Abstract
Background:
Microsatellite instability (MSI) is a molecular phenotype due to defective DNA mismatch repair (MMR) system. It is used to predict outcome of colorectal tumours and to screen tumours for Lynch syndrome (LS). A pentaplex panel composed of five mononucleotide markers has been largely recommended for determination of the MSI status. However, its sensitivity may be taken in default in occasional situations. The aim of the study was to optimise this panel for the detection of MSI.
Methods:
We developed an assay allowing co-amplification of six mononucleotide repeat markers (BAT25, BAT26, BAT40, NR21, NR22, NR27) and one polymorphic dinucleotide marker (D3S1260) in a single reaction. Performances of the new panel were evaluated on a cohort of patients suspected of LS.
Results:
We demonstrate that our assay is technically as easy to use as the pentaplex assay. The hexaplex panel shows similar performances for the identification of colorectal and non-MSH6-deficient tumours. On the other hand, the hexaplex panel has higher sensitivity for the identification of MSH6-deficient tumours (94.7% vs 84.2%) and MMR-deficient tumours other than colorectal cancer (92.9% vs 85.7%).
Conclusion:
The hexaplex panel could thus be an attractive alternative to the pentaplex panel for the identification of patients with LS.
Keywords: Lynch syndrome, microsatellite instability, DNA mismatch repair
Instability of short tandem repeats or microsatellite instability (MSI) is a molecular phenotype due to defective DNA mismatch repair (MMR) system. About 15% of sporadic colorectal cancers, about 20% of sporadic endometrial cancers, as well as a non-negligible proportion of cancers originating from other sites exhibit MSI (Ionov et al, 1993; Thibodeau et al, 1993; Salovaara et al, 2000). A MSI phenotype is also observed in most tumours from patients with Lynch syndrome (LS; Aaltonen et al, 1993; Salovaara et al, 2000; Lynch et al, 2009). At the clinical level, patients with MSI colorectal tumours are known to have better stage-adjusted prognosis and may respond differently to adjuvant chemotherapy (Gryfe et al, 2000; Ribic et al, 2003; Sargent et al, 2010).
LS (MIM# 120435, 609310), also known as hereditary non-polyposis colorectal cancer (CRC), is the most common cause of inherited CRC. It is caused by germline mutations in MMR genes, not only affecting mainly MLH1 and MSH2 (∼40% each) but also MSH6 and PMS2 (∼10% and 5%, respectively). Patients with LS are at high risk of developing early-onset CRC. Patients with LS are also at risk of developing other primary cancers involving endometrium, ovary, urinary tract, stomach, small intestine, hepatobiliary tract, skin, and brain (Vasen et al, 2007; Lynch et al, 2009; Bonadona et al, 2011). The young age of disease onset (average before 45 years) highlights the importance of identifying these patients. Tumour testing for the presence of MMR deficiency has been recognised to be a relevant screening method to identify patients with MMR germline mutations (Umar et al, 2004).
Various repeat markers have been proposed to determine MSI. In 1997, an international meeting at the National Cancer Institute recommended the use of a panel of five markers (three dinucleotide and two mononucleotide repeats), known as the Bethesda panel (Boland et al, 1998). However, some limitations appeared, primarily due to the use of dinucleotide markers that showed lower sensitivity and specificity compared with mononucleotide markers (Perucho, 1999; Suraweera et al, 2002). In 2002, Hamelin and collaborators proposed a new panel of five quasi-monomorphic mononucleotide markers, known as the pentaplex panel, which was shown to allow accurate identification of MSI tumours without the need of matched normal DNA (Suraweera et al, 2002). This panel showed increased sensitivity and specificity compared with the Bethesda panel and was thus recommended for MSI testing (Buhard et al, 2004; Umar et al, 2004; Wong et al, 2006; Xicola et al, 2007; Goel et al, 2010; You et al, 2010). Nevertheless, in our experience, the pentaplex panel still failed detecting some tumours from patients with LS, especially tumours from patients with a germline MSH6 mutation and/or tumours other than CRC.
The aim of the present study was to optimise the panel of microsatellite markers for detection of MSI and to evaluate the performances of this new panel for the identification of MMR-deficient tumours.
Materials and Methods
Tissue specimens
A total of 148 tumours from patients with suspicion of LS were included in the study. Tumours and matched normal tissues when available (n=120) were obtained from Pathology Centers from North of France. These included 71 CRC and 7 colorectal adenomas, 31 endometrial carcinomas, 12 upper urinary tract carcinomas, 12 ovarian carcinomas, 7 skin tumours (6 sebaceous carcinomas and one adenoma), 6 gastric carcinomas, and 2 cerebral tumours.
Among these tumours, 77 were with defective MMR (dMMR) and 71 were with proficient MMR (pMMR) on the basis of MMR protein expression (n=138) and/or the presence of somatic MLH1 hypermethylation or germline MMR mutation (n=61). The MSI status of the tumours was established using the original pentaplex panel (Suraweera et al, 2002). Fifty-eight tumours were found MSI-high (MSI-H) with instability at ⩾3 of the five markers. Fifteen additional tumours were found MSI-low (MSI-L) with instability at only one (n=7) or two (n=8) markers after comparison to normal DNA. Methods used were as described previously (Suraweera et al, 2002; Aissi-Ben Moussa et al, 2009; Crepin et al, 2011). Only patients with unambiguous results were included in the study. Full informed consent for the study was obtained from all the patients.
MSI analysis with the new panel
DNA was extracted from archival formalin-fixed paraffin-embedded (FFPE) tissue samples using the EZ1 DNA Tissue tissue kit with the BioRobot EZ1 (Qiagen, Courtaboeuf, France).
DNA samples isolated from tumours and corresponding normal tissues were tested for MSI using a set of six mononucleotide repeat markers: BAT25, BAT26, NR21, NR22 selected from the original pentaplex panel (Suraweera et al, 2002), NR27 selected from the modified pentaplex panel (Buhard et al, 2006), and BAT40, a polyT located on chromosome 1, which was shown to be particularly sensitive in both CRC and extra-colonic tumours (Hartmann et al, 2002; Kuismanen et al, 2002; Hendriks et al, 2004). The NR24 marker was not kept in the hexaplex panel because of its lower sensitivity (Goel et al 2010, and personal data). One highly polymorphic dinucleotide repeat marker (D3S1260) was added to the hexaplex as an internal control to check the correspondence between tumour DNA and matched normal DNA.
Primers and fluorescent markers were designed to allow efficient amplification of DNA extracted from FFPE tumours and resolution of different amplified products. Primer sequences and fluorescent markers are given in Table 1.
