Abstract
The signal transducer and activator of transcription (STAT) family of transcription factors have a spectrum of functions in mammary gland development. In some cases these roles parallel those of STATs in other organ systems, while in other instances the function of individual STATs in the mammary gland is specific to this tissue. In the immune system, STAT6 is associated with differentiation of T helper cells, while in the mammary gland, it has a fundamental role in the commitment of luminal epithelial cells to the alveolar lineage. STAT5A is required for the production of luminal progenitor cells from mammary stem cells and is essential for the differentiation of milk producing alveolar cells during pregnancy. By contrast, the initiation of regression following weaning heralds a dramatic and specific activation of STAT3, reflecting its pivotal role in the regulation of cell death and tissue remodeling during mammary involution. Although it has been demonstrated that STAT1 is regulated during a mammary developmental cycle, it is not yet determined whether it has a specific, non-redundant function. Thus, the mammary gland constitutes an unusual example of an adult organ in which different STATs are sequentially activated to orchestrate the processes of functional differentiation, cell death and tissue remodeling.
Keywords: mammary gland, STAT, apoptosis, differentiation, mouse, gene targeting, microarray
Introduction
The JAK-STAT field recently celebrated its 20th anniversary. In 1991, Andrew Wilks at the Ludwig Institute for Cancer Research in Melbourne1 described kinases that were named JAK1 and JAK2, for Janus kinase, after the Roman god of gates and doorways. Using complementation of mutagenized cell lines, George Stark and Ian Kerr at the ICRF in London identified other components of the interferon response pathway.2 In the same year, a factor highly expressed in the mammary glands of sheep and cows was described, although it was not until the following year that the true identity of this transcription factor was revealed. Originally named MPBF (milk protein binding factor)3 or MGF (mammary gland factor)4 this STAT was subsequently named STAT5 following the cloning of components of the ISGF3 complex in Darnell’s laboratory5 and coining of the term STAT.6
We now know that there are seven signal transducer and activator of transcription (STAT) family members in mammals, located in pairs on three different chromosomes, with the exception of STAT5A and STAT5B, which probably arose by a gene duplication event, which occupy the same locus as STAT3.7 Several members of the STAT family of transcription factors are essential for mammary gland development. This is perhaps surprising as STATs generally transduce signals from cytokines (Fig. 1). However, their roles in this tissue highlight the distinct function of each STAT as a signal transduction molecule. In addition to STAT5, other members of the STAT family are expressed during development of the adult mammary gland8 and several play important roles in mammopoeisis, lactogenesis and post-lactational regression.9 In this article, we will first outline the principal stages of mammary gland development and then discuss in detail the roles of the four STATs that are differentially regulated at specific developmental times in the mammary gland.
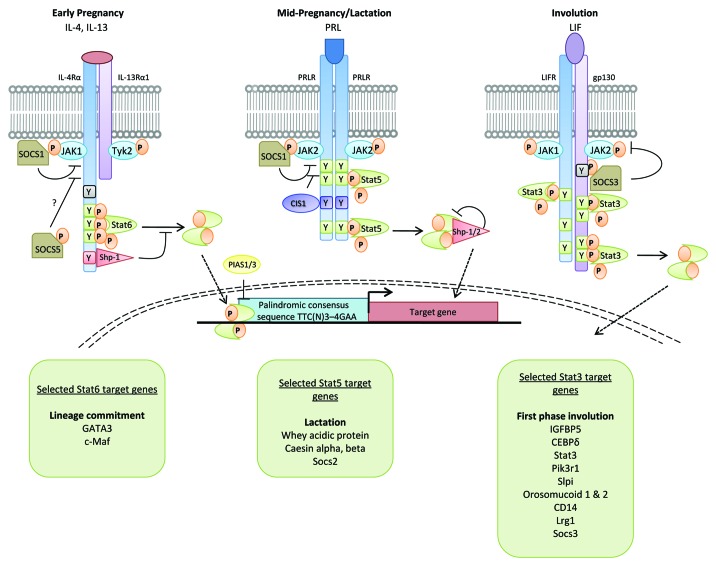
Figure 1. Cytokine signaling through the JAK-STAT pathway. Signaling components for STAT3, STAT5 and STAT6 that are utilized in mammary gland in response to IL-4/IL-13, prolactin and LIF. A subset of target genes for each of these STATs, at specific times in the developmental cycle, are indicated in the boxes. Socs proteins, which are direct transcriptional targets of STATs, are negative regulators of STATs, providing an exquisite level of regulation of STAT signaling. For a discussion of Socs proteins in mammary gland development, see Watson and Neoh.9 The phenotype of mice with deletions of selected STAT target genes is beyond the scope of this review, but is discussed elsewhere in the case of GATA318,19 and whey acidic protein.59 For a discussion of the involution phenotype of selected transgenic mice, see Radisky and Hartmann.60
Mammary Gland Development
The mammary gland is an unusual tissue since most of its growth and development occurs in the adult. It is composed of a network of ducts and branches that consist of a polarized bi-layered epithelium of luminal and basal cells that arise from a common multipotent stem cell.10,11
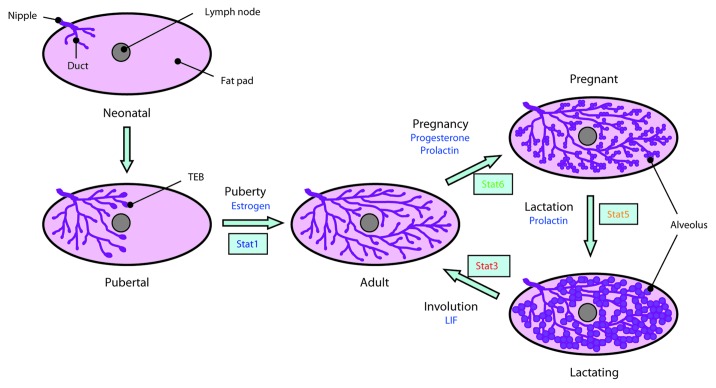
