Abstract
High-risk types of human papilloma virus (HPV) are increasingly associated with oropharyngeal squamous cell carcinoma (OPSCC). Strikingly, patients with HPV-positive OPSCC are highly curable with ionizing radiation and have better survival compared with HPV-negative patients, but the underlying molecular mechanisms remain poorly understood. We applied an array-based approach to monitor global changes in CpG island hypermethylation between HPV-negative and HPV-positive OPSCCs and identified a specific pattern of differentially methylated regions that critically depends on the presence of viral transcripts. HPV-related alterations were confirmed for the majority of candidate gene promoters by mass spectrometric, quantitative methylation analysis. There was a significant inverse correlation between promoter hypermethylation of ALDH1A2, OSR2, GATA4, GRIA4, and IRX4 and transcript levels. Interestingly, Kaplan-Meier analysis revealed that a combined promoter methylation pattern of low methylation levels in ALDH1A2 and OSR2 promoters and high methylation levels in GATA4, GRIA4, and IRX4 promoters was significantly correlated with improved survival in 3 independent patient cohorts. ALDH1A2 protein levels, determined by immunohistochemistry on tissue microarrays, confirmed the association with clinical outcome. In summary, our study highlights specific alterations in global gene promoter methylation in HPV-driven OPSCCs and identifies a signature that predicts the clinical outcome in OPSCCs.
Introduction
Head and neck squamous cell carcinoma (HNSCC) is one of the most prevalent and lethal cancers worldwide (1, 2). Although improvements have been achieved in early detection, surgical techniques, and radiation and chemotherapeutic regimens, the survival rate has only marginally improved over the past decades (3). Accordingly, appropriate therapy and stratification of patients at high risk of treatment failure remains a major challenge for translational and personalized head and neck oncology.
The principal risk factors for HNSCC are tobacco and alcohol use (4). However, the escalating incidence of oropharyngeal squamous cell carcinoma (OPSCC) in the absence of a parallel rise in smoking and alcohol consumption suggests the involvement of additional nontraditional behavioral and environmental factors (5, 6). Indeed, infection by high-risk types of the human papilloma virus (HPV), most frequently type HPV16, has been found to be etiologically associated with an increasing number of patients with OPSCC (7–9). Although the majority of patients with HPV-positive OPSCC presents with an advanced stage of the disease at the time of diagnosis, these patients show a favorable prognosis independent of the therapeutic treatment regimen (10, 11). HPV-positive OPSCCs show differences in viral load and viral oncogene expression that are closely associated with clinical outcome (8, 10, 12, 13). In particular, it has been demonstrated that specific, cervical carcinoma–like viral transcript patterns were superior to viral DNA status alone in defining OPSCC with an active HPV16 involvement (HPV-driven OPSCC) (12).
Despite the growing evidence that HPV infection represents a major risk factor for OPSCC and may serve as an important predictor for patient survival and treatment decision making, knowledge of the underlying molecular mechanisms responsible for differences in the clinical behavior of patients with HPV-driven and non–HPV-driven tumors remains rather limited (1, 2, 7, 14).
Aberrant DNA hypermethylation within gene promoters is a hallmark in various human malignancies, including HNSCC (15–17), and usually results in downregulation of the associated genes. However, gene silencing via gene promoter methylation is still a novel concept to explain the development of HNSCC; in particular, its effect on the pathogenesis of HPV-driven OPSCC as well as the improved clinical outcome of affected patients remains obscure (14). To date, the vast majority of published data did not provide a strong association between patterns of DNA hypermethylation and overall survival (OS) (18), and only a few studies considered the HPV status (19–22).
A better understanding of molecular principles underlying the more favorable prognosis of HPV-driven OPSCC patients will certainly enable better classification of treatment responders and patients at high risk for treatment failure, thereby supporting therapeutic decision making. Additionally, it will result in innovative concepts for individualized treatment of both HPV-driven and non–HPV-driven OPSCCs, based on the etiology and biology of these tumors. Consequently, we focused our attention on HPV-dependent alterations in the gene promoter methylome: its chemically stable nature makes this epigenetic modification an attractive novel molecular biomarker for disease staging and prognosis, while its physiological reversibility makes it a potent druggable target for translational head and neck oncology. Here, we describe the identification of a signature for HPV-related gene promoter methylation by a genome-wide, array-based approach. The signature was confirmed by quantitative, gene-specific methylation analysis in 3 larger independent patient cohorts. Furthermore, we demonstrate that gene promoter hypermethylation inversely correlates with transcript and protein levels of affected candidate genes and present a methylation signature score that reliably predicts the clinical outcome of OPSCC patients.
Results
HPV-related alterations in global DNA hypermethylation in OPSCC.
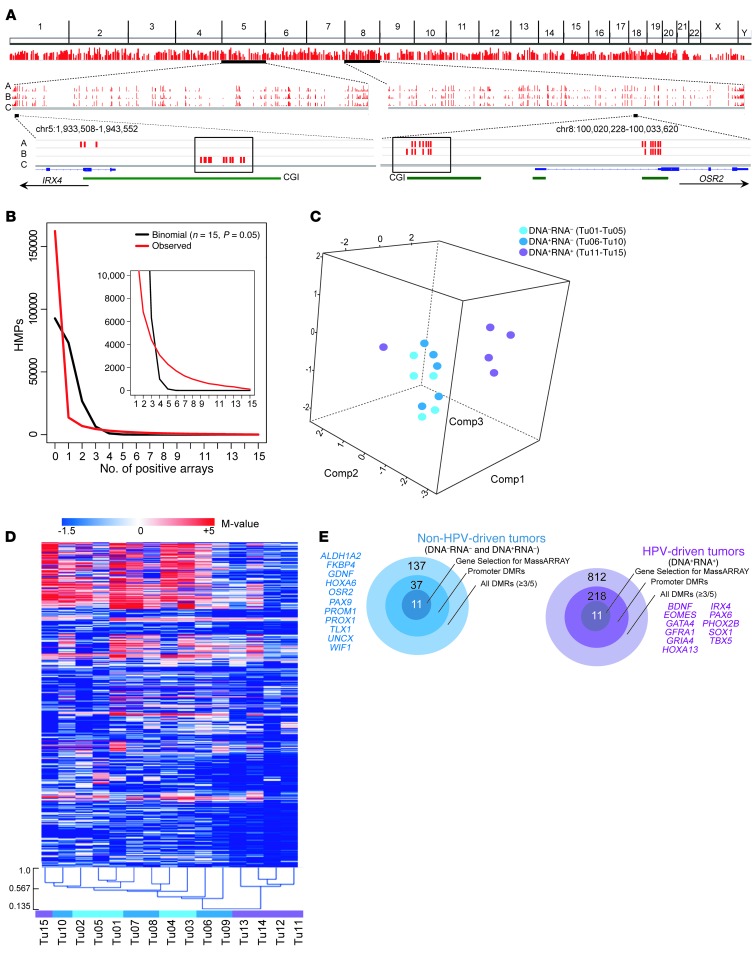
We enriched highly methylated genomic DNA by methyl-CpG IP (MCIp) from a healthy mucosa control and 3 OPSCC patient subgroups categorized by HPV status: group A was DNA–RNA– (OPSCC patients 1–5, corresponding to tumor DNA samples Tu01–Tu05); group B was DNA+RNA– (patients 6–10 and Tu06–Tu10); and group C was DNA+RNA+ (patients 11–15 and Tu11–Tu15). All 15 patients, together designated as cohort 1, were male with a median age of 57 years (range, 47–70 years; Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI67010DS1). Individual enriched tumor DNA samples were cohybridized with the enriched control on 244K CpG island arrays. The array-derived M values were used in a stepwise selection process (see “Candidate gene selection” in Methods) to identify differentially methylated regions (DMRs) being hypermethylated in the individual OPSCC subgroups (Figure 1A and Supplemental Table 2). Nonrandom occurrence of the top 5% differentially methylated CpG island microarray probes among the 15 OPSCC samples was observed (Figure 1B), indicative of association of aberrant DNA hypermethylation patterns with the disease. Overall, 4,219, 5,002, and 3,573 DMRs were identified in groups A, B, and C, respectively (Supplemental Tables 3–5); moreover, 892, 987, and 838 DMRs were found in annotated gene promoters in the respective groups. Common among all 3 groups were 2,161 DMRs, of which 520 were located in annotated gene promoters (Supplemental Tables 6–21).
Figure 1. Global HPV-related CpG island methylation changes in OPSCC samples.
(A) Top: Whole-genome distribution of DMRs commonly hypermethylated and represented at least once in all 3 groups. Line height corresponds to sample number (minimum, 3; maximum, 15). Middle: Exemplary distribution of DMRs on chromosome 5 and chromosome 8 of group A (HPV DNA–RNA–), group B (HPV DNA+RNA–), and group C (HPV DNA+RNA+). Line height corresponds to sample number (minimum, 1; maximum, 5). Bottom: Enlarged view of the promoter CpG island regions of IRX4 and OSR2. Boxed red bars represent promoter CpG island array probes hypermethylated in at least 3 of 5 samples from patient group A, B, or C. (B) Distribution of OPSCC-related hypermethylated probes (HMPs). The number of common top 5% HMPs (y axis) observed in increasing OPSCC sample numbers is plotted against the number of samples (x axis). The black curve shows the binomial distribution expected under the null hypothesis and the red curve shows the observed distribution. Inset: Enlargement (x axis coordinates, 1–15; y axis coordinates, 0–10,000). (C) PCA using all array-derived M values of the 15 OPSCC samples (Tu01–Tu15). (D) Unsupervised cluster analysis of hypermethylated DMRs. (E) Target diagrams summarizing the number of hypermethylated DMRs specific for the non–HPV-driven and HPV-driven OPSCC samples (see also Supplemental Tables 22 and 23). The candidate genes selected for confirmation are indicated.