Table 1. Primer sequences and allele sizes.
| Marker | Sense primer (5′→3′) | Antisense primer (5′→3′) | Label | Allele sizea (bp) |
|---|---|---|---|---|
| BAT25 |
TCGCCTCCAAGAATGTAAGT |
TCTGCATTTTAACTATGGCTC |
HEX |
122–124 |
| BAT26 |
TGACTACTTTTGACTTCAGCC |
AACCATTCAACATTTTTAACCC |
6-FAM |
116–117 |
| BAT40 |
AGTCCATTTTATATCCTCAAGC |
GTAGAGCAAGACCACCTTG |
NED |
142–143 and 145–146 |
| NR21 |
TAAATGTATGTCTCCCCTGG |
ATTCCTACTCCGCATTCACA |
HEX |
98–99 |
| NR22 |
GAGGCTTGTCAAGGACATAA |
AATTCGGATGCCATCCAGTT |
6-FAM |
138–140 |
| NR27 |
AACCATGCTTGCAAACCACT |
CGATAATACTAGCAATGACC |
6-FAM |
86–87 |
| D3S1260 | CTACCAGGGAAGCACTGTAG | CATGTACCTGAGCACCTACTG | 6-FAM | 171–189 |
Most common alleles.
The seven markers were co-amplified in one tube using the Multiplex Master Mix (Qiagen). PCR was carried out in a 25-μl final volume containing 1 × Multiplex Master Mix, 0.24 μℳ of each primer pair, except for NR27 (0.12 μℳ) and BAT40 (0.48 μℳ), and about 50 ng of DNA. The PCR conditions are described in details in Supplementary Methods. The amplified products were separated on an ABI Prism 3130XL analyser and analysed using GeneMapper analysis software (Applied Biosystems, Courtaboeuf, France). A microsatellite marker was considered unstable when its size differed by at least 2 bp compared with the germline DNA (matched normal DNA or quasi-monomorphic variation range obtained from normal tissues). All tumours with ⩾1 unstable markers were considered as having some degree of instability and designated as MSI. Tumours with instability at ⩾3 of the six mononucleotide markers compared with the germline DNA were defined as MSI-H. Tumours with instability at one or two markers compared with germline DNA were defined as MSI-L. The cutoff for classification was chosen on the basis of the threshold of about 30–40% that is commonly used to distinguish MSI-H and MSI-L samples (Tomlinson et al, 2002).
Statistical analyses
The performances of the different microsatellite markers for MSI detection was compared by evaluating the sensitivity, specificity, positive predictive (PPV), and negative predictive values (NPVs), which were calculated using standard definition. The statistical software GraphPad InStat, version 3.10 (La Jolla, CA, USA) was used to calculate the 95% confidence intervals.
Allele size variations were summarised using means and s.ds. and compared in groups by Mann–Whitney U test.
The cutoff distinguishing MSI tumours from MSS tumours was ⩾1 unstable marker except otherwise indicated.
A P-value <0.05 was considered as statistically significant. Statistical analyses were performed using the SPSS software version 15.0 (SPSS, Chicago, IL, USA).
Results
Fluorescent heptaplex PCR
The panel consisted of six mononucleotide repeat markers: BAT25, BAT26, BAT40, NR21, NR22, and NR27, and a highly polymorphic dinucleotide repeat marker: D3S1260. Primers were designed to be less sensitive to DNA quality, and PCR conditions were defined to allow co-amplification of the seven microsatellite markers in a single heptaplex PCR.
Most common sizes for BAT25, BAT26, BAT40, NR21, NR22, and NR27, as observed with normal DNA samples, were 122–124 bp, 116–117 bp, 145–146 bp, 98–99 bp, 138–139 bp, and 86–87 bp, respectively. These sizes were particularly short, allowing efficient amplification of the six mononucleotide markers of interest from all the samples. Examples of profiles are given in Figure 1.
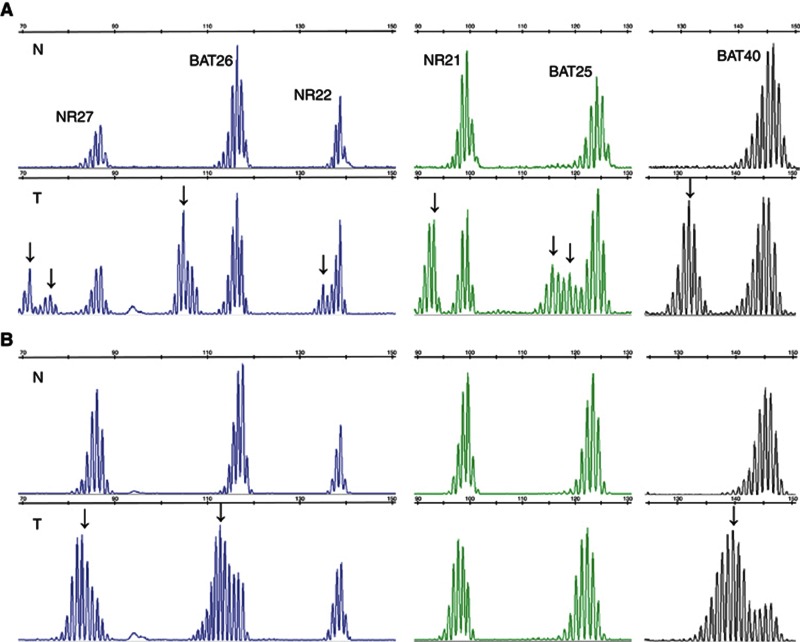
Figure 1.
Representative examples of MSI profiles obtained with the hexaplex panel. MSI profiles in (A) colorectal and (B) endometrial dMMR tumours and matched normal tissues. (A) CRC from a patient with a MLH1 germline mutation and loss of MLH1 and PMS2 expression, showing instability for the six microsatellite markers. (B) Endometrial tumour from a patient with a MLH1 germline mutation and loss of MLH1 and PMS2 expression, showing instability for NR27, BAT26, NR21, BAT25, and BAT40. Additional alleles are indicated (arrows). Note the shorter allelic shifts in the endometrial tumour compared with the CRC. Abbreviations: N=normal tissue; T=tumour tissue.
Size variations, as observed with normal DNA, were very infrequent for BAT25 (1.30%), BAT26 (0.85%), NR21 (1.28%), NR22 (0.42%), and NR27 (0.42%), indicating that these markers are highly monomorphic in our population. By contrast, BAT40 was more polymorphic. Alleles ranged from 122 bp to 150 bp, 142–143 pb and 145–146 pb being the most common alleles (28.7% and 59.1% of the observed alleles, respectively).
