Abstract
Prior to activation of the embryonic genome, the initiating events of mammalian development are under maternal control and include fertilization, the block to polyspermy and processing sperm DNA. Following gamete union, the transcriptionally inert sperm DNA is repackaged into the male pronucleus which fuses with the female pronucleus to form a 1-cell zygote. Embryonic transcription begins during the maternal to zygotic transfer of control in directing development. This transition occurs at species-specific times after one or several rounds of blastomere cleavage and is essential for normal development. However, even after activation of the embryonic genome, successful development relies on stored maternal components without which embryos fail to progress beyond initial cell divisions. Better understanding of the molecular basis of maternal to zygotic transition including fertilization, the activation of the embryonic genome and cleavage-stage development will provide insight into early human development that should translate into clinical applications for regenerative medicine and assisted reproductive technologies.
Keywords: maternal to zygotic transition (MZT), mouse fertilization, gamete recognition, DNA methylation, histone modification, zygotic genome activation (ZGA), maternal effect genes, subcortical maternal complex (SCMC)
1. INTRODUCTION
Mammalian development begins when the haploid sperm fuses with the haploid egg at fertilization to form the 1-cell diploid zygote (Fig. 1A). Both the maternal and paternal genomes are required for successful development (Mcgrath and Solter, 1984; Surani et al., 1984). At gamete fusion, the fertilizing sperm DNA is packaged with protamines and mature sperm are transcriptionally inert. Although the sperm provides DNA for the male pronucleus and is essential for egg activation, the mitochondria, the microtubule-organizing center precursors and the stored cellular components in the sperm play minor roles in fertilization and early embryogenesis (Saunders et al., 2002; Schatten et al., 1985; Shitara et al., 1998; Sutovsky and Schatten, 2000). Thus, it is incumbent on the egg to provide suitable environment for sperm-egg recognition, prevention of polyspermy, paternal genome remodeling and embryonic genome activation, ensuring a successful transition from maternal control to a shared responsibility with the male genome in directing early development (Fig. 1B).
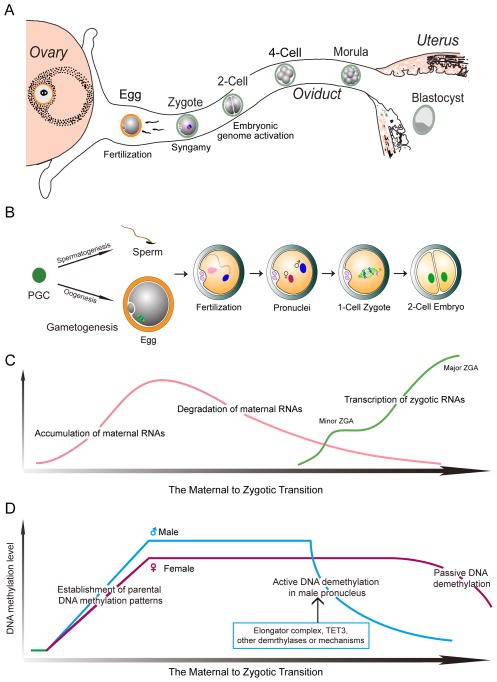
Figure 1.
The maternal to zygotic transition in mice.
(A) Ovulated eggs are fertilized in the ampulla of the oviduct to form 1-cell zygotes. After three cell divisions (cleavage stage development), the embryo undergoes compaction to form the morula. A fluid filled blastocoel forms in the blastocyst that escapes from the zona pellucida to implant on the wall of the uterus. (B) During gametogenesis, primordial germ cells (PGCs) develop into haploid sperm and oocytes after two rounds of meiotic division. The maternal to zygotic transition (MZT) is initiated when sperm and egg fuse at the time of fertilization. Each haploid gamete forms a pronucleus and after syngamy develops into a 1-cell zygote that divides to form the 2-cell embryo. (C) During oogenesis, oocytes accumulate a large pool of maternal RNAs essential for fertilization, the maternal to zygotic transition and preimplantation embryogenesis. After fertilization, maternal RNAs are gradually degraded in early embryonic stages. Transcription of the embryonic genome is initiated at the late 1-cell stage (minor ZGA) and robustly activated at 2- and 4-cell stages (major ZGA). (D) During gametogenesis, the genome of the sperm and oocyte are methylated de novo to obtain sex-specific patterns in which DNA methylation is slightly higher in sperm than in oocytes (Smith et al., 2012). After fertilization and replacement of protamines with hyperacetylated maternal histones, the male pronucleus is actively demethylated and initiates zygotic gene transcription. Following syngamy, the zygotic genome undergoes passive demethylation in a DNA replication dependent manner, but preserves the methylation status of imprinting regions.
At birth, the ovary contains a full complement of germ cells and after puberty, cohorts enter into a two week growth phase that culminates in meiotic maturation and ovulation into the oviduct. In mice, the female genome is transcriptionally and translationally active as oocytes grow to ~70 μm and the enlarging oocyte serves as a storehouse for maternal proteins (Fig. 1C) (De Leon et al., 1983). These proteins are critically important because transcription stops during meiotic maturation prior to ovulation and the embryonic genome is not robustly activated until the 2-cell stage (Fig. 1C). Thus, the maternal to zygotic transition (MZT) depends heavily on stored maternal components to form structures necessary to initiate development, eliminate redundant maternal materials and activate the embryonic genome to reprogram gene expression during the MZT (Tadros and Lipshitz, 2009).
Microarray and proteomic analyses have defined global patterns of gene expression in early development and transgenesis provides insight into the role of specific genes during the MZT. Genetic studies have shown that maternal effect genes affect multiple processes including pronuclear formation and fusion (Philipps et al., 2008; Wu et al., 2003), the first cell division (Burns et al., 2003; Tang et al., 2007), embryonic gene transcription (Bultman et al., 2006; Ramos et al., 2004) and cleavage-stage embryogenesis (Li et al., 2008a; Ma et al., 2006; Payer et al., 2003; Roest et al., 2004; Tong et al., 2000). Limited data from these studies suggest the important role of individual maternal effect genes on embryonic development, but the function and mechanisms of these genes in early embryogenesis are largely unknown.
In this review, we focus on mice as a model to highlight molecular events in the MZT in mammals. Our discussion begins with fertilization of the transcriptionally inert gametes, a process dependent on stored maternal proteins. We then examine the processing and activation of the embryonic genomes as a transition phase from maternal to zygotic control and finish with the continued role of maternal components in cleavage stage embryogenesis. Finally we briefly address future challenges associated with the MZT and the medical implications of understanding this process.
2. MATERNAL CONTRIBUTION TO THE ONSET OF DEVELOPMENT
2.1 Fertilization
Fertilization heralds the onset of development in most species, including mammals. For successful embryonic development, sperm need to penetrate outer investments to fuse with the egg and there must be an effective post-fertilization block to polyspermy. The absence of the former precludes development and the absence of the latter is an embryonic lethal. Although a multi-step process, a major arbiter of successful fertilization and preimplantation development in mammals is the zona pellucida that surrounds ovulated eggs and preimplantation embryo (Fig. 2A, B). Encoded by maternal genes, the zona pellucida not only mediates relative taxon-specific gamete recognition and provides a definitive block to polyspermy, it also ensures passage of the early embryo down the oviduct prior to implantation in the uterus.
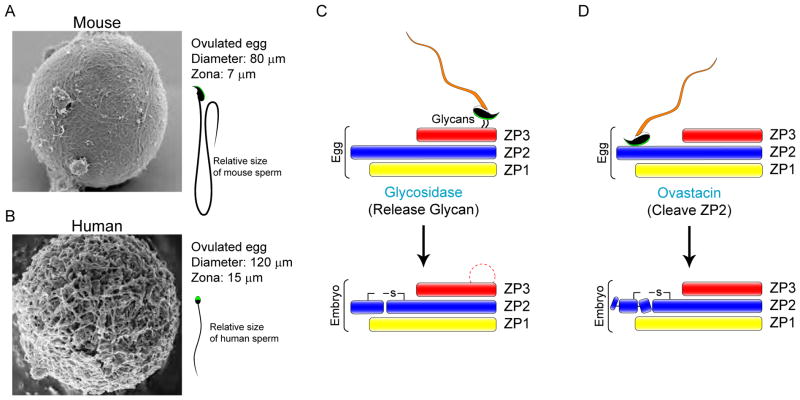
Figure 2.
Fertilization, the initiation of development.
(A) The ~7 μm wide mouse zona pellucida surrounds a ~80 μm egg. As observed by scanning electron microscopy, the zona matrix has multiple pores which may facilitate sperm penetration (Familiari et al., 2008). Mouse sperm are ~125 μm long with a thin acrosome overlying a distinctive falciform (hook-like) head. (B) The thicker (~ 15 μm) human zona pellucida surrounds a larger (~120 μm) egg and yet its three-dimensional structure is similar to mouse (Familiari et al., 2006). Human sperm are half as long (~ 60 μm) as mouse sperm with a smaller, flattened, spatulate head. Of note, human sperm are fastidious and will not bind to mouse eggs, although mouse sperm bind to human (and most other) eggs. (C) Glycan release models postulate a carbohydrate ligand attached to a zona pellucida protein that interacts with a sperm surface receptor. Although N-glycans have been proposed, most attention has been focused on O-glycans attached to ZP3. Following fertilization, the removal of the glycan by a cortical granule glycosidase would account for the inability of sperm to bind to 2-cell embryos. However, recent biochemical investigations and genetic ablation studies have not confirmed the candidacy of proposed glycan ligands or receptors as essential for sperm-egg recognition. (D) The ZP2 cleavage model proposes that sperm bind with taxon-specificity to the N-terminus of ZP2 and the cleavage status of ZP2 is crucial for sperm-zona recognition and binding. Following fertilization, ovastacin, a metalloendoprotease, is exocytosed from egg cortical granules to cleave the N-terminus of ZP2 and prevent post-fertilization sperm binding. Implicit in the model is the presence of a cognate receptor(s) on the sperm surface which has not as yet been molecularly defined.
2.2 Zona Pellucida
The genomes of eutherian mammals (including human, rat and mouse) contain four loci (Zp1, Zp2, Zp3, Zp4) on separate, but syntenic chromosomes that potentially encode zona proteins. Although all four proteins are detected in the zona pellucida of human (Lefievre et al., 2004) and rat (Hoodbhoy et al., 2005), only ZP1, ZP2 and ZP3 are present in mouse (Bleil and Wassarman, 1980b; Boja et al., 2003) because of multiple stop codons in transcripts expressed at the Zp4 locus (Lefievre et al., 2004). Each mouse gene is single copy in the genome (Chamberlin and Dean, 1989; Epifano et al., 1995; Kinloch et al., 1988; Liang et al., 1990) and mouse lines with null mutations for Zp1, Zp2 and Zp3 have been established. Mice lacking ZP1 form a zona pellucida to which sperm bind and these mice are fertile, albeit with decreased fecundity (Rankin et al., 1999). The absence of either ZP2 or ZP3 precludes formation of a stable zona pellucida around eggs which are resorbed after ovulation and these mice are sterile (Liu et al., 1996; Rankin et al., 1996; Rankin et al., 2001). However, this phenotype can be rescued by transgenic expression of homologous human proteins, suggesting that human ZP2 and ZP3 are structurally comparable to the corresponding mouse proteins (Rankin et al., 2003; Rankin et al., 1998).
2.3 Models of Sperm-zona Recognition
The molecular basis of mammalian gamete recognition has perplexed investigators for decades. Candidate glycans and proteins that were initially proposed based on biochemical or cell biology assays have not proven to be essential for fertility when ablated in transgenic mouse models.
Glycan-release Models
The most widely embraced models for sperm-egg recognition have been based on zona glycan ligands binding to a sperm surface receptor (Fig. 2C). In these models, the post-fertilization release of a glycosidase from the egg’s cortical granules is hypothesized to cleave the candidate glycan and account for the inability of sperm to bind to the zona surrounding 2-cell embryos. Mouse and human taxon-specific binding is ascribed to differences in the glycosylation of their respective zona proteins.
ZP3, initially described as a sperm ‘receptor’, but more seemingly a ‘ligand’, was first implicated in sperm-egg recognition based on the ability of gel-purified, re-natured, soluble ZP3 to inhibit (~80%) sperm binding to ovulated eggs in vitro (Bleil and Wassarman, 1980a). This model was extended to specify O-glycans on ZP3 as mediators of sperm-egg recognition (Florman and Wassarman, 1985), specifically a terminal α1,3 galactose residue (Bleil and Wassarman, 1988), and further refined to implicate ZP3 Ser332 and Ser334 as attachment sites for the glycan ligand (Chen et al., 1998a). An independent line of investigation identified a second candidate glycan, N-acetylglucosamine, as the essential ZP3 ligand that was bound by sperm surface β1,4 galactosyl transferase acting as a lectin (Miller et al., 1992). In this model, the subsequent release by cortical granule N-acetylglucosaminidase accounted for the inability of sperm to bind to 2-cell embryos (Miller et al., 1993).
However, subsequent biochemical and genetic data have not supported these early models. In particular, genetically modified mice lacking α1,3 galactose (Liu et al., 1997; Thall et al., 1995) or N-acetylglucosamine (Williams et al., 2007) remained fertile and the absence of the candidate sperm receptor, β1,4 galactosyl transferase minimally affects fertility (Asano et al., 1997; Lu and Shur, 1997). Turning to the putative attachment sites, mass spectrometric analysis, sensitive to low femtomole levels, did not detect glycosylation on either ZP3 Ser332 or Ser334 (Boja et al., 2003). Moreover, specific genetic mutation of the implicated serine residues to preclude attachment of O-glycans did not affect sperm recognition or fertility in transgenic mice (Liu et al., 1995) even when crossed into Zp3 null mice to reconstitute a zona pellucida in which ZP3Mut completely replaces endogenous mouse ZP3. Sperm bound to ovulated eggs with a zona pellucida containing mutant ZP3 in the absence of normal ZP3 and the otherwise normal appearing Zp3Mut female mice were fertile (Gahlay et al., 2010). Thus, current biochemical and genetic data are not consistent with ‘glycan-release’ models in which O-glycans attached to ZP3 Ser332 or Ser334 play an essential role in sperm-egg recognition.
ZP2 Cleavage Model
In a series of loss of function assays, sperm binding and fertility was assessed in Zp1, Zp2 or Zp3 null mice. Mice lacking ZP1 form a zona matrix with ZP2 and ZP3 that is structurally abnormal with increased porosity when viewed by electron microscopy. Sperm bind and fertilize Zp1 null eggs, but exhibit decreased fecundity with litter sizes half that of normal mice. This continued fertility documents that ZP1 is not essential for gamete recognition (Rankin et al., 1999). Mice lacking either ZP2 or ZP3 have a more striking phenotype. No zona pellucida surrounds ovulated eggs in the absence of either protein and the zona-free eggs are quickly resorbed into the epithelial lining of the oviduct. Although Zp2 and Zp3 null mice are sterile (Liu et al., 1996; Rankin et al., 1996; Rankin et al., 2001), no assessment of the role of either ZP2 or ZP3 in sperm binding is possible in the absence of a zona matrix.
