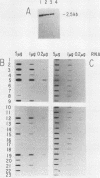
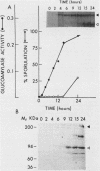
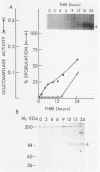
Abstract
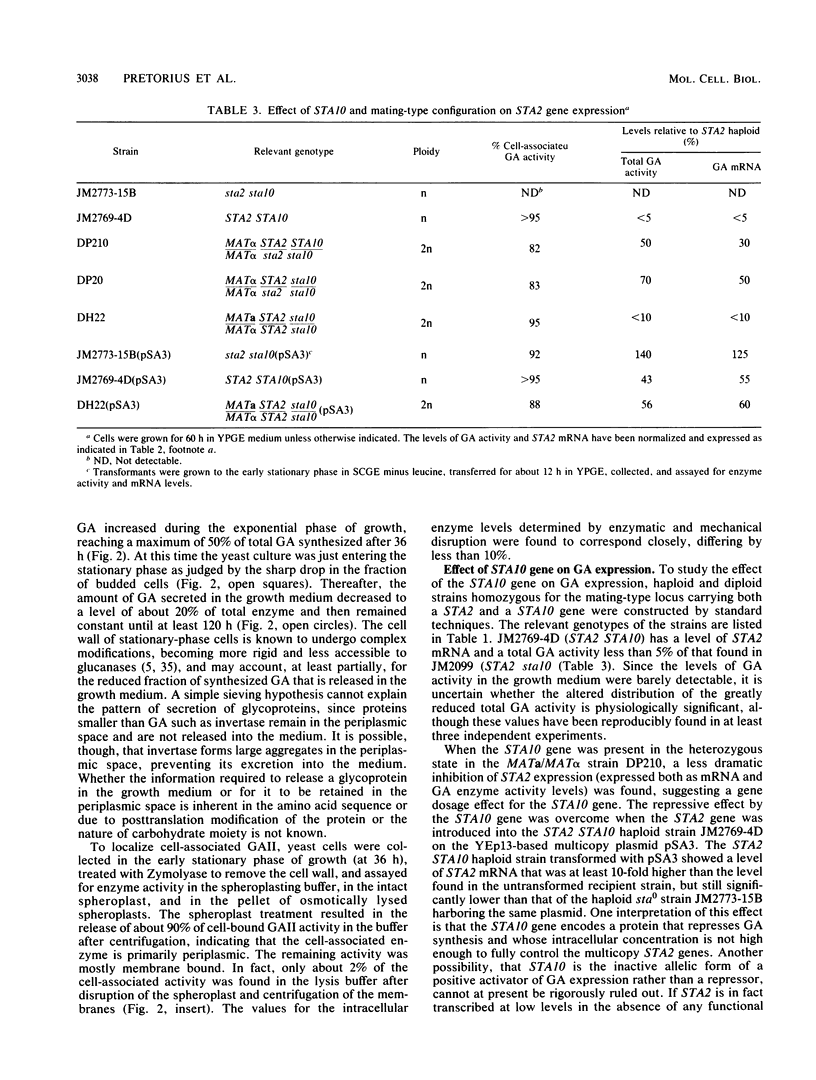
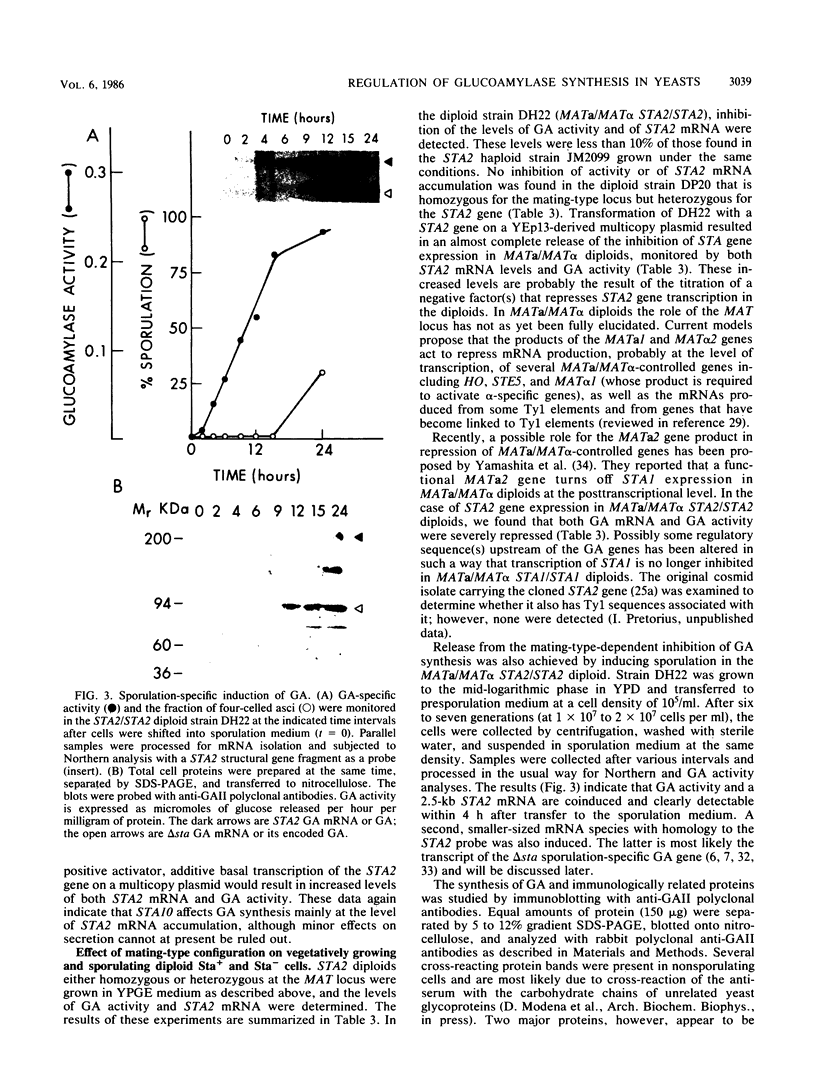
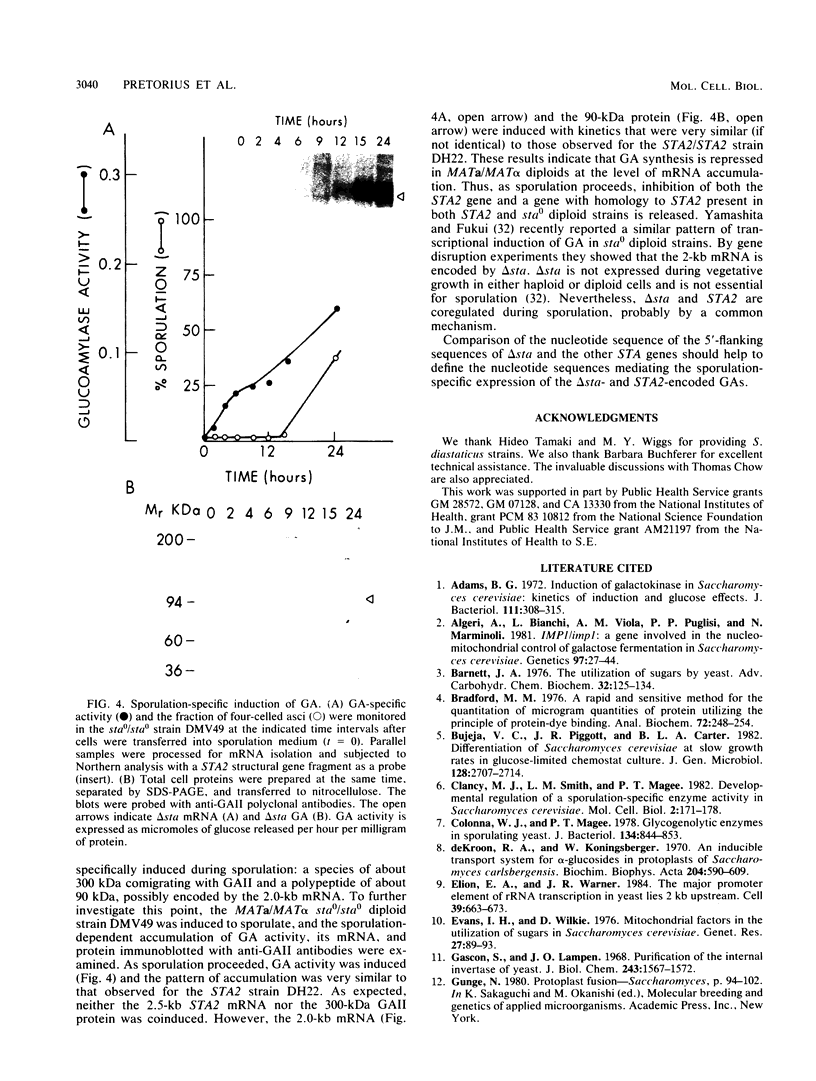
Three unlinked, homologous genes, STA1, STA2, and STA3, encode the extracellular glycosylated glucoamylase isozymes I, II, and III, respectively, in Saccharomyces species. S. cerevisiae, which is sta0 (absence of functional STA genes in haploids), does carry a glucoamylase gene, delta sta, expressed only during sporulation (W. J. Colonna and P. T. Magee, J. Bacteriol. 134:844-853, 1978; I. Yamashita and S. Fukui, Mol. Cell. Biol. 5:3069-3073, 1985). In this study we examined some of the physiological and genetic factors that affect glucoamylase expression. It was found that STA2 strains grown in synthetic medium produce glucoamylase only in the presence of either Maltrin M365 (a mixture of maltooligosaccharides) or starch. Maximal levels of glucoamylase activity were found in cells grown in rich medium supplemented with glycerol plus ethanol, starch, or Maltrin. When various sugars served as carbon sources they all supported glucoamylase synthesis, although at reduced levels. In any given growth medium glucoamylase isozyme II synthesis was modulated by functionality of the mitochondria. Synthesis of glucoamylase is continuous throughout the growth phases, with maximal secretion taking place in the early stationary phase. In the various regimens, the differences in enzyme accumulation are accounted for by differences in the levels of glucoamylase mRNA. Both glucoamylase mRNA and enzyme activity were drastically and coordinately inhibited in MATa/MAT alpha diploids and by the presence of the regulatory gene STA10. Both effects were partially overcome when the STA2 gene was present on a multicopy plasmid. The STA2 mRNA and glucoamylase were coinduced in sporulating STA2/STA2 diploids. A smaller, coinduced RNA species was also detected by Northern blotting with a STA2 probe. The same mRNA species was detected in sporulating sta0 diploids and is likely to encode the sporulation-specific glucoamylase.