Abstract
Our previous studies demonstrated that lysine-specific demethylase 1 (LSD1) and histone deacetylases (HDACs) closely interact in controlling growth of breast cancer cells. However, the underlying mechanisms are largely unknown. In this study, we showed that knockdown of LSD1 expression (LSD1-KD) by RNAi decreased mRNA levels of HDAC isozymes in triple-negative breast cancer (TNBC) cells. Small interfering RNA (siRNA)-mediated depletion of HDAC5 expression induced the most significant accumulation of H3K4me2, a specific substrate of LSD1. Combined treatment with LSD1 inhibitor, pargyline, and HDAC inhibitor, SAHA (Vorinostat), led to superior growth inhibition and apoptotic death in TNBC cells, but exhibited additive or antagonistic effect on growth inhibition in non-TNBC counterparts or non-tumorigenic breast cells. Additionally, LSD1-KD enhanced SAHA-induced reexpression of a subset of aberrantly silenced genes, such as NR4A1, PCDH1, RGS16, BIK, and E-cadherin whose reexpression may be tumor suppressive. Genome-wide microarray study in MDA-MB-231 cells identified a group of tumor suppressor genes whose expression was induced by SAHA and significantly enhanced by LSD1-KD. We also showed that concurrent depletion of RGS16 by siRNA reduced overall cytotoxicity of SAHA and blocked the reexpression of E-cadherin, CDKN1C and ING1 in LSD1-deficient MDA-MB-231 cells. Furthermore, cotreatment with RGS16 siRNA reversed the downregulation of nuclear factor-kappaB expression induced by combined inhibition of LSD1 and HDACs, suggesting a crucial role of RGS16 in controlling key pathways of cell death in response to combination therapy. Taken together, these results provide novel mechanistic insight into the breast cancer subtype-dependent role of LSD1 in mediating HDAC activity and therapeutic efficacy of HDAC inhibitor.
Introduction
Abnormally enhanced activity of histone deacetylases (HDACs) in cancer cells may lead to the anomalous loss of expression of genes that are important in curbing tumor growth. Attempts to relieve this transcriptional repression have led to clinical trials using HDAC inhibitors (HDACi) in cancer therapy (1,2). Preclinical data suggest a role for HDACi as a potential new treatment in several tumor types including breast cancer (3,4). Two leading HDACis, vorinostat and romidepsin (FK-228), have been approved by the US FDA for the clinical treatment of cutaneous T-cell lymphoma. Despite the promising results produced by HDACi in treatment of hematological cancers, little clinical evidence exists to indicate that HDACi work effectively as a monotherapy against solid tumors including breast cancer, although most trials are still in early stages (5–8). A paucity of knowledge about HDAC biology and the action of HDACi in breast cancer has led to an empirical approach to testing HDACi, which is slowing the progress of future clinical application of these drugs. To conquer these obstacles, it is necessary to better understand the mechanisms by which HDAC activity is regulated in breast cancer. It appears that HDACis are more effective in tumor growth inhibition when they are used in combination with other epigenetic or chemotherapeutic agents (9–11). It is critically important to develop effective combination strategies to improve the efficacy of HDACi and reduce the side effects by targeting, more specifically, the small regions of chromatin and the subset of genes that are associated with most prominent alterations in the breast cancer genome.
Our recent work showed that a previously unrecognized histone demethylase, LSD1, possesses great potential as a target in cancer therapy (12–15). LSD1, also known as AOF2 or KDM1A, is the first identified histone demethylase capable of specifically demethylating mono- and dimethylated lysine 4 of histone H3 (H3K4me1 and H3K4me2) (16,17). LSD1 has been typically found in association with a transcriptional repressor complex that includes HDAC1/2, CoREST and BHC80 (16). The activity of the LSD1/HDACs complex has been implicated in tumorigenesis (18–20). Our most recent work provided novel insights into molecular mechanisms by which LSD1 and HDACs interact in breast cancer cells (14). We have shown that interaction at the chromatin level between LSD1 and HDACs is dysregulated in breast cancer cells, leading to abnormal gene expression patterns that could promote breast tumorigenesis (14). However, the exact mechanism(s) underlying the interactions between LSD1 and HDACs in breast cancer is still largely unclear.
In this study, we addressed the following important issues: (i) What are the mechanisms underlying the regulation of HDAC activity by LSD1 in breast cancer? (ii) How does LSD1 activity mediate the therapeutic efficacy of HDAC inhibitors in breast cancer? (iii) What are the unique target genes and pathways that are regulated by LSD1 and HDAC crosstalk in breast cancer? To answer these questions, we define in depth the mechanisms of the functional link between histone demethylase and deacetylase in chromatin remodeling and gene transcription. The results from these studies suggest that LSD1 and HDACs closely cooperate to mediate important signaling pathways of cell death in a breast cancer subtype-dependent manner.
Materials and methods
Reagents and cell culture conditions
SAHA (suberoylanilide hydroxamic acid) (Vorinostat) was purchased from Cayman Chemical (Ann Arbor, MI). Pargyline was obtained from Sigma–Aldrich (St Louis, MO). Breast cancer cell lines were obtained from American Type Culture Collection and from the Integrative Cancer Biology Program (ICBP) 45 breast cancer cell line kit (ICBP45) of the National Cancer Institute. MDA-MB-231 and MCF-7 cells were maintained in Dulbecco’s modified Eagle’s medium, BT-474 cells were maintained in RPMI 1640 medium and MDA-MB-468 cells were cultured in Improved Minimum Essential medium, each supplemented with 5% fetal bovine serum. SK-BR-3 cells were cultured in McCoy’s modified 5A medium with 10% fetal bovine serum. MCF-10A cells were cultured in growth medium as described previously (21).
shRNA treatment and stable cell line generation
Scramble control and LSD1-specific shRNA lentiviral particles (Santa Cruz Biotechnology, Santa Cruz, CA) were infected into cells according to manufacturer’s protocol. Briefly, cells were seeded at 50% confluence on day of infection. Polybrene was added to complete medium at a final concentration of 5 μg/ml. Selection with 1 μg/ml puromycin began at 72h postinfection and continued until all negative control cells were killed. Single-cell colonies were expanded and assayed for expression of LSD1 via quantitative PCR (qPCR).
