Abstract
Purpose
To describe the clinical and genetic findings in a Chinese family with autosomal dominant cone dystrophy (adCOD).
Methods
One family was examined clinically, and genomic DNA was extracted from venous blood of all participants. Genotyping and haplotyping analysis was performed on the known genetic loci for adCOD and autosomal dominant cone-rod dystrophies (adCORD) with a panel of polymorphic markers in this family. All coding exons of the AIPL1, PTTPNM3, and GUCY2D gene were directly sequenced. Allele-specific PCR was used to validate a substitution in all available family members and 100 normal controls. Bioinformatics analysis was done using the Garnier-Osguthorpe-Robson method to predict the effect of the variants detected on the secondary structure of the GUCY2D protein.
Results
Clinical examination and pedigree analysis revealed a three-generation family with four members diagnosed with adCOD. Through genotyping, the disease-causing genes were mapped to chromosomes 17p13.1–2 (AIPL1, PITPNM3, and GUCY2D gene). A novel A->G transition at position 2545 (p.T849A) of the cDNA sequence was identified in the GUCY2D gene. No mutation was detected in the AIPL1 and PITPNM3 genes. This missense mutation co-segregated with the disease phenotype of the family but was not found in the 100 normal controls.
Conclusions
A novel missense mutation of the GUCY2D gene was identified in this study. Our results further confirm that the dimerization zone of RetGC-1 is the mutational hot region for COD and CORD.
Introduction
Inherited cone dystrophies (CODs) and cone-rod dystrophies (CORDs) are a subgroup of inherited retinal degenerative diseases [1]. Characterized by the degeneration of cones with the relative preservation of rod function, CODs cause an early loss of visual acuity and color discrimination in the first decade of life. In contrast, CORDs are characterized by the progressive loss of cone photoreceptor function, followed by the progressive loss of rod photoreceptor function [1]. Both conditions are genetically heterogeneous and can be inherited in autosomal dominant, recessive, or X-linked patterns. To date, 10 genes have been identified as being responsible for adCOD and adCORD, namely, semaphorin 4A (SEMA4A) on chromosome 1q22, prominin 1 (PROM1) on chromosome 4p15.32, guanylate cyclase activator 1A (GUCA1A) and peripherin 2 (PRPH2) on chromosome 6p21.1, regulating synaptic membrane exocytosis 1 (RIMS1) on chromosome 6p13, guanylate cyclase 2D (GUCY2D), arylhydrocarbon receptor interacting protein-like 1 (AIPL1), and PITPNM family member 3 (PITPNM3) on chromosome 17p13.1–2, unc-119 homolog (UNC119) on chromosome 17q11.2, and cone-rod homeobox (CRX) on chromosome 19q13.32 (RetNet).
The GUCY2D gene, located on chromosome 17p13.1, encodes a 1103 amino acid membrane-bound retinal guanylyl cyclase-1 protein (RetGC-1), which is expressed in both the cone and rod photoreceptors but predominantly in the cone outer segment [2-5]. RetGC-1 is one member of a pair of membrane-bound guanylate cyclases, RetGC-1 and RetGC-2, which synthesize cyclic 3′, 5′-guanosine monophosphate (cGMP) from guanosine triphosphate in mammalian photoreceptor cells. RetGC-1 and its associated activator proteins are responsible for the Ca2+-sensitive restoration of cGMP levels after light activation of the phototransduction cascade. RetGC-1 consists of an extracellular or intradiskal domain, a transmembrane segment, a kinase homology domain, a dimerization domain, and a catalytic domain [6]. Heterozygous mutations in the GUCY2D gene have been shown to cause adCOD and adCORD [2-4]; however, homozygous or compound heterozygous mutations cause autosomal recessively inherited Leber Congenital Amaurosis (LCA), the most severe form of inherited retinopathy, with total blindness or greatly impaired vision recognized at birth or in early infancy [7,8].
In this study, we investigated a Chinese family with cone dystrophy. After linkage analysis, we mapped the disease-causing gene to regions 17p13.1–17p13.2 where the GUCY2D, AIPL1, and PITPNM3 genes are located and found a novel missense mutation of the GUCY2D gene.
