Abstract
Single-cell level measurements are necessary to characterize the intrinsic biological variability in a population of cells. In this study, we demonstrate that, with the microarrays for mass spectrometry platform, we are able to observe this variability. We monitor environmentally (2-deoxy-d-glucose) and genetically (ΔPFK2) perturbed Saccharomyces cerevisiae cells at the single-cell, few-cell, and population levels. Correlation plots between metabolites from the glycolytic pathway, as well as with the observed ATP/ADP ratio as a measure of cellular energy charge, give biological insight that is not accessible from population-level metabolomic data.
Keywords: single-cell measurements, MALDI mass spectrometry, baker's yeast
Even genetically identical cells present in the same microenvironment can express different phenotypes, for a number of reasons: cell-to-cell heterogeneity can stem from differences in the cell age and differences in the cell cycle stage, and stochastic effects together with feedback mechanisms can lead to distinctively different phenotypes, too (1–6). As population-level measurement techniques inherently average out such cell-to-cell differences, biochemical mechanisms underlying a studied system cannot be deduced from such measurements. Thus, to detect and exploit this heterogeneity, new analytical platforms with a sensitivity at the single-cell level and the ability to perform quantitative analyses must be developed and validated.
Motivated by advances of mass spectrometry (MS) in metabolomics, the analytical chemistry community has stepped up its efforts toward realizing MS-based single-cell metabolomics (1, 2). A number of analytical approaches were developed with detection limits low enough for single-cell metabolite analyses [e.g., nanostructured surfaces (7, 8), postionization techniques (9, 10), modified laser optics (11), the use of microsampling tools (12, 13), microarrays for MS measurements (14, 15), etc.]. Until now, however, most MS studies targeting single-cell metabolite analysis have only shown the analytical capabilities, but have not demonstrated that true biological information can be retrieved from studying the metabolism of single cells.
Here, using the unicellular eukaryotic model organism Saccharomyces cerevisiae, we present an analytical validation of a single-cell metabolite analysis using the microarrays for mass spectrometry (MAMS) platform. This validation concerns both the analytical methodology and the biological information, by monitoring expected cellular responses upon an environmental and a genetic perturbation. Furthermore, we present examples of biological insight that are only accessible with a platform such as MAMS. Specifically, we unravel metabolite–metabolite correlations, and visualize coexisting subpopulations in an isogenic cell culture. This technology can now be used to reveal metabolic differences in cells of isogenic cell populations, such as differences caused by cell cycles stages, cell ages, or stochastically induced phenotypic differences.
Results and Discussion
MAMS represent a type of substrate for matrix-assisted laser desorption/ionization–mass spectrometry (MALDI-MS) recently introduced by our group (14, 15). MAMS uses a combination of hydrophilic reservoirs surrounded by an omniphobic surface (SI Text) for massively parallel, automated sample spotting.
Briefly, cells are taken from a liquid culture, quenched using cold solvents (to stop any metabolic activity), after which the supernatant is removed and cells are washed to remove salts. The cell suspension is then spread onto the MAMS substrate (Fig. 1). Applying the cell suspension onto the MAMS surface will result in an automated aliquoting of the cell suspension into the hydrophilic reservoirs, without the need for a microspotter. Dependent on the cell concentration used, the number of cells on each hydrophilic reservoir can be between zero and hundreds. The transparency of the MAMS substrate allows for microscopic analysis to determine the number of cells in each reservoir while the cellular metabolism remains quenched because the entire MAMS chip is kept cold in a cryochamber.
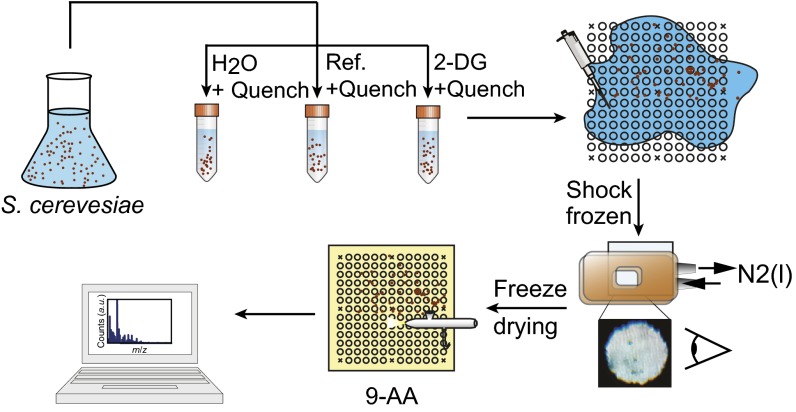
Fig. 1.
