Background: Dentin phosphophoryn (DPP) is an acidic protein involved in the nucleation of hydroxyapatite.
Results: The acidic domain of DPP is responsible for its non-receptor-mediated endocytosis.
Conclusion: The acidic DSS domain can be used for targeted delivery of proteins into cells.
Significance: The DSS domain can function as a cell penetrating peptide to deliver proteins for therapeutic applications.
Keywords: Drug Delivery System, Endocytosis, Extracellular Matrix Proteins, Nanotechnology, Nuclear Translocation, Dentin Phosphophoryn, Quantum Dots
Abstract
Dentin phosphophoryn is nature's most acidic protein found predominantly in the dentin extracellular matrix. Its unique amino acid composition containing Asp-Ser (DS)-rich repeats makes it highly anionic. It has a low isoelectric point (pI 1.1) and, therefore, tends to be negatively charged at physiological pH. Phosphophoryn is normally associated with matrix mineralization as it can bind avidly to Ca2+. It is well known that several macromolecules present in the extracellular matrix can be internalized and localized to specific intracellular compartments. In this study we demonstrate that dentin phosphophoryn (DPP) is internalized by several cell types via a non-conventional endocytic process. Utilizing a DSS polypeptide derived from DPP, we demonstrate the repetitive DSS-rich domain facilitates that endocytosis. As a proof-of-concept, we further demonstrate the use of this polypeptide as a protein delivery vehicle by delivering the osteoblast transcription factor Runx2 to the nucleus of mesenchymal cells. The functionality of the endocytosed Runx2 protein was demonstrated by performing gene expression analysis of Runx2 target genes. Nuclear localization was also demonstrated with the fusion protein DSS-Runx2 conjugated to quantum dots in two- and three-dimensional culture models in vitro and in vivo. Overall, we demonstrate that the DSS domain of DPP functions as a novel cell-penetrating peptide, and these findings demonstrate new opportunities for intracellular delivery of therapeutic proteins and cell tracking in vivo.
Introduction
Dentin phosphophoryn (DPP)2 is an acidic, highly phosphorylated protein that is a ubiquitous component of the dentin extracellular matrix (1, 2). The phosphorylated protein possesses a strong affinity for calcium ions and thus serves as a nucleator of calcium phosphate polymorphs during mineralized matrix formation (3–5). DPP is a cleavage product of the larger dentin sialophosphoprotein, a member of the SIBLING (small integrin binding ligand) family of proteins (6–10). Mutations identified in the dentin sialophosphoprotein (DSPP) gene and knock-out mouse models show that lack of functional DSPP results in dentinogenesis imperfecta, a disease characterized by mineralization defects (11–16).
DPP contains numerous aspartic and phosphorylated serine residues that render this protein as nature's most acidic protein. The molecule is called thus as it is considered to be a “phosphate carrier” (17–19). A common feature in this protein is the presence of (DSS)n (Asp-Ser-Ser) repeats distributed throughout the molecule. The principal post-translational modification is the phosphorylation of 85–90% of the serine groups in the molecule. Phosphorylation of all potential serine results in a highly negatively charged molecule and is a virtual sink for the binding of calcium ions in vivo.
Apart from its role in mineralization, DPP has also been shown to actively participate in cell signaling events leading to differentiation of precursor mesenchymal cells (20–22). We recently showed that the RGD domain in DPP induces integrin-mediated signaling events (22). Other published reports have demonstrated activation of the MAP kinase signaling pathway when human mesenchymal cells were stimulated with DPP (20, 21).
In this study we demonstrate that several cell types from different lineages can internalize DPP. Therefore, we sought to investigate the mechanism by which endocytosis of DPP is facilitated. In mammalian cells, endocytosis occurs through two major pathways that have been characterized extensively. The first one is the clathrin-mediated pathway that primarily targets its cargo to the endosomes (23). The clathrin-independent pathway is subclassified into dynamin-dependent and dynamin-independent pathways. Of these, the most prevalent one is the caveolae or lipid raft-mediated endocytosis. This pathway has been shown to be involved in the trafficking of receptors, toxins, proteins, and viruses (24–26).
In this study our observations indicate that the DSS repeat-rich sequence in DPP is responsible for its endocytosis. However, neither clathrin nor the caveolae-mediated pathways is responsible for its internalization. Finally, we provide evidence that the DSS polypeptide derived from DPP can be used as a carrier for targeted protein delivery. This approach was demonstrated by the delivery of Runx2, an osteoblast specific transcription factor into the nuclei of mesenchymal cells. We envision that the DSS-containing polypeptide could be a valuable tool for targeted intracellular protein delivery and, therefore, have therapeutic potentials.
EXPERIMENTAL PROCEDURES
Cell Culture
Several cell types were used in this study. They are dental pulp stem cells (DPSC; a gift from Dr. Songatao Shi, University of Southern California), human marrow stromal cells (HMSC; Tulane NIH Cancer Center), T4-4 rat preodontoblasts, MC3T3-E1 mouse calvarial preosteoblasts, primary mouse odontoblasts (isolated from 3-day-old wild type pups), C3H10T1/2 mouse embryonic mesenchymal cells, HAT-7 dental epithelial cells (a kind gift from Dr. Harada Japan), and human umbilical vein endothelial cells (ATCC). DPSC, HMSC, and odontoblasts were cultured in α minimum essential medium (MEM, Invitrogen) supplemented with 20% fetal bovine serum (FBS, Invitrogen), 1% l-glutamine, and 1% antibiotic and antimycotic solution (Invitrogen). T4-4, HAT-7, and MC3T3-E1 cells were cultured in DMEM/F12 (1:1) medium with 10% FBS and 1% antibiotic and antimycotic solution. C3H10T1/2 cells were cultured in Basal Medium Eagle media containing 10% FBS and 1% antibiotic and antimycotic solution.
Expression of Recombinant Proteins
Recombinant DPP was expressed as a GST fusion protein in Escherichia coli and purified as published previously (22). The 244-residue C-terminal domain of rat DPP, containing the aspartic acid-serine repeats (18) (DSS) was cloned into PGEX4T-3 GST vector and expressed as a GST-DSS fusion protein. Protein purification was carried out as per published protocols. supplemental Fig. 1A shows an image of a Stains-All stained gel showing the purified DSS polypeptide. For experiments with GST-DSS, the GST was not cleaved, and the fusion protein was eluted using glutathione. Runx2-DSS (R2DSS) fusion protein was obtained by cloning the Runx2 coding sequence (obtained as a gift in from Dr. Monetcino, Univercidad de Concepcion, Chile in PGEX4T-1 GST fusion vector) upstream of the DSS coding sequence in the PGEX4T-3 vector. The fusion protein was then purified as published (22). The purity of all recombinant proteins was verified by SDS-PAGE analysis.
