Background: Mechanism of CD36 inhibition by sulfo-N-succinimidyl oleate (SSO) was explored using mass spectrometry and mutagenesis.
Results: SSO binds lysine 164 and inhibits uptake of fatty acids and oxLDL.
Conclusion: Lysine 164 is important for CD36-mediated fatty acid uptake and Ca2+ signaling.
Significance: Fatty acids and oxLDL bind to the same site within a hydrophobic pocket of CD36 shared by several lipid ligands.
Keywords: Fatty Acid, Glycerophospholipid, Lipids, Lipid Transport, Metabolic Regulation, CD36, SSO, Fatty Acid Uptake
Abstract
FAT/CD36 is a multifunctional glycoprotein that facilitates long-chain fatty acid (FA) uptake by cardiomyocytes and adipocytes and uptake of oxidized low density lipoproteins (oxLDL) by macrophages. CD36 also mediates FA-induced signaling to increase intracellular calcium in various cell types. The membrane-impermeable sulfo-N-hydroxysuccinimidyl (NHS) ester of oleate (SSO) irreversibly binds CD36 and has been widely used to inhibit CD36-dependent FA uptake and signaling to calcium. The inhibition mechanism and whether SSO modification of CD36 involves the FA-binding site remain unexplored. CHO cells expressing human CD36 were SSO-treated, and the protein was pulled down, deglycosylated, and resolved by electrophoresis. The CD36 band was extracted from the gel and digested for analysis by mass spectrometry. NHS derivatives react with primary or secondary amines on proteins to yield stable amide or imide bonds. Two oleoylated peptides, found only in SSO-treated samples, were identified with high contribution and confidence scores as carrying oleate modification of Lys-164. Lysine 164 lies within a predicted CD36 binding domain for FA and oxLDL. CHO cells expressing CD36 with mutated Lys-164 had impaired CD36 function in FA uptake and FA-induced calcium release from the endoplasmic reticulum, supporting the importance of Lys-164 for both FA effects. Furthermore, consistent with the importance of Lys-164 for oxLDL binding, SSO inhibited oxLDL uptake by macrophages. In conclusion, SSO accesses Lys-164 in the FA-binding site on CD36, and initial modeling of this site is presented. The data suggest competition between FA and oxLDL for access to the CD36 binding pocket.
Introduction
FAT/CD36 is a multifunctional transmembrane glycoprotein homologous to the scavenger receptor class B, type I. CD36 facilitates uptake of long chain fatty acids (FA)2 by heart and adipose tissues in rodents (1, 2) and humans (3–5). The protein is also a receptor for oxidized low density lipoproteins (oxLDL) (6, 7), thrombospondin and collagen (8), hexarelin (9), apoptotic cells (10), and erythrocytes infected with Plasmodium falciparum (11). CD36 has signal transduction capabilities and is involved in a number of pathways related to immune responses, inflammation, and the development of atherosclerosis (12). CD36 was recently implicated in the regulation of store-operated calcium channels, phospholipase activation, and eicosanoid production (13). These pathways mediate taste perception of dietary fatty acids (14, 15) and FA-induced release of gut peptides (16). In humans, variants in the CD36 gene associate with sensitivity to fat taste perception (17), with blood lipids (18) and platelet reactivity (19). CD36 variants also influence susceptibility to complications of obesity such as the metabolic syndrome, stroke, etc. (20).
Our understanding of the structure-function relationships of CD36 remains limited. There is no crystal structure available, but the protein is predicted to have a large, heavily glycosylated extracellular domain and two transmembrane spanning segments. Three disulfide bridges are present in the carboxyl-terminal half of the protein. The amino-terminal half is proposed to contain binding domains for hexarelin (9), fatty acids (21), oxidized LDL or phospholipids (6), thrombospondin (8), and P. falciparum-infected erythrocytes (11). A hydrophobic sequence (residues 186–204) that might be part of a binding pocket is present in the amino-terminal half and may dip into the outer leaflet of the membrane (1, 2). The cytoplasmic tails of the protein are short but active in signal transduction via association to a number of tyrosine kinases (13, 22, 23). Several post-translational modifications, including phosphorylation, glycosylation, palmitoylation, or ubiquitination modulate levels or trafficking of CD36 and consequently its metabolic functions (24, 25).
CD36 function in FA uptake and signaling can be irreversibly inhibited by NHS esters of long-chain FA. Esters of palmitate, myristate, and oleate were shown as effective inhibitors of FA uptake and were used to identify CD36 as a membrane FA receptor (26). The most widely used derivative is the oleate ester or SSO, which inhibits CD36-mediated FA uptake (27–30) and FA signaling (13–16, 31, 32) in a variety of cell types. The molecular mechanism underlying the inhibitory effect of SSO remains unclear and understanding it would provide insight into the structural determinants of CD36 function. N-Hydroxysuccinimidyl derivatives, which are widely used as protein labeling or cross-linking reagents (33–35), react with primary or secondary amines (nucleophiles) in neutral pH to yield stable amide or imide bond and NHS. Primary amino-terminal and ϵ-amino groups in lysine side chains of proteins are the preferred targets. Sulfo-NHS esters contain a negatively charged sulfonate group on the N-hydroxysuccinimide ring, which increases water solubility and restricts membrane permeability without affecting reactivity.
In this study, we identify the target of SSO as lysine 164 situated within a binding pocket shared by other CD36 lipid ligands, notably oxLDL. We document the importance of Lys-164 for CD36-mediated FA uptake and FA-induced signaling.
EXPERIMENTAL PROCEDURES
Materials
Mini protease inhibitor mixture tablets (cOmplete) were from Roche Applied Science, FLAG immunoprecipitation kit, fatty acid-free bovine serum albumin fraction IV (BSA), and other chemicals from Sigma. SSO was synthesized as described previously (36), and purity was determined to be greater than 99% by NMR.
Cell Culture and Transfection
Chinese hamster ovary (CHO) cells (American Type Culture Collection, Manassas, VA) were maintained in Ham's F-12 medium containing 10% fetal bovine serum and penicillin/streptomycin. Carboxyl-terminal FLAG-tagged human CD36 was cloned in the pcDNA3 expression vector (37). For generating CD36 with mutated lysine 164, site-directed mutagenesis was used to substitute Lys-164 with alanine, and the mutant cDNAs were verified by sequencing. CHO cells were transfected using Lipofectamine 2000 (Invitrogen), and positive clones expressing wild type (CD36WT) or alanine 164 CD36 (CD36K164A) were selected in hygromycin (50 μg/ml) as described previously (13, 37). THP-1 (ATCC) monocyte cells were maintained in RPMI 1640 medium containing 10% FBS, 0.05 mm 2-mercaptoethanol, and penicillin/streptomycin. Macrophage differentiation was induced by phorbol 12-myristate 13-acetate (PMA, 50 ng/μl) for 24 h.