Graphical summary of the work flow used to prepare the samples for single-cell MALDI-MS analysis. Cellular metabolism is quenched by adding cold solvent. After centrifugation and discarding the supernatant, the pellet is reconstituted, and the cell suspension is spread onto a cold MAMS chip. This generates many uniform, liquid droplets trapped in the hydrophilic reservoirs on the MAMS chip that each contain ensemble of cells. The exact number of cells within a reservoir is given by a Poisson distribution and depends on the density of the cell suspension. After counting the cells under a microscope, a MALDI matrix (9-aminoacridine) is applied with an airbrush, and each reservoir on the plate is analyzed with MALDI-MS. Signals of metabolites from a reservoir are normalized by a unsaturated matrix background signal and by the number of cells found in the respective reservoir (containing between 1 and 15 cells) to reduce artifacts associated with differences in the cell size (biomass) and obtain the equivalent of a single-cell signal. See SI Text for details.
The MALDI matrix used (9-aminoacridine) and the possibility of cocrystallizing cells with the matrix within the defined reservoirs allow us to boost the sensitivity beyond that of traditional MALDI mass spectrometry (SI Text). Using the MAMS platform, we can reach the level of 100 amol to 10 fmol—a range on the order of the metabolite levels in a single yeast cell (14, 15).
MAMS Is Capable to Detect Biological Information from Single/Few Cells.
In principle, the MAMS platform is ready to be used to resolve cell-to-cell metabolic heterogeneity. However, because MS-based methods are destructive, we cannot make multiple measurements of the same sample to assess the measurement uncertainty. However, our samples (i.e., the S. cerevisiae cells) have an intrinsic biological variability. Thus, replicate measurements on different cells will inherently result in variable metabolite levels, although it is expected that the overall metabolism would present certain similarities. Therefore, for validating our single-cell metabolomics platform, we need to show that the signals acquired represent the biological variability and not just measurement noise (i.e., instrumental variability).
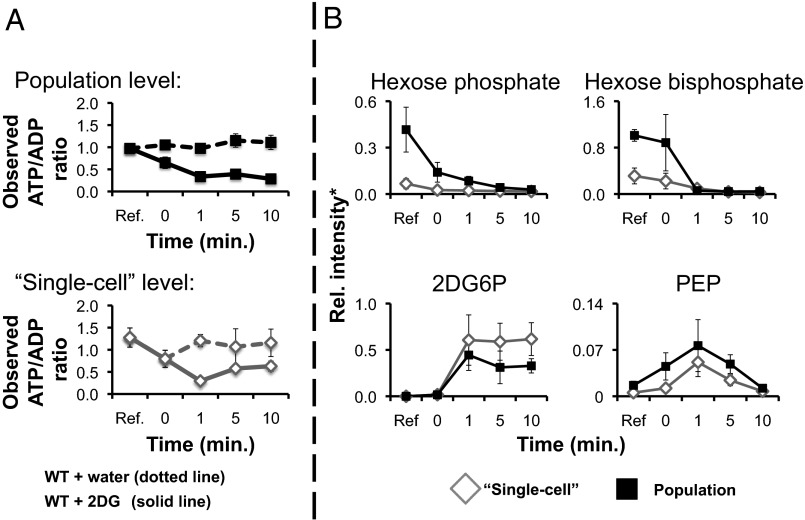
To demonstrate that MAMS is indeed able to retrieve biological information from a single or a small ensemble of yeast cells positioned in the MAMS reservoirs, we perturbed them with 2-deoxy-d-glucose (2DG), a drug that blocks glycolysis (16, 17). We dynamically monitored the response both with MAMS on the single-cell and near–single-cell level, and with traditional MALDI-MS on the population level, and compared the response of the two measurements. We indeed found that the addition of 2% (wt/vol) 2DG to the growth medium generates similar trends in both the population-level as well as in few-cell level analyses made using the MAMS platform (Fig. 2). Specifically, the quantitative change in glycolytic metabolites and the drop in the energy charge (represented by the observed ATP/ADP ratio) 1 min after the addition of 2DG to the growth medium is consistent with earlier findings on the population level (Fig. 2A) (18, 19). Furthermore, we found that, in both 2DG-treated samples (i.e., MAMS and population-level samples), we can detect 2-deoxy-d-glucose-6-phosphate (2DG6P). 2DG6P, the product of the uptake and metabolism of 2DG, could not be detected in samples that were not drug treated (Fig. 2B). Together, this is a first indication that MAMS is actually monitoring biological processes.