Small interfering RNA treatment
Target gene and control small interfering RNAs (siRNAs) (Santa Cruz Biotec hnology) were transfected into cells according to manufacturer’s protocol. Briefly, cells were seeded at 2 × 105 in 6-well plates prior to transfection. siRNA complexes were prepared in transfection medium (sc-36868) with transfection reagent (sc-29528) and added to the medium. Cells were incubated at 37°C for 5h when 1ml normal growth medium containing 2× serum was added to each well. Cells were harvested 48h posttransfection and total RNA and nuclear extracts were prepared.
RNA extraction and qPCR
Total RNA samples were extracted using RNeasy kit (Qiagen, Valencia, CA) or TRIZOL reagents (Life Technologies, Grand Island, NY). cDNA was made using 3 μg total RNA via M-MLV Reverse Transcriptase (Life Technologies, Grand Island, NY) according to manufacturer’s recommendation. Quantitative real-time PCR was performed on the StepOne real-time PCR system (Life Technologies) using TaqMan® Gene Expression Assays.
Nuclear extract preparation and western blotting
Whole and nuclear proteins were extracted as described previously (13,22). Fifty microgram of whole-cellular protein or 30 µg of nuclear protein was separated on acrylamide gels and transferred onto polyvinylidene difluoride membranes. Membranes were blocked in Odyssey® Blocking buffer and then incubated with primary antibody in Odyssey Blocking buffer supplemented with 0.1% Tween 20. Primary antibodies against H3K4me2 and LSD1 were from EMD Millipore (Billerica, MA) and the H3 antibody was purchased from Abcam (Cambridge, MA). The caspase-3, Bax, Bcl-2, cyclin D1, CDKN1A, regulator of G-protein signaling 16 (RGS16), nuclear factor-kappaB (NF-κB), proliferating cell nuclear antigen and actin antibodies were obtained from Santa Cruz Biotechnology. Nuclear receptor subfamily 4, group A, member 1 (NR4A1) antibody was purchased from Active Motif (Carlsbad, CA).
MTT growth inhibition and drug combination index analysis
Cells were seeded at 5000 cells per well in a 96-well plate and treated with appropriate concentrations of drugs. Following incubation, 3-(4,5-dimethylthiazole-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assays were performed using the method as described previously (22). The median effects (IC50) were determined by using CalcuSyn software from Biosoft (Cambridge, UK). The Chou–Talalay median effect/combination index (CI) model was used to determine synergy, additivity or antagonism of combination treatments with pargyline and HDAC inhibitors (23).
Chromatin immunoprecipitation
To crosslink proteins, 2×106 cells were exposed to 1% formaldehyde, and chromatin immunoprecipitation (ChIP) assay was performed as described previously (13). Primary antibodies against LSD1, H3K4me2 and acetyl-H3K9 (EMD Millipore) were used as indicated for immunoprecipitation of protein–DNA complexes. PCR primer sets used for amplification of precipitated fragments were as follows: NR4A1 sense, 5′-GTTCAGCAGAACAGGTGCAA; antisense, 5′-TCCCATATTGGGCT- TGGATA; PCDH1 sense, 5′-ACACACTCGGGGAGCGGGAG; antisense, 5′-CTGGGCTGTGCCTCAGGTGC. Quantitative ChIP by qPCR was performed using SYBR Green Gene Expression Assays (Life Technologies). Input DNA was used for normalization.
Determination of internucleosomal DNA cleavage
After treatment with SAHA for 48h, MDA-MB-231 cells were collected and DNA ladder fragments were prepared as described previously (22). DNA samples were analyzed by electrophoresis using a 2% agarose gel containing 0.2 μg/ml ethidium bromide and visualized under UV illumination.
Microarray analysis of gene expression
MDA-MB-231 cells were treated with 5 µM SAHA for 24h. Total RNA samples from three independent biological replicates were extracted using Qiagen RNeasy kit. The array study was performed at Cancer Biomarkers Shared Facility of University of Pittsburgh Cancer Institute (UPCI) using the Affymetrix GeneChip U133A 2.0 array platform, which contains 20928 probes representing all functionally characterized genes in the human genome to date. The data from all of the arrays were processed as RMA files (Affymetrix Robust Multi-Array Average) in which the raw intensity data were background corrected, log2 transformed and then quantile normalized according to Affymetrix recommendations. Probes were selected that were ≥1.25-fold above or ≤0.75 of the control values.
Statistical analysis
A one-way analysis of variance or Student’s t-test was used to determine the statistical differences between various experimental and control groups using GraphPad Prism 5.0 or Excel software. The cytotoxic potential of the combination treatments was evaluated by CalcuSyn analysis (Biosoft). Microarray statistical tests were performed using Significance Analysis of Microarrays software (SAM version 3.09c), which is designed to reduce the risk of Type 1 errors due to multiple testing (24).
Results
LSD1 and HDACs functionally interact in breast cancer
To better dissect the role of LSD1 in regulation of histone acetylation in breast cancer, we stably knocked down LSD1 mRNA expression in MDA-MB-231 cells with LSD1 shRNA lentiviral particles. LSD1-targeting shRNA reduced endogenous LSD1 protein expression by >90% and LSD1-KD-15 and LSD1-KD-16 were chosen for future studies (Figure 1A). Depletion of LSD1 hindered growth of both LSD1-KD clones, indicating a growth-promoting role for LSD1 in breast cancer cells (Figure 1B). In both LSD1-KD-15 and LSD1-KD-16 clones, loss of LSD1 repressed mRNA expression of most of the HDAC isozymes including class I HDACs (1, 2, 3 and 8), class II HDACs (6 and 10) and class IV HDAC (11). HDAC9 mRNA expression was induced by LSD1-KD in both clones. This finding suggests that LSD1 acts as an important regulator in mediating mRNA expression of HDAC isozymes in breast cancer cells.
Fig. 1.