FITC Labeling of Recombinant Proteins
FITC labeling of DSS was performed as published previously (27). Supplemental Fig. 1, B and C shows images of the FITC-labeled protein acquired after UV illumination of the SDS gel.
Quantum Dot (QD) Labeling of Recombinant Proteins
QD conjugation of the recombinant proteins was performed according to previously published protocols (28, 29).
Binding and Endocytosis
Binding of biotinylated DSS to T4-4 cells at 4 °C was performed as published previously (27). Endocytosis experiments were performed using FITC-labeled proteins or quantum dot-labeled proteins. For the FITC-labeled proteins, 10 μg/ml protein (DPP, DSS, R2DSS, GST-DSS, or BSA) were incubated with the cells grown on coverslips for various time points. The cells were then fixed with 4% paraformaldehyde, mounted on slides, and imaged using a Zeiss LSM 510 Meta or LSM 710 confocal microscope. When required, the cells were immunostained with the appropriate primary and secondary antibodies. The following primary antibodies used in this study were mouse monoclonal anti-tubulin antibody (Sigma, 1/1000), rabbit polyclonal anti-caveolin-1 antibody (Santa Cruz Biotechnology, 1/75), rabbit polyclonal anti-clathrin antibody (Santa Cruz Biotechnology, 1/100), and rabbit polyclonal anti-EEA-1 FITC-labeled antibody (BD Biosciences, 1/50).
Dose response curves for DSS, GST-DSS, and R2DSS were obtained by incubating MC3T3-E1 cells (50,000cells/well) seeded in 24-well plates with increasing amounts of the FITC-labeled proteins. The cells were incubated with the proteins at 37 °C for 2 h. They were then lysed using 100 μl of MPER (Thermo Scientific) protein extraction reagent per well, and green fluorescence was measured using a BioTek microtiter plate reader using appropriate filter sets. All experiments were performed in triplicate.
For endocytosis blocking experiments, T4-4 cells were pretreated for 60 min with different inhibitors and then treated for 15 min with 10 μg/ml FITC DSS. Wherever time points are not mentioned, the cells were incubated with the fluorescently labeled proteins for 15 min. The following inhibitors were used in this study: hyper-osmotic sucrose (0.45 m), ikarugamycin (2 μm), methyl β-cyclodextrin (1%), colchicine (20 μm), and cytochalasin D (1 μm). The experiments were also performed under acidic and basic conditions. Adding concentrated HCl solution to the media increased the acidity of the media (pH 4). To increase the alkalinity of the media, NaOH (1 m stock) was added (pH 10). For these experiments, T4-4 cells were pretreated for 30 min in acidic or alkaline media, and the experiment was performed for 15 min under the same conditions at 37 °C. Endocytosis experiments were also performed at 4 °C and at room temperature. For these experiments, the cells (MC3T3-E1) were incubated in a refrigerator or at room temperature for 30 min followed by treatment with FITC-labeled proteins (DSS, GST-DSS) for 1 h. After incubation, the cells were fixed in 4% paraformaldehyde, permeabilized, and immunostained as required. The cells were then imaged using a Zeiss LSM 510 META confocal microscope using appropriate lasers and emission filters. All imaging conditions for comparative experiments were maintained constant. To quantify the fluorescence, the green intensities in cells were measured using the Zeiss Axiovision Software and expressed as mean intensity per condition ± S.E.
Endocytosis experiments with proteins conjugated with QDs were performed by incubating cells cultured on coverslips or embedded in collagen scaffolds (30) with 25 μl of the conjugates (estimation of protein concentration was not possible). However, before conjugation, the protein concentration was 1 mg/ml, the concentration of the protein used for conjugation was maintained the same each time, and the suitable amount of conjugate required to get a good signal for the indicated time points was empirically determined. The cells were fixed in 4% paraformaldehyde, mounted on to slides, and imaged using the Zeiss LSM 510 Meta confocal microscope (the cells were excited using a UV laser, and emission was observed in the red spectrum).
Gene Expression Analysis
Quantitative real-time PCR was used to analyze gene expression levels of Runx2 targets upon treatment with Runx2-DSS fusion protein. HMSC were treated with either 2 μg of the DSS-Runx2 fusion protein or 2 μg of DSS peptide alone for 24 and 48 h. At the end of these time points, the RNA from the cells was extracted using the Qiagen kit as per the manufacturer's protocol. cDNA synthesis was then performed, and the expression levels of three Runx2 target genes, namely, alkaline phosphatase, SMAD 4, and DMP1, were analyzed using gene-specific primers. The data were normalized to GAPDH, and changes in expression levels were calculated using the comparative ΔΔCT method.
In Vivo Implantation
DPSC were seeded onto biomimetic scaffolds as described previously. 24 h post seeding, the scaffolds were incubated with 25 μl of DSS-Runx2 QD conjugates for 24 h. Scaffolds were prepared in triplicate and then implanted subcutaneously into 1-month-old athymic nude male mice (Charles River Laboratories) for a period of 2 weeks. Scaffolds without the QD conjugates served as controls. All experiments were performed using approved University of Illinois at Chicago animal care protocols (Assurance no A3460-01). After 1- and 2-week post-implantation, the animals were euthanized and imaged using an IVIS Spectrum animal imager. The samples were excited using UV light, and emission was observed in the red spectrum. The scaffolds were then removed, fixed in 4% formalin, and embedded in paraffin, and 5-μm thick sections were obtained and stained with DAPI nuclear stain and imaged using a Zeiss LSM 710 confocal microscope. The slides were excited using UV laser, and emission was observed in the blue spectrum for DAPI nuclear fluorescence and in the red spectrum to observe the QDs.
Endocytosis of (DSS)6 Peptide
To identify the minimum number of DSS repeats responsible for endocytosis, we synthesized a short peptide containing six DSS amino acid repeats followed by a FLAG tag (DSSDSSDSSDSSDSSDSSDYKDDDDK). 50 μg of this peptide was incubated with 20 μg of affinity-purified rabbit polyclonal FLAG antibody (Sigma) for 1 h at room temperature. The mixture was then added to MC3T3-E1 cells seeded on a cover glass (25 mm) and incubated at 37 °C for 1 h. 20 μg of FLAG antibody added to cells served as control. After incubation, the cells were washed, fixed, permeabilized, and fluorescently immunostained with anti-rabbit secondary antibody (FITC) to visualize the FLAG antibody. The cells were also immunostained with mouse monoclonal tubulin antibody (Sigma) followed by a TRITC-conjugated anti-mouse secondary antibody. The cells were imaged using a Zeiss LSM 710 confocal microscope.