Therefore, a series of gain-of-function assays were established to take advantage of the observation that, although individual proteins are conserved among mammals, human sperm do not bind to the mouse zona pellucida (Bedford, 1977). Using transgenesis, mouse lines were established in which human ZP1, ZP2, ZP3 and ZP4 replaced (or added to) endogenous mouse proteins (Rankin et al., 1999; Rankin et al., 2003; Rankin et al., 1998; Yauger et al., 2011). In a taxon-specific sperm binding assay, only human ZP2 supported human sperm binding after which sperm penetrated the ‘humanized’ zona pellucida and accumulated in the perivitelline space unable to fuse with mouse eggs (Baibakov et al., 2012). Normally ZP2 is cleaved after fertilization and sperm will not bind to the zona pellucida (Bauskin et al., 1999; Bleil et al., 1981). The cleavage site has been biochemically defined in mice (166LA↓DE169) and, when mutated to prevent cleavage, sperm bind to the zona pellucida after fertilization and cortical granule exocytosis (Gahlay et al., 2010). Taken together, these observations support a model for gamete recognition in which sperm bind to ZP2 and the post-fertilization cleavage of ZP2 accounts for the inability of sperm to bind to 2-cell embryos (Fig. 2D).
The sperm binding site on ZP2 was further defined in a bead binding assay in which human sperm bound well to recombinant human ZP239-154 or ZP239-267 peptides, but poorly to beads coated with the homologous mouse peptides. Reduction of disulfide bonds significantly decreased human sperm binding to the human peptides indicating a dependence on secondary structures. The binding of human sperm to huZP2 rescued eggs was inhibited by excess huZP239-154, but not moZP235-149 peptides, which corroborated the specificity of the binding site on ZP2. Although fewer in number, mouse sperm bound comparably to mouse and human ZP2 peptide beads consistent with its observed lack of taxon specificity in binding to mouse and human eggs (Bedford, 1977). From these results, it was concluded that the N-terminus of human ZP2 plays an important role in gamete recognition on the surface of the zona pellucida (Baibakov et al., 2012).
In this rapidly changing landscape, a significant amount of investigation will be required to obtain wide-spread consensus for the molecular basis of gamete recognition. For glycan-release models to be revived, a specific glycan ligand(s) and sperm-surface receptor will need to be identified that, when genetically ablated, prevents fertilization. In addition, expression of the human glycan ligand in transgenic mice should break taxon-specific, sperm-egg recognition. The ZP2 cleavage model requires further validation in transgenic mice expressing chimeric mouse-human and truncated forms of mouse ZP2. The identification of a cognate sperm surface receptor with taxon-specificity that could be manipulated in transgenic mouse models would be particularly compelling. The incompleteness of our understanding of this critical event should inspire renewed research into the molecular basis of sperm-egg recognition that is required for fertilization and the onset of development.
2.4 The Post-fertilization Block to Polyspermy
Polyspermy is lethal for early embryos and at least three post-fertilization blocks to gamete interactions have evolved in mice. The first two occur rapidly after fertilization and prevent additional sperm from fusing with the egg’s plasma membrane or penetrating the extracellular zona pellucida surrounding eggs and preimplantation embryos (Sato, 1979; Stewart-Savage and Bavister, 1988). The third and definitive block occurs over several hours and ensures that sperm do not bind to the surface of the zona pellucida (Baibakov et al., 2007; Inoue and Wolf, 1975). The molecular basis of the first two blocks remains largely unknown, and the third correlates with egg cortical granule exocytosis (Barros and Yanagimachi, 1971).
As described above, ZP2 is cleaved and sperm do not bind to the zona pellucida surrounding 2-cell embryos after fertilization (Bauskin et al., 1999; Bleil et al., 1981). Ovastacin, a member of the astacin family of metalloendoproteases (Quesada et al., 2004), is released from cortical granules following fertilization and normally cleaves ZP2 (Burkart et al., 2012) over several hours (Baibakov et al., 2007). The ensuing proteolysis effectively precludes sperm binding to the zona pellucida (Burkart et al., 2012; Greenhouse et al., 1999). Either mutation of the ZP2 cleavage site (Gahlay et al., 2010) or ablation of the gene encoding ovastacin (Burkart et al., 2012) prevents proteolysis (Fig. 2D). Thus, when ZP2 remains intact, sperm will bind to the zona pellucida surrounding the early embryo independent of fertilization and cortical granule exocytosis.
3. EPIGENETIC REGULATION OF THE MATERNAL TO ZYGOTIC TRANSITION
During gametogenesis, mouse primordial germ cells (PGCs) establish sex-specific epigenetic marks with distinct DNA methylation patterns that can occur globally and in specific imprinted regions (Fig. 1D) (Hayashi and Surani, 2009; Lees-Murdock and Walsh, 2008). As gametes mature, the haploid male and female genomes, packaged respectively with protamines and histones, become transcriptionally quiescent. At fertilization, the highly differentiated, haploid maternal egg and paternal sperm fuse to establish the totipotent 1-cell zygote in which both genomes undergo dynamic changes in DNA methylation (Fig. 1D) and are repackaged with histones in a specific manner to initiate embryonic development. There is substantial research that documents the critical importance of epigenetic modifications during the MZT.
3.1 Epigenetics
Epigenetics refers to heritable changes in gene expression or cellular phenotypes that occur without alterations in DNA sequence, but rather through DNA methylation, histone modification and regulation by non-coding RNAs (e.g. miRNA, siRNA, piRNA) (Berger et al., 2009). By adding a methyl group to the fifth carbon of cytosine residues in the context of CpG dinucleotides (Holliday and Pugh, 1975), DNA methylation plays vital roles in various cellular processes including regulation of gene expression, establishment of imprinting (Li et al., 1993), X chromosome inactivation in females (Robertson and Wolffe, 2000), silencing of parasitic retrotransposons (Yoder et al., 1997) and maintaining the stability and integrity of the genome (Chen et al., 1998b). The patterns of DNA methylation can be divided into two categories: de novo DNA methylation and maintenance DNA methylation, each with a set of well-characterized DNA methyltransferases (DNMTs) that are conserved in both animals and plants (Goll and Bestor, 2005; Law and Jacobsen, 2010). Enzymes responsible for de novo DNA methylation are DNMT3 family members and include DNMT3a, DNMT3b and DNMT3L (Aapola et al., 2000; Okano et al., 1999; Okano et al., 1998). DNMT3L lacks the key methyltransferase motifs and has no enzymatic activity, but acts as a regulator of both DNMT3a and DNMT3b in de novo DNA methylation (Gowher et al., 2005; Suetake et al., 2004). DNMT1 methylates DNA with a strong preference for hemimethylated target sites and maintains methylated DNA during DNA replication (Hermann et al., 2004). In general, the status of DNA methylation is associated with gene transcription in which hypermethylation indicates repression and hypomethylation specifies activation (Reik et al., 2001; Santos and Dean, 2004).
In the nucleus, DNA is packaged with histones to form core nucleosomes (two each of H2A, H2B, H3, and H4) which are separated by linker histones (H1) (Luger et al., 1997; Turner, 2002) and further arranged into higher-order chromatin structures. Post-translational modifications of individual histones (i.e., the histone code) in their amino termini include acetylation, phosphorylation, ubiquitylation, ADP-ribosylation and methylation in which specific lysine or arginine residues can be modified by mono-, di- and trimethylation. These modifications afford greater subtlety than DNA methylation alone and are essential in regulating cellular and molecular events including chromatin remodeling and gene expression (Berger, 2002; Kouzarides, 2007; Peterson and Laniel, 2004). More recent investigations of histone variants are providing additional insight into the histone code, making these modulatory epigenetic events ever more intriguing (Kamakaka and Biggins, 2005; Sarma and Reinberg, 2005).
3.2 Demethylation of DNA in the Paternal Genome
Just prior to ovulation, oocytes undergoes nuclear envelope breakdown and complete the first meiotic division. Ovulated eggs are arrested at metaphase of the second meiotic division (MII) with their DNA packaged with maternal histones. In contrast, the haploid sperm DNA is tightly packaged with protamines which must be removed after gamete fusion. The mechanisms of this removal during sperm nuclear decondensation are incompletely understood, but are accompanied by wide-spread demethylation (Abdalla et al., 2009; Mayer et al., 2000; Oswald et al., 2000) and followed by repackaging with hyperacetylated maternal histones to form the male pronucleus (Fig. 1D) (Adenot et al., 1997; Santos et al., 2002). Following syngamy of the two pronuclei, the zygotic genome undergoes passive demethylation in a DNA replication dependent manner until the morula stage (Fig. 1D). Thereafter, de novo methylation patterns are established to sustain successful cell lineage differentiation (Corry et al., 2009; Rougier et al., 1998; Santos et al., 2002).
DNA demethylation of the paternal genome is rapidly completed within 4–6 hours after fertilization and occurs before the commencement of the first round of DNA replication (Fig. 1D) (Santos and Dean, 2004; Santos et al., 2002). This indicates an active process (Abdalla et al., 2009; Mayer et al., 2000; Oswald et al., 2000) in which the methyl group from 5-methyl cytosine is enzymatically removed (Wu and Zhang, 2010). One candidate, MBD2 (methyl-CpG-binding domain protein 2) was reported to have demethylase activity and to directly remove methyl groups from 5-methylcytosine in vitro (Bhattacharya et al., 1999). However, Mbd2 null mice are viable, fertile and lack abnormal DNA methylation patterns (Hendrich et al., 2001; Santos et al., 2002) and, thus, other candidates must be sought.
Using a siRNA-mediated knockdown screening strategy coupled with live cell imaging of mouse zygote, ELP3, a submit of the elongator complex (composed of ELP1 to ELP6), was reported to play important roles in paternal genome demethylation (Okada et al., 2010). ELP3 knockdown prevented paternal DNA demethylation as shown by cytosine-5 methylation (5mC) staining and bisulphite sequencing. The same effect was observed with knockdowns of ELP1 and ELP4 which suggests that the entire elongator complex may be involved in demethylation (Okada et al., 2010). ELP3 contains a histone acetyltransferase (HAT) domain and a cysteine-rich Fe–S radical S-adenosylmethionine (SAM) domain which can catalyze paternal DNA demethylation as shown by dominant-negative mutants using domain-specific mRNAs (Okada et al., 2010). Based on the known catalytic mechanism of radical SAM domains (Wang and Frey, 2007), it is postulated that ELP3-mediated DNA demethylation is accomplished by a series of radical reactions starting from the generation of a powerful oxidizing agent, the 5′-deoxyadenosyl (5′-dA) radical from SAM, and ending with the production of 5-hydroxymethyl-cytosine (5hmC) which would be further resolved into cytosine (Wu and Zhang, 2010).
Recently, the Tet family has been discovered to mediate DNA demethylation via an oxidative process catalyzing the conversion of 5mC to 5hmC (Ito et al., 2010; Tahiliani et al., 2009; Veron and Peters, 2011). The Tet family includes Tet1, Tet2 and Tet3 in mice. Tet1 and Tet2 play a role in embryonic stem cell self-renewal and human myeloid malignancies, respectively, and their perturbation is accompanied by defects in DNA methylation (Ito et al., 2010; Nolte and Hofmann, 2008; Xu et al., 2011). Elevation of 5hmC and reduction of 5mC is associated with enrichment of TET3 in the mammalian paternal pronucleus, raising the possibility of a role of TET3 in active DNA demethylation of the paternal genome (Gu et al., 2011; Iqbal et al., 2011; Wossidlo et al., 2011). Indeed, knocking down maternal Tet3 with specific siRNA (Wossidlo et al., 2011) or through transgenesis in murine oocytes and zygotes (Gu et al., 2011) leads to aberrant methylation patterns in the male genome. The observed decrease in 5hmC and increase in 5mC levels in the late male pronucleus indicates that disruption of 5mC to 5hmC conversion causes abnormal DNA demethylation. However, there is little perturbation in the maternal genome when Tet3 is disrupted which suggests specific targeting to the male (Gu et al., 2011). 5hmC can be further oxidized to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), and the latter could be excised by thymine-DNA glycosylase (TDG)–mediated base excision repair (He et al., 2011; Ito et al., 2011). However, whether 5hmC is an intermediate of demethylation or an end-product in the male pronucleus remains to be determined. The observation of replication-dependent dilution of 5hmC during preimplantation mouse development indicates that TET proteins may also facilitate passive demethylation of the embryonic genome after active demethylation of the male pronucleus (Inoue and Zhang, 2011).
Active demethylation of the paternal genome has been observed in many mammalian embryos including those of mouse, rat, pig, bovine and human (Abdalla et al., 2009; Dean et al., 2001; Fulka et al., 2004; Mayer et al., 2000) which indicates a biological imperative in early development. One hypothesis is that active DNA demethylation de-represses the paternal genome. This could enable minor transcriptional activation of the paternal genome at the late 1-cell stage and prime the robust transcription essential for the MZT. In support of this hypothesis, BrUTP incorporation, indicative of de novo gene transcription, is initially detected in the male pronucleus 5 hours after pronuclear formation and is always higher than that observed in the female pronucleus (Aoki et al., 1997). Alternatively, active demethylation may be required for reprogramming paternal chromatin into a totipotent state, as has been observed during somatic nuclear transferred into enucleated oocyte (Dean et al., 2003; Reik et al., 2001). However, neither hypothesis adequately explains why only the paternal genome undergoes active demethylation and in such a narrow developmental window. Based on the kinship theory of imprinting (Wilkins and Haig, 2003), demethylation of the paternal genome by maternal factors during the protamine-histone exchange (Reik and Walter, 2001) could maximize the female genome fitness by reactivating genes in paternal genome (Wilkins and Haig, 2002). This evolutionary conflict based notion is consistent with the absence of active DNA demethylation in species without genomic imprinting, such as zebrafish (Macleod et al., 1999) and Xenopus (Stancheva et al., 2002).
Global and active DNA demethylation of the paternal genome in the one-cell zygotes is widely considered to occur shortly after fertilization. However, a recent study suggested that the weak staining of 5meC in the male nucleus during the first cell cycle may resulted from a progressive acid-resistant antigenic masking of 5meC (Li and O’Neill, 2012). When antigen retrieval was performed using tryptic digestion, no loss of DNA methylation in male pronuclei was observed. Rather, there was persistence of 5meC in both pronuclei, an observation that was supported by the almost equal staining with MBD1 (methyl binding domain 1 protein), a methyl group binding protein (Li and O’Neill, 2012). Considering the difficulty in identifying a specific demethylase or mechanism to account for active DNA demethylation in the male pronucleus, this general paradigm might need to be reconsidered and further investigated.
3.3. Maternal and Paternal Imprinting
While there is active DNA demethylation of the male pronucleus shortly after fertilization followed by passive, replication dependent DNA demethylation of the entire genome (Santos and Dean, 2004; Santos et al., 2002), the methylation status of imprinted regions in maternal and paternal genome remains constant. The genomic imprinting was discovered thirty years ago when investigators using pronuclear transplantation demonstrated that diploid mouse embryos with two pronuclei originating from the same parent (gynogenones or androgenones) failed to develop to term (Mcgrath and Solter, 1984; Surani et al., 1984). This was the first documentation that, although each pronucleus carried the same genetic information, maternal and paternal alleles are not functionally equivalent in mammalian development.
