Abstract
Iron refractory iron deficiency anemia is a hereditary recessive anemia due to a defect in the TMPRSS6 gene encoding Matriptase-2. This protein is a transmembrane serine protease that plays an essential role in down-regulating hepcidin, the key regulator of iron homeostasis. Hallmarks of this disease are microcytic hypochromic anemia, low transferrin saturation and normal/high serum hepcidin values. The anemia appears in the post-natal period, although in some cases it is only diagnosed in adulthood. The disease is refractory to oral iron treatment but shows a slow response to intravenous iron injections and partial correction of the anemia. To date, 40 different Matriptase-2 mutations have been reported, affecting all the functional domains of the large ectodomain of the protein. In vitro experiments on transfected cells suggest that Matriptase-2 cleaves Hemojuvelin, a major regulator of hepcidin expression and that this function is altered in this genetic form of anemia. In contrast to the low/undetectable hepcidin levels observed in acquired iron deficiency, in patients with Matriptase-2 deficiency, serum hepcidin is inappropriately high for the low iron status and accounts for the absent/delayed response to oral iron treatment. A challenge for the clinicians and pediatricians is the recognition of the disorder among iron deficiency and other microcytic anemias commonly found in pediatric patients. The current treatment of iron refractory iron deficiency anemia is based on parenteral iron administration; in the future, manipulation of the hepcidin pathway with the aim of suppressing it might become an alternative therapeutic approach.
Introduction
Iron deficiency anemia is a major health problem worldwide. Iron deficiency of nutritional origin is the most frequent cause of microcytic hypochromic anemia, but other conditions such as bleeding, gastro-intestinal malabsorption or Helicobacter pylori infection can lead to iron deficiency and anemia.1 Iron restricted erythropoiesis underlies the anemia of chronic diseases, although several other mechanisms such as suppressed erythropoiesis and poor response to erythropoietin also contribute to this form of anemia. A new cause of hereditary anemia has recently been described called iron refractory iron deficiency anemia or IRIDA (OMIM #206200, ORPHA209981), due to mutations in the TMPRSS6 gene (mapping to chromosome 22q12-q13), encoding Matriptase-2 (MT-2).2 The prevalence of this condition is not known but it has certainly been under-diagnosed up to now and should be taken into consideration when all other known causes of iron deficiency anemia have been ruled out. This disorder was recognized as a new entity after hepcidin was identified as the key regulator of systemic iron homeostasis. For a better understanding of IRIDA, the regulation of hepcidin is first discussed.
Regulation of hepcidin expression
Iron availability for erythropoiesis and cellular functions is determined by the amount of iron that circulates in the plasma. Iron is bound to transferrin [Fe(III)-Tf] and can be readily taken up by all cell types via the ubiquitously expressed transferrin receptor 1 (TfR1). Iron homeostasis is maintained by the liver-expressed peptide hormone hepcidin that regulates intestinal iron absorption, macrophage-mediated iron recycling from senescent erythrocytes, and iron mobilization from hepatic stores. Hepcidin down-regulates iron export by binding to the iron exporter ferroportin expressed on the surface of iron-releasing cells, triggering its degradation and hence reducing plasma iron levels. Hepcidin levels are regulated by systemic iron availability, iron demand for erythropoiesis, hypoxia and inflammation.3
The study of mechanisms that underlie frequent iron-related disorders, such as hereditary hemochromatosis, iron-loading anemia (e.g. thalassemia) or the anemia of chronic diseases, provided insight into hepcidin regulation. Iron balance is disrupted in the autosomal recessive disorder hereditary hemochromatosis (HH) that is hallmarked by excessive iron absorption from the diet and iron accumulation within parenchymal cells. Different HH disease subtypes are caused by mutations in the HFE,4TFR2,5HFE2 (encoding hemojuvelin HJV)6 or HAMP (hepcidin) gene7 and are characterized by inappropriately low hepcidin levels, reflecting the fact that the membrane proteins HFE, TfR2 and HJV contribute to hepcidin regulation.
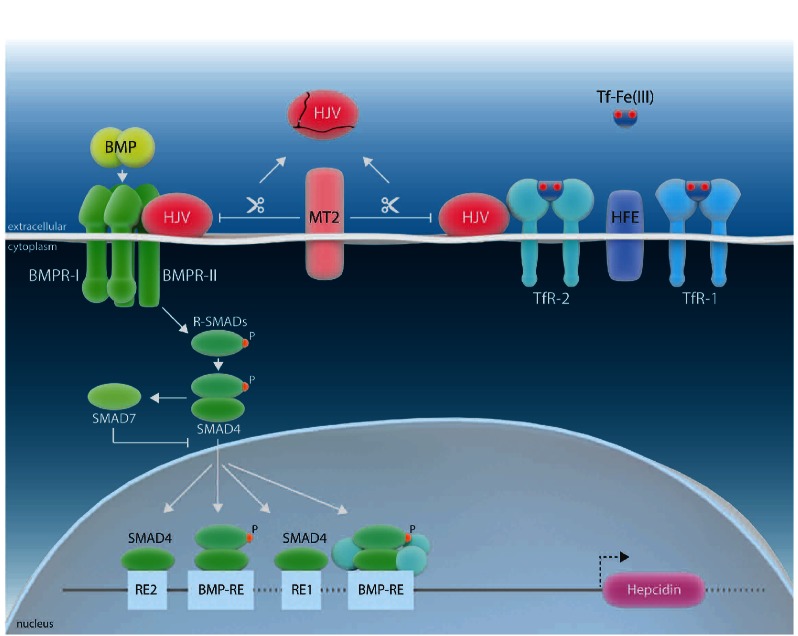
HJV is a glycophosphatidylinositol (GPI)-anchored protein that acts as a bone-morphogenetic protein (BMP) co-receptor, driving hepcidin transcription via the BMP-SMAD signaling cascade (Figure 1).8 Disease-associated mutations in HJV cause a juvenile form of HH with a severe phenotype of iron overload, indicating that the HJV/BMP pathway plays a critical role in maintaining basal hepcidin levels. It has been suggested that BMP6, which is activated by intracellular iron, is the endogenous ligand for HJV.13,14 Based on biochemical evidence, a model was proposed that suggests that HFE, TfR2 and HJV interact with each other to form a hepatocyte ‘iron-sensing complex’.10 If serum Fe(III)2-Tf levels increase, HFE is displaced from TfR1 to permit its interaction with TfR2, activating the transcription of the HAMP gene. TfR2 thus acts as a sensor for Tf saturation.15–20
Figure 1.
