Significance
Nonhomologous end joining (NHEJ) is a major DNA-break repair pathway in eukaryotes and prokaryotes but is assumed to be absent in archaea. This study establishes that a functionally homologous pathway is present in archaea. We have reconstituted archaeal NHEJ repair in vitro, demonstrating that it is closely related to the bacterial apparatus and preferentially repairs breaks using RNA intermediates. We identify a role for a functionally unascribed nuclease in preventing the accumulation of genotoxic repair intermediates produced by strand displacement. This study has important implications for our understanding of the mechanisms of DNA-break repair by NHEJ and the evolution of end-joining pathways.
Abstract
Nonhomologous end-joining (NHEJ) pathways repair DNA double-strand breaks (DSBs) in eukaryotes and many prokaryotes, although it is not reported to operate in the third domain of life, archaea. Here, we describe a complete NHEJ complex, consisting of DNA ligase (Lig), polymerase (Pol), phosphoesterase (PE), and Ku from a mesophillic archaeon, Methanocella paludicola (Mpa). Mpa Lig has limited DNA nick-sealing activity but is efficient in ligating nicks containing a 3′ ribonucleotide. Mpa Pol preferentially incorporates nucleoside triphosphates onto a DNA primer strand, filling DNA gaps in annealed breaks. Mpa PE sequentially removes 3′ phosphates and ribonucleotides from primer strands, leaving a ligatable terminal 3′ monoribonucleotide. These proteins, together with the DNA end-binding protein Ku, form a functional NHEJ break-repair apparatus that is highly homologous to the bacterial complex. Although the major roles of Pol and Lig in break repair have been reported, PE’s function in NHEJ has remained obscure. We establish that PE is required for ribonucleolytic resection of RNA intermediates at annealed DSBs. Polymerase-catalyzed strand-displacement synthesis on DNA gaps can result in the formation of nonligatable NHEJ intermediates. The function of PE in NHEJ repair is to detect and remove inappropriately incorporated ribonucleotides or phosphates from 3′ ends of annealed DSBs to configure the termini for ligation. Thus, PE prevents the accumulation of abortive genotoxic DNA intermediates arising from strand displacement synthesis that otherwise would be refractory to repair.
DNA double-strand breaks (DSBs) are one of the most lethal forms of damage encountered by cells (1). Nonhomologous end joining (NHEJ) is the primary pathway for repairing DSBs in higher eukaryotes (2). NHEJ does not require the presence of a sister chromatid, in contrast to homologous recombination, and therefore can operate in quiescent cells. The direct repair and ligation of DSBs by NHEJ is considered to be more error prone because breaks are mended without the assistance of an intact DNA template.
The higher eukaryotic NHEJ complex is composed primarily of the ligase IV, X-ray repair cross-complementing protein 4 (XRCC4), and XRCC4-like factor (XLF) complex (the LXX complex), DNA-dependent protein kinase, catalytic subunit (DNA-PKcs), and Ku70/80 (1, 3). Ku and DNA-PKcs both assist in bridging the gap between the broken termini by promoting end synapsis. NHEJ repair of nonhomologous breaks requires remodeling of the DNA termini by processing enzymes, including polymerases (Pol λ and μ) and nucleases [Artemis and flap endonuclease 1 (Fen-1)], to prepare them for ligation (4–7). The LXX complex is recruited by Ku, which enables the ligation of the DNA ends.
Most bacterial organisms also possess an NHEJ complex, composed of a more limited set of proteins, including ligase D (LigD) and Ku (8, 9), which is required for DSB repair in stationary phase (10). The mycobacterial LigD gene encodes a multifunctional enzyme which, along with Ku, is capable of remodeling and ligating a broad variety of DSBs (8, 11). In many bacteria, LigD is found as a fusion of DNA ligase (Lig), polymerase (Pol), and phosphoesterase (PE) domains in a variety of configurations. The NHEJ Lig or ligase domain (LigDom) has a strong propensity for ligating nicks with a ribonucleotide on the 3′ position of the break (12), and the Pol domain (PolDom) preferentially incorporates nucleoside triphosphates (NTPs), rather than the expected deoxynucleoside triphosphates (dNTPs) (12, 13). Together, these findings establish that NHEJ repair requires Pol insertion of ribonucleotides at the DNA termini for efficient ligation. The PE or nuclease domain (NucDom) of LigD, originally shown to have 3′ exonuclease activity (8), preferentially removes ribonucleotides and phosphates from the 3′ termini strand, by sequential phosphodiesterase and phosphomonoesterase activities (14). Although PE’s biochemical activities are known, the specific role of this nuclease in DSBs repair remains unclear, with the nucleolytic activities of PE appearing to be antagonistic to those of Pol. Indeed, no obvious alterations to repair efficiency or fidelity were observed in plasmid-based DSB repair assays that excluded PE (15).
Archaea make up the third major domain of life, and although these organisms are among the most abundant in the biosphere, relatively little is known about how they repair their DNA. Many archaeal species survive in some of the most extreme environments on the planet, and therefore it is important to investigate how they maintain genome stability to understand how life evolved in the hostile conditions present on the primordial earth and how these organisms continue to thrive in extreme terrestrial niches. Although some NHEJ-like genes have been identified in archaea (9, 16–18), a complete DNA-break repair apparatus has not been reported. Therefore, it is unclear if a related DSB-repair pathway operates in archaea. Here, we describe the identification and characterization of a complete NHEJ complex, composed of Lig, Pol, PE, and Ku, in the archaeon Methanocella paludicola (Mpa). This study establishes that a functional NHEJ apparatus exists in archaea that is highly homologous to the bacterial break-repair complex. Furthermore, we exploited the biochemically tractability of this NHEJ apparatus to identify and characterize an unexpected function of the PE nuclease activity in coordinating end joining. We demonstrate that the PE is required for ribonucleolytic resection of Pol-incorporated RNA intermediates, resulting from strand-displacement synthesis, at annealed DSBs. This unusual 3′ resection activity configures nicked NHEJ intermediates for optimal end joining, thus preventing the accumulation of potentially lethal nonligatable intermediates that can arise from strand-displacement synthesis by NHEJ Pols.
Results and Discussion
Identification of a Putative NHEJ Repair Complex in Archaea.
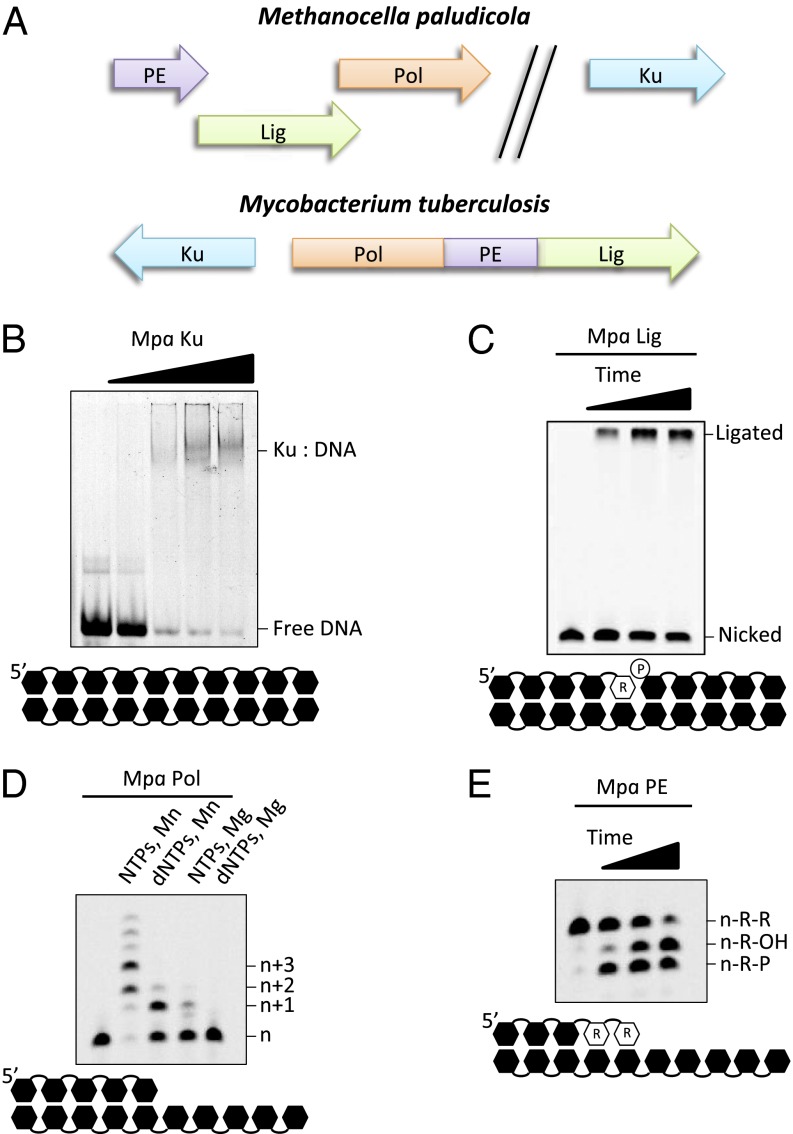
NHEJ pathways operate in both eukaryotes and prokaryotes, but whether a related DSB-repair system exists in the third domain of life, archaea, is unclear. Although a few NHEJ-like genes have been identified in archaea, a complete repertoire of factors has not been discovered in any one species, suggesting that a functional end-joining repair pathway may not exist in this kingdom. To address this question, mycobacterial NHEJ protein sequences (Ku and LigD) were used to search iteratively for related sequences in archaeal genomes. We found that the genome of the mesophillic archaeon Mpa (19) possesses an apparently intact NHEJ apparatus. Mpa encodes NHEJ Lig (Mcp 2126), Pol (Mcp 2125), and PE (Mcp 2127) genes, located in an operonic arrangement, that share significant homology with the three distinct catalytic domains of mycobacterial LigD (Fig. 1A and Fig. S1). However, unlike LigD, the different putative activities are encoded on individual genes rather than fused together into a single LigD-like gene. Mpa also encodes the DSB end-binding protein, Ku (Mcp 0581), a “hallmark” gene for the presence of a bona fide NHEJ pathway. Together, these Mpa repair protein orthologs represent of a complete NHEJ complex in an archaeal organism.