RESULTS
Internalization of DPP
Fig. 1 shows representative confocal micrographs of three different mesenchymal cell types (T4-4 pre-odontoblasts, C3H10T1/2 embryonic mesenchymal cells, and DPSC treated with FITC-labeled DPP. Results show that all these cell types were able to internalize FITC-DPP when incubated for 15 min at 37 °C.
FIGURE 1.
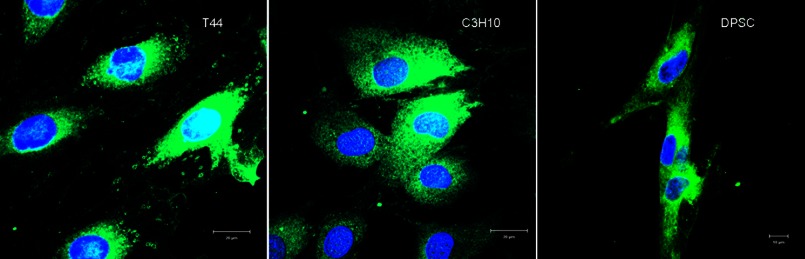
Endocytosis of DPP. Shown are representative confocal micrographs of FITC-labeled DPP endocytosed by T44 preodontoblast cells, C3H10T1/2 pluripotent mesenchymal cells, and DPSC.
Internalization of the DSS Domain Lacking RGD
DPP contains a conserved RGD domain in several species; therefore, we first verified if cell surface integrins mediated the uptake of DPP. Fig. 2, A and B, shows representative confocal images of FITC-labeled DSS polypeptide (lacking the RGD domain) endocytosed in T4-4 (A) and primary pulp cells (B) at different time points. Extensive uptake of the DSS polypeptide was observed even within 2 min. Additionally, the polypeptide was also endocytosed by several other cell types (Fig. 2C). FITC-labeled BSA was used as a negative control (Fig. 2D). These results confirmed that DPP internalization is not mediated through integrin receptors.
FIGURE 2.
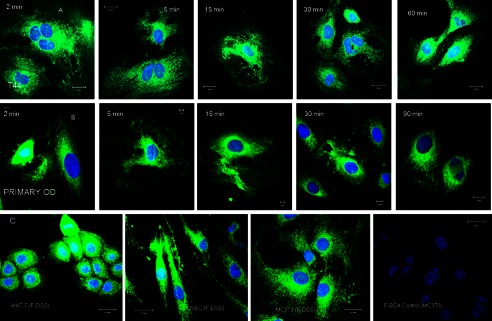
Time course of DSS polypeptide endocytosis. A and B, representative confocal micrographs show snapshots of the location of the endocytosed FITC labeled DSS peptide (F-DSS) in T44 cells (A) and primary mouse pulp cells (B) at the indicated time points. C, representative confocal images show endocytosis of the DSS peptide by epithelial cells (HAT 7), endothelial cells (human umbilical vein endothelial cells), and osteoblast cells (MC3T3). D, shown is a negative control with FITC-labeled BSA.
Endocytosis Does Not Occur via the Clathrin- or Caveolae-mediated Endocytic Pathways
We then examined the involvement of classical endocytic mechanisms for the internalization of DSS. Inhibitors for clathrin- and caveolae-mediated endocytosis were employed to identify the endocytic mechanism. Clathrin endocytic pathway was blocked using the inhibitor ikarugamycin or by adding 0.45 m sucrose to the culture medium. Ikarugamycin is also an inhibitor of receptor-mediated endocytosis. Caveolar endocytosis was blocked using methyl β-cyclodextrin. Results in Fig. 3, B, C, D, and G, show that these inhibitors had no effect on DSS uptake. Immunofluorescence colocalization experiments were also performed to observe colocalization of the endocytic vesicles with clathrin or caveolin. Results in Fig. 3, E and F, shows that there was no colocalization of the endocytic vesicles with both caveolin-1 (caveolae marker, Fig. 3E) and clathrin (Fig. 3F). Thus, both clathrin-coated pits and caveolae/lipid rafts did not partake in the endocytic process. Additionally, a dose-response endocytosis experiment showed that endocytosis was not saturable (Fig. 3G) but showed a linear increase with an increase in dosage indicating the absence of specific cell surface binding. Binding experiments were also performed with biotinylated DSS to check if DSS bound to the plasma membrane before endocytosis. No specific binding of DSS to T4-4 and MC3T3-E1 cells was observed (data not shown).
FIGURE 3.
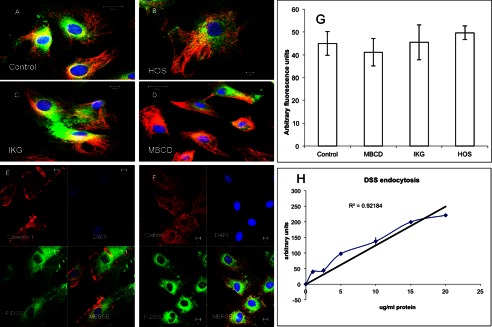
Effect of endocytic inhibitors on the endocytosis of DSS polypeptide. A, a representative confocal image shows endocytosis of the FITC labeled DSS peptide (F-DSS) by T44 cells. B, C, and D, endocytosis of the DSS peptide by T44 cells after pretreatment with hyperosmotic sucrose (HOS), an inhibitor of clathrin-mediated endocytosis (B), ikarugamycin (IKG), an inhibitor of receptor-mediated endocytosis (C), and methyl β-cyclodextrin (MBCD), an inhibitor of lipid raft and caveolae mediated endocytosis (D). Red staining in all images indicates immunofluorescent labeling of the microtubules. E and F, representative confocal images show the absence of co-localization of the endocytosed DSS (green) with caveolin-1 (E, red stain) and clathrin (F, red stain). G, quantitation of fluorescence for DSS endocytosis is shown. Data represent the mean fluorescent signal ± S.E. (n = 9). H, dose-response curve for DSS endocytosis by MC3T3-E1 cells show a linear response.
Disruption of Microtubules and Microfilaments Does Not Affect Endocytosis of DSS
Endocytic vesicles are transported from the plasma membrane through microtubules and microfilaments. We investigated if disruption of the intracellular transportation mechanisms affected the endocytic process. Results show that disruption of the microtubules with colchicine (Fig. 4A) or microfilaments with cytochalasin D (Fig. 4B) did not prevent the endocytosis of the DSS polypeptide.
FIGURE 4.