In mammals, genomic imprinting gives rise to parental-specific monoallelic expression of particular genes that are only expressed from either the maternal or the paternal allele. Since the first two paternally imprinted genes, Igf2r and H19, and the first maternally imprinted gene, Igf2, were reported (Barlow et al., 1991; Bartolomei et al., 1991; Dechiara et al., 1991), more than 150 imprinted genes have been identified in mice (http://www.mousebook.org/catalog.php?catalog=imprinting). Most imprinted genes are organized in clusters that can span more than 1Mb and are scattered throughout the genome (Verona et al., 2003). The parent-of-origin expression pattern of these genes results from different DNA methylation in cis-acting regulatory elements termed imprinting control regions (ICRs) functioning through differentially methylated regions (DMRs), in which only one allele, either maternal or paternal, is methylated. Notably, dysfunction of ICRs leads to loss of proper expression of imprinted genes in the cluster (Fitzpatrick et al., 2002; Thorvaldsen et al., 1998) and improper expression of imprinted genes can affect embryogenesis leading to growth defects and, in some cases, severe disease including cancer. The low efficiency in obtaining SCNT (somatic cell nuclear transfer) derived clones and the unpredictable abnormalities in survivors are likely a result of aberrant genomic imprinting (Humpherys et al., 2001). In humans, several malformation syndromes, including Prader-Willi, Angelman, Beckwith-Wiedemann and Silver-Russell as well as cancers such as neuroblastoma (maternal chromosome 1p36 and paternal chromosome 2), Wilms’ tumour (maternal chromosome 11p15.5) and acute myeblastic leukemia (paternal chromosome 7), have documented associations with imprinting defects (Butler, 2009; Lim and Maher, 2009; Walter and Paulsen, 2003). Clinical studies have reported that human malformations associated with imprinting disorders occur with greater frequency in babies obtained by assisted reproductive technology (ART) (Cox et al., 2002; DeBaun et al., 2003; Gicquel et al., 2003). Because of these impacts on mammalian development, patterns of genomic imprinting must be sustained faithfully throughout embryogenesis and on into adulthood (Li and Sasaki, 2011).
DNA methyltransferases have been shown to control the methylation status of imprinted regions (Cirio et al., 2008; Hirasawa et al., 2008; Howell et al., 2001; Kurihara et al., 2008). Depletion of DNMT1o, which is present in the oocyte and translocates to the nucleus of the 8-cell embryo, results in abnormal, allele-specific expression and half of the ICR methylation is lost (Howell et al., 2001). DNMT1s are associated with MII oocyte chromatin and are present in the nucleus throughout preimplantation development. Inhibiting DNMT1s activity with antibodies or RNAi (RNA interference) knockdowns decreases DNA methylation in specific repetitive sequences and imprinted loci. Thus, DNMT1s is also responsible for methylation maintenance (Cirio et al., 2008; Kurihara et al., 2008). Deletion of both maternal and zygotic DNMT1 results in complete loss of methylation at imprinted regions, suggesting that both maternal and zygotic DNMT1 are required to maintain DNA methylation patterns at imprinted loci (Hirasawa et al., 2008).
However, DNA methyltransferases are not sufficient for to maintain imprints. Stella (official name, Dppa3 and also known as Pgc7), first characterized as a maternal effect gene (Payer et al., 2003), plays important roles in maintaining methylation of most of the maternal genome and two paternally imprinted genes, H19 and Rasgrf1 (Nakamura et al., 2007). STELLA is present in both maternal and paternal pronuclei, but seems to preferentially bind to a specific ‘architecture’ in maternal chromatin (Nakamura et al., 2012), namely that provided by dimethylated histone H3 lysine 9 (H3K9me2). When the H3K9me2 mark was removed with ectopic expression of an H3K9 methylation/dimethylation-specific demethylase (Jhdm2a, official name Kdm3a) in mouse zygotes, STELLA staining was no longer observed in the maternal pronucleus and maternal DNA methylation was not maintained (Nakamura et al., 2012). In addition, the protection of DNA methylation patterns in paternally imprinted H19 and Rasgrf1 DMRs was associated with enrichment of H3K9me2 at these two loci, further supporting a role for H3K9me2 in mediating protection of DNA methylation by STELLA (Nakamura et al., 2012). DNA demethylation is accompanied by the TET3-mediated conversion of 5mC to 5hmC in the paternal genome, but not in the maternal genome (Gu et al., 2011; Iqbal et al., 2011). However, strong staining of TET3 anomalously occurred in maternal pronucleus in Stella null zygotes. Thus, the binding of STELLA to H3K9me2 containing chromatin seems to inhibit the binding of TET3 to its chromatin targets, which prevents the conversion of 5mC to 5hmC and protects DNA methylation in normal mouse zygotes (Nakamura et al., 2012).
ZFP57, a KRAB-zinc finger protein, also has been shown to play a role in maintenance of both parental imprints (Li et al., 2008b). While zygotic depletion of ZFP57 results in partial lethality and partial disruption of several imprinted loci, lack of both maternal and zygotic ZFP57 leads to a more severe phenotype with greater embryonic lethality and complete loss of methylation at multiple loci (Snrpn, Peg1, Peg3, Peg5, and Dlk1 DMRs) (Li et al., 2008b). In addition, ZFP57 plays a role in establishing maternal imprints at the Snrpn locus and maternal loss of ZFP57 precludes establishment of Snrpn imprints in mouse oocyte (Li et al., 2008b). Consistent with these observations, a clinical study reported that ZFP57 mutations were frequently found in patients with transient neonatal diabetes, a disease caused by hypomethylation of the promoter of the imprinted gene Plagl1, coupled with abnormal hypomethylation at multiple other imprinted loci (Mackay et al., 2008). Recently, Trim28 (also known as KAP-1 or Tif1β), which is highly expressed in the nuclei of oocytes and preimplantation embryos, was found to be involved in methylation maintenance at imprinted loci (Messerschmidt et al., 2012). When Trim28 was depleted in oocytes, no viable offspring were obtained, although embryos could successfully develop into blastocysts (Messerschmidt et al., 2012). Upon further analysis, it was proposed that this severe phenotype might be a result of dysregulation of genomic imprinting. Notably, the ICR of the paternally imprinted H19 was hypomethylated with up-regulation of H19 expression in maternal Trim28 mutants (Messerschmidt et al., 2012), indicating the importance of maternal TRIM28 in safeguarding paternal H19 imprints from aberrant demethylation.
Taken together, these data suggest that multiple mechanisms have evolved to counteract global DNA demethylation and thus maintain proper imprints at specific genetic loci at particular times in development.
3.4. Histone Modification
Histone modifications are dramatically different in maternal and paternal pronuclei of zygotes and undergo dynamic changes during the MZT. After fertilization, hyperacetylated histones are incorporated into the paternal genome and may account for its relatively higher transcriptional activity compared to the maternal pronucleus (Aoki et al., 1997). H3.3 is a replication-independent variant of H3 and indicative of a transcriptionally permissive state. Following fertilization, this variant is preferentially incorporated into the paternal pronucleus and enriched in paternal chromatin by means of a specific histone chaperone, HIRA (Torres-Padilla et al., 2006; van der Heijden et al., 2005). On the other hand, while H3 di- and tri- K9 and K27 methylation are readily detected in the maternal pronucleus and may provide protection of the maternal genome from the active DNA demethylation, they are not present in the paternal pronucleus (Arney et al., 2002; Feil, 2009; Reik et al., 2003; Santos et al., 2005). Trimethylation of H3K4 is readily observed in the maternal pronucleus before pronuclear stage PN4, but the extent of this modification becomes indistinguishable between the two pronuclei by PN5 (Lepikhov and Walter, 2004). Considering the complexity of histone modifications, no single set of rules can be followed to explain these phenomena and deciphering the detailed dynamics of various histone modifications during the MZT remains an intriguing challenge.
A further challenge is the need to coordinate histone modifications with DNA methylation for proper regulation of gene expression in early embryonic development. For example, transcriptionally repressive H3K9 methyltransferases G9a (official name, EHMT2), SUV39H1 and SETDB1 (also known as ESET) interact with DNA methyltransferases, including DNMT1, DNMT3A and DNMT3B. This cooperation establishes particular methylation patterns and can regulate gene expression at specific time and sites in early development (Cedar and Bergman, 2009; Saitou et al., 2012). It seems likely that similar coordination between histone modifications and DNA methylation is involved in regulation of additional genes in mammalian development. To date this remains an under-investigated area of research, but high-throughput ChIP experiments should provide insight into the relationship between histone modification and the MZT.
4. ACTIVATION OF THE EMBYRONIC GENOME
Transcription of the embryonic genome (zygotic genome activation, ZGA) occurs after fertilization and is critical for normal embryonic development. The initiation of ZGA varies depending on the species. In mouse, ZGA is observed as early as the S/G2 phase in the male pronucleus of the 1-cell zygote and becomes robust at the 2-cell stage (Fig. 1C) (Bouniol et al., 1995; Latham et al., 1991; Ram and Schultz, 1993); in human (Davis, 1985) and pig (Braude et al., 1988) the ZGA occur and at 4-8-cell stage; and in cow (Frei et al., 1989) and sheep (Crosby et al., 1988) it is delayed until the 8-16-cell stage. Because of its essential role in the onset of development, there has been considerable interest in the regulation of ZGA and changes in gene expression profiles during the MZT.
4.1 Transcriptome Profile of the Maternal to Zygotic Transition
Conventional approaches including RT-PCR (reverse transcription; polymerase chain reaction), quantitative RT-RCR and northern blot analyses have been used to examine gene expression during the MZT (Christians et al., 1995; Davis et al., 1996). In addition, mRNA differential display (Zimmermann and Schultz, 1994), expressed sequence tag (EST) based analysis (Ko et al., 2000) and suppression subtractive hybridization (Zeng and Schultz, 2003) have been used to dissect stage specific gene expression during the MZT and preimplantation mouse development. In recent years, large-scale gene expression profiles from microarrays and deep RNA sequencing have systematically provided enormous data sets of global gene expression during the MZT and preimplantation development in various mammals, including mice, cow, pigs and humans (Hamatani et al., 2004; Misirlioglu et al., 2006; Sirard et al., 2005; Vallee et al., 2008; Wang et al., 2004; Whitworth et al., 2005; Zeng et al., 2004; Zeng and Schultz, 2005). Of particular note is the dynamic wave of gene expression corresponding to the MZT, during which maternal transcripts are degraded and zygotic genes are activated (Fig. 1C).
Three groups separately have reported large-scale gene expression patterns in mouse oocytes and preimplantation embryos using different microarray platforms (Hamatani et al., 2004; Wang et al., 2004; Zeng et al., 2004; Zeng and Schultz, 2005). Employing NIA 22K 60-mer oligo microarrays, 12,179 out of 21,939 genes were observed to exhibit significant change (Hamatani et al., 2004). These genes were classified into 9 groups (clusters 1–9) and, using a k-means nonhierarchical clustering method, were further partitioned into 3 groups according to their expression patterns. The first group (clusters 7 and 9) comprises 1,589 genes highly enriched in oocytes, but progressively degraded in early development; the second group (clusters 1, 4, 5, and 8) contains 5,126 genes that are initially transcribed from the embryonic genome; and the third group (clusters 2, 3 and 6) is composed of 5,464 genes that are both inherited as maternally stored transcripts and transcribed from the embryonic genome after ZGA at the 1-2-cell stage. Maternal transcripts that are gradually degraded around ovulation and after fertilization are mainly represented in clusters 3, 6, 7 and 9, which include a total of 4,063 genes. Among these four clusters, only genes in cluster 3 are reactivated after ZGA. Gene expression patterns corresponding to the ZGA are grouped in clusters 1, 5 and 8, with genes in cluster 1 increasing throughout preimplantation development and those in cluster 5 and 8 peaking at 2-cell and 4-cell stage, respectively, and then declining as development progresses. Intriguingly, a set of genes in cluster 2 are firstly dramatically up-regulated from the 4-cell to 8-cell stage before compaction and then significantly decreased later in development. This novel gene expression pattern is designated “mid-preimplantation gene activation” (MGA), which might play crucial roles in cell polarity and the first cell lineage specification.
In a second study using Affymetrix Murine Genome Array U74Av2, more than 12,000 genes, over 1/3 of which exhibited ≧5-fold change in their expression, were identified in the MZT (Wang et al., 2004). It was found that 737 genes increased by ≧3 fold, while 1,082 genes decreased by ≧3-fold during meiotic maturation, indicating that accumulation of transcripts is coupled with degradation and/or deadenylation of many maternal transcripts (Bachvarova, 1985; Paynton et al., 1988). After fertilization and initiation of ZGA, embryonic RNA increase in complexity because multiple genes are activated. The genes in which the 12 time points were examined from GV oocyte to blastocyst stage were clustered into two large temporal groups, one of which, the “oocyte-to-embryo” phase, documents the dramatic changes of gene expression during the MZT (Wang et al., 2004).
In a third study based on MOE430 GeneChips, 14,119 genes were identified in mouse oocytes and early embryos including 1-cell, 2-cell, 8-cell and blastocyst embryos (Zeng et al., 2004). Among all the identified genes, 13,378 genes were found differentially expressed in at least one stage and were further grouped into 6 classes (‘maternal’, ‘maternal-to-zygotic’, ‘1-cell transient’, ‘2-cell transient’, ‘8-cell transient’, and ‘blastocyst’) according to their temporal expression patterns. Members in the “maternal” group, such as Mos and Zp3, represent gene products accumulated in oogenesis that progressively vanish in later development. “Maternal-to-zygotic” genes usually are expressed in both oocytes and early embryos having first been degraded and then replaced by zygotic transcripts. Expression Analysis Systematic Explorer (EASE) analysis of the 809 genes transiently expressed in the 2-cell embryos documented that these genes are involved in transcription and RNA metabolism, indicating potential roles in the MZT. Genes associated with chromatin assembly and disassembly are enriched in 1-cell and 2-cell embryos and may be critical for chromatin structure changes observed during the ZGA and the MZT (Zeng et al., 2004). Pharmacological studies documented that 1,819 genes (~17% of genes detected in the 2-cell embryos) are α-amanitin sensitive (inhibits RNA Polymerase II and III), indicating their initial expression after ZGA (Zeng and Schultz, 2005). EASE analysis of these α-amanitin sensitive 2-cell transcripts revealed that bioprocesses, such as ribosome biogenesis and assembly, protein synthesis, RNA metabolism and transcription, are overrepresented, which further substantiates the selective and biased nature of gene expression patterns at ZGA (Zeng et al., 2004; Zeng and Schultz, 2005). Moreover, the observed α-amanitin sensitive 2-cell transcripts are concordant with the NIA data (Hamatani et al., 2004), suggesting the reproducibility and validity of the data obtained in the two different platform.
In recent years, investigative attention has turned to analyzing profiles of small, non-coding RNAs including miRNAs (microRNA), endo–siRNAs (small interfering RNA) and piRNAs (Piwi-interacting RNA) during the MZT (Ma et al., 2010; Suh et al., 2010; Svoboda and Flemr, 2010; Tam et al., 2008; Tang et al., 2007; Watanabe et al., 2008). The expression pattern of miRNAs are divided into three classes: maternal, maternal-to-zygote and zygote patterns (Tang et al., 2007). Maternally inherited miRNAs (e.g. the let-7 family members) are degraded and de novo miRNAs synthesis starts at the 2-cell stage (e.g. the mir-290 cluster). However, maternal miRNA appears necessary for early development. Embryos derived from Dicer null oocytes in which almost all miRNAs are depleted, lack a functional meiotic spindle and fail to develop beyond the first cell division (Tang et al., 2007). In other model organisms, including Drosophila (Bushati et al., 2008), zebrafish (Giraldez et al., 2006) and Xenopus (Lund et al., 2009), miRNAs that mediate silencing pathways deplete maternal mRNAs and are critical for the MZT. However, global suppression of miRNA pathways in growing oocytes and preimplantation embryos has limited effect on the MZT in mice (Ma et al., 2010; Suh et al., 2010).
Endo–siRNAs in mouse oocytes are derived from naturally occurring dsRNAs (double strand RNAs) that are produced by inverted repeat structures, bidirectional transcription, antisense transcripts and some expressed pseudogenes (Tam et al., 2008; Watanabe et al., 2008). Reduction of siRNA from the loss of DICER or AGO2 in oocytes leads to increase of mobile and repetitive sequences and complementary endogenous transcripts (Watanabe et al., 2008). In addition, many up-regulated transcripts in Dicer1−/−oocytes have complementary sequences to endo-siRNAs (Ma et al., 2010). Given the minimal affect observed after suppression of the miRNA pathways in mouse oocyte and preimplantation embryos, the siRNA pathway might serve as the dominant RNA silencing mechanism controlling the MZT.