Methods
Patients and DNA samples collection
This study was performed according to the tenets of the Declaration of Helsinki for research involving human subjects. This study was approved by the Beijing Tongren Hospital Joint Committee on Clinical Investigation. After informed consent was obtained, all participants underwent full ophthalmic examinations, which included bilateral best-corrected visual acuity using E decimal charts, detailed examination of the anterior segment by slit-lamp biomicroscopy, fundus examination with dilated pupils, and a color discrimination test using pseudoisochromatic plates. The proband underwent visual field testing, an electroretinogram, and optical coherence tomography examination.
Genotyping and haplotyping analysis
Genotyping was performed with 24 microsatellite markers from the autosomes for the known adCOD and adCORD loci in this family (Appendix 1). The fine mapping primer sequences were obtained from the Human Genome Database. Pedigree and haplotype maps were constructed using Cyrillic v. 2.0 software.
Mutation screening of the GUCY2D, AIPL1, and PITPNM3 genes
Mutation screening was performed in the family using direct DNA sequence analysis. All coding regions of the GUCY2D, AIPL1, and PITPNM3 genes were amplified by PCR from the genomic DNA. Primers for all coding exons and exon-intron boundaries of the three genes (18 exons for the GUCY2D, 5 exons for the AIPL1, and 20 exons for the PITPNM3) were designed by the Primer3 program (Appendix 2). For direct sequencing, the PCR products were purified (Shenneng Bocai PCR purification kit; Shenneng, Shanghai, China). The purified PCR products were sequenced using an automatic fluorescence DNA sequencer (ABI, Prism 373A; Perkin Elmer, Foster City, CA) according to the manufacturer’s instructions. All PCR products were sequenced in both forward and reverse directions, and the nucleotide sequences were compared with the published DNA sequences of the GUCY2D, AIPL1, and PITPNM3 genes (GenBank accession number NM_000180, NM_014336, and NM_031220, respectively) using DNAssit version 1.0. For the three genes, cDNA numbering +1 corresponded to A in the ATG translation initiation codon of GUCY2D, AIPL1, and PITPNM3, respectively.
Allele-specific PCR analysis
To confirm the variation found in the sequencing, allele-specific PCR analysis (AS-PCR) was performed in the available family members and in 100 normal controls [9]. An allele-specific forward primer in Exon 13 of the GUCY2D gene was designed: 5′-GGA GCT GGA AAA GCA GAA GG-3′, where C is the mutation-specific nucleotide. The AS-PCR fragment was amplified with the forward allele specific primer and the normal reverse primer of the exon13 of the GUCY2D gene.