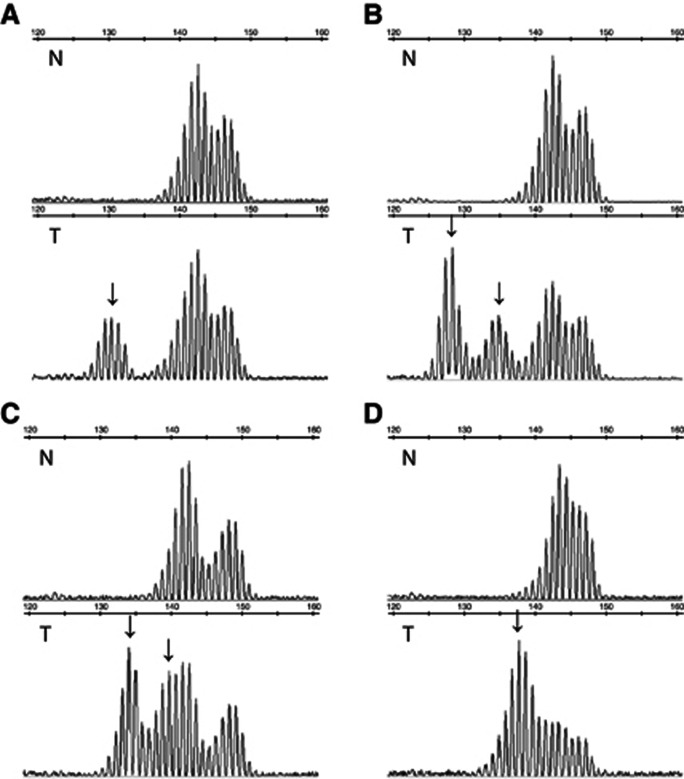
Out of the 148 tumours included in the study, 76 (51.4%) showed instability at ⩾1 markers. Sixty-six tumours (44.6%) were MSI-H with instability at ⩾3 markers and 10 (6.8%) were MSI-L with instability at 1 (n=4) or 2 (n=6) markers. Matched normal DNA was available for 120 tumours (81.1%). Of the 28 tumours without matched normal DNA, six showed uninterpretable markers due to the presence of uncommon alleles: one for BAT26, one for NR21, one for NR27, and three for BAT40. None of the 28 tumours displayed >1 uninterpretable marker. Notably, BAT40 profile was profoundly modified in most unstable tumours (Figures 1 and 2). Blinded analysis of BAT40 without reference to matched normal DNA revealed that all tumours but one (147/148, 99.3%) would have been correctly classified in the absence of normal tissue due to the presence of >2 alleles (Figure 2A–C) or to an abnormal pattern of slippage that is not observed with germline DNA (Figure 2 D).
Figure 2.
Representative examples of MSI profiles obtained with BAT40. MSI profiles in four dMMR tumours and matched normal tissues. Patients are heterozygous for BAT40. Profiles are profoundly modified in tumours compared with the germline DNA with one or multiple additional alleles (arrows). Abbreviations: N=normal tissue; T=tumour tissue.
Screening performance of the hexaplex panel
To determine the performance of our assay, we investigated a series of 148 tumours from patients suspected of LS, which consisted of 77 dMMR tumours and 71 pMMR tumours based on the results of immunohistochemistry for MMR protein expression (n=138), MLH1 methylation analysis, and/or genetic testing (n=61). Among dMMR tumours, 24 were MLH1-deficient (i.e., with combined loss of MLH1 and PMS2, somatic hypermethylation of the MLH1 promoter, and/or germline mutation in MLH1), 30 were MSH2-deficient (with combined loss of MSH2 and MSH6 and/or germline mutation in MSH2), 19 were MSH6-deficient (with selective loss of MSH6 and/or germline mutation in MSH6), and 4 were PMS2-deficient (with selective loss of PMS2 and germline mutation in PMS2). Characteristics of the tumours are summarised in Table 2.
Table 2. Characteristics of tumours (n=148).
| Tumours | dMLH1 (n=24) | dMSH2 (n=30) | dMSH6 (n=19) | dPMS2 (n=4) | dMMR (n=77) | pMMR (n=71) |
|---|---|---|---|---|---|---|
| CRC (n=71) |
18 |
8 |
7 |
2 |
35 |
36 |
| Endometrial cancer (n=31) |
4 |
10 |
5 |
2 |
21 |
10 |
| Urothelial cancer (n=12) |
1 |
4 |
1 |
0 |
6 |
6 |
| Ovary cancer (n=12) |
0 |
2 |
3 |
0 |
5 |
7 |
| Skin cancer (n=7) |
0 |
3 |
2 |
0 |
5 |
2 |
| Gastric cancer (n=6) |
1 |
0 |
0 |
0 |
1 |
5 |
| Colorectal adenoma (n=7) |
0 |
2 |
1 |
0 |
3 |
4 |
| Brain cancer (n=2) | 0 | 1 | 0 | 0 | 1 | 1 |
Abbreviations: CRC=colorectal cancer; dMMR=defective DNA mismatch repair; pMMR=proficient DNA mismatch repair.Bold faced numbers correspond to global effectives. dMMR corresponds to the sum of dMLH1, dMSH2, dMSH6 and dPMS2.
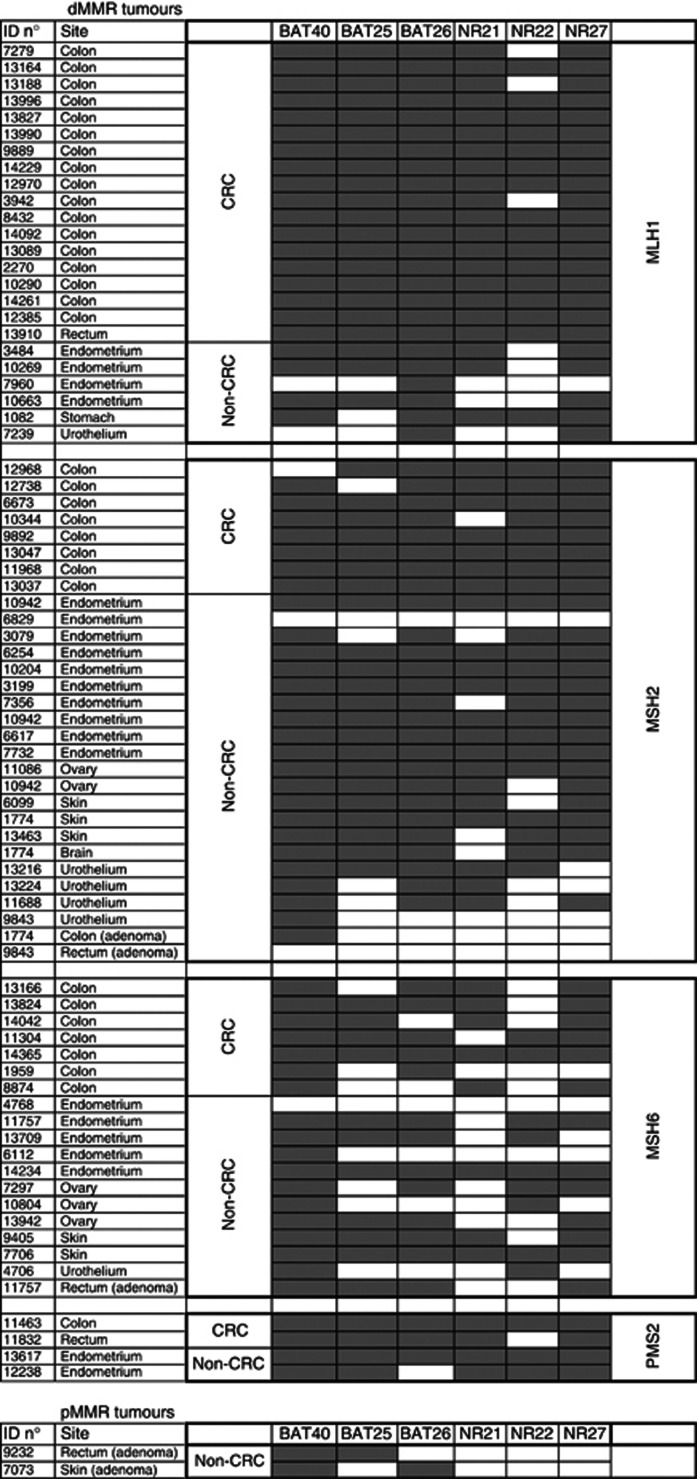
We evaluated the performance characteristics of the six mononucleotide repeat markers for identification of dMMR tumours. The panel displayed 85.7% sensitivity, 100% PPV, 100% specificity, and 86.6% NPV when instability at ⩾3 markers defined MSI. Corresponding values were 96.1% sensitivity and 97.4% PPV with 97.2% specificity and 95.8% NPV for a cutoff at one or more unstable markers (Table 3). Of the 77 dMMR tumours, 66 (85.7%) showed instability at ⩾3 markers and 8 (10.4%) showed instability at 1 (n=4) or 2 (n=4) markers (Figure 2). Tumours with low instability included one CRC, two endometrial tumours, three urothelial tumours, one ovary tumour, and one rectal adenoma. Characteristics of the tumours are summarised in Table 4 and Figure 3. Three tumours did not show instability at any of the six markers. These included two endometrial cancers, one of which was shown to contain <10% of viable tumour cells and one rectal adenoma with low-grade dysplasia. Notably, two out of the three dMMR adenomas tested showed some instability (with 4 out of 6 unstable markers in one case and 1 out of 6 in the other case). Of the 71 pMMR-classified tumours, 2 showed some instability using the hexaplex panel: 1 rectal adenoma and 1 sebaceous adenoma, both with instability at 2 markers (Table 4).