With the large scale and global gene expression data during the MZT, the next step will be screening specific factors that potentially play critical roles in the MZT, such as the degradation of maternal factors, fertilization, the initiation of ZGA and even the whole process of preimplantation development, and deciphering the molecular regulatory mechanisms of these functional genes.
4.2. Proteome Profile of the Maternal to Zygotic Transition
Although RNA profiles provide essential data, they may not accurately reflect abundance of cognate proteins because some RNAs may be un-coupled (non-coding RNAs) or not immediately coupled (delayed translation) with protein synthesis (Nothias et al., 1996). Therefore, high-resolution 1-dimentional or 2-dimensional protein gel electrophoresis, polysomal mRNA microarray and protein microarray, coupled with mass spectrometry and bioinformatic analyses have been used to inventory proteins during the MZT (Flach et al., 1982; Latham et al., 1991; Potireddy et al., 2006; Wang et al., 2010; Yurttas et al., 2010).
Growing oocytes accumulate large pools of stored maternal RNAs and, although many are degraded after fertilization, some are stable and persist in early development (Potireddy et al., 2006; Yurttas et al., 2010; Zhang et al., 2009). During the MZT, maternal mRNAs are recruited for translation in support of particular developmental events that occur before ZGA. To identify these mRNAs, polysomal mRNAs in MII stage oocytes and late 1-cell zygotes were identified on microarrays (Potireddy et al., 2006). Maternal mRNAs recruited for translation depended on cis-regulatory elements within the transcripts and changed significantly during transition from oocyte to embryo. While most polysomal maternal mRNA isolated from oocyte are associated with cellular homeostasis, those over-represented in late 1-cell embryos are related to biosynthesis, especially protein biosynthesis (Potireddy et al., 2006) which is consistent with data obtained by analyzing total mRNA (Zeng et al., 2004). Of note, one class of proteins encoded by genes recruited for translation in late 1-cell embryos is associated with gene transcription, which may contribute to de novo zygotic gene transcription at 2-cell stage.
Using semi-quantitative mass spectrometry, 2,781, 2,973, and 2,082 proteins were respectively identified in germinal vesicle (GV), MII oocytes and zygotes (Wang et al., 2010). Proteins relating to metabolism and transport that may support oocyte maturation were over-represented in GV oocytes and those associated with cell cycle, epigenetic modification, DNA repair and pluripotency regulation were over-represented in MII oocytes. Unexpectedly, zygotes were highly enriched in proteins of the ubiquitination pathway including ubiquitin B (UBB), ubiquitin C (UBC) and proteasome proteins. This may account for the observed decrease in peptides from MII oocytes (185,643) and to zygotes (85,369) and suggest a role for the ubiquitin-proteasome pathway in active degradation of maternal proteins after fertilization.
Although important strides have been made in cataloging the proteome, de novo zygotic protein synthesis during the ZGA remains inadequately understood. Thus, the next challenge seemingly lies in developing high-resolution, high-throughput protein analyses from the limited biomaterials available from early embryos. Once catalogued, advances can be anticipated in deciphering the potential molecular functions of maternal proteins that have been degraded and, more importantly, zygotic proteins that have been synthesized de novo and play critical roles in the MZT.
4.3. Regulation of Zygotic Genome Activation
ZGA is indispensable for preimplantation development and was investigated in the early 1970s using α-amanitin, a specific inhibitor of RNA polymerase II and III (Golbus et al., 1973; Warner and Versteegh, 1974). When α-amanitin was added to the medium at the 1–2 cell stage, mouse embryos could not develop beyond 2-cells. In addition, fertilized eggs cultured in vitro tend to arrest at the 2-cell stage, a phenomenon termed the “2-cell block” (Goddard and Pratt, 1983) which may reflected delayed ZGA (Qiu et al., 2003) and depends on culture condidtions (Summers et al., 1995). Given the crucial roles of ZGA in embryonic development, multiple molecular mechanisms and levels of control may exist including maternal effect genes, chromatin remodeling and DNA replication (Minami et al., 2007; Schultz, 1993).
Maternal factors, including Mater (official name, Nlrp5) (Tong et al., 2000), Padi6 (Yurttas et al., 2008), Hsf1 (Christians et al., 2000), Zar1 (Wu et al., 2003), Npm2 (Burns et al., 2003), Ctcf (Wan et al., 2008), Zfp36l2 (Ramos et al., 2004), Atg5 (Tsukamoto et al., 2008), Ago2 (official name, Eif2c2) (Lykke-Andersen et al., 2008), Basonuclin (Ma et al., 2006), Zag1 (official name, N4bp2l2) (Matsuoka et al., 2008), Zar1l (Hu et al., 2010), Granzyme G (official name, Gzmg) (Tsai et al., 2010), Ring1, Rnf2 (Posfai et al., 2012), the maternal pluripotency transcription factors Oct4 (official name, Pou5f1) (Foygel et al., 2008) and Sox2 (Pan and Schultz, 2011) have been implicated in regulation of ZGA. Ablation of the gene encoding any one of these proteins results in embryonic arrest at cleavage-stage development and is reminiscent of cycloheximide inhibition of the initiation of ZGA (Cho et al., 2002; Li et al., 2010; Minami et al., 2007; Wang and Latham, 1997).
Chromatin remodeling has long been considered to play a critical role in the MZT by ensuring specific patterns of gene expression (Cho et al., 2002; Kanka, 2003; Thompson et al., 1998). Several lines of evidence have documented that a chromatin mediated transcriptionally repressive state is superimposed on the ZGA at the onset of mouse development. For example, enhancers are needed to express exogenous genes injected into the maternal pronucleus of 1-cell zygotes as well as the nuclei of 2-cell embryos (Henery et al., 1995; Wiekowski et al., 1991, 1993), and treatment with butyrate, an inhibitor of histone deacetylase, significantly increases exogenous gene expression in early embryos (Wiekowski et al., 1993). The repressive transcriptional state of the chromatin structure might serve to selectively modulate promoter activity to ensure proper spatiotemporal expression of genes that are time-critical in triggering normal developmental while repressing non-required or deleterious genes (Schultz, 2002).
A chromatin mediated repressive state could account for the observation that genes involved in transcription and RNA processing are preferentially expressed at the 2-cell stage of development (Zeng et al., 2004). Employing in situ DNase I sensitivity assays (indicative of open chromatin structures) and in vitro transcription analysis (indicative of the total transcriptional activity), a direct association between the chromatin structure and the ZGA is observed (Cho et al., 2002). By and large, increased DNase I sensitivity corresponds with increased transcriptional activity from the late 1-cell to the late 2-cell stage while both are decreased from the early to late 2-cell stage. However, DNase I sensitivity and transcriptional activity exhibited opposite changes from early 1-cell to late 1-cell where initial transcriptional activity is associated with decreased DNase I sensitivity. Thus, it seems that the involvement of chromatin structure in the regulation of ZGA begins at the late 1-cell stage, and that maternal proteins (Wang and Latham, 1997), rather than chromatin structure, may account for the initiation of ZGA and gene expression patterns at the early 1-cell stage.
Given that chromatin structure is critical for the ZGA, it is reasonable to speculate that chromatin remodeling factors are involved in the MZT. Indeed, abnormality of factors involved in chromatin remodeling tends to result in prevention of ZGA. BRG1, a catalytic subunit of SWI/SNF chromatin remodeling complex, has been shown to be involved in this transition (Bultman et al., 2006). Maternal mutation of Brg1 blocks embryonic development at early cleavage stages and reduces transcription for ~30% of genes. In addition, the depletion of Brg1 reduces the level of dimethyl H3K4, a mark for transcriptionally active chromatin (Bultman et al., 2006). Another factor, TIF1α (transcription intermediary factor 1 α), also modulates gene expression during the first wave of transcription activation (Torres-Padilla and Zernicka-Goetz, 2006). TIF1α has been shown to translocate from the cytoplasm to the pronucleus and accumulate in specific regions that are enriched with chromatin remodelers including SNF2H and BRG1. Ablation of TIF1α, using either RNAi or anti-TIF1α antibody, results in arrest of embryos at the 2-4-cell stage, misallocation of RNA polymerase II, SNF2H and BRG1 as well as dysregulation of a subset of genes necessary for proper ZGA (Torres-Padilla and Zernicka-Goetz, 2006).
The observation that embryonic gene expression is initially detected as early as mid-S-phase in 1-cell embryos (Aoki et al., 1997) and the subtle correlation between DNA replication and active or repressive transcription (Wolffe, 1991) raises the possibility that DNA replication might take part in the regulation of ZGA. The inhibition of the first round of DNA synthesis does not prevent initiation of ZGA which is consistent with an internal “zygotic clock” regulating ZGA independent of embryonic progression to the 2-cell stage (Wiekowski et al., 1991). However, failure of the first round of DNA replication leads to a pronounced reduction in abundance of two transiently expressed genes in 2-cell embryos, TRC (Transcription Required Complex) and eIF-1A (Davis et al., 1996), and inhibits incorporation of BrUTP by ~35% (Aoki et al., 1997). The inhibition of the second round of DNA replication in 2-cell embryos prevents the expression of Hsp70 (Christians et al., 1995), the TRC and elF-1A (Davis et al., 1996). Furthermore, BrUTP incorporation in aphidicolin (blocks DNA replication) treated 2-cell embryos at G2 of the cell cycle is ~4-fold more than controls (Aoki et al., 1997). This suggests that the second round of DNA replication could exert influence on ZGA by establishing a transcriptionally repression architecture. Thus, it seems that both the first and second round of DNA replication are involved in the establishment of repressive chromatin structure, which would further regulate gene expression patterns during ZGA (Cho et al., 2002).
5. PERSISTENT MATERNAL EFFECTS IN EARLY DEVLOPMENT
In additional to individual proteins, complexes of maternal proteins assembled during oogenesis can persist and play critical roles in preimplantation development. For example, the zona pellucida that surrounds ovulated eggs to mediate fertilization also protects the dividing embryo as it passes through the oviduct prior to implantation in the uterus. If the zona pellucida is removed at the cleavage stage of development either biochemically (Bronson and McLaren, 1970; Modlinski, 1970) or by ablation of Zp2 (Rankin et al., 2001) or Zp3 (Liu et al., 1996; Rankin et al., 1996), the zona-free embryos are resorbed into the epithelial lining of the oviduct and no offspring are observed. More recently a second maternal structure has been identified and designated the subcortical maternal complex (SCMC). Like the extracellular zona matrix, the intracellular SCMC matrix is assembled during oogenesis and is essential for successful preimplantation development (Fig. 3).
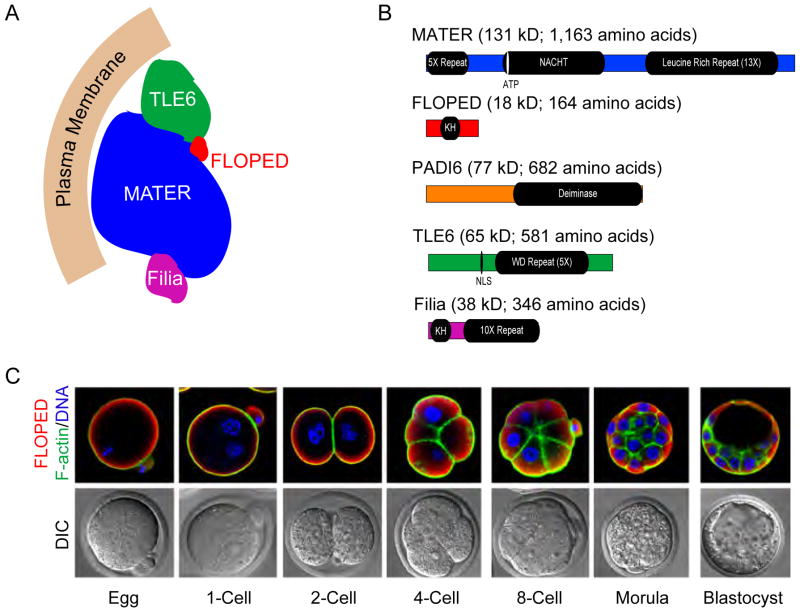
Figure 3.
The Subcortical Maternal Complex.
(A) A model of the SCMC (subcortical maternal complex). The SCMC includes at least four proteins: MATER, FLOPED, TLE6, Filia and, most likely, PADI6. The first three proteins interacts each other, while Filia only binds to MATER (modified from (Li et al., 2008a). (B) Protein domains in the members of the SCMC. MATER includes a novel 5X N- terminal repeat, a NACHT domain and a 13-fold leucine-rich domain. FLOPED contains an atypical KH domain and PADI6 has a deiminase domain that converts arginine to citrulline. TLE6 belongs to the Groucho co-repressor family and has a nuclear localization signal (NLS) and a WD domain, but lacks the N-terminal Q domain required for DNA binding. Filia has an atypical KH domain in its N- terminal and a novel 23 amino acid 10-fold repeat. (C) Localization of the SCMC in egg and early embryos. Mouse eggs and early embryos were stained with phalloidin labeled with fluorescence (F-actin, green), DAPI (DNA, blue) and rabbit anti-FLOPED (red) and imaged by confocal microscopy (modified from Li et al., 2008a).
5.1 Proteins of the Subcortical Maternal Complex
The founding member of the SCMC is FLOPED, a maternal protein that was first identified as a downstream target of FIGLA and defined by its localization in the subcortex of the egg and 1-cell zygote (Li et al., 2008a). In an earlier investigation, FIGLA had been identified as a germ-cell specific, basic helix-loop-helix transcription factor that coordinated expression of Zp1, Zp2 and Zp3 (Liang et al., 1997). The further observation that FIGLA was required for neonatal formation of primordial follicles (Soyal et al., 2000) suggested a role in the activation of multiple downstream oocyte-specific genes. Using microarrays and SAGE to compare the transcriptome of newborn normal and FiglaNull ovaries, additional potential gene targets were identified (Joshi et al., 2007).
Floped (official name, Ooep), characterized as a maternal effect gene from this screen (Li et al., 2008a), was also identified in a proteomic screen of ovulated eggs and by digital differential display analysis of pre-existing cDNA libraries (Herr et al., 2008; Pierre et al., 2007). To discover potential binding partners, ovarian lysates were immunoprecipitated with antibodies to FLOPED and analyzed by tandem mass spectrometry. Numerous candidate proteins, including those encoded by Mater, Filia, Padi6 and Tle6 were detected. The interactions of these proteins were confirmed by co-expression of Myc- or HA-tagged recombinant proteins in heterologous cell lines (Li et al., 2008a). Mater (official name, Nlrp5) had been characterized as a gene encoding an antigen associated with a mouse model of autoimmune oophoritis (Tong and Nelson, 1999). Unexpectedly, its ablation in female (but not male) mice prevented embryonic progression past cleavage-stage development and resulted in female sterility(Tong et al., 2000). In further functional investigations of this maternal effect gene, Filia (official name, Khdc3), was identified as a binding partner of MATER (Ohsugi et al., 2008).
The phenotype of the MaterNull prompted a search for additional maternal effect genes in these data sets. To date, multiple independent approaches now have identified at four protein components in the SCMC (Fig. 3B) and based on its subcortical localization, immunoprecipitation with antibodies to FLOPED and a similar phenotype in gene-targeted mice, it is likely that PADI6 also participates in the SCMC (Li et al., 2008a; Yurttas et al., 2008). Sizing by gel filtration revealed that the SCMC has a molecular mass between ~669 kDa and ~2000 kDa, which is much larger than the total mass (~325 kDa) of the known proteins (Li et al., 2008a) (Fig. 3A). This discrepancy raises several possibilities regarding to the organization and composition of the SCMC. First, additional proteins other than the aforementioned are present in the SCMC. Second, components in the SCMC oligomerize or polymerize with each other to conjointly form the ~MDa complex, which is supported by recent crystallographic observation that the N-terminal fragment of Filia (Filia-N) forms a stable dimer in both solution and crystal (Wang et al., 2012). Third, both possibilities may contribute to the supramolecular organization of the SCMC.
