Abstract
Background
Formation of the epicardium requires several cellular processes including migration, transformation, invasion and differentiation in order to give rise to fibroblast, smooth muscle, coronary endothelial and myocyte cell lineages within the developing myocardium. Snai1 is a zinc finger transcription factor that plays an important role in regulating cell survival and fate during embryonic development and under pathological conditions. However, its role in avian epicardial development has not been examined.
Results
Here we show that Snai1 is highly expressed in epicardial cells from as early as the proepicardial cell stage and its expression is maintained as proepicardial cells migrate and spread over the surface of the myocardium and undergo epicardial-to-mesenchymal transformation in the generation of epicardial-derived cells. Using multiple in vitro assays we show that Snai1 overexpression in chick explants enhances proepicardial cell migration at Hamburger Hamilton Stage (HH St.) 16, and epicardial-to-mesenchymal transformation, cell migration and invasion at HH St. 24. Further, we demonstrate that Snai1-mediated cell migration requires matrix metalloproteinase activity, and MMP15 is sufficient for this process.
Conclusion
Together our data provides new insights into the multiple roles that Snai1 has in regulating avian epicardial development.
Keywords: Snai1, epicardium, chicken embryo
Introduction
The mammalian four-chamber heart is composed of multiple cell lineages with distinct developmental origins. In addition to the primary and secondary heart fields, an additional population of epicardial-derived cells contributes to cardiac myocytes and non-muscle cell types (Schlueter and Brand, 2012). The epicardium is an epithelial cell layer that covers the outermost layer of the heart and development begins with formation of the proepicardium (PE). This extracardiac structure is composed of a cluster of mesothelial cells that overlies the septum transversum in the mouse and sinus venosus in the chick. At around Hamburger Hamilton (HH) stage 18–19 in the chicken and embryonic day (E) 9.5 in the mouse, the proepicardial cells attach to the myocardial surface and migrate over the surface of the developing heart (Rodgers et al., 2008; Ishii et al., 2010) Following migration, a subset of these cells undergo epicardial-to-mesenchymal transformation (EpMT) and invade the underlying subepicardial space and myocardium as epicardially-derived cells (EPDCs). Subsequently EPDCs differentiate into fibroblast and smooth muscle cell lineages and additionally contribute to coronary endothelial cells and cardiac myocytes (Dettman et al., 1998; Gittenberger-de Groot et al., 1998; Cai et al., 2008; Zhou et al., 2008; Smart et al., 2011; Katz et al., 2012). Previous studies have demonstrated that alterations in PE formation, cell migration, EpMT and EPDC differentiation affect myocardial maturation (Kwee et al., 1995; Yang et al., 1995; Gittenberger-de Groot et al., 2000; Lavine et al., 2006; Lin et al., 2010; Martinez-Estrada et al., 2010; Combs et al., 2011; Smith et al., 2011; von Gise et al., 2011; Braitsch et al., 2012), however the molecular mechanisms regulating these processes are not fully understood.
The zinc-finger transcriptional factor Snai1 plays important roles during cardiogenesis (Timmerman et al., 2004; Lomeli et al., 2009; Schlueter and Brand, 2009; Martinez-Estrada et al., 2010; Bax et al., 2011; Chen et al., 2012) and we have previously demonstrated its requirement for endothelial-to-mesenchymal transformation (EMT) and cell motility during endocardial cushion formation (Tao et al., 2011). In addition to heart valves, Snai1 has also been implicated in epicardial development. During early stages Snai1 signaling is required for asymmetric development of the proepicardium on the right side of the chick embryo (Schlueter and Brand, 2009). While later, Snai1 is highly expressed in murine epicardial cells and EPDCs (Casanova et al., 2012), however its function in epicardial cells is not fully understood. A study by Martinez-Estrada et al., shows that Snai1 is a direct target of Wilms’ Tumor 1 (Wt1), a key regulator of epicardial development, and Snai1 is sufficient to rescue EpMT defects associated with Wt1-null phenotypes (Martinez-Estrada et al., 2010). However, a more recent report describes normal cardiovascular development in mice with epicardial-specific deletion of Snai1 (Casanova et al., 2012). While these controversial studies in mice have provided insights into Snai1 function in the mouse, studies focused on epicardial development in the chick are limited.
In this study, we determined the role of Snai1 in avian epicardial development using established in vitro systems. We show that Snai1 is sufficient to enhance PE cell migration in Hamburger Hamilton Stage (HH St.) 16 explants and induce EpMT in epicardial cells derived from HH St. 24 chicks. In addition, we demonstrate that Snai1 increases invasion of cells from the outermost layer of the heart into the underlying myocardium at HH St. 24, and this process requires matrix metalloproteinase (MMP) activity. More specifically, we report that overexpression of MMP15 a known downstream target of Snai1 (Tao et al., 2011), is sufficient to recapitulate increased cell invasion phenotypes observed by Snai1 overexpression. These results suggest that Snai1 plays a role during multiple steps of avian epicardial development.
Results
Snai1 is expressed throughout epicardial development of the chick
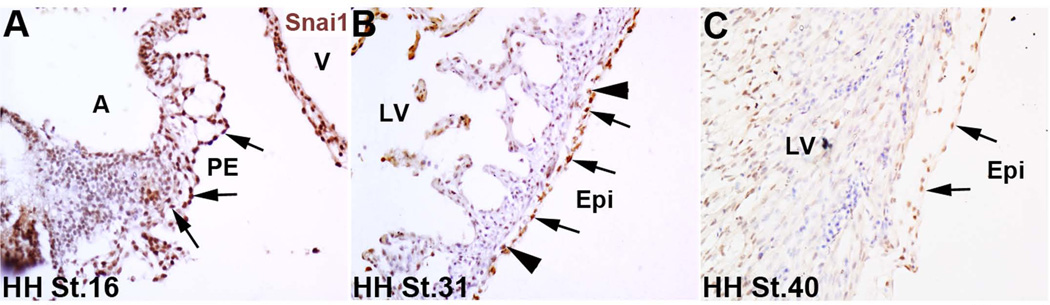
A previous study has described the role of Snai1 during early stages of proepicardial formation in the chick (Schlueter and Brand, 2009), however its expression pattern has not been described. To examine this, immunohistochemistry was performed. At HH St. 16, Snai1 is highly expressed in the majority of mesothelial cells within the proepicardium (PE) (Figure 1A). Snai1 is maintained during stages of epicardial cell migration and high levels of expression are observed throughout the epicardium, as well as in cells within the subepicardial space at HH St. 31 (Figure 1B). By HH St. 40 (embryonic day 14), Snai1 expression has decreased but remains detectable in the maturing epicardium (Figure 1C). These expression studies demonstrate that similar to the mouse (Casanova et al., 2012), Snai1 is highly expressed in the developing epicardium of the chick.
Figure 1. Snai1 is highly expressed during avian epicardial development.
Immunohistochemistry was used to detect Snai1 expression in the proepicardium (PE) (arrows) at HH St. 16 (A), and in the epicardial cell layer (Epi) covering the myocardium (arrows) in addition to cells within the sub-epicardial space (arrowheads) at HH St. 31 (B). (C) By HH St. 40, Snai1 expression has decreased but levels are detectable in the epicardium. A, atria; V, ventricle; LV, left ventricle.