To examine whether the methylation data reflected the HPV status of the 15 OPSCC samples, we applied principal component analysis (PCA), using all array-probe M values from samples Tu01–Tu15 as the measure of enrichment. Components 1, 2, and 3 with the largest variances distinguished 2 groups, Tu01–Tu10 and Tu11–Tu14, while Tu15 behaved as an outlier (Figure 1C). Unsupervised cluster analysis of differentially methylated probes confirmed the grouping of Tu01–Tu10 and Tu11–Tu14, while Tu15 clustered with Tu01–Tu10, yet was the least related (Figure 1D). Among patients 11–15, with identical HPV status (DNA+RNA+), patient 15 was the only one who was not event-free with regard to progression-free survival (PFS). Based on HPV RNA status, PCA, and clustering data, we therefore combined groups A and B (both HPV RNA–; Tu01–Tu10) in a single group designated non–HPV-driven, while group C (HPV RNA+; Tu11–Tu15, Tu15 still included because of being HPV RNA+) was designated HPV-driven. In total, 137 hypermethylated DMRs were enriched in the non–HPV-driven OPSCCs (present in at least 3 of 5 patients per group and in less than 3 of 5 samples within the group of comparison), of which 37 DMRs were located in annotated gene promoters. Conversely, 812 hypermethylated DMRs were enriched in HPV-driven tumors, and 218 of these DMRs were located in annotated gene promoters (Figure 1E and Supplemental Tables 22 and 23).
Confirmation of gene promoter DMRs by quantitative DNA methylation analysis in 2 independent OPSCC patient cohorts.
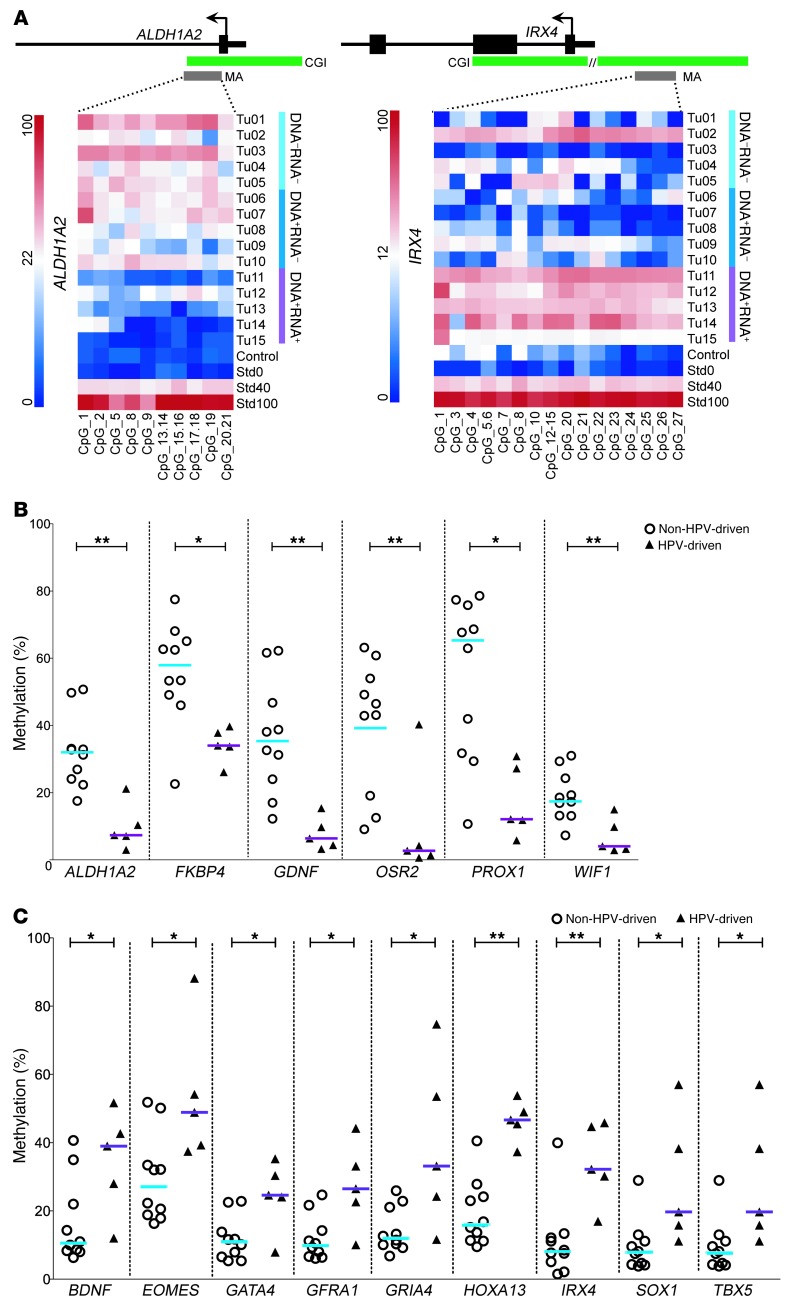
We applied STRING network analysis (23) for all genes with promoter DMRs identified per HPV status (37 and 218 for non–HPV-driven and HPV-driven, respectively) and selected 22 among those with physical and/or functional interactions that were hypermethylated in HPV-driven (n = 11) or non–HPV-driven (n = 11) tumors for further analysis (Figure 1E and Supplemental Figure 1). We confirmed the array-based methylation data of all gene promoter DMRs in cohort 1 by mass spectrometric analysis (MassARRAY), excepting the DMR of PAX6 (Figure 2 and Supplemental Figures 2 and 3). Moreover, methylation levels of 15 DMRs were significantly different between HPV-driven and non–HPV-driven tumors (Figure 2, B and C).
Figure 2. Confirmation of DMRs by quantitative mass spectrometric analysis.
(A) Exemplary heatmaps of promoter DMRs of ALDH1A2 and IRX4 with single CpG units (lines). ALDH1A2 proved to be hypermethylated in the non–HPV-driven tumors (Tu01–Tu10), IRX4 in the HPV-driven tumors (Tu11–Tu15). A healthy mucosa sample served as control; standards (Std) for 0%, 40%, and 100% methylation were included. Schemes above the heatmaps display the transcriptional start site (arrow) of the genes (black bars), the relative location of the CpG islands, and the MassARRAY amplicons (MA). (B) Significantly different hypermethylation in non–HPV-driven versus HPV-driven samples for ALDH1A2, FKBP4, GDNF, OSR2, PROX1, and WIF1. (C) Significantly different hypermethylation in HPV-driven versus non–HPV-driven samples for BDNF, EOMES, GATA4, GFRA1, GRIA4, HOXA13, IRX4, SOX1, and TBX5. Bars denote medians. **P < 0.01, *P < 0.05, Mann-Whitney U test.
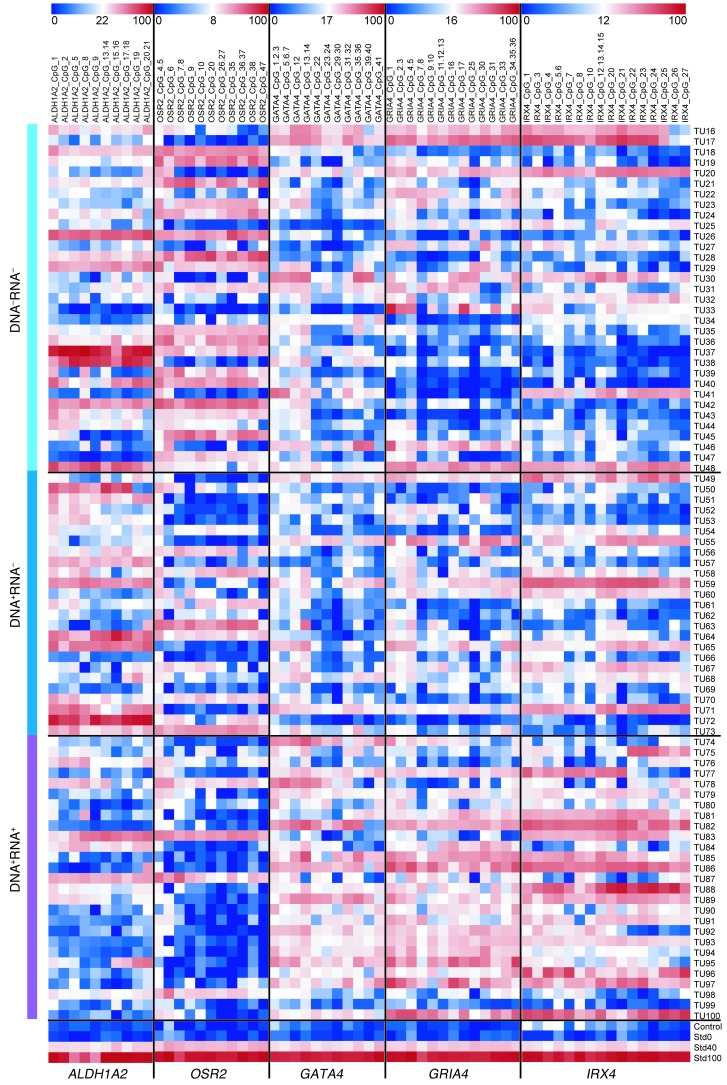
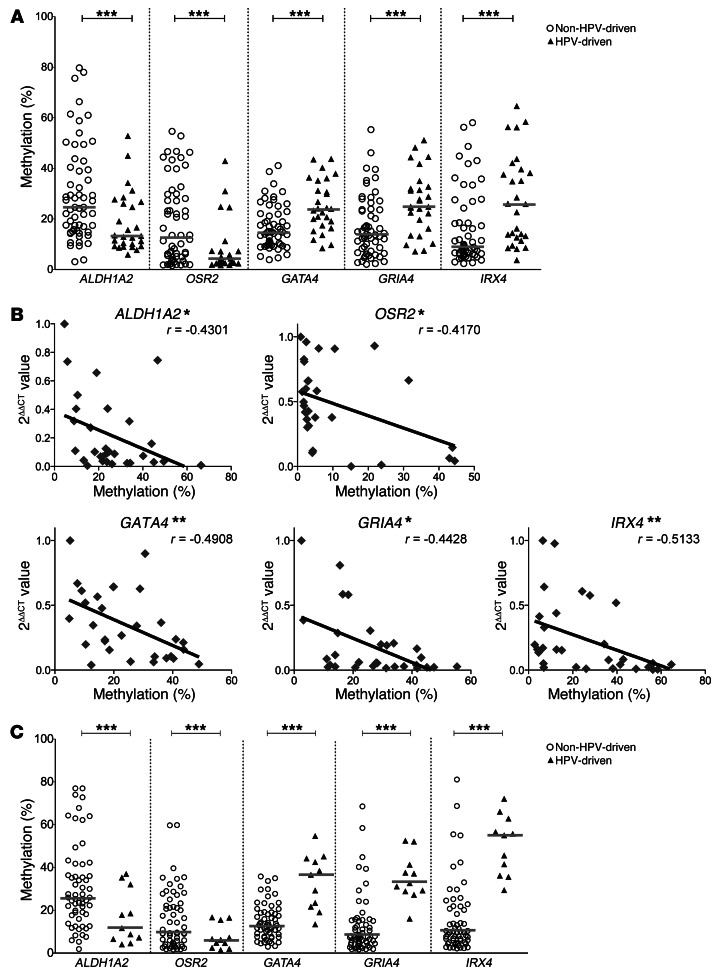
To further corroborate the methylation state of HPV-associated promoter DMRs, we analyzed ALDH1A2, OSR2, GATA4, GRIA4, and IRX4 by MassARRAY in 2 independent, larger cohorts of OPSCC patients with known HPV status. Cohort 2 (n = 85; Supplemental Table 1) included patients that were treated at Heidelberg University Hospital (n = 33 HPV DNA–RNA–; n = 25 HPV DNA+RNA–; n = 27 HPV DNA+RNA+), and cohort 3 (n = 70; Supplemental Table 1) included patients that were treated at the Leipzig University Hospital (n = 41 HPV DNA–RNA–; n = 18 HPV DNA+RNA–; n = 11 HPV DNA+RNA+). The genes were chosen because of their robust methylation differences observed within cohort 1 and the inverse correlation between methylation state and transcript level (see below). In agreement with our data from cohort 1, DMR methylation states were significantly different between the HPV-driven and the non–HPV-driven OPSCC cases in both cohorts (P < 0.001, Mann-Whitney U test; Figure 3 and Figure 4, A and C).