5.2 Formation of the Subcortical Maternal Complex
Mater (Chr7:24170908-24226941 bp), Floped (Chr9:78223916-78226532 bp), Padi6 (Chr4:140283270-140298558 bp), Tle6 (Chr10:81053649-81063818 bp) and Filia (Chr9:72950268-72952220 bp) are conserved, single copy genes located on four different chromosomes in the mouse genome and yet their expression is coordinately regulated during oogenesis to establish the SCMC (Li et al., 2008a; Ohsugi et al., 2008). Accumulation of transcripts of each gene is specific or enhanced in growing oocytes and, based on microarray and SAGE comparison of normal and null newborn ovaries, at least four out of these five genes are regulated by FIGLA. The regulation of Filia was indeterminate in these assays which lacked an appropriate element on the microarray or a tag in the SAGE libraries for Figla transcripts (Joshi et al., 2007).
MATER, FLOPED, PADI6, TLE6 and Filia are synthesized during oogenesis, and co-localize in the subcortex of growing oocytes, ovulated eggs and one-cell zygotes (Fig. 3C). As embryos completes their first divisions, these proteins are excluded from the region of cell-cell contact and segregate to the outer cells of the morula and blastocyst where they are asymmetrically restricted to the apical cortex. This exclusion from cell-cell contact (Fig. 3C) is reversible (Herr et al., 2008; Li et al., 2008a; Ohsugi et al., 2008) and suggests a dynamic equilibrium between a cytoplasmic pool of preassembled complex (or component parts) and the subcortical complex. Cell divisions non-parallel to the apical-basal axis of the polarized blastomeres result in cell populations marked by the presence or absence of the SCMC (Fig. 3C). These observations are consistent with a model in which the early embryo, although subject to regulative development, differentially accumulates a maternally expressed protein complex in topologically distinct blastomeres. The progeny of those containing the SCMC complex preferentially form the trophectoderm; those without the complex preferentially become the inner cell mass of the blastocyst (Fig. 3C).
5.3 The Function of the Subcortical Maternal Complex
Ablation of the genes encoding individual components of the SCMC documented that each had maternal effect on early embryogenesis. For example, adult male and female MaterNull mice appeared normal and, although males were fertile, females produced no offspring. MaterNull mice have normal ovarian histology and ovulate eggs that can be fertilized in vitro and in vivo, but embryos progress poorly past the first cell division and uniformly perish prior to implantation (Tong et al., 2000). A similar phenotype is observed in FlopedNull and Padi6Null mice, but a milder phenotype with delayed embryonic progression and decreased fecundity was reported for FiliaNull female mice (Li et al., 2008a; Tong et al., 2000; Yurttas et al., 2008; Zheng and Dean, 2009). The observed 2-cell arrest might be a consequence of incompletion of ZGA, as de novo RNA and protein synthesis are significantly decreased in 2-cell embryos lacking the SCMC and the marker of ZGA, the TRC, is significantly reduced (Tong et al., 2000; Yurttas et al., 2008). Nonetheless, how the SCMC might modulate the initiation of ZGA is unknown. Although there is no obvious evidence that the SCMC is involved in the clearance of maternal factors, the potential roles of Filia (Wang et al., 2012) and FLOPED (Li et al., 2010; Pierre et al., 2007) in RNA interaction raise the possibility that the SCMC might participate in RNA metabolism, localization and regulation of transcription and translation.
Thus, the SCMC joins the zona pellucida as a maternal complex that is vital in preimplantation mouse development, but our understanding of its function is far from complete. Each component of the SCMC has a human homologue with human and mouse MATER (1200 aa) sharing 46% identity, FLOPED (149 aa) sharing 39% identity, Filia (217) sharing 41% identity, TLE6 (449 aa) sharing 44% identity and PADI6 (694 aa) sharing 67% (Li et al., 2010). Thus, exploring the molecular function of the SCMC in mouse models systems may provide therapeutic insight to causes of human infertility and recurrent spontaneous abortions.
6. CONCLUSIONS
Developmental control of the early mammalian embryo is gradually transferred from stored maternal proteins to the newly activated embryonic genome through a series of carefully orchestrated steps. There appear to be multiple threads in this gradual transfer rather than an abrupt change and maternal proteins can participate at least until the blastocyst stage. Development is initiated by gamete recognition, fertilization and the initial processing of the sperm genome, all of which depend heavily on stored maternal components. With intricate modulation of genetic and epigenetic regulators, the maternal to zygotic transition is concomitant with a dramatic reprogramming of gene expression and a series of cellular and molecular events, including degradation of maternal factors, DNA demethylation, changes of histone modification and eventual zygotic genome activation.
Despite significant progress, comprehensive understanding of the molecular basis and the corresponding consequences of the mammalian MZT remains a yet to be attained goal. Particularly intriguing among the unanswered questions are: what are the downstream effects triggered by fertilization that facilitate processing of the male genome; what processes control degradation of maternal RNAs and proteins and the initiation of ZGA; how and why is DNA actively and globally demethylated in the paternal pronucleus while specific regions in both parent genomes avoid demethylation to preserve imprinting; how does the histone code function in the two pronuclei and what is its effect on zygotic genome activation? To answer these questions, more powerful technologies and systemic analyses, including high-throughput sequencing of transcriptomes and epigenomes (e.g. bisulphite genomic sequencing and ChIP experiments) are required to investigate qualitative and quantitative changes in oocytes and early embryos. Considering the persistence of maternal proteins during the MZT and into preimplantation development, specific protein knockout technologies, which relies on the conserved eukaryotic ubiquitin pathway and has been used in cultured cell lines and Drosophila (Caussinus et al., 2012; Zhang et al., 2003), need to be developed in oocytes and early embryos to determine specific protein functions during the MZT.
Beyond its intrinsic importance, the maternal to zygotic transition also represents a paradigm of reverting terminally differentiated cells (gametes) to totipotency (1-cell zygotes) under physiological conditions which could provide insight into regenerative medicine. It is likely that improved understanding of the molecular and systematic basis of the maternal to zygotic transition also will provide useful diagnostic and therapeutic information that can be translated to the clinical with implications for improved medical technologies and reproductive choices for infertile couples.
Acknowledgments
We thank Professor Giuseppe Familiari for the scanning electron microscopy images of the mouse and human zonae pellucidae as well as our scientific colleagues for the many discussions that helped shape this review. This work is supported by the National Basic Research Program of China (2012CB944401, 2011CB944501), the National Natural Science Foundation of China (30971655, 31171382), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA01010103), the Hundred Talents Program of Chinese Academy of Sciences and the Intramural Research Program of the National Institutes of Health, NIDDK.
Abbreviations
- MZT
maternal to zygotic transition
- DNMTs
DNA methyltransferases
- 5fC
5-formylcytosine
- 5caC
5-carboxylcytosine
- TDG
thymine-DNA glycosylase
- ICR
imprinting control regions
- DMR
differentially methylated regions
- SCNT
somatic cell nuclear transfer
- ART
assisted reproductive technology
- RNAi
RNA interference
- ZGA
zygotic genome activation
- TRC
transcription required complex
- SCMC
subcortical maternal complex
- miRNA
microRNA
- endo–siRNA
endo-small interfering RNA
- piRNA
Piwi-interacting RNA
- dsRNA
double strand RNA
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aapola U, Kawasaki K, Scott HS, Ollila J, Vihinen M, Heino M, Shintani A, Minoshima S, Krohn K, Antonarakis SE, Shimizu N, Kudoh J, Peterson P. Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics. 2000;65 (3):293–298. doi: 10.1006/geno.2000.6168. [DOI] [PubMed] [Google Scholar]
- Abdalla H, Yoshizawa Y, Hochi S. Active demethylation of paternal genome in mammalian zygotes. J Reprod Dev. 2009;55 (4):356–360. doi: 10.1262/jrd.20234. [DOI] [PubMed] [Google Scholar]
- Adenot PG, Mercier Y, Renard JP, Thompson EM. Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development. 1997;124 (22):4615–4625. doi: 10.1242/dev.124.22.4615. [DOI] [PubMed] [Google Scholar]
- Aoki F, Worrad DM, Schultz RM. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol. 1997;181 (2):296–307. doi: 10.1006/dbio.1996.8466. [DOI] [PubMed] [Google Scholar]
- Arney KL, Bao S, Bannister AJ, Kouzarides T, Surani MA. Histone methylation defines epigenetic asymmetry in the mouse zygote. Int J Dev Biol. 2002;46 (3):317–320. [PubMed] [Google Scholar]
- Asano M, Furukawa K, Kido M, Matsumoto S, Umesaki Y, Kochibe N, Iwakura Y. Growth retardation and early death of beta-1,4-galactosyltransferase knockout mice with augmented proliferation and abnormal differentiation of epithelial cells. EMBO J. 1997;16 (8):1850–1857. doi: 10.1093/emboj/16.8.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachvarova R. Gene expression during oogenesis and oocyte development in mammals. Dev Biol (N Y 1985) 1985;1:453–524. doi: 10.1007/978-1-4615-6814-8_11. [DOI] [PubMed] [Google Scholar]
- Baibakov B, Boggs NA, Yauger B, Baibakov G, Dean J. Human sperm bind to the N-terminal domain of ZP2 in humanized zonae pellucidae in transgenic mice. J Cell Biol. 2012;197 (7):897–905. doi: 10.1083/jcb.201203062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baibakov B, Gauthier L, Talbot P, Rankin TL, Dean J. Sperm binding to the zona pellucida is not sufficient to induce acrosome exocytosis. Development. 2007;134 (5):933–943. doi: 10.1242/dev.02752. [DOI] [PubMed] [Google Scholar]
- Barlow DP, Stoger R, Herrmann BG, Saito K, Schweifer N. The Mouse Insulin-Like Growth-Factor Type-2 Receptor Is Imprinted and Closely Linked to the Tme Locus. Nature. 1991;349 (6304):84–87. doi: 10.1038/349084a0. [DOI] [PubMed] [Google Scholar]
- Barros C, Yanagimachi R. Induction of zona reaction in golden hamster eggs by cortical granule material. Nature. 1971;233 (5317):268–269. doi: 10.1038/233268a0. [DOI] [PubMed] [Google Scholar]
- Bartolomei MS, Zemel S, Tilghman SM. Parental Imprinting of the Mouse H19 Gene. Nature. 1991;351 (6322):153–155. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- Bauskin AR, Franken DR, Eberspaecher U, Donner P. Characterization of human zona pellucida glycoproteins. Mol Hum Reprod. 1999;5 (6):534–540. doi: 10.1093/molehr/5.6.534. [DOI] [PubMed] [Google Scholar]
- Bedford JM. Sperm/egg interaction: the specificity of human spermatozoa. Anat Rec. 1977;188 (4):477–487. doi: 10.1002/ar.1091880407. [DOI] [PubMed] [Google Scholar]
- Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev. 2002;12 (2):142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23 (7):781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397 (6720):579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- Bleil JD, Beall CF, Wassarman PM. Mammalian sperm-egg interaction: fertilization of mouse eggs triggers modification of the major zona pellucida glycoprotein, ZP2. Dev Biol. 1981;86 (1):189–197. doi: 10.1016/0012-1606(81)90329-8. [DOI] [PubMed] [Google Scholar]
- Bleil JD, Wassarman PM. Mammalian sperm-egg interaction: identification of a glycoprotein in mouse egg zonae pellucidae possessing receptor activity for sperm. Cell. 1980a;20 (3):873–882. doi: 10.1016/0092-8674(80)90334-7. [DOI] [PubMed] [Google Scholar]
- Bleil JD, Wassarman PM. Structure and function of the zona pellucida: identification and characterization of the proteins of the mouse oocyte’s zona pellucida. Dev Biol. 1980b;76 (1):185–202. doi: 10.1016/0012-1606(80)90371-1. [DOI] [PubMed] [Google Scholar]
- Bleil JD, Wassarman PM. Galactose at the Nonreducing Terminus of O-Linked Oligosaccharides of Mouse Egg Zona Pellucida Glycoprotein Zp3 Is Essential for the Glycoproteins Sperm Receptor Activity. P Natl Acad Sci USA. 1988;85 (18):6778–6782. doi: 10.1073/pnas.85.18.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boja ES, Hoodbhoy T, Fales HM, Dean J. Structural characterization of native mouse zona pellucida proteins using mass spectrometry. J Biol Chem. 2003;278 (36):34189–34202. doi: 10.1074/jbc.M304026200. [DOI] [PubMed] [Google Scholar]
- Bouniol C, Nguyen E, Debey P. Endogenous transcription occurs at the 1-cell stage in the mouse embryo. Exp Cell Res. 1995;218 (1):57–62. doi: 10.1006/excr.1995.1130. [DOI] [PubMed] [Google Scholar]
- Braude P, Bolton V, Moore S. Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature. 1988;332 (6163):459–461. doi: 10.1038/332459a0. [DOI] [PubMed] [Google Scholar]
- Bronson RA, McLaren A. Transfer to the mouse oviduct of eggs with and without the zona pellucida. J Reprod Fertil. 1970;22 (1):129–137. doi: 10.1530/jrf.0.0220129. [DOI] [PubMed] [Google Scholar]
- Bultman SJ, Gebuhr TC, Pan H, Svoboda P, Schultz RM, Magnuson T. Maternal BRG1 regulates zygotic genome activation in the mouse. Genes Dev. 2006;20 (13):1744–1754. doi: 10.1101/gad.1435106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkart AD, Xiong B, Baibakov B, Jimenez-Movilla M, Dean J. Ovastacin, a cortical granule protease, cleaves ZP2 in the zona pellucida to prevent polyspermy. J Cell Biol. 2012;197 (1):37–44. doi: 10.1083/jcb.201112094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KH, Viveiros MM, Ren YS, Wang P, DeMayo FJ, Frail DE, Eppig JJ, Matzuk MM. Roles of NPM2 in chromatin and nucleolar organization in oocytes and embryos. Science. 2003;300 (5619):633–636. doi: 10.1126/science.1081813. [DOI] [PubMed] [Google Scholar]
- Bushati N, Stark A, Brennecke J, Cohen SM. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr Biol. 2008;18 (7):501–506. doi: 10.1016/j.cub.2008.02.081. [DOI] [PubMed] [Google Scholar]