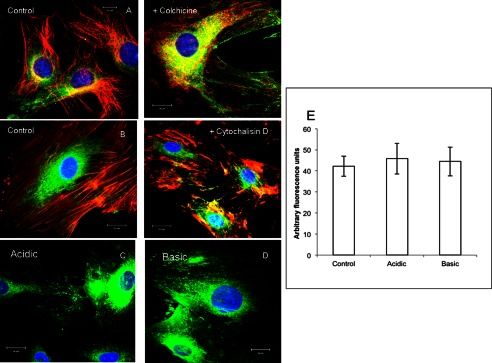
Disruption of microtubules and microfilaments and the presence of acidic or basic medium does not inhibit endocytosis of DSS by T44 cells. A and B, FITC-tagged DSS peptide endocytosis (green) is not inhibited by disruption of microtubules (A) and microfilaments (B). In both images red staining indicates microtubules and microfilaments respectively. C and D, DSS peptide endocytosis is not inhibited in the presence of acidic (C) and basic cell culture media (B). E, quantitation of fluorescence for DSS endocytosis in the presence of acidic and basic cell culture medium indicates the absence of endocytosis blocking. Data represent mean fluorescent signal ± S.E. (n = 9).
Effect of pH on the Endocytosis of DSS
Perturbations in the cytoplasmic and extracellular pH are known to inhibit the endocytosis of cell surface proteins (31, 32). Therefore, we investigated if acidic or alkaline environment had an effect on the internalization of DSS. Results indicated that neither acidic pH (Fig. 4, C and E) nor alkaline pH (Fig. 4, D and E) affected the endocytic process.
Temperature Dependence of DSS Endocytosis
Results from endocytosis experiments performed at 4 °C and at room temperature indicated that the endocytosis of DSS was reduced but not abrogated by changes in temperature (Fig. 5, A and B). Results also show that the vesicles containing DSS were not observed on the microtubules, suggesting random diffusion of the protein within the cytoplasm. This indicates that the DSS polypeptide requires ATP for regulated intracellular transportation.
FIGURE 5.
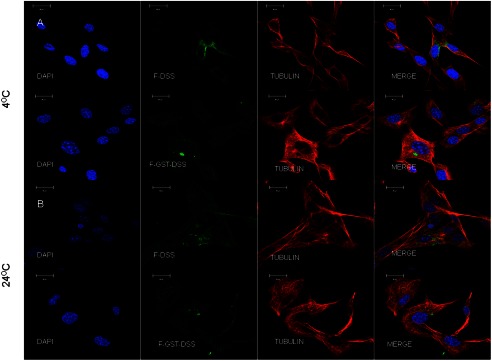
Endocytosis at 4 °C and room temperature. Representative confocal images show endocytosis of FITC-labeled DSS (F-DSS) and GST-DSS (F-GST-DSS) at 4 °C (A) and room temperature (B). Note that the endocytosed vesicles (green) do not co-localize on the microtubules (red).
Use of the DSS Polypeptide as a Vehicle for Protein Delivery
With the DSS peptide showing the ability to cross the plasma membrane of several cell types via a non-classical, non-receptor mediated pathway, we tested the possibility of using this peptide as a protein delivery agent. To visualize the delivery process, GST-DSS fusion protein was FITC-labeled, and the uptake was examined in T4-4 and MC3T3-E1 cells to observe if the DSS domain possessed the ability to carry the GST protein across the plasma membrane. Fig. 6A shows representative confocal micrographs of the GST-DSS fusion protein endocytosed by these two cell types. GST protein alone was used as control and was not endocytosed by either of the cell types. A dose response for the endocytosis of this fusion protein by MC3T3-E1 cells indicated a linear profile similar to the endocytosis of the DSS polypeptide (Fig. 6B).
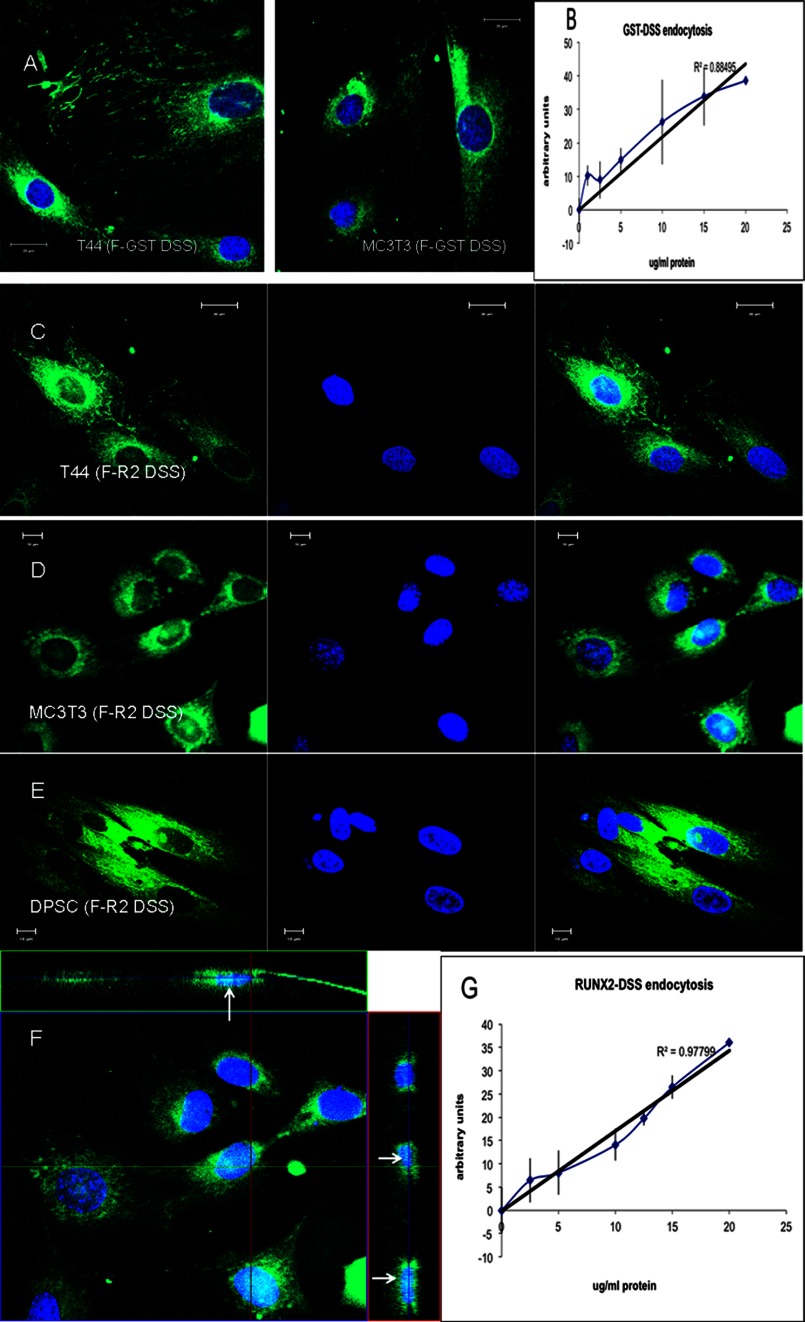
FIGURE 6.
