Abstract
Organelles inside cells move to position themselves at the right place at the right time. A mechanism for generating active force exists for each of such directed organelle movements. In our recent study on cytoplasmic streaming in the Caenorhabditis elegans one-cell embryo, we demonstrated that an anterior-directed force generated by myosin could drive not only anterior-directed cortical flow but also posterior-directed cytoplasmic flow. This coupling of flows in opposing directions is mediated by the hydrodynamic properties of the cytoplasm. This work provided a good example of an active force generation mechanism that drives organelle movements in two opposite directions inside the cell, just as a funicular moves up and down a slope. Interestingly, the funicular-like coupling of intracellular movements is also seen in our recent studies on centrosome positioning in the C. elegans embryo and on interkinetic nuclear movement during mouse neurogenesis. Thus, funicular-like coupling may be a general strategy used repeatedly in cells. The use of the funicular-like coupling seems advantageous because it is efficient, as one active force generation mechanism can drive movements in two directions, and also because the two movements can be coordinated to have similar speeds.
Keywords: actomyosin, centrosome centration, cytoplasmic streaming, dynein, hydrodynamics, interkinetic nuclear migration, microtubule
Organelle Movement and Force Generation
The cell is highly organized in its architecture, with precise positioning of organelles and other structures at the right place and the right time. Appropriate positioning of the organelles is critical for cell function. For example, the positioning of the centrosome and nucleus at the center of the cell is important for symmetric cell division. To achieve such positioning, active force generators, typically the cytoskeleton and molecular motors, transport organelles to appropriate locations in the cell. A straightforward mechanism of organelle transport is direct cargo transport along the cytoskeleton by molecular motors.1 Alternatively, motors may be anchored at structures inside the cell (e.g., the cell cortex), and pull the cytoskeleton and organelles bound to the cytoskeleton.2 Elongation of an elastic cytoskeleton between an organelle and the cell cortex may push the organelle away from the cortex.3 Contraction of the cortical cytoskeletal network may cause cortical flow, which globally moves organelles inside the cell.4 Previous studies on organelle transport mainly focused on cases wherein a single active force generating mechanism is responsible for the movement of an organelle toward one direction. In these cases, there is a one-to-one relationship between force generation and directional transport.
Funicular-Like Coupling for Organelle Movements: One Active Force Generation Drives Two Directional Movements
The above-described one-to-one relationship may not always be the only mechanism for organelle transport in a cell. One active force generation mechanism in a cell may drive organelle movements in two directions at the same time, like a funicular. A funicular is a transport system located on slopes in which a pair of vehicles is attached by a cable (Fig. 1A). A force that pulls one vehicle up results in the downward movement of the other vehicle. The movements of the two vehicles are coupled and a single active force generation mechanism drives the movement of the two vehicles in opposite directions. Three independent studies using Caenorhabditis elegans embryos and embryonic mouse brain cells suggest the existence of such funicular-like coupled movements of organelles inside cells. Hence, we propose the existence of a “cellular funicular,” in which one active force generation mechanism drives organelle movements in two opposite directions; this may apply to many intracellular movements, and therefore, it potentially comprises a common mechanism in cell construction.
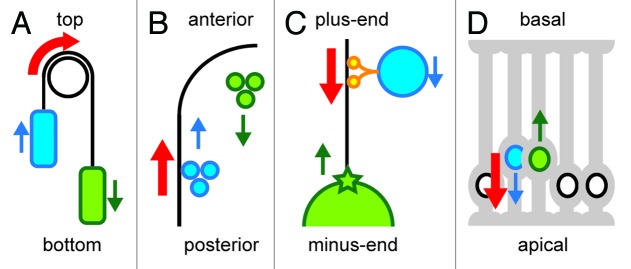
Figure 1. A funicular and cellular funiculars. The directions of active force generation are indicated in red. Blue and green indicate each of the two components moving in opposite directions (the directions are shown in arrows). The blue components are more directly linked to the active force generators, while the green components are moved more passively. (A) A funicular. Blue and green vehicles are linked by a cable. By applying a force to rotate the pulley at the top, the blue vehicle moves up, and the green vehicle moves down. (B) Cytoplasmic streaming in C. elegans. The molecular motor myosin generates forces at the cell cortex (black line) to move the proteins and granules toward the anterior (cortical flow, blue). The hydrodynamic property of the cytoplasm transmits the force to the inner components to move them posteriorly (cytoplasmic flow, green). (C) Centrosome centration in C. elegans. Organelles (blue) move toward the minus-end of a microtubule (black line) driven by the molecular motor dynein (orange). These movements generate a reactionary force that pulls the microtubule and associated centrosome (green star) and nucleus (green circle) toward the plus-end. (D) Interkinetic nuclear movement during mouse neurogenesis. Inside the developing brain, neural progenitor cells (gray cells with processes at their apical and basal surfaces) are densely packed. The nucleus in a G2-phase cell (blue) moves toward the apical surface using the force generated by the microtubule motor dynein. As the apical side gets crowded with the nuclei, the nucleus in the G1-phase cell (green) is pushed out toward the basal side.