- Butler MG. Genomic imprinting disorders in humans: a mini-review. J Assist Reprod Genet. 2009;26 (9–10):477–486. doi: 10.1007/s10815-009-9353-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caussinus E, Kanca O, Affolter M. Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat Struct Mol Biol. 2012;19 (1):117–121. doi: 10.1038/nsmb.2180. [DOI] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10 (5):295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- Chamberlin ME, Dean J. Genomic organization of a sex specific gene: the primary sperm receptor of the mouse zona pellucida. Dev Biol. 1989;131 (1):207–214. doi: 10.1016/s0012-1606(89)80052-1. [DOI] [PubMed] [Google Scholar]
- Chen J, Litscher ES, Wassarman PM. Inactivation of the mouse sperm receptor, mZP3, by site-directed mutagenesis of individual serine residues located at the combining site for sperm. P Natl Acad Sci USA. 1998a;95 (11):6193–6197. doi: 10.1073/pnas.95.11.6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RZ, Pettersson U, Beard C, Jackson-Grusby L, Jaenisch R. DNA hypomethylation leads to elevated mutation rates. Nature. 1998b;395 (6697):89–93. doi: 10.1038/25779. [DOI] [PubMed] [Google Scholar]
- Cho T, Sakai S, Nagata M, Aoki F. Involvement of chromatin structure in the regulation of mouse zygotic gene activation. Animal Science Journal. 2002;73 (2):113–122. [Google Scholar]
- Christians E, Campion E, Thompson EM, Renard JP. Expression of the Hsp-70.1 Gene, a Landmark of Early Zygotic Activity in the Mouse Embryo, Is Restricted to the First Burst of Transcription. Development. 1995;121 (1):113–122. doi: 10.1242/dev.121.1.113. [DOI] [PubMed] [Google Scholar]
- Christians E, Davis AA, Thomas SD, Benjamin IJ. Maternal effect of Hsf1 on reproductive success. Nature. 2000;407 (6805):693–694. doi: 10.1038/35037669. [DOI] [PubMed] [Google Scholar]
- Cirio MC, Ratnam S, Ding F, Reinhart B, Navara C, Chaillet JR. Preimplantation expression of the somatic form of Dnmt1 suggests a role in the inheritance of genomic imprints. BMC Dev Biol. 2008;8:9. doi: 10.1186/1471-213X-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corry GN, Tanasijevic B, Barry ER, Krueger W, Rasmussen TP. Epigenetic regulatory mechanisms during preimplantation development. Birth Defects Res C Embryo Today. 2009;87 (4):297–313. doi: 10.1002/bdrc.20165. [DOI] [PubMed] [Google Scholar]
- Cox GF, Burger J, Lip V, Mau UA, Sperling K, Wu BL, Horsthemke B. Intracytoplasmic sperm injection may increase the risk of imprinting defects. Am J Hum Genet. 2002;71 (1):162–164. doi: 10.1086/341096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby IM, Gandolfi F, Moor RM. Control of Protein-Synthesis during Early Cleavage of Sheep Embryos. Journal of Reproduction and Fertility. 1988;82 (2):769–775. doi: 10.1530/jrf.0.0820769. [DOI] [PubMed] [Google Scholar]
- Davis DL. Culture and storage of pig embryos. J Reprod Fertil Suppl. 1985;33:115–124. [PubMed] [Google Scholar]
- Davis W, DeSousa PA, Schultz RM. Transient expression of translation initiation factor eIF-4C during the 2-cell stage of the preimplantation mouse embryo: Identification by mRNA differential display and the role of DNA replication in zygotic gene activation. Dev Biol. 1996;174 (2):190–201. doi: 10.1006/dbio.1996.0065. [DOI] [PubMed] [Google Scholar]
- De Leon V, Johnson A, Bachvarova R. Half-lives and relative amounts of stored and polysomal ribosomes and poly(A) + RNA in mouse oocytes. Dev Biol. 1983;98 (2):400–408. doi: 10.1016/0012-1606(83)90369-x. [DOI] [PubMed] [Google Scholar]
- Dean W, Santos F, Reik W. Epigenetic reprogramming in early mammalian development and following somatic nuclear transfer. Semin Cell Dev Biol. 2003;14 (1):93–100. doi: 10.1016/s1084-9521(02)00141-6. [DOI] [PubMed] [Google Scholar]
- Dean W, Santos F, Stojkovic M, Zakhartchenko V, Walter J, Wolf E, Reik W. Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos. Proc Natl Acad Sci U S A. 2001;98 (24):13734–13738. doi: 10.1073/pnas.241522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBaun MR, Niemitz EL, Feinberg AP. Association of in vitro fertilization with Beckwith-Wiedemann syndrome and epigenetic alterations of LIT1 and H19. Am J Hum Genet. 2003;72 (1):156–160. doi: 10.1086/346031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechiara TM, Robertson EJ, Efstratiadis A. Parental Imprinting of the Mouse Insulin-Like Growth Factor-Ii Gene. Cell. 1991;64 (4):849–859. doi: 10.1016/0092-8674(91)90513-x. [DOI] [PubMed] [Google Scholar]
- Epifano O, Liang LF, Dean J. Mouse Zp1 encodes a zona pellucida protein homologous to egg envelope proteins in mammals and fish. J Biol Chem. 1995;270 (45):27254–27258. doi: 10.1074/jbc.270.45.27254. [DOI] [PubMed] [Google Scholar]
- Familiari G, Heyn R, Relucenti M, Sathananthan H. Structural changes of the zona pellucida during fertilization and embryo development. Front Biosci. 2008;13:6730–6751. doi: 10.2741/3185. [DOI] [PubMed] [Google Scholar]
- Familiari G, Relucenti M, Heyn R, Micara G, Correr S. Three-dimensional structure of the zona pellucida at ovulation. Microsc Res Tech. 2006;69 (6):415–426. doi: 10.1002/jemt.20301. [DOI] [PubMed] [Google Scholar]
- Feil R. Epigenetic asymmetry in the zygote and mammalian development. International Journal of Developmental Biology. 2009;53 (2–3):191–201. doi: 10.1387/ijdb.082654rf. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick GV, Soloway PD, Higgins MJ. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat Genet. 2002;32 (3):426–431. doi: 10.1038/ng988. [DOI] [PubMed] [Google Scholar]
- Flach G, Johnson MH, Braude PR, Taylor RA, Bolton VN. The transition from maternal to embryonic control in the 2-cell mouse embryo. EMBO J. 1982;1 (6):681–686. doi: 10.1002/j.1460-2075.1982.tb01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florman HM, Wassarman PM. O-linked oligosaccharides of mouse egg ZP3 account for its sperm receptor activity. Cell. 1985;41 (1):313–324. doi: 10.1016/0092-8674(85)90084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foygel K, Choi B, Jun S, Leong DE, Lee A, Wong CC, Zuo E, Eckart M, Pera RAR, Wong WH, Yao MWM. A Novel and Critical Role for Oct4 as a Regulator of the Maternal-Embryonic Transition. PLoS One. 2008;3(12) doi: 10.1371/journal.pone.0004109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei RE, Schultz GA, Church RB. Qualitative and Quantitative Changes in Protein-Synthesis Occur at the 8-16-Cell Stage of Embryogenesis in the Cow. Journal of Reproduction and Fertility. 1989;86 (2):637–641. doi: 10.1530/jrf.0.0860637. [DOI] [PubMed] [Google Scholar]
- Fulka H, Mrazek M, Tepla O, Fulka J., Jr DNA methylation pattern in human zygotes and developing embryos. Reproduction. 2004;128 (6):703–708. doi: 10.1530/rep.1.00217. [DOI] [PubMed] [Google Scholar]
- Gahlay G, Gauthier L, Baibakov B, Epifano O, Dean J. Gamete recognition in mice depends on the cleavage status of an egg’s zona pellucida protein. Science. 2010;329 (5988):216–219. doi: 10.1126/science.1188178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gicquel C, Gaston V, Mandelbaum J, Siffroi JP, Flahault A, Le Bouc Y. In vitro fertilization may increase the risk of Beckwith-Wiedemann syndrome related to the abnormal imprinting of the KCNQ1OT gene. Am J Hum Genet. 2003;72 (5):1338–1341. doi: 10.1086/374824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312 (5770):75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Goddard MJ, Pratt HP. Control of events during early cleavage of the mouse embryo: an analysis of the ‘2-cell block’. J Embryol Exp Morphol. 1983;73:111–133. [PubMed] [Google Scholar]
- Golbus MS, Calarco PG, Epstein CJ. The effects of inhibitors of RNA synthesis (alpha-amanitin and actinomycin D) on preimplantation mouse embryogenesis. J Exp Zool. 1973;186 (2):207–216. doi: 10.1002/jez.1401860211. [DOI] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Gowher H, Liebert K, Hermann A, Xu G, Jeltsch A. Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J Biol Chem. 2005;280 (14):13341–13348. doi: 10.1074/jbc.M413412200. [DOI] [PubMed] [Google Scholar]
- Greenhouse S, Castle PE, Dean J. Antibodies to human ZP3 induce reversible contraception in transgenic mice with ‘humanized’ zonae pellucidae. Human Reproduction. 1999;14 (3):593–600. doi: 10.1093/humrep/14.3.593. [DOI] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, Iqbal K, Shi YG, Deng Z, Szabo PE, Pfeifer GP, Li J, Xu GL. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477 (7366):606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- Hamatani T, Carter MG, Sharov AA, Ko MS. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell. 2004;6 (1):117–131. doi: 10.1016/s1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Surani MA. Resetting the epigenome beyond pluripotency in the germline. Cell Stem Cell. 2009;4 (6):493–498. doi: 10.1016/j.stem.2009.05.007. [DOI] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang QY, Ding JP, Jia YY, Chen ZC, Li L, Sun Y, Li XX, Dai Q, Song CX, Zhang KL, He C, Xu GL. Tet-Mediated Formation of 5-Carboxylcytosine and Its Excision by TDG in Mammalian DNA. Science. 2011;333 (6047):1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Guy J, Ramsahoye B, Wilson VA, Bird A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev. 2001;15 (6):710–723. doi: 10.1101/gad.194101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henery CC, Miranda M, Wiekowski M, Wilmut I, Depamphilis ML. Repression of Gene-Expression at the Beginning of Mouse Development. Dev Biol. 1995;169 (2):448–460. doi: 10.1006/dbio.1995.1160. [DOI] [PubMed] [Google Scholar]
- Hermann A, Goyal R, Jeltsch A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J Biol Chem. 2004;279 (46):48350–48359. doi: 10.1074/jbc.M403427200. [DOI] [PubMed] [Google Scholar]
- Herr JC, Chertihin O, Digilio L, Jha KN, Vemuganti S, Flickinger CJ. Distribution of RNA binding protein MOEP19 in the oocyte cortex and early embryo indicates pre-patterning related to blastomere polarity and trophectoderm specification. Dev Biol. 2008;314 (2):300–316. doi: 10.1016/j.ydbio.2007.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirasawa R, Chiba H, Kaneda M, Tajima S, Li E, Jaenisch R, Sasaki H. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev. 2008;22 (12):1607–1616. doi: 10.1101/gad.1667008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187 (4173):226–232. [PubMed] [Google Scholar]
- Hoodbhoy T, Joshi S, Boja ES, Williams SA, Stanley P, Dean J. Human sperm do not bind to rat zonae pellucidae despite the presence of four homologous glycoproteins. J Biol Chem. 2005;280 (13):12721–12731. doi: 10.1074/jbc.M413569200. [DOI] [PubMed] [Google Scholar]
- Howell CY, Bestor TH, Ding F, Latham KE, Mertineit C, Trasler JM, Chaillet JR. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell. 2001;104 (6):829–838. doi: 10.1016/s0092-8674(01)00280-x. [DOI] [PubMed] [Google Scholar]
- Hu JJ, Wan FC, Zhu XQ, Yuan Y, Ding MX, Ga SR. Mouse ZAR1-Like (XM_359149) Colocalizes With mRNA Processing Components and Its Dominant-Negative Mutant Caused Two-Cell-Stage Embryonic Arrest. Dev Dynam. 2010;239 (2):407–424. doi: 10.1002/dvdy.22170. [DOI] [PubMed] [Google Scholar]
- Humpherys D, Eggan K, Akutsu H, Hochedlinger K, Rideout WM, 3rd, Biniszkiewicz D, Yanagimachi R, Jaenisch R. Epigenetic instability in ES cells and cloned mice. Science. 2001;293 (5527):95–97. doi: 10.1126/science.1061402. [DOI] [PubMed] [Google Scholar]
- Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334 (6053):194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue M, Wolf DP. Fertilization-associated changes in the murine zona pellucida: a time sequence study. Biol Reprod. 1975;13 (5):546–551. doi: 10.1095/biolreprod13.5.546. [DOI] [PubMed] [Google Scholar]
- Iqbal K, Jin SG, Pfeifer GP, Szabo PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. P Natl Acad Sci USA. 2011;108 (9):3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466 (7310):1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet Proteins Can Convert 5-Methylcytosine to 5-Formylcytosine and 5-Carboxylcytosine. Science. 2011;333 (6047):1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S, Davies H, Sims LP, Levy SE, Dean J. Ovarian gene expression in the absence of FIGLA, an oocyte-specific transcription factor. BMC Dev Biol. 2007;7:67. doi: 10.1186/1471-213X-7-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamakaka RT, Biggins S. Histone variants: deviants? Genes Dev. 2005;19 (3):295–310. doi: 10.1101/gad.1272805. [DOI] [PubMed] [Google Scholar]
- Kanka J. Gene expression and chromatin structure in the pre-implantation embryo. Theriogenology. 2003;59 (1):3–19. doi: 10.1016/s0093-691x(02)01267-0. [DOI] [PubMed] [Google Scholar]
- Kinloch RA, Roller RJ, Fimiani CM, Wassarman DA, Wassarman PM. Primary structure of the mouse sperm receptor polypeptide determined by genomic cloning. Proc Natl Acad Sci U S A. 1988;85 (17):6409–6413. doi: 10.1073/pnas.85.17.6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko MS, Kitchen JR, Wang X, Threat TA, Hasegawa A, Sun T, Grahovac MJ, Kargul GJ, Lim MK, Cui Y, Sano Y, Tanaka T, Liang Y, Mason S, Paonessa PD, Sauls AD, DePalma GE, Sharara R, Rowe LB, Eppig J, Morrell C, Doi H. Large-scale cDNA analysis reveals phased gene expression patterns during preimplantation mouse development. Development. 2000;127 (8):1737–1749. doi: 10.1242/dev.127.8.1737. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128 (4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Kawamura Y, Uchijima Y, Amamo T, Kobayashi H, Asano T, Kurihara H. Maintenance of genomic methylation patterns during preimplantation development require’s the somatic form of DNA methyltransferase 1. Dev Biol. 2008;313 (1):335–346. doi: 10.1016/j.ydbio.2007.10.033. [DOI] [PubMed] [Google Scholar]
- Latham KE, Garrels JI, Chang C, Solter D. Quantitative-Analysis of Protein-Synthesis in Mouse Embryos .1. Extensive Reprogramming at the One-Cell and 2-Cell Stages. Development. 1991;112 (4):921–932. doi: 10.1242/dev.112.4.921. [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11 (3):204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees-Murdock DJ, Walsh CP. DNA methylation reprogramming in the germ line. Epigenetics. 2008;3 (1):5–13. doi: 10.4161/epi.3.1.5553. [DOI] [PubMed] [Google Scholar]
- Lefievre L, Conner SJ, Salpekar A, Olufowobi O, Ashton P, Pavlovic B, Lenton W, Afnan M, Brewis IA, Monk M, Hughes DC, Barratt CL. Four zona pellucida glycoproteins are expressed in the human. Hum Reprod. 2004;19 (7):1580–1586. doi: 10.1093/humrep/deh301. [DOI] [PubMed] [Google Scholar]