Bioinformatics analysis
Garnier-Osguthorpe-Robson software was used to predict the effect of the mutation on the secondary structure of GUCY2D (Biotools) [10]. This method infers the secondary structure of a sequence by calculating the probability for each of the four structure classes (helix, sheet, turn, and loop) based on the central residue and its neighbors from the calculated matrices [10]. The PolyPhen2 (Polymorphism Phenotyping 2) program was used to predict the potential functional impact of an amino acid change [11].
Results
Clinical findings
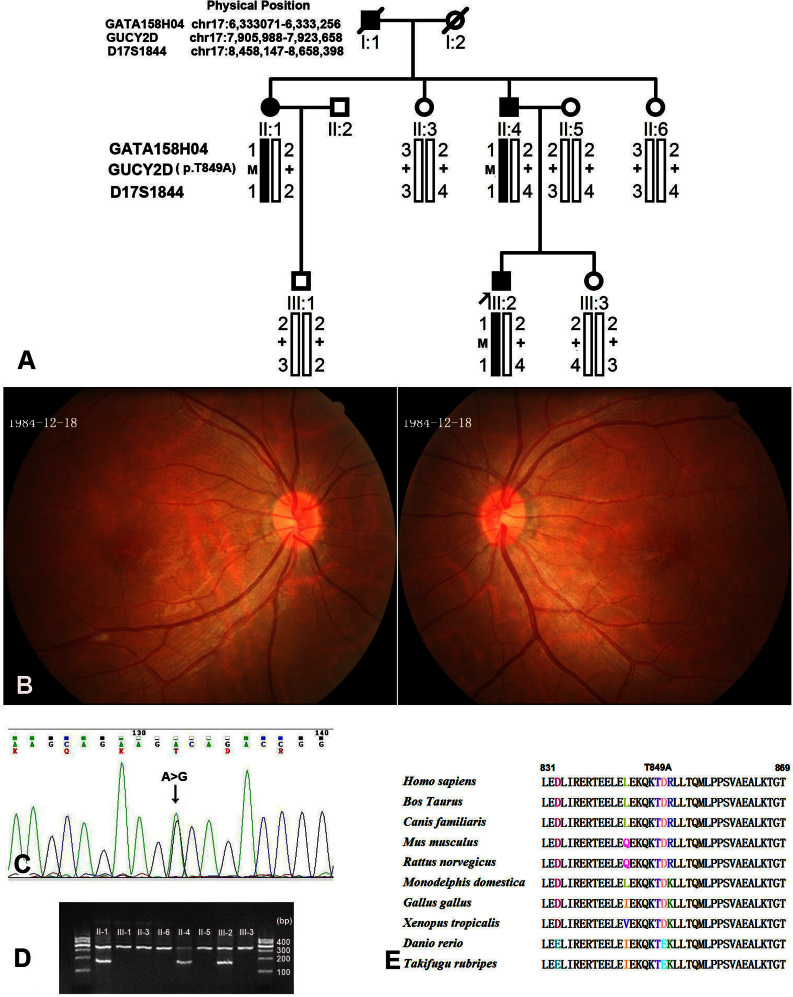
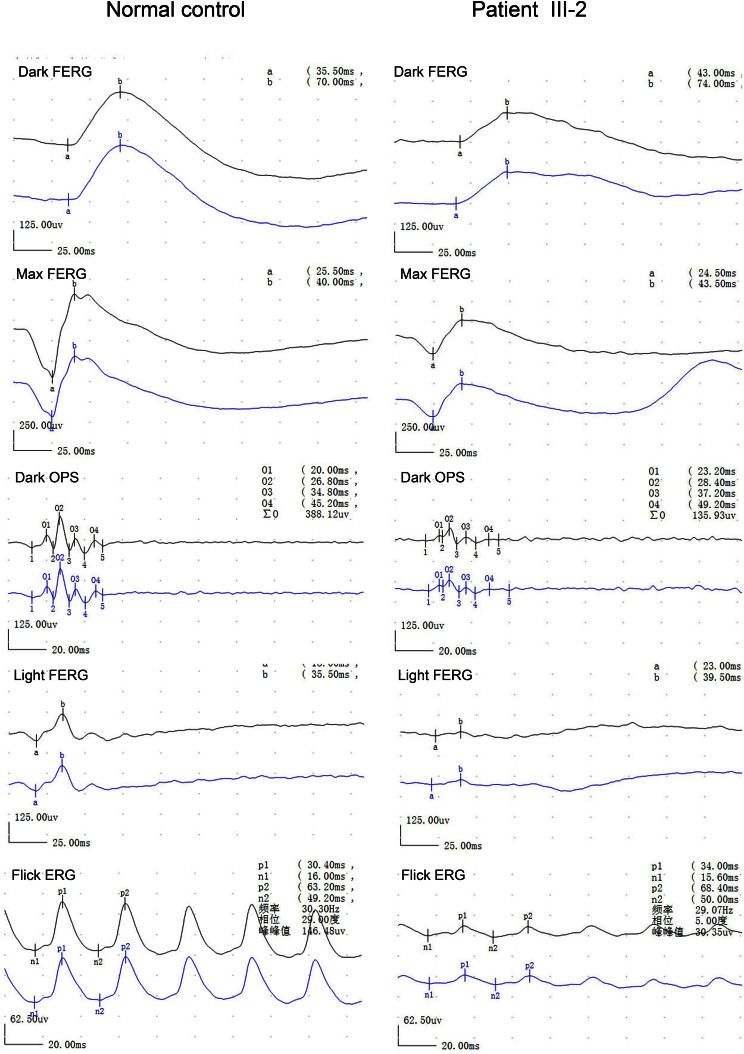
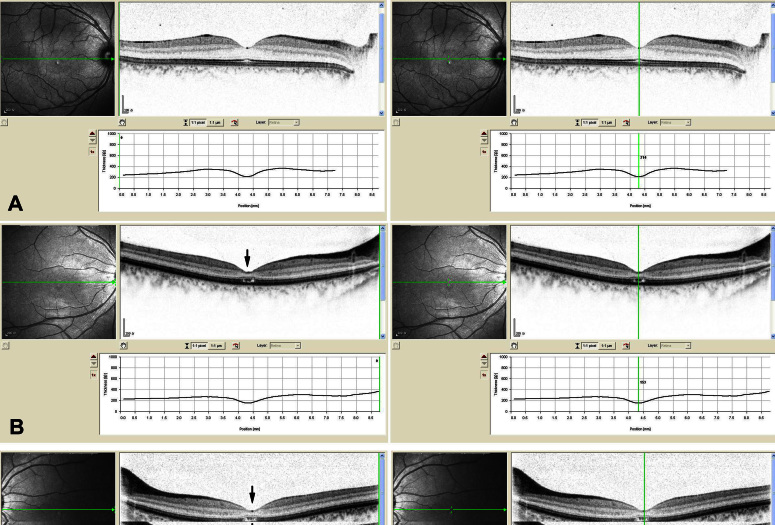
We identified a three-generation family consisting of four patients diagnosed with cone dystrophy (Figure 1A). All patients had experienced bilateral visual acuity impairment and marked photophobia in their early childhood. No patients, including the two patients aged over 50 years, had peripheral field loss or a nyctalopia complaint. Slit-lamp examination showed the anterior segments were normal with the exception of mild cataracts in both eyes of one patient (II-1). Fundus examinations revealed subtle RPE granular abnormalities in the macular area and normal appearance of the peripheral retina. A pseudoisochromatic plates test showed red-green color weakness. The proband was examined in 2008 at age 24 years and again in 2011 at age 27 years. His best-corrected visual acuity was 0.5 in both eyes, and there was no significant deterioration during the three following years. Fundus examination showed almost normal fundus appearance with the exception of subtle mottling of the RPE in the macular area (Figure 1B). Electroretinographic testing revealed a significant reduction in cone responses and normal rod responses (Figure 2). Optical coherence tomography showed thinning of the retina and loss of the photoreceptor inner segment (IS) and photoreceptor outer segment (OS) in the macular area (Figure 3). The detailed clinical features are summarized in Table 1.
Figure 1.