Studying Cellular Funiculars by Combining Quantitative Measurements and Numerical Modeling
Although the idea of a funicular-like coupled movement is intuitive and reasonable, it may be difficult to demonstrate the existence and function of such coupled movements through conventional molecular genetics alone. Manipulation of genes responsible for active force generation should affect both movements equally, thus making it difficult to precisely dissect the relationship among the three components of this system (one force and two movements). Thus, in addition to utilizing molecular genetics, we use methods to quantify the movements of cellular structures from time-lapse microscopy images using image processing, and computer simulation to validate different possible models. Our approach, namely, a combination of quantitative measurement and simulation analyses, is an effective strategy for characterizing the role of cellular funiculars.
Cytoplasmic Streaming in C. elegans: Anterior-Directed Myosin Drives Both Anterior-Directed Cortical Flow and Posterior-Directed Cytoplasmic Flow
Cytoplasmic flow and cortical flow
The timing and direction of cytoplasmic streaming in the C. elegans zygote are developmentally programmed. Upon fertilization, the location of the sperm entry point specifies the posterior of the embryo. Cortical and cytoplasmic flow initiates around the S-phase of female and male pronuclei, in an actomyosin-dependent manner (Fig. 2A).5-7 Cortical flow is the anterior-directed flow near the cell surface, while cytoplasmic flow is the posterior-directed flow in the central region of the cell. Gradually decreasing the flow velocity, actomyosin-dependent streaming continues until the two pronuclei meet at a slightly posterior region of the central part of the cell in microtubule-dependent manner.
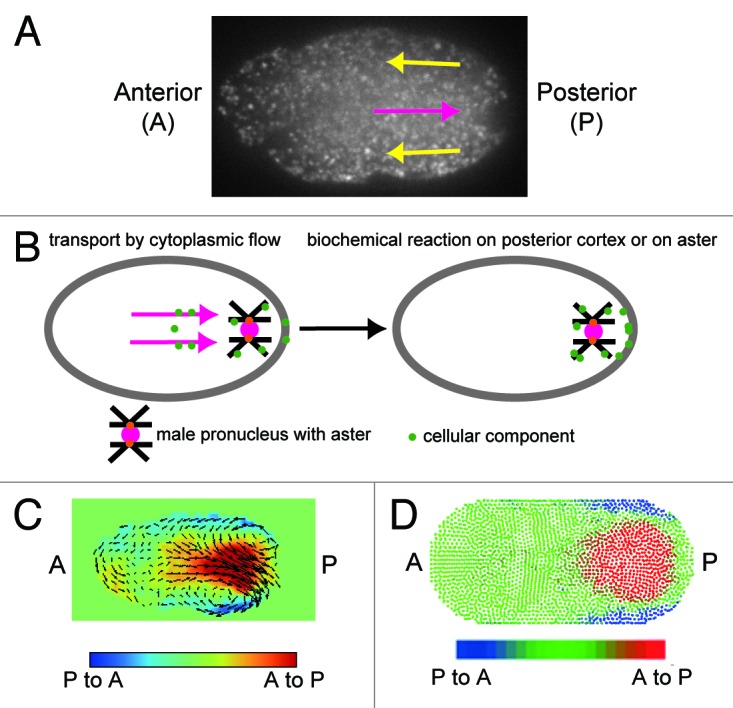
Figure 2. Cytoplasmic streaming in C. elegans. (A) Cytoplasmic streaming visualized using GFP-labeled yolk granules. Anterior-directed cortical flow and posterior-directed cytoplasmic flow are indicated by yellow and magenta arrows, respectively. (B) Our model for the possible role of cytoplasmic flow. The flow may enhance the probability of cytoplasmic material attaching to the aster or the posterior cortex. (C) Velocity distribution of cytoplasmic streaming was measured using the PIV method and visualized with vectors and colors.13 In the red region, the flow is posterior-directed, while in the blue region, the flow is anterior-directed. (D) Velocity distribution of streaming reproduced with computer simulation utilizing the MPS method.13 In the red region, the flow is posterior-directed, while in the blue region, the flow is anterior-directed.