- Lepikhov K, Walter J. Differential dynamics of histone H3 methylation at positions K4 and K9 in the mouse zygote. BMC Dev Biol. 2004;4:12. doi: 10.1186/1471-213X-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366 (6453):362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- Li L, Baibakov B, Dean J. A subcortical maternal complex essential for preimplantation mouse embryogenesis. Dev Cell. 2008a;15 (3):416–425. doi: 10.1016/j.devcel.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zheng P, Dean J. Maternal control of early mouse development. Development. 2010;137 (6):859–870. doi: 10.1242/dev.039487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XJ, Ito M, Zhou F, Youngson N, Zuo XP, Leder P, Ferguson-Smith AC. A Maternal-Zygotic Effect Gene, Zfp57, Maintains Both Maternal and Paternal Imprints. Developmental Cell. 2008b;15 (4):547–557. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, O’Neill C. Persistence of cytosine methylation of DNA following fertilisation in the mouse. PLoS One. 2012;7 (1):e30687. doi: 10.1371/journal.pone.0030687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YF, Sasaki H. Genomic imprinting in mammals: its life cycle, molecular mechanisms and reprogramming. Cell Res. 2011;21 (3):466–473. doi: 10.1038/cr.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Soyal SM, Dean J. FIGalpha, a germ cell specific transcription factor involved in the coordinate expression of the zona pellucida genes. Development. 1997;124 (24):4939–4947. doi: 10.1242/dev.124.24.4939. [DOI] [PubMed] [Google Scholar]
- Liang LF, Chamow SM, Dean J. Oocyte-specific expression of mouse Zp-2: developmental regulation of the zona pellucida genes. Mol Cell Biol. 1990;10 (4):1507–1515. doi: 10.1128/mcb.10.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DH, Maher ER. Human imprinting syndromes. Epigenomics. 2009;1 (2):347–369. doi: 10.2217/epi.09.24. [DOI] [PubMed] [Google Scholar]
- Liu C, Litscher ES, Mortillo S, Sakai Y, Kinloch RA, Stewart CL, Wassarman PM. Targeted disruption of the mZP3 gene results in production of eggs lacking a zona pellucida and infertility in female mice. Proc Natl Acad Sci U S A. 1996;93 (11):5431–5436. doi: 10.1073/pnas.93.11.5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CY, Litscher ES, Wassarman PM. Transgenic Mice with Reduced Numbers of Functional Sperm Receptors on Their Eggs Reproduce Normally. Mol Biol Cell. 1995;6 (5):577–585. doi: 10.1091/mbc.6.5.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DY, Baker HW, Pearse MJ, d’Apice AJ. Normal sperm-zona pellucida interaction and fertilization in vitro in alpha-1,3-galactosyltransferase gene knockout mice. Mol Hum Reprod. 1997;3 (11):1015–1016. doi: 10.1093/molehr/3.11.1015. [DOI] [PubMed] [Google Scholar]
- Lu Q, Shur BD. Sperm from beta 1,4-galactosyltransferase-null mice are refractory to ZP3-induced acrosome reactions and penetrate the zona pellucida poorly. Development. 1997;124 (20):4121–4131. doi: 10.1242/dev.124.20.4121. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 angstrom resolution. Nature. 1997;389 (6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Lund E, Liu MZ, Hartley RS, Sheets MD, Dahlberg JE. Deadenylation of maternal mRNAs mediated by miR-427 in Xenopus laevis embryos. Rna. 2009;15 (12):2351–2363. doi: 10.1261/rna.1882009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen K, Gilchrist MJ, Grabarek JB, Das P, Miska E, Zernicka-Goetz M. Maternal Argonaute 2 Is Essential for Early Mouse Development at the Maternal-Zygotic Transition. Mol Biol Cell. 2008;19 (10):4383–4392. doi: 10.1091/mbc.E08-02-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Flemr M, Stein P, Berninger P, Malik R, Zavolan M, Svoboda P, Schultz RM. MicroRNA Activity Is Suppressed in Mouse Oocytes. Curr Biol. 2010;20 (3):265–270. doi: 10.1016/j.cub.2009.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Zeng FY, Schultz RM, Tseng H. Basonuclin: a novel mammalian maternal-effect gene. Development. 2006;133 (10):2053–2062. doi: 10.1242/dev.02371. [DOI] [PubMed] [Google Scholar]
- Mackay DJG, Callaway JLA, Marks SM, White HE, Acerini CL, Boonen SE, Dayanikli P, Firth HV, Goodship JA, Haemers AP, Hahnemann JMD, Kordonouri O, Masoud AF, Oestergaard E, Storr J, Ellard S, Hattersley AT, Robinson DO, Temple IK. Hypomethylation of multiple imprinted loci in individuals with transient neonatal diabetes is associated with mutations in ZFP57. Nature Genetics. 2008;40 (8):949–951. doi: 10.1038/ng.187. [DOI] [PubMed] [Google Scholar]
- Macleod D, Clark VH, Bird A. Absence of genome-wide changes in DNA methylation during development of the zebrafish. Nat Genet. 1999;23 (2):139–140. doi: 10.1038/13767. [DOI] [PubMed] [Google Scholar]
- Matsuoka T, Sato M, Tokoro M, Shin SW, Uenoyama A, Ito K, Hitomi S, Amano T, Anzai M, Kato H, Mitani T, Saeki K, Hosoi Y, Iritani A, Matsumoto K. Identification of ZAG1, a novel protein expressed in mouse prei rn plantation, and its putative roles in zygotic genome activation. J Reprod Develop. 2008;54 (3):192–197. doi: 10.1262/jrd.20008. [DOI] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403 (6769):501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- Mcgrath J, Solter D. Completion of Mouse Embryogenesis Requires Both the Maternal and Paternal Genomes. Cell. 1984;37 (1):179–183. doi: 10.1016/0092-8674(84)90313-1. [DOI] [PubMed] [Google Scholar]
- Messerschmidt DM, de Vries W, Ito M, Solter D, Ferguson-Smith A, Knowles BB. Trim28 Is Required for Epigenetic Stability During Mouse Oocyte to Embryo Transition. Science. 2012;335 (6075):1499–1502. doi: 10.1126/science.1216154. [DOI] [PubMed] [Google Scholar]
- Miller DJ, Gong XH, Decker G, Shur BD. Egg Cortical Granule N-Acetylglucosaminidase Is Required for the Mouse Zona Block to Polyspermy. Journal of Cell Biology. 1993;123 (6):1431–1440. doi: 10.1083/jcb.123.6.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DJ, Macek MB, Shur BD. Complementarity between Sperm Surface Beta-1,4-Galactosyl-Transferase and Egg-Coat Zp3 Mediates Sperm Egg Binding. Nature. 1992;357 (6379):589–593. doi: 10.1038/357589a0. [DOI] [PubMed] [Google Scholar]
- Minami N, Suzuki T, Tsukamoto S. Zygotic gene activation and maternal factors in mammals. J Reprod Dev. 2007;53 (4):707–715. doi: 10.1262/jrd.19029. [DOI] [PubMed] [Google Scholar]
- Misirlioglu M, Page GP, Sagirkaya H, Kaya A, Parrish JJ, First NL, Memili E. Dynamics of global transcriptome in bovine matured oocytes and preimplantation embryos. Proc Natl Acad Sci U S A. 2006;103 (50):18905–18910. doi: 10.1073/pnas.0608247103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modlinski JA. The role of the zona pellucida in the development of mouse eggs in vivo. J Embryol Exp Morphol. 1970;23 (3):539–547. [PubMed] [Google Scholar]
- Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, Sekimoto T, Ikawa M, Yoneda Y, Okabe M, Tanaka S, Shiota K, Nakano T. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol. 2007;9 (1):64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Liu YJ, Nakashima H, Umehara H, Inoue K, Matoba S, Tachibana M, Ogura A, Shinkai Y, Nakano T. PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos. Nature. 2012;486 (7403):415–419. doi: 10.1038/nature11093. [DOI] [PubMed] [Google Scholar]
- Nolte F, Hofmann WK. Myelodysplastic syndromes: molecular pathogenesis and genomic changes. Ann Hematol. 2008;87 (10):777–795. doi: 10.1007/s00277-008-0502-z. [DOI] [PubMed] [Google Scholar]
- Nothias JY, Miranda M, DePamphilis ML. Uncoupling of transcription and translation during zygotic gene activation in the mouse. EMBO J. 1996;15 (20):5715–5725. [PMC free article] [PubMed] [Google Scholar]
- Ohsugi M, Zheng P, Baibakov B, Li L, Dean J. Maternally derived FILIA-MATER complex localizes asymmetrically in cleavage-stage mouse embryos. Development. 2008;135 (2):259–269. doi: 10.1242/dev.011445. [DOI] [PubMed] [Google Scholar]
- Okada Y, Yamagata K, Hong K, Wakayama T, Zhang Y. A role for the elongator complex in zygotic paternal genome demethylation. Nature. 2010;463 (7280):554–558. doi: 10.1038/nature08732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99 (3):247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19 (3):219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10 (8):475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- Pan H, Schultz RM. SOX2 Modulates Reprogramming of Gene Expression in Two-Cell Mouse Embryos. Biol Reprod. 2011;85 (2):409–416. doi: 10.1095/biolreprod.111.090886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B, Saitou M, Barton SC, Thresher R, Dixon JPC, Zahn D, Colledge WH, Carlton MBL, Nakano T, Surani MA. Stella is a maternal effect gene required for normal early development in mice. Curr Biol. 2003;13 (23):2110–2117. doi: 10.1016/j.cub.2003.11.026. [DOI] [PubMed] [Google Scholar]
- Paynton BV, Rempel R, Bachvarova R. Changes in state of adenylation and time course of degradation of maternal mRNAs during oocyte maturation and early embryonic development in the mouse. Dev Biol. 1988;129 (2):304–314. doi: 10.1016/0012-1606(88)90377-6. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Laniel MA. Histones and histone modifications. Curr Biol. 2004;14 (14):R546–R551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Philipps DL, Wigglesworth K, Hartford SA, Sun F, Pattabiraman S, Schimenti K, Handel M, Eppig JJ, Schimenti JC. The dual bromodomain and WD repeat-containing mouse protein BRWD1 is required for normal spermiogenesis and the oocyte-embryo transition. Dev Biol. 2008;317 (1):72–82. doi: 10.1016/j.ydbio.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre A, Gautier M, Callebaut I, Bontoux M, Jeanpierre E, Pontarotti P, Monget P. Atypical structure and phylogenomic evolution of the new eutherian oocyte-and embryo-expressed KHDC1/DPPA5/ECAT1/OOEP gene family. Genomics. 2007;90 (5):583–594. doi: 10.1016/j.ygeno.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Posfai E, Kunzmann R, Brochard V, Salvaing J, Cabuy E, Roloff TC, Liu ZC, Tardat M, van Lohuizen M, Vidal M, Beaujean N, Peters AHFM. Polycomb function during oogenesis is required for mouse embryonic development. Gene Dev. 2012;26 (9):920–932. doi: 10.1101/gad.188094.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potireddy S, Vassena R, Patel BG, Latham KE. Analysis of polysomal mRNA populations of mouse oocytes and zygotes: dynamic changes in maternal mRNA utilization and function. Dev Biol. 2006;298 (1):155–166. doi: 10.1016/j.ydbio.2006.06.024. [DOI] [PubMed] [Google Scholar]
- Qiu JJ, Zhang WW, Wu ZL, Wang YH, Qian M, Li YP. Delay of ZGA initiation occurred in 2-cell blocked mouse embryos. Cell Res. 2003;13 (3):179–185. doi: 10.1038/sj.cr.7290162. [DOI] [PubMed] [Google Scholar]
- Quesada V, Sanchez LM, Alvarez J, Lopez-Otin C. Identification and characterization of human and mouse ovastacin: a novel metalloproteinase similar to hatching enzymes from arthropods, birds, amphibians, and fish. J Biol Chem. 2004;279 (25):26627–26634. doi: 10.1074/jbc.M401588200. [DOI] [PubMed] [Google Scholar]
- Ram PT, Schultz RM. Reporter Gene-Expression in G2 of the 1-Cell Mouse Embryo. Dev Biol. 1993;156 (2):552–556. doi: 10.1006/dbio.1993.1101. [DOI] [PubMed] [Google Scholar]
- Ramos SB, Stumpo DJ, Kennington EA, Phillips RS, Bock CB, Ribeiro-Neto F, Blackshear PJ. The CCCH tandem zinc-finger protein Zfp36l2 is crucial for female fertility and early embryonic development. Development. 2004;131 (19):4883–4893. doi: 10.1242/dev.01336. [DOI] [PubMed] [Google Scholar]
- Rankin T, Familari M, Lee E, Ginsberg A, Dwyer N, Blanchette-Mackie J, Drago J, Westphal H, Dean J. Mice homozygous for an insertional mutation in the Zp3 gene lack a zona pellucida and are infertile. Development. 1996;122 (9):2903–2910. doi: 10.1242/dev.122.9.2903. [DOI] [PubMed] [Google Scholar]
- Rankin T, Talbot P, Lee E, Dean J. Abnormal zonae pellucidae in mice lacking ZP1 result in early embryonic loss. Development. 1999;126 (17):3847–3855. doi: 10.1242/dev.126.17.3847. [DOI] [PubMed] [Google Scholar]
- Rankin TL, Coleman JS, Epifano O, Hoodbhoy T, Turner SG, Castle PE, Lee E, Gore-Langton R, Dean J. Fertility and taxon-specific sperm binding persist after replacement of mouse sperm receptors with human homologs. Dev Cell. 2003;5 (1):33–43. doi: 10.1016/s1534-5807(03)00195-3. [DOI] [PubMed] [Google Scholar]
- Rankin TL, O’Brien M, Lee E, Wigglesworth K, Eppig J, Dean J. Defective zonae pellucidae in Zp2-null mice disrupt folliculogenesis, fertility and development. Development. 2001;128 (7):1119–1126. doi: 10.1242/dev.128.7.1119. [DOI] [PubMed] [Google Scholar]
- Rankin TL, Tong ZB, Castle PE, Lee E, Gore-Langton R, Nelson LM, Dean J. Human ZP3 restores fertility in Zp3 null mice without affecting order-specific sperm binding. Development. 1998;125 (13):2415–2424. doi: 10.1242/dev.125.13.2415. [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293 (5532):1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Reik W, Santos F, Mitsuya K, Morgan H, Dean W. Epigenetic asymmetry in the mammalian zygote and early embryo: relationship to lineage commitment? Philos Trans R Soc Lond B Biol Sci. 2003;358(1436):1403–1409. doi: 10.1098/rstb.2003.1326. discussion 1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W, Walter J. Evolution of imprinting mechanisms: the battle of the sexes begins in the zygote. Nat Genet. 2001;27 (3):255–256. doi: 10.1038/85804. [DOI] [PubMed] [Google Scholar]
- Robertson KD, Wolffe AP. DNA methylation in health and disease. Nat Rev Genet. 2000;1 (1):11–19. doi: 10.1038/35049533. [DOI] [PubMed] [Google Scholar]
- Roest HP, Baarends WM, de Wit J, van Klaveren JW, Wassenaar E, Hoogerbrugge JW, van Cappellen WA, Hoeijmakers JH, Grootegoed JA. The ubiquitin-conjugating DNA repair enzyme HR6A is a maternal factor essential for early embryonic development in mice. Mol Cell Biol. 2004;24 (12):5485–5495. doi: 10.1128/MCB.24.12.5485-5495.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougier N, Bourc’his D, Gomes DM, Niveleau A, Plachot M, Paldi A, Viegas-Pequignot E. Chromosome methylation patterns during mammalian preimplantation development. Genes Dev. 1998;12 (14):2108–2113. doi: 10.1101/gad.12.14.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M, Kagiwada S, Kurimoto K. Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development. 2012;139 (1):15–31. doi: 10.1242/dev.050849. [DOI] [PubMed] [Google Scholar]
- Santos F, Dean W. Epigenetic reprogramming during early development in mammals. Reproduction. 2004;127 (6):643–651. doi: 10.1530/rep.1.00221. [DOI] [PubMed] [Google Scholar]
- Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241 (1):172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- Santos F, Peters AH, Otte AP, Reik W, Dean W. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev Biol. 2005;280 (1):225–236. doi: 10.1016/j.ydbio.2005.01.025. [DOI] [PubMed] [Google Scholar]
- Sarma K, Reinberg D. Histone variants meet their match. Nat Rev Mol Cell Bio. 2005;6 (2):139–149. doi: 10.1038/nrm1567. [DOI] [PubMed] [Google Scholar]
- Sato K. Polyspermy-preventing mechanisms in mouse eggs fertilized in vitro. J Exp Zool. 1979;210 (2):353–359. doi: 10.1002/jez.1402100219. [DOI] [PubMed] [Google Scholar]