Family structure, proband fundus appearance, DNA sequence chromatograms, and co-segregation analysis of the p.T849A mutation with the disease phenotype in a Chinese family with cone dystrophy. A: The pedigree and haplotype analysis of the family with cone dystrophy showed segregation with three microsatellite markers on chromosome 17 listed in rising order from the telomere end. Squares indicate males; circles indicate females; slashed symbols indicate deceased; solid symbols indicate affected; open symbols indicate unaffected; M indicates mutant; and + indicates wild-type. B: Fundus appearance of the proband shows the subtle mottling of the RPE in the macula. C: Heterozygote sequence (sense strand) shows an A/G transition in codon 849 that changed threonine to alanine. D: Allele-specific PCR analysis presents the amplified products of the mutation allele (184 bp) co-segregated with patients in this family. The fragments (325 bp), which are the parts of exon3 of the MYOC gene, were used as the internal control in the allele-specific PCR analysis. E: The sequence alignment portion of the dimerization domain spanning the p.T849 of the GUCD2Y of the human with other species.
Figure 2.
Electroretinography of the proband and a normal control. Electroretinography of the proband shows reduced photopic and 30 Hz responses and normal scotopic responses.
Figure 3.
Macular optical coherence tomography images from a visually normal subject and the proband of this family with cone dystrophy. A: The macular optical coherence tomography images of the right eye from a normal individual show organization of retinal microstructures with a well defined photoreceptor inner/outer segment layer and normal thickness (214 μm). B and C: The macular optical coherence tomography images of both eyes from the proband exhibit loss of inner/outer segment layer and thinning of the retina in the macular area (151 μm of the right eye, 153 μm of the left eye).
Table 1. Clinical features of the patients of this family with adCOD.
| Patient | Age | Best corrected visual acuity (R/L) | Onset age of photophobia | Night blindess | Refraction (diopters) | Fundus appearance | Color vision | ERG |
|---|---|---|---|---|---|---|---|---|
| II-1 |
56 |
0.1/0.2 |
EC |
NO |
−2.75–1.0X180, −2.25–0.75X180 |
RPE granular abnormalities at the fovea |
red-green defect |
N/A |
| II-4 |
53 |
0.1/01 |
EC |
NO |
−5.0–1.25X175,-4.5–1.50X180 |
RPE granular abnormalities at the fovea |
red-green defect |
N/A |
| III-6 | 28 | 0.5/0.5 | EC | NO | −0.5X180,-0.5X175 | RPE granular abnormalities at the fovea | red-green defect | reduction in cone responses and normal rod responses |
Abbreviations: R, right eye; L, left eye; EC, early childhood, N/A, data not available.