Fig. 2.
(A) Dynamic population-level measurements with a traditional bulk-level MALDI-MS metabolomic study and an average of multiple single-cell level metabolomic measurements with MAMS after applying 2DG. Reference, untreated cells; control, cells treated with doubly deionized water; treated, cells treated with 2DG. The spike of water and 2DG occurs at time = 0 min. (B) Metabolite time study for 2DG-treated samples. Here, we present the changes observed in time for selected metabolites of the glycolytic pathway. Some of the assayed glycolytic metabolites are isobaric (i.e., they have the same molecular weight); thus, they cannot be differentiated using mass spectrometry alone. For example, glucose-6-phosphate (G6P) and fructose-6-phosphate (F6P) are therefore summarized as hexose phosphate. *The relative ion signal intensity for each plot metabolite is additionally normalized, in the case of the single-cell level measurement (alone), by the number of cells found in the MAMS reservoir (SI Text). The cell number distributions for the MAMS samples are as follows: (i) reference = 62 measurements (1.694 ± 1.521); (ii) control (t = 1 min) = 77 measurements (2.364 ± 1.356); (iii) control (t = 1 min) = 77 measurements (2.364 ± 1.356); (iv) control (t = 5 min) = 38 measurements (1.763 ± 1.651); (v) control (t = 10 min) = 76 measurements (4.500 ± 3.048); (vi) treated (t = 1 min) = 94 measurements (2.500 ± 1.435); (vii) treated (t = 5 min) = 24 measurements (10.542 ± 2.843); (viii) treated (t = 10 min) = 23 measurements (1.763 ± 1.651). The population-level data are as follows (cell number = ±20): (i) reference = 110 measurements; (ii) control (t = 1 min) = 8 measurements; (iii) control (t = 5 min) = 8 measurements; (iv) control (t = 10 min) = 8 measurements; (v) treated (t = 1 min) = 119 measurements; (vi) treated (t = 5 min) = 132 measurements; (vii) treated (t = 10 min) = 132 measurements. For detailed analytical instrumental conditions, see SI Text.
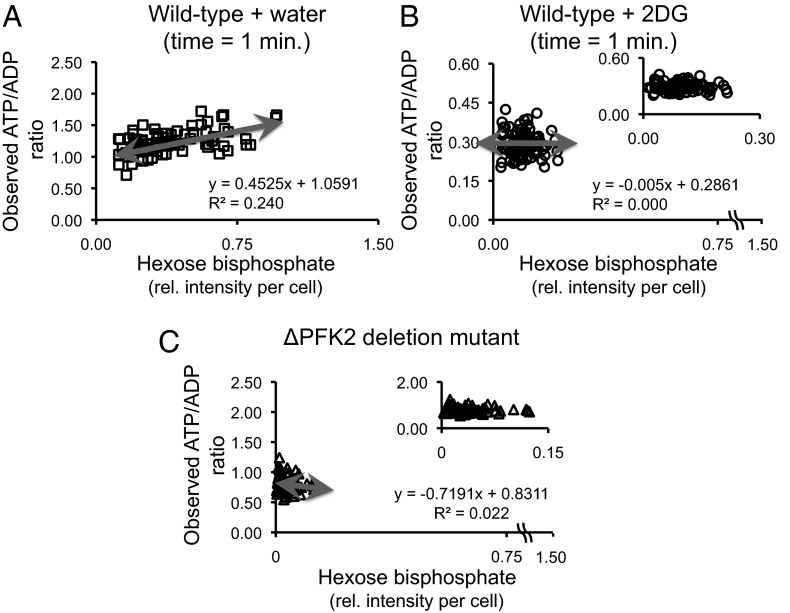
Focusing in on the metabolite signals that we retrieved from individual MAMS reservoirs (containing between 1 and 15 cells), we sought further evidence that the different signals reflect the biological variability rather than just the technical or analytical variability. Before drawing any correlation plots, the metabolite signals were normalized by a nonsaturated matrix signal and by the number of cells found in the respective reservoir (SI Text); then, we plotted the metabolite data retrieved from single reservoirs against each other. We observed a number of correlations between different metabolite signals (Fig. 3). Specifically, we found that fructose-1,6-bisphosphate (F16BP) and the observed ATP/ADP ratio are correlated for cells treated with water but not with 2DG (Fig. 3 A and B). This is expected, because 2DG6P inhibits the glycolysis pathway upstream of the phosphofructokinase (Pfk) enzyme, which generates F16BP (16, 17), rendering the production of ATP (and thus the observed ATP/ADP ratio) independent of the amount of F16BP present in the cell.