Mammary gland development occurs in three specific stages: in the embryo, during puberty and during gestation. Similar processes are involved in all mammals although the number of glands varies considerably. The first evidence of mammary gland development occurs in the mouse at embryonic day 10.5 when the two mammary (or milk) lines appear. These ridges of ectoderm are derived from embryonic skin and run in an anteroposterior direction from the fore- to the hind-limb buds. The cells in the milk lines are thought to migrate at E11.5 to form five pairs of placodes that are not only symmetrically positioned but appear at predictable locations.12 However, there are specific genetic mutations that can alter both the number and the position of the placodes. By E13.5, epithelial buds form and then sink into the underlying dermis and induce mammary mesenchyme to develop, followed by elongation of the buds to give mammary sprouts. Limited branching then forms a rudimentary mammary gland embedded within a subdermal fat pad at E18.5 after which development is arrested.
Initially postnatal development reflects overall body growth. Following puberty, terminal end buds (TEBs) develop at the ductal tips and in response to estrogen start to invade the fat pad, forming an elongated ductal network. When the limits of the fat pad have been reached at approximately 10 weeks of age, growth ceases and the TEBs regress. No further changes take place unless pregnancy ensues, whereupon, in response primarily to progesterone and prolactin signaling, tertiary branching is initiated and alveolar buds form. These lobuloalveolar structures produce milk during lactation and alveolar epithelial cells are a distinct lineage from the luminal epithelial cells present in the ducts of non-pregnant mammals.
Remarkably, with each pregnancy, the mammary gland undergoes a new cycle of alveologenesis. This is required as, following lactation, the milk-producing cells become superfluous and are removed by programmed cell death coupled with extensive tissue remodeling. This process of regression, called involution, is one of the most dramatic examples of physiologically regulated cell death that occurs in an adult tissue. One particularly interesting aspect of involution is that is occurs in two discrete phases.13 In the mouse, cell death can be halted by returning the pups to the mother and re-commencing lactation. This reversible phase is approximately 48 h in the mouse although it is considerably longer in larger mammals such as cows.14 Reversibility is lost after this time window and the architecture of the gland changes dramatically with loss of the alveolar epithelium and its replacement by re-differentiated adipocytes. These stages of mammary gland development are depicted schematically in Figure 2.
Figure 2. A schematic overview of postnatal mammary gland development in the mouse. A mammary gland developmental cycle from mature non-pregnant adult through pregnancy and involution is indicated on the right of the diagram. Abbreviations: TEB, terminal end bud; LIF, leukemia inhibitory factor. Based on a figure from Wiseman and Werb61 and reproduced from reference 9 with permission.
Expression and Activation of STATs during a Mammary Developmental Cycle
With the exception of STAT3, genetic deletion of STAT genes has little impact on mammary gland development during embryogenesis since mammary glands form normally in their absence. Thus we can conclude that STATs 1, 2, 4, 5 and 6 are not required for embryonic and early post-natal development. STAT3 gene deletion results in early embryonic lethality thereby precluding any investigation of its role in mammary gland. It is possible that STAT3 could be important in mammary stem cell self-renewal or maintenance, as it is for embryonic stem cells15 but this has not been directly investigated.
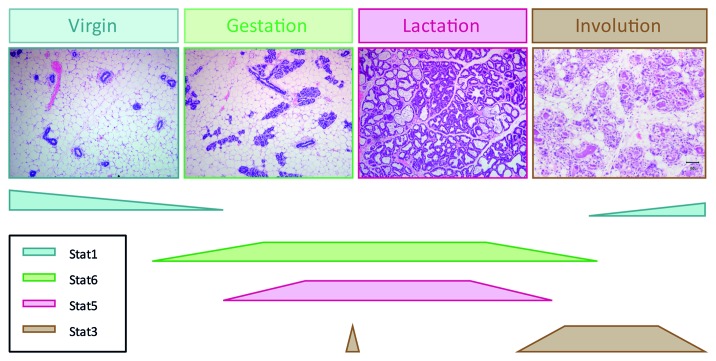
It is however during pregnancy that the function of individual STATs is revealed. Studies of expression levels showed that STATs 1, 3, 5 and 6 were expressed during gestation, lactation and involution while very low levels of STAT4 were detected and it was not determined whether this was restricted to mammary epithelium or was present in stromal cells. STAT2 has not been detected.8 Interestingly, tyrosine phosphorylation patterns are distinct albeit over-lapping. The highest levels of pSTAT1 are found in virgin gland and in late involution, during remodeling of the gland, while STAT3 activity is restricted to the day of birth and the first 10 d of involution. Despite the observation that STATs 1 and 3 can form heterodimers, these STATs have reciprocal patterns of activity during a mammary developmental cycle. Low levels of pSTAT6 are found in virgin glands where a small number of cells stain positively with pSTAT6 antibodies.16 However, during early gestation, most alveolar cells become pSTAT6+. The pattern of pSTAT5 is similar to pSTAT6 but appears later in gestation and reaches a peak during lactation. These developmentally regulated windows of STAT activity (Fig. 3) are highly suggestive of specific roles for individual STAT factors in the adult mammary developmental cycle. Genetic studies using knockout mice has revealed these roles which are discussed in the following sections.