Genotyping results
This family was genotyped with 24 polymorphic markers around known adCOD and adCORD loci. The mapping results excluded the other known adCOD and adCORD loci with the exception of a locus, 17p13.1–2, where the GUCY2D, AIPL1, and PITPNM3 genes are found.
Mutation analysis
Sequencing of the three genes (GUCY2D, AIPL1, and PITPNM3) revealed one novel heterozygous mutation c.2545 A> G (p.T849A) in the GUCY2D gene. Using AS-PCR analysis, this mutation co-segregated with the adCOD phenotype in this family and was not detected in the unaffected members or 100 normal controls (Figure 1C,D).
In addition to the pathogenic mutation detected in the GUCY2D gene, six nonpathogenic sequence variants were also identified in this study. Table 2 summarizes these variants based on their nature.
Table 2. Presumed nonpathogenic variants found in this study.
| Gene | Exon | Nucleotide change | Codon | SNP |
|---|---|---|---|---|
| AIPL1 |
Exon3 |
c.276–10 A>C |
rs12453262 |
|
| c.300A>G |
p. L100L |
rs8075035 |
||
| PITPNM3 |
Exon4 |
IVS3+56G>T |
rs11656015 |
|
| Exon6 |
c.477C>T |
p.S159S |
rs145362623 |
|
| GUCY2D |
Exon3 |
c.741C>T |
p.H243H |
rs3829789 |
| Exon10 | c.2100C>T | p.P700P | rs34598902 |
Bioinformatics analysis
Using the Garnier-Osguthorpe-Robson method, the results of the secondary structure prediction suggested that the mutant GUCY2D 849A replaced one helix, “H,” with one β sheet, “E,” at position 852. Through PolyPhen-2 program analysis, p.T849A was predicted to be potentially damaging.
Discussion
In this study, we identified one novel missense mutation, p.T849A, in the GUCY2D gene in a small family with adCOD. The mutation co-segregated with the disease phenotype but was absent in the unaffected family members and 100 normal controls.
RetGC-1 is essential for the recovery of the dark state after the excitation process of the photoreceptor cells by light stimulation [6]. To date, more than 120 mutations of the GUCY2D gene have been identified as being responsible for retinal degeneration [2-5,7,8,12-23]. Most of them were found in the autosomal recessively inherited LCA [7,8]. The mutations found in COD and CORD were mainly clustered in codon 838 or the two adjacent codons, 837 and 839 [2-5,12-15]. Codon 838 is a mutational hot spot with five disease-causing sequence variations (R → C/G/H/P/S) [2-5,12-15,20]. The most frequent mutations, p.R838C and p.R838H, have been identified in different ethnicities, such as the Caucasian, Spanish, Japanese, and Chinese populations [2-5,12-15,18,22,23]. Unlike the mutations detected in LCA, which are mainly located in the catalytic and kinase-like domains of the RetGC-1 [7,8], most of the mutations identified in COD or CORD are located in the putative dimerization domain, which extends from amino acid 817 to 857 [2-5,12-15,17,18,22,23]. The Thr849 residue located in the dimerization domain is fully conserved in the different species (Figure 1E). The complex missense mutations, p.Q847L and p.K848Q, which were identified in a Japanese family with COD, are just adjacent to the novel mutation p.T849A [17]. Our results further confirm that the dimerization zone of RetGC-1 is the mutational hot region for COD or CORD, and a heterozygous mutation of GUCY2D not involving codon 838 can also be linked to COD and CORD.
In the clinical phenotype of the affected members of the family with the mutation p.T849A, the visual acuity of the proband was 0.5 with almost normal fundus. The male subject’s electroretinograms demonstrated reduced cone function and nearly normal rod function. Two elder patients (over 50 years old) had preserved peripheral visual fields and no complaints of night blindness. These findings are similar to the previous descriptions of the phenotypes associated with the mutations p.R838C and p.R838H [2-5]. Since the mutation p.R838C has been identified, the detailed clinical phenotypes with similar or different mutations have been reported by several studies [2-5,14,16,17,21,22]. Usually, patients with the mutations p.R838C and p.R838H have relatively similar clinical features of COD, which include the marked dysfunction of the cones from a young age while rod dysfunction appears later or does not present until a later stage [2-5,15,17,18,22,23].