Fig. 3.
Relationship between the abundance of hexose bisphosphate and observed cellular ATP/ADP ratio. The correlation between the abundance of hexose bisphosphate [most probable fructose-1,6-bisphosphate (F16BP)] vs. the observed cellular ATP/ADP ratio is shown 1 min after applying two different stress conditions to the growth media: (A) plus doubly deionized water (control); (B) plus 2-deoxy-d-glucose (treated) (SI Text). In C, a different perturbation (genetic) is used to simulate part of the effects of 2DG on the S. cerevisiae wild-type cells. The metabolite abundance is based on the value of the relative signal intensity of the metabolite in the MS spectra divided by an unsaturated matrix background signal and the number of cells in the respective MAMS reservoir (see SI Text for more details). Changes in the slope of the linear regression (red line) between the ”control” and ”treated” MAMS samples is due to the glycolytic inhibitor action of 2-deoxy-d-glucose in the yeast cells (for more details, see text). The cell number distributions for the MAMS samples are as follows: (i) wild type plus water (t = 1 min) = 77 measurements (2.364 ± 1.356); (ii) wild type plus 2DG (t = 1 min) = 94 measurements (2.5 ± 1.435); (iii) ΔPFK2 deletion mutant = 73 measurements (2.836 ± 2.255).
To further validate that our observations are of biological origin, we generated a deletion mutant of the gene pfk2, which encodes an isoenzyme of Pfk. This mutant has reduced phosphofructokinase activity and is known to partially mimic the effect of 2DG (20). We also found a correlation between the observed ATP/ADP ratio and F16BP (Fig. 3C) that resembled the wild type treated with 2DG (Fig. 3B) rather than the wild-type control (Fig. 3A). Together, these findings indicate that MAMS is indeed able to retrieve signals from individual and small ensemble of cells, and that the data contain valid biological information, because averaged correlation plots made from random data would not show any significant biological trend. Thus, this is clearly not a measurement artifact.
MAMS Uncovers Metabolite–Metabolite Correlations.
Next, equipped with this confidence, we used MAMS to gain insight into the yeast metabolism. Above, we have shown a metabolite–metabolite correlation between hexose bisphosphate (most probably F16BP) and the observed ATP/ADP ratio (Fig. 3). Such a correlation is a consequence of the underlying metabolic and regulatory mechanisms, reflecting the system’s properties (21). Previously, metabolite–metabolite correlations have been reported with population-level measurements on samples that had quite different origin, i.e., different plant tissues (21–24). Unlike what was done with tissues, we exploit the intrinsic biological variability on the single-cell level to uncover such metabolite–metabolite correlations.
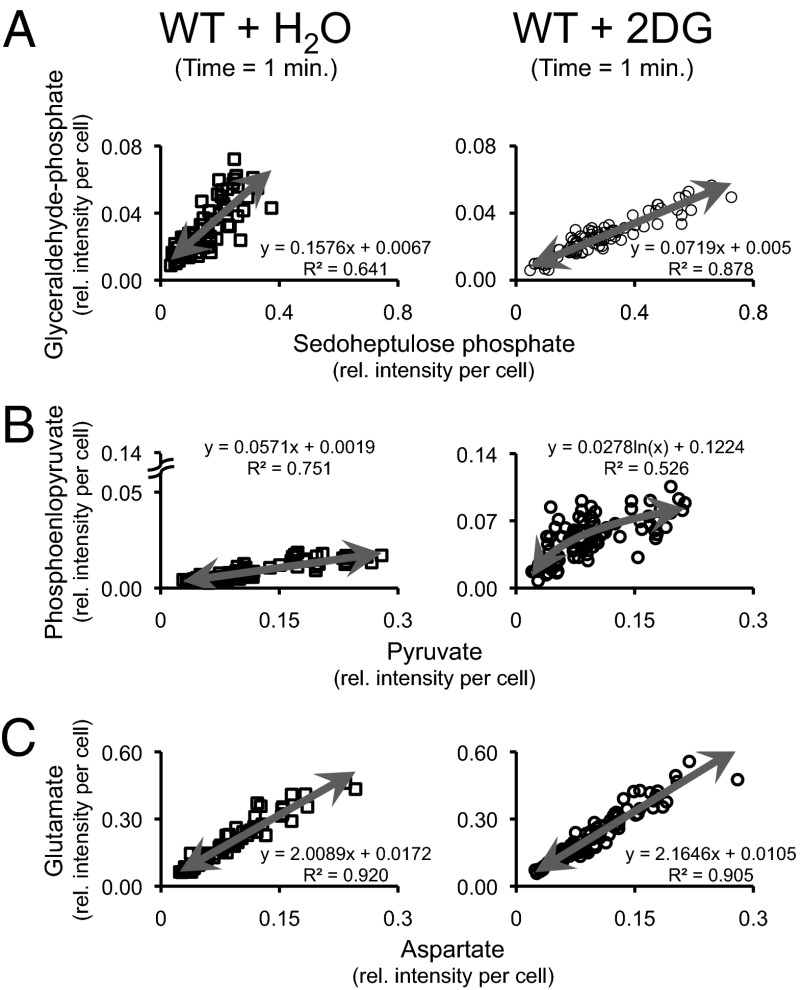