- Saunders CM, Larman MG, Parrington J, Cox LJ, Royse J, Blayney LM, Swann K, Lai FA. PLC zeta: a sperm-specific trigger of Ca(2+) oscillations in eggs and embryo development. Development. 2002;129 (15):3533–3544. doi: 10.1242/dev.129.15.3533. [DOI] [PubMed] [Google Scholar]
- Schatten G, Simerly C, Schatten H. Microtubule configurations during fertilization, mitosis, and early development in the mouse and the requirement for egg microtubule-mediated motility during mammalian fertilization. Proc Natl Acad Sci U S A. 1985;82 (12):4152–4156. doi: 10.1073/pnas.82.12.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz RM. Regulation of zygotic gene activation in the mouse. Bioessays. 1993;15 (8):531–538. doi: 10.1002/bies.950150806. [DOI] [PubMed] [Google Scholar]
- Schultz RM. The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Hum Reprod Update. 2002;8 (4):323–331. doi: 10.1093/humupd/8.4.323. [DOI] [PubMed] [Google Scholar]
- Shitara H, Hayashi JI, Takahama S, Kaneda H, Yonekawa H. Maternal inheritance of mouse mtDNA in interspecific hybrids: segregation of the leaked paternal mtDNA followed by the prevention of subsequent paternal leakage. Genetics. 1998;148 (2):851–857. doi: 10.1093/genetics/148.2.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirard MA, Dufort I, Vallee M, Massicotte L, Gravel C, Reghenas H, Watson AJ, King WA, Robert C. Potential and limitations of bovine-specific arrays for the analysis of mRNA levels in early development: preliminary analysis using a bovine embryonic array. Reprod Fertil Dev. 2005;17 (1–2):47–57. doi: 10.1071/rd04113. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, Gu HC, Gnirke A, Regev A, Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484 (7394):339–U374. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyal SM, Amleh A, Dean J. FIGalpha, a germ cell-specific transcription factor required for ovarian follicle formation. Development. 2000;127 (21):4645–4654. doi: 10.1242/dev.127.21.4645. [DOI] [PubMed] [Google Scholar]
- Stancheva I, El-Maarri O, Walter J, Niveleau A, Meehan RR. DNA methylation at promoter regions regulates the timing of gene activation in Xenopus laevis embryos. Dev Biol. 2002;243 (1):155–165. doi: 10.1006/dbio.2001.0560. [DOI] [PubMed] [Google Scholar]
- Stewart-Savage J, Bavister BD. A cell surface block to polyspermy occurs in golden hamster eggs. Dev Biol. 1988;128 (1):150–157. doi: 10.1016/0012-1606(88)90277-1. [DOI] [PubMed] [Google Scholar]
- Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. Journal of Biological Chemistry. 2004;279 (26):27816–27823. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- Suh N, Baehner L, Moltzahn F, Melton C, Shenoy A, Chen J, Blelloch R. MicroRNA Function Is Globally Suppressed in Mouse Oocytes and Early Embryos. Curr Biol. 2010;20 (3):271–277. doi: 10.1016/j.cub.2009.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers MC, Bhatnagar PR, Lawitts JA, Biggers JD. Fertilization in vitro of mouse ova from inbred and outbred strains: complete preimplantation embryo development in glucose-supplemented KSOM. Biol Reprod. 1995;53 (2):431–437. doi: 10.1095/biolreprod53.2.431. [DOI] [PubMed] [Google Scholar]
- Surani MA, Barton SC, Norris ML. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature. 1984;308 (5959):548–550. doi: 10.1038/308548a0. [DOI] [PubMed] [Google Scholar]
- Sutovsky P, Schatten G. Paternal contributions to the mammalian zygote: fertilization after sperm-egg fusion. Int Rev Cytol. 2000;195:1–65. doi: 10.1016/s0074-7696(08)62703-5. [DOI] [PubMed] [Google Scholar]
- Svoboda P, Flemr M. The role of miRNAs and endogenous siRNAs in maternal-to-zygotic reprogramming and the establishment of pluripotency. Embo Rep. 2010;11 (8):590–597. doi: 10.1038/embor.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136 (18):3033–3042. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324 (5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, Hannon GJ. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453 (7194):534–U538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Kaneda M, O’Carroll D, Hajkova P, Barton SC, Sun YA, Lee C, Tarakhovsky A, Lao K, Surani MA. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007;21 (6):644–648. doi: 10.1101/gad.418707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thall AD, Maly P, Lowe JB. Oocyte Gal-Alpha-1,3gal Epitopes Implicated in Sperm Adhesion to the Zona-Pellucida Glycoprotein Zp3 Are Not Required for Fertilization in the Mouse. Journal of Biological Chemistry. 1995;270 (37):21437–21440. doi: 10.1074/jbc.270.37.21437. [DOI] [PubMed] [Google Scholar]
- Thompson EM, Legouy E, Renard JP. Mouse embryos do not wait for the MBT: Chromatin and RNA polymerase remodeling in genome activation at the onset of development. Dev Genet. 1998;22 (1):31–42. doi: 10.1002/(SICI)1520-6408(1998)22:1<31::AID-DVG4>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Thorvaldsen JL, Duran KL, Bartolomei MS. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998;12 (23):3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong ZB, Gold L, Pfeifer KE, Dorward H, Lee E, Bondy CA, Dean J, Nelson LM. Mater, a maternal effect gene required for early embryonic development in mice. Nat Genet. 2000;26 (3):267–268. doi: 10.1038/81547. [DOI] [PubMed] [Google Scholar]
- Tong ZB, Nelson LM. A mouse gene encoding an oocyte antigen associated with autoimmune premature ovarian failure. Endocrinology. 1999;140 (8):3720–3726. doi: 10.1210/endo.140.8.6911. [DOI] [PubMed] [Google Scholar]
- Torres-Padilla ME, Bannister AJ, Hurd PJ, Kouzarides T, Zernicka-Goetz M. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. International Journal of Developmental Biology. 2006;50 (5):455–461. doi: 10.1387/ijdb.052073mt. [DOI] [PubMed] [Google Scholar]
- Torres-Padilla ME, Zernicka-Goetz M. Role of TIF1 alpha as a modulator of embryonic transcription in the mouse zygote. Journal of Cell Biology. 2006;174 (3):329–338. doi: 10.1083/jcb.200603146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai TC, Lin W, Yang SH, Cheng WTK, Cheng EH, Lee MS, Chong KY, Chen CM. Granzyme G is expressed in the two-cell stage mouse embryo and is required for the maternal-zygotic transition. Bmc Developmental Biology. 2010:10. doi: 10.1186/1471-213X-10-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto S, Kuma A, Mizushima N. The role of autophagy during the oocyte-to-embryo transition. Autophagy. 2008;4 (8):1076–1078. doi: 10.4161/auto.7065. [DOI] [PubMed] [Google Scholar]
- Turner BM. Cellular memory and the histone code. Cell. 2002;111 (3):285–291. doi: 10.1016/s0092-8674(02)01080-2. [DOI] [PubMed] [Google Scholar]
- Vallee M, Aiba K, Piao Y, Palin MF, Ko MSH, Sirard MA. Comparative analysis of oocyte transcript profiles reveals a high degree of conservation among species. Reproduction. 2008;135 (4):439–448. doi: 10.1530/REP-07-0342. [DOI] [PubMed] [Google Scholar]
- van der Heijden GW, Dieker JW, Derijck AAHA, Muller S, Berden JHM, Braat DDM, van der Vlag J, de Boer P. Asymmetry in Histone H3 variants and lysine methylation between paternal and maternal chromatin of the early mouse zygote. Mech Develop. 2005;122 (9):1008–1022. doi: 10.1016/j.mod.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Veron N, Peters AH. Epigenetics: Tet proteins in the limelight. Nature. 2011;473 (7347):293–294. doi: 10.1038/473293a. [DOI] [PubMed] [Google Scholar]
- Verona RI, Mann MR, Bartolomei MS. Genomic imprinting: intricacies of epigenetic regulation in clusters. Annu Rev Cell Dev Biol. 2003;19:237–259. doi: 10.1146/annurev.cellbio.19.111401.092717. [DOI] [PubMed] [Google Scholar]
- Walter J, Paulsen M. Imprinting and disease. Semin Cell Dev Biol. 2003;14 (1):101–110. doi: 10.1016/s1084-9521(02)00142-8. [DOI] [PubMed] [Google Scholar]
- Wan LB, Pan H, Hannenhalli S, Cheng Y, Ma J, Fedoriw A, Lobanenkov V, Latham KE, Schultz RM, Bartolomei MS. Maternal depletion of CTCF reveals multiple functions during oocyte and preimplantation embryo development. Development. 2008;135 (16):2729–2738. doi: 10.1242/dev.024539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Xu M, Zhu K, Li L, Liu X. The N-terminus of FILIA forms an atypical KH domain with a unique extension involved in interaction with RNA. PLoS One. 2012;7 (1):e30209. doi: 10.1371/journal.pone.0030209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QT, Piotrowska K, Ciemerych MA, Milenkovic L, Scott MP, Davis RW, Zernicka-Goetz M. A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Developmental Cell. 2004;6 (1):133–144. doi: 10.1016/s1534-5807(03)00404-0. [DOI] [PubMed] [Google Scholar]
- Wang QX, Latham KE. Requirement for protein synthesis during embryonic genome activation in mice. Mol Reprod Dev. 1997;47 (3):265–270. doi: 10.1002/(SICI)1098-2795(199707)47:3<265::AID-MRD5>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Wang SC, Frey PA. S-adenosylmethionine as an oxidant: the radical SAM superfamily. Trends Biochem Sci. 2007;32 (3):101–110. doi: 10.1016/j.tibs.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Wang SF, Kou ZH, Jing ZY, Zhang Y, Guo XZ, Dong MQ, Wilmut I, Gao SR. Proteome of mouse oocytes at different developmental stages. P Natl Acad Sci USA. 2010;107 (41):17639–17644. doi: 10.1073/pnas.1013185107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner CM, Versteegh LR. In vivo and in vitro effect of alpha-amanitin on preimplantation mouse embryo RNA polymerase. Nature. 1974;248 (450):678–680. doi: 10.1038/248678a0. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, Surani MA, Sakaki Y, Sasaki H. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453 (7194):539–U539. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Whitworth KM, Agca C, Kim JG, Patel RV, Springer GK, Bivens NJ, Forrester LJ, Mathialagan N, Green JA, Prather RS. Transcriptional profiling of pig embryogenesis by using a 15-K member unigene set specific for pig reproductive tissues and embryos. Biol Reprod. 2005;72 (6):1437–1451. doi: 10.1095/biolreprod.104.037952. [DOI] [PubMed] [Google Scholar]
- Wiekowski M, Miranda M, Depamphilis ML. Regulation of Gene-Expression in Preimplantation Mouse Embryos - Effects of the Zygotic Clock and the 1st Mitosis on Promoter and Enhancer Activities. Dev Biol. 1991;147 (2):403–414. doi: 10.1016/0012-1606(91)90298-h. [DOI] [PubMed] [Google Scholar]
- Wiekowski M, Miranda M, Depamphilis ML. Requirements for Promoter Activity in Mouse Oocytes and Embryos Distinguish Paternal Pronuclei from Maternal and Zygotic Nuclei. Dev Biol. 1993;159 (1):366–378. doi: 10.1006/dbio.1993.1248. [DOI] [PubMed] [Google Scholar]
- Wilkins JF, Haig D. Parental modifiers, antisense transcripts and loss of imprinting. Proc Biol Sci. 2002;269 (1502):1841–1846. doi: 10.1098/rspb.2002.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins JF, Haig D. What good is genomic imprinting: The function of parent-specific gene expression. Nature Reviews Genetics. 2003;4 (5):359–368. doi: 10.1038/nrg1062. [DOI] [PubMed] [Google Scholar]
- Williams SA, Xia L, Cummings RD, McEver RP, Stanley P. Fertilization in mouse does not require terminal galactose or N-acetylglucosamine on the zona pellucida glycans. J Cell Sci. 2007;120 (Pt 8):1341–1349. doi: 10.1242/jcs.004291. [DOI] [PubMed] [Google Scholar]
- Wolffe AP. Implications of DNA replication for eukaryotic gene expression. J Cell Sci. 1991;99 (Pt 2):201–206. doi: 10.1242/jcs.99.2.201. [DOI] [PubMed] [Google Scholar]
- Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, Walter J. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun. 2011:2. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11 (9):607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XM, Viveiros MM, Eppig JJ, Bai YC, Fitzpatrick SL, Matzuk MM. Zygote arrest 1 (Zar1) is a novel maternal-effect gene critical for the oocyte-to-embryo transition. Nature Genetics. 2003;33 (2):187–191. doi: 10.1038/ng1079. [DOI] [PubMed] [Google Scholar]
- Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S, Min J, Nicholson T, Chen T, Xu G, Shi Y, Zhang K, Shi YG. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell. 2011;42 (4):451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauger B, Boggs NA, Dean J. Human ZP4 is not sufficient for taxon-specific sperm recognition of the zona pellucida in transgenic mice. Reproduction. 2011;141 (3):313–319. doi: 10.1530/REP-10-0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13 (8):335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- Yurttas P, Morency E, Coonrod SA. Use of proteomics to identify highly abundant maternal factors that drive the egg-to-embryo transition. Reproduction. 2010;139 (5):809–823. doi: 10.1530/REP-09-0538. [DOI] [PubMed] [Google Scholar]
- Yurttas P, Vitale AM, Fitzhenry RJ, Cohen-Gould L, Wu W, Gossen JA, Coonrod SA. Role for PADI6 and the cytoplasmic lattices in ribosomal storage in oocytes and translational control in the early mouse embryo. Development. 2008;135 (15):2627–2636. doi: 10.1242/dev.016329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng F, Baldwin DA, Schultz RM. Transcript profiling during preimplantation mouse development. Dev Biol. 2004;272 (2):483–496. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Zeng F, Schultz RM. Gene expression in mouse oocytes and preimplantation embryos: use of suppression subtractive hybridization to identify oocyte- and embryo-specific genes. Biol Reprod. 2003;68 (1):31–39. doi: 10.1095/biolreprod.102.007674. [DOI] [PubMed] [Google Scholar]
- Zeng FY, Schultz RM. RNA transcript profiling during zygotic gene activation in the preimplantation mouse embryo. Dev Biol. 2005;283 (1):40–57. doi: 10.1016/j.ydbio.2005.03.038. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zheng N, Zhou P. Exploring the functional complexity of cellular proteins by protein knockout. Proc Natl Acad Sci U S A. 2003;100 (24):14127–14132. doi: 10.1073/pnas.2233012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Ni X, Guo Y, Guo X, Wang Y, Zhou Z, Huo R, Sha J. Proteomic-based identification of maternal proteins in mature mouse oocytes. BMC Genomics. 2009;10:348. doi: 10.1186/1471-2164-10-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P, Dean J. Role of Filia, a maternal effect gene, in maintaining euploidy during cleavage-stage mouse embryogenesis. Proc Natl Acad Sci U S A. 2009;106 (18):7473–7478. doi: 10.1073/pnas.0900519106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann JW, Schultz RM. Analysis of gene expression in the preimplantation mouse embryo: use of mRNA differential display. Proc Natl Acad Sci U S A. 1994;91 (12):5456–5460. doi: 10.1073/pnas.91.12.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]