Purification and Deglycosylation of SSO-labeled CD36
CHO cells were grown to confluency in 100-mm dishes, washed three times with phosphate-buffered saline (PBS: 138 NaCl, 5.4 KCl, 7 Na2HPO4, 1.4 KH2PO4, 5.5 glucose in mm, pH 7.4), and incubated with or without 100 μm SSO in PBS for 30 min on ice. SSO was freshly dissolved in DMSO and mixed with PBS just before application onto cells. Labeling was quenched by F-12 medium containing 0.2% BSA, washed three times with ice-cold PBS, and the cells were scraped into lysis buffer (PBS, 0.1% octylglucoside, protease inhibitors), homogenized on ice by sonication (10 cycles, amplitude 60, Hielscher, Ultrasound Technology, Germany), left at 4 °C for 30 min, centrifuged (12,000 × g, 10 min, 4 °C), and the supernatant harvested and incubated with FLAG immunoprecipitation slurry overnight. The resin was washed five times with PBS containing 0.01% octyl glucoside to remove nonspecific protein binding, and CD36 was eluted using FLAG peptide. The eluted samples were denatured (70 °C, 10 min) and deglycosylated (37 °C, 60 min) with peptide:N-glycosidase F (New England Biolabs), and sample volume was reduced to 30 μl (SpeedVac) before SDS-PAGE and Coomassie staining.
Mass Spectrometry of CD36
The protein bands were excised from the gel, reduced with DTT, and alkylated with iodoacetamide before digestion with chymotrypsin overnight. Analysis of samples dissolved in 0.1% formic acid was performed on UltiMate 3000 RSLCnano system (Dionex/Thermo Scientific) coupled to a TripleTOF 5600 mass spectrometer with a NanoSpray III source (AB Sciex, Framingham, MA). The instrument was operated with Analyst TF 1.6 (AB Sciex). After injection, the samples were trapped and desalted with 2% acetonitrile in 0.1% formic acid at flow rate of 5 μl/min on Acclaim® PepMap100 column (5 μm, 2 cm × 100 μm inner diameter, Thermo Scientific). Eluted peptides were separated using Acclaim® PepMap100 analytical column (3 μm, 15 cm × 75 μm inner diameter, Thermo Scientific). The 60-min elution gradient at a constant flow of 300 nl/min was set to 5% of phase B (0.1% formic acid, 99.9% acetonitrile; phase A, 0.1% formic acid) for the first 10 min, then stepped from 5 to 30% B over 25 min, from 30 to 50% B over 5 min, from 50 to 90% B over 2 min, from 90 to 5% over 9 min, and then remaining at 5% B for 9 min. An information-dependent acquisition method was utilized with total cycle time of 2.3 s. Maximum 25 MS/MS spectra per cycle were acquired, and former target ions were excluded for 15 s after two occurrences. TOF MS mass range was set to 350–1500 m/z. In the MS/MS mode, the instrument acquired fragmentation spectra with m/z ranging from 100 to 2000. Data files were analyzed using the ProteinPilot 4.5beta software (38) (revision 1656, Paragon Algorithm, 4.5.0.0.1654, AB Sciex) and plotted using mMass 5.3.0 (39). The following sample parameters were used: detected protein threshold 0.05; ProtScore 2; identification; iodoacetamide cysteine alkylation; chymotrypsin digestion; biological modifications and amino acid substitutions; thorough search effort; acetylation emphasis; database UniProtKB 2013_03, March 6, 2013, and custom database, UniProt P16671 CD36_HUMAN sequence prolonged with FLAG tag -DYKDDDK). Custom modification of the software was made to search for oleoylated peptides based on the manufacturer's guidelines (new biological modification, target, lysine; TS, 255; Fma, C18H33O; RpF, H; Chg, 0; Prob, 0.1).
Biotinylation of Cell Surface Proteins
Cell surface proteins were labeled with biotin using EZ-Link Sulfo-NHS-LC-Biotin (Pierce) and purified as described (37). The amounts of CD36 protein on the cell surface and in total cell lysates were evaluated by immunoblots.
Fatty Acid Uptake and SSO Treatment
Uptake of [3H]oleate or [3H]palmitate was measured as before (37).
Intracellular Calcium Transients
Calcium transients were measured as described previously (13). Briefly, cells were loaded with the FURA-2AM Ca2+ dye and exposed to 30 μm linoleic acid complexed to BSA (4:1), and the cell response was monitored by microscopy (Cell∧R imaging system based on an Olympus IX-81 inverted microscope).
Oxidized LDL Uptake
THP-1 monocytes were differentiated into macrophages for 24 h using 50 ng/μl PMA and switched back to growth medium following differentiation. Two days later, the cells were serum-starved overnight before incubation with or without 25 μm SSO for 15 min at 37 °C in HEPES/saline buffer (20 HEPES, 138 NaCl, 1 MgCl2, 6 KCl in mm, pH 7.4). This was followed by washing and then by incubation with 50 μg/ml Dil-OxLDL (Kalen Biomedical, Montgomery Village, MD) for 2 h at 37 °C. Cells were harvested using a nonenzymatic cell dissociation solution (Sigma), stained with allophycocyanin-labeled anti-CD36 antibody (Clone CB38, BD Biosciences), and fixed with formaldehyde. Fixed cells were kept in the dark at 4 °C until assayed by flow cytometry (FACSCalibur, BD Biosciences), and results were analyzed by FlowJo (Tree Star, OR).
Bioinformatics
Statistical analysis used SigmaStat, and a p ≤ 0.05 was considered significant. Prediction of the secondary structure of CD36 (UniProt P16671) was performed using the I-TASSER platform (40).
RESULTS
Identification of the Binding Site of SSO in CD36
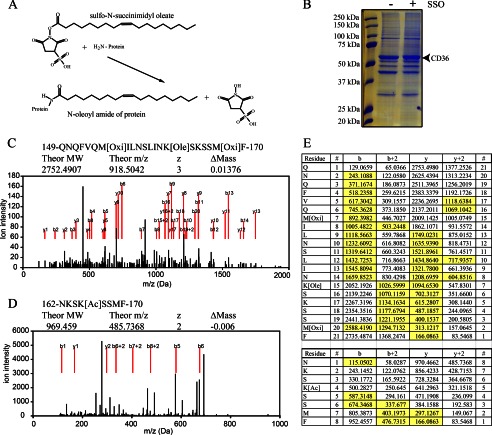
Harmon and Abumrad (26) showed that CD36 is irreversibly inhibited by a short treatment with SSO, and this associates with covalent binding of [3H]oleate to the CD36 protein. Based on the known reactivity of NHS-derived reagents (35), the expected covalent modification is an N-oleoyl amide of free amine groups on CD36 (Fig. 1A).
FIGURE 1.