Table 3. Performance characteristics of the hexaplex and pentaplex panels for the identification of dMMR tumoursa (n=148).
| Panel | No. of unstable markers | Sensitivity (95% CI) | Specificity (95% CI) | PPV (95% CI) | NPV (95% CI) |
|---|---|---|---|---|---|
| Hexaplex |
⩾3 |
85.7 (75.9–92.6) |
100.0 (94.9–100.0) |
100.0 (94.6–100.0) |
86.6 (77.3–93.1) |
| |
⩾2 |
90.9 (81.9–96.2) |
97.2 (90.2–99.7) |
97.2 (90.2–99.7) |
90.7 (81.0–95.6) |
| |
⩾1 |
96.1 (89.0–99.2) |
97.2 (90.2–99.7) |
97.4 (90.8–99.7) |
95.8 (88.3–99.1) |
| Pentaplex |
⩾3 |
75.3 (64.1–84.4) |
100.0 (94.9–100.0) |
100.0 (93.8–100.0) |
78.9 (69.0–86.8) |
| |
⩾2 |
85.7 (75.9–92.6) |
100.0 (94.9–100.0) |
100.0 (94.6–100.0) |
86.6 (77.3–93.1) |
| ⩾1 | 92.2 (83.8–97.1) | 97.2 (90.2–99.7) | 97.3 (90.5–99.7) | 92.0 (83.4–97.0) |
Abbreviations: CI=confidence interval; dMMR=defective DNA mismatch repair; NPV=negative predictive value; PPV=positive predictive value.
Results are expressed as percentages, with 95% CIs in brackets.
Table 4. Clinical characteristics of dMMR tumours with low or absence of microsatellite instability and pMMR tumours with instability.
| Group | Site | Histology | Age (years)a | Criteriab | No. of unstable markers (type) | MMR protein expression | MMR germline/somatic mutation | Remarks |
|---|---|---|---|---|---|---|---|---|
| dMMR |
Colon |
ADC |
39 |
Beth |
2/6 (BAT26, BAT40) |
MSH6 |
MSH6 c.2150_2153del (p.Val717Alafs) |
+Synchronous colon cancer |
| |
Endometrium |
ADC |
49 |
Ams-II |
1/6 (BAT26) |
MLH1 PMS2 |
MLH1 somatic hypermethylation |
|
| |
Endometrium |
ADC |
46 |
EC |
1/6 (BAT40) |
MSH6 |
MSH6 c.3268_3272del (p.Glu1090Lysfs) |
|
| |
Endometrium |
ADC |
58 |
Ams-II |
0/6 |
MSH6 |
MSH6 c.3261delC p.Phe1088Serfs |
|
| |
Endometrium |
ADC |
57 |
Ams-I |
0/6 |
N |
MLH1 c.1165C>A (p.Arg389*) |
|
| |
Ovary |
ADC |
49 |
Ams-II |
2/6 (BAT40, NR22) |
MSH6 |
MSH6 c.2277_2281dup (p.Arg761Lysfs) |
|
| |
Urothelium |
Carcinoma |
48 |
Ams-I |
2/6 (BAT26, NR27) |
MLH1 PMS2 |
MLH1 c.2117_2130del (p.Gly706Valfs) |
|
| |
Urothelium |
Carcinoma |
53 |
EC |
2/6 (BAT40, NR22) |
N |
MSH6 c.3080dupT (p.Ser1028Ilefs) |
+Endometrial carcinoma at 53 |
| |
Urothelium |
Carcinoma |
56 |
Ams-I |
1/6 (BAT40) |
MSH2 MSH6 |
MSH2 c.1022T>C (p.Leu341Pro)c |
+Colorectal adenoma at 56 |
| |
Rectum |
Adenoma |
51 |
— |
1/6 (BAT40) |
MSH2 MSH6 |
MSH2 c.793-2A>C (p.Val265_Gln314del) |
+Multiple sebaceous carcinomas at 51 |
| |
Rectum |
Adenoma |
56 |
Ams-I |
0/6 |
N |
MSH2 c.1022T>C (p.Leu341Pro)c |
+Urothelial carcinoma at 56 |
| pMMR |
Skin |
Sebaceous adenoma |
54 |
EC |
2/6 (BAT26, BAT40) |
N |
ND |
+Multiple sebaceous adenomas at <54+past history of ovary carcinoma at 44 |
| Rectum | Adenoma | 48 | — | 2/6 (BAT25, BAT40) | N | ND |
Abbreviations: ADC=adenocarcinoma; dMMR=defective DNA mismatch repair; EC=extended criteria; N=normal expression of the four DNA mismatch repair (MMR) proteins; ND=not determined; pMMR=proficient MMR.
Age at diagnosis.
Ams, Amsterdam criteria: Ams-I ( Vasen et al., 1991), Ams-II (Vasen et al., 1999); Beth, revised Bethesda criteria (Umar et al., 2004); EC Lynch Syndrome-related cancer <60 years (Olschwang et al., 2004; French National Cancer Institute guidelines).