Fig. 1.
Biochemical activities associated with Mpa NHEJ repair proteins. (A) Comparison of gene arrangement in NHEJ operons from Mpa and Mtu. Mpa NHEJ factors are expressed as discrete proteins, whereas Mtu LigD is a multidomain single polypeptide. Ku is present in both genomes and is operonic with LigD in Mtu. (B) In Ku DNA-binding assays, 100 nM 5′-fluorescein–labeled 33mer dsDNA was incubated with 0, 200, 400, 800, and 1,600 nM Mpa Ku protein. (C) Ligase reactions contained 30 nM 5′-fluorescein–labeled 16mer DNA-RNA (D15R1) annealed to template DNA and a 5′-phosphorylated D-strand to create a nicked substrate and 300 nM Mpa Lig protein. The ligated product is 35 bases in length. (D) Pol primer extension reactions contained 30 nM 5′-fluorescein–labeled 16mer DNA annealed to template DNA and 300 nM Mpa Pol protein. Incubations included 250 µM NTPs/dNTPs and 5 mM MnCl2 or MgCl2 where indicated. (E) PE reactions contained 30 nM 5′-fluorescein–labeled 16mer DNA/RNA (D14R2) annealed to template DNA and 300 nM Mpa PE protein. n-R-R, unmodified substrate; n-R-P, removal of the terminal ribonucleotide monophosphate to yield a terminal phosphate group; n-R-OH, product after phosphate removal leaving a terminal OH group. All reactions were performed in the presence of manganese (5 mM).
Mpa NHEJ Proteins Possess Activities Equivalent to Those of Prokaryotic Orthologs.
To allow biochemical characterization of the putative Mpa NHEJ proteins, the genes fused to an N-terminal hexahistidine tag were individually overexpressed in Escherichia coli. The proteins were purified to homogeneity in high yields using immobilized metal affinity chromatography (IMAC), ion exchange chromatography, and gel filtration chromatography (Methods).
Mpa Ku gene encodes a 252-aa polypeptide that shares significant sequence homology with the prokaryotic and viral Ku proteins. After purification, Mpa Ku was observed to migrate at ∼30 kD by SDS/PAGE (Fig. S2) and eluted as an apparent homodimer from gel filtration columns, as is consistent with other Ku proteins (9, 20). Increasing titration of Mpa Ku reduced the electrophoretic mobility dsDNA (Fig. 1B), confirming that it binds to dsDNA, as is consistent with other Ku proteins (13).
The Mpa DNA Pol (Mpa Pol) gene encodes a 295-aa protein (∼34 kD) (Fig. S2) containing the three archaeo-eukaryotic primase (AEP) catalytic motifs present in other LigD-associated primase Pols (Fig. S3). In primer-template extension assays, Mpa Pol incorporated either dNTPs or NTPs, in the presence of manganese, opposite a DNA template of a 5′ overhang (Fig. 1C). Similar to other bacterial NHEJ Pols (13), Mpa Pol displayed a marked preference for insertion of NTPs rather than dNTPs, with optimal activity observed in the presence of manganese ions rather than magnesium (Fig. S2).
The Mpa PE gene encodes a 198-aa protein migrating ∼24 kD in SDS/PAGE (Fig. S2). Mpa PE represents a full-length, stand-alone, ortholog of the LigD PE domain. Mpa PE cleaved 3′ ribonucleotides and 3′ phosphates from a recessed DNA–RNA primer strand, leaving a single 3′ ribonucleotide containing a 3′ hydroxyl (OH) group (Fig. 1E), in common with the orthologous ribonucleolytic activity reported for other 3′ PEs (14, 18). As previously reported, this ability to remove all but the last ribonucleotide from a primer strand indicates the need for the PE to recognize a 2′-OH ribose moiety on the penultimate base. Mpa PE required either manganese or cobalt as a cofactor for efficient catalytic activities. Copper, cadmium, and magnesium all elicited moderate ribonuclease and phosphatase activities (Fig. S4).
The Mpa DNA Lig (Mpa Lig) gene encodes a 334-aa protein, which migrated at ∼40 kD in SDS/PAGE (Fig. S2). Mpa Lig possesses all the conserved motifs found in members of the Lig nucleotidyltransferase superfamily (Fig. S5) (21, 22). Notably, Mpa Lig had limited ligation activity on nicked DNA substrates (Fig. S6). In contrast, it preferentially catalyzed the ligation of nicked DNA containing at least one ribonucleotide on the 3′-OH side of a nick (Fig. 1C). Ligation was optimal in the presence of manganese, with only limited end joining in the presence of magnesium (Fig. S6). Mpa Lig was able to catalyze the ligation of nicks with a 5′ phosphate and a 3′ monoribonucleotide in the absence of ATP, suggesting that some of the recombinant protein was preadenylated, as observed previously (23).
Together, these data verify that the Mpa NHEJ proteins possess biochemical activities similar to those of their bacterial counterparts in vitro and suggest that they perform equivalent roles in NHEJ-mediated DSB repair processes in this species, implying that this DSB repair pathway also operates in archaea. This NHEJ apparatus provides a tractable in trans model system to study how components of the archaeo-prokaryotic (AP) NHEJ apparatus coordinate the repair of DSBs in vitro.
Mpa Ku Is Required for DNA End Joining in Vitro.