Schematic representation of the regulation of HAMP gene expression by systemic iron availability, modified from 9. HFE is displaced from TfR1 by high concentrations of the transferrin-iron complex [Tf-Fe(III)] to promote its interaction with transferrin receptor 2 (TfR2). HFE and TfR2 bind the BMP co-receptor hemojuvelin (HJV),10 and activate HAMP transcription via bone morphogenetic protein (BMP)/SMAD signaling. Additionally, this involves type I and type II BMP receptors (BMPR) at the plasma membrane to induce phosphorylation of receptor-activated SMAD (R-SMAD) proteins. The subsequent formation of active transcriptional complexes involves the co-SMAD factor SMAD4. MT-2 interacts with HJV and causes HJV fragmentation. SMAD7 interferes with SMAD4-controlled hepcidin activation.11 Response Elements (RE) critical for SMAD-mediated control of the HAMP promoter are shown.12
Secondary iron overload associated with ineffective erythropoiesis is also due to hepcidin misregulation, at least in the absence of blood transfusions. In this case, soluble factors secreted from erythroid cells have been proposed to attenuate hepatic hepcidin levels by interfering with the BMP/SMAD signaling pathway. However, the molecular details remain elusive.9 Finally, increased hepcidin levels are the hallmark of the anemia of chronic diseases that is caused by cytokines produced in response to chronic inflammatory and infectious disorders or cancer.21 In this case, hepcidin is activated by inflammatory cytokines, such as IL-6 and IL-1β, via the Jak/Stat3 signaling cascade.22
Matriptase-2: gene identification and lessons from two mouse models, the mask mouse and the Tmprss6 knock-out mouse
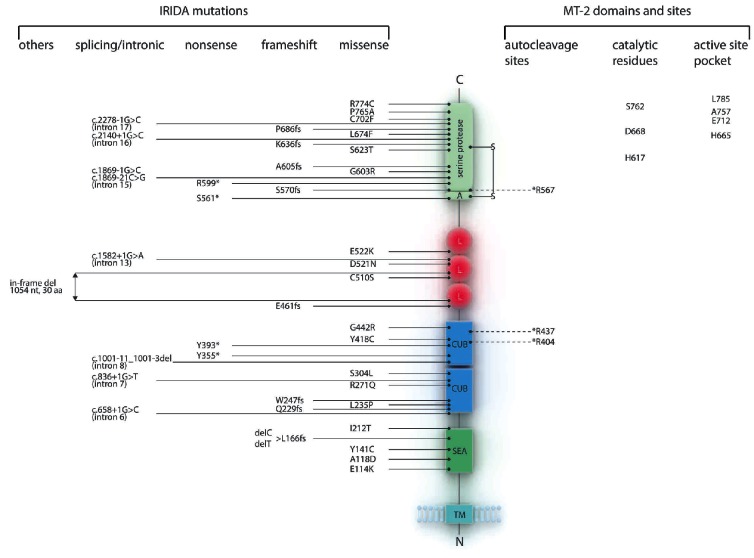
MT-2 is a type II trans-membrane serine protease (TTSP) first identified by a genome-wide in silico screen.23 It presents all the expected features of trans-membrane serine proteases,24 including a large ectodomain with a SEA (Sea urchin sperm protein, Enteropeptidase, Agrin) region, two CUB (Complement factor C1s/C1r, Urchin embryonic growth factor, Bone morphogenic protein) domains, three Low Density Lipoprotein Receptor (LDLR) domains and a C terminal serine protease domain with the conserved Ser, Asp and His residues required for the catalytic activity (Figure 2).
Figure 2.
IRIDA mutations reported in the literature and schematic representation of the predicted domain structure of the MT-2. The provided numbers refer to the amino acid position in the pre-proenzyme. Mutations are classified as nonsense, frameshift, missense, splicing/intronic and others. Domains are shown as: serine protease domain (serine protease), activation domain (A), LDL (Low Density Lipoprotein) receptor class A domain (L), CUB (Cls/Clr, Urchin embryonic growth fator, Bone morphogenic protein 1) domain, SEA (Sea urchin sperm protein, Enteropeptidase, Agrin) domain and transmembrane (TM) domain. The conserved disulphide bond linking the pro- and catalytic domain is shown as S-S. Residues that undergo autocleavage including the proteolytic activation site25 (R567) are marked with a discontinuous line and an asterisk. The three conserved catalytic residues, histidine (H617), aspartatic acid (D668) and serine (S762) and residues that constitute the active site pocket are also shown.26
MT-2 is highly homologous to Matriptase-1/MT-1/ST14. In contrast to MT-1, which is expressed in most epithelial cells and plays essential roles in the establishment and maintenance of epithelial integrity,27 MT-2 is expressed predominantly in hepatocytes.28 Like MT-1, MT-2 undergoes a complex activation process including several cleavage steps (Figure 2): one in the SEA region, two in the CUB domains (at amino acid positions 404 and 437), and one in the conserved activation site (amino acid position 567).25 Overall, these proteolytic cleavages release several MT-2 fragments that can be detected in the culture media of transfected cells. However, in the absence of specific antibodies detecting endogenous MT-2, it is still not known whether these fragments are present in the serum.
Following the cloning of MT-2, Velasco and collaborators showed that the recombinant protein exerts proteolytic activity against several synthetic substrates as well as against proteins of the cell matrix such as type I collagen or fibronectin,23 although the function of MT-2 remained unclear. Six years later, the analysis of Tmprss6 knock-out mice by the same group revealed an anemia and iron deficiency phenotype.29 However, the role of MT-2 in controlling hepcidin expression had just been discovered with the description of the mask mice.30 These mice were generated by ENU mutagenesis and were hallmarked by a progressive loss of body but not of facial hair (hence their name) and infertility of homozygous female mice. When maintained on a standard laboratory diet, these animals developed microcytic anemia, with depletion of iron in spleen macrophages. Interestingly, these mice failed to suppress Hamp expression when placed on an iron deficient diet. A gene mapping strategy led to the identification of a splicing defect in the Tmprss6 gene, encoding a MT-2 protein lacking the serine protease domain. The phenotype of the Tmprss6-/- mice was very similar to that of the mask mice, with progressive alopecia of the trunk, post-natal onset of iron deficiency and microcytic hypochromic anemia.29 This phenotype was attributed to a marked upregulation of liver hepcidin mRNA and reduced ferroportin expression on the baso-lateral side of duodenal enterocytes, as well as iron accumulation in these cells. Altogether, these data strongly suggested that MT-2 was a novel regulator of hepcidin expression, required for downregulation of HAMP expression following iron deprivation. Nevertheless, the sensing mechanism that controls and regulates MT-2 zymogen expression and activation is still unknown. Recently, the Kunitz-type serine protease inhibitor HAI-2 was shown to form a complex with MT-2 at the cell surface and to inhibit its proteolytic activity, thereby inducing HAMP expression.31 However, the role of HAI-2 in the iron sensing pathway is not known. The TMPRSS6 gene is transcriptionally up-regulated by Hypoxia-Inducible Factor 1 (HIF-1α) and acute iron deprivation,28,32,33 through a Hypoxia Responsive Element (HRE) present in the promoter region, thus providing an additional functional link between hypoxia and iron homeostasis. In addition, TMPRSS6 mRNA is also increased by BMP6/iron-dependent Id1 activation, as part of a negative feedback mechanism controlling excessive hepcidin upregulation in iron overload.34 How these transcriptional responses contribute to MT-2 surface expression and how these regulatory mechanisms are linked to zymogen activation and hepcidin regulation is not known.