Both actin and myosin are concentrated near the cell cortex6 and appear to form the network of the foci. At the initiation of cortical and cytoplasmic flow, the network of actomyosin covers the entire cell cortex. As cortical and cytoplasmic flows occur, the network migrates in an anterior direction. The speed of the cortical actomyosin is almost the same as that of yolk granules near the cell surface, which indicates that actomyosin and cortical materials are tightly associated during migration.6 As the consequence of migration, the network of the actomyosin foci covers the anterior half of the cell. Molecules that characterize cortical cell polarity, such as PAR-3, move in an anterior direction along with, and at the same speed as, actomyosin.6 PAR-3 is also known to form anterior cortical domain. A PAR-2 domain is formed posteriorly where PAR-3 is eliminated. Eventually, the PAR-2 domain occupies the posterior half of the cell.8 Thus, cortical flow seems to facilitate the formation of cell surface polarity.
Compared with cortical flow, the biological role of cytoplasmic flow has been obscure. Posterior-directed cytoplasmic flow was suspected to concentrate P granule, a germline granule in C. elegans, to the posterior region of the embryo, where future germ cells arise. However, cytoplasmic streaming does not change the number of the P granules distributed between the anterior and posterior sides of the embryo.9 We still believe that cytoplasmic flow may contribute to the efficient formation or robustness of the cellular polarity. For example, it may enhance the probability of P granules or other cellular components encountering the posterior cell cortex or the microtubule asters, where the biochemical reactions that impart cell polarity occur10 (Fig. 2B). Cytoplasmic streaming is proposed to play several roles in species other than C. elegans. For example, cytoplasmic streaming contributes to the appropriate positioning of the spindle in mouse oocytes.11 Similarly, cytoplasmic streaming is proposed to mix the intracellular materials in Chara.12
Mechanism of cytoplasmic flow analyzed by combining PIV and MPS methods
Recently, we analyzed the physics of cytoplasmic flow generation.13 We examined a hypothesis in which the hydrodynamic property of the cytoplasm couples posterior-directed cytoplasmic flow with anterior-directed cortical flow. In this study, the flow velocity distribution of cytoplasmic streaming in vivo was quantitatively compared with that in simulation. Particle image velocimetry (PIV) extracted the flow velocity distribution in vivo. The simulation used a moving particle semi-implicit (MPS) method to elicit a theoretical velocity distribution based on fluid mechanics.
PIV is an image processing method to quantify the velocity distribution from images. The input for PIV is the images in which the motions of probes are captured. The output of PIV is the velocity distribution in these images. We used GFP-labeled yolk granules as the probe in our study. The probe in PIV does not necessarily have to be a particle; for example, a network of myosin was used as a probe in a previous study.9 In our PIV analysis, probe flow velocity is estimated at every 8th pixel (or about 1 μm in our system).14,15 At each location (x, y), the velocity components in the x and y directions (vx, vy) is acquired by calculating the maximum cross-correlation of brightness of two square windows in two consecutive images. In addition, sub-pixel-level movement is calculated using a gradient method. In our PIV analysis in the C. elegans embryo, intense cortical and cytoplasmic flows were detected in the posterior region, with velocity peaks at about 10 μm from the posterior pole (Fig. 2C).13
MPS is relatively new simulation method that describes fluid as collection of particles.16 The calculation of Navier-Stokes equation, which describes simple viscous fluid dynamics, is modeled as an interaction between particles. The Navier-Stokes equation is Dv/Dt = μ/ρ∇2v − 1/ρ∇p, where v is velocity (a three-dimensional vector in each location), p is pressure, μ is viscosity, and ρ is mass density. The first term on the right-hand side of the equation represents the effect of viscosity. The second term on the right-hand side of the equation represents the effect of pressure. The merit of the particle method lies in its ease of use for analyzing flow in a deforming structure. In our analysis, we also simulated the flow that accompanies the movement of pronuclei, which can be considered a kind of deformation of the cytoplasm.13 Particle methods, including the MPS method, are also easy to use for the analysis of interactions between fluid flow and an elastic body. Noguchi et al. recapitulated the deformation of the red blood cells while they flow.17 We believe that particle methods present promising tools for hydrodynamic simulation inside cells and organisms. In our simulation of cytoplasmic streaming in the C. elegans embryo, we only applied forces at the cortex to move cortical particles at speeds observed in real embryos. The application of this cortical flow reproduces in simulation a cytoplasmic flow with a velocity distribution similar to that generated in vivo (Fig. 2D).13 The quantitative agreement between in vivo cytoplasmic flow and the flow generated by hydrodynamic simulation supports the hypothesis that cytoplasmic flow is indeed the hydrodynamic flow generated by cortical flow (Fig. 1B).