Loss of LSD1 suppresses HDAC mRNA expression. (A) MDA-MB-231 cells were infected with either non-targeting scramble control or LSD1-specific shRNA lentiviral particles. Three LSD1-KD clones were shown to have suppressed LSD1 protein level by immunoblots. (B) Scramble shRNA and LSD1-KD MDA-MB-231 cells were analyzed for growth by MTT assays. (C) LSD1-KD by shRNA in MDA-MB-231 cells altered mRNA expression of HDAC isozymes.Bars represent the fraction of mRNA expression in LSD1-KD cells versus scramble control (as fold 1). Data are means ± SD of three independent experiments. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 (LSD1-KD versus Scramble, Student’s t-test). (D) MDA-MB-231 cells were treated with 5 μM SAHA for 24h or transiently transfected with siRNA against individual HDAC isozymes. The western blots shown are representative of three experiments with similar results. (E) Histograms represent the mean nuclear H3K4me2 expression levels of three determinations relative to PCNA ± SD as determined by quantitative immunoblotting using infrared detection and analysis. *P ≤ 0.05, **P ≤ 0.01 (HDAC siRNA versus Scramble, Student’s t-test).
Our earlier study demonstrated that HDACi treatment induced accumulation of H3K4me1 and H3K4me2, two specific substrates of LSD1, suggesting that HDACs represent key targets through which HDACi modulates LSD1 (14). To prove this hypothesis, we transfected MDA-MB-231 cells with siRNAs against each class I, class II and class IV HDAC isozyme. class III HDACs were not included in this experiment, as our previous experiments showed that H3K4me2 level was not altered by inhibitors against class III HDACs in breast cancer cells (14). The selectivity and efficacy of the HDAC knockdown were validated by qPCR. siRNAs successfully depleted approximately ≥50% of mRNA expression for each targeted HDAC isozyme (data not shown). The effect of HDAC-KD on cellular LSD1 activity was subsequently assessed by quantifying the level of its substrate, H3K4me2. We observed that siRNA-mediated inhibition of the class II HDAC, HDAC5, led to the most significant increase of H3K4me2 (Figure 1D and E). This result indicates that a decrease in HDAC5 mRNA expression mimics SAHA effect on histone methylation and thus identifies HDAC5 as a key HDAC isozyme that functionally modulates LSD1 activity. We further determined that none of these HDAC siRNAs significantly altered LSD1 mRNA expression (Supplementary Figure S1, available at Carcinogenesis Online), suggesting that the effect of HDAC inhibition on histone demethylation is apparently due to blocking LSD1 enzymatic function rather than inhibiting its expression. These findings point to a close functional association between LSD1 and HDACs in breast cancer cells.
Inhibition of LSD1 sensitizes TNBC cells to HDACi-induced apoptosis
We have recently demonstrated that LSD1 inhibitor, pargyline, sensitized MDA-MB-231 cells to HDACi-induced cell death (14). In this study, we extended the investigation on combinatorial effect of LSD1 and HDAC inhibitors on different subtypes of breast cancer cells and non-tumorigenic breast epithelial cells. Using a concomitant 48h treatment schedule of agents, significant synergistic growth inhibition (CI < 1) between the LSD1 inhibitor, pargyline, and HDAC inhibitor, SAHA, was observed in two TNBC or basal-like MDA-MB-231 and MDA-MB-468 cells. In contrast, the combination exhibited moderate additivity (CI = 1) or antagonism (CI > 1) in non-TNBC or luminal-like MCF-7, BT-474 and SK-BR-3 cells. Strong antagonistic effect of compounds was observed in treated non-tumorigenic breast epithelial cells MCF-10A (Figure 2A).
Fig. 2.
Inhibition of LSD1 sensitizes TNBC cells to HDACi-induced apoptosis. (A) Effect of combination therapy on growth of breast cancer cells by CI. Synergy was defined as any CI < 1, additivity as CI = 1 and antagonism as any CI > 1. (B) Depletion of LSD1 sensitizes MDA-MB-231 cells to SAHA. Cells were treated with SAHA for 48h. Cell proliferation was analyzed by MTT assays. (C) Scramble control and LSD1-KD MDA-MB-231 cells were treated with5 μM SAHA for 48h. Fragmented DNA indicative of apoptosis was analyzed by electrophoresis. (D) Immunoblots with anti-caspase-3, Bcl-2, Bax and cyclin D1 antibodies were performed and analyzed using β-actin protein as a control. (E) Effect of LSD1-KD on mRNA expression of HDAC isozymes in MCF-7 and SK-BR-3 cells. (F) Effect of SAHA on growth of MCF-7 scramble control and LSD1-KD cells. Cells were treated with SAHA for 48h. The experiments were performed three times with similar results. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 (LSD1-KD versus Scramble, Student’s t-test).
The above results indicate that combination therapy targeting crosstalk between LSD1 and HDACs may represent an effective therapeutic strategy for TNBC. We tested this hypothesis using an LSD1-KD model with more specific inhibition of LSD1. Similar to pharmacological results, LSD1 depletion by RNAi sensitizes cells to SAHA-induced growth inhibition in MDA-MB-231 cells (Figure 2B). Furthermore, LSD1-KD in combination with SAHA treatment led to a significant induction of DNA fragmentation (Figure 2C) and enhanced cleavage of caspase-3, downregulated antiapoptotic Bcl-2 protein, upregulated proapoptotic Bax protein and led to inhibition of cell cycle regulator cyclin D1 (Figure 2D). These results support the hypothesis that inhibition of LSD1 sensitizes MDA-MB-231 cells to HDACi-induced apoptosis.
The effect of the pharmacological inhibition of LSD1 on HDACi efficacy was also compared with a lentiviral shRNA-mediated knockdown of LSD1 expression in MCF-7 and SK-BR-3 cells. Loss of LSD1 was unable to attenuate mRNA expression of HDAC isozymes in both cell lines (Figure 2E) and failed to change the cellular sensitivity to SAHA in MCF-7 cells (Figure 2F). These results suggest that combination therapy inhibiting LSD1 and HDACs may affect breast tumor cell growth in a cell subtype-dependent manner.