The disease
Identification of families with hereditary microcytic iron-deficient anemia
Prior to the molecular era and the identification of the TMPRSS6 gene, very few families were reported in the literature with individuals affected by iron refractory iron deficiency anemia.
In 1981, 3 siblings were described with iron unresponsive anemia, malabsorption of medicinal iron and a partial but incomplete hematologic response to parenteral iron dextran.35 Two sisters with similar findings and normal parents suggested recessive inheritance.36 In other cases with similar findings, defective iron absorption was documented by the absorption test, and acquired intestinal disorders and blood losses were excluded. The authors concluded that the patients had a rare inherited form of anemia with disordered iron metabolism only partially corrected by parenteral iron dextran.37
In 1997, an 18-month old African child with iron resistant iron deficiency anemia and severe microcytosis was reported.38 Anemia was unresponsive to oral iron supplementation and persisted after iron stores were replete. The anemic iron deficient mk mouse and the corresponding Belgrade rat were initially considered as models for this disease but were later found to have mutations in the Slc11a2 gene encoding Dmt1, the divalent metal iron transporter 1.39
An iron unresponsive microcytic anemia was further identified in several individuals of a large inbred Sardinian kindred, clearly indicating that this type of anemia had a genetic cause and facilitating the locus identification. By using genomic studies and homozygosity mapping, the locus for the hereditary microcytic anemia was mapped on chromosome 22q12-q13, definitely excluding SLC11A2 (located on 12q13) as the responsible gene.
Ferrokinetic studies in 2 probands with microcytic iron refractory anemia provided different results to those obtained in the mk mouse,40 suggesting an altered mobilization of iron into plasma from both the intestine and the reticuloendothelial cells. In 2001, the case of 2 Caucasian siblings with a similar hematologic phenotype was reported.41
Most of the reported cases were children. In those cases followed for several years, it was noted that despite anemia, growth, development and intellectual performance were normal.40,42 In 2008, when the gene responsible for this disease was identified, molecular diagnosis was performed in some of these families43 and in the large Sardinian pedigree described above.42
IRIDA biological and clinical phenotype
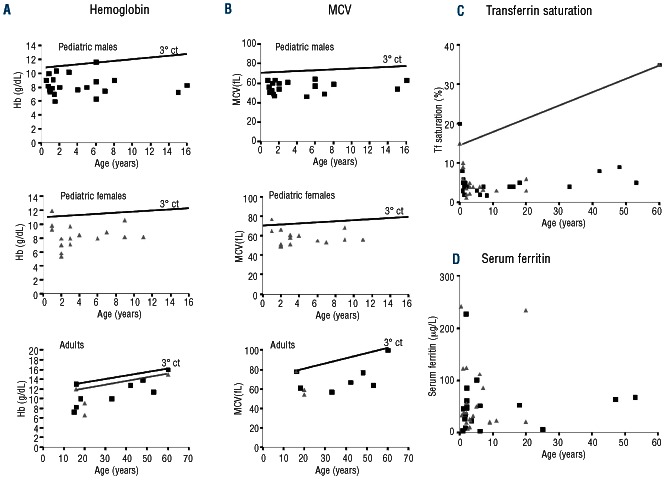
Clinically, IRIDA subjects are characterized by a hypochromic, microcytic anemia (Figure 3A and B), and very low serum iron and transferrin saturation levels (Figure 3C). However, serum ferritin levels are mostly within the normal range, or even slightly elevated following intravenous iron treatment (Figure 3D).
Figure 3.
Clinical phenotype of IRIDA subjects versus controls according to age. In all the graphs, the lines indicate values of the parameters corresponding to the 3rd percentile of healthy controls. (A) Hemoglobin (Hb, g/dL) values of pediatric male (black square), pediatric female (gray triangle) or adult (age > 16 years) IRIDA patients. (B) Mean corpuscular volume (MCV, fL) values of pediatric male (black square), pediatric female (gray triangle) or adult (age > 16 years) IRIDA patients. (C) Transferrin (Tf) saturation values of IRIDA patients (male: black square; female: gray triangle). (D) Ferritin values of IRIDA patients (control values: male <300 μg/L, female < 200 μg/L; male: black square; female: gray triangle).
The degree of anemia varies (Figure 3A), and is mostly mild, and more pronounced during childhood. However, no direct correlation has been observed between age at diagnosis and the degree of anemia (A Iolascon, personal observations, 2013). From a theoretical point of view, females of a reproductive age could be more exposed to IRIDA because of iron loss due to menses or due to pregnancies, but analysis of the literature data does not reveal a difference in sex prevalence of IRIDA.
In all the cases reported so far, as well as from the personal experience of some of our group, it has been found that anemia is not detectable at birth and that the clinical phenotype develops only after the neonatal period, suggesting that MT-2 may not be essential during fetal life. Suspicion of IRIDA usually occurs during a pediatric routine evaluation. However, in some patients, the condition is recognized only in adulthood, either because the anemia is mild or because it has been misclassified. Overall, it is likely that up till now this condition has been under-diagnosed.
How to diagnose IRIDA?
It is mandatory to differentiate IRIDA from nutritional iron deficiency and from other genetic microcytic anemias. The presence of several affected siblings in the family may suggest the existence of an inherited disorder. However, many patients are sporadic cases because of the recessive mode of transmission and the small size of many pedigrees. In general, clinical data could help establish whether iron deficiency is inherited or acquired. Acquired iron deficiency may result from blood loss or decreased iron absorption, as in celiac disease, where anemia is the most common hematologic complication.44 Duodenal atrophy resulting in a delayed response to oral iron but a good response to intravenous iron is another confounding factor with IRIDA (see below: Iron therapy). A positive anti-endomysium antibody test, and a positive response to a gluten-free diet will rule out IRIDA.45 The age of onset may contribute to diagnosis since microcytic anemia is not present at birth in contrast to other genetic conditions such as DMT1 mutations or atransferrinemia.46 Sideroblastic anemia due to either ALAS-2 or SLC25A38 deficiency can also occur during childhood,47 but iron overload is also present in sideroblastic anemia as well as in DMT1 deficiency or atransferrinemia, as indicated by elevated serum iron and transferrin saturation. Microcytosis, hypochromia and low iron stores are present in both acquired iron deficiency and IRIDA. However, RBC count tends to be higher in IRIDA whereas in true iron deficiency serum ferritin is lower. Microcytosis and low mean corpuscular hemoglobin (MCH) are also hallmarks of beta-thalassemia carriers, who have normal or slightly elevated iron parameters and increased hemoglobin (Hb)A2.
Carriers of alpha-thalassemia may have remarkable microcytosis but are not (or only mildly) anemic, and often have normal or increased iron parameters. When IRIDA patients show very low Hb levels and increased number of RBCs,48,49 a helpful marker for a differential diagnosis is the reticulocyte count, which is high in beta-thalassemia and low in IRIDA.