Identification of SSO-binding site in CD36. A, schematic representation of the SSO protein labeling reaction showing that when the sulfo-N-succinimidyl oleate ester reacts with a primary amine group on a membrane protein, an N-oleoyl amide of the target protein is formed together with the NHS group. B, Coomassie-stained gel showing the deglycosylated CD36 bands at about 53 kDa. CHO cells stably expressing wild type FLAG-tagged CD36 (CD36WT) were used. The cells were labeled with 100 μm SSO as indicated, and the CD36 protein was immunoprecipitated, deglycosylated, and resolved on a gel. The CD36 bands were extracted and subjected to mass spectrometry. C, mass spectrometry evidence of oleoylation of lysine 164. CD36 protein chymotryptic fragments were ionized into y, y2+, b, and b2+ series with m/z values detected by MS. y and b series are marked with red lines. Modification by SSO (Ole) was detected together with oxidation of methionines (Oxi), caused by ionization. Spectrum of oleoylated peptide is shown. D, mass spectrometry evidence of acetylation (Ac) of lysine 166, details as above. E, sequence of the fragments and complete b and y series ions generated by the ionization. Oleoylation of Lys-164, top table; acetylation of Lys-166, bottom table. Detected and assigned ions are highlighted in yellow.
CHO cells stably expressing human CD36, FLAG-tagged at the carboxyl terminus (CD36WT), were previously used to study CD36 functions in FA uptake (37) or signal transduction (13), and SSO treatment of these cells was effective in inhibiting the CD36-specific effects. CHO cells (CD36WT) were used for purification of SSO-labeled CD36. Confluent cells were treated with 100 μm SSO in PBS for 30 min on ice and lysed, and the CD36 protein was pulled down using FLAG immunoprecipitation. Concentrated and purified lysates were treated with peptide:N-glycosidase F to cleave N-linked oligosaccharides to decrease the complexity of the mixture created during fragmentation in mass spectrometry. The deglycosylated lysate was separated on SDS-PAGE and stained with Coomassie (Fig. 1B), and a distinct band at about 53 kDa corresponding to deglycosylated CD36 was extracted from the gel and analyzed on nano-liquid chromatography-MS/MS.
Complete chymotryptic digests of control and SSO-labeled CD36 were analyzed using mass spectrometry. Data files were searched with ProteinPilot software (38). A total of 290 unique CD36 peptides were identified with greater than 98% confidence and were scanned for N-amide oleate modification. Oleoylated peptides were only found in SSO-treated samples (Table 1). Two overlapping peptides, with high contribution (unique) and high confidence scores, were identified as carriers of oleate modification on lysine 164. Two other overlapping peptides covering a sequence containing lysine 334 were identified but with low contribution and confidence scores. The MS spectrum and table of fragments of the modified peptides are presented in Fig. 1, C and E.
TABLE 1.
SSO-modified CD36 peptides
Chymotryptic digests of CD36 from control and SSO-treated cells were analyzed using mass spectrometry and ProteinPilot software. Peptides with an oleate attached to a side chain of an amino acid residue of the CD36 backbone were found only in the SSO-treated samples. Two overlapping peptides, identified with high confidence, had the oleate attached to lysine 164. Two other peptides, mapping lysine 334 as the SSO target, were identified with lower confidence. Contribution scores range from 0 to 2, and confidence scores are shown in percent.
| Contribution <0–2> | Confidence (%) | Peptide sequence | Peptide lysine | CD36 residue | Additional peptide modifications |
|---|---|---|---|---|---|
| 1.96 | 99.00 | QNQFVQMILNSLINKSKSSMF | Lys-15 | Lys-164 | Oxidation Met-7 and Met-20 |
| 1.82 | 99.00 | NLAVAAASHIYQNQFVQMILNSLINKSKSSMF | Lys-26 | Lys-164 | Oxidation Met-18 and Met-31 |
| 0.89 | 95.31 | DISKCKEGRPVY | Lys-6 | Lys-334 | Carbamidomethyl C5 |
| 0.00 | 59.59 | KEGRPVY | Lys-1 | Lys-334 | None |
In addition to oleoylation sites, MS identified acetylation of CD36 lysines 52, 166, 231, and 403. Mass spectrum and table of fragments are presented for a peptide acetylated at Lys-166 in Fig. 1, D and E. Lysines 52, 166, 231, and 403 were previously shown to be acetylated in mouse and rat CD36 (41), so our data document that this is conserved for human CD36.
Mutation of Lysine 164 Impairs the Long-chain FA Transport Function of CD36
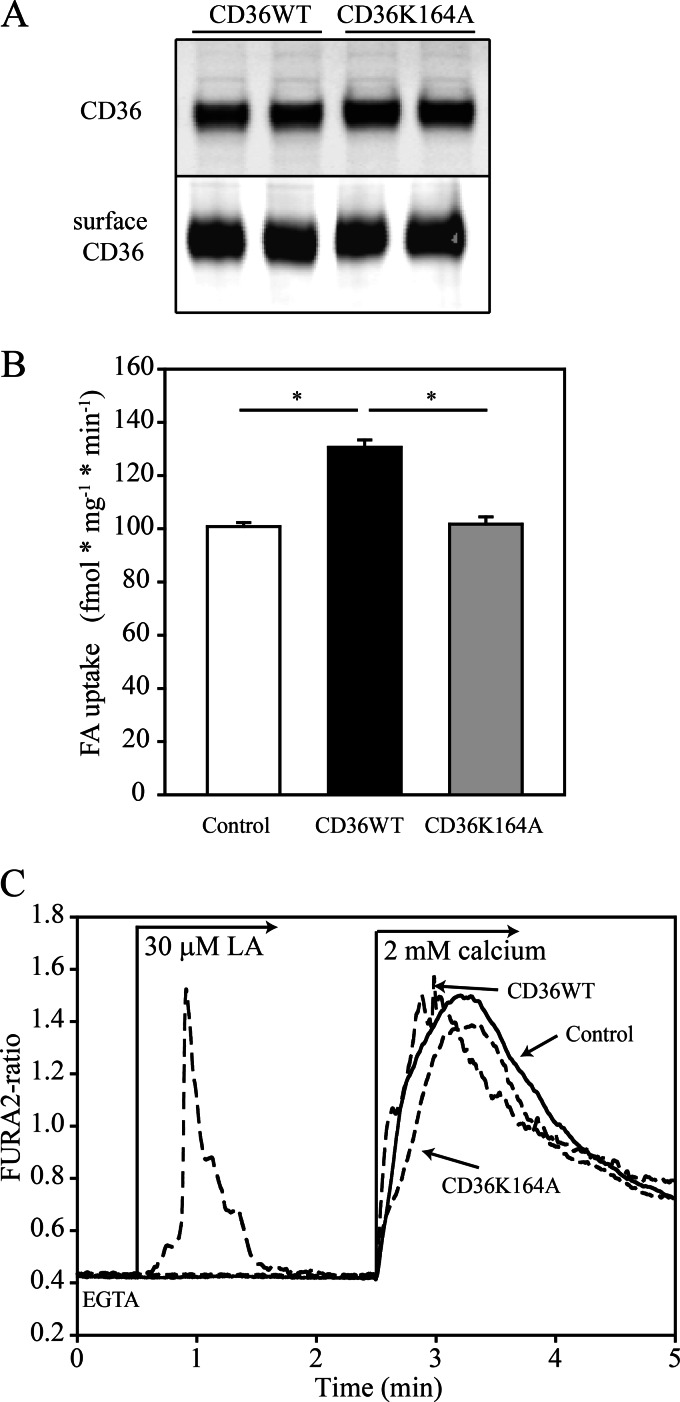
The two overlapping peptides oleoylated at lysine 164 that were identified with high confidence scores lie within the CD36 region previously predicted to contain a binding domain for fatty acids, based on simulation to the structure of the FA-binding protein (21). Lysine 164 was chosen for further characterization, and CHO cells stably expressing wild type or mutated CD36, where lysine 164 was substituted by alanine, were generated. The mutation did not interfere with CD36 expression or surface localization because total CD36 level and CD36 membrane content, analyzed by biotinylation, were similar for CD36WT and CD36K164A cells (Fig. 2A).