Functional interplay between LSD1 and HDACs is an important epigenetic signature contributing to aberrant gene silencing in TNBC
Our recent microarray assay identified a cluster of genes whose expression is uniquely upregulated by combined inhibition of LSD1 and HDACs. These genes include NR4A1, protocadherin 1 (PCDH1), RGS16 and Bcl-2-interacting killer (BIK). Importantly, activation of these genes might be tumor suppressive in breast cancer (14,25–27). Our initial comparison study in a small group of breast cancer cell lines showed that the mRNA expression level of these genes is much lower in TNBC/basal-like MDA-MB-231 and MDA-MB-468 cells in comparison with their hormone receptor-positive or HER2-amplified counterparts MCF-7, BT-474 and SK-BR-3 cells (Figure 3A). This result raised a question of whether aberrant epigenetic silencing of these genes represents a unique feature for TNBC/basal-like tumors. To better address this issue, we undertook a more comprehensive assessment of gene expression profiling in a panel of 36 human breast cancer cell lines across nearly all distinct genetic or clinical subtypes of breast tumors (Supplementary Table S1, available at Carcinogenesis Online). The gene expression level of NR4A1, PCDH1, RGS16 and BIK in each individual cell line was compared with that of MDA-MB-231 cells (set as 1) (Supplementary Figure S2, available at Carcinogenesis Online). We first assessed the expression of these genes in Basal A, Basal B and luminal breast cancer cell lines, three distinct genetic clusters of breast cancers, that were defined according to previously published standards (28). The baseline expression of all these genes is lowest in the Basal B group with a significantly reduced mRNA expression of NR4A1, RGS16 and BIK in Basal B group compared with luminal counterparts (Figure 3B). Because the Basal B cluster mirrors the clinical TNBC subtype, we further compared the gene expression in TNBC cells versus non-TNBC cells. The analysis clearly indicates that TNBC cells express significantly lower levels of mRNA for all four tested genes with statistical significance for NR4A1, RGS16 and BIK (Figure 3C). These findings point to a possible association of epigenetic silencing for a unique subset of genes within a specific subtype of breast tumors. Importantly, no difference in the mRNA expression of LSD1 or HDAC5 was found between distinct subtypes of breast cancer cells (Supplementary Figure S3, available at Carcinogenesis Online), suggesting that abnormal gene silencing likely occurs through the enhanced crosstalk between LSD1 and HDACs rather than amplified gene expression of histone enzymes.
Fig. 3.
Comparison of gene expression in distinct subtypes of breast cancer cells. (A) mRNA expression of indicated genes in different subtypes of breast cancer cells. Data are means ± SD of three independent experiments. (B) Comparison of mRNA levels of NR4A1, PCDH1, RGS16 and BIK in 36 human breast cancer cell lines representing different subtypes as indicated in Supplementary Table 1, available at Carcinogenesis Online. (C) Expression of indicated genes in TNBC versus non-TNBC cell lines. mRNA expression of each cell line was compared with that of MDA-MB-231 as fold 1. Data were analyzed by one-way analysis of variance, posttest of analysis of variance and Student’s t-test. *P ≤ 0.05, **P ≤ 0.01 (TNBC versus non-TNBC).
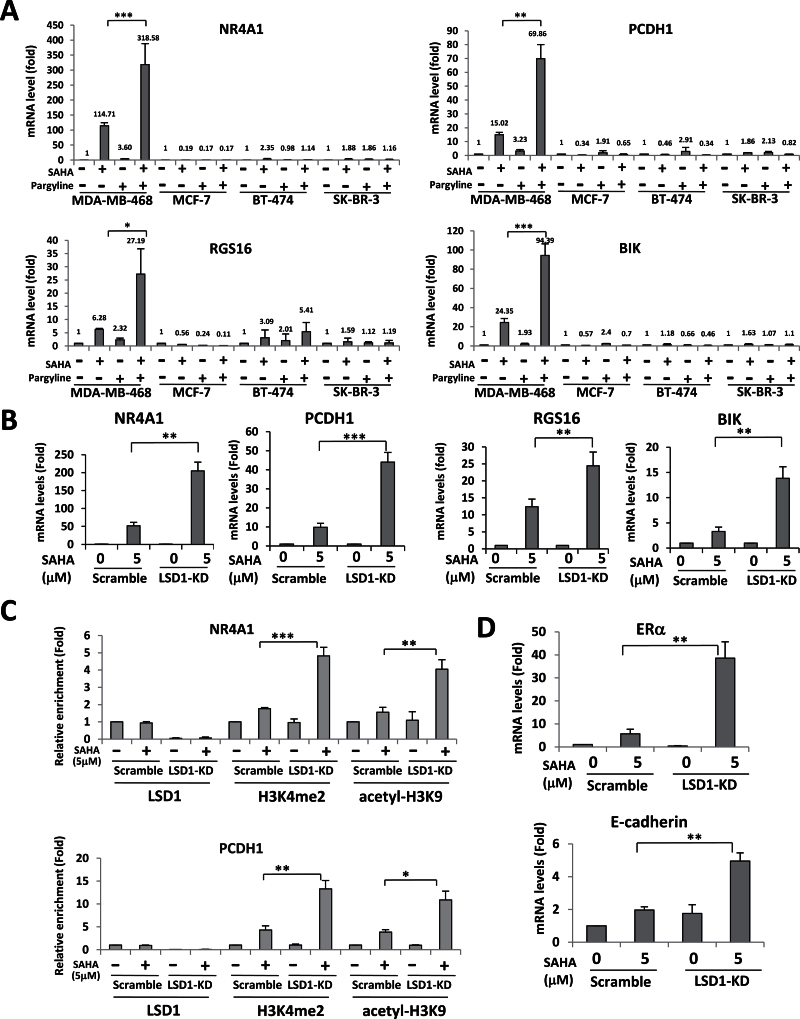
The above data have signaled a possible role of these genes as a unique panel of therapeutic biomarkers to indicate a potential response to combination therapy targeting LSD1 and HDACs in breast cancer. To validate this hypothesis, we examined the effect of combined treatment with LSD1 and HDAC inhibitors on gene expression in distinct subtypes of breast cancer cells. Similar to published results in MDA-MB-231 cells (14), the mRNA expression of all four genes exhibited a striking increase upon combination treatment with pargyline and SAHA in MDA-MB-468 cells (Figure 4A). However, combination treatment exerted little or no effect on gene activation in MCF-7, BT-474 and SK-BR-3 cells (Figure 4A). These data clearly indicate that the expression of these genes is indeed abnormally silenced as a result of ‘transcriptional corepression’ by interaction of LSD1 and HDACs in TNBC cells. To validate this conclusion, we further compared the effect of pharmacological inhibition of LSD1 with LSD1 RNAi on gene expression via qPCR. As shown in Figure 4B, loss of LSD1 significantly enhanced SAHA-induced gene reexpression in LSD1-KD MDA-MB-231 cells, indicating that pharmacologic inhibition and RNAi exert similar effects on reexpression of silenced genes.