Finally, despite a lack of harmonization among the hepcidin assays currently available,50 normal/high serum hepcidin levels characterize IRIDA due to MT-2 mutations (Online Supplementary Table S1), on the contrary to iron-deficiency in which hepcidin levels are very low,51 and to thalassemia or other hereditary microcytic anemia in which ineffective erythropoiesis suppresses hepcidin synthesis and results in increased intestinal iron absorption.52 Hepcidin levels are also high in anemia of chronic diseases since pro-inflammatory cytokines, especially IL-6,53 stimulate hepcidin synthesis and contribute to the onset of anemia. In inflammatory conditions, elevated hepcidin levels induce iron retention in macrophages, normal/high serum ferritin levels and low serum iron values,21 a biological pattern very similar to what is observed in IRIDA patients. However, the anemia is normo- or moderately microcytic as opposed to the marked microcytosis in IRIDA.
To date, the most widely used method for screening candidate IRIDA patients for MT-2 mutations is based on the sequencing of exons and exon-intron boundaries of the TMPRSS6 gene. Two alternative cost-effective methods have been recently described for detecting TMPRSS6 mutations. One is a SNaPshot assay design for screening several iron-related gene mutations54 and the other is a combined Polymerase chain reaction (PCR) and (high resolution melting (HRM) assay.55 However, in the near future, it is likely that next-generation sequencing methods will allow direct sequencing of a panel of candidate genes for rare iron-related anemia, including TMPRSS6.
TMPRSS6 mutation heterogeneity
The molecular basis of IRIDA in anemic patients was first identified in 2008.43 Up to now, 32 IRIDA families with 50 patients of different ethnic origin have been reported, accounting for 40 different mutations in the TMPRSS6 gene (18 missense, 4 nonsense, 9 frameshift, one large in-frame deletion, and 8 intronic mutations probably affecting the splicing) (Online Supplementary Table S2). As shown in Figure 2, MT-2 mutations are spread throughout the entire protein, affecting all extra-cellular domains.
The most frequent mutation is S304L, described in 6 patients from four different families. However, neither frequent mutations with a greater contribution to the phenotype nor mutations with a founder effect have been identified. As for the genotype-phenotype relationship, we observed a tendency towards lower hemoglobin, MCV and transferrin saturation, in patients with two nonsense mutations, but not in patients with two missense or one missense and one nonsense mutation.
The increasing number of cases that are being regularly diagnosed will make it easier to study possible genotype-phenotype correlations. Several common, uncommon and rare polymorphisms, including the non-synonymous polymorphisms K225E, K253E, G228D, R446W, V736A and V795I, have been described in TMPRSS6.56
IRIDA phenotype with a single or no MT-2 mutation: the case for a heterogeneous disorder?
As expected for a recessive disorder, bi-allelic mutations of TMPRSS6 are usually found in IRIDA patients. However, in the first description of IRIDA,43 single heterozygous mutations were found in 2 of the 7 probands, although the possibility of a gene deletion was not excluded. The first genomic deletion of TMPRSS6 resulting in IRIDA was described in 2009.57 In a French cohort of IRIDA patients, 5 out of 23 patients were found with a single heterozygous deleterious mutation of TMPRSS6 (B Grandchamp, personal communication, 2013). Furthermore, for 3 of these patients, exonic deletions were excluded by custom CGH array. The phenotype was milder in these patients than in typical IRIDA, but serum hepcidin levels were abnormally elevated. The phenotype in patients with a single mutated allele could be modified by environmental factors, the presence of deep intronic mutations or mutations in regulatory regions missed by the sequencing strategy, digenic inheritance, or associations with a hypomorphic allele. The presence of only one deleterious TMPRSS6 allele might be sufficient for the onset of anemia in young females who are more at risk of developing iron deficiency. In animal models, haploinsufficiency of Tmprss6 confers susceptibility to iron deficiency.58,59 The association between a deleterious mutation and a low expressed allele has been found in several inherited disorders such as erythropoietic protoporphyria,60 spherocytosis61 or microcytic anemia due to STEAP3 mutations.62 This might be a rather frequent situation since recent reports have demonstrated that preferential/specific allele expression may concern 5–20% of the genes.63,64 Finally, hypomorphic TMPRSS6 alleles have been found to contribute to the IRIDA phenotype, despite an apparent normal biological activity in functional in vitro assay.56,65
Furthermore, common TMPRSS6 SNPs have been associated with increased risk of iron deficiency and iron deficiency anemia in a Chinese population.66
Finally, a large Serbian family was recently described with a severe IRIDA phenotype responding only to intravenous iron and with no TMPRSS6 mutation.67 Based on SNP genotyping, the authors proposed gene-gene interaction between TMPRSS6 and SCL11A2, although this was not formally proven. However, it is tempting to speculate that another monogenic defect might contribute to the microcytic anemia phenotype in the reported children, especially since hepcidin levels were low. In our European cohort of microcytic iron deficiency anemia, some patients have no TMPRSS6 mutation and also have very low hepcidin levels, strongly suggesting another pathophysiological mechanism. Whole exome sequencing approaches are currently underway to identify the underlying gene.
Functional studies of the normal and mutated MT-2 proteins
Putative substrates of MT-2 include MT-2 itself, which undergoes cis and trans auto-proteolytic processing during zymogen activation,25 and hemojuvelin. In vitro, MT-2 interacts and cleaves membrane HJV,68 a finding compatible with the inappropriately high serum hepcidin levels observed in IRIDA patients and in TMPRSS6 deficient mice. Inactivation of HJV (or BMP6) in Tmprss6 knock-out mice completely rescued the IRIDA phenotype, confirming the functional interaction of MT-2 with the BMP-SMAD pathway in mice.58,69 To study the modulation of hepcidin by MT-2, several functional assays have been developed. These assays are based on the measurements of: 1) the luciferase activity driven by a HAMP promoter after overexpression of wild-type or mutated MT-2 in cells in the presence of HJV;68 2) the release of the serine pro-tease domain in the culture media of transfected cells to assess the proteolytic activity of MT-2;43,49,70–73 3) the release of the chromogenic molecule p-nitroaniline based on the ability of the serine protease domain to cleave the artificial substrate N-(tert-butoxycarbonyl)-Gln-Ala-Arg-p-nitroanilide (chromogenic assay developed by Sisay and collaborators).34,73,74
Therefore, a MT-2 variant may be considered as pathogenic on the basis of the following findings: 1) a lack of release of the MT-2 S/P domain in the culture media of transfected cells; 2) a defective ability to repress the HAMP promoter, using the luciferase assay; or 3) no detection of membrane-derived HJV fragments, using cells co-transfected with HJV and MT-2.68
Treatment of IRIDA patients: intravenous iron and future treatments
Iron therapy
In most patients, oral iron is ineffective in correcting the anemia and patients must receive intravenous iron. The response to parenteral administration of iron is variable but generally leads to a progressive increase in Hb levels. Correction of the anemia is partial and much slower than in patients with acquired iron deficiency. In addition, Hb levels rarely normalize, microcytosis persists, and transferrin saturation remains below normal values. By contrast, serum ferritin increases following iron injections, somehow in a dose-dependant manner. These features can be easily explained based on the hepcidin-ferroportin interactions.75 Indeed, hepcidin levels in these patients prevent increasing intestinal iron absorption in response to iron deficiency, or following oral iron therapy. However, they are probably not sufficient to fully suppress iron release from macrophages. Serum ferritin levels are also within the normal range, indicating absence of macrophage iron retention. Intravenous iron preparations usually consist of a colloidal suspension of large iron-sucrose complexes that are phagocytosed and metabolized by macrophages before iron is released to plasma transferrin. Adult patients usually require no or reduced iron supplementation compared to young subjects, probably as a consequence of the spontaneous reduction in iron requirement in adulthood.42
Although the term Iron Refractory Iron Deficiency Anemia (IRIDA) implies that oral iron is completely inefficient, in at least 4 IRIDA cases from three unrelated families sustained administration of oral iron partially corrected the anemia and Hb raised to acceptable levels without intravenous iron administration.71,73 Nevertheless, the partial correction of the anemia required an unusually long treatment. Concomitant administration of iron and ascorbate might be beneficial. It was recently reported that ascorbic acid (30 mg/day) supplementation at the time of ferrous sulfate administration had improved Hb and iron status in an infant patient.76 Some of us have observed a similar response in a young patient for whom Hb levels were maintained around 12 g/dL under a long-term therapy with ferrous ascorbate, while anemia reappeared when the treatment was discontinued (B Grandchamp, personal observation, 2013).