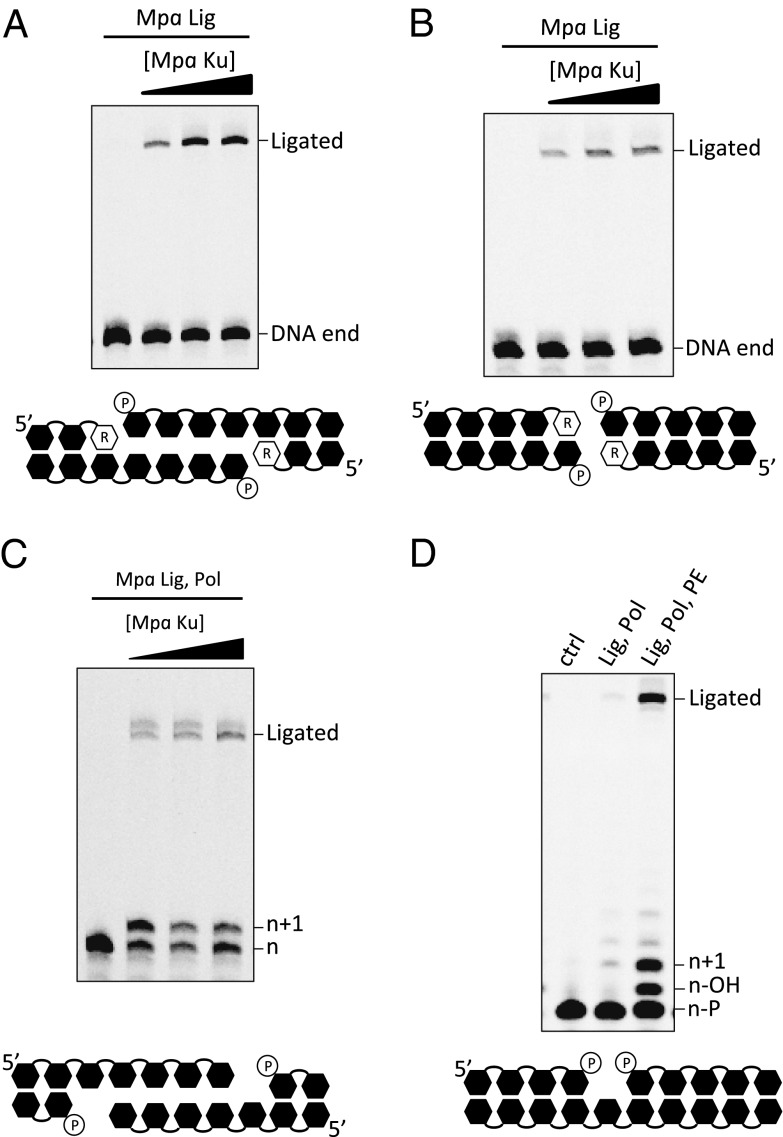
Ku’s proposed cellular function is to promote end synapsis and recruit other NHEJ repair factors to the break. We therefore assayed the necessity for Mpa Ku to facilitate end joining by Mpa Lig. The initial assay was performed on dsDNA containing a 3′ monoribonucleotide and a four-base 5′ DNA overhang with a self-annealing sequence to form a microhomology, thus creating a doubly nicked duplex (Fig. 2A). Mpa Lig alone was not sufficient to ligate these breaks (Fig. 2A); however, incubation with Ku allowed successful ligation. The amount of product was proportional to the concentration of Ku added to the reactions. Mpa Lig’s end-joining activity also was assayed using blunt-end dsDNA, containing a 3′ monoribonucleotide and a 5′ phosphate (Fig. 2B). Again, Mpa Lig alone was unable to ligate this substrate, but Ku stimulated its end-joining activity. Ligation of the blunt-ended DNA was not as efficient as that of the “synapsed” DNA, as would be expected given the increased stability imparted by the presence of a region of microhomology.
Fig. 2.
Mpa NHEJ complexes repair a diverse range of DNA breaks in vitro. (A) End-joining reaction mixtures contained 30 nM 5′-fluorescein–labeled 36mer DNA–RNA (D35R1) annealed to 5′-phosphorylated 40mer template DNA to create a self-complementary 4mer overhang, with 300 nM Mpa Lig and 200–800 nM Mpa Ku proteins. Ligated products were 76 bases in length. (B) End-joining reaction mixtures contained 30 nM 5′-fluorescein–labeled 36mer DNA–RNA (D35R1) annealed to 5′-phosphorylated 36mer template DNA to create blunt-ended 5′-phosphorylated DNA, with 300 nM Mpa Lig and 200–800 nM Mpa Ku proteins. Ligated products were 72 bases in length. (C) End-joining reaction mixtures contained 30 nM 5′-fluorescein–labeled 42mer DNA annealed with 5′-phosphorylated 37mer template DNA to create a 5mer 3′-overhang DNA, four bases of which are self-complementary, with 300 nM Mpa Lig and Pol proteins and 200–800 nM Mpa Ku proteins. Ligated products were 79 bases in length. (D) 3′-phosphatase, gap-filling, and ligation reaction mixtures contained 30 nM 5′-fluorescein–labeled 16mer with a 3′-phosphate annealed to template DNA and 5′-phosphorylated D-strand, with 300 nM Mpa Lig, Pol, and PE where indicated. n-P, unmodified substrate; n-OH, removal of 3′-phosphate to reveal 3′-OH which then can receive incoming NTPs for n+1. All reactions were performed in the presence of manganese (5 mM).
Although it had been determined that Mpa Pol was not necessary for the physical act of connecting DNA ends to achieve ligation, a terminal ribonucleotide remained essential for ligation (Fig. S6). To ascertain whether Mpa Pol could efficiently fill a gap to promote ligation after break annealing, we designed a DNA terminus with a five-base 3′ DNA overhang with a self-annealing sequence of four bases (Fig. 2C). The noncomplementary base requires gap-filling, in a template-dependent manner (see Fig. S7 A and C), subsequent to break annealing, before ligation can occur. These results indicate nucleotide incorporation by Mpa Pol and subsequent ligation of RNA/DNA nicks. The successful ligation of this substrate was again dependent on the presence of Ku for end joining. Together, these data verify that Mpa Pol, Lig, and Ku are bona fide NHEJ repair proteins and establish that a functional NHEJ complex exists in an archaeal species.
PE Removal of 3′ Phosphate Assists DNA Gap-Filling and Ligation.
As discussed, Mpa PE possesses both phosphodiesterase and monophosphatase activities. PE is capable of removing a 3′ phosphate group from any DNA primer strand configuration (Fig. S8). This ability confirms that, unlike its ribonuclease activity, monophosphoesterase activity does not require a 2′-OH ribose moiety on the penultimate nucleotide. PE’s ability to remove 3′ phosphates, in a context that is different from that of its ribonuclease activity, suggests that PE’s phosphatase activity may participate in distinct processing events during end joining. The likely role of the phosphatase is to facilitate 3′ phosphate removal at damaged termini to expose a free 3′ OH, thus allowing the DNA termini to be receptive to subsequent modifications by the Pol and Lig whose activities are dependent on the availability of an OH group.
To scrutinize this possibility further, we incubated a DNA substrate with a single-nucleotide gap and containing both 3′ and 5′ phosphate groups with and without Mpa Pol, Lig, and PE (Fig. 2D). dsDNA with gaps frequently are used as substrates for NHEJ enzymes because they mimic annealed breaks formed during DSB repair (13, 24). As predicted, the presence of a 3′ phosphate prevented NTP incorporation by Mpa Pol (Fig. 2D), because of the lack of a 3′ OH required for nucleophilic attack during extension. Mpa Lig also was unable to ligate this DNA, because ligation mechanisms also require a free 3′ OH. This impediment was overcome by the addition of PE, which catalyzed removal of the 3′ phosphate, producing a phosphate shift (annotated as n-OH; Fig. 2D) followed by subsequent gap filling and ligation. These data demonstrate that Mpa PE performs additional DNA-modification tasks in NHEJ, aside from its ribonuclease function, thus acting as a phosphatase that can operate at early stages of DSB repair to facilitate further processing and ligation of the break termini.
Mpa Pol, a Nonprocessive Pol with Strand-Displacement Activity.