Figure 3. DNA methylation patterns of gene promoter DMRs for ALDH1A2, OSR2, GATA4, GRIA4, and IRX4 in an independent and larger cohort.
Methylation heatmaps from MassARRAY analysis of cohort 2 with 85 OPSCC patients (Tu16–Tu100; Heidelberg University Hospital). DNA–RNA–, n = 33 (Tu16–Tu48); DNA+RNA–, n = 25 (Tu49–Tu73); DNA+RNA+, n = 27 (Tu74–Tu10r0). A healthy mucosa sample served as control; standards for 0%, 40%, and 100% methylation are also depicted.
Figure 4. DNA methylation levels of gene promoter DMRs and transcript levels of ALDH1A2, OSR2, GATA4, GRIA4, and IRX4.
(A) Confirmation of HPV-related gene promoter methylation for ALDH1A2, OSR2, GATA4, GRIA4, and IRX4 in the combined cohort 1/2 (Heidelberg) (n = 100) by MassARRAY. Each symbol represents the mean of all CpG units in a sample amplicon. Bars denote medians. ***P < 0.001, Mann-Whitney U test. (B) Correlation between gene promoter methylation (mean of all CpG units) and transcript levels of 30 samples from cohort 2. Statistical nonparametric comparison for correlation between methylation and expression values was performed by Spearman’s Rho method (r is indicated). *P < 0.05, **P < 0.01, 2-tailed Spearman’s Rho test. (C) Confirmation of the discriminating methylation signature of 5 gene promoters in validation cohort 3 (Leipzig) (n = 70). Each symbol represents the mean of all CpG units in a sample amplicon; bars denote medians. ***P < 0.001, Mann-Whitney U test.
Gene promoter hypermethylation inversely correlates with transcript levels.
It is well established that gene promoter hypermethylation may downregulate transcription of the affected gene (24). We performed quantitative RT-PCR (qRT-PCR) analysis with cDNA from the 15 tumor samples of cohort 1 and observed a significant inverse correlation between gene promoter hypermethylation and relative transcript levels for ALDH1A2 and OSR2, which were hypermethylated in non–HPV-driven tumors, and for GRIA4, IRX4, GATA4, which were hypermethylated in HPV-driven tumors (Supplemental Figure 4). The same inverse correlation was also observed for ALDH1A2, OSR2, GATA4, GRIA4, and IRX4 in analyses of cDNA from 30 tumor samples of cohort 2 (n = 6 HPV DNA–RNA–; n = 11 HPV DNA+RNA–; n = 13 HPV DNA+RNA+; Figure 4B). Taken together, our data suggest that ALDH1A2, OSR2, GATA4, GRIA4, and IRX4 gene transcription is regulated by gene promoter hypermethylation, which in turn correlates with HPV status in OPSCC tumors.
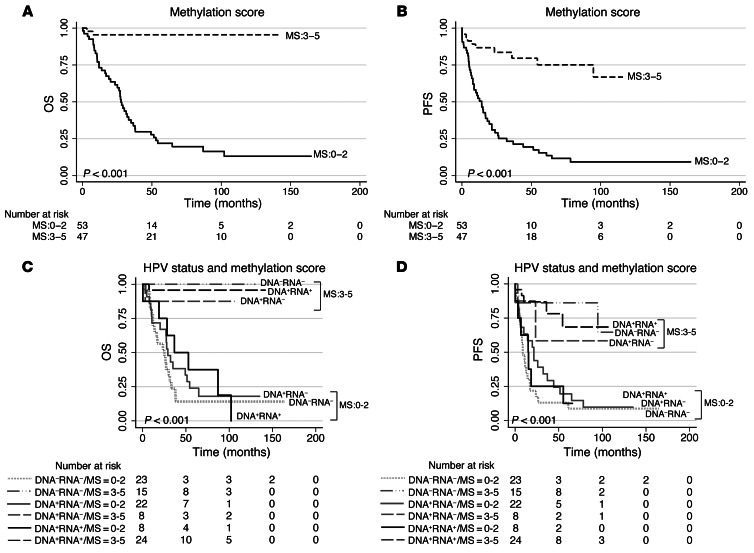
HPV-related promoter methylation states correlate with the clinical outcome of OPSCC patients.
We investigated whether the promoter methylation states of selected candidate genes ALDH1A2, OSR2, GATA4, GRIA4, and IRX4 correlate with the clinical outcome of OPSCC patients, using Heidelberg cohorts 1 and 2 combined (n = 100; referred to hereafter as cohort 1/2). We defined 2 methylation states, high and low, corresponding to above and below the median methylation value of all patients in the combined cohort, respectively. PFS and OS revealed a highly significant correlation (P < 0.001, log-rank/Mantel-Cox test) between states ALDH1A2lo, OSR2lo, GATA4hi, GRIA4hi, or IRX4hi and improved clinical outcome (Supplemental Figure 5). Furthermore, a strong pairwise correlation for methylation states of all 5 genes was observed (Spearman rho correlation, P < 0.001; r ranging from 0.505 to 0.718 for positive correlations and –0.303 to –0.465 for inverse correlations) and prompted us to define a score ranging 0–5 with respect to the methylation signature ALDH1A2lo, OSR2lo, GATA4hi, GRIA4hi, IRX4hi for further analysis (see Methods for details). Indeed, a high score of 3–5 significantly correlated with a good clinical outcome of OPSCC patients (P < 0.001, log-rank/Mantel-Cox test), whereas a low score of 0–2 correlated with a worse clinical outcome (Figure 5, A and B). Strikingly, the prognostic power of the methylation signature score concerning PFS and OS even appeared superior to that of HPV status (as defined by both DNA and RNA viral status, as previously described; Figure 5, C, and D, Supplemental Figure 6A, and ref. 12) or that of HPV DNA and p16 status (Supplemental Figure 6B and Supplemental Tables 25 and 26). Moreover, the strong correlation of the methylation signature score with clinical outcome was evident after stratification according to distinct HPV RNA patterns, HPV DNA viral load, and first-line treatment modality (Supplemental Figure 7, A–D, Supplemental Tables 25 and 26, and ref. 12). Multivariate models (Cox regression) for subgroups analyses were performed in order to assess the prognostic value of methylation score groups on both OS and PFS and showed highly significant differences for HPV status (P < 0.0001), as defined by both HPV DNA and RNA consideration, HPV DNA viral load, RNA patterns, and DNA/p16 status (Supplemental Table 26).
Figure 5. A signature of 5-gene promoter DMRs is associated with the clinical outcome of OPSCC patients.
Kaplan-Meier plot for OS (A) and PFS (B) with regard to methylation signature score (MS) based on the 5 promoter DMRs in cohort 1/2 (Heidelberg): ALDH1A2, OSR2, GATA4, GRIA4, and IRX4. Kaplan-Meier plot for OS (C) and PFS (D) with regard to the methylation signature score stratified for HPV status in cohort 1/2. P values were derived by log-rank/Mantel-Cox test.
Methylation signature score correlates with clinical outcome of OPSCC patients in 2 independent validation cohorts.