In rare cases, intravenous iron is inefficient, badly tolerated or made difficult by the lack of adequate venous access, and other therapies must be considered. In 2 patients, recombinant erythropoietin was added to the iron therapy in order to counteract excessive liver iron deposition caused by large doses of parenteral iron that were necessary to correct the anemia.70 This led the authors to suggest that it might be appropriate to target a sub-normal concentration of Hb to avoid the risk of iron overload
Future treatments
In the future, other treatments aimed at lowering plasma hepcidin levels might become available. These innovative therapies are currently being developed with the aim of correcting the anemia of chronic diseases but they could represent an interesting alternative to intravenous iron treatment in other conditions of iron-restricted erythropoiesis, especially in IRIDA. Anti-hepcidin antibodies were shown to correct the anemia of chronic diseases in a mouse model based on injections of heat-killed Brucella abortus.77 Another promising approach is the development of Anticalins® (Pieris AG, Germany) which are genetically modified lipocalins that can target almost any desired molecule with a high affinity. One Anticalin® has been developed that is highly specific for hepcidin and is proposed to antagonize hepcidin for the treatment of anemia of chronic diseases.78 An interesting alternative for the sequestration of hepcidin comes from the Spiegelmer® technology (Noxxon Pharma) with a PEGylated anti-hepcidin L-RNA oligonucleotide, now starting in phase I/IIa clinical trials in cancer patients.
Finally, the reduction of HAMP mRNA by siRNA technology (Alnyam, USA) has also proven pre-clinical efficacy79 and the same company also plans to apply this technology to the silencing of other molecules in the hepcidin regulatory pathway.
Conclusion
Iron refractory iron deficiency anemia is a new disease entity that must be considered whenever contemplating a diagnosis of microcytic anemia. The biological hallmarks of the disease are microcytic hypochromic anemia, low transferrin saturation and normal/high serum hepcidin values. Refractoriness to oral iron, and a slow and partial response to intravenous iron injections are features of this disease. The prevalence of the disease is still unknown, but it is worth noting that in GWAS studies, common SNPs in the TMPRSS6 gene have been found to be associated with Hb,80 or with iron status and erythrocyte volume.81 The strongest TMPRSS6 association with serum iron, transferrin saturation and Hb values was found with the non-synonymous coding SNP A736V (rs855791), while the intronic SNP NM_153609.2:c.1468+426A>G (rs2413450) was found to be strongly associated with hematocrit, MCV and MCH values. This suggests that TMPRSS6 might have a role in common disorders in which it could act as a gene modifier. For instance, in a large meta-analysis of around 50,000 non-diabetic adults of European descent, TMPRSS6 was one of the loci found to influence HbA1c levels. However, this association was likely indirect, occurring via the association between TMPRSS6 and erythrocyte biology.82 Considering the function of MT-2 as a negative regulator of HAMP expression, it is possible that TMPRSS6 could also be a modifier gene for hemochromatosis. Indeed, this has been shown in mouse models.83 However, from the limited number of studies available, TMPRSS6 SNPs do not seem to modulate iron accumulation in hemochromatosis patients.
Further studies are still required to fully understand the sensing pathway leading to MT-2 activation in response to iron deficiency and the role of this protein in rare anemia or common diseases. Finally, there is also evidence to suggest that, as with other members of the TTSPs, MT-2 may have a role in cancer development and progression.84 Although the evidence currently available points to a role for MT-2 only in prostate and breast tumor development, the link between MT-2 and liver tumor is a challenging avenue to explore, especially considering the well-known relationship between iron overload and hepatocellular carcinoma.85
Acknowledgments
The authors are grateful to Bernard Grandchamp, Claire Oudin and Flavia Guillem at Hopital Bichat (Paris, France) and Erica Morán at IMPPC (Barcelona, Spain) for helpful discussions. MUM acknowledges support of the Dietmar Hopp Stiftung.
Footnotes
The online version of this article has a Supplementary Appendix.
Funding
This work was supported by the eRARE HMA-IRON funding to MUM, CC and CB and by grants: PS09/00341 from “Instituto de Salud Carlos III”, SAF2012-40106 from Spanish Secretary of Research, Development and Innovation (MINECO) and CIVP16A1857 from “Ayudas a proyectos de Investigación en Ciéncias de la Vida - Fundación Ramón Areces” to MS. M.S. held a research contract under the Ramón y Cajal program from the Spanish Ministry of Science and Innovation (RYC-2008-02352).