Mutation c.1022T>C is classified as pathogenic (UMD MSH2 mutations database: http://www.umd.be/MSH2/).
Figure 3.
Performance characteristics of hexaplex markers in dMMR tumours and MSI-positive pMMR tumours. Black scares indicate unstable markers; white scares indicate stable markers.
We next examined the performance characteristics of individual markers, with special focus on BAT40. The sensitivity and NPV were high for all markers, ranging from 64.1% and 71.4%, respectively, for NR22, to 92.2% and 91.7%, respectively, for BAT40 (Table 5). Six dMMR tumours showed stable BAT40, three of which were stable for all markers (Figure 3). Notably, one tumour with stable BAT40 showed instability at the five other markers. This tumour from a patient of African origin was homozygous for BAT40 with two unusual short 122 bp alleles.
Table 5. Performance characteristics of individual markers for the identification of dMMR tumoursa (n=148).
| Marker | Sensitivity (95% CI) | Specificity (95% CI) | PPV (95% CI) | NPV (95% CI) |
|---|---|---|---|---|
| BAT25 |
74.4 (63.6–82.8) |
98.6 (91.6–100.0) |
98.3 (90.2–100.0) |
77.5 (67.8–85.0) |
| BAT26 |
84.6 (74.9–91.1) |
98.6 (91.5–100.0) |
98.5 (91.3–100.0) |
85.0 (75.4–91.4) |
| BAT40 |
92.2 (83.7–96.7) |
97.1 (89.3–99.8) |
97.3 (90.0–99.8) |
91.7 (82.7–96.4) |
| NR21 |
70.1 (59.1–79.2) |
100.0 (93.7–100.0) |
100.0 (92.1–100.0) |
75.0 (65.2–82.8) |
| NR22 |
64.1 (53.0–73.9) |
100.0 (93.8–100.0) |
100.0 (91.5–100.0) |
71.4 (61.8–79.5) |
| NR27 | 82.1 (72.0–89.1) | 100.0 (93.7–100.0) | 100.0 (93.2–100.0) | 83.1 (73.5–89.8) |
Abbreviations: CI=confidence interval; dMMR=defective DNA mismatch repair; NPV=negative predictive value; PPV=positive predictive value.
Results are expressed as percentages, with 95% CIs in brackets.
The performance characteristics of the hexaplex panel were better for CRC than for non-CRC tumours regardless of the cutoff used for the MSI definition, with 100% sensitivity for CRC vs 92.9% for non-CRC tumours when ⩾1 unstable markers defined MSI (Table 6 and Supplementary Figure 1). As expected, the mean number of unstable markers per dMMR tumour was significantly reduced in non-CRC tumours (4.3 vs 5.4, P=0.001) with a lower allelic shift when compared with matched normal DNA (mean total variation, 22.5 vs 39.5 bp, P<0.0001) (Figure 3 and Supplementary Table 1). These differences were bigger for BAT40, NR27, and BAT26, contributing to the higher sensitivity of these markers in identifying non-CRC dMMR tumours.
Table 6. Performance characteristics of the hexaplex and pentaplex panels for the identification of non-CRC-dMMR and MSH6-deficient tumoursa, b (n=148).
| |
Hexaplex |
Pentaplex |
||
|---|---|---|---|---|
| Tumour | Sensitivity (95% CI) | Specificity (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) |
| CRC |
100.0 (90.0–100.0) |
100.0 (90.3–100.0) |
100.0 (90.0–100.0) |
100.0 (90.3–100.0) |
| Non-CRC |
92.9 (80.5–98.5) |
94.3 (80.9–99.3) |
85.7 (71.5–94.6) |
94.3 (80.9–99.3) |
| Non-MSH6 |
96.6 (88.1–99.6) |
— |
94.8 (85.6–98.9) |
— |
| MSH6 |
94.7 (74.0–99.9) |
— |
84.2 (60.4–96.6) |
— |
| All | 96.1 (89.0–99.2) | 97.2 (90.2–99.7) | 92.2 (83.8–97.1) | 97.2 (90.2–99.7) |
Abbreviations: CI=confidence interval; CRC=colorectal cancer; dMMR=defective DNA mismatch repair.
Results are expressed as percentages, with 95% CIs in brackets
Tumours showing instability for ⩾1 markers were considered as MSI.
The performance characteristics of the panel were very similar for MLH1-, MSH2-, or PMS2-deficient tumours and MSH6-deficient tumours, with 96.6% and 94.7% sensitivity, respectively, when ⩾1 unstable markers defined MSI (Table 6 and Supplementary Figure 1). However, the mean number of unstable markers per dMMR tumour was significantly smaller in MSH6-deficient tumours compared with other MMR-deficient tumours (3.9 vs 5.3, P=0.001) (Figure 3). Accordingly, the sensitivity of the panel was lesser for MSH6-deficient tumours compared with non-MSH6-deficient tumours (73.7% vs 88.1%) when the cutoff for MSI definition was increased to three unstable markers (Supplementary Figure 1). Notably, allele size variations were bigger for BAT40, BAT26, BAT25, and NR27, suggesting higher sensitivity of these markers in identifying MSH6-deficient tumours (Supplementary Table 1).
Comparison of screening performance between the hexaplex and pentaplex panels
We compared the data obtained with our panel and with the original pentaplex panel composed of BAT25, BAT26, NR21, NR22, and NR24 markers on the same tumours (n=148). As shown in Table 6 and Supplementary Figure 1, performances of the two panels were comparable for CRC-dMMR and non-MSH6-deficient tumours. But interestingly, the sensitivity of the hexaplex was higher for non-CRC-dMMR tumours and MSH6-deficient tumours, irrespective of the cutoff used for MSI definition (92.9% vs 85.7% and 94.7% vs 84.2%, respectively, when ⩾1 unstable markers defined MSI).
Discussion
Determination of the MSI status is routinely performed as a screening test for LS. A pentaplex panel has been recognised as very sensitive and specific for determination of MSI status and also easy to use (Buhard et al, 2004; Wong et al, 2006; Xicola et al, 2007; Goel et al, 2010; You et al, 2010). This led to its widespread recommendation and adoption (Umar et al, 2004; Xicola et al, 2007; Goel et al, 2010). However, a lower sensitivity of MSI testing has been described for some extra-colonic tumours and some MSH6-deficient tumours (Wu et al, 1999; Hartmann et al, 2002; Kuismanen et al, 2002; Hendriks et al, 2004). This contributes to an under-diagnosis of LS. We further optimised this panel for detection of MMR deficiency by adding a sixth mononucleotide marker, BAT40, which appeared to be particularly sensitive in extra-colonic tumours (Hartmann et al, 2002; Kuismanen et al, 2002; Hendriks et al, 2004). One highly polymorphic dinucleotide repeat marker was also added to the panel as an internal control to check the correspondence between tumour DNA and matched normal DNA when the latter is available. Primers and PCR conditions were determined to allow amplification of all markers in a single multiplex PCR, rendering the panel as simple to use as the pentaplex panel. BAT25, BAT26, NR21, NR22, and NR27 were highly monomorphic in germline DNA, in accordance with previous data obtained from a wide spectrum of population worldwide (Buhard et al, 2006). BAT40 was more polymorphic, with two common alleles (142–143 bp and 145–146 bp) accounting for 87.8% of the observed alleles. This implies that matched normal DNA would be necessary in a non-negligible proportion of cases for its interpretation. However, even in cases of heterozygosity without matched normal DNA for comparison, its profile is per se a good indicator of the real presence/absence of instability. Thus, matched normal DNA would remain not mandatory in most cases.