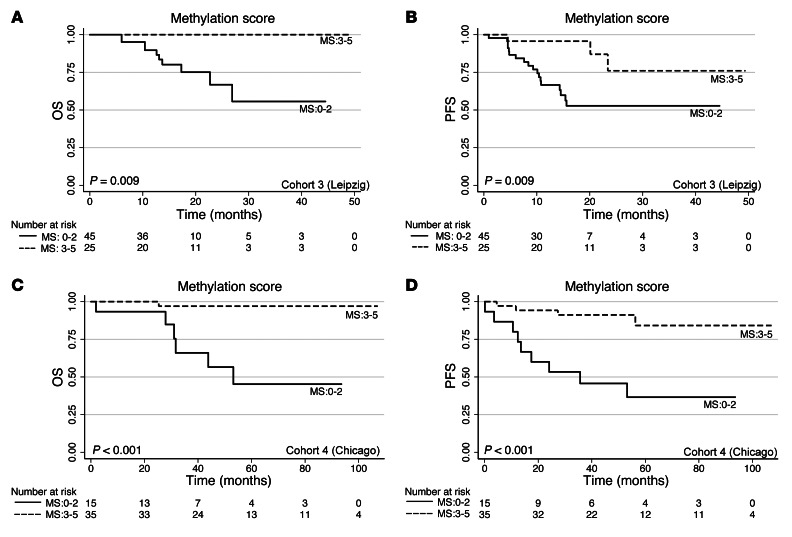
To confirm the prognostic value of the methylation signature score, we performed quantitative MassARRAY and survival analysis for 2 independent OPSCC patient cohorts. The first, cohort 3 (Leipzig), consisted of patients of a current prospective study with a median follow-up of 14.5 months (range, 1–49 months; n = 70). Analyses of PFS and OS with respect to promoter methylation pattern of individual candidate genes revealed similar findings as those of cohort 1/2 (Supplemental Table 27), and the methylation signature score reliably predicted the clinical outcome (P = 0.009; Figure 6, A and B), even after stratification according to HPV status (Supplemental Figure 6C).
Figure 6. The 5-gene (ALDH1A2, OSR2, GATA4, GRIA4, and IRX4) methylation signature score is associated with the clinical outcome of OPSCC patients in validation cohorts 3 and 4.
(A and B) Kaplan-Meier plots for OS (A) and PFS (B) with regard to the methylation signature score based on the 5 promoter DMRs for cohort 3 (Leipzig). (C and D) Kaplan-Meier plots for OS (C) and PFS (D) with regard to the methylation signature score based on the 5 promoter DMRs for cohort 4 (Chicago). P values were derived by log-rank/Mantel-Cox test.
The second validation cohort, cohort 4 (Chicago), represented patients with chemoradiation as first-line treatment (n = 50); again, Kaplan-Meier analysis confirmed the strong predictive power of the methylation signature score with respect to clinical outcome (P < 0.001; Figure 6, C and D, and Supplemental Table 28).
Correlation between ALDH1A2 protein level and clinical outcome.
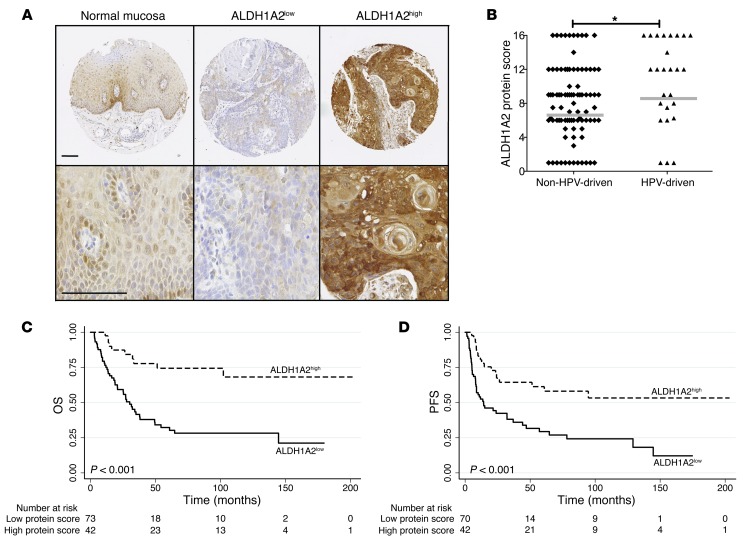
The candidate gene ALDH1A2 encodes aldehyde dehydrogenase family 1 member A2 (ALDH1A2; also known as RALDH2), a putative tumor suppressor and key player in retinoid metabolism and signaling (25–28). We determined ALDH1A2 protein levels by immunohistochemical staining of tissue microarrays generated from primary tumors of 115 OPSCC patients with known HPV status (n = 89 non–HPV-driven; n = 26 HPV-driven; Supplemental Table 29). Immunohistochemical staining revealed low to moderate ALDH1A2 protein levels (combined as ALDH1A2low) in keratinocytes of healthy mucosa as well as 73 (63.5%) primary tumors, and high ALDH1A2 protein levels in 42 (36.5%) primary tumors (Figure 7A). In line with our previous finding that a low ALDH1A2 promoter methylation state is common in HPV-driven OPSCCs accompanied with increased transcript levels, we found significantly higher ALDH1A2 protein levels in HPV-driven than non–HPV-driven tumors (Figure 7B). Moreover, PFS and OS strongly correlated with the ALDH1A2 protein expression level (OS, P < 0.001; PFS, P = 0.001; log-rank/Mantel-Cox test; Figure 7, C and D).
Figure 7. Correlation of ALDH1A2 protein expression and clinical outcome of OPSCC patients.
(A) Representative images at different power of ALDH1A2 expression on tissue microarrays of normal mucosa and exemplary tumors with low and high protein levels (brown signal). Counterstaining with hematoxylin. Scale bars: 100 μm. (B) Lower ALDH1A2 expression score in non–HPV-driven (n = 89) versus HPV-driven (n = 26) tumors. Bars denote medians. *P < 0.05, 2-tailed Mann-Whitney U test. Kaplan-Meier plots for OS (C) and PFS (D) with regard to ALDH1A2 protein levels in n = 115 OPSCC patients. P values were derived by log-rank/Mantel-Cox test.
Discussion
Epidemiological and clinical studies highlighted an unexpected heterogeneity for HNSCC concerning etiology as well as environmental, cellular, and molecular features, hampering accurate prognosis, treatment planning, and identification of causative genes that may serve as molecular drug targets (1, 2). Several groups used comparative genomic hybridization and whole exome sequencing to identify genomic imbalance and somatic gene mutations in HNSCCs, including samples of HPV-positive tumors (29–34). These studies unraveled an obvious difference in the genetic landscape, with significantly more mutations in HPV-negative compared with HPV-positive tumors, supporting the hypothesis that better clinical outcome of patients with HPV-positive tumors is simply a reflection of less genetic aberrations at the time of treatment. In this study, we screened the CpG island methylome of 15 OPSCC tumors and identified specific methylation signatures (corroborated by 3 larger and independent cohorts of OPSCC patients) that discriminated between HPV-driven and non–HPV-driven cases and strongly correlated with patient outcome. Our data demonstrated that, in addition to genetic aberrations, epigenetic alterations may also critically contribute to histopathological and clinical differences between HPV-driven and non–HPV-driven tumors.
In line with previous findings that heterogeneity — with respect to biological and clinical behavior of OPSCC cases typed positive for HPV DNA — is a reflection of viral load and viral oncogene expression (12, 13, 35–44), our HPV DNA+RNA– OPSCC cases were closer to HPV-negative than were the HPV DNA+RNA+ cases with respect to aberrant DNA methylation changes. Our data suggest that differences in DNA methylation are mediated by active viral transcription and not merely by the presence of viral DNA. Indeed, expression of HPV16 oncoproteins E6 and E7 has been shown to modulate the expression and function of key regulators in DNA methylation, such as DNMT1 and DNMT3B (45–47), but the underlying molecular mechanisms by which HPV components contribute to DNA methylation changes of the host cell genome remain to be addressed.
Despite the relatively small number of OPSCC cases in the initial array-based methylome screen (i.e., cohort 1), we confirmed selected DMRs by quantitative methylation analysis in 3 much larger, independent OPSCC cohorts. This high reliability of our initial screen may be attributed to both the stringent selection criteria applied and to the stable nature and specificity of aberrant DNA methylation in HPV-driven OPSCCs. We selected 5 genes for a more detailed analysis, based on their robust difference in HPV-dependent gene promoter hypermethylation in larger OPSCC patient cohorts and their highly significant and inverse correlation between gene promoter hypermethylation and transcript levels in tumor samples. We identified a promoter methylation signature of 5 genes — ALDH1A2lo, OSR2lo, GATA4hi, GRIA4hi, and IRX4hi — that not only characterized HPV-driven tumors, but also highly correlated with (and, hence, may serve as reliable predictor for) better OPSCC patient clinical outcome. Aberrant promoter hypermethylation of ALDH1A2, OSR2, GATA4, and GRIA4 as such is not a novel finding and has already been described in established human tumor cell lines or in tumor samples (27, 48–56). A striking novelty, however, was the robust correlation of the methylation signature score with the clinical outcome of OPSCC patients, which was confirmed in 3 independent cohorts, independent of HPV status or first-line treatment modality.
Considering the current debate in the field of head and neck oncology on de-escalating the treatment regimen of HPV-driven tumors to reduce toxicity and morbidity (57), our 5-gene methylation signature may enable a refined classification for appropriate treatment options of OPSCC patients, regardless of their HPV status. Accordingly, validation of the methylation signature score in a prospective and multicenter study including OPSCC, but also HNSCCs of other locations, is warranted.
So far, studies on the association between HPV and aberrant gene promoter methylation in HNSCC are merely descriptive and lack causal explanation for differences in clinical behavior (19, 20, 58, 59). Only one study (22) identified a potential causal link among HPV, promoter hypermethylation of SMG1 (encoding a PI3K-related kinase involved in the maintenance of genome integrity via genotoxic stress response pathways; refs. 60, 61), and enhanced radiation sensitivity of cultivated cells.