Physical properties of the cytoplasm
Our study also has implications on how we can approximate the physical property of complex cytoplasm. Viscosity and elasticity are two major physical signatures of the cytoplasm. Interestingly, the contribution of each of these signatures differs from one biological system to another. In C. elegans one-cell embryos, viscosity appears to be the dominant force when compared with the elasticity, as normal diffusion of the micro-beads was observed in vivo,18 and because our simulation, assuming a simple fluid, agrees quite well with in vivo cytoplasmic streaming.13 Thus, an approximation assuming that the cytoplasm is a viscous fluid works well in C. elegans embryo. Similarly, in Xenopus laevis egg extract, viscosity seems to dominate over elasticity.19 By contrast, there is a report that elasticity is strong in cultured mouse fibroblast cells.20
Centrosome Centering in C. elegans: The Minus-End-Directed Microtubule Motor, Dynein, Drives Both Minus-End-Directed Organelle Movements and a Plus-End-Directed Centrosome Pull
Coupling between two intracellular migrations in opposite directions with a single force generating mechanism was also implicated in our study on centrosome positioning.21,22 The centrosome is a major microtubule-organizing center in animal cells. Centrosomes position themselves at the geometric center of cells.23,24 Several lines of observations imply that microtubules elongating from centrosomes are pulled throughout the cytoplasm in a microtubule-length dependent manner.24-28 However, until recently, the nature of the hypothetical structure that anchors microtubule motors to pull the centrosome throughout the cytoplasm have been unclear; thus, the model lacked a critical mechanical basis. We proposed that the process of organelle transport along microtubules provides the microtubule length-dependent force to pull centrosomes.21 This proposal was based on our experimental observation of a strong link between organelle transport and centrosome centration in C. elegans embryos. A dynein subunit (dyrb-1) required selectively for centrosome centration was also required for organelle transport. In addition, reducing the organelle transport significantly and selectively reduces the speed of centration. Based on these observations, we proposed that an organelle transported along a microtubule by the dynein motor protein toward the minus-end of the microtubule produces a concomitant “reaction” force that pulls the microtubule and the centrosome toward the plus-end. This mechanism is another example of the cellular funicular, in which a force generated by dynein drives both minus-end-directed organelle movements and plus-end-directed centrosome pull (Fig. 1C).
As organelle transport is a general process constantly occurring in all eukaryotic cells, our model bears universal implications. Each event during organelle transport along the cytoskeleton should generate a reaction force that pulls the cytoskeleton and its associated structures. As the cytoskeleton network is spread across most of the cell, a local force that is generated for organelle transport may be transmitted throughout the cell. Although, the effect produced by single organelle transport should not be so large, multiple pairs of cellular funiculars may be further coupled inside the cell in order to accomplish collective movements of the organelles or the entire cell. However, if the cell requires movement of only a single component without affecting the others, then a funicular-like movement can be disadvantageous. In this case, the cell may use certain mechanisms to prevent such funicular-like movements.
Interkinetic Nuclear Migration During Mouse Neurogenesis: Dynein Drives Both Apical and Basal Nuclear Migrations
The interkinetic migration of the nucleus during mouse neurogenesis is an example of a cellular funicular found outside C. elegans cells. In this case, two organelle movements in different cells are coupled. Interkinetic nuclear migration, also known as “elevator movement,” is a hallmark of neurogenesis in the vertebrate brain, in which the positions of nuclei oscillate along the apical-basal axis in polarized neural progenitor cells synchronized by cell cycle progression.29 Apical migration is driven by an apical force directed by dynein motors and microtubules. The microtubule-associated protein Tpx2 localizes to apical processes in the G2 phase, contributing to G2-phase-specific apical migration.30 By contrast, little is known about the mechanism of basal migration during G1-phase. Kosodo et al. hypothesized that the basal migration of the nuclei is a passive consequence of crowding due to the active migration of nuclei toward the apical region, because of the tight packing of the neural progenitors within the tissue. We constructed a computer simulation model based on an excluded volume effect to test this hypothesis. Our simulation recapitulated the behavior of interkinetic nuclear migration. The apical movements of the G2-phase nuclei cause the basal movement of the nuclei in neighboring cells. Because the M-phase nuclei/spindles are anchored onto the apical surface, the G1-phase nuclei are the primary nuclei that are excluded from the apical side and move toward the basal side. The S-phase nuclei are already located at the basal region because of the movements in the G1 phase, and do not show much further movement toward the basal side. Importantly, the simulation predicted different distributions of nuclei depending on the duration of the G1-phase, which was then experimentally confirmed. These analyses collectively support the hypothesis that the basal migration of nuclei is driven as a by-product of an apical-directed microtubule-based nuclear migration (Fig. 1D).30 This model explains how the robustness of epithelial architecture is maintained despite the massive neural progenitor divisions that occur during brain development.