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams B. G. Induction of galactokinase in Saccharomyces cerevisiae: kinetics of induction and glucose effects. J Bacteriol. 1972 Aug;111(2):308–315. doi: 10.1128/jb.111.2.308-315.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Algeri A. A., Bianchi L., Viola A. M., Puglisi P. P., Marmiroli N. IMP1/imp1: a gene involved in the nucleo-mitochondrial control of galactose fermentation in Saccharomyces cerevisiae. Genetics. 1981 Jan;97(1):27–44. doi: 10.1093/genetics/97.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett J. A. The utilization of sugars by yeasts. Adv Carbohydr Chem Biochem. 1976;32:125–234. doi: 10.1016/s0065-2318(08)60337-6. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Clancy M. J., Smith L. M., Magee P. T. Developmental regulation of a sporulation-specific enzyme activity in Saccharomyces cerevisiae. Mol Cell Biol. 1982 Feb;2(2):171–178. doi: 10.1128/mcb.2.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna W. J., Magee P. T. Glycogenolytic enzymes in sporulating yeast. J Bacteriol. 1978 Jun;134(3):844–853. doi: 10.1128/jb.134.3.844-853.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion E. A., Warner J. R. The major promoter element of rRNA transcription in yeast lies 2 kb upstream. Cell. 1984 Dec;39(3 Pt 2):663–673. doi: 10.1016/0092-8674(84)90473-2. [DOI] [PubMed] [Google Scholar]
- Evans I. H., Wilkie D. Mitochondrial factors in the utilization of sugars in Saccharomyces cerevisiae. Genet Res. 1976 Feb;27(1):89–93. doi: 10.1017/s001667230001627x. [DOI] [PubMed] [Google Scholar]