Several in vitro studies described functional interference by HPV16 with retinoid signaling in keratinocyte differentiation and transformation (62–67). Whether this interference contributes to HPV-related pathogenesis is unclear, yet it may offer novel treatment options. A common feature of the 5 methylation signature genes identified in our study is their involvement in tissue development and regeneration, and 4 of them, ALDH1A2, OSR2, GATA4, and IRX4, are related to retinoid metabolism and signaling (25, 26, 68–74). Since retinoic acids exert potent effects on cell growth, differentiation, and apoptosis, targeting one or a combination of the 4 genes and thereby modulating retinoid metabolism and signaling may offer novel avenues in successful treatment of OPSCC (75, 76). Interestingly, a recent study demonstrated that a retinoic acid–rich tumor microenvironment is required for cytotoxic T cell accumulation accompanied by protective antitumor immunity (77), and it is worth mentioning that an improved clinical outcome of OPSCC patients has been shown to be associated with increased amounts of peripheral and tumor infiltrating T cells (78, 79).
The protein ALDH1A2 belongs to a large family of aldehyde dehydrogenases and represents the rate-limiting enzyme in the production of retinoic acid from retinaldehyde (25, 80). Although ALDH1A2 expression has been observed in normal and tumor stem cells (28), several reports support its role as a putative tumor suppressor gene in prostate and ovarian cancer (27, 68, 81). In line with these findings, we observed a strong correlation between ALDH1A2 protein levels and favorable clinical outcome of OPSCC patients.
In summary, we identified an HPV-related promoter methylation signature of 5 genes with strong correlation to and strong predictive power for clinical outcome of OPSCC patients. Concerning clinical application, detection of the newly identified methylation signature score at the time of first diagnosis offers a novel strategy of stratification to discriminate patients at high risk for treatment failure, who will urgently need multimodal therapy, and those who will benefit from treatment de-escalation to avoid toxic side effects and improve quality of life. Moreover, encoded proteins that are affected by gene promoter methylation or other key players in the retinoic acid signaling pathway may represent promising drug targets for innovative and more efficient cancer therapy.
Methods
Patient samples
Tumor samples of cohorts 1 and 2 as well as specimens for tissue microarray analysis were obtained from OPSCC patients treated at the University Hospital Heidelberg between 1990 and 2008. Clinical and follow-up data were provided by the Department of Otolaryngology, Head and Neck Surgery, at the University Hospital Heidelberg (Supplemental Tables 1, 25, 26, and 29). Tissue specimens from OPSCC patients were provided by the tissue bank of the National Center for Tumor Disease (Institute of Pathology, University Hospital Heidelberg) after approval by the local institutional review board (ethic vote 206/2005).
DNA and RNA from tumors of reference cohort 3 were obtained from OPSCC patients treated at the University Hospital Leipzig between 2007 and 2011. Samples were collected upon patient’s consent as approved by the local institutional review board (Medical Faculty of University Leipzig ethic votes 201-10-12072010 and 202-10-1207210). Clinical data were provided by the Department of Otolaryngology, Head and Neck Surgery, at the University Hospital Leipzig (Supplemental Tables 1 and 27).
DNA from tumors of an additional validation cohort, cohort 4 (Supplemental Tables 1 and 28), from The University of Chicago Medicine were also included in the present study. Frozen OCT tumor samples were obtained from the University of Chicago Head and Neck Cancer tissue bank (IRB approved protocol UCCCC#8980). Specimens were obtained prior to treatment with concurrent chemoradiotherapy.
DNA and RNA isolation
Cohort 1/2.
2–10 mg of 16-μm tissue cryosections were homogenized in liquid nitrogen and stored at –80°C. H&E staining of adjacent sections indicated a tumor cell content of at least 70%. Genomic DNA was isolated by the QIAamp DNA Mini Kit (Qiagen). Total RNA was isolated using the RNeasy mini kit including the on-column DNase I digestion according to the manufacturer’s instructions (Qiagen). Quantity and quality of DNA and RNA were determined using the NanoDrop 2000 spectrophotometer (Thermo Scientific).
Cohort 3.
Samples of the Leipzig cohort were put into TRIzol (Invitrogen) and stored at –80°C. The frozen samples in TRIzol were homogenized for 80 seconds at 160 g using the Minilys benchtop homogenizer (Peqlab Biotechnologie) followed by isolation of RNA and DNA according the Chomczynski protocol (82). The aqueous phase was used for RNA preparation by use of the Ambion Pure Link RNA Mini Kit (Invitrogen) including the on-column DNase I digestion (Qiagen) according to the manufacturer’s instructions. Genomic DNA was prepared from the organic phase according to the standard TRIzol protocol. After ethanol precipitation, DNA was resolved in water and used without further cleanup. Quantity and quality of DNA and RNA were determined using the NanoDrop 2000 spectrophotometer.
Cohort 4.
H&E slides were prepared by cutting and staining a section from each OCT frozen block and reviewed for their tumor content by an expert HNC pathologist. Samples or fractions of samples with a tumor content of at least 60% were extracted. Tissue was cut from the OCT blocks, pulverized using CryoPrep (Covaris), and homogenized in lysis buffer from an AllPrepRNA/DNA/Protein Mini kit (Qiagen) using an Ultrasonicator (Covaris). DNA, RNA, and protein were isolated from each sample following the manufacturer’s protocol. Quantity and quality of the purified genomic DNA were determined with Quant-iT Picogreen (Invitrogen) and NanoDrop 2000c spectrophotometer, respectively.
HPV status analysis and definition
The HPV DNA status and viral transcript pattern were determined as previously described by multiplex HPV genotyping and viral RNA pattern analysis on fresh-frozen OPSCC specimens (12). Tumors from cohort 1/2 (Heidelberg) and cohort 3 (Leipzig) were classified based on HPV DNA and RNA pattern analysis (12) into DNA–RNA– (group A), DNA+RNA– (group B), and DNA+RNA+ (group C). Groups A and B are referred to herein as non–HPV-driven, and group C is referred to as HPV-driven. DNA viral load and RNA patterns for HPV16 were as follows: RNA pattern 1, high E6I/E1^E4; RNA pattern 2, high E1C/L1. RNA positivity was defined by the combination of the 2 RNA patterns (Supplemental Table 25). In cohort 2, 2 cases were positive for HPV33 and HPV35. We considered the HPV16 DNA+ pattern as HPV DNA+, and RNA patterns concerning HPV16 were as previously described (12). For cohort 4 (Chicago), DNA HPV status was assessed performing an HPV E6 nested multiplex PCR using AmpliTaq Gold (Applied Biosystems) and subsequent methodology as described previously (83). SCC2 (HPV16) and HeLa (HPV18) cell lines were used as positive controls, whereas SCC151 and water were used as negative and no-template controls, respectively. PCR products were analyzed on a 2% agarose gel.
MCIp and CpG island microarray analysis
2.5 μg genomic DNA were sonicated with the Bioruptor NextGen (Diagenode) to fragments of 200–800 bp controlled by agarose gel electrophoresis. Highly methylated DNA was enriched by MCIp (84, 85) using the SX-8G IP-Star robot (Diagenode) and 60 μg methyl-CpG–binding domain–Fc protein coupled to 40 μl protein A–coated magnetic beads (Diagenode). DNA eluates with increasing salt concentrations (fraction A, 300 mM; fraction B, 400 mM; fraction C, 500 mM; fraction D, 550 mM; fraction E, 1,000 mM), and methylation degree were desalted with MinElute columns (Qiagen) and controlled for enrichment of methylated DNA by real-time PCR analyzing the imprinted gene SNRPN. The nonmethylated SNRPN allele was eluted at low-salt concentration, and the methylated allele was eluted at high-salt concentration.
Highly methylated DNA from fraction E was labeled with Alexa 5 (tumor) or Alexa 3 (healthy control), and labeled tumor and control samples were cohybridized to a human CpG island tiling array (244K; Agilent Technologies) that carries 237,220 oligonucleotide probes (45–60 bp length) representing 27,800 CpG islands. Probes were annotated according to Agilent (NCBI Build 36.1; March 2006). Microarrays were scanned with a DNA microarray scanner (Agilent), and scanning TIF files were analyzed using Feature Extraction Software 10.5 (Agilent) with the ChIP protocol setting. Raw data were analyzed using R statistical package (version 2.13.1) as described previously, with background correction and log2 ratio transformation according to the NormExp method with offset of 50 (86, 87). Variation between cohybridized samples was reduced by intensity-based LOESS normalization on rank-invariant probes and negative controls (M values, calculated as log2[Alexa 5/Alexa 3]). For between-array normalization, log intensity ratios (M values) and log intensity averages (A values) were scaled to have the same median absolute value across all arrays. SAM test was done with scaled M values all ranging between –5 and +5 (88). This normalization was done using the R statistical environment (version 2.14; package samr).
Candidate gene selection
To narrow down the list of DMRs displaying tumor type–specific hypermethylation, we chose the following stepwise criteria: (a) a “region” consisted of a coherent sequence with more than 1 probe, and in which 2 vicinal probes were separated by at least 500 bp; (b) per tumor sample (array), only the top 5% hypermethylated probes were considered; (c) a DMR of a tumor sample was represented by at least 2 vicinal top 5% probes, allowing a gap of a single non–top 5% probe; (d) a DMR was positive in at least 3 of 5 samples from the same tumor type. Furthermore, we applied network analysis (STRING; ref. 23) and mainly selected genes included in networks either in the original confidence view or with 1-step enhancement of the network with intermediate predicted functional partners (score, >0.997). Also included were 2 genes, HOXA6 and HOXA13, that were presented as disconnected nodes in the network. To visualize genomic location of DMRs and CpG island array probes (Figure 1A), custom tracks were generated and loaded into the Integrated Genomics Viewer (IGV version 2.1.; ref. 89).
Quantitative DNA methylation analysis
Percent DNA methylation was determined by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MassARRAY; Sequenom) as previously described (90). Amplicons were designed according to positive regions derived from microarray results (Supplemental Table 24). Methylation standards (0%, 20% 40%, 60%, 80%, and 100% methylated genomic DNA) were used to control for the full dynamic range of measurements. Heatmaps were generated to visualize methylation levels for each CpG unit or averages of amplicons using R Bioconductor scripts library (gplots).