Authorship and Disclosures
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Hershko C, Hoffbrand AV, Keret D, Souroujon M, Maschler I, Monselise Y, et al. Role of autoimmune gastritis, Helicobacter pylori and celiac disease in refractory or unexplained iron deficiency anemia. Haematologica. 2005;90(5):585–95 [PubMed] [Google Scholar]
- 2.Finberg KE. Iron-refractory iron deficiency anemia. Semin Hematol. 2009;46(4):378–86 [DOI] [PubMed] [Google Scholar]
- 3.Darshan D, Anderson GJ. Interacting signals in the control of hepcidin expression. Biometals. 2009;22(1):77–87 [DOI] [PubMed] [Google Scholar]
- 4.Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, Basava A, et al. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13(4):399–408 [DOI] [PubMed] [Google Scholar]
- 5.Camaschella C, Roetto A, Cali A, De Gobbi M, Garozzo G, Carella M, et al. The gene TFR2 is mutated in a new type of haemochromatosis mapping to 7q22. Nat Genet. 2000;25(1):14–5 [DOI] [PubMed] [Google Scholar]
- 6.Papanikolaou G, Samuels ME, Ludwig EH, MacDonald ML, Franchini PL, Dube MP, et al. Mutations in HFE2 cause iron overload in chromosome 1q-linked juvenile hemochromatosis. Nat Genet. 2004;36(1):77–82 [DOI] [PubMed] [Google Scholar]
- 7.Roetto A, Papanikolaou G, Politou M, Alberti F, Girelli D, Christakis J, et al. Mutant antimicrobial peptide hepcidin is associated with severe juvenile hemochromatosis. Nat Genet. 2003;33(1):21–2 [DOI] [PubMed] [Google Scholar]
- 8.Babitt JL, Huang FW, Wrighting DM, Xia Y, Sidis Y, Samad TA, et al. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet. 2006;38(5):531–9 [DOI] [PubMed] [Google Scholar]
- 9.Hentze MW, Muckenthaler MU, Galy B, Camaschella C. Two to tango: regulation of Mammalian iron metabolism. Cell. 2010;142(1):24–38 [DOI] [PubMed] [Google Scholar]
- 10.D’Alessio F, Hentze MW, Muckenthaler MU. The hemochromatosis proteins HFE, TfR2, and HJV form a membrane-associated protein complex for hepcidin regulation. J Hepatol. 2012;57(5):1052–60 [DOI] [PubMed] [Google Scholar]
- 11.Mleczko-Sanecka K, Casanovas G, Ragab A, Breitkopf K, Muller A, Boutros M, et al. SMAD7 controls iron metabolism as a potent inhibitor of hepcidin expression. Blood. 2010;115(13):2657–65 [DOI] [PubMed] [Google Scholar]
- 12.Casanovas G, Mleczko-Sanecka K, Altamura S, Hentze MW, Muckenthaler MU. Bone morphogenetic protein (BMP)-responsive elements located in the proximal and distal hepcidin promoter are critical for its response to HJV/BMP/SMAD. J Mol Med (Berl). 2009;87(5):471–80 [DOI] [PubMed] [Google Scholar]
- 13.Andriopoulos B, Jr, Corradini E, Xia Y, Faasse SA, Chen S, Grgurevic L, et al. BMP6 is a key endogenous regulator of hepcidin expression and iron metabolism. Nat Genet. 2009;41(4):482–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meynard D, Kautz L, Darnaud V, Canonne-Hergaux F, Coppin H, Roth MP. Lack of the bone morphogenetic protein BMP6 induces massive iron overload. Nat Genet. 2009;41(4):478–81 [DOI] [PubMed] [Google Scholar]
- 15.Feder JN, Penny DM, Irrinki A, Lee VK, Lebron JA, Watson N, et al. The hemochromatosis gene product complexes with the transferrin receptor and lowers its affinity for ligand binding. Proc Natl Acad Sci U S A. 1998;95(4):1472–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lebron JA, West AP, Jr, Bjorkman PJ. The hemochromatosis protein HFE competes with transferrin for binding to the transferrin receptor. J Mol Biol. 1999;294(1):239–45 [DOI] [PubMed] [Google Scholar]
- 17.Goswami T, Andrews NC. Hereditary hemochromatosis protein, HFE, interaction with transferrin receptor 2 suggests a molecular mechanism for mammalian iron sensing. J Biol Chem. 2006;281(39):28494–8 [DOI] [PubMed] [Google Scholar]
- 18.Chen J, Chloupkova M, Gao J, Chapman-Arvedson TL, Enns CA. HFE modulates transferrin receptor 2 levels in hepatoma cells via interactions that differ from transferrin receptor 1-HFE interactions. J Biol Chem. 2007;282(51):36862–70 [DOI] [PubMed] [Google Scholar]
- 19.Schmidt PJ, Toran PT, Giannetti AM, Bjorkman PJ, Andrews NC. The transferrin receptor modulates Hfe-dependent regulation of hepcidin expression. Cell Metab. 2008;7(3):205–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao J, Chen J, Kramer M, Tsukamoto H, Zhang AS, Enns CA. Interaction of the hereditary hemochromatosis protein HFE with transferrin receptor 2 is required for transferrin-induced hepcidin expression. Cell Metab. 2009;9(3):217–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiss G, Goodnough LT. Anemia of chronic disease. N Engl J Med. 2005;352(10):1011–23 [DOI] [PubMed] [Google Scholar]
- 22.Verga Falzacappa MV, Vujic Spasic M, Kessler R, Stolte J, Hentze MW, Muckenthaler MU. STAT3 mediates hepatic hepcidin expression and its inflammatory stimulation. Blood. 2007;109(1):353–8 [DOI] [PubMed] [Google Scholar]
- 23.Velasco G, Cal S, Quesada V, Sanchez LM, Lopez-Otin C. Matriptase-2, a membrane-bound mosaic serine proteinase predominantly expressed in human liver and showing degrading activity against extracellular matrix proteins. J Biol Chem. 2002; 277(40):37637–46 [DOI] [PubMed] [Google Scholar]
- 24.Ramsay AJ, Hooper JD, Folgueras AR, Velasco G, Lopez-Otin C. Matriptase-2 (TMPRSS6): a proteolytic regulator of iron homeostasis. Haematologica. 2009;94(6):840–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stirnberg M, Maurer E, Horstmeyer A, Kolp S, Frank S, Bald T, et al. Proteolytic processing of the serine protease matriptase-2: identification of the cleavage sites required for its autocatalytic release from the cell surface. Biochem J. 2010;430(1):87–95 [DOI] [PubMed] [Google Scholar]
- 26.Maurer E, Sisay MT, Stirnberg M, Steinmetzer T, Bajorath J, Gutschow M. Insights into matriptase-2 substrate binding and inhibition mechanisms by analyzing active-site-mutated variants. ChemMedChem. 2012;7(1):68–72 [DOI] [PubMed] [Google Scholar]
- 27.Yuan C, Chen L, Meehan EJ, Daly N, Craik DJ, Huang M, et al. Structure of catalytic domain of Matriptase in complex with Sunflower trypsin inhibitor-1. BMC Struct Biol. 2011;11:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang AS, Anderson SA, Wang J, Yang F, DeMaster K, Ahmed R, et al. Suppression of hepatic hepcidin expression in response to acute iron deprivation is associated with an increase of matriptase-2 protein. Blood. 2011;117(5):1687–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Folgueras AR, de Lara FM, Pendas AM, Garabaya C, Rodriguez F, Astudillo A, et al. Membrane-bound serine protease matriptase-2 (Tmprss6) is an essential regulator of iron homeostasis. Blood. 2008;112(6):2539–45 [DOI] [PubMed] [Google Scholar]