Figure 3. STAT activity during a mammary gland developmental cycle. Although STATs are expressed throughout the cycle, their activation by tyrosine phosphorylation is strictly regulated and occurs at specific stages in the cycle. This pattern of activity reflects the requirement for each STAT as determined using genetic deletion of each individual factor.
The Function of Individual STATs in Mammary Gland
We will consider each STAT in turn, in the order in which they are activated, starting with the gestational time points of the developmental cycle.
STAT6 regulates commitment to the alveolar lineage
STAT6 has a clearly demonstrated role in immune cells and in particular as a regulator of the differentiation of naive T helper (Th) cells to the Th2 lineage whereby the cytokines IL-4 and IL-13 activate STAT6. This is associated with changes in chromatin structure and with transcriptional upregulation of the Th2 transcription factors Gata-3, c-Maf and NFAT1/NFATc2.17
A role for both IL-4/IL-13 and STAT6 in alveologenesis has been shown using mice deficient for either STAT6 or for both of these cytokines.16 An early defect in proliferation of alveolar cells during gestation and a 70% reduction in the numbers of alveoli was observed in knockout mice. As pregnancy progressed, the differences between knockout and control glands diminished and lactation was unaffected. In contrast, precocious alveologenesis was observed in SOCS5 knockout glands as predicted from the role of SOCS5 as a negative regulator of STAT6 signaling. Increased expression of IL-4Rα and Gata-3 occurs concomitantly with STAT6 activity at day 5 gestation while levels of pSTAT5 are decreased in the absence of STAT6. Interestingly, Gata-3 is important for maintenance of differentiated alveolar cells as conditional deletion of Gata-3 during gestation results in death of the alveolar cells and lactation failure.18,19 Thus, the IL-4/IL-13/STAT6 pathway is a regulator of mammary gland development adding a further complexity to this process which has long been associated with progesterone and prolactin (PRL) signaling.20
Studies in HC11 mammary epithelial cells in culture revealed that STAT6 can regulate expression of β-casein21 although, as discussed below, this is a task more usually performed by STAT5. KIM-2 mammary epithelial cells can be induced to undergo a program of differentiation in response to treatment with lactogenic hormones that mimics differentiation of alveolar cells during pregnancy.22 Treatment of KIM-2 cells with IL-4 or IL-13 induced tyrosine phosphorylation of STAT6 and Gata-3 expression. Interestingly, the type-1 cytokines IL-12α, IFNγ and TNF, which activate STAT1, are secreted by undifferentiated KIM-2 cells but there is a switch to secretion of the type-2 cytokines IL-4, IL-13 and IL-5 when these cells are induced to differentiate.16 This suggests that differentiation requires autocrine or paracrine signaling by type 2 cytokines.
STAT5A is essential for lobuloalveolar development and the expression of milk protein genes
STAT5 was initially shown to be an essential regulator of milk protein gene expression. The promoter of the sheep β-lactoglobulin (BLG) gene has three STAT5 binding sites in the proximal 406 bp promoter region.3 Using in vivo transgenic studies, it was demonstrated that these motifs are essential for maximal expression of BLG although mutation of these sites did not affect tissue specificity.23 Similarly, the whey acidic protein (WAP) promoter has STAT5 binding motifs that are required for expression of WAP in mammary gland24 and STAT5 is also important for β-casein expression in HC11 mammary epithelial cells.25 Clearly, lack of STAT5 would seriously impair milk production.
