. Gene-specific oligonucleotides used for EMSA.
Supplemental Table S2. Gene-specific oligonucleotides used for qRT-PCR.
Supplemental Table S3. Gene-specific oligonucleotides used for ChIP.
Supplemental Table S4. Gene-specific oligonucleotides used for GUS assay.
Acknowledgments
We thank the Arabidopsis Biological Resource Center for providing seed stocks.
Glossary
- CPD
cyclobutane pyrimidine dimer
- RNAi
RNA interference
- RBR
retinoblastoma-related
- EMSA
electrophoretic mobility shift assay
- qRT-PCR
quantitative reverse transcription-PCR
- ChIP
chromatin immunoprecipitation
- HA
hemagglutinin
- DSB
double-strand break
- DAS
d after sowing
- TBS
Tris-buffered saline
- PMSF
phenylmethylsulfonyl fluoride
- qPCR
quantitative PCR
References
- Adkins MW, Howar SR, Tyler JK. (2004) Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol Cell 14: 657–666 [DOI] [PubMed] [Google Scholar]
- Adkins MW, Williams SK, Linger J, Tyler JK. (2007) Chromatin disassembly from the PHO5 promoter is essential for the recruitment of the general transcription machinery and coactivators. Mol Cell Biol 27: 6372–6382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avvakumov NA, Nourani A, Côté J. (2011) Histone chaperones: modulators of chromatin marks. Mol Cell 41: 502–514 [DOI] [PubMed] [Google Scholar]
- Bieza K, Lois R. (2001) An Arabidopsis mutant tolerant to lethal ultraviolet-B levels shows constitutively elevated accumulation of flavonoids and other phenolics. Plant Physiol 126: 1105–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray CM, West CE. (2005) DNA repair mechanisms in plants: crucial sensors and effectors for the maintenance of genome integrity. New Phytol 168: 511–528 [DOI] [PubMed] [Google Scholar]
- Britt AB. (1996) DNA damage and repair in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 75–100 [DOI] [PubMed] [Google Scholar]
- Burnette WN. (1981) “Western blotting”: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem 112: 195–203 [DOI] [PubMed] [Google Scholar]
- Campi M, D’Andrea L, Emiliani J, Casati P. (2012) Participation of chromatin-remodeling proteins in the repair of ultraviolet-B-damaged DNA. Plant Physiol 158: 981–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P, Campi M, Chu F, Suzuki N, Maltby D, Guan S, Burlingame AL, Walbot V. (2008) Histone acetylation and chromatin remodeling are required for UV-B-dependent transcriptional activation of regulated genes in maize. Plant Cell 20: 827–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P, Walbot V. (2004a) Crosslinking of ribosomal proteins to RNA in maize ribosomes by UV-B and its effects on translation. Plant Physiol 136: 3319–3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P, Walbot V. (2004b) Rapid transcriptome responses of maize (Zea mays) to UV-B in irradiated and shielded tissues. Genome Biol 5: R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano MM, del Pozo JC, Ramirez-Parra E, Brown S, Gutierrez C. (2001) Expression and stability of Arabidopsis CDC6 are associated with endoreplication. Plant Cell 13: 2671–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Desvoyes B, Ramirez-Parra E, Xie Q, Chua NH, Gutierrez C. (2006) Cell type-specific role of the retinoblastoma/E2F pathway during Arabidopsis leaf development. Plant Physiol 140: 67–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donham DC, II, Scorgie JK, Churchill ME. (2011) The activity of the histone chaperone yeast Asf1 in the assembly and disassembly of histone H3/H4-DNA complexes. Nucleic Acids Res 39: 5449–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberharter A, Becker PB. (2002) Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep 3: 224–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehsan H, Reichheld JP, Durfee T, Roe JL. (2004) TOUSLED kinase activity oscillates during the cell cycle and interacts with chromatin regulators. Plant Physiol 134: 1488–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco AA, Lam WM, Burgers PM, Kaufman PD. (2005) Histone deposition protein Asf1 maintains DNA replisome integrity and interacts with replication factor C. Genes Dev 19: 1365–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W (1995) DNA Damage. ASM Press, Washington, DC [Google Scholar]
- Gallagher TF, Ellis RJ. (1982) Light-stimulated transcription of genes for two chloroplast polypeptides in isolated pea leaf nuclei. EMBO J 1: 1493–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt KE, Wilson MI, Greenberg BM. (1999) Tryptophan photolysis leads to a UVB-induced 66 kDa photoproduct of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) in vitro and in vivo. Photochem Photobiol 70: 49–56 [Google Scholar]
- Gutierrez C, Ramirez-Parra E, Castellano MM, del Pozo JC. (2002) G(1) to S transition: more than a cell cycle engine switch. Curr Opin Plant Biol 5: 480–486 [DOI] [PubMed] [Google Scholar]