Correlations (and in particular positive correlations), however, could simply also emerge from variability in cell size, with larger cells generating higher metabolite signals. If this would be the case, then all metabolite–metabolite correlations would show the same profile. That this is not the case becomes obvious when comparing metabolite–metabolite correlations obtained after different environmental perturbations: although control samples and 2DG-treated samples show differences in some correlations, others are identical (Fig. 4), indicating that the correlations must report on metabolic system properties, because if it were a mere cell size effect, the effect of the perturbation would have been the same for all pairs.
Fig. 4.
Pairwise metabolite–metabolite correlations can be used to unravel slight differences in the metabolic network regulation after the addition of a drug (2-deoxyglucose). (A) Glyceraldehyde-3-phosphate is solely produced by the pentose phosphate pathway (represented by sedoheptulose-7-phosphate). (B) Low glycolytic fluxes in the cell due to the presence of 2DG increase the amount of phosphoenolpyruvate, which itself activates the pentose phosphate pathway (previously reported in refs. 24 and 25). (C) Glutamate is a key compound in cellular metabolism, whose biosynthesis (from its precursor aspartate) is apparently unmodified by the presence of 2DG in the growth medium. Glutamate can thus be used as a control for A and B. The cell number distributions for the MAMS samples are as follows: (i) wild type plus water (t = 1 min) = 77 measurements (2.364 ± 1.356); (ii) wild type plus 2DG (t = 1 min) = 94 measurements (2.5 ± 1.435).
One prominent correlation is the one between glyceraldehyde-3-phosphate and sedoheptulose-7-phosphate (Fig. 4A). Glyceraldehyde-3-phosphate is produced via the glycolysis or via the pentose phosphate pathway (PPP), to which the metabolite sedoheptulose-7-phosphate belongs. After blocking the glycolysis-related synthesis of glyceraldehyde-3-phosphate with 2DG, the correlation between sedoheptulose-7-phosphate and glyceraldehyde-3-phosphate becomes more obvious (Fig. 4A), indicating that the synthesis of glyceraldehyde-3-phosphate is dependent of sedoheptulose-7-phosphate and the PPP. Another correlation is found between phosphoenolpyruvate (PEP) and pyruvate (PYR) (Fig. 4B)—two intermediates of glycolysis. Although PYR and PEP have a linear relationship for the control samples, the 2DG-treated samples show that the amount of PEP present seems to level off at higher PYR concentrations (Fig. 4B). Finally, we found a strong correlation between glutamic acid and aspartic acid (Fig. 4C). This correlation can be considered as an analytical/biological control for the previous presented correlations, because neither the addition of 2DG nor water affected the correlation between these metabolites during the time frame covered in our experiments (10 min), because these two metabolites are too far away from the entry point of the added 2DG.
MAMS Uncovers Different Phenotypes.