Fig. 4.
Combined inhibition of LSD1 and HDACs reactivates silenced genes. (A) MDA-MB-468, MCF-7, BT-474 and SK-BR-3 cells were treated with 5 μM SAHA or 2.5mM pargyline or both for 24h. qPCR analysis of gene expression of NR4A1, PCDH1, RGS16 and BIK was performed. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 (combination versus SAHA, Student’s t-test). (B) MDA-MB-231 scramble or LSD1-KD cells were treated with 5 μM SAHA for 24h. Total RNA was extracted for qPCR analysis of NR4A1, PCDH1, BIK and RGS16 (LSD1-KD versus Scramble). (C) MDA-MB-231 scramble or LSD1-KD cells were treated with 5 μM SAHA for 24h. Quantitative ChIP analysis was used to determine the occupancy of the indicated promoters by LSD1, H3K4me2 and acetyl-H3K9. (D) MDA-MB-231 scramble or LSD1-KD cells were treated with 5 μM SAHA for 24h. Total RNA was extracted for quantitative RT–PCR analysis ofERα and E-cadherin expression. Actin is included as an internal control. In all panels, the quantified results are the means of three independent experiments ± SD *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 (LSD1-KD versus Scramble, Student’s t-test).
To examine if gene silencing is caused by enhanced cooperation of LSD1 and HDACs activities at gene promoters, quantitative ChIP analysis was performed to measure the level of regulatory histone marks at promoters of NR4A1 and PCDH in LSD1-KD MDA-MB-231 cells. ChIP assays revealed that LSD1-KD abolished LSD1 occupancy from both NR4A1 and PCDH gene promoters. We observed that SAHA treatment in LSD1-KD cells significantly enhances the level of both H3K4me2 and acetyl-H3K9, the key histone substrate for HDACs, at the promoter region of both NR4A1 and PCDH1 (Figure 4C), suggesting that the derepression of these genes lies exclusively in the cooperation between histone demethylation and deacetylation at key active marks.
HDAC inhibitors have been shown to reexpress methylated genes without changing DNA CpG methylation in breast cancer cells. For example, HDACi reactivates silenced estrogen receptor alpha (ERα) without loss of DNA hypermethylation (29). Inhibition of the class III HDAC, SIRT1, restored expression of epigenetically silenced tumor suppressive SFRP1, SFRP2 and E-cadherin genes in human breast cancer cells without altering CpG methylation at promoters (4). We investigated if combined inhibition of histone demethylation and deacetylation could result in superior reactivation of these heavily methylated genes. We examined a panel of methylated genes including ERα, members of the secreted frizzle-related protein family, SFRP1, SFRP2, E-cadherin and retinoic acid (RA) receptor beta (RARb), etc. Of these, SAHA-induced reexpression of silenced genes ERα and E-cadherin was enhanced by LSD1 inhibition (Figure 4D). Interestingly, we have found that LSD1-KD in combination with SAHA treatment has no effect on global or gene-specific CpG methylation (data not shown), suggesting that histone modification alone may be potent enough as activating marks to produce some reexpression of even heavily methylated genes.
LSD1 is extensively involved in regulation of expression of HDAC-targeted tumor suppressor genes
We have recently carried out a comprehensive genome-wide microarray analysis to define the effect of SAHA on gene expression in MDA-MB-231 cells. It showed that 416 genes were upregulated and 255 genes were downregulated with 1.5-fold or greater (P < 0.01, Supplementary Tables S2 and S3, available at Carcinogenesis Online). Among these affected genes, we identified a group of tumor suppressor genes (TSGs) whose expression levels were induced by SAHA treatment in MDA-MB-231 cells. These genes include CDKN1A, CDKN1C, CRABP2, TNFAIP3, TP53TG1 and ING1. The microarray results of gene expression were validated by qPCR in MDA-MB-231 cells and similar gene regulation pattern was observed in MDA-MB-468 cells treated with SAHA (Figure 5A). We have examined the protein expression of CDKN1A to validate if mRNA induction leads to concordant increase of protein level. Great induction of both cytoplasmic and nuclear CDKN1A proteins by SAHA treatment in MDA-MB-231 cells was observed, suggesting that induction of mRNA by SAHA indeed leads to steady increase of protein expression (Supplementary Figure S4A, available at Carcinogenesis Online).
Fig. 5.
LSD1 regulates SAHA-induced expression of TSGs. (A) Validation of TSG mRNA expression altered by SAHA in MDA-MB-231 and MDA-MB-468 cells. All cells were treated with 5 μM SAHA for 24h. (B) Effect of LSD1 depletion on SAHA-induced expression of TSGs in MDA-MB-231, MCF-7 and SK-BR-3 cells. Total RNA was extracted for qPCR analysis of expression of indicated genes. Actin is included as an internal control. *P ≤ 0.05, **P ≤ 0.01 (fold changes by SAHA in LSD1-KD cells/fold changes by SAHA in scramble control cells).
Next, we investigated whether activity of LSD1 is involved in regulation of expression of these TSGs. As shown in Figure 5B, loss of LSD1 in MDA-MB-231 cells enhances SAHA-induced expression of CRABP2, TP53TG1, CDKN1C and ING1. We also compared the effect of LSD1-KD on TSG expression in different subtypes of breast cancer cells. Loss of LSD1 in MCF-7 cells led to a modest increase of expression of CDKN1A and TNFAIP3. In SK-BR-3 LSD1-KD cells, TP53TG1 is the only gene whose expression was promoted by LSD1-KD (Figure 5B). These data indicate that LSD1 plays a more influential role in mediating expression of HDAC-targeted TSGs in TNBC cells.
Reexpression of RGS16 plays an important role in mediating activity of crosstalk between LSD1 and HDACs
The recognition of the important role of epigenetic regulation in breast cancer has led to active efforts to develop strategies to restore the genes of interest to a transcriptionally active state (30,31). Among our candidate target genes, NR4A1 and RGS16 have been demonstrated to act as important growth factors and signaling transducers in tumorigenesis. NR4A1 acts as a critical nuclear transcription factor that functions as a growth factor, but it can become a potent killer when certain death stimuli induce its migration to mitochondria, where it binds to Bcl-2 and conformationally converts it to a killer that triggers cytochrome c release and apoptosis (25,32,33). RGS16 interacts with G proteins and inhibits the G-protein-coupled mitogenic signal transduction of mitogen-activated protein kinase-signaling cascade in tumor cell proliferation and transformation (34,35).