FIGURE 2.
Fatty acid uptake and FA-induced Ca2+ signaling are impaired in the CD36 K164A mutant CD36 protein. A, surface expression of wild type (CD36WT) and CD36 K164A expressed in CHO cells. Upper panel, total CD36; lower panel, surface-exposed CD36. CHO cells stably expressing FLAG-tagged CD36WT, mutated CD36 with alanine substitution at lysine 164, or transfected with an empty vector were labeled on ice with biotin and lysed, and CD36 was immunoprecipitated using FLAG. B, fatty acid uptake. CHO cells were serum-starved overnight (16 h) and [3H]palmitate uptake was measured for 4 min at room temperature. Data are representative of five different experiments ± S.E., *, p < 0.05. C, representative FA-induced release of intracellular Ca2+. CHO cells were treated with 30 μm linoleic acid (LA) in the absence of extracellular Ca2+ and then Ca2+ was added to the medium, as indicated. Only CD36WT cells were able to recognize linoleic acid and trigger endoplasmic reticulum release of Ca2+. The data are representative of three different experiments.
CHO cells expressing CD36WT, CD36K164A, or empty vector were serum-starved overnight and assayed for uptake of [3H]palmitate. A time course of the uptake was determined (data not shown), and 4-min incubations were selected from the linear part of the time course for comparison of uptake rates. Palmitate uptake was enhanced in CD36-expressing cells by ∼25% in agreement with previous data (37). This enhancement is reproducible but modest reflecting the limited capacity of CHO cells for lipid accumulation due to lack of key proteins of triglyceride turnover (42, 43). The enhancement of FA uptake resulting from CD36 expression was absent in CD36K164A cells where uptake was comparable with cells transfected with empty vector (Fig. 2B). Similar results were obtained with oleate (data not shown).
Mutation of Lysine 164 Impairs the Signaling Function of CD36
Long chain polyunsaturated FA interaction with CD36 induces an increase in cytosolic Ca2+ from intracellular stores that is inhibited by SSO (14, 16). We tested whether the K164A mutation modulates the ability of CD36 to mediate the effect of FA to signal for an increase in cytosolic Ca2+. CHO cells were loaded with Ca2+-sensitive dye and Ca2+ transients monitored after stimulation with linoleic acid. Linoleic acid addition to CD36WT cells maintained in the absence of medium Ca2+ induced a rapid and transient rise in intracellular Ca2+ that was completely blunted in CD36K164A cells (Fig. 2C). However, Ca2+ influx into the cell after addition of Ca2+ to the extracellular medium was similar in both cell types indicating that Lys-164 is critical for CD36-mediated FA-induced release of intracellular Ca2+.
SSO Inhibits Oxidized LDL Uptake by Macrophages
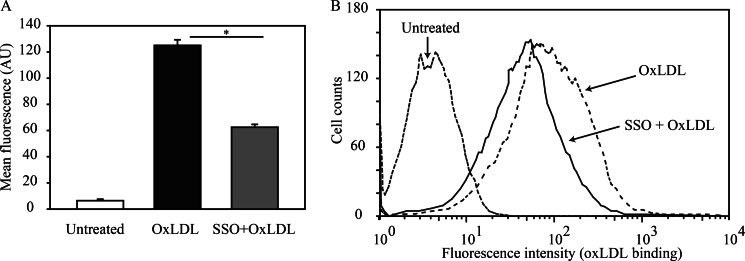
It was previously shown that neutralization of Lys-164 in CD36 reduces the protein's ability to recognize oxidized LDL and oxidized phosphatidylcholine (oxPC) (6). Because SSO was identified to bind to Lys-164, we examined whether it would inhibit oxLDL uptake, which to our knowledge had not been tested before. THP-1 macrophages were pretreated for 15 min with 25 μm SSO, washed, and assayed for uptake of Dil-OxLDL by flow cytometry. Under these conditions, SSO inhibited oxLDL uptake by macrophages by about 50% as shown in Fig. 3 (A and B).
FIGURE 3.
SSO inhibits uptake of oxLDL by macrophages. A, uptake of Dil-OxLDL by THP-1 macrophages was determined by flow cytometry. B, histogram of cells sorted for fluorescence using the FlowJo software. THP-1 monocytes were differentiated into macrophages using PMA. The cells were serum-starved overnight before incubation with or without 25 μm SSO followed by washing and before incubation with Dil-OxLDL. Harvested cells were stained with allophycocyanin-labeled anti-CD36 antibody and measured by flow cytometry. Data are means ± S.E., n = 3–4, representative of two separate experiments; *, p < 0.05.
DISCUSSION
This study identified the amino acid residue targeted by the CD36 inhibitor SSO and documented its functional relevance to FA uptake and FA signaling to Ca2+. This residue was previously implicated in oxLDL binding, and we showed that SSO also inhibits oxLDL uptake. The findings support FA and oxLDL binding to the same site on CD36 and the potential of direct competition between the two ligands.
The NHS ester of oleic acid SSO was generated by Harmon et al. (26, 36) to test the possibility that a plasma membrane protein facilitates FA uptake by adipocytes. SSO inhibited FA uptake and bound covalently to an 88-kDa protein identified as CD36. Inhibition by SSO of adipocyte FA uptake occurred in absence of FA metabolism and without effects on the uptake of other substrates such as retinoic acid or glucose (36). Furthermore, SSO retained its ability to inhibit long-chain FA uptake by cardiac giant membrane vesicles (27). These findings resulted in the use of SSO in a wide variety of cell types as a specific inhibitor of CD36-facilitated FA uptake (27, 28, 44, 45). More recently, SSO was shown to also potently inhibit CD36-mediated FA signaling to calcium (13–16). The findings in this study validate the assumption that SSO is a specific inhibitor of FA interaction with CD36 by documenting that SSO binding to a specific residue on CD36 is the molecular mechanism underlying its inhibitory effects. Using mass spectrometry, we identified lysine 164 as the major amino acid targeted by SSO. Site-specific mutation and stable expression in CHO cells showed that lysine 164 is critical for the functions of CD36 in facilitating FA uptake and FA-induced signaling to activate Ca2+ transients. Lysine 164 is localized within a putative binding pocket in the amino-terminal half of CD36 where binding sequences for oxLDL and oxPC have been previously identified (6). A sequence for binding FA had been predicted within the same pocket based on simulation analysis with the FA-binding site of the fatty acid-binding protein for which the crystal structure was available (21).