In conclusion, we identified a novel mutation, p.T849A, in a Chinese family with COD. Our results further suggest that the dimerization zone of RetGC-1 is the mutational hot region for COD or CORD.
Acknowledgments
We thank the patients and their family for participating in this study. The study was supported by the National Natural Science Foundation of China (No 81170878).
Appendix 1.
Markers used in known autosomal dominant cone dystrophy and autosomal dominant rod dystrophy genotyping. To access the data, click or select the words “Appendix 1.”
Appendix 2.
Primer information for the AIPL1, PITPNM2, and GUCY2D gene sequence. To access the data, click or select the words “Appendix 2.”
References
- 1.Hamel CP. Cone rod dystophies. Orphanet J Rare Dis. 2007;2:7. doi: 10.1186/1750-1172-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelsell RE, Gregory-Evans K, Payne A, Perrault I, Kaplan J, Yang RB, Garbers DL, Bird AC, Moore AT, Hunt DM. Mutations in the retinal guanylate cyclase (RETGC-1) gene in dominant cone-rod dystrophy. Hum Mol Genet. 1998;7:1179–84. doi: 10.1093/hmg/7.7.1179. [DOI] [PubMed] [Google Scholar]
- 3.Gregory-Evans K, Kelsell R, Gregory-Evans C, Downes SM, Fitzke FM, Holder GR, Simunovic M, Mollon JD, Taylor R, Hunt DM, Bird AC, Moore AT. Autosomal dominant cone rod retinal dystrophy (CORD6) from heterozygous mutation of GUCY2D, which encodes retinal guanylate cyclase. Ophthalmology. 2000;107:55–61. doi: 10.1016/s0161-6420(99)00038-x. [DOI] [PubMed] [Google Scholar]
- 4.Payne AM, Morris AG, Downes SM, Johnson S, Bird AC, Moore AT, Bhattacharya SS, Hunt DM. Clustering and, frequency of mutations in the retinal guanylate cyclase (GUCY2D) gene in patients with dominant cone-rod dystrophies. J Med Genet. 2001;38:611–4. doi: 10.1136/jmg.38.9.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kitiratschky VB, Wilke R, Renner AB, Kellner U, Vadala M, Birch DG, Wissinger B, Zrenner E, Kohl S. Mutation analysis identifies GUCY2D as the major gene responsible for autosomal dominant progressive cone degeneration. Invest Ophthalmol Vis Sci. 2008;49:5015–23. doi: 10.1167/iovs.08-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunt DM, Buch P, Michaelides M. Guanylate cyclases and associated activator proteins in retinal disease. Mol Cell Biochem. 2010;334:157–68. doi: 10.1007/s11010-009-0331-y. [DOI] [PubMed] [Google Scholar]
- 7.Cremers FPM, van den Hurk JAJM, den Hollander AI. Molecular genetics of Leber congenital amaurosis. Hum Mol Genet. 2002;11:1169–76. doi: 10.1093/hmg/11.10.1169. [DOI] [PubMed] [Google Scholar]
- 8.den Hollander AI, Roepman R, Koenekoop RK, Cremers FPM. Leber congenital amaurosis: Genes, proteins and disease mechanisms. Prog Retin Eye Res. 2008;27:391–419. doi: 10.1016/j.preteyeres.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 9.Ruano G, Kidd KK. Direct haplotying of chromosomal segments from multiple heterozygous via allele-specific PCR amplification. Nucleic Acids Res. 1989;17:8392. doi: 10.1093/nar/17.20.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garnier J, Gibrat JF, Robson B. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 1996;266:540–53. doi: 10.1016/s0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]