To study the role of NR4A1 and RGS16 in mediating activity of crosstalk between LSD1 and HDACs, we first examined whether mRNA reexpression of these genes led to protein reexpression. Combined use of pargyline and SAHA resulted in detectable reexpression of NR4A1 and RGS16 proteins in MDA-MB-231 cells (Figure 6A). Most reexpressed NR4A1 protein was observed in the cytoplasm, suggesting a potential nuclear export and proapoptotic activity of this reexpressed gene in response to therapy (Figure 6A). Similar results were observed in MDA-MB-468 cells (Supplementary Figure S4B, available at Carcinogenesis Online). To determine if restoration of these key silenced genes can be a potential mechanism for antitumor activity of vorinostat, MDA-MB-231 cells were transfected with siRNA to genetically reverse the gene reexpression to assess their roles in drug-induced cell death. Proliferation studies using MTT assay showed that simultaneous treatment with NR4A1 siRNA failed to rescue the growth inhibition by SAHA in LSD1-KD cells. However, concurrent siRNA depletion of RGS16 significantly reduces the sensitivity of LSD1-KD cells to SAHA at higher concentrations (>1 μM), which are required to reexpress RGS16 in LSD1-KD cells (Figure 6B).
Fig. 6.
RGS16 mediates activity of crosstalk between LSD1 and HDACs. (A) Effect of combination therapy on reexpression of NR4A1 and RGS16 proteins. MDA-MB-231 cells were treated with 5 μM SAHA or 2.5mM pargyline or both for 24h. Cytoplasmic or nuclear protein was extracted for immunoblotting with anti-NR4A1 antibody. Whole cellular lysates were subjected to immunoblot with anti-RGS16 antibody. β-Actin protein was blotted as a loading control for whole cellular or cytoplasmic lysate and PCNA was used as a loading control for nuclear lysate. (B) Scramble and LSD1-KD MDA-MB-231 cells were transiently transfected with scramble, NR4A1 or RGS16 siRNA and simultaneously treated with different concentrations of SAHA for 48h. Cell proliferationwas analyzed by MTT assays. The quantified results are the means of three independent experiments ± SD. *P ≤ 0.05, **P ≤ 0.01 (LSD1-KD treated with scramble siRNA and SAHA versus LSD1-KD treated with RGS16 siRNA and SAHA, Student’s t-test). (C) LSD1-KD MDA-BM-231 cells were transiently transfected with scramble or RGS16 siRNA and simultaneously treated with 5 μM SAHA for 48h. qPCR was performed to analyze mRNA expression of RGS16, E-cadherin, CDKN1C and ING1. The quantified results are the means of three independent experiments ± SD. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 (LSD1-KD treated with scramble siRNA and SAHA versus LSD1-KD treated with RGS16 siRNA and SAHA, Student’s t-test). (D) Scramble and LSD1-KD MDA-MB-231 cells were treated with 5 μM SAHA for 48h. Western blot was performed to examine NF-κB protein expression. β-Actin protein was blotted as a control. (E) LSD1-KD MDA-MB-231 cells were transiently transfected with scramble or RGS16 siRNA and simultaneously treated with 5 μM SAHA for 48h. Western blot was performed to examine NF-κB (p65) protein expression. β-Actin protein was blotted as a control. (F) Proposed model of role of LSD1 in mediating HDAC inhibitor-induced gene expression and cell death in breast cancer.
Next, we examined if RGS16 mediates drug cytotoxicity through regulation of the activity of key growth regulatory genes whose expression was induced by combination treatment using the strategy as stated above. qPCR results indicate that simultaneous reduction of SAHA-induced RGS16 reexpression by siRNA in LSD1-KD MDA-MB-231 cells diminished the mRNA reexpression of CDKN1C, ING1 and E-cadherin (Figure 6C).
Our recent microarray study revealed that combined treatment with pargyline and SAHA suppressed expression of a group of important prosurvival growth factors such as NF-κB, EpCAM (epithelial cell adhesion molecule), MKI67 (antigen identified by monoclonal antibody Ki-67) (14). The downregulation of these genes by drugs is thought to be a downstream effect of combination therapy. We demonstrated that SAHA treatment in combination with LSD1-KD remarkably downregulated NF-κB (p65) protein expression (Figure 6D), which was reversed by concurrent treatment with RGS16 siRNA in MDA-MB-231 cells (Figure 6E). This result suggests that NF-κB is an important downstream target of RGS16 in mediating the cytotoxic effect of combination treatment.
Discussion
In this study, as depicted in Figure 6F, our findings provided supportive evidence that an orchestrated interplay between LSD1 and HDACs is a fundamental epigenetic mechanism contributing to aberrant gene silencing. Combination therapy targeting crosstalk between these two critical histone posttranslational modification systems may represent a novel and effective approach to curb the growth of breast cancer, especially the clinically aggressive TNBC.
We demonstrated for the first time that inhibition of LSD1 downregulated the mRNA expression of important HDAC family members, which may enhance susceptibility of breast cancer cells to HDACi-induced apoptotic death. This critical finding provides us a previously unknown mechanistic insight into the pivotal role of LSD1 in governing HDAC mRNA expression. Because LSD1 serves mainly as a transcription repressor, one possible effect is that inhibition of LSD1 increases H3K4 methylation which in turn induces the expression of one or multiple transcription factors that repress HDAC mRNA synthesis. In this study, we also showed that inhibition of HDAC5 mimics the effect of HDAC inhibitor on nuclear expression of H3K4me2. From a clinical perspective, this finding has significance for design and development of isozyme-selective HDAC inhibitors to improve the specificity of combination therapy in breast cancer cells. However, the role of HDAC5 in mediating histone demethylation activity is still largely unknown. One recent study reported that LSD1 interacts with HDAC5 through the nuclear receptor, TLX (36). Further study will be necessary to probe the exact mechanism by which these two enzymes are linked.