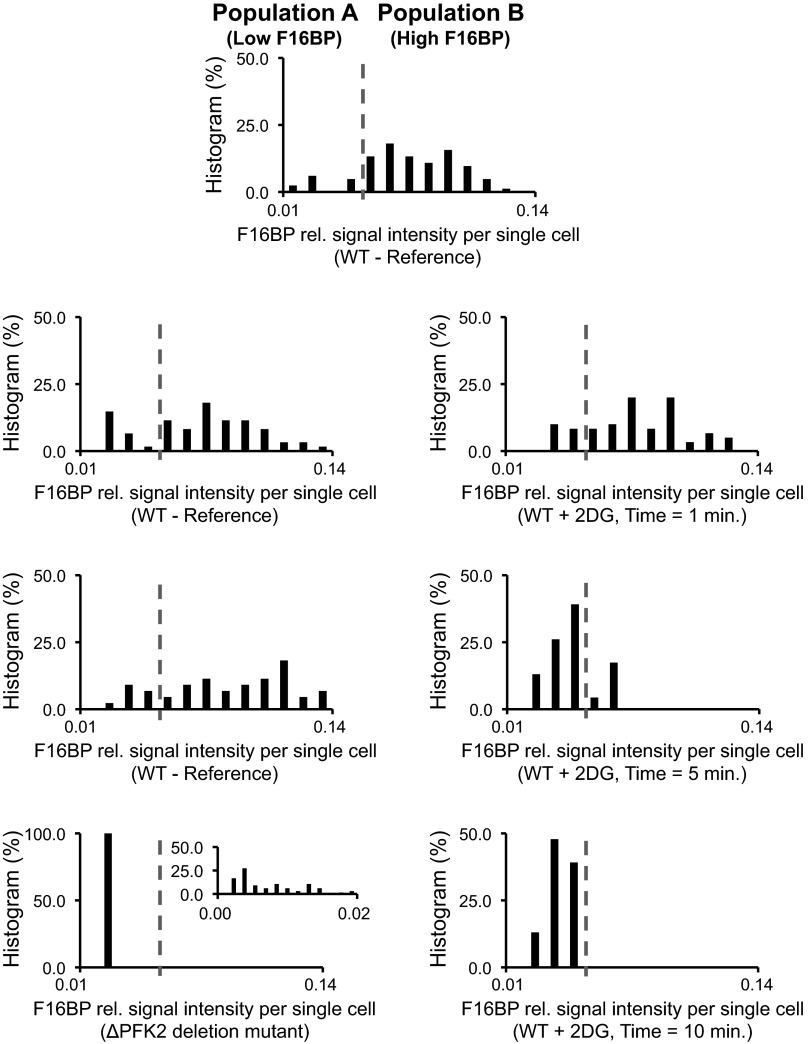
Next, we focused on the glycolytic metabolite F16BP. The MAMS platform revealed evidence for the presence of two different phenotypes present in the cell population: one phenotype characterized by low levels of F16BP and one with high levels (Fig. 5). Independent replicate experiments demonstrated that this bimodal distribution is not a measurement artifact (Fig. 5, left panels), but rather a robust feature of heterogeneous single-cell behavior that is now uncovered with the MAMS platform.
Fig. 5.
The F16BP signal intensity frequency distributions using an equal number of bins (between 0.0083 and 0.01439) for S. cerevisiae cells detected using the MAMS platform at the ”single-cell” level under different environmental conditions (reference, treated samples with 2DG, and the ΔPFK2 deletion mutant). The histogram of the latter is blown as an Inset (using the same number of bins as in the other histograms between the values of 0.0006 and 0.0186). The cell number distributions for the MAMS samples are as follows: (i) reference (1) = 62 measurements (1.694 ± 1.521); (ii) reference (2) = 83 measurements (1.337 ± 0.859); (iii) reference (3) = 42 measurements (1.452 ± 0.942); (iv) ΔPFK2 deletion mutant = 73 measurements (2.836 ± 2.255); (v) treated (t = 1 min) = 94 measurements (2.500 ± 1.435); (vi) treated (t = 5 min) = 24 measurements (10.542 ± 2.843); (vii) treated (t = 10 min) = 23 measurements (1.763 ± 1.651). For detailed analytical instrumental conditions, see SI Text.
Measurements on the population level revealed that F16BP levels correlate with the magnitude of glycolytic flux (25). Using this information, we speculated that the two populations found here are characterized by glycolytic fluxes of two distinct magnitudes. Indeed, when we followed the F16BP levels upon the addition of 2DG—a treatment blocking glycolysis upstream of F16BP—the phenotype with the high F16BP levels (high flux, population B) disappears. Consistently, the phenotype at high F16BP levels is also not present in the ΔPFK2 deletion mutant (Fig. 5), which is a mutation that only realizes a low glycolytic flux.
We do not know the cause for the presence of these two distinct phenotypes. Although it is tempting to speculate that it might be cell cycle related, with cells in the G1 phase having high and cells in the S phase, low F16BP levels (26), only future studies on the metabolism of single cells—now enabled by the MAMS platform—will reveal the true cause.
Conclusions
In this work, we addressed one of the most difficult challenges associated with single-cell level analysis of metabolites, validation. We reduced the technical (or analytical) variability to be able to observe the naturally occurring cell-to-cell heterogeneity. Furthermore, we presented a qualitative biological study based on a comprehensive dataset that shows the feasibility of monitoring Saccharomyces cerevisiae metabolism at the single-cell and few-cell level.