DSS mediated endocytosis of GST-DSS and Runx2-DSS fusion proteins. A, representative confocal images show endocytosis of FITC labeled GST-DSS fusion protein (F-GST-DSS) by T44 and MC3T3 cells. B, a dose-response curve for GST-DSS fusion protein endocytosis by MC3T3-E1 cells shows a linear response. C, D, and E, representative confocal images show the endocytosis of the FITC labeled DSS-tagged Runx2 (F-R2-DSS) transcription factor by T44 (C) and MC3T3 (D) cells and DPSC (E). Note the nuclear localization of the endocytosed protein. F, shown is an orthogonal view of a z-stack of confocal images for the endocytosis of the DSS-tagged Runx2 protein by MC3T3 cells. Arrows point to nuclear localization of the endocytosed protein in the x-z and the y-z planes. G, a dose-response curve for Runx2-DSS fusion protein endocytosis by MC3T3-E1 cells shows a linear response similar to that of DSS and GST-DSS proteins.
Targeted Nuclear Delivery of the Osteoblast Transcription Factor Runx2 in Various Mesenchymal Cell Types
As internalization of DSS in eukaryotic cells occurs instantaneously, we investigated its potential for targeted nuclear delivery of Runx2. FITC-labeled fusion protein containing the DSS polypeptide with Runx2 was used in this study. Runx2 contains a nuclear localization signal, and therefore, fluorescence microscopy was used to visualize endocytosed protein and subsequent nuclear localization. Results in Fig. 6, C–E, shows that the FITC-labeled DSS-Runx2 fusion protein was internalized and nuclear localized in T4-4 (C), MC3T3-E1 cells (D), and DPSC (E). Nuclear localization was confirmed by performing a z-stack confocal imaging. Fig. 6F is an orthogonal representation of the Z-stack imaging of the area shown in Fig. 6D (MC3T3-E1). The arrows in Fig. 6F represent areas of nuclear localization observed in the y-z and the x-z planes. Negative controls with FITC-labeled Runx2 protein alone indicated that Runx2 is not endocytosed by itself. Additionally, similar to the DSS and the GST-DSS proteins, a linear dose-response curve was obtained for the endocytosis of the FITC-labeled DSS-Runx2 fusion protein (Fig. 6G), indicating that the addition of a cargo protein does not alter the endocytic mechanism.
The DSS-tagged Runx2 Retains Its Intracellular Functionality
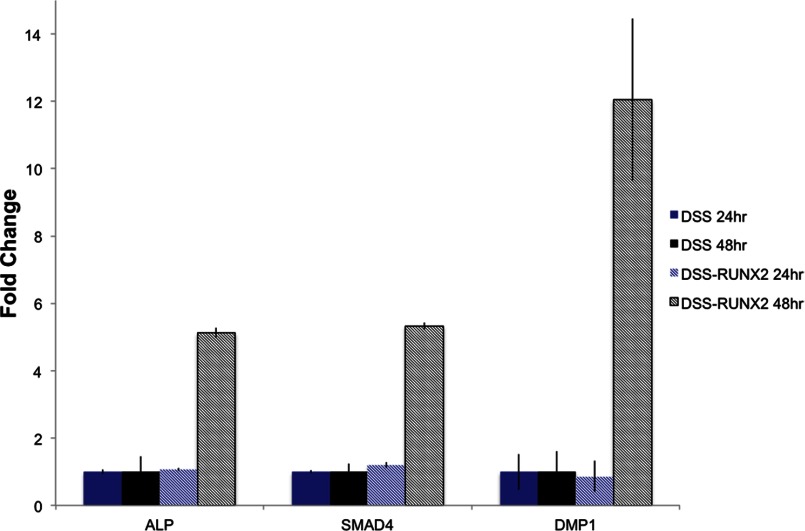
We next investigated if the endocytosed transcription factor is functionally active. To test this, we treated HMSC with the DSS-Runx2 fusion protein for a period of 24 and 48 h and observed the expression levels of DMP1, alkaline phosphatase and SMAD4 that are three of the many downstream target genes for Runx2. No change in gene expression was observed at the 24-h time point. However, all of these genes showed increased expression levels at 48 h, indicating the effectiveness of the transcription factor (Fig. 7). Additionally, the DSS peptide alone (carrier control) did not increase the expression levels of these genes.
FIGURE 7.
Endocytosed Runx2 retains its functionality after being expressed as a fusion protein with DSS. The graph shows the changes in expression pattern of three Runx2 downstream target genes upon treatment of HMSC with Runx2-DSS fusion protein for 24 and 48 h. No change was observed after 24 h post treatment. However, the expression levels of the three genes tested increased after 48 h of treatment. ALP, alkaline phosphatase.
Endocytosis of Runx2-DSS Fusion Protein Conjugated to Nanocrystals/QDs
QDs are resistant to photobleaching and can serve as tools for tracking the internalized proteins or as a marker for implanted cells. We, therefore, analyzed if Runx2-DSS fusion protein conjugated to QDs could be used to track the vehicle and the cargo proteins. We were able to observe internalization of the QD-tagged Runx2-DSS fusion protein (supplemental Fig. 2, A and B). Quantum dots are notorious for being trapped in endosomes after endocytosis. Therefore, we analyzed if the internalized Runx2-DSS-QD conjugates co-localized with the early endosome antigen 1 (EEA-1), an endosomal marker. Results in Fig. 8, A and B, demonstrate that the Runx2-DSS-QD signals did not co-localize with EEA-1 after 4 h (Fig. 8A) and 24 h (Fig. 8B). Fig. 8B also shows nuclear localization of the QD-tagged fusion protein. Results in Fig. 8, C and D, show three-dimensional rendering of z-stack confocal images of the endocytosed Runx2-DSS-QD conjugates after 24 h in MC3T3-E1 cells. Fig. 8, C and D, are three-dimensional reconstructions of the same z-stack section. In Fig. 8C, the DAPI signal was turned off to demonstrate the presence of the Runx2-DSS-QD conjugate in the nucleus. Fig. 8D shows the same image with the DAPI signal (white pseudo color). BSA-tagged quantum dots and quantum dots alone were used as negative controls and did not show any internalization (supplemental Fig. 2C).
FIGURE 8.
Endocytosis of the QD-tagged Runx2-DSS protein. A and B, representative confocal images show endocytosis of the QD-tagged Runx2-DSS (R2DSS QD) fusion protein (red) and its non-colocalization with the endosome marker EEA-1 in MC3T3 cells after 4 h (A) and 24 h (B). Note the nuclear localization of the endocytosed protein. C and D, three-dimensional reconstruction of z-stack confocal images of the MC3T3 cell-endocytosed QD-tagged R2 DSS fusion protein shows nuclear localization after 24 h of incubation in the absence (C) and presence (D) of DAPI-stained nuclear signal.