STAT5 is predominantly activated by PRL during gestation. The phenotype of PRL receptor knockout mice, which fail to undergo alveologenesis, indicates that STAT5 may have a more extensive role than just a regulator of milk protein gene expression and could be involved in regulating proliferation and/or differentiation of alveolar epithelial cells.26
STAT5 is encoded by two different genes, STAT5A and STAT5B.27 Mice in which the STAT5A gene was disrupted showed incomplete lobuloalveolar development at late pregnancy time points and failed to lactate and nurse their pups.28 In contrast, STAT5B deficient mice had growth defects.29 Combined deletion of STAT5A and STAT5B showed that, although there is some redundancy between these two proteins, they mediate virtually all growth hormone and prolactin functions.30 Subsequent generation of complete null alleles of STAT5A/B by conditional gene targeting at two different stages of development (virgin and mid-pregnancy) revealed that STAT5 is indeed required for alveolar development in pregnancy and showed also that loss of STAT5 from differentiated alveolar cells results in rapid cell death.31 The phenotype of conditional JAK2 knockout mice, an upstream regulator of STAT5A, essentially recapitulates that of the STAT5A deficient mice.32
More recently, STAT5A has been shown to be required for the generation and/or expansion of alveolar luminal progenitor cells from mammary stem cells.33 The mechanism by which STAT5 gene expression is regulated is not clear as the Ets transcription factor Elf5 has been shown to bind to the proximal STAT5A gene promoter in late pregnancy. Furthermore, in Elf5 knockout mammary epithelial cells, levels of STAT5A are downregulated.34 However, it also appears that STAT5A/5B can regulate Elf5 expression since STAT5A/5B-null luminal progenitor cells do not express Elf5. Furthermore, the distal region of the Elf5 gene promoter contains multiple STAT binding sites33 indicating that there could be a positive transcriptional regulatory loop between Elf5 and STAT5.
STAT3 is a mediator of cell death and inflammatory signaling in involution
The profile of STAT3 activation via tyrosine phosphorylation suggests a potential function in involution and, indeed, STAT3 activity has been demonstrated to have a pivotal role in this process. Interestingly, this role encompasses both control of epithelial cell death, and modulation of the inflammatory environment of the gland.
As previously mentioned, deletion of STAT3 results in early embryonic lethality.35 Consequently a murine mammary conditional deletion of STAT3 has been developed using the Cre-lox recombination system where Cre recombinase is under the control of a mammary-specific promoter36 such as whey acidic protein (WAP-Cre)37 or β-lactoglobulin (BLG-Cre).38 Although the latter is a whey protein not naturally present in rodent milk,39 Cre expression under the control of the ovine BLG promoter can target transgenes efficiently to secretory mammary epithelial cells of rodents.40
Using the BLG promoter to drive Cre expression in STAT3fl/fl mice, it was shown that STAT3 is essential for the initiation of cell death and remodeling following induction of synchronous weaning.38 Such mice, with a mammary-specific conditional deletion of STAT3, exhibited a notable delay in involution of at least 3 d and reduced levels of cell death. This delay in cell death and tissue remodeling was associated with the downregulation of IGFBP5, a negative regulator of IGF survival signaling which is considered to have a pro-apoptotic role in the mammary gland41 and the upregulation of p53 and p21. An independent study using WAP-Cre to drive conditional deletion of STAT3 yielded similar results and revealed an extension of the reversible phase of involution by 4 d in the absence of STAT3.37 These studies confirmed that STAT3 has a cell autonomous role in inducing cell death in mammary epithelium.
A recent study42 has shifted the paradigm that cell death during first phase of involution is via classical apoptotic pathways. Given that mammary gland regression progressed unabated both in caspase 3/caspase 6 doubly deficient mice, and in transgenic mice overexpressing the viral caspase inhibitor p35 under the control of the mouse mammary tumor virus promoter, it was inferred that cell death in early involution must be independent of executioner caspases. Interestingly, it was demonstrated that during involution mammary epithelial lysosomes undergo lysosomal membrane permeabilization and that STAT3 upregulates the expression of the lysosomal proteolytic enzymes cathepsins B and L while downregulating their inhibitor Spi2A. Correspondingly, delayed involution was observed in mice administered with cathepsin B inhibitor, and tellingly, the phenotypic delay correlated with the extent of inhibition of cathepsin B. Thus, it is now considered that cell death during reversible involution is regulated by STAT3 activity coordinating cell death via a lysosomal-mediated pathway rather than apoptosis.42
Studies using leukemia inhibitory factor (LIF) deficient mice43 and implantable LIF containing pellets44 demonstrated that the initial activator of STAT3 in the mammary gland following weaning is LIF. TGFβ3 also regulates STAT3 activity in involution.45 By contrast, IL-6, which is known to activate STAT3 in other contexts, does not play a role in control of STAT3 activity during this process.46 During the irreversible phase STAT3 is activated by the IL-6 cytokine family member oncostatin M (OSM) and its receptor (OSMR).47 OSMR itself is regulated by LIF induced STAT3, in a positive feedback loop acting to propagate continued activation of STAT3 as LIF levels decline.38,47,48
It has previously been noted that the onset of involution heralds a dramatic upregulation of genes connected with the acute phase response and inflammation,49-51 some of which have a microarray expression profile that mirrors the phosphorylation profile of STAT3.49,50 A wider role for epithelial STAT3 in modulating the inflammatory environment of the gland during involution has also been recently demonstrated showing STAT3 is a key transcriptional regulator of genes associated with innate immunity in first phase involution, and wound healing in second phase involution.52 Furthermore, epithelial STAT3 activity influences the influx of macrophages and mast cells during involution and promotes polarization of macrophages toward an alternatively activated (M2) phenotype. In the absence of STAT3, the gland retains a phenotype associated with classically activated (M1) macrophages, evidenced by significantly increased expression of iNOS (an M1 marker) and decreased expression of arginase-1 and Ym1 (M2 markers). Given the significant decrease in IL-4Rα expression in STAT3 deleted glands at 96 h involution, it seems probable that IL-4 signaling has a role in this polarization.52
In spite of the significant advances in understanding of STAT3 signaling during mammary gland involution made in the last decade, several key questions remain relatively unexplored. Investigation of the role of STAT3 activity in the stroma will require conditional deletion in stromal cells. This is a potentially exciting area for future research, particularly given the increasing recognition of the intricate levels of interplay between epithelial, immune cell and stromal compartments.