These advances were feasible thanks to the use of the MAMS platform. The possibility of carrying out a single-cell level metabolic analysis, which takes advantage of the naturally occurring cell-to-cell variability and exploits it to directly visualize two different phenotypes (both phenotypes characterized by different levels of F16BP), gives us insight into metabolic networks as well as regulation processes without the need of applying a nonnatural perturbation (e.g., genetic modification that favors one particular phenotype).
We believe that in the near future the use of the MAMS platform will allow us to isolate data from subgroups of cells to investigate, in more depth, either bistability or metabolite/pathway interactions. The analytical variability that is still present in our measurements should still be further decreased. An increase in the number of data points for more extensive statistical studies of one particular subpopulation (e.g., multivariate data processing of metabolic profiles) would also be highly desirable. The number of reservoirs on the MAMS chips used here was limited to 160 per time point. However, available microfabrication techniques allow us to increase the density of the array to 250,000 wells on a single glass slide. This will be relevant for visualizing system-level interactions of metabolic pathways by observing metabolites that show a bimodal or higher order distribution.
Materials and Methods
All raw MS data collected for this publication have been uploaded to a European Molecular Biology Organization database. The MS data and metadata are available to the reader at MetaboLights (www.ebi.ac.uk/metabolights/), under the study identifier MTBLS29. The reader may also contact the corresponding authors for additional details of the experiments presented here.
Additional information, such as MAMS chip fabrication, cell culture and handling conditions, MALDI-MS measurement parameters, and data treatment can be found in SI Text.
Supplementary Material
Acknowledgments
A.I. gratefully acknowledges Dr. Madina Mansurova for her advice and thoughtful insights, the help of Guille Zampar for statistical interpretation of the data, the support of Robert Steinhoff, Jasmin Krismer, and Martin Pabst, and financial support by the Seventh European Community Framework through the program Marie Curie Intra-European Fellowship (PIEF-GA-2011-271895-DiSC-MS).
Footnotes
Conflict of interest statement: A.I., S.R.F., P.L.U., K.J., and R.Z. are developing a commercial product based on MAMS technology.
This article is a PNAS Direct Submission.
Database deposition: The MS data and metadata are available at MetaboLights, www.ebi.ac.uk/metabolights (study identifier: MTBLS29).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209302110/-/DCSupplemental.
References
- 1.Heinemann M, Zenobi R. Single cell metabolomics. Curr Opin Biotechnol. 2011;22(1):26–31. doi: 10.1016/j.copbio.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 2.Walling MA, Shepard JRE. Cellular heterogeneity and live cell arrays. Chem Soc Rev. 2011;40(7):4049–4076. doi: 10.1039/c0cs00212g. [DOI] [PubMed] [Google Scholar]
- 3.Raj A, van Oudenaarden A. Nature, nurture, or chance: Stochastic gene expression and its consequences. Cell. 2008;135(2):216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dubnau D, Losick R. Bistability in bacteria. Mol Microbiol. 2006;61(3):564–572. doi: 10.1111/j.1365-2958.2006.05249.x. [DOI] [PubMed] [Google Scholar]
- 5.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305(5690):1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 6.Sumner ER, Avery SV. Phenotypic heterogeneity: Differential stress resistance among individual cells of the yeast Saccharomyces cerevisiae. Microbiology. 2002;148(Pt 2):345–351. doi: 10.1099/00221287-148-2-345. [DOI] [PubMed] [Google Scholar]
- 7.O’Brien , et al. 2013 Monitoring metabolic responses to chemotherapy in single cells and tumors using nanostructure-initiator mass spectrometry (NIMS) imaging. Cancer Metab 1:4. [Google Scholar]
- 8.Amantonico A, Flamigni L, Glaus R, Zenobi R. Negative mode nanostructure-initiator mass spectrometry for detection of phosphorylated metabolites. Metabolomics. 2009;5:346–353. [Google Scholar]