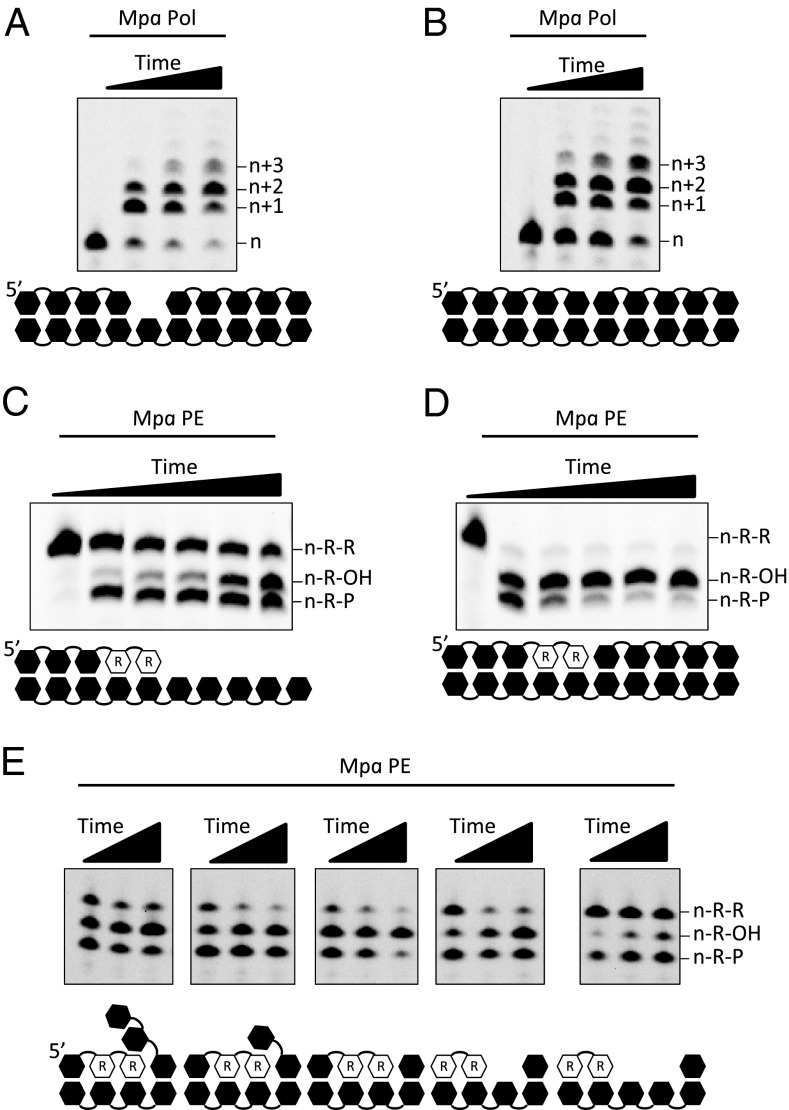
As discussed, Mpa Pol incorporated NTPs onto a primer strand in a template-dependent but nonprocessive manner (Fig. 1D). Mpa Pol inserted only three or four NTPs opposite a template strand (19 bases long), and efficiency was reduced even further with dNTPs, as noted above. This limited extension may result from the formation of DNA/RNA hybrids, switching the DNA conformation from B to A form, thus encouraging Pol dissociation (25). Mpa Pol also is capable of catalyzing the incorporation of nontemplated NTP on blunt-ended DNA, another activity shared with bacterial NHEJ Pols (Fig. S9) (13).
To determine if Mpa Pol displays strand-displacement activity after gap-filling (13, 26), we measured incorporation on DNA substrate with a single-nucleotide gap. Although Mpa Pol filled in the gap efficiently, it also synthesized additional longer products, incorporating up to three nucleotides (Fig. 3A). This activity suggests that, after gap-filling, Mpa Pol proceeds to incorporate further NTPs, presumably by displacing the downstream (D) strand. Notably, when Mpa Pol was assayed with a nicked DNA substrate (Fig. 3B) containing no gap, it also inserted up to three nucleotides. Even with nicked DNA, the NTP was incorporated in a template-dependent manner (Fig. S7B). Strand-displacement synthesis also is intrinsic to the mycobacterial NHEJ Pols [the Mycobacterium tuberculosis (Mtu) PolDom; Fig. S10] and has been described in other DNA-repair pathways (27). It is unclear what precise role displacement synthesis plays during NHEJ repair, at least in the absence of a 5′ exonuclease that could resect the displaced DNA flap. However, in addition to gap-filling, these Pols also play important roles in end synapsis by directly facilitating the annealing of break termini (24). Therefore, Pol may maintain intrinsic displacement activity to enable it to open up breaks with limited complementarity, e.g., blunt ends. Pol may ingress into the termini to uncover regions of microhomology that then can be annealed together to facilitate end synapsis (24). It also may enable these enzymes to locate an internal 5′ phosphate, if a terminal one is not present, to allow it to bind more securely to a DSB (24, 28). However, although it may be beneficial for some purposes, strand displacement can lead to some undesirable consequences, as discussed below.
Fig. 3.
Characterization of Pol strand displacement and PE activities. (A and B) Gap-filling reaction mixtures contained 30 nM 5′-fluorescein–labeled 16mer DNA annealed to template DNA and D-strands to create DNA with a one-nucleotide gap (A) or nicked DNA (B), respectively, and 300 nM Mpa Pol protein. (C–E) PE reaction mixtures contained 5′-fluorescein–labeled DNA-RNA (D14R2) annealed to a template with no D-strand (C), with a D-strand (D), or with D-strands producing two- and one-base nucleotide 5′ flaps and one- and two-nucleotide gaps with respect to the DNA-RNA primer (E) and 300 nM Mpa PE. Products are as described in Fig. 1E. All reactions were performed in the presence of manganese (5 mM).
PE Removes Ribonucleotides from Annealed DNA Breaks.
Although PE resects terminal ribonucleotides from recessed 3′ primer strands (Fig. 1D), the specific role of this nucleolytic activity in processing NHEJ intermediates has remained elusive. Given that Pol incorporation of NTPs is the likely source for the presence of small tracts of RNA at sites of DSBs, we asked if PE’s ribonuclease activity is required for processing these DNA–RNA intermediates. To address this question, we assayed PE’s ability to resect an annealed DNA-break substrate containing RNA, a diribonucleotide at the 3′ end of the primer strand, and a D-strand. Notably, PE readily removed nucleoside monophosphates (NMPs) from the 3′ DNA–RNA primer strand on this “annealed” break, leaving a single 3′ ribonucleotide. Significantly, PE’s resection activity was far more efficient than on an equivalent 3′ primer lacking an adjacent D-strand (Fig. 3 C and D).
Given the striking stimulation of PE’s resecting activity in the presence of a D-strand, we assayed a variety of different D-strands [ranging from a two-base 5′ flap (+2) to a two-base gap (−2)] to identify PE’s preferred substrate (Fig. 3E). A nicked substrate was the most efficiently processed intermediate, with almost 100% resection observed. A one-base 5′ flap (+1) and gapped substrate was processed less efficiently but considerably better than the remaining substrates (Fig. 3E). Notably, the activity of PE decreased drastically if the gap was larger than +1, whereas a two-base 5′ flap did not greatly suppress activity. These data demonstrate that PE is most efficient at resecting RNA incorporation at annealed breaks with little or no gap, even preferring to remove RNA that is displacing DNA, rather than from a gap of two or more bases.
Together, these data establish that PE preferentially resects RNA from the 3′ primer strand when a D-strand is present, and not from a recessed 3′ strand of a free end, as previously reported (14, 29, 30). This observed preference for resecting 3′ RNA from nicked DNA supports a model in which PE specifically operates not on break termini but after DSB synapsis and gap filling has already occurred.
PE Regulates Incorporation of RNA at Annealed Breaks.