STAT1 has no apparent role in development but influences mammary tumorigenesis
The pattern of STAT1 phosphorylation is unlike the other STATs in that it tends to be high when other STATs are low. Thus, pSTAT1 is highest in virgin and late involuting glands but is low during gestation and lactation.9,38 Although STAT1 knockout mice have been available for over 15 years, no analysis of mammary development in STAT1 deficient mice has been published. Given the normal growth of pups nursed by STAT1−/− dams, it is unlikely that STAT1 has a major role in mammary gland development. STAT1 can be phosphorylated by IFNγ in mammary epithelial cells in vitro, and IRF1, a primary downstream target of STAT1 has been shown to play a minor role in involution.53 This is interesting in the context of the second wave of STAT 1 phosphorylation that is observed around day 3 to 4 of involution, although the role of STAT1 signaling in second phase involution remains poorly characterized.
The enhanced and precocious activation of STAT1 via tyrosine phosphorylation observed in the absence of epithelial STAT3 at 24 h involution38 demonstrates that STAT1 and STAT3 may both be activated via the common signal transduction subunit gp13054 and that STAT3 preferentially binds to this receptor chain. STAT1 could adopt a compensatory role in the eventual commencement of involution observed in STAT3 deleted mammary glands.38 However, it is not known whether the downstream effects of STAT1 in this situation are mediated through the same targets as STAT3. It is worth noting that microarray analysis of STAT3 knockout glands at 24 h involution revealed that most of the highly upregulated genes in the absence of STAT3 are STAT1 target genes (Hughes and Watson, unpublished).
Recently, there has been some interest in determining a function for STAT1 in breast tumorigenesis. Using MMTV-Cre mediated deletion of floxed STAT alleles to ablate STAT1 specifically in mammary epithelium resulted in an increased ErbB2/Neu-induced tumor burden.55 On the other hand, STAT1 was shown to suppress Neu mediated tumorigenesis through both immune cell regulatory mechanisms and tumor specific effects.56
Conclusion
Development of the adult mammary gland is critically dependent on several members of the STAT family of transcription factors. STAT5 and STAT6 are sequentially activated to promote the development of the alveolar lineage. STAT5 is also required to regulate the expression of milk protein genes, and Elf5, a master regulator of alveologenesis. STAT6 induces expression of the cytokines IL-4 and IL-13 and the transcription factor Gata-3, which is required for the maintenance of the luminal alveolar cells. Following lactation, the involution process requires STAT3, which regulates a lysosomal pathway of cell death and controls the balance of inflammatory and anti-inflammatory signaling to effect tissue remodeling.
In addition to their role in transducing cytokine signals, STATs have been shown to act as functional repressors57 where they could suppress the expression of genes associated with a different cell fate. With the advent of technologies that allow genome-wide mapping of STAT binding, additional roles for STATs as mediators of epigenetic modifications are emerging. Interestingly, a comparison of ChIP-seq data for STAT5 and STAT6 in TH2 cells showed that some of their target genes overlap.57 It is likely that this would apply also to mammary alveolar cells where STAT5 and STAT6 are both involved in establishing the alveolar lineage. In contrast, STAT3 and STAT5 can have opposing functions in regulating IL-17 expression58 and this resonates well with their opposing roles in mammary gland development. Further work is needed to delineate and understand the landscape of STAT target genes in mammary gland.
It is intriguing that multiple members of the STAT family of transcription factors have essential roles in the development of the mammary gland. These STATs all have specific functions in other tissues and we can speculate that, as a late evolving tissue, the mammary gland has hijacked programs of gene expression controlled by individual STATs to drive such diverse events as differentiation and milk production, programmed cell death, and immune cell mediated tissue remodeling.
Acknowledgments
K.H. is funded by a Wellcome Trust Research Training Fellowship (WT086672MA).