- 11.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weigell-Weber M, Fokstuen S, Torok B, Niemeyer G, Schinzel A, Hergersberg M. Codons 837 and 838 in the retinal guanylate cyclase gene on chromosome 17p: hot spots for mutations in autosomal dominant cone-rod dystrophy? Arch Ophthalmol. 2000;118:300. doi: 10.1001/archopht.118.2.300. [DOI] [PubMed] [Google Scholar]
- 13.Downes SM, Payne AM. Kelsel REl, Fitzke FW, Holder GE, Hunt DM, Moore AT, Bird AC. Autosomal Dominant Cone-Rod dystrophy with mutations in the guanylate cyclase 2D gene encoding Retinal Guanylate Cyclase-1. Arch Ophthalmol. 2001;119:1667–73. doi: 10.1001/archopht.119.11.1667. [DOI] [PubMed] [Google Scholar]
- 14.Udar N, Yelchits S, Chalukya M, Yellore V, Nusinowitz S, Silva-Garcia R, Vrabec T, Hussles Maumenee I, Donoso L, Small KW. Identification of GUCY2D gene mutations in CORD5 families and evidence of incomplete penetrance. Hum Mutat. 2003;21:170–1. doi: 10.1002/humu.9109. [DOI] [PubMed] [Google Scholar]
- 15.Ito S, Nakamura M, Nuno Y, Ohnishi Y, Nishida T, Miyake Y. Autosomal dominant cone-rod dystrophy with R838H and R838C mutations in the GUCY2D gene in Japanese patients. Jpn J Ophthalmol. 2004;48:228–35. doi: 10.1007/s10384-003-0050-y. [DOI] [PubMed] [Google Scholar]
- 16.Ito S, Nakamura M, Nuno Y, Ohnishi Y, Nishida T, Miyake Y. Novel complex GUCY2D mutation in Japanese family with cone-rod dystrophy. Invest Ophthalmol Vis Sci. 2004;45:1480–5. doi: 10.1167/iovs.03-0315. [DOI] [PubMed] [Google Scholar]
- 17.Yoshida S, Yamaji Y, Yoshida A, Kuwahara R, Yamamoto K, Kubata T, Ishibashi T. Novel triple missense mutations of GUCY2D gene in Japanese family with cone-rod dystrophy: possible use of genotyping microarray. Mol Vis. 2006;12:1558–64. [PubMed] [Google Scholar]
- 18.Smith M, Whittock N, Searle A, Croft M, Brewer C, Cole M. Phenotype of autosomal dominant cone-rod dystrophy due to the R838C mutation of the GUCY2D gene encoding retinal guanylate cyclase-1. Eye (Lond) 2007;21:1220–5. doi: 10.1038/sj.eye.6702612. [DOI] [PubMed] [Google Scholar]
- 19.Small KW, Silva-Garcia R, Udar N, Nguyen EV, Heckenlively JR. New mutation, P575L, in the GUCY2D gene in a family with autosomal dominant progressive cone degeneration. Arch Ophthalmol. 2008;126:397–403. doi: 10.1001/archopht.126.3.397. [DOI] [PubMed] [Google Scholar]
- 20.Auz-Alexandre CL, Vallespin E, Aguirre-Lamban J, Cantalapiedra D, Avila-Fernandez A, Villaverde-Montero C, Ainse E, Trujillo-Tiebas MJ, Ayuso C. Novel human pathological mutations. Gene symbol: GUCY2D. Disease: Leber congenital amaurosis. Hum Genet. 2009;125:349. [PubMed] [Google Scholar]
- 21.Ugur Iseri SA, Durlu YK, Tolun A. A novel recessive GUCY2D mutation causing cone-rod dystrophy and not Leber's congenital amaurosis. Eur J Hum Genet. 2010;18:1121–6. doi: 10.1038/ejhg.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Hoyos M, Auz-Alexandre CL, Almoguera B, Cantalapiedra D, Riveiro-Alvarez R, Lopez-Martinez MA, Gimenez A, Blanco-Kelly F, Avila-Fernandez A. Trujillo- Tiebas MJ, Garcia-Sandoval B, Ramos C, Ayuso C. Mutation analysis at codon 838 of the Guanylate Cyclase 2D gene in Spanish families with autosomal dominant cone, cone-rod, and macular dystrophies. Mol Vis. 2011;17:1103–9. [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao X, Guo X, Jia X, Li S, Wang P, Zhang Q. A recurrent mutation in GUCY2D associated with autosomal dominant cone dystrophy in a Chinese family. Mol Vis. 2011;17:3271–8. [PMC free article] [PubMed] [Google Scholar]