Lysines are positively charged residues suggesting that electrostatic interactions might play a role in FA binding by CD36. The negatively charged long-chain FA carboxylic group would be a preferential partner for such interactions. When oleate is esterified to sulfo-NHS, the resulting SSO molecule gets a highly charged negative sulfonate group. This combined with the uncharged alkyl chain would make SSO an excellent ligand for the CD36 binding pocket. Based on the preponderance of hydrophobic residues in the binding pocket, we propose a model whereby the alkyl chain plays an important role in directing the FA to the binding site in CD36. This interpretation is consistent with our earlier finding that a shorter chain NHS derivative such as sulfo-N-succinimidyl myristate bound to several proteins in adipocyte membranes, whereas SSO was recovered predominantly on CD36 (26, 36). Once the alkyl chain positions SSO into the hydrophobic pocket, the negative sulfonate-containing headgroup can interact with positively charged amines, notably with the Lys-164 amine group to yield an amide bond. In the case of the native FA positioned in the pocket, its carboxyl group would form electrostatic interactions with Lys-164, which would stabilize the binding and possibly initiate a conformational change in the protein to promote FA uptake and or FA-induced signaling. Hydrophobic interactions contributed by the alkyl chain and electrostatic interactions involving the charged carboxyl terminus of the FA and lysine residues of the protein have been described for FA binding to albumin (46, 47). FA interaction with lysines in the binding pocket of the intestinal FA-binding protein was shown to influence FA transfer from intestinal fatty acid-binding protein to model membranes (48, 49).
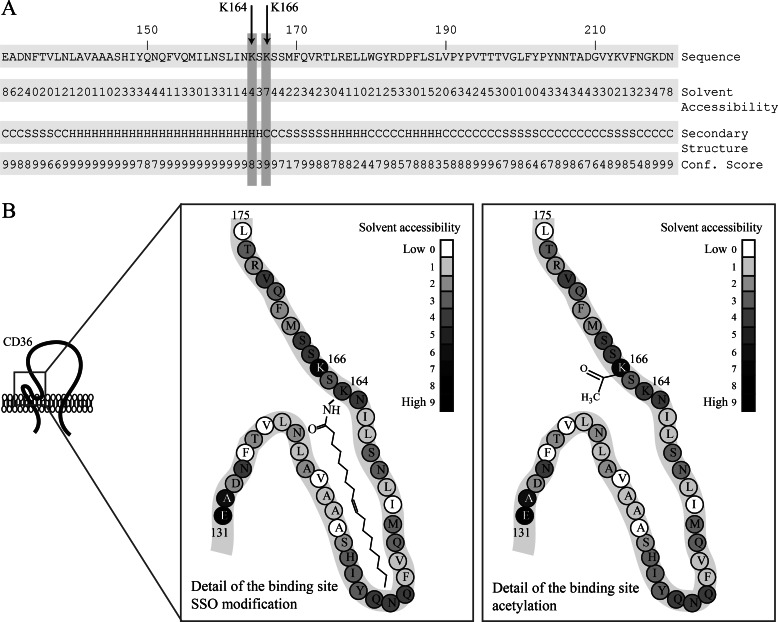
The binding pocket of CD36 contains two lysines, Lys-164 and Lys-166, but SSO was only recovered bound to Lys-164. We believe this reflects the fact that Lys-166 is acetylated, which neutralizes its charge. Indeed, mass spectral analysis documented acetylation of four different lysines in CD36, including Lys-166 (Fig. 4). These data with human CD36 are in agreement with the previously reported acetylation of rat CD36 (41). Acetylation of Lys-166 increases hydrophobicity of the binding pocket and might contribute to positioning the FA by limiting the interaction of the carboxyl group to Lys-164. It is unknown whether acetylation of Lys-166 is subject to regulation and whether its deacetylation can occur under certain conditions and influence FA binding. Acetylation of proteins, notably those involved in metabolism of glucose and fatty acids, has been shown to be a common mode of regulating function (41, 50).
FIGURE 4.
Structural prediction of the SSO-binding site in CD36. A, prediction of the secondary structure of the CD36 sequence (UniProt P16671) containing the SSO-binding site using the I-TASSER platform. Predicted secondary structures: C, coil; S, sheet; H, helix; confidence scores range from 0 (low) to 10 (high). Predicted solvent accessibility of amino acid residues: 0 (buried) 9 (exposed). B, schematic model of the CD36-binding site for long-chain FA. Lysine 166 is predicted to be solvent-exposed, although Lys-164 is predicted to be less solvent-accessible. Gray scale is based on I-TASSER predictions. Lys-164 can be modified by SSO to yield N-oleoyl amide, which occupies the putative hydrophobic pocket (left panel). Lys-166 can be acetylated, and this modification might modify accessibility of the pocket (right panel).
Secondary structure prediction using the I-TASSER platform (40) suggests that CD36 lysines 164 and 166 are localized on a turn between helical and sheet structures on the edge of the hydrophobic pocket. Although Lys-164 is predicted to be rather hidden inside the structure (Fig. 4B), Lys-166 would be exposed to solvent and could act in fatty acyl docking into the hydrophobic pocket. Schematic representation of the binding site shows that Lys-166 and its modification by acetylation could control ligand access to the binding site/pocket (Fig. 4B). We were not able to detect peptides concurrently acetylated on Lys-166 and oleoylated on Lys-164, suggesting that Lys-166 acetylation might restrict access to the binding site.
The FA-binding site in CD36 overlaps with that of oxLDL and of oxPC. Interestingly, binding for both oxLDL and oxPC was proposed to involve interactions with lysines 164 and 166 (6, 51). Consistent with the above data, we found that a short incubation with a low concentration of SSO was effective in inhibiting oxLDL uptake by macrophages. Thus, SSO might be useful as an inhibitor of CD36-mediated oxLDL uptake.