- 30.Du X, She E, Gelbart T, Truksa J, Lee P, Xia Y, et al. The serine protease TMPRSS6 is required to sense iron deficiency. Science. 2008;320(5879):1088–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maurer E, Gutschow M, Stirnberg M. Hepatocyte growth factor activator inhibitor type 2 (HAI-2) modulates hepcidin expression by inhibiting cell surface protease matriptase-2. Biochem J. 2013;450(3):585–93 [DOI] [PubMed] [Google Scholar]
- 32.Lakhal S, Schodel J, Townsend AR, Pugh CW, Ratcliffe PJ, Mole DR. Regulation of type II transmembrane serine proteinase TMPRSS6 by hypoxia-inducible factors: new link between hypoxia signaling and iron homeostasis. J Biol Chem. 2011;286(6):4090–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maurer E, Gutschow M, Stirnberg M. Matriptase-2 (TMPRSS6) is directly up-regulated by hypoxia inducible factor-1: identification of a hypoxia-responsive element in the TMPRSS6 promoter region. Biol Chem. 2012;393(6):535–40 [DOI] [PubMed] [Google Scholar]
- 34.Meynard D, Vaja V, Sun CC, Corradini E, Chen S, Lopez-Otin C, et al. Regulation of TMPRSS6 by BMP6 and iron in human cells and mice. Blood. 2011;118(3):747–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buchanan GR, Sheehan RG. Malabsorption and defective utilization of iron in three siblings. J Pediatr. 1981;98(5):723–8 [DOI] [PubMed] [Google Scholar]
- 36.Brown AC, Lutton JD, Pearson HA, Nelson JC, Levere RD, Abraham NG. Heme metabolism and in vitro erythropoiesis in anemia associated with hypochromic microcytosis. Am J Hematol. 1988;27(1):1–6 [DOI] [PubMed] [Google Scholar]
- 37.Hartman KR, Barker JA. Microcytic anemia with iron malabsorption: an inherited disorder of iron metabolism. Am J Hematol. 1996;51(4):269–75 [DOI] [PubMed] [Google Scholar]
- 38.Andrews NC. Iron deficiency: lessons from anemic mice. Yale J Biol Med. 1997;70(3):219–26 [PMC free article] [PubMed] [Google Scholar]
- 39.Fleming MD, Trenor CC, 3rd, Su MA, Foernzler D, Beier DR, Dietrich WF, et al. Microcytic anaemia mice have a mutation in Nramp2, a candidate iron transporter gene. Nat Genet. 1997;16(4):383–6 [DOI] [PubMed] [Google Scholar]
- 40.Pearson HA, Lukens JN. Ferrokinetics in the syndrome of familial hypoferremic microcytic anemia with iron malabsorption. J Pediatr Hematol Oncol. 1999;21(5):412–7 [DOI] [PubMed] [Google Scholar]
- 41.Mayo MM, Samuel SM. Iron deficiency anemia due to a defect in iron metabolism: a case report. Clin Lab Sci. 2001;14(3):135–8 [PubMed] [Google Scholar]
- 42.Melis MA, Cau M, Congiu R, Sole G, Barella S, Cao A, et al. A mutation in the TMPRSS6 gene, encoding a transmembrane serine pro-tease that suppresses hepcidin production, in familial iron deficiency anemia refractory to oral iron. Haematologica. 2008;93(10):1473–9 [DOI] [PubMed] [Google Scholar]
- 43.Finberg KE, Heeney MM, Campagna DR, Aydinok Y, Pearson HA, Hartman KR, et al. Mutations in TMPRSS6 cause iron-refractory iron deficiency anemia (IRIDA). Nat Genet. 2008;40(5):569–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Halfdanarson TR, Litzow MR, Murray JA. Hematologic manifestations of celiac disease. Blood. 2007;109(2):412–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gujral N, Freeman HJ, Thomson AB. Celiac disease: Prevalence, diagnosis, pathogenesis and treatment. World J Gastroenterol. 2012;18(42):6036–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iolascon A, De Falco L, Beaumont C. Molecular basis of inherited microcytic anemia due to defects in iron acquisition or heme synthesis. Haematologica. 2009;94(3):395–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Camaschella C. Hereditary sideroblastic anemias: pathophysiology, diagnosis, and treatment. Semin Hematol. 2009;46(4):371–7 [DOI] [PubMed] [Google Scholar]
- 48.De Falco L, Totaro F, Nai A, Pagani A, Girelli D, Silvestri L, et al. Novel TMPRSS6 mutations associated with iron-refractory iron deficiency anemia (IRIDA). Hum Mutat. 2010;31(5):E1390–405 [DOI] [PubMed] [Google Scholar]
- 49.Guillem F, Lawson S, Kannengiesser C, Westerman M, Beaumont C, Grandchamp B. Two nonsense mutations in the TMPRSS6 gene in a patient with microcytic anemia and iron deficiency. Blood. 2008;112(5):2089–91 [DOI] [PubMed] [Google Scholar]
- 50.Kroot JJ, van Herwaarden AE, Tjalsma H, Jansen RT, Hendriks JC, Swinkels DW. Second round robin for plasma hepcidin methods: First steps toward harmonization. Am J Hematol. 2012;87(10):977–83 [DOI] [PubMed] [Google Scholar]
- 51.Ganz T, Olbina G, Girelli D, Nemeth E, Westerman M. Immunoassay for human serum hepcidin. Blood. 2008;112(10):4292–7 [DOI] [PubMed] [Google Scholar]
- 52.Rivella S. The role of ineffective erythropoiesis in non-transfusion-dependent thalassemia. Blood Rev. 2012;26 Suppl 1:S12–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nemeth E, Rivera S, Gabayan V, Keller C, Taudorf S, Pedersen BK, et al. IL-6 mediates hypoferremia of inflammation by inducing the synthesis of the iron regulatory hormone hepcidin. J Clin Invest. 2004;113(9):1271–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bertoncini S, Blanco-Rojo R, Baeza C, Arroyo-Pardo E, Vaquero MP, Lopez-Parra AM. A novel SNaPshot assay to detect genetic mutations related to iron metabolism. Genet Test Mol Biomarkers. 2011;15(3):173–9 [DOI] [PubMed] [Google Scholar]
- 55.Wu HM, Li L, Yuan XW, Zhou YQ, Xiao QZ, Liu WY, et al. Rapid, accurate detection of TMPRSS6 gene causative mutations with a high-resolution melting assay. Blood Cells Mol Dis. 2011;47(3):198–204 [DOI] [PubMed] [Google Scholar]
- 56.Beutler E, Van Geet C, te Loo DM, Gelbart T, Crain K, Truksa J, et al. Polymorphisms and mutations of human TMPRSS6 in iron deficiency anemia. Blood Cells Mol Dis. 2010;44(1):16–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tchou I, Diepold M, Pilotto PA, Swinkels D, Neerman-Arbez M, Beris P. Haematologic data, iron parameters and molecular findings in two new cases of iron-refractory iron deficiency anaemia. Eur J Haematol. 2009;83(6):595–602 [DOI] [PubMed] [Google Scholar]