- Hayashi R, Goto Y, Tanaka R, Oonogi K, Hisasue M, Yoshida K. (2007) Transcriptional regulation of human chromatin assembly factor ASF1. DNA Cell Biol 26: 91–99 [DOI] [PubMed] [Google Scholar]
- Hays JB. (2002) Arabidopsis thaliana, a versatile model system for study of eukaryotic genome-maintenance functions. DNA Repair (Amst) 1: 579–600 [DOI] [PubMed] [Google Scholar]
- Hennig L, Menges M, Murray JA, Gruissem W. (2003) Arabidopsis transcript profiling on Affymetrix GeneChip arrays. Plant Mol Biol 53: 457–465 [DOI] [PubMed] [Google Scholar]
- Jansen MAK, Gaba V, Greenberg BM. (1998) Higher plants and UV-B radiation: balancing damage, repair and acclimation. Trends Plant Sci 3: 131–135 [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Seeger K, Lautrette A, Gaubert A, Mousson F, Guerois R, Mann C, Ochsenbein F. (2012) Surprising complexity of the Asf1 histone chaperone-Rad53 kinase interaction. Proc Natl Acad Sci USA 109: 2866–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Sakaguchi K. (2006) DNA repair in plants. Chem Rev 106: 753–766 [DOI] [PubMed] [Google Scholar]
- Lammens T, Boudolf V, Kheibarshekan L, Zalmas LP, Gaamouche T, Maes S, Vanstraelen M, Kondorosi E, La Thangue NB, Govaerts W, et al. (2008) Atypical E2F activity restrains APC/CCCS52A2 function obligatory for endocycle onset. Proc Natl Acad Sci USA 105: 14721–14726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lario LD, Ramirez-Parra E, Gutierrez C, Casati P, Spampinato CP. (2011) Regulation of plant MSH2 and MSH6 genes in the UV-B-induced DNA damage response. J Exp Bot 62: 2925–2937 [DOI] [PubMed] [Google Scholar]
- Le S, Davis C, Konopka JB, Sternglanz R. (1997) Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast 13: 1029–1042 [DOI] [PubMed] [Google Scholar]
- Lincker F, Roa H, Lang J, Sanchez-Calderon L, Smetana O, Cognat V, Keller M, Mediouni C, Houlne G, Chaboute ME (2008) Plant E2F Factors in Cell Cycle, Development and DNA Damage Response. Research Signpost, Trivandrum, India [Google Scholar]
- Mariconti L, Pellegrini B, Cantoni R, Stevens R, Bergounioux C, Cella R, Albani D. (2002) The E2F family of transcription factors from Arabidopsis thaliana: novel and conserved components of the retinoblastoma/E2F pathway in plants. J Biol Chem 277: 9911–9919 [DOI] [PubMed] [Google Scholar]
- Masumoto H, Hawke D, Kobayashi R, Verreault A. (2005) A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436: 294–298 [DOI] [PubMed] [Google Scholar]
- Mazza CA, Boccalandro HE, Giordano CV, Battista D, Scopel AL, Ballaré CL. (2000) Functional significance and induction by solar radiation of ultraviolet-absorbing sunscreens in field-grown soybean crops. Plant Physiol 122: 117–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minard LV, Williams JS, Walker AC, Schultz MC. (2011) Transcriptional regulation by Asf1: new mechanistic insights from studies of the DNA damage response to replication stress. J Biol Chem 286: 7082–7092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K, Pennaneach V, Kats ES, Kolodner RD. (2003) Saccharomyces cerevisiae chromatin-assembly factors that act during DNA replication function in the maintenance of genome stability. Proc Natl Acad Sci USA 100: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naouar N, Vandepoele K, Lammens T, Casneuf T, Zeller G, van Hummelen P, Weigel D, Rätsch G, Inzé D, Kuiper M, et al. (2009) Quantitative RNA expression analysis with Affymetrix Tiling 1.0R arrays identifies new E2F target genes. Plant J 57: 184–194 [DOI] [PubMed] [Google Scholar]
- Osada S, Sutton A, Muster N, Brown CE, Yates JR, III, Sternglanz R, Workman JL. (2001) The yeast SAS (something about silencing) protein complex contains a MYST-type putative acetyltransferase and functions with chromatin assembly factor ASF1. Genes Dev 15: 3155–3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R, Müller A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, Bender J, Mount DW, Jorgensen RA. (2002) Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res 30: 5036–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YJ, Luger K. (2008) Histone chaperones in nucleosome eviction and histone exchange. Curr Opin Struct Biol 18: 282–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfluger J, Wagner D. (2007) Histone modifications and dynamic regulation of genome accessibility in plants. Curr Opin Plant Biol 10: 645–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado F, Cortés-Ledesma F, Aguilera A. (2004) The absence of the yeast chromatin assembly factor Asf1 increases genomic instability and sister chromatid exchange. EMBO Rep 5: 497–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qüesta JI, Walbot V, Casati P. (2010) Mutator transposon activation after UV-B involves chromatin remodeling. Epigenetics 5: 352–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramey CJ, Howar S, Adkins M, Linger J, Spicer J, Tyler JK. (2004) Activation of the DNA damage checkpoint in yeast lacking the histone chaperone anti-silencing function 1. Mol Cell Biol 24: 