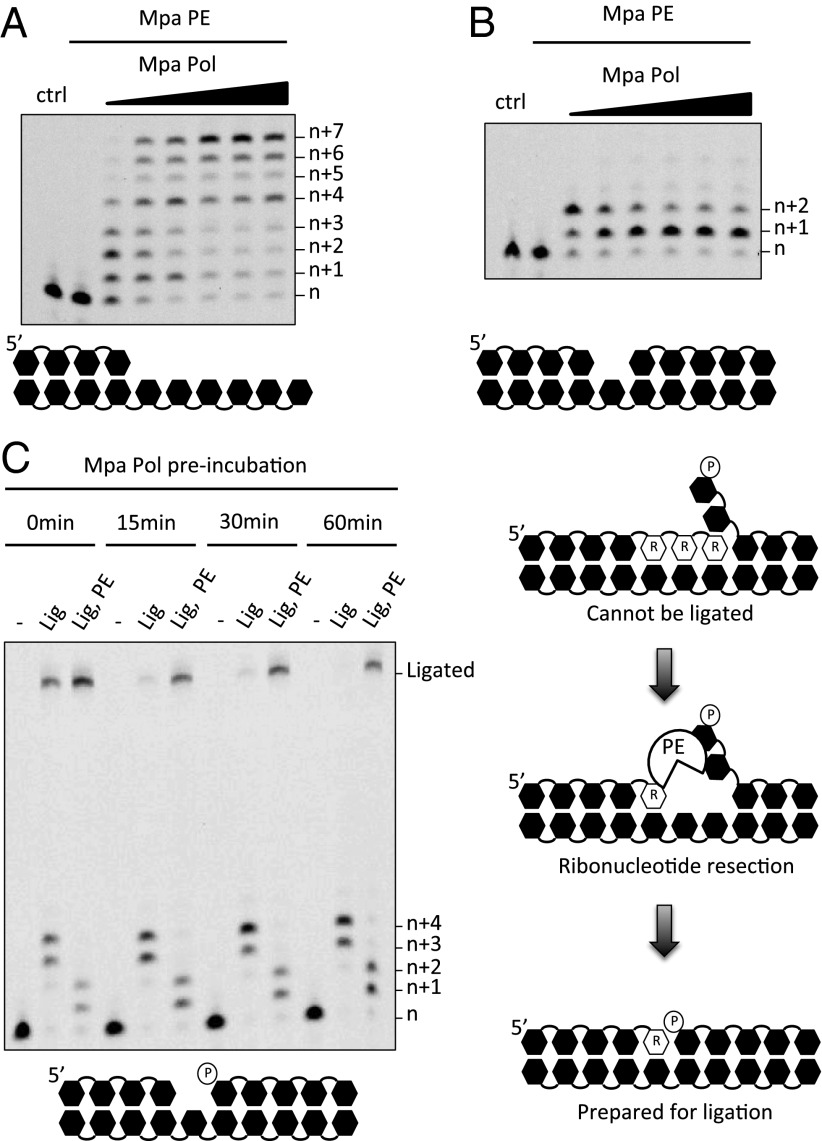
The roles of NHEJ Pol and PE are antagonistic, because one inserts ribonucleotides at DSBs and the other one removes them. Therefore it is critical to understand the context and coordination of these opposing end-processing activities to delineate how they cooperate to repair DSBs. As shown, PE prefers to resect RNA from the 3′ end of primer strands at annealed breaks, indicating that it may be required to modulate the length of extension products incorporated during gap-filling. To test this model, a fixed concentration of PE was incubated with an increasing concentration of Pol and two different DNA substrates (with or without a D-strand) containing a one-nucleotide gap to assess the effects, if any, on NTP incorporation (Fig. 4 A and B). Notably, PE limited the amount of NTP incorporation, at approximately equimolar concentrations, on both substrates. PE significantly limited NTP incorporation in the presence of a D-strand, with the major product at +1, even at saturating Pol concentrations (Fig. 4B). These data establish that PE directly modulates the length of extension of primer strands at annealed DSBs by resecting the 3′ RNA products incorporated by NHEJ Pols.
Fig. 4.
PE resection of strand-displacement intermediates is required for NHEJ repair. (A and B) Polymerase and PE reaction mixtures contained 5′-fluorescein–labeled 16mer DNA primers annealed to a template with no D-strand (A) or with a one-nucleotide gap (B), and 300 nM Mpa PE and 0.1–1.4 µM Mpa Pol. (C) (Left) Gap-filling, PE, and ligation reaction mixtures contained 5′-fluorescein–labeled 16mer DNA annealed to template DNA and a D-strand producing a single-nucleotide gap, and 300 nM Mpa Lig, Pol, and PE where indicated were preincubated with Mpa Pol at 37 °C for 0, 15, 30, and 60 min before the addition of Mpa Lig, and PE and incubation at 37 °C for 1 h. All reactions were performed in the prsence of manganese (5 mM). (Right) Rescue of abortive NHEJ repair intermediates that cannot be ligated. Downstream DNA displaced by incorporation of NTPs prevents ligation; however, Mpa PE resects the RNA tract and leaves a monoribonucleotide. Downstream DNA then can reanneal, and the substrate is ready for ligation with the 5′ phosphate opposite the 3′-OH group.
PE Resection of Strand-Displacement Intermediates During NHEJ Repair.
Although NHEJ Pols are proficient at filling in gaps formed at annealed breaks (24), they also catalyze, less desirably, strand-displacement synthesis on gapped substrates. This ability is a particularly dangerous one to possess, because it can lead to the formation of unligatable intermediates containing a 5′ flap that are refractory to repair. A mechanism therefore is required to minimize the production of such abortive genotoxic intermediates, allowing end ligation to proceed in a favorable way.
Because PE regulates the amount of nucleotides incorporated by Pol into gapped NHEJ substrates, we assessed if PE provides a mechanism for modulating strand-displacement synthesis, thus preventing the formation of unligatable DNA intermediates (Fig. 4C, Left). A single-nucleotide gapped substrate was preincubated with Mpa Pol (for 0, 15, 30, and 60 min), with manganese and NTPs, before the addition of Mpa PE and Lig. Although NTP incorporation by Pol remained at +3 or +4 across the time course, the amount of ligated product reduced dramatically over time. For example, after 15 min Mpa Pol had catalyzed almost total strand-displacement synthesis, rendering most of the DNA inert to ligation (Fig. 4C, Left). However, the addition of Mpa PE (after 15, 30, or 60 min) rescued a considerable proportion of the nonrepairable DNA (Fig. 4C, Left), presenting Mpa Lig with a nicked substrate optimized for ligation. Indeed, the addition of PE directly after the addition of Pol (0 min; Fig. 4C) yielded a much greater quantity of ligated substrates than was present in reactions lacking it. Similar results also were observed for Mtu PolDom (Fig. S11). End-joining substrates with 3′ overhangs also were assayed as described above (Fig. S12), and the results are consistent with the gap-filling and ligation assays in Fig. 4C, with PE counteracting Pol’s downstream displacement activity. Together, these data robustly support a model in which PE is required for optimal resecting of RNA-containing NHEJ intermediates, particularly those produced by strand-displacement synthesis, to ensure the remodeled annealed break is amenable to ligation (Fig. 4C, Right). The availability of this editing mechanism may be particularly important for ensuring the repair of all DSBs, because even a single unrepaired DNA break can be lethal.
Concluding Remarks.
The potentially lethal nature of DNA DSBs has ensured that all organisms have evolved repair pathways to mend these potent lesions effectively. Although recombination-based repair mechanisms are conserved across all living organisms, NHEJ break-repair pathways were considered to operate exclusively in eukaryotes. However, in recent years it has been recognized that most bacterial species also use a closely related NHEJ repair apparatus to mend breaks. In contrast, there was limited evidence to support the existence of NHEJ repair pathways in the third major domain of life, archaea. This absence is particularly surprising because many archaeal species thrive in some of the harshest environmental niches, e.g., deep sea thermal vents, putting their genomes at significantly increased risk of breakage. This current study resolves this apparent discrepancy with the identification of a fully functional NHEJ repair apparatus in archaea, thus establishing the conservation of this pathway across all three domains of life.
Although many archaeal DNA metabolism pathways (e.g., DNA replication) share many more common genes with eukaryotes rather than with bacteria, archaeal NHEJ is much more homologous to the bacterial DSB-repair complexes, particularly in terms of its conservation of the key repair proteins and their common intrinsic biochemical activities. Therefore, these closely related break-repair pathways can be grouped together as AP-NHEJ to differentiate them from the more divergent eukaryotic pathway. Notably, a complete LigD AP-NHEJ apparatus is present in some plants (e.g., castor bean), suggesting that this pathway diverged quite late in evolution. The discovery of a shared end-joining repair apparatus in both archaeal and prokaryotic organisms further supports the hypothesis that a primordial NHEJ repair pathway arose early in evolution and was maintained and evolved further in lower eukaryotes before eventually becoming the predominant DSB repair pathway in mammalian cells.