Snai1 is sufficient to enhance avian PE cell migration in vitro
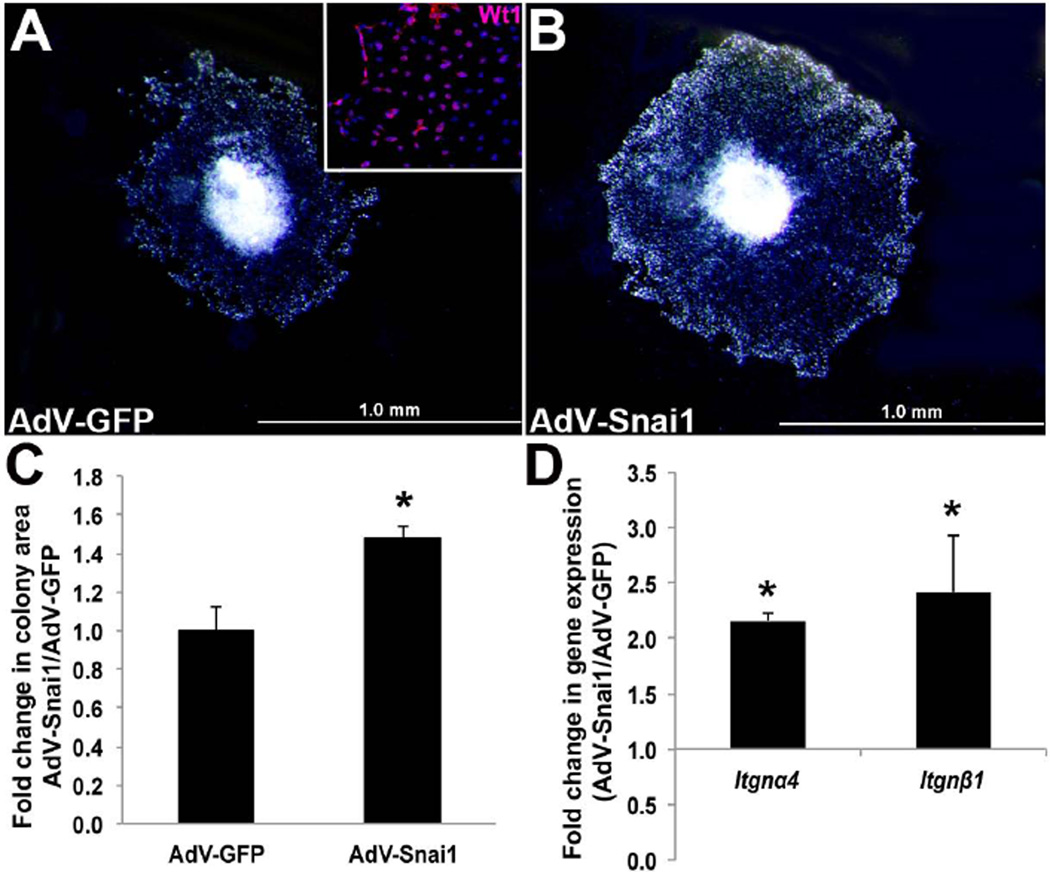
Our lab has previously shown that Snai1 is required for migration of mesenchyme cells during stages of endocardial cushion formation (Tao et al., 2011). As migration is also essential for proepicardial cell outgrowth and ‘spreading’ over the myocardium (Kwee et al., 1995; Yang et al., 1995), we tested the hypothesis that Snai1 plays a similar role in this process. To do this, HH St. 16 PE explants were cultured and migrating cells were infected with adenovirus (AdV) expressing full-length GFP-tagged mouse Snai1 (AdV-Snai1) (Tao et al., 2011) or AdV-GFP that served as a control. Wt1 immunostaining was performed to confirm the migration of proepicardial cells from the PE explants over the culture plate (inset, Figure 2A). As indicated in Figure 1C, AdV-Snai1 significantly increased the colony area covered by migrating proepicardial cells compared to AdV-GFP. Cell proliferation was not significantly altered in AdV-Snai1 infected explants as indicated by insignificant changes in Cdk1, Ccne1, pCNA and Ccnd1 expression (data not shown). Previous studies have shown that PE migration over the myocardial surface requires integrins (Kwee et al., 1995; Yang et al., 1995; Pae et al., 2008), and therefore fold changes in Integrin-α4 (Itgnα4) and -β1 (Itgnβ1) expression were examined. In AdV-Snai1 treated explants, both Itgnα4 and Itgnβ1 were significantly increased (Figure 2D), consistent with increased colony area (Figure 2C). These studies suggest that Snai1 plays a positive role in enhancing PE cell migration in the developing avian heart.
Figure 2. Snai1 enhances avian proepicardial cell migration in vitro.
The colony area of migrating, Wt1-positive (inset) epicardial cells was determined in HH St. 16 proepicardium (PE) explants infected with adenovirus to overexpress full-length Snai1 (AdV-Snai1) (B) or AdV-GFP (A) for 24 hours. (C) Fold change in colony area of AdV-Snai1 infected PE explants over AdV-GFP controls (n=5). (D) qPCR analysis to show fold changes in Integrin-α4 (Itgnα4) and -β1 (Itgnβ1) gene expression in AdV-Snai1 infected PE explants compared to AdV-GFP (n=4). * =p<0.05.
Snai1 overexpression induces EpMT in avian epicardial cells in vitro
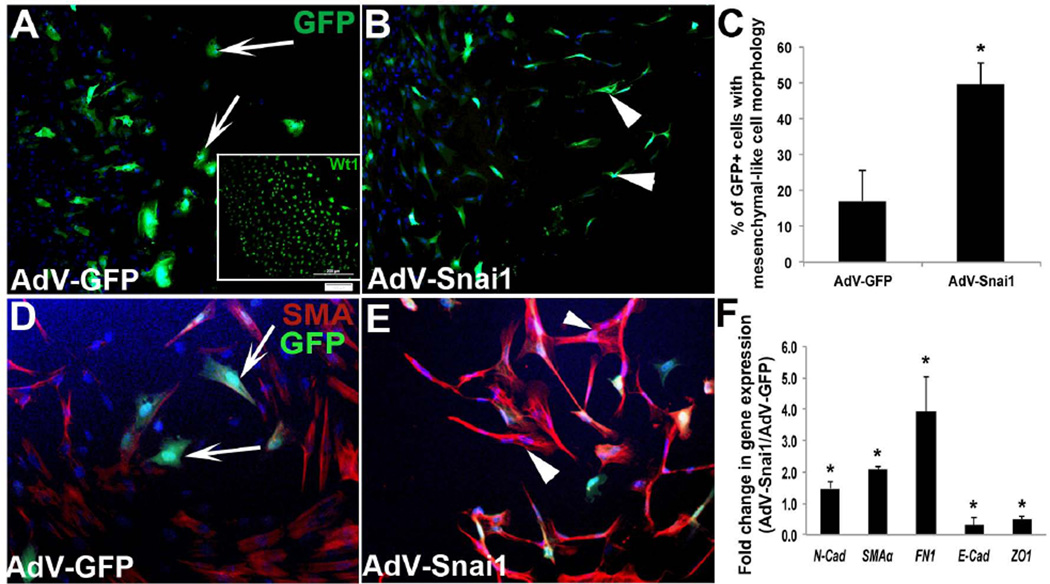
In many developmental and pathological systems, Snai1 has been described as a master regulator of EMT (Batlle et al., 2000; Cano et al., 2000; Carver et al., 2001) and we have shown that Snai1 is important for EMT during early stages of valve development (Tao et al., 2011). In epicardial cells, transformation to a mesenchymal cell fate referred to as epicardial-to-mesenchymal transformation (EpMT), is essential for the generation of EPDCs. This cell population serves as precursors to mature fibroblasts, smooth muscle cells, coronary endothelial cells and possibly cardiac myocytes within the myocardium (Dettman et al., 1998; Gittenberger-de Groot et al., 1998; Cai et al., 2008; Zhou et al., 2008; Smart et al., 2011; Katz et al., 2012). Studies in mice have shown that defects in EpMT lead to underdevelopment of the myocardium and early lethality (Martinez-Estrada et al., 2010; Smith et al., 2011; von Gise et al., 2011; Acharya et al., 2012; Baek and Tallquist, 2012). To determine the sufficiency of Snai1 to promote EpMT in the chick, HH St. 24 whole heart explants were plated and epicardial cell outgrowths were infected with AdV-Snai1 or AdV-GFP. Using qPCR, levels of murine Snai1 overexpression in AdV-Snai1 assays were observed at 116822±53795-fold compared to AdV-GFP controls, while endogenous chicken Snai1 levels were not significantly different. At this time during chick development the epicardium has formed and EpMT has been initiated. In cell outgrowths infected with AdV-GFP, GFP+ cells were largely rounded (Figure 3A), while AdV-Snai1 treatment resulted in less rounded and more mesenchymal shaped cells (Figure 3B, C). In support of Snai1 enhancing EpMT, we examined changes in mesenchyme markers SMA, Fibronectin1 (FN1) and N-cadherin (N-cad) (Martinez-Estrada et al., 2010; Zhou et al., 2010), in addition to epicardial cell junction markers E-cadherin (E-cad) (Zhou et al., 2010; von Gise et al., 2011) and ZO-1 (Austin et al., 2008). Further, SMA, N-cad and FN1 were significantly increased (Figure 3D-F), while E-cad was decreased following AdV-Snai1 infection (Figure 3F). These observations suggest that Snai1 is sufficient to enhance mesenchyme cell transformation of epicardial cells during avian cardiac development.