10313–10327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Parra E, Gutierrez C. (2000) Characterization of wheat DP, a heterodimerization partner of the plant E2F transcription factor which stimulates E2F-DNA binding. FEBS Lett 486: 73–78 [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra E, Gutierrez C. (2007) E2F regulates FASCIATA1, a chromatin assembly gene whose loss switches on the endocycle and activates gene expression by changing the epigenetic status. Plant Physiol 144: 105–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Parra E, López-Matas MA, Fründt C, Gutierrez C. (2004) Role of an atypical E2F transcription factor in the control of Arabidopsis cell growth and differentiation. Plant Cell 16: 2350–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386 [DOI] [PubMed] [Google Scholar]
- Sapountzi V, Logan IR, Robson CN. (2006) Cellular functions of TIP60. Int J Biochem Cell Biol 38: 1496–1509 [DOI] [PubMed] [Google Scholar]
- Schulz LL, Tyler JK. (2006) The histone chaperone ASF1 localizes to active DNA replication forks to mediate efficient DNA replication. FASEB J 20: 488–490 [DOI] [PubMed] [Google Scholar]
- Sen SP, De Benedetti A. (2006) TLK1B promotes repair of UV-damaged DNA through chromatin remodeling by Asf1. BMC Mol Biol 7: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W-H. (2002) The plant E2F-Rb pathway and epigenetic control. Trends Plant Sci 7: 505–511 [DOI] [PubMed] [Google Scholar]
- Singer MS, Kahana A, Wolf AJ, Meisinger LL, Peterson SE, Goggin C, Mahowald M, Gottschling DE. (1998) Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 150: 613–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squatrito M, Gorrini C, Amati B. (2006) Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol 16: 433–442 [DOI] [PubMed] [Google Scholar]
- Stevens R, Mariconti L, Rossignol P, Perennes C, Cella R, Bergounioux C. (2002) Two E2F sites in the Arabidopsis MCM3 promoter have different roles in cell cycle activation and meristematic expression. J Biol Chem 277: 32978–32984 [DOI] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Price BD. (2010) Tip60: connecting chromatin to DNA damage signaling. Cell Cycle 9: 930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. (2004) Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116: 51–61 [DOI] [PubMed] [Google Scholar]
- Tang Y, Poustovoitov MV, Zhao K, Garfinkel M, Canutescu A, Dunbrack R, Adams PD, Marmorstein R. (2006) Structure of a human ASF1a-HIRA complex and insights into specificity of histone chaperone complex assembly. Nat Struct Mol Biol 13: 921–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT. (1999) The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402: 555–560 [DOI] [PubMed] [Google Scholar]
- Ulm R, Baumann A, Oravecz A, Máté Z, Adám E, Oakeley EJ, Schäfer E, Nagy F. (2004) Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci USA 101: 1397–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant I, Paszkowski J. (2007) Role of histone and DNA methylation in gene regulation. Curr Opin Plant Biol 10: 528–533 [DOI] [PubMed] [Google Scholar]
- Vandepoele K, Vlieghe K, Florquin K, Hennig L, Beemster GTS, Gruissem W, Van de Peer Y, Inzé D, De Veylder L. (2005) Genome-wide identification of potential plant E2F target genes. Plant Physiol 139: 316–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbsky ML, Richards EJ. (2001) Chromatin remodeling in plants. Curr Opin Plant Biol 4: 494–500 [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu J, Xia R, Wang J, Shen J, Cao R, Hong X, Zhu J-K, Gong Z. (2007) The protein kinase TOUSLED is required for maintenance of transcriptional gene silencing in Arabidopsis. EMBO Rep 8: 77–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Zhang K, Grunstein M. (2005) Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 121: 375–385 [DOI] [PubMed] [Google Scholar]
- Yamane K, Mizuguchi T, Cui B, Zofall M, Noma K, Grewal SIS. (2011) Asf1/HIRA facilitate global histone deacetylation and associate with HP1 to promote nucleosome occupancy at heterochromatic loci. Mol Cell 41: 56–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan G, Zhu B. (2012) Histone variants and epigenetic inheritance. Biochim Biophys Acta 1819: 222–229 [DOI] [PubMed] [Google Scholar]
- Zhang R, Poustovoitov MV, Ye X, Santos HA, Chen W, Daganzo SM, Erzberger JP, Serebriiskii IG, Canutescu AA, Dunbrack RL, et al. (2005) Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev Cell 8: 19–30 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Dong A, Shen W-H. (2012) Histone variants and chromatin assembly in plant abiotic stress responses. Biochim Biophys Acta 1819: 343–348 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Weng M, Yang Y, Zhang C, Li Z, Shen WH, Dong A. (2011) Arabidopsis homologues of the histone chaperone ASF1 are crucial for chromatin replication and cell proliferation in plant development. Plant J 66: 443–455 [DOI] [PubMed] [Google Scholar]