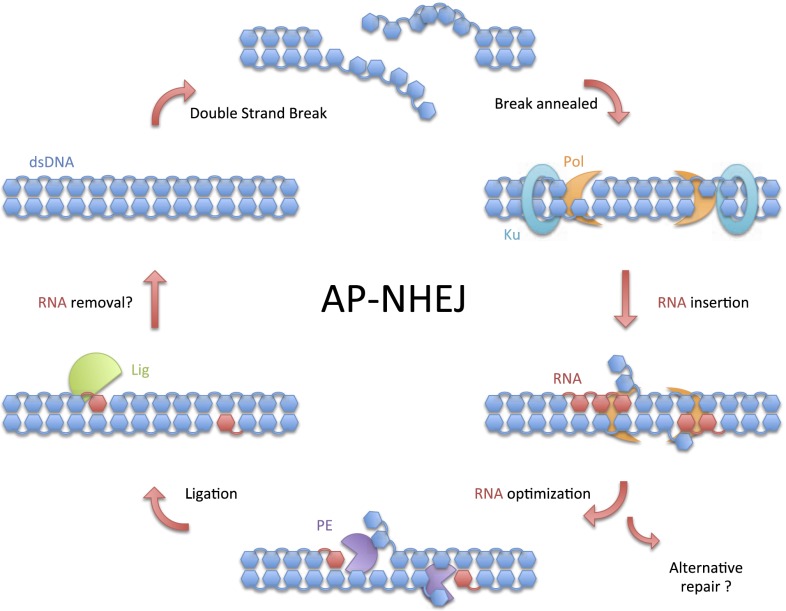
Although bacterial LigD has proven invaluable in providing insights into AP-NHEJ repair mechanisms, it also has some major experimental limitations. However, unlike LigD, the Mpa archaeon end-joining machinery is not fused into a single protein. This physical disconnectivity of the NHEJ activities has provided a tractable in trans repair apparatus that allows dissection of the interplay between the end-processing enzymes to elucidate how they cooperate to coordinate break repair. Although the AP-NHEJ PE family was identified more than a decade ago, its specific role in break repair has remained elusive. The modularity of the archaeal end-joining complex has enabled us to delineate a major function for PE in break repair. We demonstrate that the propensity of NHEJ Pols to insert excessive ribonucleotides into gaps by strand-displacement synthesis after end synapsis is strongly counteracted by the resection activity of PE. NHEJ PEs limit excessive ribonucleotide incorporation by using a ribonucleolytic resection mechanism, thus ensuring that nicked DNA substrates are available for ligation, preventing the accumulation of potentially genotoxic NHEJ intermediates (Fig. 5). The principle of strand displacement in DNA repair is not novel and, in fact, is well established in the eukaryotic base-excision repair (BER) pathway (27). Pol β actively displaces downstream damaged DNA as an interim repair step of BER, before FEN-1 endonucleolytically resects the 5′ damaged flap. The PE activity associated with AP-NHEJ offers an alternative resolution of strand-displaced intermediates. Resection of the newly synthesized ingressing DNA strand by PE facilitates reannealing of the displaced strand to produce a nicked template that is optimal for end ligation.
Fig. 5.
Model of AP-NHEJ using RNA-repair intermediates. The initial step in AP-NHEJ repair is the binding of Ku homodimers to broken ends and bringing them into close proximity. Ku and Pol promote microhomology-mediated synapsis of overhanging termini, if present, to anneal the break. Pol fills in any resulting gaps at either end of the annealed break by a template-dependent RNA synthesis. Pol may displace several bases of downstream DNA using its distinct ability to mobilize and direct DNA strands, offering a greater flexibility to repair a variety of configurations of damaged DNA. Strand-displacement RNA synthesis by Pol is regulated by PE, which can remove unnecessary NMPs and allow the displaced DNA to realign with the template. PE always leaves a single terminal 3′ ribonucleotide in place in resected breaks, presenting Lig with an optimized nicked substrate for ligation. Although the broken DNA is reconnected, repair may not be fully completed, because several RNA bases still reside in the duplex. RNA removal may be performed by additional enzymes, such as RNase HII, before other repair Pols fill in the gaps with DNA.
Replicative DNA Pols possess a 3′ proofreading exonuclease (Exo) function to maintain fidelity. AP-NHEJ Pols also maintain an associated 3′ Exo activity but do so to limit displacement synthesis rather than to maintain fidelity. This functional association of these atypical nucleases with Pol and Lig is supported by the genomic distribution of PE genes, which are found almost exclusively in an operonic arrangement with Pol and Lig genes, indicating that a selective genetic requirement maintains PE in association with these NHEJ-processing enzymes. PE-like genes also exist in some yeast species, although their role is unknown.
An intriguing feature of AP-NHEJ repair pathways is the preferential insertion, removal, and ligation of ribonucleotides at DSBs by the break-repair enzymes. Utilization of ribonucleotides during break repair is receiving growing recognition, with reports of the preferential incorporation of ribonucleotides during eukaryotic NHEJ (31, 32) suggesting that RNA–DNA intermediates are not limited to AP-NHEJ (33). There are several reasons why it may be favorable to incorporate RNA during DSB end joining. NHEJ is the DSB-repair pathway of choice for nondividing cells (10), where NTPs are much more abundant than dNTPs in the available nucleotide pools (33). Another distinct possibility, driven by the availability of ribonucleotides, is that NHEJ repair pathways evolved to incorporate RNA rather than DNA selectively into repaired DSBs to demarcate the boundaries of any alterations made to the repaired sites. This “labeling” of the repaired break with ribonucleotides may be useful in a number of ways. First, it prevents the loss of genetic material at sites of DSBs by preventing resection of the DNA termini by the ribocentric NHEJ repair machinery. Significantly in this regard, PE cannot resect DNA ends and prefers to resect short RNA tracts incorporated by the Pol. Secondly, efficient ligation of breaks also is predicated on the incorporation of a terminal 3′ ribonucleotide that potentially acts as a molecular signal to indicate when a nick is ready for ligation, thus preventing aberrant end joining. A major consequence of AP-NHEJ repair is the incorporation of at least one ribonucleotide into ligated DSB junctions, and it is likely that subsequently this RNA is excised from the repaired breaks (33, 34). Studies in murine systems have revealed that RNase HII performs an essential role in removing ribonucleotides from genomic DNA (35). Although this role is thought primarily to be protection from RNA misincorporated during replication, RNase HII (or a similar enzyme) also may resolve the final complication of AP-NHEJ repair. However, this role remains to be established.
Methods
Purification of Mpa Lig, Pol, PE, and Ku Proteins.
All Mpa NHEJ genes were cloned into pET 28A, the histidine-tagged proteins expressed in B834S (DE3), and were purified using the same chromatography protocols, except where noted. Pelleted cells containing the appropriate plasmid were grown in Terrific Broth, and pelleted cultures were resuspended in lysis buffer [50 mM Tris⋅HCl (pH 7.5), 500 mM NaCl, 30 mM imidazole, 10% (vol/vol) glycerol, 17 mg/mL PMSF, 34 μg/mL benzamidine]. Cells were disrupted by sonication, and soluble proteins were isolated by centrifugation. The supernatant was applied to a 25-mL NTA (Qiagen) column equilibrated in IMAC buffer A (the lysis buffer) and eluted in IMAC buffer B (buffer A with 300 mM imidazole). Mpa Ku supernatant was subjected to an extended wash period in IMAC buffer C (buffer A with 1M NaCl) to remove excess DNA bound to Ku proteins. Protein-containing fractions were loaded onto 5-mL fast-flow Q and S (Lig and Pol), or Q (PE and Ku) columns (GE Healthcare) pre-equilibrated in IEx buffer A [50 mM Tris⋅HCl (pH 7.5), 10% (vol/vol) glycerol] for separation by ionic charge. Lig, Pol, and PE were eluted in the flowthrough without binding the column; Ku was eluted in 10% (vol/vol) IEx buffer B [50 mM Tris⋅HCl (pH7.5), 2M NaCl, 10% (vol/vol) glycerol]. Any remaining contaminating proteins were removed by gel filtration on a Superdex S200 column (GE Healthcare) equilibrated in GF buffer [25 mM Tris (pH 7.5), 500 mM NaCl, 10% (vol/vol) glycerol]. Eluted proteins were analyzed for purity using SDS/PAGE, and concentrations were ascertained by spectrophotometry of samples at 280 nm.
EMSA.
Fluorescein labeled DNA primer (5′-CATATCCGTGTCGCCCCTTATTCCGATAGTGACTACA) was annealed to DNA template (5′-TGTAGTCACTATCGGAATAAGGGGCGACACGGATATG) creating oligo 20 for DNA-binding assays. DNA (60 nM) was incubated with 0.2–1.6 µM Mpa Ku in 50 mM Tris (pH 7.5) and 5% (vol/vol) glycerol for 30 min at room temperature in a volume of 20 µL. Samples were separated by electrophoresis on a native 5% (vol/vol) polyacrylamide gel in 0.5× Tris/borate/EDTA (TBE) buffer for 2 h. Fluorescently labeled DNA oligomers were detected by scanning using a Fujifilm FLA-5100 fluorescent image analyzer.