Figure 3. Snai1 overexpression enhances epicardial-to-mesenchymal transformation in avian epicardial explants.
Epicardial cell outgrowths from HH St. 24 whole heart explants were infected with AdV-GFP (A, D) or AdV-Snai1 (B, E) for 48 hours and differences in cell morphology (A, B), protein (D, E) and gene (F) expression were analyzed. (A) GFP-positive spherical epicardial cells were observed in cultures infected with AdV-GFP (arrows, A), while fibroblast-like mesenchyme cells (arrowheads) were apparent in AdV-Snai1 infected epicardial cells (B). Inset in (A) shows Wt1 immunohistochemistry to indicate the epicardial phenotype of these cells prior to treatment. (C) Quantitation of the percentage of GFP+ cells that display fibroblast-like mesenchyme cell morphology in AdV-GFP and AdV-Snai1 infected cells. (D) Representative immunohistochemistry to detect SMA expression in AdV-GFP (D) and AdV-Snai1 (E) infected epicardial cells. Arrows in D indicate rounded cells types, and arrowheads in E highlight mesenchymal shaped cell types. (F) qPCR to examine changes in expression levels of N-cad (N-cadherin), SMAα (Smooth muscle α-actin), FN1 (Fibronectin-1), E-cad (E-cadherin) and ZO-1 in AdV-Snai1 infected cells compared to AdV-GFP (n=3). * =p<0.05.
Snai1 plays an important role in enhancing cell invasion through activation of MMPs
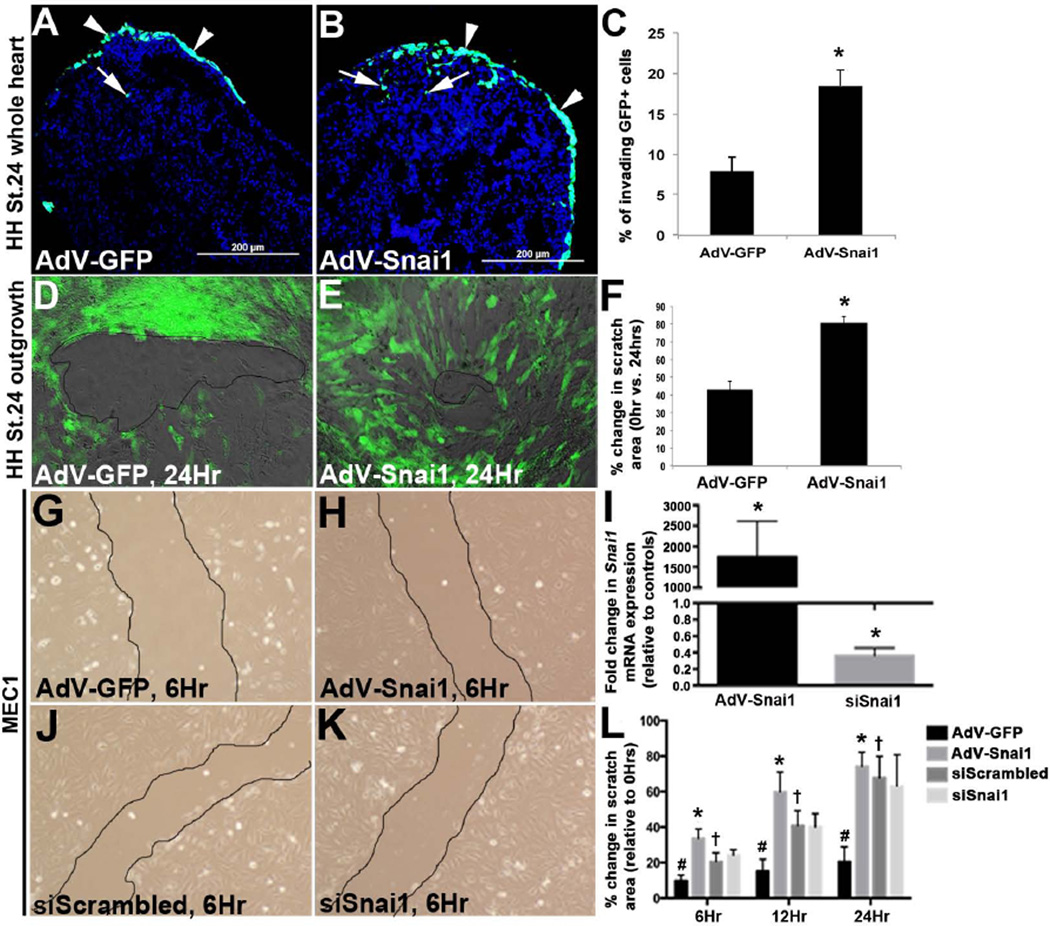
Following EpMT, newly transformed mesenchyme cells invade the myocardium as EPDCs where they differentiate into several myocardial lineages. Using an approach similar to that described by others (Dokic and Dettman, 2006; Nesbitt et al., 2009), HH St. 24 whole hearts were infected with AdV-GFP and the GFP-tagged AdV-Snai1 to label the outermost layer of cells consistent with the epicardium at an infection efficiency of ~75%. After 48 hours, tissue sections were prepared from treated whole hearts and the fate of GFP+ cells was determined. As shown, cells infected with AdV-GFP largely remained on the outermost layer of the heart (Figure 4A), while cells treated with AdV-Snai1 had invaded deep into the myocardium (Figure 4B, C). To further support the hypothesis that Snai1 promotes epicardial cell motility, scratch-healing assays were performed on epicardial cell outgrowths from HH St. 24 whole heart explants (Figures 4D, E, H) and a primary mouse epicardial cell line (MEC1) (Li et al., 2011) (Figure 4G-L) following gain and loss of Snai1 function. In avian epicardial cell outgrowths, cultures infected with AdV-Snai1 showed a greater percent decrease in scratched area at 24 hours compared to 0 hours, relative to AdV-GFP controls. Similar observations were observed in AdV-Snai1 infected MEC1 cells (Figure 4G, H) at 6, 12 and 24 hours (Figure 4L). However, significant changes in scratched area were not detected following successful Snai1 knockdown in MEC1 cells at each time point (Figure 4I-L). Therefore suggesting that Snai1 is sufficient, but not required for epicardial cell motility.