We validated the utility of the hexaplex for MSI detection in a large series of MMR-deficient and -proficient tumours. Our series was voluntarily enriched with non-CRC tumours as well as MSH6-deficient tumours that have been reported to be associated with a lower proportion of unstable markers and a smaller size of allelic shifts resulting in some false-negative results (Wu et al, 1999; Hartmann et al, 2002). We also chose to include adenomas in our study, because benign tumours are known to present a lower frequency and degree of instability when compared with malignant tumours, which may therefore be challenging to detect (Ferreira et al, 2009).
Our assay demonstrated high sensitivity with instability in 74 out of 77 (96.1%) dMMR cases and high instability (⩾3 markers) in 66 (85.7%) cases. Of note, three tumours (one endometrium, one urothelium, and one colonic adenoma) would have been missed and five additional tumours with a MSI-low phenotype (two colon, one ovary, and two urothelium) would have been also considered as MSS if the BAT40 marker had not been tested. This demonstrates the interest of this marker to identify dMMR tumours. In addition, allele size variations were bigger for BAT40, and therefore easier to detect, which should facilitate the interpretation in cases with slight slippage.
At the same time, our assay demonstrated two apparently ‘false-positive' cases. These included one rectal adenoma and one sebaceous adenoma with two out of six unstable markers and maintained expression of MLH1, MSH2, MSH6, and PMS2 proteins. Of note, these tumours were also positive (with 1 out of 5 unstable marker) using the pentaplex panel. Unfortunately, blood samples were not available for MMR gene analysis for these patients. Such analysis may have revealed a defect of IHC in detecting MMR deficiency, as observed for two other cases in our series. The first one was an urothelial cancer that showed low MSI and maintained expression of the four MMR proteins and was subsequently found associated with a MSH6 pathogenic germline mutation (c.3080dupT); the second one was an endometrial cancer that showed high MSI and maintained MMR protein expression but was associated to a PMS2 germline mutation (c.1831dupA).
Cutoffs regarding the number of unstable markers for the classification of MSI have been suggested to minimise false-positive results due to non-specific slippage or heterozygosity when matched normal DNA is missing. LS has been excluded or considered very unlikely in MSI-L cases (Umar et al, 2004). In addition, only MSI-H tumours are believed to have distinctive clinical features, including better prognosis (Halford et al, 2002; Laiho et al, 2002). Nevertheless, MSI-L has been shown to occur as a real phenomenon in colorectal, endometrial, and ovarian cancers, which are all LS-related cancers (Halford et al, 2003). Moreover, it has been shown recently that defining tumours showing instability at ⩾2 markers as MSI (instead of ⩾3 out of 5) considerably enhanced the screening performance of the pentaplex panel for MSH6-deficient tumours (Goel et al, 2010). In our series, the statistical analysis of the hexaplex panel performances revealed even better sensitivity when ⩾1 unstable markers defined MSI, with no loss of specificity. Eight dMMR tumours showed instability at <3 markers, including four tumours with instability at only one marker. Seven out of them were from patients carrying a pathogenic germline MMR mutation (4 MSH6, 2 MSH2, and 1 MLH1), indicating that the detection of this class of tumours is useful for LS identification.
It is particularly true for MSH6-deficient tumours that are expected to display lower instability due to its biological function. Indeed, MSH6 is preferentially involved in repair of single base pair mismatches and small insertion/deletion loops. Moreover, its repair function can partially be compensated by MSH3 (Drummond et al, 1995; Palombo et al, 1995; Acharya et al, 1996). This phenomenon has also been well demonstrated in MSH6-mutant mice (Edelmann et al, 1997).
It is also particularly relevant for extra-colonic tumours such as endometrial tumours that have been described to follow a specific tumourigenic pathway, leading to a lower proportion of unstable markers with shorter allelic shifts, and thus to a frequent MSI-low or MSS phenotype (Wijnen et al, 1999; Wu et al, 1999; Kuismanen et al, 2002; Wong et al, 2006). Of note, endometrial cancer is the most common extra-colonic cancer in LS, with an estimated cumulative risk of 35–60% for female mutation carriers, that may reach up to 71% for MSH6 mutation carriers at 70 years of age (Hendriks et al, 2004; Vasen et al, 2007; Bonadona et al, 2011). This should be taken in consideration in the context of LS screening. Similarly, other tumours from patients with LS, such as urothelial tumours, brain tumours, or colorectal adenomas have been described to be frequently associated with a MSI-low or MSS phenotype (Hartmann et al, 2002; Gylling et al, 2008; Ferreira et al, 2009; Giunti et al, 2009). Because of the smaller size of allelic shifts observed in these tumours, we recommend to maintain analysis of matched germline DNA for routine-practice MSI screening of these cancer types.
In conclusion, this study further confirms the utility of routine molecular screening for LS in every type of potentially LS-associated tumours. Moreover, given the comparable advantages of our hexaplex panel in terms of rapidity and simplicity to the pentaplex one and its superiority in identifying some non-CRC dMMR and some MSH6-deficient tumours, we think that it is a good alternative screening test to identify patients with LS.
Acknowledgments
We thank all clinicians and pathologists who refer tumour samples through the Platform of Molecular Biology of Cancers of the ‘Centre Régional de Référence en Cancérologie' (C2RC) of Lille. This work was supported by the ‘Institut National du Cancer' (INCa).