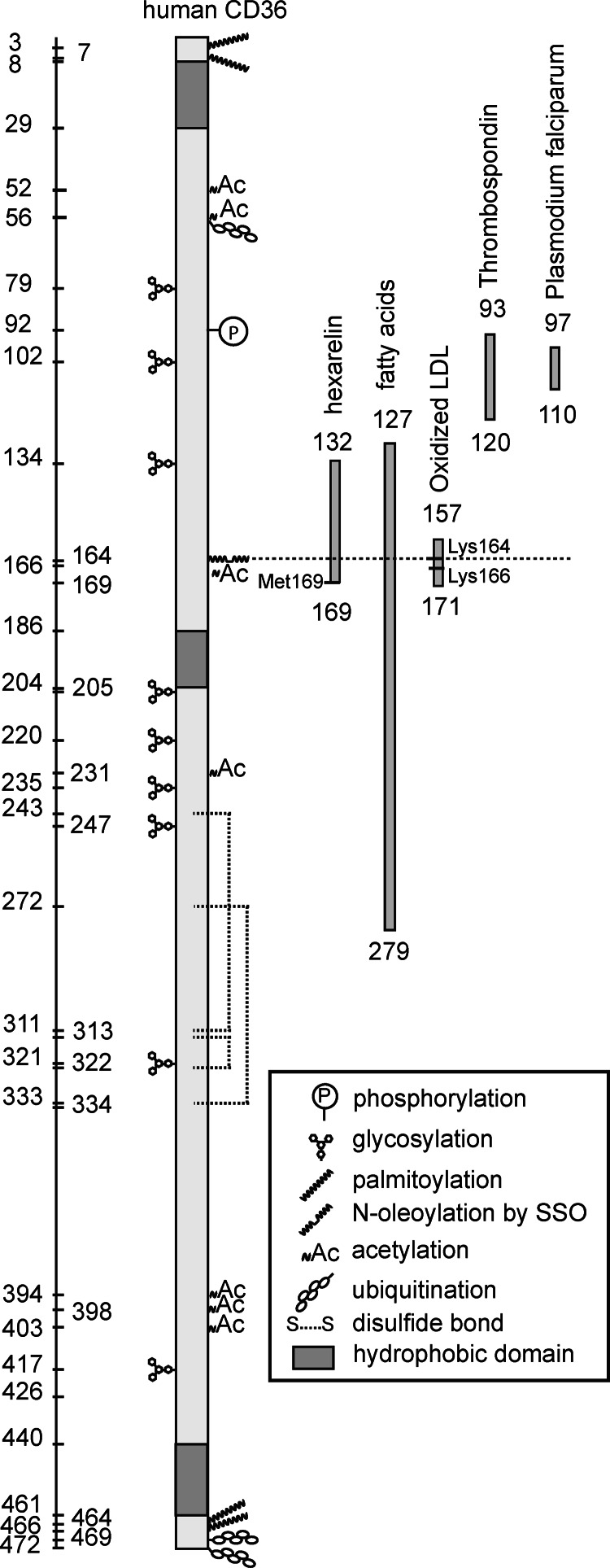
Our understanding of CD36 structure-function remains incomplete, but as summarized in Fig. 5 current knowledge supports existence of at least two major regions for ligand interaction. The first binding domain was identified for oxLDL, oxPC and hexarelin recognition (6, 9) and as shown in this study it now includes FA. The second binding domain is involved in recognition of thrombospondin and the malaria parasite P. falciparum (8, 11). A number of post-translational modifications of CD36 that include ubiquitination, palmitoylation, acetylation, etc. (Fig. 5) have now been identified, and these might be involved in ligand uptake or signaling functions. Of interest is ubiquitination of the two lysines 469 and 472, which was found important for CD36 signaling to Ca2+. Mutation of the two lysines interfered with CD36's ability to induce Ca2+ influx via the store-operated membrane channels, although release of calcium from intracellular stores appeared normal (13, 22, 23). In contrast, mutation of Lys-164 primarily abolished CD36-mediated calcium release from intracellular stores in response to linoleic acid (Fig. 2). These mutations should be helpful in dissecting the molecular steps involved in CD36 regulation of Ca2+ signaling.
FIGURE 5.
Schematic representation of CD36 structure, binding domains, and known or predicted post-translational modifications. CD36 has two short intracellular domains at the carboxyl- and amino-terminal ends, two transmembrane domains, and a large globular extracellular portion with a hydrophobic sequence. The extracellular domain is heavily glycosylated (53) and cross-linked with three disulfide bridges (54, 55), and Thr-92 was shown to be phosphorylated (8). Both the amino and carboxyl termini contain two palmitoylation sites (56). The carboxyl terminus also has two lysines, which can be ubiquitinated (37), and another ubiquitination was localized to Lys-56 (57). CD36 contains at least seven lysines, which can be acetylated (41), and phosphorylation of Tyr-62 and Thr-323 and acetylation of Lys-298 was reported at PhosphoSitePlus. Binding sequences for fatty acids, hexarelin, oxLDL, thrombospondin, and P. falciparum are aligned to the backbone of CD36. The SSO-binding site Lys-164 is highlighted with a dotted line.
In summary, we have shown that one amino acid residue, lysine 164, is the primary target of SSO bound to CD36 and that this lysine is important for FA uptake and FA-induced signaling to Ca2+. Lysine 164 is situated in a hydrophobic pocket shared by a number of CD36 ligands and was previously shown to mediate binding of oxLDL and oxPC. Consistent with this, we showed that SSO inhibits oxLDL uptake by macrophages. The data suggest that direct interaction between the various CD36 lipid ligands (oxLDL, oxPC, and FA) competing for binding to the same amino acid residue could occur under physiological conditions and influence CD36 function. Interestingly, one study previously reported that oleic acid competed for the binding of ox-LDL to CD36-transfected 3T3 cells (52).
This work was supported, in whole or in part, by National Institutes of Health Grants DK033301, DK60022, and P01 HL057278. This work was also supported by Czech Science Foundation Grant 13-04449P, EC Project DIABAT HEALTH-F2-2011-278373, and Academy of Sciences of the Czech Republic Grant RVO 61388963.
- FA
- fatty acid
- NHS
- N-hydroxysuccinimidyl
- oxLDL
- oxidized low density lipoprotein
- PMA
- phorbol 12-myristate 13-acetate
- SSO
- sulfo-N-succinimidyl oleate
- Dil-OxLDL
- 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate-oxidized LDL
- oxPC
- oxidized phosphatidylcholine.