For methylation score calculation, the mean value of all CpG units per amplicon was used. Samples were divided into 2 groups (high and low methylation), based on the median of the methylation values of the cohort (above and below median). At gene level, a favorable prognostic methylation pattern (ALDH1A2lo, OSR2lo, GATA4hi, GRIA4hi, and IRX4hi) scored 1 per gene, whereas an unfavorable prognostic methylation pattern (ALDH1A2hi, OSR2hi, GATA4lo, GRIA4lo, and IRX4lo) scored 0 per gene. The total of all 5 scores for the 5 genes per patient was calculated (score values, 0–5), and 2 groups were used for survival functions: (a) methylation score 0–2 (0–2 genes with favorable prognostic pattern) and (b) methylation score 3–5 (3–5 genes with favorable prognostic pattern). Methylation cutoff values from training cohort 1/2 (above/below median) were used for defining the methylation signature score in validation cohorts 3 and 4.
cDNA preparation and qRT-PCR
First-strand cDNA was synthesized from 2 μg total RNA using the Superscript II First strand synthesis system (Invitrogen) according to the manufacturer’s protocol. The cDNA was diluted 1:5 for use in qRT-PCR. The qRT-PCR reaction consisted of 2 μl template cDNA, 0.5 μl primer mix (10 μM), 0.05 μl Taqman probe (RealTime ready Universal ProbeLibrary; Roche Applied Science), and 2.5 μl Master Mix (Roche Applied Science) in a total volume of 5 μl. See Supplemental Table 24 for primers, amplicon sequences, and probe IDs. qRT-PCR reaction was performed in a Lightcycler 480 (Roche Applied Science) with the following protocol: denaturation at 95°C for 10 minutes; followed by 45 cycles of 95°C for 10 seconds, 55°C for 20 seconds, and 72°C for 1 minute; and cooling at 40°C for 10 seconds. 3 housekeeping genes were included in the analysis (ACTB, GAPDH, and HPRT). Results for Ct values were obtained with LightCycler 480 software (version 1.5.0; Roche Applied Science) and Abs Quant/2nd Derivative Max analysis. 2ΔΔCt values were calculated with the mean of the Ct of the 3 housekeeping genes as reference and the Ct of healthy mucosa sample as calibrator.
Tissue microarrays and immunohistochemical staining
2 tissue microarrays containing multiple spots from 115 primary OPSCC patients (Supplemental Table 28) were generated as previously described (91). Tissue microarrays included 61 of 100 patients from cohort 1/2 (Heidelberg) (Supplemental Table 29). Briefly, H&E-stained sections were cut from each donor block to define representative tumor regions. Small tissue cylinders with a diameter of 0.6 mm were taken from selected areas of each donor block using a tissue chip microarrayer (Beecher Instruments) and transferred to a recipient paraffin block. This block was cut in 2-μm paraffin sections using standard techniques. Immunohistochemistry was performed with the anti-human ALDH1A2 antibody (HPA010022, Sigma-Aldrich). Tissue microarrays were scanned using the Nanozoomer HT Scan System (Hamamatsu Photonics). 3 experienced observers analyzed scanned slides by using the NDP Viewer software (version 1.1.27). Semiquantitative analysis was performed according to the number of stained tumor cells (score A; 1, no positive cells; 2, less than 33% positive cells; 3, between 34% and 66% positive cells; 4, more than 66% positive cells), and according to the staining intensity (score B; 1, no staining; 2, weak staining; 3, moderate staining; 4, high staining) (Supplemental Table 25). The multiple score for number of stained tumor cells (score A) and staining intensity (score B) was used for defined subgroups (final expression scores ranged 1–16). For p16 expression, immunohistochemistry was performed as previously described (12). p16 status was available for 91 of 100 patients from cohort 1/2 (Heidelberg) (Supplemental Tables 25 and 26).
Statistical analyses and survival functions
To investigate global methylation patterns, we noted for each differentially methylated CpG island microarray probe (DMP) the number of samples in which the probe was among the top 5% probes. This distribution was then compared with the expected null distribution if randomly selected probes were used instead of those identified in the top 5%. The expected distribution was a binomial distribution with parameters sample number n = 15 and P = 0.05 (fraction of top 5% probes). For PCA, R Bioconductor software (prcom and pcaMethods packages) was used. Hierarchical cluster analysis was performed with R (pvclust and gplots packages) and used Euclidian distance (average linkage). The analysis was run repeatedly by decreasing the number of differently methylated probes from 100% to 10% to validate cluster stability.
Statistical analysis of methylation data was performed for grouped data by Mann-Whitney U test, and 2-tailed P values were calculated in Graphpad Prism (version 5.0c; GraphPad Software). Spearman’s Rho test (Spearman’s rank correlation coefficient) was used for coefficiency check of methylation status (mean of all CpG units) versus relative expression data (2ΔΔCt values), and Spearman’s Rho test and estimation of Spearman’s rank correlation coefficient (r value) as well as Rho 2-tailed P value were calculated in Graphpad Prism (version 5.0c; GraphPad Software).
OS was measured from registration date until date of OPSCC-related death, censoring patients alive at last follow-up and non–OPSCC-related deaths. PFS was measured from registration date until date of progression or death, censoring patients without a documented progression at last follow-up. Local recurrence, lymph node or distant metastasis, second primary carcinoma, or date of OPSCC-related death within the follow-up period were considered progression events. Estimation of OS and PFS distributions was performed by the Kaplan-Meier method (SPSS version 20, Stata version 12). Differences between groups were determined by log-rank/Mantel-Cox test. Multivariate Cox regression was performed to assess independent effect of methylation signature score groups using R software (version 2.15).
Study approval
The studies presented herein received approval from the ethics committee of the Medical Faculty of Heidelberg University (ethic vote 206/2005), the ethics committee of the Medical Faculty of the University Leipzig (ethic votes 201-10-12072010 and 202-10-1207210), and the IRB of University of Chicago (approved protocol no. UCCCC#8980). Samples were collected upon patient consent, as approved by the ethics committee of the Medical Faculty of the University Leipzig.
Data access
The global methylation data from the present study were submitted to NCBI GEO (accession no. GSE41152; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE41152).
Supplementary Material
Acknowledgments
We gratefully acknowledge Nataly Henfling, Antje Schuhmann, Ines Kaden, and Marion Bähr for excellent technical assistance; Oliver Mücke for support with MassARRAY experiments; and Manuela Zucknick for critical feedback on biostatistics. We thank the tissue bank of the National Center for Tumor Disease (Institute of Pathology, University Hospital Heidelberg) for providing tumor specimens of OPSCC patients. This work was supported by the Dietmar Hopp Stiftung (to J. Hess and P.K. Plinkert) and the Stiftung Tumorforschung Kopf-Hals (to J. Hess). E. Kostareli was supported by the Postdoc Fellowship of the Medical Faculty of Heidelberg University. D. Holzinger was supported by a PhD grant of the German Research Foundation (DFG), Graduiertenkolleg 793: “Epidemiology of communicable and chronic, non-communicable diseases and their interrelationships.” This study was funded in part by European Commission grant HPV-AHEAD (FP7-HEALTH-2011-282562; to M. Pawlita and D. Holzinger)
Footnotes
Conflict of interest: Michael Pawlita has received research support through cooperation contracts of DKFZ with Roche and Qiagen in the field of development of HPV diagnostics.
Citation for this article: J Clin Invest. 2013;123(6):2488–2501. doi:10.1172/JCI67010.