- 9.Shrestha B, Nemes P, Vertes A. Ablation and analysis of small cell populations and single cells by consecutive laser pulses. Appl Phys A Mater Sci Process. 2010;101:121–126. [Google Scholar]
- 10.Shrestha B, Vertes A. In situ metabolic profiling of single cells by laser ablation electrospray ionization mass spectrometry. Anal Chem. 2009;81(20):8265–8271. doi: 10.1021/ac901525g. [DOI] [PubMed] [Google Scholar]
- 11.Bouschen W, Schulz O, Eikel D, Spengler B. Matrix vapor deposition/recrystallization and dedicated spray preparation for high-resolution scanning microprobe matrix-assisted laser desorption/ionization imaging mass spectrometry (SMALDI-MS) of tissue and single cells. Rapid Commun Mass Spectrom. 2010;24(3):355–364. doi: 10.1002/rcm.4401. [DOI] [PubMed] [Google Scholar]
- 12.Hölscher D, et al. Matrix-free UV-laser desorption/ionization (LDI) mass spectrometric imaging at the single-cell level: Distribution of secondary metabolites of Arabidopsis thaliana and Hypericum species. Plant J. 2010;60:907–918. doi: 10.1111/j.1365-313X.2009.04012.x. [DOI] [PubMed] [Google Scholar]
- 13.Rubakhin SS, Sweedler JV. Characterizing peptides in individual mammalian cells using mass spectrometry. Nat Protoc. 2007;2(8):1987–1997. doi: 10.1038/nprot.2007.277. [DOI] [PubMed] [Google Scholar]
- 14.Urban PL, et al. Carbon-13 labelling strategy for studying the ATP metabolism in individual yeast cells by micro-arrays for mass spectrometry. Mol Biosyst. 2011;7(10):2837–2840. doi: 10.1039/c1mb05248a. [DOI] [PubMed] [Google Scholar]
- 15.Urban PL, et al. High-density micro-arrays for mass spectrometry. Lab Chip. 2010;10(23):3206–3209. doi: 10.1039/c0lc00211a. [DOI] [PubMed] [Google Scholar]
- 16.Pelicano H, Martin DS, Xu RH, Huang P. Glycolysis inhibition for anticancer treatment. Oncogene. 2006;25(34):4633–4646. doi: 10.1038/sj.onc.1209597. [DOI] [PubMed] [Google Scholar]
- 17.Walther T, et al. Control of ATP homeostasis during the respiro-fermentative transition in yeast. Mol Syst Biol. 2010;6:344. doi: 10.1038/msb.2009.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ralser M, et al. A catabolic block does not sufficiently explain how 2-deoxy-D-glucose inhibits cell growth. Proc Natl Acad Sci USA. 2008;105(46):17807–17811. doi: 10.1073/pnas.0803090105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Randez-Gil F, Blasco A, Prieto JA, Sanz P. DOGR1 and DOGR2: Two genes from Saccharomyces cerevisiae that confer 2-deoxyglucose resistance when overexpressed. Yeast. 1995;11(13):1233–1240. doi: 10.1002/yea.320111303. [DOI] [PubMed] [Google Scholar]
- 20.Pearce AK, et al. Pyruvate kinase (Pyk1) levels influence both the rate and direction of carbon flux in yeast under fermentative conditions. Microbiology. 2001;147(Pt 2):391–401. doi: 10.1099/00221287-147-2-391. [DOI] [PubMed] [Google Scholar]
- 21.Steuer R. Review: On the analysis and interpretation of correlations in metabolomic data. Brief Bioinform. 2006;7(2):151–158. doi: 10.1093/bib/bbl009. [DOI] [PubMed] [Google Scholar]
- 22.Morgenthal K, Weckwerth W, Steuer R. Metabolomic networks in plants: Transitions from pattern recognition to biological interpretation. Biosystems. 2006;83(2–3):108–117. doi: 10.1016/j.biosystems.2005.05.017. [DOI] [PubMed] [Google Scholar]
- 23.Camacho D, de la Fuente A, Mendes P. The origin of correlations in metabolomics data. Metabolomics. 2005;1:53–63. [Google Scholar]
- 24.Steuer R, Kurths J, Fiehn O, Weckwerth W. Observing and interpreting correlations in metabolomic networks. Bioinformatics. 2003;19(8):1019–1026. doi: 10.1093/bioinformatics/btg120. [DOI] [PubMed] [Google Scholar]
- 25.Huberts DHEW, Niebel B, Heinemann M. A flux-sensing mechanism could regulate the switch between respiration and fermentation. FEMS Yeast Res. 2012;12(2):118–128. doi: 10.1111/j.1567-1364.2011.00767.x. [DOI] [PubMed] [Google Scholar]
- 26.Wittmann C, Hans M, van Winden WA, Ras C, Heijnen JJ. Dynamics of intracellular metabolites of glycolysis and TCA cycle during cell-cycle-related oscillation in Saccharomyces cerevisiae. Biotechnol Bioeng. 2005;89(7):839–847. doi: 10.1002/bit.20408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.