Our studies have revealed a unique panel of tumor suppressive or growth regulatory genes whose expression is exclusively upregulated by combined inhibition of histone demethylation and deacetylation in a breast cancer subtype-dependent manner. It is now axiomatic that breast cancer is a heterogeneous disease and shows different patterns of transcriptomic, genomic and epigenomic abnormality. We determined that the expression of a novel set of genes is more profoundly suppressed in TNBC cells. Clinically, TNBC is the most aggressive subtype and one of the major clinical hurdles encountered in the development of specific treatment of this disease is lack of effective targeted therapy due to the loss of important hormonal receptors or TSGs. Based on our findings, we speculate that silencing of these genes is closely associated with aggressive phenotype of TNBC and that reactivation of these genes holds hope for enhancing antitumor efficacy of reagents targeting crosstalk between LSD1 and HDACs.
Our study suggests that RGS16 acts as a tumor suppressor by transduction of growth suppressive signaling evoked by inhibition of crosstalk activity between LSD1 and HDACs. RGS16 was recently identified as a breast cancer susceptibility gene. A detailed mapping study in human breast carcinomas revealed that the promoter region of RGS16 was found to be methylated in 10% of the breast tumors (37). Results from our study suggest that RGS16 plays a key role in determining the susceptibility of tumor cells to cytotoxic effect of combination therapy. Several important downstream growth regulatory factors were identified as RGS16 target genes such as NF-κB, E-cadherin, CDKN1C and ING1. Among these genes, NF-κB is a key regulator of tumor cell proliferation and survival and holds potential therapeutic application in cancer. However, the precise mechanism underlying the regulation of RGS16 on NF-κB has not been elucidated. A recent study showed that RGS16 limits proliferation of breast cancer cells through regulation of PI3K/Akt signaling (27). Activation of the PI3K/Akt pathway has been reported to stimulate activation of NF-κB (38–40). Future work is needed to determine if combination therapy inhibits NF-κB through RGS16/PI3K/Akt axis.
This study also identified a group of nuclear receptors or their related factors whose expression is upregulated by combined inhibition of LSD1 and HDACs. Nuclear export of NR4A1 by drug treatment has been proposed to be involved in induction of apoptosis through conversion of Bcl-2 into a proapoptotic factor (41,42). Our observation that combination therapy induced cytoplasmic localization of NR4A1 and significantly decreased Bcl-2 protein and increased Bax protein supports a proapoptotic role of NR4A1 in response to drug treatment. However, simultaneous treatment with NR4A1 siRNA failed to rescue LSD1-KD enhanced SAHA cytotoxicity, suggesting that manipulation of NR4A1 expression alone may not suffice to alter combination therapy-induced growth inhibition. Future investigation using concurrent siRNA depletion on multiple NR4A1-associated factors may help understand its precise role in regulating activity of crosstalk between LSD1 and HDACs. Reactivation of another important nuclear hormone receptor ERα was also found in this study, perhaps predisposing to a therapeutic intervention in the treatment of ER-negative breast cancers through restoration of sensitivity to antiestrogen therapy. CRABP2 is a protein that transports RA to the nucleus and regulates the access of RA to nuclear retinoic acid receptors. Overexpression of CRABP2 has been reported to greatly enhance the sensitivity of breast cancer cells to RA-induced growth arrest (43). Thus, restoration of CRABP2 by targeting histone abnormality may have clinical significance in promoting the efficacy of RA in breast cancer therapy.
In sum, the data from these studies provide important insight into the mechanisms underlying histone demethylation as it relates to HDACi-mediated gene regulation and tumor growth inhibition. These studies will be instrumental in designing more effective schedules and combination strategies that could have the potential to improve the treatment of breast cancer.
Supplementary material
Supplementary Tables 1–3 and Figures 1–4 can be found at http://carcin.oxfordjournals.org/
Funding
National Institutes of Health (CA88843); the Breast Cancer Research Foundation; Samuel and Emma Winters Foundation and Competitive Medical Research Fund of the University of Pittsburgh Medical Center Health System.
Supplementary Material
Acknowledgement
We thank UPCI Cancer Biomarker Facility. University of Pittsburgh Cancer Institute shared resources supported by NCI P30CA047904.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- BIK
Bcl-2-interacting killer
- ChIP
chromatin immunoprecipitation
- CI
combination index
- ER
estrogen receptor
- HDAC
histone deacetylases
- HDACi
HDAC inhibitors
- KD
knockdown
- LSD1
lysine-specific demethylase 1
- MTT
3-(4,5-dimethylthiazole-2-yl)-2,5-diphenyl tetrazolium bromide
- NF-κB
nuclear factor-kappaB
- PCDH1
protocadherin 1
- qPCR
quantitative PCR
- RA
retinoic acid
- RGS16
regulator of G-protein signaling 16
- siRNA
small interfering RNA
- TNBC
triple-negative breast cancer
- TSG
tumor suppressor genes.