References
- 1.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11(1):9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 2.Rothenberg MS, Ellisen LW. The molecular pathogenesis of head and neck squamous cell carcinoma. J Clin Invest. 2012;122(6):1951–1957. doi: 10.1172/JCI59889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masuda M, Wakasaki T, Toh S, Shimizu M, Adachi S. Chemoprevention of head and neck cancer by green tea extract: EGCG-the role of EGFR signaling and “Lipid Raft”. J Oncol. 2011;2011:540148. doi: 10.1155/2011/540148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D’Souza G, et al. Case-control study of human papillomavirus and oropharyngeal cancer. N Engl J Med. 2007;356(19):1944–1956. doi: 10.1056/NEJMoa065497. [DOI] [PubMed] [Google Scholar]
- 5.Ernster JA, et al. Rising incidence of oropharyngeal cancer and the role of oncogenic human papilloma virus. Laryngoscope. 2007;117(12):2115–2128. doi: 10.1097/MLG.0b013e31813e5fbb. [DOI] [PubMed] [Google Scholar]
- 6.Pai SI, Westra WH. Molecular pathology of head and neck cancer: implications for diagnosis, prognosis, and treatment. Annu Rev Pathol. 2009;4:49–70. doi: 10.1146/annurev.pathol.4.110807.092158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gillison ML, et al. Distinct risk factor profiles for human papillomavirus type 16-positive and human papillomavirus type 16-negative head and neck cancers. J Natl Cancer Inst. 2008;100(6):407–420. doi: 10.1093/jnci/djn025. [DOI] [PubMed] [Google Scholar]
- 8.Ang KK, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363(1):24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marur S, D’Souza G, Westra WH, Forastiere AA. HPV-associated head and neck cancer: a virus-related cancer epidemic. Lancet Oncol. 2010;11(8):781–789. doi: 10.1016/S1470-2045(10)70017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fakhry C, et al. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst. 2008;100(4):261–269. doi: 10.1093/jnci/djn011. [DOI] [PubMed] [Google Scholar]
- 11.Vu HL, Sikora AG, Fu S, Kao J. HPV-induced oropharyngeal cancer, immune response and response to therapy. Cancer Lett. 2010;288(2):149–155. doi: 10.1016/j.canlet.2009.06.026. [DOI] [PubMed] [Google Scholar]
- 12.Holzinger D, Schmitt M, Dyckhoff G, Benner A, Pawlita M, Bosch FX. Viral RNA patterns and high viral load reliably define oropharynx carcinomas with active HPV16 involvement. Cancer Res. 2012;72(19):4993–5003. doi: 10.1158/0008-5472.CAN-11-3934. [DOI] [PubMed] [Google Scholar]
- 13.Jung AC, et al. Biological and clinical relevance of transcriptionally active human papillomavirus (HPV) infection in oropharynx squamous cell carcinoma. Int J Cancer. 2010;126(8):1882–1894. doi: 10.1002/ijc.24911. [DOI] [PubMed] [Google Scholar]
- 14.Kostareli E, Holzinger D, Hess J. New concepts for translational head and neck oncology: Lessons from HPV-related oropharyngeal squamous cell carcinomas. Front Oncol. 2012;2:36. doi: 10.3389/fonc.2012.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez-Ramirez I, Garcia-Cuellar C, Sanchez-Perez Y, Granados-Garcia M. DNA methylation in oral squamous cell carcinoma: molecular mechanisms and clinical implications. Oral Dis. 2011;17(8):771–778. doi: 10.1111/j.1601-0825.2011.01833.x. [DOI] [PubMed] [Google Scholar]
- 16.Stephen JK, Chen KM, Havard S, Harris G, Worsham MJ. Promoter methylation in head and neck tumorigenesis. Methods Mol Biol. 2012;863:187–206. doi: 10.1007/978-1-61779-612-8_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Worsham MJ, et al. Cancer Lett. Delineating an epigenetic continuum in head and neck cancer [published online ahead of print March 21, 2012]. doi: 10.1016/j.canlet.2012.02.018 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poage GM, et al. Identification of an epigenetic profile classifier that is associated with survival in head and neck cancer. Cancer Res. 2012;72(11):2728–2737. doi: 10.1158/0008-5472.CAN-11-4121-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marsit CJ, McClean MD, Furniss CS, Kelsey KT. Epigenetic inactivation of the SFRP genes is associated with drinking, smoking and HPV in head and neck squamous cell carcinoma. Int J Cancer. 2006;119(8):1761–1766. doi: 10.1002/ijc.22051. [DOI] [PubMed] [Google Scholar]
- 20.Sartor MA, et al. Genome-wide methylation and expression differences in HPV(+) and HPV(–) squamous cell carcinoma cell lines are consistent with divergent mechanisms of carcinogenesis. Epigenetics. 2011;6(6):777–787. doi: 10.4161/epi.6.6.16216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiss D, Basel T, Sachse F, Braeuninger A, Rudack C. Promoter methylation of cyclin A1 is associated with human papillomavirus 16 induced head and neck squamous cell carcinoma independently of p53 mutation. Mol Carcinog. 2011;50(9):680–688. doi: 10.1002/mc.20798. [DOI] [PubMed] [Google Scholar]
- 22.Gubanova E, et al. Downregulation of SMG-1 in HPV-positive head and neck squamous cell carcinoma due to promoter hypermethylation correlates with improved survival. Clin Cancer Res. 2012;18(5):1257–1267. doi: 10.1158/1078-0432.CCR-11-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szklarczyk D, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39(Database issue):D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 25.Gudas LJ. Emerging roles for retinoids in regeneration and differentiation in normal and disease states. Biochim Biophys Acta. 2012;1821(1):213–221. doi: 10.1016/j.bbalip.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffman LM, et al. BMP action in skeletogenesis involves attenuation of retinoid signaling. J Cell Biol. 2006;174(1):101–113. doi: 10.1083/jcb.200604150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim H, et al. The retinoic acid synthesis gene ALDH1a2 is a candidate tumor suppressor in prostate cancer. Cancer Res. 2005;65(18):8118–8124. doi: 10.1158/0008-5472.CAN-04-4562. [DOI] [PubMed] [Google Scholar]
- 28.Muzio G, Maggiora M, Paiuzzi E, Oraldi M, Canuto RA. Aldehyde dehydrogenases and cell proliferation. Free Radic Biol Med. 2012;52(4):735–746. doi: 10.1016/j.freeradbiomed.2011.11.033. [DOI] [PubMed] [Google Scholar]
- 29.Dahlgren L, et al. Comparative genomic hybridization analysis of tonsillar cancer reveals a different pattern of genomic imbalances in human papillomavirus-positive and -negative tumors. Int J Cancer. 2003;107(2):244–249. doi: 10.1002/ijc.11371. [DOI] [PubMed] [Google Scholar]
- 30.Smeets SJ, et al. Genome-wide DNA copy number alterations in head and neck squamous cell carcinomas with or without oncogene-expressing human papillomavirus. Oncogene. 2006;25(17):2558–2564. doi: 10.1038/sj.onc.1209275. [DOI] [PubMed] [Google Scholar]
- 31.Klussmann JP, et al. Genetic signatures of HPV-related and unrelated oropharyngeal carcinoma and their prognostic implications. Clin Cancer Res. 2009;15(5):1779–1786. doi: 10.1158/1078-0432.CCR-08-1463. [DOI] [PubMed] [Google Scholar]
- 32.Wilting SM, et al. Genomic profiling identifies common HPV-associated chromosomal alterations in squamous cell carcinomas of cervix and head and neck. BMC Med Genomics. 2009;2:32. doi: 10.1186/1755-8794-2-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agrawal N, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333(6046):1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stransky N, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paz IB, Cook N, Odom-Maryon T, Xie Y, Wilczynski SP. Human papillomavirus (HPV) in head and neck cancer. An association of HPV 16 with squamous cell carcinoma of Waldeyer’s tonsillar ring. Cancer. 1997;79(3):595–604. doi: 10.1002/(SICI)1097-0142(19970201)79:3<595::AID-CNCR24>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 36.Andl T, et al. Etiological involvement of oncogenic human papillomavirus in tonsillar squamous cell carcinomas lacking retinoblastoma cell cycle control. Cancer Res. 1998;58(1):5–13. [PubMed] [Google Scholar]
- 37.Klussmann JP, et al. Prevalence, distribution, and viral load of human papillomavirus 16 DNA in tonsillar carcinomas. Cancer. 2001;92(11):2875–2884. doi: 10.1002/1097-0142(20011201)92:11<2875::AID-CNCR10130>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 38.van Houten VM, et al. Biological evidence that human papillomaviruses are etiologically involved in a subgroup of head and neck squamous cell carcinomas. Int J Cancer. 2001;93(2):232–235. doi: 10.1002/ijc.1313. [DOI] [PubMed] [Google Scholar]
- 39.Mellin H, et al. Human papillomavirus type 16 is episomal and a high viral load may be correlated to better prognosis in tonsillar cancer. Int J Cancer. 2002;102(2):152–158. doi: 10.1002/ijc.10669. [DOI] [PubMed] [Google Scholar]
- 40.Wiest T, Schwarz E, Enders C, Flechtenmacher C, Bosch FX. Involvement of intact HPV16 E6/E7 gene expression in head and neck cancers with unaltered p53 status and perturbed pRb cell cycle control. Oncogene. 2002;21(10):1510–1517. doi: 10.1038/sj.onc.1205214. [DOI] [PubMed] [Google Scholar]
- 41.Braakhuis BJ, et al. Genetic patterns in head and neck cancers that contain or lack transcriptionally active human papillomavirus. J Natl Cancer Inst. 2004;96(13):998–1006. doi: 10.1093/jnci/djh183. [DOI] [PubMed] [Google Scholar]
- 42.Lindquist D, et al. Human papillomavirus is a favourable prognostic factor in tonsillar cancer and its oncogenic role is supported by the expression of E6 and E7. Mol Oncol. 2007;1(3):350–355. doi: 10.1016/j.molonc.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reimers N, et al. Combined analysis of HPV-DNA, p16 and EGFR expression to predict prognosis in oropharyngeal cancer. Int J Cancer. 2007;120(8):1731–1738. doi: 10.1002/ijc.22355. [DOI] [PubMed] [Google Scholar]
- 44.Attner P, et al. The role of human papillomavirus in the increased incidence of base of tongue cancer. Int J Cancer. 2010;126(12):2879–2884. doi: 10.1002/ijc.24994. [DOI] [PubMed] [Google Scholar]
- 45.Leonard SM, et al. Oncogenic human papillomavirus imposes an instructive pattern of DNA methylation changes which parallel the natural history of cervical HPV infection in young women. Carcinogenesis. 2012;33(7):1286–1293. doi: 10.1093/carcin/bgs157. [DOI] [PubMed] [Google Scholar]