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Aaltonen LA, Peltomaki P, Leach FS, Sistonen P, Pylkkanen L, Mecklin JP, Jarvinen H, Powell SM, Jen J, Hamilton SR, Petersen GM, Kinzler KW, Vogelstein B, de la Chapelle A. Clues to the pathogenesis of familial colorectal cancer. Science. 1993;260 (5109:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- Acharya S, Wilson T, Gradia S, Kane MF, Guerrette S, Marsischky GT, Kolodner R, Fishel R. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc Natl Acad Sci USA. 1996;93 (24:13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aissi-Ben Moussa S, Moussa A, Lovecchio T, Kourda N, Najjar T, Ben Jilani S, El Gaaied A, Porchet N, Manai M, Buisine MP. Identification and characterization of a novel MLH1 genomic rearrangement as the cause of HNPCC in a Tunisian family: evidence for a homologous Alu-mediated recombination. Fam Cancer. 2009;8 (2:119–126. doi: 10.1007/s10689-008-9215-7. [DOI] [PubMed] [Google Scholar]
- Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58 (22:5248–5257. [PubMed] [Google Scholar]
- Bonadona V, Bonaiti B, Olschwang S, Grandjouan S, Huiart L, Longy M, Guimbaud R, Buecher B, Bignon YJ, Caron O, Colas C, Nogues C, Lejeune-Dumoulin S, Olivier-Faivre L, Polycarpe-Osaer F, Nguyen TD, Desseigne F, Saurin JC, Berthet P, Leroux D, Duffour J, Manouvrier S, Frebourg T, Sobol H, Lasset C, Bonaiti-Pellie C. Cancer risks associated with germline mutations in MLH1, MSH2, and MSH6 genes in Lynch syndrome. JAMA. 2011;305 (22:2304–2310. doi: 10.1001/jama.2011.743. [DOI] [PubMed] [Google Scholar]
- Buhard O, Cattaneo F, Wong YF, Yim SF, Friedman E, Flejou JF, Duval A, Hamelin R. Multipopulation analysis of polymorphisms in five mononucleotide repeats used to determine the microsatellite instability status of human tumors. J Clin Oncol. 2006;24 (2:241–251. doi: 10.1200/JCO.2005.02.7227. [DOI] [PubMed] [Google Scholar]
- Buhard O, Suraweera N, Lectard A, Duval A, Hamelin R. Quasimonomorphic mononucleotide repeats for high-level microsatellite instability analysis. Dis Markers. 2004;20 (4-5:251–257. doi: 10.1155/2004/159347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crepin M, Dieu MC, Lejeune S, Escande F, Boidin D, Porchet N, Morin G, Manouvrier S, Mathieu M, Buisine MP. Evidence of constitutional MLH1 epimutation associated to transgenerational inheritance of cancer susceptibility. Hum Mutat. 2011;33 (1:180–188. doi: 10.1002/humu.21617. [DOI] [PubMed] [Google Scholar]
- Drummond JT, Li GM, Longley MJ, Modrich P. Isolation of an hMSH2-p160 heterodimer that restores DNA mismatch repair to tumor cells. Science. 1995;268 (5219:1909–1912. doi: 10.1126/science.7604264. [DOI] [PubMed] [Google Scholar]
- Edelmann W, Yang K, Umar A, Heyer J, Lau K, Fan K, Liedtke W, Cohen PE, Kane MF, Lipford JR, Yu N, Crouse GF, Pollard JW, Kunkel T, Lipkin M, Kolodner R, Kucherlapati R. Mutation in the mismatch repair gene Msh6 causes cancer susceptibility. Cell. 1997;91 (4:467–477. doi: 10.1016/s0092-8674(00)80433-x. [DOI] [PubMed] [Google Scholar]
- Ferreira AM, Westers H, Sousa S, Wu Y, Niessen RC, Olderode-Berends M, van der Sluis T, Reuvekamp PT, Seruca R, Kleibeuker JH, Hollema H, Sijmons RH, Hofstra RM. Mononucleotide precedes dinucleotide repeat instability during colorectal tumour development in Lynch syndrome patients. J Pathol. 2009;219 (1:96–102. doi: 10.1002/path.2573. [DOI] [PubMed] [Google Scholar]
- Giunti L, Cetica V, Ricci U, Giglio S, Sardi I, Paglierani M, Andreucci E, Sanzo M, Forni M, Buccoliero AM, Genitori L, Genuardi M. Type A microsatellite instability in pediatric gliomas as an indicator of Turcot syndrome. Eur J Hum Genet. 2009;17 (7:919–927. doi: 10.1038/ejhg.2008.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel A, Nagasaka T, Hamelin R, Boland CR. An optimized pentaplex PCR for detecting DNA mismatch repair-deficient colorectal cancers. PLoS One. 2010;5 (2:e9393. doi: 10.1371/journal.pone.0009393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryfe R, Kim H, Hsieh ET, Aronson MD, Holowaty EJ, Bull SB, Redston M, Gallinger S. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N Engl J Med. 2000;342 (2:69–77. doi: 10.1056/NEJM200001133420201. [DOI] [PubMed] [Google Scholar]
- Gylling AH, Nieminen TT, Abdel-Rahman WM, Nuorva K, Juhola M, Joensuu EI, Jarvinen HJ, Mecklin JP, Aarnio M, Peltomaki PT. Differential cancer predisposition in Lynch syndrome: insights from molecular analysis of brain and urinary tract tumors. Carcinogenesis. 2008;29 (7:1351–1359. doi: 10.1093/carcin/bgn133. [DOI] [PubMed] [Google Scholar]
- Halford S, Sasieni P, Rowan A, Wasan H, Bodmer W, Talbot I, Hawkins N, Ward R, Tomlinson I. Low-level microsatellite instability occurs in most colorectal cancers and is a nonrandomly distributed quantitative trait. Cancer Res. 2002;62 (1:53–57. [PubMed] [Google Scholar]
- Halford SE, Sawyer EJ, Lambros MB, Gorman P, Macdonald ND, Talbot IC, Foulkes WD, Gillett CE, Barnes DM, Akslen LA, Lee K, Jacobs IJ, Hanby AM, Ganesan TS, Salvesen HB, Bodmer WF, Tomlinson IP, Roylance RR. MSI-low, a real phenomenon which varies in frequency among cancer types. J Pathol. 2003;201 (3:389–394. doi: 10.1002/path.1453. [DOI] [PubMed] [Google Scholar]
- Hartmann A, Zanardo L, Bocker-Edmonston T, Blaszyk H, Dietmaier W, Stoehr R, Cheville JC, Junker K, Wieland W, Knuechel R, Rueschoff J, Hofstaedter F, Fishel R. Frequent microsatellite instability in sporadic tumors of the upper urinary tract. Cancer Res. 2002;62 (23:6796–6802. [PubMed] [Google Scholar]
- Hendriks YM, Wagner A, Morreau H, Menko F, Stormorken A, Quehenberger F, Sandkuijl L, Moller P, Genuardi M, Van Houwelingen H, Tops C, Van Puijenbroek M, Verkuijlen P, Kenter G, Van Mil A, Meijers-Heijboer H, Tan GB, Breuning MH, Fodde R, Wijnen JT, Brocker-Vriends AH, Vasen H. Cancer risk in hereditary nonpolyposis colorectal cancer due to MSH6 mutations: impact on counseling and surveillance. Gastroenterology. 2004;127 (1:17–25. doi: 10.1053/j.gastro.2004.03.068. [DOI] [PubMed] [Google Scholar]
- Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363 (6429:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- Kuismanen SA, Moisio AL, Schweizer P, Truninger K, Salovaara R, Arola J, Butzow R, Jiricny J, Nystrom-Lahti M, Peltomaki P. Endometrial and colorectal tumors from patients with hereditary nonpolyposis colon cancer display different patterns of microsatellite instability. Am J Pathol. 2002;160 (6:1953–1958. doi: 10.1016/S0002-9440(10)61144-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laiho P, Launonen V, Lahermo P, Esteller M, Guo M, Herman JG, Mecklin JP, Jarvinen H, Sistonen P, Kim KM, Shibata D, Houlston RS, Aaltonen LA. Low-level microsatellite instability in most colorectal carcinomas. Cancer Res. 2002;62 (4:1166–1170. [PubMed] [Google Scholar]
- Lynch HT, Lynch PM, Lanspa SJ, Snyder CL, Lynch JF, Boland CR. Review of the Lynch syndrome: history, molecular genetics, screening, differential diagnosis, and medicolegal ramifications. Clin Genet. 2009;76 (1:1–18. doi: 10.1111/j.1399-0004.2009.01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olschwang S, Bonaïti C, Feingold J, Frébourg T, Grandjouan S, Lasset C, Laurent-Puig P, Lecuru F, Millat B, Sobol H, Thomas G, Eisinger F. Identification and management of HNPCC syndrome (hereditary non polyposis colon cancer), hereditary predisposition to colorectal and endometrial adenocarcinomas. Bull Cancer. 2004;91 (4:303–315. [PubMed] [Google Scholar]
- Palombo F, Gallinari P, Iaccarino I, Lettieri T, Hughes M, D'Arrigo A, Truong O, Hsuan JJ, Jiricny J. GTBP, a 160-kilodalton protein essential for mismatch-binding activity in human cells. Science. 1995;268 (5219:1912–1914. doi: 10.1126/science.7604265. [DOI] [PubMed] [Google Scholar]
- Perucho M. Correspondence ref.: C.R. Boland et al, A National Cancer Institute workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res., 58: 5248–5257, 1998. Cancer Res. 1999;59 (1:249–256. [PubMed] [Google Scholar]
- Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, Hamilton SR, Laurent-Puig P, Gryfe R, Shepherd LE, Tu D, Redston M, Gallinger S. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349 (3:247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salovaara R, Loukola A, Kristo P, Kaariainen H, Ahtola H, Eskelinen M, Harkonen N, Julkunen R, Kangas E, Ojala S, Tulikoura J, Valkamo E, Jarvinen H, Mecklin JP, Aaltonen LA, de la Chapelle A. Population-based molecular detection of hereditary nonpolyposis colorectal cancer. J Clin Oncol. 2000;18 (11:2193–2200. doi: 10.1200/JCO.2000.18.11.2193. [DOI] [PubMed] [Google Scholar]
- Sargent DJ, Marsoni S, Monges G, Thibodeau SN, Labianca R, Hamilton SR, French AJ, Kabat B, Foster NR, Torri V, Ribic C, Grothey A, Moore M, Zaniboni A, Seitz JF, Sinicrope F, Gallinger S. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28 (20:3219–3226. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraweera N, Duval A, Reperant M, Vaury C, Furlan D, Leroy K, Seruca R, Iacopetta B, Hamelin R. Evaluation of tumor microsatellite instability using five quasimonomorphic mononucleotide repeats and pentaplex PCR. Gastroenterology. 2002;123 (6:1804–1811. doi: 10.1053/gast.2002.37070. [DOI] [PubMed] [Google Scholar]
- Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260 (5109:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- Tomlinson I, Halford S, Aaltonen L, Hawkins N, Ward R. Does MSI-low exist. J Pathol. 2002;197 (1:6–13. doi: 10.1002/path.1071. [DOI] [PubMed] [Google Scholar]
- Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Ruschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R, Hamilton SR, Hiatt RA, Jass J, Lindblom A, Lynch HT, Peltomaki P, Ramsey SD, Rodriguez-Bigas MA, Vasen HF, Hawk ET, Barrett JC, Freedman AN, Srivastava S. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96 (4:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasen HF, Mecklin JP, Khan PM, Lynch HT. The International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC) Dis Colon Rectum. 1991;34 (5:424–425. doi: 10.1007/BF02053699. [DOI] [PubMed] [Google Scholar]
- Vasen HF, Moslein G, Alonso A, Bernstein I, Bertario L, Blanco I, Burn J, Capella G, Engel C, Frayling I, Friedl W, Hes FJ, Hodgson S, Mecklin JP, Moller P, Nagengast F, Parc Y, Renkonen-Sinisalo L, Sampson JR, Stormorken A, Wijnen J. Guidelines for the clinical management of Lynch syndrome (hereditary non-polyposis cancer) J Med Genet. 2007;44 (6:353–362. doi: 10.1136/jmg.2007.048991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasen HF, Watson P, Mecklin JP, Lynch HT. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology. 1999;116 (6:1453–1456. doi: 10.1016/s0016-5085(99)70510-x. [DOI] [PubMed] [Google Scholar]
- Wijnen J, de Leeuw W, Vasen H, van der Klift H, Moller P, Stormorken A, Meijers-Heijboer H, Lindhout D, Menko F, Vossen S, Moslein G, Tops C, Brocker-Vriends A, Wu Y, Hofstra R, Sijmons R, Cornelisse C, Morreau H, Fodde R. Familial endometrial cancer in female carriers of MSH6 germline mutations. Nat Genet. 1999;23 (2:142–144. doi: 10.1038/13773. [DOI] [PubMed] [Google Scholar]
- Wong YF, Cheung TH, Lo KW, Yim SF, Chan LK, Buhard O, Duval A, Chung TK, Hamelin R. Detection of microsatellite instability in endometrial cancer: advantages of a panel of five mononucleotide repeats over the National Cancer Institute panel of markers. Carcinogenesis. 2006;27 (5:951–955. doi: 10.1093/carcin/bgi333. [DOI] [PubMed] [Google Scholar]
- Wu Y, Berends MJ, Mensink RG, Kempinga C, Sijmons RH, van Der Zee AG, Hollema H, Kleibeuker JH, Buys CH, Hofstra RM. Association of hereditary nonpolyposis colorectal cancer-related tumors displaying low microsatellite instability with MSH6 germline mutations. Am J Hum Genet. 1999;65 (5:1291–1298. doi: 10.1086/302612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xicola RM, Llor X, Pons E, Castells A, Alenda C, Pinol V, Andreu M, Castellvi-Bel S, Paya A, Jover R, Bessa X, Giros A, Duque JM, Nicolas-Perez D, Garcia AM, Rigau J, Gassull MA. Performance of different microsatellite marker panels for detection of mismatch repair-deficient colorectal tumors. J Natl Cancer Inst. 2007;99 (3:244–252. doi: 10.1093/jnci/djk033. [DOI] [PubMed] [Google Scholar]
- You JF, Buhard O, Ligtenberg MJ, Kets CM, Niessen RC, Hofstra RM, Wagner A, Dinjens WN, Colas C, Lascols O, Collura A, Flejou JF, Duval A, Hamelin R. Tumours with loss of MSH6 expression are MSI-H when screened with a pentaplex of five mononucleotide repeats. Br J Cancer. 2010;103 (12:1840–1845. doi: 10.1038/sj.bjc.6605988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.