DNA Ligation Assay.
Fluorescein-labeled DNA primer (5′-CTATGAGCGAATCGCrC) was annealed to DNA template (5′-AGTCGCATAGTGTAGTCGGGGCGATTCGCTCATAG) with a downstream sequence (5′-P-CGACTACACTATGCGACT) creating oligo 60. DNA (30 nM) was incubated with 300 nM Mpa Lig in 50 mM Tris⋅HCl (pH 7.5), 5 mM MnCl2 for 30, 60, and 90 min at 37 °C in a volume of 20 µL. The reactions were stopped by the addition of Stop buffer [95% (vol/vol) formamide, 0.09% xylene cyanol] and the mixtures were boiled at 95 °C for 10 min. Samples were separated by electrophoresis on an 8-M urea, 15% polyacrylamide gel in 1× TBE buffer for 2 h. Fluorescently labeled oligos were detected by scanning using a Fujifilm FLA-5100 fluorescent image analyzer.
DNA Extension Assay.
Fluorescein-labeled DNA primer (5′-CTATGAGCGAATCGCC) was annealed to a DNA template (5′-AGTCGCATAGTGTAGTCGGGGCGATTCGCTCATAG) creating oligo 9 for extension. Further downstream sequences (5′-CCGACTACACTATGCGAC and 5′-P-CGACTACACTATGCGACT) were annealed to this DNA oligo, creating oligos 11 and 37 for gap-filling assays. DNA (30 nM) was incubated with 300 nM Mpa Pol in 50 mM Tris⋅HCl (pH 7.5), 5 mM MnCl2, and 250 µM NTP mix for 1 h at 37 °C in a volume of 20 µL. The reactions were stopped by the addition of Stop buffer (95% formamide, 0.09% xylene cyanol) and the mixtures were boiled at 95 °C for 10 min. Samples were separated by electrophoresis on an 8-M urea, 15% polyacrylamide gel in 1× TBE buffer for 2 h. Fluorescently labeled oligos were detected by scanning using a Fujifilm FLA-5100 fluorescent image analyzer.
Ribonuclease and Phosphatase Assay.
Fluorescein-labeled DNA–RNA primer (5′-CTATGAGCGAATCGrCrC) was annealed to a DNA template (5′-AGTCGCATAGTGTAGTCGGGGCGATTCGCTCATAG), creating oligo 3 for PE assays. Further downstream sequences (5′-CTACCGACTACACTATGCGACT, 5′-CCCGACTACACTATGCGACT, 5′-CCGACTACACTATGCGAC, 5′-P-CGACTACACTATGCGACT, and 5′- GACTACACTATGCGACT) were annealed to this oligo, creating oligos 25, 62, 2, 38, and 43, respectively. DNA (30 nM) was incubated with 300 nM Mpa PE in 50 mM Tris (pH 7.5) and 5 mM MnCl2 at 37 °C for varying time points across 90 min. The reactions were quenched by the addition of Stop buffer (95% formamide, 0.09% xylene cyanol) and the mixtures were boiled at 95 °C for 10 min. Samples were separated by electrophoresis on an 8 M urea, 15% polyacrylamide gel in 1× TBE buffer for 2 h. Fluorescently labeled oligos were detected by scanning using a Fujifilm FLA-5100 fluorescent image analyzer.
DNA End-Joining Assay.
Fluorescein-labeled DNA and DNA-RNA primers (5′-CATATCCGTGTCGCCCCTTATTCCGATAGTGACTArC, 5′- CATATCCGTGTCGCCCCTTATTCCGATAGTGACTACrA, and 5′-CATATCCGTGTCGCCCCTTATTCCGATAGTGACTACAACGCG) were annealed to DNA templates (5′-P-CATGGTAGTCACTATCGGAATAAGGGGCGACACGGATAT and 5′-P- TGTAGTCACTATCGGAATAAGGGGCGACACGGATATG) to form oligos 45, 68, and 76, respectively, for DNA end-joining assays. Mpa Lig (300 nM) was incubated with 200–800 nM Mpa Ku and with 300 nM Mpa Pol where indicated in 50 mM Tris (pH 7.5), 5 mM MnCl2, 1 mM DTT, 5% glycerol, 0.1 mg/mL BSA, 250 µM NTPs (Pol assay only) at 37 °C in a final volume of 2 µL. The reactions were quenched by the addition of Stop buffer (95% formamide, 0.09% xylene cyanol) and the mixtures were boiled at 95 °C for 10 min. Samples were separated by electrophoresis on an 8 M urea, 15% polyacrylamide gel in 1× TBE buffer for 2 h. Fluorescently labeled oligos were detected by scanning using a Fujifilm FLA-5100 fluorescent image analyzer.
Phosphatase, Gap-Filling, and Ligation Assay.
Fluorescein-labeled DNA primer (5′-CTATGAGCGAATCGCC-P) was annealed to a DNA template (5′-AGTCGCATAGTGTAGTCGGGGCGATTCGCTCATAG) and downstream strand (5′-P-CGACTACACTATGCGACT) creating oligo 63 for phosphatase, gap-filling, and ligation. Mpa Lig, Pol, and PE (300 nM) were incubated where indicated with 50 m M Tris⋅HCl (pH 7.5), 5 mM MnCl2, 250 µM NTP mix for 1 h at 37 °C in a volume of 20 µL. The reactions were quenched by the addition of Stop buffer (95% formamide, 0.09% xylene cyanol) and the mixtures were boiled at 95 °C for 10 min. Samples were separated by electrophoresis on an 8 M urea, 15% polyacrylamide gel in 1× TBE buffer for 2 h. Fluorescently labeled oligos were detected by scanning using a Fujifilm FLA-5100 fluorescent image analyzer.
Polymerase DNA Extension, Gap-Filling, and PE Assay.
Oligos 9, 11, and 37, as described above, were used for DNA extension and PE assays. DNA (30 nM) was incubated with 300 nM Mpa Pol and PE where indicated in 50 mM Tris (pH 7.5) and 5 mM MnCl2 at 37 °C for varying time points across 90 min. The reactions were quenched by the addition of Stop buffer (95% formamide, 0.09% xylene cyanol) and the mixtures were boiled at 95 °C for 10 min. Samples were separated by electrophoresis on an 8 M urea, 15% polyacrylamide gel in 1× TBE buffer for 2 h. Fluorescently labeled oligos were detected by scanning using a Fujifilm FLA-5100 fluorescent image analyzer.
DNA Gap-Filling, PE, and Ligation Assay.
Oligo 37, as described above, was used for gap-filling, PE, and ligation assays. DNA (30 nM) was incubated with 300 nM Mpa Pol in 50 mM Tris (pH 7.5) and 5 mM MnCl2 at 37 °C for 0, 15, 30, and 60 min. Mpa Lig and PE (300 nM) then were added, and incubation continued for a further 60 min. The reactions were quenched by the addition of Stop buffer (95% formamide, 0.09% xylene cyanol) and the mixtures were boiled at 95 °C for 10 min. Samples were separated by electrophoresis on an 8 M urea, 15% polyacrylamide gel in 1× TBE buffer for 2 h. Fluorescently labeled oligos were detected by scanning using a Fujifilm FLA-5100 fluorescent image analyzer.
Supplementary Material
Acknowledgments
We thank Dr. H. Imachi for providing M. paludicola genomic DNA. Work in the A.J.D. laboratory is supported by grants from the Biotechnology and Biological Sciences Research Council (BBSRC) and indirectly by a centre grant from the Medical Research Council. E.J.B. was supported by a BBSRC PhD studentship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1302616110/-/DCSupplemental.