- 58.Finberg KE, Whittlesey RL, Fleming MD, Andrews NC. Down-regulation of Bmp/Smad signaling by Tmprss6 is required for maintenance of systemic iron homeostasis. Blood. 2010;115(18):3817–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nai A, Pagani A, Silvestri L, Camaschella C. Increased susceptibility to iron deficiency of Tmprss6-haploinsufficient mice. Blood. 2010;116(5):851–2 [DOI] [PubMed] [Google Scholar]
- 60.Gouya L, Martin-Schmitt C, Robreau AM, Austerlitz F, Da Silva V, Brun P, et al. Contribution of a common single-nucleotide polymorphism to the genetic predisposition for erythropoietic protoporphyria. Am J Hum Genet. 2006;78(1):2–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Delaunay J. Molecular basis of red cell membrane disorders. Acta Haematol. 2002;108(4):210–8 [DOI] [PubMed] [Google Scholar]
- 62.Grandchamp B, Hetet G, Kannengiesser C, Oudin C, Beaumont C, Rodrigues-Ferreira S, et al. A novel type of congenital hypochromic anemia associated with a nonsense mutation in the STEAP3/TSAP6 gene. Blood. 2011;118(25):6660–6 [DOI] [PubMed] [Google Scholar]
- 63.Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452(7186):423–8 [DOI] [PubMed] [Google Scholar]
- 64.Serre D, Gurd S, Ge B, Sladek R, Sinnett D, Harmsen E, et al. Differential allelic expression in the human genome: a robust approach to identify genetic and epigenetic cis-acting mechanisms regulating gene expression. PLoS Genet. 2008; 4(2):e1000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pellegrino RM, Coutinho M, D’Ascola D, Lopes AM, Palmieri A, Carnuccio F, et al. Two novel mutations in the tmprss6 gene associated with iron-refractory iron-deficiency anaemia (irida) and partial expression in the heterozygous form. Br J Haematol. 2012;158(5):668–72 [DOI] [PubMed] [Google Scholar]
- 66.An P, Wu Q, Wang H, Guan Y, Mu M, Liao Y, et al. TMPRSS6, but not TF, TFR2 or BMP2 variants are associated with increased risk of iron-deficiency anemia. Hum Mol Genet. 2012;21(9):2124–31 [DOI] [PubMed] [Google Scholar]
- 67.Kloss-Brandstatter A, Erhart G, Lamina C, Meister B, Haun M, Coassin S, et al. Candidate gene sequencing of SLC11A2 and TMPRSS6 in a family with severe anaemia: common SNPs, rare haplotypes, no causative mutation. PLoS One. 2012;7(4):e35015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Silvestri L, Pagani A, Nai A, De Domenico I, Kaplan J, Camaschella C. The serine protease matriptase-2 (TMPRSS6) inhibits hepcidin activation by cleaving membrane hemojuvelin. Cell Metab. 2008;8(6):502–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lenoir A, Deschemin JC, Kautz L, Ramsay AJ, Roth MP, Lopez-Otin C, et al. Iron-deficiency anemia from matriptase-2 inactivation is dependent on the presence of functional Bmp6. Blood. 2011;117(2):647–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ramsay AJ, Quesada V, Sanchez M, Garabaya C, Sarda MP, Baiget M, et al. Matriptase-2 mutations in iron-refractory iron deficiency anemia patients provide new insights into protease activation mechanisms. Hum Mol Genet. 2009;18(19):3673–83 [DOI] [PubMed] [Google Scholar]
- 71.Silvestri L, Guillem F, Pagani A, Nai A, Oudin C, Silva M, et al. Molecular mechanisms of the defective hepcidin inhibition in TMPRSS6 mutations associated with iron-refractory iron deficiency anemia. Blood. 2009;113(22):5605–8 [DOI] [PubMed] [Google Scholar]
- 72.Altamura S, D’Alessio F, Selle B, Muckenthaler MU. A novel TMPRSS6 mutation that prevents protease auto-activation causes IRIDA. Biochem J. 2010; 431(3):363–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guillem F, Kannengiesser C, Oudin C, Lenoir A, Matak P, Donadieu J, et al. Inactive matriptase-2 mutants found in IRIDA patients still repress hepcidin in a transfection assay despite having lost their serine protease activity. Hum Mutat. 2012;33(9):1388–96 [DOI] [PubMed] [Google Scholar]
- 74.Sisay MT, Steinmetzer T, Stirnberg M, Maurer E, Hammami M, Bajorath J, et al. Identification of the first low-molecular-weight inhibitors of matriptase-2. J Med Chem. 2010;53(15):5523–35 [DOI] [PubMed] [Google Scholar]
- 75.Ganz T. Hepcidin and iron regulation, 10 years later. Blood. 2011;117(17):4425–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cau M, Galanello R, Giagu N, Melis MA. Responsiveness to oral iron and ascorbic acid in a patient with IRIDA. Blood Cells Mol Dis. 2012;48(2):121–3 [DOI] [PubMed] [Google Scholar]
- 77.Sasu BJ, Cooke KS, Arvedson TL, Plewa C, Ellison AR, Sheng J, et al. Antihepcidin antibody treatment modulates iron metabolism and is effective in a mouse model of inflammation-induced anemia. Blood. 2010;115(17):3616–24 [DOI] [PubMed] [Google Scholar]
- 78.Hohlbaum AM, Trentman S, Gille H, Allersdorfer A, Belaiba RS, Huelsmeyer M, et al. Discovery and Preclinical Characterization of a Novel Hepcidin Antagonist with Tunable PK/PD Properties for the Treatment of Anemia in Different Patient Populations. ASH Annual Meeting Abstracts. 2011;118(21):687 [Google Scholar]
- 79.Akinc A, Chan-Daniels A, Sehgal A, Foster D, Bettencourt BR, Hettinger J, et al. Targeting the Hepcidin Pathway with RNAi Therapeutics for the Treatment of Anemia. ASH Annual Meeting Abstracts. 2011;118(21):688 [Google Scholar]
- 80.Ganesh SK, Zakai NA, van Rooij FJ, Soranzo N, Smith AV, Nalls MA, et al. Multiple loci influence erythrocyte phenotypes in the CHARGE Consortium. Nat Genet. 2009;41(11):1191–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Benyamin B, Ferreira MA, Willemsen G, Gordon S, Middelberg RP, McEvoy BP, et al. Common variants in TMPRSS6 are associated with iron status and erythrocyte volume. Nat Genet. 2009;41(11):1173–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Soranzo N, Sanna S, Wheeler E, Gieger C, Radke D, Dupuis J, et al. Common variants at 10 genomic loci influence hemoglobin A(1)(C) levels via glycemic and nonglycemic pathways. Diabetes. 2010;59(12):3229–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Finberg KE, Whittlesey RL, Andrews NC. Tmprss6 is a genetic modifier of the Hfe-hemochromatosis phenotype in mice. Blood. 2011;117(17):4590–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sanders AJ, Webb SL, Parr C, Mason MD, Jiang WG. The type II transmembrane serine protease, matriptase-2: Possible links to cancer?. Anticancer Agents Med Chem. 2010; 10(1):64–9 [DOI] [PubMed] [Google Scholar]
- 85.Deugnier Y, Turlin B. Iron and hepatocellular carcinoma. J Gastroenterol Hepatol. 2001; 16(5):491–4 [DOI] [PubMed] [Google Scholar]