Future Perspectives
The above-discussed research in C. elegans and mouse models suggests the existence of the cellular funiculars, i.e., the coupling of two movements in opposite directions using a single active force generating mechanism. The idea of funicular-like coupling provides a fresh perspective on cellular and developmental processes. We believe that the cellular funicular can be used in many cellular movements, and thus, it represents a common principle in constructing a cell- and tissue-architectures. Cellular funicular mechanisms can confer some distinct advantages on the cells. First, this mechanism is efficient; one active force generating mechanism drives movements in two opposing directions. Second, in a cellular funicular, the two movements generated by a single force are roughly equivalent and highly coordinated. Further characterization of the mechanism of funicular-like coupling between cellular movements should provide clues on the biological significance of such coupling.
Acknowledgments
The ideas presented in this manuscript were obtained during our collaboration with Drs Kyosuke Shinohara, Yoichi Kosodo, Fumio Matsuzaki and Kenji Kimura. We thank Drs. Yoichi Kosodo, Kenji Kimura, Ritsuko Arai and Takeshi Sugawara for their comments on the manuscript. Studies in the Kimura lab are supported by a grant from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and by the Transdisciplinary Research Integration Center of the Research Organization of Information and Systems, Japan.
Glossary
Abbreviations:
- PIV
particle image velocimetry
- MPS
moving particle semi-implicit
- GFP
green fluorescent protein
Footnotes
Previously published online: www.landesbioscience.com/journals/worm/article/19039
References
- 1.Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10:682–96. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- 2.Bloom K. Nuclear migration: cortical anchors for cytoplasmic dynein. Curr Biol. 2001;11:R326–9. doi: 10.1016/S0960-9822(01)00176-2. [DOI] [PubMed] [Google Scholar]
- 3.Tran PT, Marsh L, Doye V, Inoue S, Chang F. A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J Cell Biol. 2001;153:397–412. doi: 10.1083/jcb.153.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayer M, Depken M, Bois JS, Jülicher F, Grill SW. Anisotropies in cortical tension reveal the physical basis of polarizing cortical flows. Nature. 2010;467:617–21. doi: 10.1038/nature09376. [DOI] [PubMed] [Google Scholar]
- 5.Hird SN, White JG. Cortical and cytoplasmic flow polarity in early embryonic cells of Caenorhabditis elegans. J Cell Biol. 1993;121:1343–55. doi: 10.1083/jcb.121.6.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munro E, Nance J, Priess JR. Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev Cell. 2004;7:413–24. doi: 10.1016/j.devcel.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 7.Golden A.Cytoplasmic flow and the establishment of polarity in C. elegans 1-cell embryos. Curr Opin Genet Dev 2000; 10:414-20 [DOI] [PubMed] [Google Scholar]
- 8.Cheeks RJ, Canman JC, Gabriel WN, Meyer N, Strome S, Goldstein B. C. elegans PAR proteins function by mobilizing and stabilizing asymmetrically localized protein complexes. Curr Biol. 2004;14:851–62. doi: 10.1016/j.cub.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 9.Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–32. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- 10.Motegi F, Zonies S, Hao Y, Cuenca AA, Griffin E, Seydoux G. Microtubules induce self-organization of polarized PAR domains in Caenorhabditis elegans zygotes. Nat Cell Biol. 2011;13 doi: 10.1038/ncb2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yi K, Unruh JR, Deng M, Slaughter BD, Rubinstein B, Li R. Dynamic maintenance of asymmetric meiotic spindle position through Arp2/3-complex-driven cytoplasmic streaming in mouse oocytes. Nat Cell Biol. 2011;13:1252–8. doi: 10.1038/ncb2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldstein RE, Tuval I, van de Meent JW. Microfluidics of cytoplasmic streaming and its implications for intracellular transport. Proc Natl Acad Sci USA. 2008;105:3663–7. doi: 10.1073/pnas.0707223105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niwayama R, Shinohara K, Kimura A. Hydrodynamic property of the cytoplasm is sufficient to mediate cytoplasmic streaming in the Caenorhabiditis elegans embryo. Proc Natl Acad Sci USA. 2011;108:11900–5. doi: 10.1073/pnas.1101853108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sugii Y, Nishio S, Okuno T, Okamoto K. A highly accurate iterative PIV technique using a gradient method. Meas Sci Technol. 