Footnotes
Previously published online: www.landesbioscience.com/journals/jak-stat/article/19691
References
- 1.Wilks AF, Harpur AG, Kurban RR, Ralph SJ, Zürcher G, Ziemiecki A. Two novel protein-tyrosine kinases, each with a second phosphotransferase-related catalytic domain, define a new class of protein kinase. Mol Cell Biol. 1991;11:2057–65. doi: 10.1128/mcb.11.4.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.John J, McKendry R, Pellegrini S, Flavell D, Kerr IM, Stark GR. Isolation and characterization of a new mutant human cell line unresponsive to alpha and beta interferons. Mol Cell Biol. 1991;11:4189–95. doi: 10.1128/mcb.11.8.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watson CJ, Gordon KE, Robertson M, Clark AJ. Interaction of DNA-binding proteins with a milk protein gene promoter in vitro: identification of a mammary gland-specific factor. Nucleic Acids Res. 1991;19:6603–10. doi: 10.1093/nar/19.23.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmitt-Ney M, Doppler W, Ball RK, Groner B. Beta-casein gene promoter activity is regulated by the hormone-mediated relief of transcriptional repression and a mammary-gland-specific nuclear factor. Mol Cell Biol. 1991;11:3745–55. doi: 10.1128/mcb.11.7.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schindler C, Fu XY, Improta T, Aebersold R, Darnell JE., Jr Proteins of transcription factor ISGF-3: one gene encodes the 91-and 84-kDa ISGF-3 proteins that are activated by interferon alpha. Proc Natl Acad Sci U S A. 1992;89:7836–9. doi: 10.1073/pnas.89.16.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shuai K, Stark GR, Kerr IM, Darnell JE., Jr A single phosphotyrosine residue of Stat91 required for gene activation by interferon-gamma. Science. 1993;261:1744–6. doi: 10.1126/science.7690989. [DOI] [PubMed] [Google Scholar]
- 7.Miyoshi K, Cui Y, Riedlinger G, Robinson P, Lehoczky J, Zon L, et al. Structure of the mouse Stat 3/5 locus: evolution from Drosophila to zebrafish to mouse. Genomics. 2001;71:150–5. doi: 10.1006/geno.2000.6433. [DOI] [PubMed] [Google Scholar]
- 8.Philp JA, Burdon TG, Watson CJ. Differential activation of STATs 3 and 5 during mammary gland development. FEBS Lett. 1996;396:77–80. doi: 10.1016/0014-5793(96)01069-1. [DOI] [PubMed] [Google Scholar]
- 9.Watson CJ, Neoh K. The Stat family of transcription factors have diverse roles in mammary gland development. Semin Cell Dev Biol. 2008;19:401–6. doi: 10.1016/j.semcdb.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 10.Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–8. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 11.Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–7. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 12.Veltmaat JM, Mailleux AA, Thiery JP, Bellusci S. Mouse embryonic mammogenesis as a model for the molecular regulation of pattern formation. Differentiation. 2003;71:1–17. doi: 10.1046/j.1432-0436.2003.700601.x. [DOI] [PubMed] [Google Scholar]
- 13.Watson CJ. Involution: apoptosis and tissue remodelling that convert the mammary gland from milk factory to a quiescent organ. Breast Cancer Res. 2006;8:203. doi: 10.1186/bcr1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noble MS, Hurley WL. Effects of secretion removal on bovine mammary gland function following an extended milk stasis. J Dairy Sci. 1999;82:1723–30. doi: 10.3168/jds.S0022-0302(99)75402-0. [DOI] [PubMed] [Google Scholar]
- 15.Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–60. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khaled WT, Read EK, Nicholson SE, Baxter FO, Brennan AJ, Came PJ, et al. The IL-4/IL-13/Stat6 signalling pathway promotes luminal mammary epithelial cell development. Development. 2007;134:2739–50. doi: 10.1242/dev.003194. [DOI] [PubMed] [Google Scholar]
- 17.Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–56. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- 18.Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2007;9:201–9. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- 19.Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 2006;127:1041–55. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hennighausen L, Robinson GW. Information networks in the mammary gland. Nat Rev Mol Cell Biol. 2005;6:715–25. doi: 10.1038/nrm1714. [DOI] [PubMed] [Google Scholar]
- 21.Moriggl R, Berchtold S, Friedrich K, Standke GJ, Kammer W, Heim M, et al. Comparison of the transactivation domains of Stat5 and Stat6 in lymphoid cells and mammary epithelial cells. Mol Cell Biol. 1997;17:3663–78. doi: 10.1128/mcb.17.7.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gordon KE, Binas B, Chapman RS, Kurian KM, Clarkson RW, Clark AJ, et al. A novel cell culture model for studying differentiation and apoptosis in the mouse mammary gland. Breast Cancer Res. 2000;2:222–35. doi: 10.1186/bcr57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burdon TG, Maitland KA, Clark AJ, Wallace R, Watson CJ. Regulation of the sheep beta-lactoglobulin gene by lactogenic hormones is mediated by a transcription factor that binds an interferon-gamma activation site-related element. Mol Endocrinol. 1994;8:1528–36. doi: 10.1210/me.8.11.1528. [DOI] [PubMed] [Google Scholar]