Visualization of the DSS-Runx2-QD Conjugates in Vivo
We tested the possibility of tracking cells treated with the DSS-Runx2-QD conjugates in vivo. We implanted biomimetic scaffolds containing DPSC pretreated for 24 h with the QD conjugates subcutaneously into nude mice. The mice were euthanized at 1- and 2-week time points and imaged. Fig. 9, A and B, show representative images of mice containing scaffold treated with QD conjugates (Fig. 9A) on one side and the control scaffold on the other (Fig. 9B) after 2 weeks of implantation. The white arrow in Fig. 9A represents the signal from the scaffold. We speculate that the trail of signal observed could be from migrating cells. The red arrow in Fig. 9, A and B, indicates nonspecific signal. Sections from the removed implants were also imaged for the presence of DSS-Runx2-QD conjugates. Fig. 9, C and D, shows representative images of 1- and 2-week-old scaffold implant sections with the DSS-Runx2-QD conjugates in the nucleus. No signal was observed from the control scaffold sections (data not shown). Collectively, these results indicate that it is possible to observe the QD-tagged cells in vivo in the animal and in vitro after removal of the implant.
FIGURE 9.
In vivo tracking of QD conjugated Runx2-DSS fusion protein. A, a representative image shows fluorescence from QD-tagged fusion protein containing cells embedded within biomimetic scaffolds after 2 weeks of implantation. The white arrow points to signal from the cells. B, shown is representative image shows fluorescence from control cells embedded within biomimetic scaffolds after 2 weeks of implantation. Note the absence of specific fluorescence signal. Red arrows in both images point to nonspecific signal. C and D, shown are representative confocal images from implant sections containing QD-tagged Runx2-DSS fusion protein after 1 week (C) and 2 weeks (D) of implantation. In both images, the red color indicates signal from the QDs, and blue color indicates DAPI nuclear stain. Note the nuclear localization of the fusion protein.
Endocytosis of (DSS)6-FLAG Peptide
We performed experiments with 6×DSS repeats to observe if they were enough to achieve endocytosis. Results from endocytosis experiments with the 6-DSS-FLAG peptide indicated that the FLAG antibody, when pre-bound to the peptide, could be endocytosed (Fig. 10, B and B1). The yellow arrows in Fig. 10B1 point to the intracellular presence of the FLAG antibody-peptide conjugate. However, unbound antibody was not endocytosed and instead bound to the cell surface and the cover glass non-specifically (Fig. 10, A and A1). The white arrows in Fig. 10A1 point to non-specifically bound FLAG antibody.
FIGURE 10.

Endocytosis of the (DSS)6- FLAG peptide. A and B, representative three-dimensional z-stack reconstructions of confocal images show that the FLAG antibody bound non-specifically to the cell surface and the cover glass when MC3T3 cells were incubated with it at 37 °C for 1 h (A) and that it is present in the cytoplasm when pre-bound to the (DSS)6-FLAG peptide. Images A and B show the reconstruction in the x-y plane, and images A1 and B1 show the reconstruction in the x-z plane. White arrows in A1 point to nonspecific binding of the FLAG antibody to the cell surface and to the cover glass. Yellow arrows in B1 point to intracellular presence of the antibody.
DISCUSSION
Cell penetrating peptides are currently being used for delivery of cargo through the cell membrane. In this study we demonstrate that the DSS domain in DPP can be internalized in a wide variety of cell types. Therefore, internalization pathway of the anionic DSS polypeptide was first determined. DPP contains an RGD domain, and it is known that RGD-containing proteins and peptides can be internalized as they recognize integrin receptors that mediate the interaction of cells with the ECM components. The absence of RGD domain in the DSS polypeptide confirmed that integrins were not involved in the internalization process. We tested the effect of Ikarugamycin, an inhibitor of receptor-mediated endocytosis on the internalization of DSS. Uptake of the DSS polypeptide was not hindered in the presence of ikarugamycin. As the internalization process was extremely rapid in all cell types studied and showed a linear dose response, it is most likely that DPP internalization is not receptor-mediated. This begs the question regarding the mechanism by which the DSS peptide is endocytosed. Classical endocytic pathways have been suggested as endocytosis mechanisms for cellular uptake of cell penetrating peptides. Interestingly, inhibitors for the classical endocytic pathways did not prevent endocytosis. Microtubules, actin filaments, and their associated molecular motors are known to drive the movement of endocytic vesicles. Disruption of the microtubules that are involved in long-range transport or microfilaments that provides short-range transport did not abort the internalization of DSS polypeptide. Even changes in pH did not affect endocytosis. At present, the mechanism responsible for the translocation of the DSS peptide is not known. However, it is possible that the charged residues of the DSS peptide can translocate across the hydrophobic core of the membrane through passive diffusion.
The above-mentioned attributes are the most sought-after properties for carrier proteins and cell-penetrating peptides. The use of therapeutic drugs and protein delivery is severely limited by the fact that most of these proteins are not permeable through the plasma membrane. T-cell receptors are considered to be an attractive therapeutic target; however, these are not applicable to all cell types. This can be remedied by the use of peptides or proteins that can function as carriers for therapeutic targets. One of the cell-penetrating peptides that has been extensively studied and is currently considered as a promising candidate is the TAT peptide from HIV. The TAT peptide can deliver biologically active target proteins in vivo and is considered therapeutically important (33–37). However, a limitation with these peptides is that the target proteins should be cross-linked with the peptides. This frequently requires denaturing of the target proteins and in other circumstances results in a conformational change that might render the target protein or the carrier ineffective. Another existing drawback is that after cellular uptake, there is a possibility that the carrier can be routed to endosomes and then lysosomes for degradation. Therefore, an efficient peptide for targeted intracellular delivery should possess the ability to translocate across the plasma membrane, have access to specific organelles, and bypass the lysosomal degradation pathway.
We investigated the possibility of utilizing the DSS peptide as a delivery vehicle for intracellular delivery of proteins. To demonstrate that the DSS polypeptide would qualify as a universal carrier for protein delivery, we tested its uptake in several cell types. Endocytosis was observed in all cell types studied (epithelial, mesenchymal, and endothelial cells). This suggests that the endocytosis of the DSS peptide is not a cell type-specific event. We further show that the DSS polypeptide could act as a carrier for other proteins by demonstrating the endocytosis of GST-tagged DSS polypeptide.
To test the ability of the DSS polypeptide to deliver a target protein to a specific intracellular location, we designed a fusion protein of the DSS polypeptide with the osteoblast “master” transcription factor Runx2. Runx2 contains a nuclear localization signal domain that directs nuclear localization. However, Runx2 by itself does not get endocytosed. Results from this study demonstrate the internalization and nuclear localization of the DSS-Runx2 fusion protein. The endocytosed Runx2 retained its activity as a transcription factor as shown by the up-regulation of its target genes. These results support our finding that the DSS peptide could be used as a vehicle for site-specific delivery.