Figure 4. Snai1 enhances cell invasion from the epicardial cell layer into the myocardium.
(A-C) HH St. 24 whole heart explants were infected with AdV-GFP or AdV-Snai1 for 48 hours to label the outermost layer of the cells over the myocardium. (A, B) Tissue sections of infected whole hearts to show fate of GFP+ cells following infection with AdV-GFP (A) or GFP-tagged AdV-Snai1 (B) on the inside (arrows) and outside (arrowheads) of the developing heart. (C) Percent of invading GFP+ cells within the myocardium, over GFP+ cells on the outermost layer of the heart. (D-L) Scratch-healing assay of epicardial cell outgrowths from HH St. 24 whole heart cultures (D-F) and MEC1 cells (G-L) infected with AdV-GFP (D, G), AdV-Snai1 (E, H), siScrambled siRNA (J) or siSnai1 (K). (F) Percent change in scratch area in HH St.24 epicardial cell outgrowths following AdV treatments at 24 hours, compared to 0 hours (n=3, * =p<0.05). (I) qPCR of Snai1 expression in MEC1 cells following AdV-Snai1 and siSnai1 treatments compared to AdV-GFP and siScrambled controls respectively (n=3, * =p<0.05). (L) Percent change in scratch area in MEC1 cells following AdV-GFP, AdV-Snai1, siScrambled and siSnai1 treatments at 6, 12 and 24 hours, compared to 0 hours. #=p<0.05 AdV-GFP infections at 6, 12, and 24 hours compared to 0 hours, *=p<0.05 AdV-Snai1 at 6, 12 and 24 hours compared to AdV-GFP, †=p<0.05 siScrambled at 6, 12, and 24 hours compared to 0 hours.
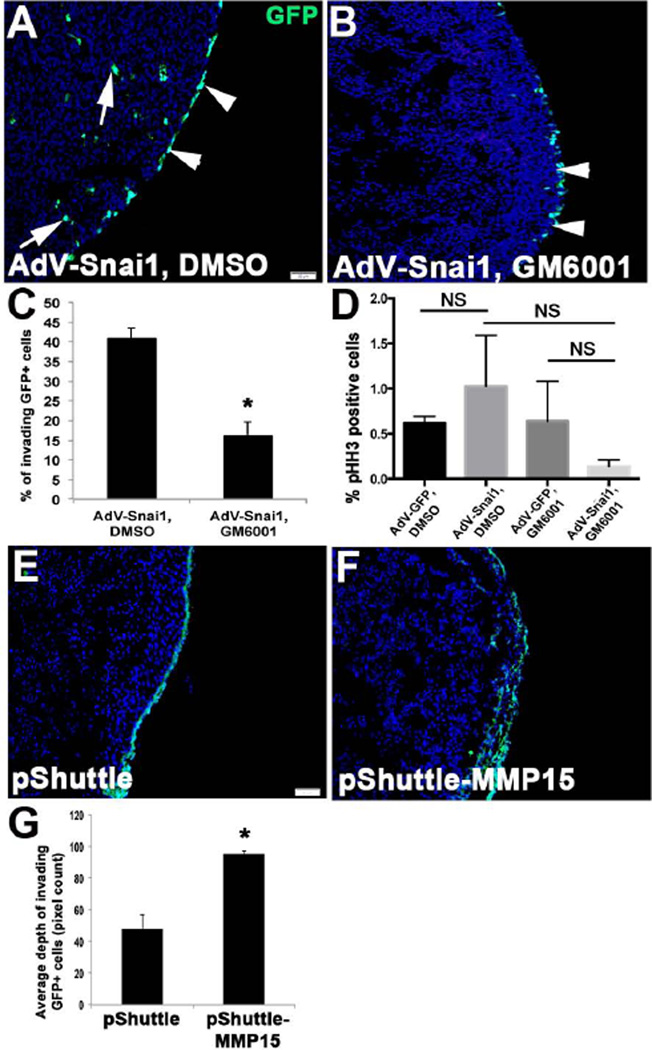
We have previously shown that MMPs, notably MMP15, are required for Snai1-mediated cell migration and invasion during endocardial cushion development (Tao et al., 2011). Therefore, we examined if a similar mechanism is active in motile epicardial cells by co-treating AdV-Snai1 infected HH St. 24 whole hearts with the pan-MMP inhibitor, GM6001 (Figure 5A, B). As quantitated in Figure 5C, significantly less GFP+ labeled cells invade the underlying myocardium in AdV-Snai1 infected hearts treated with GM6001 (Figure 5B) compared to AdV-Snai1, DMSO vehicle (Figure 5A) and AdV-GFP, GM6001 (data not shown) controls. This significant change in cell invasion was not accompanied by changes in cell proliferation (Figure 5D). To support a role for MMP15 in this system HH St. 24 whole hearts were transfected with pShuttle-MMP15, a known downstream target of Snai1 (Tao et al., 2011). pShuttle-MMP15 treatment increased the number of invading GFP+ cells within the underlying myocardium compared to empty pShuttle controls in which GFP+ cells are observed on the outermost layer of the heart (Figure 5E-G). Therefore suggesting that Snai1 can mediate avian epicardial cell invasion into the myocardium through MMPs.
Figure 5. Snai1-mediated epicardial cell invasion requires MMPs.
HH St. 26 whole heart explants were infected with AdV-Snai1 (A, B) and treated with DMSO (A) or the MMP inhibitor GM6001 (B) and the percentage of GFP+ cells within the myocardium was determined compared to GFP+ that remained on the outside of the heart (C) (n=4). (D) Quantitation of pHH3 immunostaining to detect proliferating cells in treated explants (NS, not significant). (E-G) Similar analysis was performed on explants transfected with empty pShuttle vector (E) or p-Shuttle containing the full length mouse MMP15 (pShuttle-MMP15) (F). (G) Quantitation of the average depth of GFP+ invading cells (n=5). * =p<0.05.
Discussion
In this study we show that Snai1 is highly expressed in the proepicardium during early stages of avian epicardial development and expression is maintained in the epicardium during stages of EpMT. At these early stages our in vitro data suggests that Snai1 is important for proepicardial cell migration of HH St. 16 explants, a process associated with formation of the epicardium over the surface of the myocardium. At later stages, Snai1 is sufficient to enhance EpMT through mechanisms similar to other EMT systems including downregulation of cell adhesion molecules and increased expression of mesenchymal cell markers. In addition to enhancing EpMT at HH St. 24, overexpression of Snai1 increases invasion of cells consistent with the epicardium into the underlying myocardium. Further, we show that this process requires MMPs, and likely MMP15 as gain of function studies recapitulate cell invasion phenotypes observed following Snai1 overexpression. Together our studies are the first to describe the function of Snai1 during avian epicardial development.