2000;11:1666–73. doi: 10.1088/0957-0233/11/12/303. [DOI] [Google Scholar]
- 15.Shinohara K, Sugii Y, Aota A, Hibara A, Tokeshi M, Kitamori T, et al. High-speed micro-PIV measurements of transient flow in microfluidic devices Meas. Sci Tech (Paris) 2004;15:1965–70. [Google Scholar]
- 16.Koshizuka S, Oka Y. Moving-Particle Semi-implicit Method for Fragmentation of Incompressible Fluid. Nucl Sci Eng. 1996;123:421–34. [Google Scholar]
- 17.Noguchi H, Gompper G. Shape transitions of fluid vesicles and red blood cells in capillary flows. Proc Natl Acad Sci USA. 2005;102:14159–64. doi: 10.1073/pnas.0504243102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daniels BR, Masi BC, Wirtz D. Probing single-cell micromechanics in vivo: the microrheology of C. elegans developing embryos. Biophys J. 2006;90:4712–9. doi: 10.1529/biophysj.105.080606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valentine MT, Perlman ZE, Mitchison TJ, Weitz DA. Mechanical properties of Xenopus egg cytoplasmic extracts. Biophys J. 2005;88:680–9. doi: 10.1529/biophysj.104.048025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kole TP, Tseng Y, Huang L, Katz JL, Wirtz D. Rho kinase regulates the intracellular micromechanical response of adherent cells to rho activation. Mol Biol Cell. 2004;15:3475–84. doi: 10.1091/mbc.E04-03-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimura K, Kimura A. Intracellular organelles mediate cytoplasmic pulling force for centrosome centration in the Caenorhabditis elegans early embryo. Proc Natl Acad Sci USA. 2011;108:137–42. doi: 10.1073/pnas.1013275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimura K, Kimura A. A novel mechanism of microtubule length-dependent force to pull centrosomes toward the cell center. BioArchitecture. 2011;1:74–9. doi: 10.4161/bioa.1.2.15549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson EB. The Cell in Development and Heredity. New York: Macmillan, 1925. [Google Scholar]
- 24.Minc N, Burgess D, Chang F. Influence of cell geometry on division-plane positioning. Cell. 2011;144:414–26. doi: 10.1016/j.cell.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamaguchi MS, Hiramoto Y. Analysis of the role of astral rays in pronuclear migration in sand dollar eggs by the colcemid-UV method. Develop Growth and Differ 1986; 28:143-56 [DOI] [PubMed] [Google Scholar]
- 26.Reinsch S, Gönczy P. Mechanisms of nuclear positioning. J Cell Sci. 1998;111:2283–95. doi: 10.1242/jcs.111.16.2283. [DOI] [PubMed] [Google Scholar]
- 27.Kimura A, Onami S. Computer simulations and image processing reveal length-dependent pulling force as the primary mechanism for C. elegans male pronuclear migration. Dev Cell. 2005;8:765–75. doi: 10.1016/j.devcel.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 28.Wühr M, Tan ES, Parker SK, Detrich HW, 3rd, Mitchison TJ. A model for cleavage plane determination in early amphibian and fish embryos. Curr Biol. 2010;20:2040–5. doi: 10.1016/j.cub.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taverna E, Huttner WB. Neural progenitor nuclei IN motion. Neuron. 2010;67:906–14. doi: 10.1016/j.neuron.2010.08.027. [DOI] [PubMed] [Google Scholar]
- 30.Kosodo Y, Suetsugu T, Suda M, Mimori-Kiyosue Y, Toida K, Baba SA, et al. Regulation of interkinetic nuclear migration by cell cycle-coupled active and passive mechanisms in the developing brain. EMBO J. 2011;30:1690–704. doi: 10.1038/emboj.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]