- Gascón S., Lampen J. O. Purification of the internal invertase of yeast. J Biol Chem. 1968 Apr 10;243(7):1567–1572. [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan N. A. Suppression of maltose-negative phenotype by a specific nuclear gene (PMU1) in the petite cells of the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1982;186(1):40–43. doi: 10.1007/BF00422909. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lutstorf U., Megnet R. Multiple forms of alcohol dehydrogenase in Saccharomyces cerevisiae. I. Physiological control of ADH-2 and properties of ADH-2 and ADH-4. Arch Biochem Biophys. 1968 Sep 10;126(3):933–944. doi: 10.1016/0003-9861(68)90487-6. [DOI] [PubMed] [Google Scholar]
- Ng R., Abelson J. Isolation and sequence of the gene for actin in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3912–3916. doi: 10.1073/pnas.77.7.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunberg J. H., Meade J. H., Cole G., Lawyer F. C., McCabe P., Schweickart V., Tal R., Wittman V. P., Flatgaard J. E., Innis M. A. Molecular cloning and characterization of the glucoamylase gene of Aspergillus awamori. Mol Cell Biol. 1984 Nov;4(11):2306–2315. doi: 10.1128/mcb.4.11.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottolenghi P. Some properties of five non-allelic -D-fructofuranosidases (invertases) of Saccharomyces. C R Trav Lab Carlsberg. 1971;38(13):213–221. [PubMed] [Google Scholar]
- Pearson N. J., Fried H. M., Warner J. R. Yeast use translational control to compensate for extra copies of a ribosomal protein gene. Cell. 1982 Jun;29(2):347–355. doi: 10.1016/0092-8674(82)90151-9. [DOI] [PubMed] [Google Scholar]
- Post-Beittenmiller M. A., Hamilton R. W., Hopper J. E. Regulation of basal and induced levels of the MEL1 transcript in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Jul;4(7):1238–1245. doi: 10.1128/mcb.4.7.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pretorius I. S., Chow T., Modena D., Marmur J. Molecular cloning and characterization of the STA2 glucoamylase gene of Saccharomyces diastaticus. Mol Gen Genet. 