- 24.Li S, Rosen JM. Nuclear factor I and mammary gland factor (STAT5) play a critical role in regulating rat whey acidic protein gene expression in transgenic mice. Mol Cell Biol. 1995;15:2063–70. doi: 10.1128/mcb.15.4.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Happ B, Groner B. The activated mammary gland specific nuclear factor (MGF) enhances in vitro transcription of the beta-casein gene promoter. J Steroid Biochem Mol Biol. 1993;47:21–30. doi: 10.1016/0960-0760(93)90053-Y. [DOI] [PubMed] [Google Scholar]
- 26.Ormandy CJ, Naylor M, Harris J, Robertson F, Horseman ND, Lindeman GJ, et al. Investigation of the transcriptional changes underlying functional defects in the mammary glands of prolactin receptor knockout mice. Recent Prog Horm Res. 2003;58:297–323. doi: 10.1210/rp.58.1.297. [DOI] [PubMed] [Google Scholar]
- 27.Liu X, Robinson GW, Gouilleux F, Groner B, Hennighausen L. Cloning and expression of Stat5 and an additional homologue (Stat5b) involved in prolactin signal transduction in mouse mammary tissue. Proc Natl Acad Sci U S A. 1995;92:8831–5. doi: 10.1073/pnas.92.19.8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X, Robinson GW, Wagner KU, Garrett L, Wynshaw-Boris A, Hennighausen L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997;11:179–86. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- 29.Udy GB, Towers RP, Snell RG, Wilkins RJ, Park SH, Ram PA, et al. Requirement of STAT5b for sexual dimorphism of body growth rates and liver gene expression. Proc Natl Acad Sci U S A. 1997;94:7239–44. doi: 10.1073/pnas.94.14.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teglund S, McKay C, Schuetz E, van Deursen JM, Stravopodis D, Wang D, et al. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell. 1998;93:841–50. doi: 10.1016/S0092-8674(00)81444-0. [DOI] [PubMed] [Google Scholar]
- 31.Cui Y, Riedlinger G, Miyoshi K, Tang W, Li C, Deng CX, et al. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol Cell Biol. 2004;24:8037–47. doi: 10.1128/MCB.24.18.8037-8047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner KU, Krempler A, Triplett AA, Qi Y, George NM, Zhu J, et al. Impaired alveologenesis and maintenance of secretory mammary epithelial cells in Jak2 conditional knockout mice. Mol Cell Biol. 2004;24:5510–20. doi: 10.1128/MCB.24.12.5510-5520.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamaji D, Na R, Feuermann Y, Pechhold S, Chen W, Robinson GW, et al. Development of mammary luminal progenitor cells is controlled by the transcription factor STAT5A. Genes Dev. 2009;23:2382–7. doi: 10.1101/gad.1840109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi YS, Chakrabarti R, Escamilla-Hernandez R, Sinha S. Elf5 conditional knockout mice reveal its role as a master regulator in mammary alveolar development: failure of Stat5 activation and functional differentiation in the absence of Elf5. Dev Biol. 2009;329:227–41. doi: 10.1016/j.ydbio.2009.02.032. [DOI] [PubMed] [Google Scholar]
- 35.Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, et al. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci U S A. 1997;94:3801–4. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Selbert S, Bentley DJ, Melton DW, Rannie D, Lourenço P, Watson CJ, et al. Efficient BLG-Cre mediated gene deletion in the mammary gland. Transgenic Res. 1998;7:387–96. doi: 10.1023/A:1008848304391. [DOI] [PubMed] [Google Scholar]
- 37.Humphreys RC, Bierie B, Zhao L, Raz R, Levy D, Hennighausen L. Deletion of Stat3 blocks mammary gland involution and extends functional competence of the secretory epithelium in the absence of lactogenic stimuli. Endocrinology. 2002;143:3641–50. doi: 10.1210/en.2002-220224. [DOI] [PubMed] [Google Scholar]
- 38.Chapman RS, Lourenco PC, Tonner E, Flint DJ, Selbert S, Takeda K, et al. Suppression of epithelial apoptosis and delayed mammary gland involution in mice with a conditional knockout of Stat3. Genes Dev. 1999;13:2604–16. doi: 10.1101/gad.13.19.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kontopidis G, Holt C, Sawyer L. Invited review: beta-lactoglobulin: binding properties, structure, and function. J Dairy Sci. 2004;87:785–96. doi: 10.3168/jds.S0022-0302(04)73222-1. [DOI] [PubMed] [Google Scholar]
- 40.Whitelaw CB, Harris S, McClenaghan M, Simons JP, Clark AJ. Position-independent expression of the ovine beta-lactoglobulin gene in transgenic mice. Biochem J. 1992;286:31–9. doi: 10.1042/bj2860031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flint DJ, Boutinaud M, Whitelaw CB, Allan GJ, Kolb AF. Prolactin inhibits cell loss and decreases matrix metalloproteinase expression in the involuting mouse mammary gland but fails to prevent cell loss in the mammary glands of mice expressing IGFBP-5 as a mammary transgene. J Mol Endocrinol. 2006;36:435–48. doi: 10.1677/jme.1.01873. [DOI] [PubMed] [Google Scholar]
- 42.Kreuzaler PA, Staniszewska AD, Li W, Omidvar N, Kedjouar B, Turkson J, et al. Stat3 controls lysosomal-mediated cell death in vivo. Nat Cell Biol. 2011;13:303–9. doi: 10.1038/ncb2171. [DOI] [PubMed] [Google Scholar]