Targeted delivery of the DSS peptide was also visualized by conjugating with semiconductor fluorescent QDs. These are nanometer-sized functionalized particles that display unique physical properties that make them particularly well suited for visualizing and tracking molecules in cells using florescence microscopy over prolonged periods. Several different peptides have been used to deliver nanoparticles and nanocrystals to intracellular locations (38). In this study we show that the fusion protein DSS-Runx2, when conjugated to QDs retains its ability to get endocytosed. However, the timeframe for nuclear localization was prolonged when conjugated with QDs. Importantly, the QDs did not colocalize with the endosomal marker EEA-1, indicating that the endosomal pathway has been avoided. Our cells also did not show any effects of cytotoxicity during the timeframe of the studies both in vitro and in vivo. Intracellular QDs could be used as a versatile tool especially to track transplanted cells in vivo. To achieve this, the endocytosis process should be feasible in a three-dimensional culture environment in scaffolds. We, therefore, tested the ability of the QD-tagged DSS-Runx2 fusion protein to be endocytosed by mesenchymal stem cells cultured in a three-dimensional biomimetic collagen/chitosan scaffold that we had published earlier (30). Results from this study showed that the QD-tagged fusion protein could be used for intracellular delivery in both three-dimensional and two-dimensional cultures and that cells tagged with these QD conjugates can be visualized in vivo.
In an attempt to narrow down the number of DSS repeats required for successful endocytosis, we synthesized a peptide containing six DSS repeats and a FLAG tag and examined its ability to carry the FLAG antibody across the plasma membrane. Three-dimensional confocal microscopy indicated that the 6-DSS-FLAG peptide showed the ability to transport the FLAG antibody across the plasma membrane. Future work will be devoted to comparing the endocytic efficiencies of the small DSS peptide with the longer DSS domain.
In conclusion we demonstrate that the DSS polypeptide derived from DPP has a cell membrane-penetrating capability. Endocytosis of this peptide was demonstrated in eight different cell types that include three primary cell types (primary mouse odontoblasts, DPSC and HMSC). This property makes the DSS polypeptide a promising tool for delivering bioactive proteins into various cell types. The peptide could be used to deliver therapeutic proteins to specific organelles bypassing the lysosomal degradation pathway. Another advantage of the DSS polypeptide is that the target protein/peptide can be genetically engineered at the N or C terminus of the DSS nucleotide sequence, resulting in a fusion recombinant protein of the carrier and the target. Future studies will be geared toward identifying the endocytic efficiency of the short DSS peptide and also toward identifying if the non-interference with the target protein function is a universal phenomenon.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant DE 19633. This work was also supported by the Brodie Endowment Fund.

This article contains supplemental Figs. 1–3.
- DPP
- dentin phosphophoryn
- rDPP
- recombinant DPP
- DPSC
- dental pulp stem cells
- HMSC
- human marrow stromal cell
- QD
- quantum dot
- TRITC
- tetramethylrhodamine isothiocyanate
- EEA-1
- early endosome antigen 1.
REFERENCES
- 1. Sabsay B., Stetler-Stevenson W. G., Lechner J. H., Veis A. (1991) Domain structure and sequence distribution in dentin phosphophoryn. Biochem. J. 276, 699–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stetler-Stevenson W. G., Veis A. (1983) Bovine dentin phosphophoryn. Composition and molecular weight. Biochemistry 22, 4326–4335 [DOI] [PubMed] [Google Scholar]
- 3. Hao J., Zou B., Narayanan K., George A. (2004) Differential expression patterns of the dentin matrix proteins during mineralized tissue formation. Bone 34, 921–932 [DOI] [PubMed] [Google Scholar]
- 4. Veis A., Wei K., Sfeir C., George A., Malone J. (1998) Properties of the (DSS)n triplet repeat domain of rat dentin phosphophoryn. Eur. J. Oral. Sci. 106, 234–238 [DOI] [PubMed] [Google Scholar]
- 5. He G., Ramachandran A., Dahl T., George S., Schultz D., Cookson D., Veis A., George A. (2005) Phosphorylation of phosphophoryn is crucial for its function as a mediator of biomineralization. J. Biol. Chem. 280, 33109–33114 [DOI] [PubMed] [Google Scholar]
- 6. Feng J. Q., Luan X., Wallace J., Jing D., Ohshima T., Kulkarni A. B., D'Souza R. N., Kozak C. A., MacDougall M. (1998) Genomic organization, chromosomal mapping, and promoter analysis of the mouse dentin sialophosphoprotein (Dspp) gene, which codes for both dentin sialoprotein and dentin phosphoprotein. J. Biol. Chem. 273, 9457–9464 [DOI] [PubMed] [Google Scholar]
- 7. George A., Srinivasan R., Thotakura S. R., Liu K., Veis A. (1999) Rat dentin matrix protein 3 is a compound protein of rat dentin sialoprotein and phosphophoryn. Connect Tissue Res. 40, 49–57 [DOI] [PubMed] [Google Scholar]
- 8. Yamakoshi Y., Lu Y., Hu J. C., Kim J. W., Iwata T., Kobayashi K., Nagano T., Yamakoshi F., Hu Y., Fukae M., Simmer J. P. (2008) Porcine dentin sialophosphoprotein. Length polymorphisms, glycosylation, phosphorylation, and stability. J. Biol. Chem. 283, 14835–14844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yamakoshi Y., Hu J. C., Iwata T., Kobayashi K., Fukae M., Simmer J. P. (2006) Dentin sialophosphoprotein is processed by MMP-2 and MMP-20 in vitro and in vivo. J. Biol. Chem. 281, 38235–38243 [DOI] [PubMed] [Google Scholar]
- 10. Yamakoshi Y., Hu J. C., Fukae M., Zhang H., Simmer J. P. (2005) Dentin glycoprotein. The protein in the middle of the dentin sialophosphoprotein chimera. J. Biol. Chem. 280, 17472–17479 [DOI] [PubMed] [Google Scholar]
- 11. Song Y. L., Wang C. N., Fan M. W., Su B., Bian Z. (2008) Dentin phosphoprotein frameshift mutations in hereditary dentin disorders and their variation patterns in normal human population. J. Med. Genet. 45, 457–464 [DOI] [PubMed] [Google Scholar]