References
- 1.Daley JM, Palmbos PL, Wu D, Wilson TE. Nonhomologous end joining in yeast. Annu Rev Genet. 2005;39:431–451. doi: 10.1146/annurev.genet.39.073003.113340. [DOI] [PubMed] [Google Scholar]
- 2.Helleday T, Lo J, van Gent DC, Engelward BP. DNA double-strand break repair: From mechanistic understanding to cancer treatment. DNA Repair (Amst) 2007;6(7):923–935. doi: 10.1016/j.dnarep.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J. 2009;417(3):639–650. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nick McElhinny SA, et al. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol Cell. 2005;19(3):357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 5.Weinfeld M, Mani RS, Abdou I, Aceytuno RD, Glover JNM. Tidying up loose ends: The role of polynucleotide kinase/phosphatase in DNA strand break repair. Trends Biochem Sci. 2011;36(5):262–271. doi: 10.1016/j.tibs.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gu J, et al. DNA-PKcs regulates a single-stranded DNA endonuclease activity of Artemis. DNA Repair (Amst) 2010;9(4):429–437. doi: 10.1016/j.dnarep.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu X, Wilson TE, Lieber MR. A role for FEN-1 in nonhomologous DNA end joining: The order of strand annealing and nucleolytic processing events. Proc Natl Acad Sci USA. 1999;96(4):1303–1308. doi: 10.1073/pnas.96.4.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Della M, et al. Mycobacterial Ku and ligase proteins constitute a two-component NHEJ repair machine. Science. 2004;306(5696):683–685. doi: 10.1126/science.1099824. [DOI] [PubMed] [Google Scholar]
- 9.Weller GR, et al. Identification of a DNA nonhomologous end-joining complex in bacteria. Science. 2002;297(5587):1686–1689. doi: 10.1126/science.1074584. [DOI] [PubMed] [Google Scholar]
- 10.Pitcher RS, et al. NHEJ protects mycobacteria in stationary phase against the harmful effects of desiccation. DNA Repair (Amst) 2007;6(9):1271–1276. doi: 10.1016/j.dnarep.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 11.Gong C, et al. Mechanism of nonhomologous end-joining in mycobacteria: A low-fidelity repair system driven by Ku, ligase D and ligase C. Nat Struct Mol Biol. 2005;12(4):304–312. doi: 10.1038/nsmb915. [DOI] [PubMed] [Google Scholar]
- 12.Zhu H, Shuman S. Bacterial nonhomologous end joining ligases preferentially seal breaks with a 3′-OH monoribonucleotide. J Biol Chem. 2008;283(13):8331–8339. doi: 10.1074/jbc.M705476200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pitcher RS, et al. Structure and function of a mycobacterial NHEJ DNA repair polymerase. J Mol Biol. 2007;366(2):391–405. doi: 10.1016/j.jmb.2006.10.046. [DOI] [PubMed] [Google Scholar]
- 14.Zhu H, Shuman S. Novel 3′-ribonuclease and 3′-phosphatase activities of the bacterial non-homologous end-joining protein, DNA ligase D. J Biol Chem. 2005;280(28):25973–25981. doi: 10.1074/jbc.M504002200. [DOI] [PubMed] [Google Scholar]
- 15.Aniukwu J, Glickman MS, Shuman S. The pathways and outcomes of mycobacterial NHEJ depend on the structure of the broken DNA ends. Genes Dev. 2008;22(4):512–527. doi: 10.1101/gad.1631908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aravind L, Koonin EV. Prokaryotic homologs of the eukaryotic DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system. Genome Res. 2001;11(8):1365–1374. doi: 10.1101/gr.181001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nair PA, Smith P, Shuman S. Structure of bacterial LigD 3′-phosphoesterase unveils a DNA repair superfamily. Proc Natl Acad Sci USA. 2010;107(29):12822–12827. doi: 10.1073/pnas.1005830107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith P, Nair PA, Das U, Zhu H, Shuman S. Structures and activities of archaeal members of the LigD 3′-phosphoesterase DNA repair enzyme superfamily. Nucleic Acids Res. 2011;39(8):3310–3320. doi: 10.1093/nar/gkq1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakai S, et al. Methanocella paludicola gen. nov., sp. nov., a methane-producing archaeon, the first isolate of the lineage ‘Rice Cluster I’, and proposal of the new archaeal order Methanocellales ord. nov. Int J Syst Evol Microbiol. 2008;58(Pt 4):929–936. doi: 10.1099/ijs.0.65571-0. [DOI] [PubMed] [Google Scholar]
- 20.Pitcher RS, Tonkin LM, Green AJ, Doherty AJ. Domain structure of a NHEJ DNA repair ligase from Mycobacterium tuberculosis. J Mol Biol. 2005;351(3):531–544. doi: 10.1016/j.jmb.2005.06.038. [DOI] [PubMed] [Google Scholar]
- 21.Shuman S, Liu Y, Schwer B. Covalent catalysis in nucleotidyl transfer reactions: Essential motifs in Saccharomyces cerevisiae RNA capping enzyme are conserved in Schizosaccharomyces pombe and viral capping enzymes and among polynucleotide ligases. Proc Natl Acad Sci USA. 1994;91(25):12046–12050. doi: 10.1073/pnas.91.25.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doherty AJ, Suh SW. Structural and mechanistic conservation in DNA ligases. Nucleic Acids Res. 2000;28(21):4051–4058. doi: 10.1093/nar/28.21.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doherty AJ, Ashford SR, Subramanya HS, Wigley DB. Bacteriophage T7 DNA ligase. Overexpression, purification, crystallization, and characterization. J Biol Chem. 1996;271(19):11083–11089. doi: 10.1074/jbc.271.19.11083. [DOI] [PubMed] [Google Scholar]
- 24.Brissett NC, et al. Structure of a NHEJ polymerase-mediated DNA synaptic complex. Science. 2007;318(5849):456–459. doi: 10.1126/science.1145112. [DOI] [PubMed] [Google Scholar]
- 25.Zhu H, Bhattarai H, Yan H-G, Shuman S, Glickman MS. Characterization of Mycobacterium smegmatis PolD2 and PolD1 as RNA/DNA polymerases homologous to the POL domain of bacterial DNA ligase D. Biochemistry. 2012;51(51):10147–10158. doi: 10.1021/bi301202e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu H, Shuman S. Gap filling activities of Pseudomonas DNA ligase D (LigD) polymerase and functional interactions of LigD with the DNA end-binding Ku protein. J Biol Chem. 2010;285(7):4815–4825. doi: 10.1074/jbc.M109.073874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imoto S, Bransfield LA, Croteau DL, Van Houten B, Greenberg MM. DNA tandem lesion repair by strand displacement synthesis and nucleotide excision repair. Biochemistry. 2008;47(14):4306–4316. doi: 10.1021/bi7021427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brissett NC, et al. Structure of a preternary complex involving a prokaryotic NHEJ DNA polymerase. Mol Cell. 2011;41(2):221–231. doi: 10.1016/j.molcel.2010.12.026. [DOI] [PubMed] [Google Scholar]
- 29.Zhu H, Wang LK, Shuman S. Essential constituents of the 3′-phosphoesterase domain of bacterial DNA ligase D, a nonhomologous end-joining enzyme. J Biol Chem. 2005;280(40):33707–33715. doi: 10.1074/jbc.M506838200. [DOI] [PubMed] [Google Scholar]
- 30.Zhu H, Shuman S. Substrate specificity and structure-function analysis of the 3′-phosphoesterase component of the bacterial NHEJ protein, DNA ligase D. J Biol Chem. 2006;281(20):13873–13881. doi: 10.1074/jbc.M600055200. [DOI] [PubMed] [Google Scholar]
- 31.Martin MJ, Garcia-Ortiz MV, Esteban V, Blanco L. Ribonucleotides and manganese ions improve non-homologous end joining by human Polμ. Nucleic Acids Res. 2012;41(4):2428–2436. doi: 10.1093/nar/gks1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nick McElhinny SA, Ramsden DA. Polymerase mu is a DNA-directed DNA/RNA polymerase. Mol Cell Biol. 2003;23(7):2309–2315. doi: 10.1128/MCB.23.7.2309-2315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pitcher RS, Wilson TE, Doherty AJ. New insights into NHEJ repair processes in prokaryotes. Cell Cycle. 2005;4(5):675–678. doi: 10.4161/cc.4.5.1676. [DOI] [PubMed] [Google Scholar]
- 34.Rydberg B, Game J. Excision of misincorporated ribonucleotides in DNA by RNase H (type 2) and FEN-1 in cell-free extracts. Proc Natl Acad Sci USA. 2002;99(26):16654–16659. doi: 10.1073/pnas.262591699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reijns MA, et al. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell. 2012;149(5):1008–1022. doi: 10.1016/j.cell.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.