References
- 1. Ficner R. (2009). Novel structural insights into class I and II histone deacetylases. Curr. Top. Med. Chem, 9, 235–240 [DOI] [PubMed] [Google Scholar]
- 2. Marks P.A, et al. (2009). Histone deacetylase inhibitors: potential in cancer therapy. J. Cell. Biochem, 107, 600–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sharma D, et al. (2006). Restoration of tamoxifen sensitivity in estrogen receptor-negative breast cancer cells: tamoxifen-bound reactivated ER recruits distinctive corepressor complexes. Cancer Res, 66, 6370–6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pruitt K, et al. (2006). Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet, 2, e40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Traynor A.M, et al. (2009). Vorinostat (NSC# 701852) in patients with relapsed non-small cell lung cancer: a Wisconsin Oncology Network phase II study. J. Thorac. Oncol, 4, 522–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Luu T.H, et al. (2008). A phase II trial of vorinostat (suberoylanilide hydroxamic acid) in metastatic breast cancer: a California Cancer Consortium study. Clin. Cancer Res, 14, 7138–7142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Modesitt S.C, et al. (2008). A phase II study of vorinostat in the treatment of persistent or recurrent epithelial ovarian or primary peritoneal carcinoma: a Gynecologic Oncology Group study. Gynecol. Oncol, 109, 182–186 [DOI] [PubMed] [Google Scholar]
- 8. Blumenschein G.R., Jr, et al. (2008). Phase II trial of the histone deacetylase inhibitor vorinostat (Zolinza, suberoylanilide hydroxamic acid, SAHA) in patients with recurrent and/or metastatic head and neck cancer. Invest. New Drugs, 26, 81–87 [DOI] [PubMed] [Google Scholar]
- 9. Belinsky S.A, et al. (2003). Inhibition of DNA methylation and histone deacetylation prevents murine lung cancer. Cancer Res, 63, 7089–7093 [PubMed] [Google Scholar]
- 10. Lee M.J, et al. (2008). Histone deacetylase inhibitors in cancer therapy. Curr. Opin. Oncol, 20, 639–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rasheed W.K, et al. (2007). Histone deacetylase inhibitors in cancer therapy. Expert Opin. Investig. Drugs, 16, 659–678 [DOI] [PubMed] [Google Scholar]
- 12. Huang Y, et al. (2007). Inhibition of lysine-specific demethylase 1 by polyamine analogues results in reexpression of aberrantly silenced genes. Proc. Natl Acad. Sci. USA, 104, 8023–8028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang Y, et al. (2009). Novel oligoamine analogues inhibit lysine-specific demethylase 1 and induce reexpression of epigenetically silenced genes. Clin. Cancer Res, 15, 7217–7228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang Y, et al. (2012). Inhibitors of histone demethylation and histone deacetylation cooperate in regulating gene expression and inhibiting growth in human breast cancer cells. Breast Cancer Res. Treat, 131, 777–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang Y, et al. (2009). Polyamine analogues targeting epigenetic gene regulation. Essays Biochem, 46, 95–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shi Y, et al. (2004). Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell, 119, 941–953 [DOI] [PubMed] [Google Scholar]
- 17. Lee M.G, et al. (2005). An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature, 437, 432–435 [DOI] [PubMed] [Google Scholar]
- 18. Garcia-Bassets I, et al. (2007). Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell, 128, 505–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lim S, et al. (2010). Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis, 31, 512–520 [DOI] [PubMed] [Google Scholar]
- 20. Metzger E, et al. (2005). LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature, 437, 436–439 [DOI] [PubMed] [Google Scholar]
- 21. Abukhdeir A.M, et al. (2006). Physiologic estrogen receptor alpha signaling in non-tumorigenic human mammary epithelial cells. Breast Cancer Res. Treat, 99, 23–33 [DOI] [PubMed] [Google Scholar]
- 22. Huang Y, et al. (2003). A novel polyamine analog inhibits growth and induces apoptosis in human breast cancer cells. Clin. Cancer Res, 9, 2769–2777 [PMC free article] [PubMed] [Google Scholar]
- 23. Chou T.C, et al. (1984). Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul, 22, 27–55 [DOI] [PubMed] [Google Scholar]
- 24. Tusher V.G, et al. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA, 98, 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alexopoulou A.N, et al. (2010). Dissecting the transcriptional networks underlying breast cancer: NR4A1 reduces the migration of normal and breast cancer cell lines. Breast Cancer Res, 12, R51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Novak P, et al. (2008). Agglomerative epigenetic aberrations are a common event in human breast cancer. Cancer Res, 68, 8616–8625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liang G, et al. (2009). RGS16 inhibits breast cancer cell growth by mitigating phosphatidylinositol 3-kinase signaling. J. Biol. Chem, 284, 21719–21727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Neve R.M, et al. (2006). A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell, 10, 515–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou Q, et al. (2007). Histone deacetylase inhibitor LBH589 reactivates silenced estrogen receptor alpha (ER) gene expression without loss of DNA hypermethylation. Cancer Biol. Ther, 6, 64–69 [DOI] [PubMed] [Google Scholar]
- 30. Jones P.A, et al. (2007). The epigenomics of cancer. Cell, 128, 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang Y, et al. (2011). Epigenetics in breast cancer: what’s new? Breast Cancer Res, 13, 225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu Q, et al. (1997). Inhibition of trans-retinoic acid-resistant human breast cancer cell growth by retinoid X receptor-selective retinoids. Mol. Cell. Biol, 17, 6598–6608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moll U.M, et al. (2006). p53 and Nur77/TR3 - transcription factors that directly target mitochondria for cell death induction. Oncogene, 25, 4725–4743 [DOI] [PubMed] [Google Scholar]
- 34. Druey K.M, et al. (1996). Inhibition of G-protein-mediated MAP kinase activation by a new mammalian gene family. Nature, 379, 742–746 [DOI] [PubMed] [Google Scholar]
- 35. De Vries L, et al. (1999). RGS proteins: more than just GAPs for heterotrimeric G proteins. Trends Cell Biol, 9, 138–144 [DOI] [PubMed] [Google Scholar]
- 36. Sun G, et al. (2010). Histone demethylase LSD1 regulates neural stem cell proliferation. Mol. Cell. Biol, 30, 1997–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wiechec E, et al. (2008). A fragile site within the HPC1 region at 1q25.3 affecting RGS16, RGSL1, and RGSL2 in human breast carcinomas. Genes. Chromosomes Cancer, 47, 766–780 [DOI] [PubMed] [Google Scholar]
- 38. Ozes O.N, et al. (1999). NF-kappaB activation by tumour necrosis factor requires the Akt serine-threonine kinase. Nature, 401, 82–85 [DOI] [PubMed] [Google Scholar]
- 39. deGraffenried L.A, et al. (2004). NF-kappa B inhibition markedly enhances sensitivity of resistant breast cancer tumor cells to tamoxifen. Ann. Oncol, 15, 885–890 [DOI] [PubMed] [Google Scholar]
- 40. Gong L, et al. (2003). Inactivation of NF-kappaB by genistein is mediated via Akt signaling pathway in breast cancer cells. Oncogene, 22, 4702–4709 [DOI] [PubMed] [Google Scholar]
- 41. Yoon K, et al. (2011). Activation of nuclear TR3 (NR4A1) by a diindolylmethane analog induces apoptosis and proapoptotic genes in pancreatic cancer cells and tumors. Carcinogenesis, 32, 836–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mohan H.M, et al. (2012). Molecular pathways: the role of NR4A orphan nuclear receptors in cancer. Clin. Cancer Res, 18, 3223–3228 [DOI] [PubMed] [Google Scholar]
- 43. Budhu A.S, et al. (2002). Direct channeling of retinoic acid between cellular retinoic acid-binding protein II and retinoic acid receptor sensitizes mammary carcinoma cells to retinoic acid-induced growth arrest. Mol. Cell. Biol, 22, 2632–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.