- 43.Kritikou EA, Sharkey A, Abell K, Came PJ, Anderson E, Clarkson RW, et al. A dual, non-redundant, role for LIF as a regulator of development and STAT3-mediated cell death in mammary gland. Development. 2003;130:3459–68. doi: 10.1242/dev.00578. [DOI] [PubMed] [Google Scholar]
- 44.Schere-Levy C, Buggiano V, Quaglino A, Gattelli A, Cirio MC, Piazzon I, et al. Leukemia inhibitory factor induces apoptosis of the mammary epithelial cells and participates in mouse mammary gland involution. Exp Cell Res. 2003;282:35–47. doi: 10.1006/excr.2002.5666. [DOI] [PubMed] [Google Scholar]
- 45.Nguyen AV, Pollard JW. Transforming growth factor beta3 induces cell death during the first stage of mammary gland involution. Development. 2000;127:3107–18. doi: 10.1242/dev.127.14.3107. [DOI] [PubMed] [Google Scholar]
- 46.Zhao L, Melenhorst JJ, Hennighausen L. Loss of interleukin 6 results in delayed mammary gland involution: a possible role for mitogen-activated protein kinase and not signal transducer and activator of transcription 3. Mol Endocrinol. 2002;16:2902–12. doi: 10.1210/me.2001-0330. [DOI] [PubMed] [Google Scholar]
- 47.Tiffen PG, Omidvar N, Marquez-Almuina N, Croston D, Watson CJ, Clarkson RW. A dual role for oncostatin M signaling in the differentiation and death of mammary epithelial cells in vivo. Mol Endocrinol. 2008;22:2677–88. doi: 10.1210/me.2008-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clarkson RW, Boland MP, Kritikou EA, Lee JM, Freeman TC, Tiffen PG, et al. The genes induced by signal transducer and activators of transcription (STAT)3 and STAT5 in mammary epithelial cells define the roles of these STATs in mammary development. Mol Endocrinol. 2006;20:675–85. doi: 10.1210/me.2005-0392. [DOI] [PubMed] [Google Scholar]
- 49.Pensa S, Watson CJ, Poli V. Stat3 and the inflammation/acute phase response in involution and breast cancer. J Mammary Gland Biol Neoplasia. 2009;14:121–9. doi: 10.1007/s10911-009-9124-x. [DOI] [PubMed] [Google Scholar]
- 50.Clarkson RW, Wayland MT, Lee J, Freeman T, Watson CJ. Gene expression profiling of mammary gland development reveals putative roles for death receptors and immune mediators in post-lactational regression. Breast Cancer Res. 2004;6:R92–109. doi: 10.1186/bcr754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stein T, Morris JS, Davies CR, Weber-Hall SJ, Duffy MA, Heath VJ, et al. Involution of the mouse mammary gland is associated with an immune cascade and an acute-phase response, involving LBP, CD14 and STAT3. Breast Cancer Res. 2004;6:R75–91. doi: 10.1186/bcr753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hughes K, Wickenden JA, Allen JE, Watson CJ. Conditional deletion of Stat3 in mammary epithelium impairs the acute phase response and modulates immune cell numbers during post-lactational regression. J Pathol. 2012;227:106–17. doi: 10.1002/path.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chapman RS, Duff EK, Lourenco PC, Tonner E, Flint DJ, Clarke AR, et al. A novel role for IRF-1 as a suppressor of apoptosis. Oncogene. 2000;19:6386–91. doi: 10.1038/sj.onc.1204016. [DOI] [PubMed] [Google Scholar]
- 54.Heinrich PC, Behrmann I, Müller-Newen G, Schaper F, Graeve L. Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem J. 1998;334:297–314. doi: 10.1042/bj3340297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klover PJ, Muller WJ, Robinson GW, Pfeiffer RM, Yamaji D, Hennighausen L. Loss of STAT1 from mouse mammary epithelium results in an increased Neu-induced tumor burden. Neoplasia. 2010;12:899–905. doi: 10.1593/neo.10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raven JF, Williams V, Wang S, Tremblay ML, Muller WJ, Durbin JE, et al. Stat1 is a suppressor of ErbB2/Neu-mediated cellular transformation and mouse mammary gland tumor formation. Cell Cycle. 2011;10:794–804. doi: 10.4161/cc.10.5.14956. [DOI] [PubMed] [Google Scholar]
- 57.O’Shea JJ, Lahesmaa R, Vahedi G, Laurence A, Kanno Y. Genomic views of STAT function in CD4+ T helper cell differentiation. Nat Rev Immunol. 2011;11:239–50. doi: 10.1038/nri2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang XP, Ghoreschi K, Steward-Tharp SM, Rodriguez-Canales J, Zhu J, Grainger JR, et al. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat Immunol. 2011;12:247–54. doi: 10.1038/ni.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Triplett AA, Sakamoto K, Matulka LA, Shen L, Smith GH, Wagner KU. Expression of the whey acidic protein (Wap) is necessary for adequate nourishment of the offspring but not functional differentiation of mammary epithelial cells. Genesis. 2005;43:1–11. doi: 10.1002/gene.20149. [DOI] [PubMed] [Google Scholar]
- 60.Radisky DC, Hartmann LC. Mammary involution and breast cancer risk: transgenic models and clinical studies. J Mammary Gland Biol Neoplasia. 2009;14:181–91. doi: 10.1007/s10911-009-9123-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wiseman BS, Werb Z. Stromal effects on mammary gland development and breast cancer. Science. 2002;296:1046–9. doi: 10.1126/science.1067431. [DOI] [PMC free article] [PubMed] [Google Scholar]