1986 Apr;203(1):29–35. doi: 10.1007/BF00330380. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sigmund R. D., McNally M. T., Lee D. B., Free S. J. Neurospora glucamylase and a mutant affected in its regulation. Biochem Genet. 1985 Feb;23(1-2):89–103. doi: 10.1007/BF00499115. [DOI] [PubMed] [Google Scholar]
- Sprague G. F., Jr, Blair L. C., Thorner J. Cell interactions and regulation of cell type in the yeast Saccharomyces cerevisiae. Annu Rev Microbiol. 1983;37:623–660. doi: 10.1146/annurev.mi.37.100183.003203. [DOI] [PubMed] [Google Scholar]
- Yamashita I., Fukui S. Transcriptional control of the sporulation-specific glucoamylase gene in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1985 Nov;5(11):3069–3073. doi: 10.1128/mcb.5.11.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita I., Maemura T., Hatano T., Fukui S. Polymorphic extracellular glucoamylase genes and their evolutionary origin in the yeast Saccharomyces diastaticus. J Bacteriol. 1985 Feb;161(2):574–582. doi: 10.1128/jb.161.2.574-582.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita I., Takano Y., Fukui S. Control of STA1 gene expression by the mating-type locus in yeasts. J Bacteriol. 1985 Nov;164(2):769–773. doi: 10.1128/jb.164.2.769-773.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotnik H., Fernandez M. P., Bowers B., Cabib E. Saccharomyces cerevisiae mannoproteins form an external cell wall layer that determines wall porosity. J Bacteriol. 1984 Sep;159(3):1018–1026. doi: 10.1128/jb.159.3.1018-1026.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kroon R. A., Koningsberger V. V. An inducible transport system for alpha-glucosides in protoplasts of Saccharomyces carlsbergensis. Biochim Biophys Acta. 1970 Apr 15;204(2):590–609. doi: 10.1016/0005-2787(70)90178-4. [DOI] [PubMed] [Google Scholar]