In many developmental and pathological systems Snai1 serves as a key inducer of endothelial-to-mesenchymal transformation (EMT) (Carver et al., 2001; Timmerman et al., 2004; Thiery et al., 2009; Tao et al., 2011). It is known to regulate this process through direct repression of cell adhesion genes including E-cadherin (Batlle et al., 2000; Cano et al., 2000) to breakdown endothelial cell-cell interactions and allow for mesenchymal transformation and migration. Similar to EMT, EpMT also requires cell transformation and motility and many of the key inducers are shared in these processes (Thiery and Sleeman, 2006; Compton et al., 2007; Zamora et al., 2007; von Gise et al., 2011). The first indication that Snai1 may also play a role in EpMT came from studies showing decreased Snai1 and E-cadherin in Gata5-Cre;Wt1loxP/GFP mice that display EpMT defects by E16.5 (Martinez-Estrada et al., 2010). This study also showed that Wt1 directly binds Snai1 and is sufficient to rescue EpMT defects in Wt1-knockout embryoid bodies (Martinez-Estrada et al., 2010). In support, we show for the first time in this study that direct overexpression of Snai1 in avian epicardial cells enhances EpMT through decreased E-cadherin and increased N-cadherin, SMA and FN1 (Figure 3). However, it is not yet known if Wt1 functions upstream of Snai1 in the avian system as other known regulators are also highly expressed and function in the epicardium, including Tgfβ (Dokic and Dettman, 2006; Austin et al., 2008; Bax et al., 2011; Sanchez and Barnett, 2012) and Wnts (Wu et al., 2010; Horvay et al., 2011; von Gise et al., 2011). Similarly, it is considered that Snai1 is not the only effector of Wt1 during EpMT as more recent work has shown that β-catenin and retinoic acid signaling pathways function downstream and this appears to be independent of changes in E-cadherin expression (von Gise et al., 2011) and therefore, presumably Snai1. Although our study strongly suggests that Snai1 is important for avian EpMT, mice with epicardial-specific deletion of Snai1 display no overt phenotypes (Casanova et al., 2012). It is not clear why Snai1 is sufficient to enhance EpMT studies in the chick, yet not required in the mouse. However, this current study largely utilizes gain of function approaches in the avian system, as opposed to loss of function in the mouse. Therefore, the discrepancies between these two studies may be dependent on levels of Snai1 function and species. Further, it is considered that compensatory responses of other Snai family members could play a role in loss of function studies.
Our data suggests that Snai1 is sufficient to enhance migration of proepicardial cells (Figure 2), while at later stages Snai1 overexpression in cells consistent with the epicardium, leads to increased cell invasion within the underlying myocardium (Figure 4). This additional role of Snai1 in cell mobility was recently described by our group during endocardial cushion development (Tao et al., 2011), and others have shown that high expression levels in cancer cells lines correlates with cell migration and metastasis (Ota et al., 2009; Thiery et al., 2009; Wu and Zhou, 2010). From our data, we can conclude that increased cell motility observed in AdV-Snai1 treated HH St. 24 explants is not associated with cell proliferation, however we are not able to determine if this is secondary to increased EpMT. Nonetheless, our data shows that MMPs are required for Snai1-mediated cell invasion as chemical inhibition with GM6001 attenuates the effects of AdV-Snai1 treatment (Figure 5). Consistent with our previous studies in endocardial cushions,(Tao et al., 2011) MMP15 gain of function is also sufficient to promote cell motility in the epicardium (Figure 5E) and recapitulate the effects of AdV-Snai1 (Figure 5B). As our previous work has shown that Snai1 molecularly interacts with MMP15 in E13.5 whole heart lysates to repress its transcriptional activity, we anticipate that this function is conserved in both endocardial and epicardial cell populations. However, it is clear from Snai1’s more established role during EMT that MMP15 is not the only downstream target gene in these cell types. Based on its function in other systems, it is likely that MMP15 facilitates epicardial cell motility by targeting its ECM substrate type IV collagen (Hotary et al., 2000; Rebustini et al., 2009), previously shown to mark the basement membrane of the epicardium (Wu et al., 2010). Thereby facilitating cell movement from the outer layer of the heart into the underlying myocardium. While it remains unclear if Snai1-mediated effects on cell invasion are secondary to increased EpMT in our system, we predict based on our previous studies (Tao et al., 2011), that MMP15 promotes cell invasion independent of EpMT as overexpression in endocardial cushion explants was not sufficient to promote EMT. These studies suggest that Snai1 has conserved functions for cell motility in avian valve endothelial and epicardial cells.
There has been recent focus on the role of epicardial cells in repair of the damaged adult myocardium (reviewed (Schlueter and Brand, 2012; Smart et al., 2012))(Huang et al., 2012). In the uninjured adult heart, expression of embryonic epicardial markers including Wt1, Raldh2 and several FGF ligands and their receptors are not detected. However their expression is rapidly increased in epicardial cells after cryo injury in the zebrafish heart (Lepilina et al., 2006) and myocardial infarction (MI) in the mouse (Wagner et al., 2002; Limana et al., 2010; Zhou et al., 2011). In the murine MI injury model, EpMT is reactivated resulting in thickening of the epicardial layer, however resulting EPDCs do not invade the myocardium (Zhou et al., 2011). While it has been concluded in both murine and zebrafish injury models that the activated epicardium does not directly contribute to the regenerative adult ventricular myocardium (Jopling et al., 2010; Kikuchi et al., 2010; Kikuchi et al., 2011), more recent studies have shown that the process occurs by activation of developmental pathways by epicardial cells acting on resident myocardial cell types (Lepilina et al., 2006; Kikuchi et al., 2011). In the zebrafish this process is facilitated by migration of activated epicardial cells to the site of injury (Lepilina et al., 2006), an observation not noted in the mouse MI model (Zhou et al., 2011). Increased Snai1 expression has not been reported in injured heart models, but based on findings from this current study it is speculated that in lower vertebrates Snai1 could facilitate migration of activated epicardial cells to the site of injury, through MMPs to promote regeneration. In addition, Snai1 may play a role in inducing EpMT in mouse injury models in response to increased Wt1 (Zhou et al., 2011). This potential role for Snai1 in adult epicardial cells is consistent with previous reports in human cells showing increased EpMT and Snai1 expression in response to Wt1 and Tgfβ signaling (Bax et al., 2011). However, the need of EpMT in the injured mouse heart does not appear to be the same as developmental EpMT to generate a pool of mesenchyme precursor cells, as EPDCs do not directly contribute to the regenerative myocardium. Although the hypothesis that Snai1 is important for EpMT in cardiac injury is an attractive one, it is considered that epicardial-specific knockouts of Snai1 display no developmental defects (Casanova et al., 2012), however this observation in embryos does not eliminate a role in the adult heart. In closing, this study has revealed new and previously unappreciated insights into the role of Snai1 in avian epicardial development and highlights diverse roles in cell migration, transformation and invasion that could have implications in regeneration of the injured myocardium.
Experimental Procedures
Histology
Whole chicken embryos staged at Hamburger Hamilton stages (HH St.) 16, 31 and 40 were collected from White Leghorn chicken eggs (Charles River Laboratories) in 1× Phosphate Buffered Saline (PBS) and fixed in 4% paraformaldehyde (PFA) overnight at 4°C. Fixed tissues were subsequently processed for paraffin embedding and 6µm tissue sections were cut as previously described (Peacock et al., 2010). For colorimetric immunohistochemistry, fixed tissue sections were subject to antigen retrieval by boiling for 10 minutes in unmasking solution (Vector Laboratories) prior to overnight incubation at 4°C with a primary antibody against Snai1 (Abcam, 1:500). Detection using diaminobenzidine was performed according to the manufacturer’s instructions (ABC staining system, Santa Cruz Biotechnology) and slides were counterstained with hematoxylin for 5 minutes and visualized using an Olympus BX51 microscope.