REFERENCES
- 1. Coburn C. T., Abumrad N. A. (2004) in Cellular Proteins and Their Fatty Acids in Health and Disease (Duttaroy A. K., Spener F., eds) pp. 3–29, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany [Google Scholar]
- 2. Glatz J. F., Luiken J. J., Bonen A. (2010) Membrane fatty acid transporters as regulators of lipid metabolism: implications for metabolic disease. Physiol. Rev. 90, 367–417 [DOI] [PubMed] [Google Scholar]
- 3. Hirano K., Kuwasako T., Nakagawa-Toyama Y., Janabi M., Yamashita S., Matsuzawa Y. (2003) Pathophysiology of human genetic CD36 deficiency. Trends Cardiovasc. Med. 13, 136–141 [DOI] [PubMed] [Google Scholar]
- 4. Koutsari C., Ali A. H., Mundi M. S., Jensen M. D. (2011) Storage of circulating free fatty acid in adipose tissue of postabsorptive humans: quantitative measures and implications for body fat distribution. Diabetes 60, 2032–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Koutsari C., Mundi M. S., Ali A. H., Jensen M. D. (2012) Storage rates of circulating free fatty acid into adipose tissue during eating or walking in humans. Diabetes 61, 329–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kar N. S., Ashraf M. Z., Valiyaveettil M., Podrez E. A. (2008) Mapping and characterization of the binding site for specific oxidized phospholipids and oxidized low density lipoprotein of scavenger receptor CD36. J. Biol. Chem. 283, 8765–8771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Endemann G., Stanton L. W., Madden K. S., Bryant C. M., White R. T., Protter A. A. (1993) Cd36 is a receptor for oxidized low density lipoprotein. J. Biol. Chem. 268, 11811–11816 [PubMed] [Google Scholar]
- 8. Asch A. S., Liu I., Briccetti F. M., Barnwell J. W., Kwakye-Berko F., Dokun A., Goldberger J., Pernambuco M. (1993) Analysis of CD36 binding domains: ligand specificity controlled by dephosphorylation of an ectodomain. Science 262, 1436–1440 [DOI] [PubMed] [Google Scholar]
- 9. Demers A., McNicoll N., Febbraio M., Servant M., Marleau S., Silverstein R., Ong H. (2004) Identification of the growth hormone-releasing peptide binding site in CD36: a photoaffinity cross-linking study. Biochem. J. 382, 417–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Navazo M. D., Daviet L., Savill J., Ren Y., Leung L. L., McGregor J. L. (1996) Identification of a domain (155–183) on CD36 implicated in the phagocytosis of apoptotic neutrophils. J. Biol. Chem. 271, 15381–15385 [DOI] [PubMed] [Google Scholar]
- 11. Barnwell J. W., Asch A. S., Nachman R. L., Yamaya M., Aikawa M., Ingravallo P. (1989) A human 88-kDa membrane glycoprotein (CD36) functions in vitro as a receptor for a cytoadherence ligand on Plasmodium falciparum-infected erythrocytes. J. Clin. Invest. 84, 765–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Febbraio M., Hajjar D. P., Silverstein R. L. (2001) CD36: a class B scavenger receptor involved in angiogenesis, atherosclerosis, inflammation, and lipid metabolism. J. Clin. Invest. 108, 785–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kuda O., Jenkins C. M., Skinner J. R., Moon S. H., Su X., Gross R. W., Abumrad N. A. (2011) CD36 protein is involved in store-operated calcium flux, phospholipase A2 activation, and production of prostaglandin E2. J. Biol. Chem. 286, 17785–17795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dramane G., Abdoul-Azize S., Hichami A., Vögtle T., Akpona S., Chouabe C., Sadou H., Nieswandt B., Besnard P., Khan N. A. (2012) STIM1 regulates calcium signaling in taste bud cells and preference for fat in mice. J. Clin. Invest. 122, 2267–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. El-Yassimi A., Hichami A., Besnard P., Khan N. A. (2008) Linoleic acid induces calcium signaling, Src kinase phosphorylation, and neurotransmitter release in mouse CD36-positive gustatory cells. J. Biol. Chem. 283, 12949–12959 [DOI] [PubMed] [Google Scholar]
- 16. Sundaresan S., Shahid R., Riehl T. E., Chandra R., Nassir F., Stenson W. F., Liddle R. A., Abumrad N. A. (2013) CD36-dependent signaling mediates fatty acid-induced gut release of secretin and cholecystokinin. FASEB J. 27, 1191–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pepino M. Y., Love-Gregory L., Klein S., Abumrad N. A. (2012) The fatty acid translocase gene CD36 and lingual lipase influence oral sensitivity to fat in obese subjects. J. Lipid Res. 53, 561–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Love-Gregory L., Sherva R., Sun L., Wasson J., Schappe T., Doria A., Rao D. C., Hunt S. C., Klein S., Neuman R. J., Permutt M. A., Abumrad N. A. (2008) Variants in the CD36 gene associate with the metabolic syndrome and high-density lipoprotein cholesterol. Hum. Mol. Genet. 17, 1695–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ghosh A., Murugesan G., Chen K., Zhang L., Wang Q., Febbraio M., Anselmo R. M., Marchant K., Barnard J., Silverstein R. L. (2011) Platelet CD36 surface expression levels affect functional responses to oxidized LDL and are associated with inheritance of specific genetic polymorphisms. Blood 117, 6355–6366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Love-Gregory L., Abumrad N. A. (2011) CD36 genetics and the metabolic complications of obesity. Curr. Opin. Clin. Nutr. Metab. Care 14, 527–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Baillie A. G., Coburn C. T., Abumrad N. A. (1996) Reversible binding of long-chain fatty acids to purified FAT, the adipose CD36 homolog. J. Membr. Biol. 153, 75–81 [DOI] [PubMed] [Google Scholar]
- 22. Huang M. M., Bolen J. B., Barnwell J. W., Shattil S. J., Brugge J. S. (1991) Membrane glycoprotein IV (CD36) is physically associated with the Fyn, Lyn, and Yes protein-tyrosine kinases in human platelets. Proc. Natl. Acad. Sci. U.S.A. 88, 7844–7848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Silverstein R. L., Febbraio M. (2009) CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci. Signal. 2, re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Su X., Abumrad N. A. (2009) Cellular fatty acid uptake: a pathway under construction. Trends Endocrinol. Metab. 20, 72–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abumrad N. A., Davidson N. O. (2012) Role of the gut in lipid homeostasis. Physiol. Rev. 92, 1061–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Harmon C. M., Abumrad N. A. (1993) Binding of sulfosuccinimidyl fatty acids to adipocyte membrane proteins: isolation and amino-terminal sequence of an 88-kDa protein implicated in transport of long-chain fatty acids. J. Membr. Biol. 133, 43–49 [DOI] [PubMed] [Google Scholar]
- 27. Coort S. L., Willems J., Coumans W. A., van der Vusse G. J., Bonen A., Glatz J. F., Luiken J. J. (2002) Sulfo-N-succinimidyl esters of long chain fatty acids specifically inhibit fatty acid translocase (FAT/CD36)-mediated cellular fatty acid uptake. Mol. Cell. Biochem. 239, 213–219 [PubMed] [Google Scholar]
- 28. Kerkhoff C., Sorg C., Tandon N. N., Nacken W. (2001) Interaction of S100A8/S100A9-arachidonic acid complexes with the scavenger receptor CD36 may facilitate fatty acid uptake by endothelial cells. Biochemistry 40, 241–248 [DOI] [PubMed] [Google Scholar]