- 46.Au Yeung CL, Tsang WP, Tsang TY, Co NN, Yau PL, Kwok TT. HPV-16 E6 upregulation of DNMT1 through repression of tumor suppressor p53. Oncol Rep. 2010;24(6):1599–1604. doi: 10.3892/or_00001023. [DOI] [PubMed] [Google Scholar]
- 47.Burgers WA, Blanchon L, Pradhan S, de Launoit Y, Kouzarides T, Fuks F. Viral oncoproteins target the DNA methyltransferases. Oncogene. 2007;26(11):1650–1655. doi: 10.1038/sj.onc.1209950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kawai S, Amano A. Odd-skipped related 2 is epigenetically regulated in cellular quiescence. Biochem Biophys Res Commun. 2010;396(4):831–836. doi: 10.1016/j.bbrc.2010.04.159. [DOI] [PubMed] [Google Scholar]
- 49.Kawai S, Abiko Y, Amano A. Odd-skipped related 2 regulates genes related to proliferation and development. Biochem Biophys Res Commun. 2010;398(2):184–190. doi: 10.1016/j.bbrc.2010.06.054. [DOI] [PubMed] [Google Scholar]
- 50.Agnihotri S, et al. A GATA4-regulated tumor suppressor network represses formation of malignant human astrocytomas. J Exp Med. 2011;208(4):689–702. doi: 10.1084/jem.20102099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Derks S, et al. Promoter methylation precedes chromosomal alterations in colorectal cancer development. Cell Oncol. 2006;28(5–6):247–257. doi: 10.1155/2006/846251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo M, et al. Hypermethylation of the GATA gene family in esophageal cancer. Int J Cancer. 2006;119(9):2078–2083. doi: 10.1002/ijc.22092. [DOI] [PubMed] [Google Scholar]
- 53.Wakana K, Akiyama Y, Aso T, Yuasa Y. Involvement of GATA-4/-5 transcription factors in ovarian carcinogenesis. Cancer Lett. 2006;241(2):281–288. doi: 10.1016/j.canlet.2005.10.039. [DOI] [PubMed] [Google Scholar]
- 54.Hellebrekers DM, et al. GATA4 and GATA5 are potential tumor suppressors and biomarkers in colorectal cancer. Clin Cancer Res. 2009;15(12):3990–3997. doi: 10.1158/1078-0432.CCR-09-0055. [DOI] [PubMed] [Google Scholar]
- 55.Bennett LB, et al. DNA hypermethylation accompanied by transcriptional repression in follicular lymphoma. Genes Chromosomes Cancer. 2009;48(9):828–841. doi: 10.1002/gcc.20687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wen XZ, et al. Methylation of GATA-4 and GATA-5 and development of sporadic gastric carcinomas. World J Gastroenterol. 2010;16(10):1201–1208. doi: 10.3748/wjg.v16.i10.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Psyrri A, Sasaki C, Vassilakopoulou M, Dimitriadis G, Rampias T. Future directions in research, treatment and prevention of HPV-related squamous cell carcinoma of the head and neck. Head Neck Pathol. 2012;6(suppl 1):S121–S128. doi: 10.1007/s12105-012-0361-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taioli E, et al. Recurrence in oral and pharyngeal cancer is associated with quantitative MGMT promoter methylation. BMC Cancer. 2009;9:354. doi: 10.1186/1471-2407-9-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dong SM, Sun DI, Benoit NE, Kuzmin I, Lerman MI, Sidransky D. Epigenetic inactivation of RASSF1A in head and neck cancer. Clin Cancer Res. 2003;9(10 pt 1):3635–3640. [PubMed] [Google Scholar]
- 60.Brumbaugh KM, et al. The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol Cell. 2004;14(5):585–598. doi: 10.1016/j.molcel.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 61.Gewandter JS, Bambara RA, O’Reilly MA. The RNA surveillance protein SMG1 activates p53 in response to DNA double-strand breaks but not exogenously oxidized mRNA. Cell Cycle. 2011;10(15):2561–2567. doi: 10.4161/cc.10.15.16347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khan SA. Cervical cancer, human papillomavirus and vaccines. Clin Oncol (R Coll Radiol). 1993;5(6):386–390. doi: 10.1016/S0936-6555(05)80093-7. [DOI] [PubMed] [Google Scholar]
- 63.Merrick DT, Gown AM, Halbert CL, Blanton RA, McDougall JK. Human papillomavirus-immortalized keratinocytes are resistant to the effects of retinoic acid on terminal differentiation. Cell Growth Differ. 1993;4(10):831–840. [PubMed] [Google Scholar]
- 64.Creek KE, et al. Retinoic acid suppresses human papillomavirus type 16 (HPV16)-mediated transformation of human keratinocytes and inhibits the expression of the HPV16 oncogenes. Adv Exp Med Biol. 1994;354:19–35. doi: 10.1007/978-1-4899-0939-8_2. [DOI] [PubMed] [Google Scholar]
- 65.Creek KE, Geslani G, Batova A, Pirisi L. Progressive loss of sensitivity to growth control by retinoic acid and transforming growth factor-beta at late stages of human papillomavirus type 16-initiated transformation of human keratinocytes. Adv Exp Med Biol. 1995;375:117–135. doi: 10.1007/978-1-4899-0949-7_11. [DOI] [PubMed] [Google Scholar]
- 66.Choo CK, Rorke EA, Eckert RL. Retinoid regulation of cell differentiation in a series of human papillomavirus type 16-immortalized human cervical epithelial cell lines. Carcinogenesis. 1995;16(2):375–381. doi: 10.1093/carcin/16.2.375. [DOI] [PubMed] [Google Scholar]
- 67.Narayanan BA, Holladay EB, Nixon DW, Mauro CT. The effect of all-trans and 9-cis retinoic acid on the steady state level of HPV16 E6/E7 mRNA and cell cycle in cervical carcinoma cells. Life Sci. 1998;63(7):565–573. doi: 10.1016/S0024-3205(98)00307-5. [DOI] [PubMed] [Google Scholar]
- 68.Touma SE, Perner S, Rubin MA, Nanus DM, Gudas LJ. Retinoid metabolism and ALDH1A2 (RALDH2) expression are altered in the transgenic adenocarcinoma mouse prostate model. Biochem Pharmacol. 2009;78(9):1127–1138. doi: 10.1016/j.bcp.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liang D, Jia W, Li J, Li K, Zhao Q. Retinoic acid signaling plays a restrictive role in zebrafish primitive myelopoiesis. PLoS One. 2012;7(2):e30865. doi: 10.1371/journal.pone.0030865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Neto A, Mercader N, Gomez-Skarmeta JL. The Osr1 and Osr2 genes act in the pronephric anlage downstream of retinoic acid signaling and upstream of Wnt2b to maintain pectoral fin development. Development. 2012;139(2):301–311. doi: 10.1242/dev.074856. [DOI] [PubMed] [Google Scholar]
- 71.Mauney JR, Ramachandran A, Yu RN, Daley GQ, Adam RM, Estrada CR. All-trans retinoic acid directs urothelial specification of murine embryonic stem cells via GATA4/6 signaling mechanisms. PLoS One. 2010;5(7):e11513. doi: 10.1371/journal.pone.0011513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ghatpande S, Brand T, Zile M, Evans T. Bmp2 and Gata4 function additively to rescue heart tube development in the absence of retinoids. Dev Dyn. 2006;235(8):2030–2039. doi: 10.1002/dvdy.20836. [DOI] [PubMed] [Google Scholar]
- 73.Wang GF, Nikovits W, Jr, Bao ZZ, Stockdale FE. Irx4 forms an inhibitory complex with the vitamin D and retinoic X receptors to regulate cardiac chamber-specific slow MyHC3 expression. J Biol Chem. 2001;276(31):28835–28841. doi: 10.1074/jbc.M103716200. [DOI] [PubMed] [Google Scholar]
- 74.Cheng Z, et al. Two novel mutations of the IRX4 gene in patients with congenital heart disease. Hum Genet. 2011;130(5):657–662. doi: 10.1007/s00439-011-0996-7. [DOI] [PubMed] [Google Scholar]
- 75.Wirth LJ, Haddad RI, Posner MR. Progress and perspectives in chemoprevention of head and neck cancer. Expert Rev Anticancer Ther. 2003;3(3):339–355. doi: 10.1586/14737140.3.3.339. [DOI] [PubMed] [Google Scholar]
- 76.Armstrong WB, Meyskens FL. Chemoprevention of head and neck cancer. Otolaryngol Head Neck Surg. 2000;122(5):728–735. doi: 10.1016/S0194-5998(00)70205-1. [DOI] [PubMed] [Google Scholar]
- 77.Guo Y, et al. A retinoic acid-rich tumor microenvironment provides clonal survival cues for tumor-specific CD8+ T cells. Cancer Res. 2012;72(20):5230–5239. doi: 10.1158/0008-5472.CAN-12-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wansom D, et al. Correlation of cellular immunity with human papillomavirus 16 status and outcome in patients with advanced oropharyngeal cancer. Arch Otolaryngol Head Neck Surg. 2010;136(12):1267–1273. doi: 10.1001/archoto.2010.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wansom D, et al. Infiltrating lymphocytes and human papillomavirus-16–associated oropharyngeal cancer. Laryngoscope. 2012;122(1):121–127. doi: 10.1002/lary.22133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Napoli JL. Physiological insights into all-trans-retinoic acid biosynthesis. Biochim Biophys Acta. 2012;1821(1):152–167. doi: 10.1016/j.bbalip.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Williams SJ, Cvetkovic D, Hamilton TC. Vitamin A metabolism is impaired in human ovarian cancer. Gynecol Oncol. 2009;112(3):637–645. doi: 10.1016/j.ygyno.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chomczynski P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques. 1993;15(3):532–534, 536–537. [PubMed] [Google Scholar]
- 83.Sotlar K, et al. Detection and typing of human papillomavirus by e6 nested multiplex PCR. J Clin Microbiol. 2004;42(7):3176–3184. doi: 10.1128/JCM.42.7.3176-3184.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gebhard C, et al. Genome-wide profiling of CpG methylation identifies novel targets of aberrant hypermethylation in myeloid leukemia. Cancer Res. 2006;66(12):6118–6128. doi: 10.1158/0008-5472.CAN-06-0376. [DOI] [PubMed] [Google Scholar]
- 85.Schilling E, Rehli M. Global, comparative analysis of tissue-specific promoter CpG methylation. Genomics. 2007;90(3):314–323. doi: 10.1016/j.ygeno.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 86.Ritchie ME, et al. A comparison of background correction methods for two-colour microarrays. Bioinformatics. 2007;23(20):2700–2707. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- 87.Kuhmann C, Weichenhan D, Rehli M, Plass C, Schmezer P, Popanda O. DNA methylation changes in cells regrowing after fractioned ionizing radiation. Radiother Oncol. 2011;101(1):116–121. doi: 10.1016/j.radonc.2011.05.048. [DOI] [PubMed] [Google Scholar]
- 88.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ehrich M, et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci U S A. 2005;102(44):15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Roesch Ely M, et al. Transcript and proteome analysis reveals reduced expression of calgranulins in head and neck squamous cell carcinoma. Eur J Cell Biol. 2005;84(2–3):431–444. doi: 10.1016/j.ejcb.2005.01.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.