In vitro explant culture
Proepicardial organ cultures
The proepicardium (PE) was collected from HH St. 16 embryos, plated on an un-coated 2-well Lab-Tek Permanox plastic sterile chamber slide (Nunc #177437) and immediately infected with 5×107 PFU adenovirus expressing Snai1 (AdV-Snai1) or GFP (AdV-GFP). All viruses were diluted in serum-free culture media (M199 supplemented with 1% Chick Embryo Extract and 1% penicillin/streptomycin) for 2 hours. Following infection, fresh serum-free media was added and PE explants were cultured for a further 24 hours. Following culture, mRNA was extracted as described (Peacock et al., 2010) for qPCR analysis (see below), or the fold change in colony area of epicardial cell outgrowth was determined by capturing 2D images using an Olympus SZX7, and calculating the average of the total pixel count from each cultured PE explant using ImagePro Plus software (n=5). Significant differences in area between AdV-GFP and AdV-Snai1 treated explants were determined using Student’s t-test (p<0.05). Alternatively, treated PE explants were cultured for 48 hours and fixed in 4% PFA, or subjected to mRNA isolation as previously described (Peacock et al., 2010) for further qPCR analysis (see below). Fixed PE explants were treated with 0.2% TritonX-100/PBS and subject to immunohistochemistry using anti-Wt1 (Santa Cruz, 1:200, incubation of 2 hours at room temperature) and a donkey-anti-rabbit-568 Alexa-Fluor secondary antibody (Invitrogen, 1:400). Images were captured using an Olympus Fluoview F-1000 confocal microscope.
Whole heart epicardial cultures
Whole hearts were dissected from HH St. 24 embryos and plated on 2-well chamber slides coated with 0.1% rat tail type I collagen (Tao et al., 2011) with complete culture media (M199 supplemented with 10% Fetal Bovine Serum, 1% Chick Embryo Extract and 1% penicillin/streptomycin). After 24 hours, whole heart explant was removed and the remaining epicardial cell layer outgrowth was infected with 5×107 PFU of AdV-Snai1 or AdV-GFP for 48 hours. Following infection, cells were fixed for immunohistochemistry using an antibody against smooth muscle alpha Actin (SMAα) (Invitrogen, 1:500) or mRNA was isolated for qPCR analysis (see below).
Scratch Healing Assay
Primary mouse epicardial cells (MEC1), a kind gift from Dr. Henry Sucov were cultured as previously described (Li et al., 2011). MEC1 cells and HH St.24 whole heart epicardial cell outgrowths were subject to AdV-GFP and AdV-Snai1 infection for 24 hours as described above, or MEC1 cells were transfected with 5nM On-TARGEtplus SMARTpool siRNA (Thermo scientific) specific for mouse Snai1 (siSnai1), or a non-targeting pool (siScrambled) as a control using 8µl Dharmafect according to the manufacturer’s protocol. 24 hours after adenoviral infection or siRNA transfection, 50% serum media was added and treated cell monolayers were scratched with a 25½ guage dissection needle (avian) or a 0.1–10µl pipette tip (MEC1) and images were captured at 0, 6, 12 and 24 hours following scratch. Significant differences between AdV-GFP and AdV-Snai1, and siScrambled and siSnai1 treated cells was determined by measuring the scratch area at 0 hours and comparing to 6, 12 and 24 hours using ImagePro Plus and presented as a percentage change. Significant differences were determined using Student’s t-test (n=5, p<0.05).
Cell Invasion Assay
For cell invasion assays, HH St.24 hearts were placed on 0.2µm filters (Millpore) floating in serum free culture media in 30mm dishes. Explants were then directly treated with 50µl media containing 5×107 PFU AdV-Snai1 or AdV-GFP, or 10µg/mL GM6001 (Millipore) or dimethyl sulfoxide (DMSO) in the presence of AdV-GFP or AdV-Snai1 for 48 hours. Following treatment(s), explants were fixed in 4% PFA and processed for frozen sections (Tao et al., 2011) and invading GFP+ cells were visualized and captured using an Olympus BX51 microscope. The percentage of invading cells was determined by counting the number of GFP+ cells within the myocardial space, versus the number of GFP+ that remained on the surface of the heart (n=5). In addition, immunofluorescence was performed to detect phospho-histone H3 (pHH3) expression in proliferating cells (Thermo Scientific, 1:200). Significant differences in cell invasion and proliferation between AdV-GFP and AdV-Snai1 treated explants were determined using Student’s t-test (p<0.05).
Alternatively, the whole heart culture was transfected with pShuttle-IRES-hrGFP-1 vector expressing mouse MMP15. Briefly, mouse MMP15 coding region was amplified from cDNA prepared from E10.5 mouse heart using the specific primers containing restriction sites (Forward: CATCGCGGCCGCCCAACCATGGGCAGCGACCGGAGCGC ; Reverse:CATCCTCGAGCACCCACTCCTGCAGTGAGCGC). Purified cDNA fragment was digested with NotI and XhoI (New England BioLabs) and ligated into linearized pShuttle-IRES-hrGFP-1 plasmid (Agilent Technologies). 4µg plasmid was mixed with 20 µl lipofectamine (per 40 µl Opti-MEM media) as per the manufacturer’s instruction (Invitrogen). The mixture was added on top of the filter to cover the heart explants. After 48 hours, the heart explants were fixed and subjected to fluorescence microscopy as mentioned above. The maximum depth of GFP+ cell invasion was measured by ImageJ (n=4). Significant differences between MMP15-expressing vector and empty vector treated explants were determined using Student’s t-test (p<0.05).
Realtime PCR
mRNA from treated cultures was isolated using Trizol as described (Peacock et al., 2010) and 200–400ng was used to generate cDNA using the high-capacity cDNA kit (Applied Biosystems) (Tao et al., 2011). cDNA was subject to quantitative PCR amplification (StepOne Plus, Applied Biosystems) using Sybr Green fast master mix (Applied Biosystems) and specific primers targeting chicken Integrin-α4 (Forward:CGGATCTTTTGTCGCTTGTG; Reverse: GGCATGACCCATGGTTTTCT), Integrin-β1 (Forward: ATCCGCTGTCTCACTGCAAA; Reverse: CACGCCAGCTACAATGGGTA), ZO1 (Forward:CGCCTCCATCGTCTACATCA; Reverse:CGATGAGGAACCCACAGACA), N-cadherin (Forward:AAGCAGTCCCTCTCCCAACA; Reverse:TTGGGTTTCCTTCCATGTCC), SMAα (Forward:; Reverse:), Fibronectin1 (Forward:CGTTCGTCTCACTGGCTACA; Reverse:ATTAATCCCGACACGACAGC) and E-cadherin (Forward:GCAAGCCGTTTACCACATCA; Reverse:TTGTTCTCCACCGTCACCAC). Alternatively, PCR amplification was performed using pre-designed Primetime qPCR Assays (Integrated DNA Technologies) to detect chicken Cdk1, Ccne1, pCNA, Ccnd1, Snai1 and mouse Snai1. Following PCR analyses, the cycle count (Ct) was normalized to the GAPDH housekeeping gene (Forward: GGGTCTTATGACCACTGTCC; Reverse: GTAAGCTTCCCATTCAGCTCAG) to give the ΔCt value. The ΔΔCt and fold changes in experimental samples over respective controls were then determined using Power(2,−ΔΔCt).
Bullet points.