- 29. Pohl J., Ring A., Korkmaz U., Ehehalt R., Stremmel W. (2005) FAT/CD36-mediated long-chain fatty acid uptake in adipocytes requires plasma membrane rafts. Mol. Biol. Cell 16, 24–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nicholls H. T., Kowalski G., Kennedy D. J., Risis S., Zaffino L. A., Watson N., Kanellakis P., Watt M. J., Bobik A., Bonen A., Febbraio M., Lancaster G. I., Febbraio M. A. (2011) Hematopoietic cell-restricted deletion of CD36 reduces high-fat diet-induced macrophage infiltration and improves insulin signaling in adipose tissue. Diabetes 60, 1100–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Naville D., Duchampt A., Vigier M., Oursel D., Lessire R., Poirier H., Niot I., Bégeot M., Besnard P., Mithieux G. (2012) Link between intestinal CD36 ligand binding and satiety induced by a high protein diet in mice. PloS One 7, e30686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Le Foll C., Irani B. G., Magnan C., Dunn-Meynell A. A., Levin B. E. (2009) Characteristics and mechanisms of hypothalamic neuronal fatty acid sensing. Am. J. Physiol. Regul. Integr. Comp. Physiol. 297, R655–R664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Geffen I., Wessels H. P., Roth J., Shia M. A., Spiess M. (1989) Endocytosis and recycling of subunit-H1 of the asialoglycoprotein receptor is independent of oligomerization with H2. EMBO J. 8, 2855–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hare J. F., Taylor K. (1991) Mechanisms of plasma-membrane protein-degradation-recycling proteins are degraded more rapidly than those confined to the cell-surface. Proc. Natl. Acad. Sci. U.S.A. 88, 5902–5906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kalkhof S., Sinz A. (2008) Chances and pitfalls of chemical cross-linking with amine-reactive N-hydroxysuccinimide esters. Anal. Bioanal. Chem. 392, 305–312 [DOI] [PubMed] [Google Scholar]
- 36. Harmon C. M., Luce P., Beth A. H., Abumrad N. A. (1991) Labeling of adipocyte membranes by sulfo-N-succinimidyl derivatives of long-chain fatty acids: inhibition of fatty acid transport. J. Membr. Biol. 121, 261–268 [DOI] [PubMed] [Google Scholar]
- 37. Smith J., Su X., El-Maghrabi R., Stahl P. D., Abumrad N. A. (2008) Opposite regulation of CD36 ubiquitination by fatty acids and insulin: effects on fatty acid uptake. J. Biol. Chem. 283, 13578–13585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shilov I. V., Seymour S. L., Patel A. A., Loboda A., Tang W. H., Keating S. P., Hunter C. L., Nuwaysir L. M., Schaeffer D. A. (2007) The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics 6, 1638–1655 [DOI] [PubMed] [Google Scholar]
- 39. Strohalm M., Kavan D., Novák P., Volný M., Havlícek V. (2010) mMass 3: a cross-platform software environment for precise analysis of mass spectrometric data. Anal. Chem. 82, 4648–4651 [DOI] [PubMed] [Google Scholar]
- 40. Roy A., Kucukural A., Zhang Y. (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lundby A., Lage K., Weinert B. T., Bekker-Jensen D. B., Secher A., Skovgaard T., Kelstrup C. D., Dmytriyev A., Choudhary C., Lundby C., Olsen J. V. (2012) Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep. 2, 419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gros J., Gerhardt C. C., Strosberg A. D. (1999) Expression of human β3-adrenergic receptor induces adipocyte-like features in CHO/K1 fibroblasts. J. Cell Sci. 112, 3791–3797 [DOI] [PubMed] [Google Scholar]
- 43. Listenberger L. L., Han X., Lewis S. E., Cases S., Farese R. V., Jr., Ory D. S., Schaffer J. E. (2003) Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc. Natl. Acad. Sci. U.S.A. 100, 3077–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kennedy D. J., Kuchibhotla S., Westfall K. M., Silverstein R. L., Morton R. E., Febbraio M. (2011) A CD36-dependent pathway enhances macrophage and adipose tissue inflammation and impairs insulin signalling. Cardiovasc. Res. 89, 604–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Riquelme C. A., Magida J. A., Harrison B. C., Wall C. E., Marr T. G., Secor S. M., Leinwand L. A. (2011) Fatty acids identified in the Burmese python promote beneficial cardiac growth. Science 334, 528–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huang B. X., Dass C., Kim H. Y. (2005) Probing conformational changes of human serum albumin due to unsaturated fatty acid binding by chemical cross-linking and mass spectrometry. Biochem. J. 387, 695–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reed R. G. (1986) Location of long chain fatty acid-binding sites of bovine serum albumin by affinity labeling. J. Biol. Chem. 261, 15619–15624 [PubMed] [Google Scholar]
- 48. Falomir-Lockhart L. J., Laborde L., Kahn P. C., Storch J., Córsico B. (2006) Protein-membrane interaction and fatty acid transfer from intestinal fatty acid-binding protein to membranes. Support for a multistep process. J. Biol. Chem. 281, 13979–13989 [DOI] [PubMed] [Google Scholar]
- 49. Storch J., McDermott L. (2009) Structural and functional analysis of fatty acid-binding proteins. J. Lipid Res. 50, S126–S131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Xiong Y., Guan K. L. (2012) Mechanistic insights into the regulation of metabolic enzymes by acetylation. J. Cell Biol. 198, 155–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gao D., Ashraf M. Z., Kar N. S., Lin D., Sayre L. M., Podrez E. A. (2010) Structural basis for the recognition of oxidized phospholipids in oxidized low density lipoproteins by class B scavenger receptors CD36 and SR-BI. J. Biol. Chem. 285, 4447–4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nicholson A. C., Frieda S., Pearce A., Silverstein R. L. (1995) Oxidized Ldl binds to Cd36 on human monocyte-derived macrophages and transfected cell lines–Evidence implicating the lipid moiety of the lipoprotein as the binding site. Arterioscler. Thromb. Vasc. Biol. 15, 269–275 [DOI] [PubMed] [Google Scholar]
- 53. Hoosdally S. J., Andress E. J., Wooding C., Martin C. A., Linton K. J. (2009) The human scavenger receptor CD36: Glycosylation status and its role in trafficking and function. J. Biol. Chem. 284, 16277–16288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Berglund L., Petersen T. E., Rasmussen J. T. (1996) Structural characterization of bovine CD36 from the milk fat globule membrane. Biochim. Biophys. Acta 1309, 63–68 [DOI] [PubMed] [Google Scholar]
- 55. Rasmussen J. T., Berglund L., Rasmussen M. S., Petersen T. E. (1998) Assignment of disulfide bridges in bovine CD36. Eur. J. Biochem. 257, 488–494 [DOI] [PubMed] [Google Scholar]
- 56. Tao N., Wagner S. J., Lublin D. M. (1996) CD36 is palmitoylated on both N- and C-terminal cytoplasmic tails. J. Biol. Chem. 271, 22315–22320 [DOI] [PubMed] [Google Scholar]
- 57. Wagner S. A., Beli P., Weinert B. T., Schölz C., Kelstrup C. D., Young C., Nielsen M. L., Olsen J. V., Brakebusch C., Choudhary C. (2012) Proteomic analyses reveal divergent ubiquitylation site patterns in murine tissues. Mol. Cell. Proteomics 11, 1578–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]