- 12. Hart P. S., Hart T. C. (2007) Disorders of human dentin. Cells Tissues Organs 186, 70–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. MacDougall M., Dong J., Acevedo A. C. (2006) Molecular basis of human dentin diseases. Am. J. Med. Genet. A 140, 2536–2546 [DOI] [PubMed] [Google Scholar]
- 14. Thotakura S. R., Mah T., Srinivasan R., Takagi Y., Veis A., George A. (2000) The non-collagenous dentin matrix proteins are involved in dentinogenesis imperfecta type II (DGI-II). J. Dent. Res. 79, 835–839 [DOI] [PubMed] [Google Scholar]
- 15. Sreenath T., Thyagarajan T., Hall B., Longenecker G., D'Souza R., Hong S., Wright J. T., MacDougall M., Sauk J., Kulkarni A. B. (2003) Dentin sialophosphoprotein knockout mouse teeth display widened predentin zone and develop defective dentin mineralization similar to human dentinogenesis imperfecta type III. J. Biol. Chem. 278, 24874–24880 [DOI] [PubMed] [Google Scholar]
- 16. MacDougall M., Simmons D., Luan X., Gu T. T., DuPont B. R. (1997) Assignment of dentin sialophosphoprotein (DSPP) to the critical DGI2 locus on human chromosome 4 band q21.3 by in situ hybridization. Cytogenet. Cell Genet. 79, 121–122 [DOI] [PubMed] [Google Scholar]
- 17. Veis A. (1988) Phosphoproteins from teeth and bone. CIBA Found. Symp. 136, 161–177 [DOI] [PubMed] [Google Scholar]
- 18. George A., Bannon L., Sabsay B., Dillon J. W., Malone J., Veis A., Jenkins N. A., Gilbert D. J., Copeland N. G. (1996) The C-terminal domain of phosphophoryn contains unique extended triplet amino acid repeat sequences forming ordered carboxyl-phosphate interaction ridges that may be essential in the biomineralization process. J. Biol. Chem. 271, 32869–32873 [DOI] [PubMed] [Google Scholar]
- 19. Sfeir C., Veis A. (1995) Casein kinase localization in the endoplasmic reticulum of the ROS 17/2.8 cell line. J. Bone Miner. Res. 10, 607–615 [DOI] [PubMed] [Google Scholar]
- 20. Jadlowiec J., Koch H., Zhang X., Campbell P. G., Seyedain M., Sfeir C. (2004) Phosphophoryn regulates the gene expression and differentiation of NIH3T3, MC3T3-E1, and human mesenchymal stem cells via the integrin/MAPK signaling pathway. J. Biol. Chem. 279, 53323–53330 [DOI] [PubMed] [Google Scholar]
- 21. Jadlowiec J. A., Zhang X., Li J., Campbell P. G., Sfeir C. (2006) Extracellular matrix-mediated signaling by dentin phosphophoryn involves activation of the Smad pathway independent of bone morphogenetic protein. J. Biol. Chem. 281, 5341–5347 [DOI] [PubMed] [Google Scholar]
- 22. Eapen A., Ramachandran A., George A. (2012) Dentin phosphoprotein (DPP) activates integrin-mediated anchorage-dependent signals in undifferentiated mesenchymal cells. J. Biol. Chem. 287, 5211–5224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Katzmann D. J., Babst M., Emr S. D. (2001) Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106, 145–155 [DOI] [PubMed] [Google Scholar]
- 24. Anderson R. G., Kamen B. A., Rothberg K. G., Lacey S. W. (1992) Potocytosis. Sequestration and transport of small molecules by caveolae. Science 255, 410–411 [DOI] [PubMed] [Google Scholar]
- 25. Norkin L. C., Anderson H. A., Wolfrom S. A., Oppenheim A. (2002) Caveolar endocytosis of simian virus 40 is followed by brefeldin A-sensitive transport to the endoplasmic reticulum, where the virus disassembles. J. Virol. 76, 5156–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nichols B. J. (2002) A distinct class of endosome mediates clathrin-independent endocytosis to the Golgi complex. Nat. Cell Biol. 4, 374–378 [DOI] [PubMed] [Google Scholar]
- 27. Ravindran S., Narayanan K., Eapen A. S., Hao J., Ramachandran A., Blond S., George A. (2008) Endoplasmic reticulum chaperone protein GRP-78 mediates endocytosis of dentin matrix protein 1. J. Biol. Chem. 283, 29658–29670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang X., Mohandessi S., Miller L. W., Snee P. T. (2011) Efficient functionalization of aqueous CdSe/ZnS nanocrystals using small-molecule chemical activators. Chem. Commun. (Camb.) 47, 3532–3534 [DOI] [PubMed] [Google Scholar]
- 29. Shen H., Jawaid A. M., Snee P. T. (2009) Poly(ethylene glycol) carbodiimide coupling reagents for the biological and chemical functionalization of water-soluble nanoparticles. ACS Nano 3, 915–923 [DOI] [PubMed] [Google Scholar]
- 30. Ravindran S., Gao Q., Kotecha M., Magin R. L., Karol S., Bedran-Russo A., George A. (2012) Biomimetic extracellular matrix-incorporated scaffold induces osteogenic gene expression in human marrow stromal cells. Tissue Eng. Part A 18, 295–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heuser J. (1989) Effects of cytoplasmic acidification on clathrin lattice morphology. J. Cell Biol. 108, 401–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Frankel A. D., Pabo C. O. (1988) Cellular uptake of the tat protein from human immunodeficiency virus. Cell 55, 1189–1193 [DOI] [PubMed] [Google Scholar]
- 33. Mann D. A., Frankel A. D. (1991) Endocytosis and targeting of exogenous HIV-1 Tat protein. EMBO J. 10, 1733–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fawell S., Seery J., Daikh Y., Moore C., Chen L. L., Pepinsky B., Barsoum J. (1994) Tat-mediated delivery of heterologous proteins into cells. Proc. Natl. Acad. Sci. U.S.A. 91, 664–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vivès E., Brodin P., Lebleu B. (1997) A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J. Biol. Chem. 272, 16010–16017 [DOI] [PubMed] [Google Scholar]
- 36. Schwarze S. R., Ho A., Vocero-Akbani A., Dowdy S. F. (1999) In vivo protein transduction. Delivery of a biologically active protein into the mouse. Science 285, 1569–1572 [DOI] [PubMed] [Google Scholar]
- 37. Nagahara H., Vocero-Akbani A. M., Snyder E. L., Ho A., Latham D. G., Lissy N. A., Becker-Hapak M., Ezhevsky S. A., Dowdy S. F. (1998) Transduction of full-length TAT fusion proteins into mammalian cells. TAT-p27Kip1 induces cell migration. Nat. Med. 4, 1449–1452 [DOI] [PubMed] [Google Scholar]
- 38. Delehanty J. B., Boeneman K., Bradburne C. E., Robertson K., Bongard J. E., Medintz I. L. (2010) Peptides for specific intracellular delivery and targeting of nanoparticles. Implications for developing nanoparticle-mediated drug delivery. Ther. Deliv. 1, 411–433 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.