Snai1 is highly expressed throughout avian epicardial development
Snai1 is sufficient to promote proepicardial cell migration in vitro
Snai1 overexpression enhances epicardial-to-mesenchymal transformation in avian explants in vitro
Snai1 enhances epicardial cell invasion through MMPs in avian explants in vitro
Acknowledgements
We acknowledge Harriet Hammond, Blair Austin and Danielle Huk for technical assistance and Dr. Henry Sucov for sharing the MEC1 cell line. This work was supported by NHLBI R01HL091878 (JL), The Research Institute at Nationwide Children’s Hospital, and The American Heart Association Predoctoral Fellowship 10PRE4360052 (GT).
References
- Acharya A, Baek ST, Huang G, Eskiocak B, Goetsch S, Sung CY, Banfi S, Sauer MF, Olsen GS, Duffield JS, Olson EN, Tallquist MD. The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors. Development. 2012;139:2139–2149. doi: 10.1242/dev.079970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin AF, Compton LA, Love JD, Brown CB, Barnett JV. Primary and immortalized mouse epicardial cells undergo differentiation in response to TGFbeta. Dev Dyn. 2008;237:366–376. doi: 10.1002/dvdy.21421. [DOI] [PubMed] [Google Scholar]
- Baek ST, Tallquist MD. Nf1 limits epicardial derivative expansion by regulating epithelial to mesenchymal transition and proliferation. Development. 2012;139:2040–2049. doi: 10.1242/dev.074054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De Herreros A. The transcription factor snail is a repressor of Ecadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- Bax NA, van Oorschot AA, Maas S, Braun J, van Tuyn J, de Vries AA, Groot AC, Goumans MJ. In vitro epithelial-to-mesenchymal transformation in human adult epicardial cells is regulated by TGFbeta-signaling and WT1. Basic Res Cardiol. 2011;106:829–847. doi: 10.1007/s00395-011-0181-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braitsch CM, Combs MD, Quaggin SE, Yutzey KE. Pod1/Tcf21 is regulated by retinoic acid signaling and inhibits differentiation of epicardium-derived cells into smooth muscle in the developing heart. Dev Biol. 2012;368:345–357. doi: 10.1016/j.ydbio.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai CL, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, Yang L, Bu L, Liang X, Zhang X, Stallcup WB, Denton CP, McCulloch A, Chen J, Evans SM. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454:104–108. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelialmesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- Carver EA, Jiang R, Lan Y, Oram KF, Gridley T. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol Cell Biol. 2001;21:8184–8188. doi: 10.1128/MCB.21.23.8184-8188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova JC, Travisano S, de la Pompa JL. Epithelial-to-mesenchymal transition in epicardium is independent of Snail1. Genesis. 2012 doi: 10.1002/dvg.22353. [DOI] [PubMed] [Google Scholar]
- Chen IH, Wang HH, Hsieh YS, Huang WC, Yeh HI, Chuang YJ. PRSS23 is essential for the Snail-dependent endothelial to mesenchymal transition during valvulogenesis in zebrafish. Cardiovasc Res. 2012 doi: 10.1093/cvr/cvs355. [DOI] [PubMed] [Google Scholar]
- Combs MD, Braitsch CM, Lange AW, James JF, Yutzey KE. NFATC1 promotes epicardium-derived cell invasion into myocardium. Development. 2011;138:1747–1757. doi: 10.1242/dev.060996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton LA, Potash DA, Brown CB, Barnett JV. Coronary vessel development is dependent on the type III transforming growth factor beta receptor. Circ Res. 2007;101:784–791. doi: 10.1161/CIRCRESAHA.107.152082. [DOI] [PubMed] [Google Scholar]
- Dettman RW, Denetclaw W, Jr, Ordahl CP, Bristow J. Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Dev Biol. 1998;193:169–181. doi: 10.1006/dbio.1997.8801. [DOI] [PubMed] [Google Scholar]
- Dokic D, Dettman RW. VCAM-1 inhibits TGFbeta stimulated epithelialmesenchymal transformation by modulating Rho activity and stabilizing intercellular adhesion in epicardial mesothelial cells. Dev Biol. 2006;299:489–504. doi: 10.1016/j.ydbio.2006.08.054. [DOI] [PubMed] [Google Scholar]
- Gittenberger-de Groot AC, Vrancken Peeters MP, Bergwerff M, Mentink MM, Poelmann RE. Epicardial outgrowth inhibition leads to compensatory mesothelial outflow tract collar and abnormal cardiac septation and coronary formation. Circ Res. 2000;87:969–971. doi: 10.1161/01.res.87.11.969. [DOI] [PubMed] [Google Scholar]
- Gittenberger-de Groot AC, Vrancken Peeters MP, Mentink MM, Gourdie RG, Poelmann RE. Epicardium-derived cells contribute a novel population to the myocardial wall and the atrioventricular cushions. Circ Res. 1998;82:1043–1052. doi: 10.1161/01.res.82.10.1043. [DOI] [PubMed] [Google Scholar]
- Horvay K, Casagranda F, Gany A, Hime GR, Abud HE. Wnt signaling regulates Snai1 expression and cellular localization in the mouse intestinal epithelial stem cell niche. Stem Cells Dev. 2011;20:737–745. doi: 10.1089/scd.2010.0188. [DOI] [PubMed] [Google Scholar]
- Hotary K, Allen E, Punturieri A, Yana I, Weiss SJ. Regulation of cell invasion and morphogenesis in a three-dimensional type I collagen matrix by membrane-type matrix metalloproteinases 1, 2, and 3. J Cell Biol. 2000;149:1309–1323. doi: 10.1083/jcb.149.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang GN, Thatcher JE, McAnally J, Kong Y, Qi X, Tan W, Dimaio JM, Amatruda JF, Gerard RD, Hill JA, Bassel-Duby R, Olson EN. C/EBP Transcription Factors Mediate Epicardial Activation During Heart Development and Injury. Science. 2012 doi: 10.1126/science.1229765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii Y, Garriock RJ, Navetta AM, Coughlin LE, Mikawa T. BMP signals promote proepicardial protrusion necessary for recruitment of coronary vessel and epicardial progenitors to the heart. Dev Cell. 2010;19:307–316. doi: 10.1016/j.devcel.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C, Sleep E, Raya M, Marti M, Raya A, Izpisua Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz TC, Singh MK, Degenhardt K, Rivera-Feliciano J, Johnson RL, Epstein JA, Tabin CJ. Distinct compartments of the proepicardial organ give rise to coronary vascular endothelial cells. Dev Cell. 2012;22:639–650. doi: 10.1016/j.devcel.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Major RJ, Blum N, Dahn RD, Begemann G, Poss KD. Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev Cell. 2011;20:397–404. doi: 10.1016/j.devcel.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DY, Poss KD. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwee L, Baldwin HS, Shen HM, Stewart CL, Buck C, Buck CA, Labow MA. Defective development of the embryonic and extraembryonic circulatory systems in vascular cell adhesion molecule (VCAM-1) deficient mice. Development. 1995;121:489–503. doi: 10.1242/dev.121.2.489. [DOI] [PubMed] [Google Scholar]
- Lavine KJ, White AC, Park C, Smith CS, Choi K, Long F, Hui CC, Ornitz DM. Fibroblast growth factor signals regulate a wave of Hedgehog activation that is essential for coronary vascular development. Genes Dev. 2006;20:1651–1666. doi: 10.1101/gad.1411406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, Burns CG, Poss KD. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- Li P, Cavallero S, Gu Y, Chen TH, Hughes J, Hassan AB, Bruning JC, Pashmforoush M, Sucov HM. IGF signaling directs ventricular cardiomyocyte proliferation during embryonic heart development. Development. 2011;138:1795–1805. doi: 10.1242/dev.054338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limana F, Bertolami C, Mangoni A, Di Carlo A, Avitabile D, Mocini D, Iannelli P, De Mori R, Marchetti C, Pozzoli O, Gentili C, Zacheo A, Germani A, Capogrossi MC. Myocardial infarction induces embryonic reprogramming of epicardial c-kit(+) cells: role of the pericardial fluid. J Mol Cell Cardiol. 2010;48:609–618. doi: 10.1016/j.yjmcc.2009.11.008. [DOI] [PubMed] [Google Scholar]
- Lin SC, Dolle P, Ryckebusch L, Noseda M, Zaffran S, Schneider MD, Niederreither K. Endogenous retinoic acid regulates cardiac progenitor differentiation. Proc Natl Acad Sci U S A. 2010;107:9234–9239. doi: 10.1073/pnas.0910430107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomeli H, Starling C, Gridley T. Epiblast-specific Snai1 deletion results in embryonic lethality due to multiple vascular defects. BMC Res Notes. 2009;2:22. doi: 10.1186/1756-0500-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Estrada OM, Lettice LA, Essafi A, Guadix JA, Slight J, Velecela V, Hall E, Reichmann J, Devenney PS, Hohenstein P, Hosen N, Hill RE, Munoz- Chapuli R, Hastie ND. Wt1 is required for cardiovascular progenitor cell formation through transcriptional control of Snail and E-cadherin. Nat Genet. 2010;42:89–93. doi: 10.1038/ng.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbitt TL, Roberts A, Tan H, Junor L, Yost MJ, Potts JD, Dettman RW, Goodwin RL. Coronary endothelial proliferation and morphogenesis are regulated by a VEGF-mediated pathway. Dev Dyn. 2009;238:423–430. doi: 10.1002/dvdy.21847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota I, Li XY, Hu Y, Weiss SJ. Induction of a MT1-MMP and MT2-MMPdependent basement membrane transmigration program in cancer cells by Snail1. Proc Natl Acad Sci U S A. 2009;106:20318–20323. doi: 10.1073/pnas.0910962106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pae SH, Dokic D, Dettman RW. Communication between integrin receptors facilitates epicardial cell adhesion and matrix organization. Dev Dyn. 2008;237:962–978. doi: 10.1002/dvdy.21488. [DOI] [PubMed] [Google Scholar]
- Peacock JD, Levay AK, Gillaspie DB, Tao G, Lincoln J. Reduced sox9 function promotes heart valve calcification phenotypes in vivo. Circ Res. 2010;106:712–719. doi: 10.1161/CIRCRESAHA.109.213702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebustini IT, Myers C, Lassiter KS, Surmak A, Szabova L, Holmbeck K, Pedchenko V, Hudson BG, Hoffman MP. MT2-MMP-dependent release of collagen IV NC1 domains regulates submandibular gland branching morphogenesis. Dev Cell. 2009;17:482–493. doi: 10.1016/j.devcel.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers LS, Lalani S, Runyan RB, Camenisch TD. Differential growth and multicellular villi direct proepicardial translocation to the developing mouse heart. Dev Dyn. 2008;237:145–152. doi: 10.1002/dvdy.21378. [DOI] [PubMed] [Google Scholar]
- Sanchez NS, Barnett JV. TGFbeta and BMP-2 regulate epicardial cell invasion via TGFbetaR3 activation of the Par6/Smurf1/RhoA pathway. Cell Signal. 2012;24:539–548. doi: 10.1016/j.cellsig.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlueter J, Brand T. A right-sided pathway involving FGF8/Snai1 controls asymmetric development of the proepicardium in the chick embryo. Proc Natl Acad Sci U S A. 2009;106:7485–7490. doi: 10.1073/pnas.0811944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlueter J, Brand T. Epicardial progenitor cells in cardiac development and regeneration. J Cardiovasc Transl Res. 2012;5:641–653. doi: 10.1007/s12265-012-9377-4. [DOI] [PubMed] [Google Scholar]
- Smart N, Bollini S, Dube KN, Vieira JM, Zhou B, Davidson S, Yellon D, Riegler J, Price AN, Lythgoe MF, Pu WT, Riley PR. De novo cardiomyocytes from within the activated adult heart after injury. Nature. 2011;474:640–644. doi: 10.1038/nature10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart N, Dube KN, Riley PR. Epicardial progenitor cells in cardiac regeneration and neovascularisation. Vascul Pharmacol. 2012 doi: 10.1016/j.vph.2012.08.001. [DOI] [PubMed] [Google Scholar]
- Smith CL, Baek ST, Sung CY, Tallquist MD. Epicardial-derived cell epithelial-to-mesenchymal transition and fate specification require PDGF receptor signaling. Circ Res. 2011;108:e15–e26. doi: 10.1161/CIRCRESAHA.110.235531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao G, Levay AK, Gridley T, Lincoln J. Mmp15 is a direct target of Snai1 during endothelial to mesenchymal transformation and endocardial cushion development. Dev Biol. 2011;359:209–221. doi: 10.1016/j.ydbio.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- Timmerman LA, Grego-Bessa J, Raya A, Bertran E, Perez-Pomares JM, Diez J, Aranda S, Palomo S, McCormick F, Izpisua-Belmonte JC, de la Pompa JL. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 2004;18:99–115. doi: 10.1101/gad.276304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gise A, Zhou B, Honor LB, Ma Q, Petryk A, Pu WT. WT1 regulates epicardial epithelial to mesenchymal transition through beta-catenin and retinoic acid signaling pathways. Dev Biol. 2011;356:421–431. doi: 10.1016/j.ydbio.2011.05.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner KD, Wagner N, Bondke A, Nafz B, Flemming B, Theres H, Scholz H. The Wilms' tumor suppressor Wt1 is expressed in the coronary vasculature after myocardial infarction. FASEB J. 2002;16:1117–1119. doi: 10.1096/fj.01-0986fje. [DOI] [PubMed] [Google Scholar]
- Wu M, Smith CL, Hall JA, Lee I, Luby-Phelps K, Tallquist MD. Epicardial spindle orientation controls cell entry into the myocardium. Dev Cell. 2010;19:114–125. doi: 10.1016/j.devcel.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhou BP. Snail: More than EMT. Cell Adh Migr. 2010;4:199–203. doi: 10.4161/cam.4.2.10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JT, Rayburn H, Hynes RO. Cell adhesion events mediated by alpha 4 integrins are essential in placental and cardiac development. Development. 1995;121:549–560. doi: 10.1242/dev.121.2.549. [DOI] [PubMed] [Google Scholar]
- Zamora M, Manner J, Ruiz-Lozano P. Epicardium-derived progenitor cells require beta-catenin for coronary artery formation. Proc Natl Acad Sci U S A. 2007;104:18109–18114. doi: 10.1073/pnas.0702415104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Honor LB, He H, Ma Q, Oh JH, Butterfield C, Lin RZ, Melero-Martin JM, Dolmatova E, Duffy HS, Gise A, Zhou P, Hu YW, Wang G, Zhang B, Wang L, Hall JL, Moses MA, McGowan FX, Pu WT. Adult mouse epicardium modulates myocardial injury by secreting paracrine factors. J Clin Invest. 2011;121:1894–1904. doi: 10.1172/JCI45529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang D, von Gise A, Ikeda S, Chien KR, Pu WT. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, von Gise A, Ma Q, Hu YW, Pu WT. Genetic fate mapping demonstrates contribution of epicardium-derived cells to the annulus fibrosis of the mammalian heart. Dev Biol. 2010;338:251–261. doi: 10.1016/j.ydbio.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]